Abstract
Korean ginseng (Panax ginseng) and American ginseng (Panax quinquefolius) are widely used medicinal plants with similar morphology but different medicinal efficacy. Roots, flowers, and processed products of Korean and American ginseng can be difficult to differentiate from each other, leading to illegal trade in which one species is sold as the other. This study was carried out to develop convenient and reliable chloroplast genome-derived DNA markers for authentication of Korean and American ginseng in commercial processed products. One codominant marker could reproducibly identify both species and intentional mixtures of the two species. We further developed a set of species-unique dominant DNA markers. Each species-specific dominant marker could detect 1% cross contamination with other species by low resolution agarose gel electrophoresis or quantitative polymerase chain reaction. Both markers were successfully applied to evaluate the original species from various processed ginseng products purchased from markets in Korea and China. We believe that high-throughput application of this marker system will eradicate illegal trade and promote confident marketing for both species to increase the value of Korean as well as American ginseng in Korea and worldwide.
Keywords: authentication, chloroplast intergenic spacer DNA marker, Panax ginseng, Panax quinquefolius, processed ginseng products
1. Introduction
Ginseng (Panax ginseng Meyer) is one of the most important medicinal plants and is particularly prized in Asian countries [1,2]. It has been a popular medicine for thousands of years in East Asia [3]. Ginseng is a deciduous perennial herb belonging to the family Araliaceae. Most Panax species including P. ginseng are indigenous to East Asia, but two species are found in Eastern North America [4]. Among them, P. ginseng (Korean ginseng) and Panax quinquefolius (American ginseng) have been the most widely cultivated and marketed in various commercial products because of their prominent medicinal effects, including immune system stimulation [5], anticarcinogenic activity, and reduction of blood glucose levels [6]. The two species are morphologically similar even though their origins were continentally separated by the Pacific Ocean. Most P. ginseng production is centralized in Korea and Northeast China, whereas P. quinquefolius is cultivated in China, Canada, and the United States.
P. ginseng contains more than 30 kinds of triterpenoid saponin glycosides, commonly called ginsenosides, as well as other phytochemical compounds [7–9]. Korean ginseng has the best reputation and its price is higher than that of other ginseng species, which has led to fraudulent labeling. Illegal trade disguising P. quinquefolius as P. ginseng has become an increasing problem in recent years in the Korean ginseng market because roots of P. ginseng and P. quinquefolius are similar in morphological appearance. Furthermore, authentication of both species within commercial processed ginseng products is almost impossible because they are sold in the form of red ginseng, ginseng powder, shredded slices, pellets, liquid extracts, and even tea. Therefore, methods for authentication of commercial ginseng products are in urgent demand.
Authentication can be achieved using high-performance liquid chromatography [10], gas chromatography–mass spectroscopy [11], and proteome analysis. However, those applications may be limited because secondary metabolite accumulation in ginseng is significantly affected by various factors such as growth conditions, developmental stage, internal metabolism, and manufacturing process. Moreover, those methods are expensive and difficult to utilize for high-throughput analysis.
Sequence-based DNA markers have advantages for the purpose of practical authentication. DNA markers can differentiate P. ginseng from other foreign ginsengs using a small amount of sample material in a time- and cost-effective manner [12]. The method is also applicable to any plant tissue as well as to processed products, with stable and reproducible results. Various DNA markers, including nuclear genomic sequence-derived simple sequence repeat markers, can be utilized for authentication of species [13]. However, these markers show intraspecies level variation, such as variation among ginseng cultivars and individuals [14,15], which constitutes a limitation to practical application of these markers for reproducible authentication of different species.
DNA markers based on the chloroplast genome are able to classify ginseng species swiftly and reliably because of their unique features. Chloroplasts are intracellular organelles that contain their own genome and are responsible for photosynthesis in plants [16]. A plant cell can contain up to 1,000 copies of the chloroplast genome, which is >100 times greater than the number of nuclear genome copies found in plant tissues [17]. Therefore, a target region in the chloroplast genome can be more easily amplified by polymerase chain reaction (PCR) than a target region in the nuclear genome from trace amounts of genomic DNA. The chloroplast genome size ranges between 120 kbp and 216 kbp, and the structure is highly conserved across plant species [18–20]. Most gene sequences are also highly conserved, but considerable amounts of nucleotide variation have been identified in chloroplast intergenic spacer (CIS) regions at above the interspecies level and rare variations were identified at the intraspecies level [21,22].
Using the P. ginseng chloroplast genome sequence as a backbone [23], we previously identified 60 CIS regions showing single nucleotide polymorphisms (SNPs) and 40 CIS regions showing insertion and deletion (InDel) variation among 101 CIS regions of three Panax species: P. ginseng, P. quinquefolius and Panax notoginseng. We identified no polymorphism between cultivars and individuals in P. ginseng [24] at these regions, which is an important characteristic if the authentication markers are to be used to distinguish between Korean and American ginseng.
We previously identified 38 SNPs and 24 InDels between P. ginseng and P. quinquefolius. Among the 24 InDels, 18 were derived from tandem repeats longer than 5 bp. All of the polymorphic regions could potentially be utilized as targets for DNA markers identifying P. ginseng and P. quinquefolius. Here, we focused on two target regions showing large InDels in order to develop tools for practical applications and efficient and high-throughput authentication methods to distinguish between Korean and American ginseng in commercial products.
2. Materials and methods
2.1. Purchase of primary processed roots and leaf products, and DNA extraction
Three-to-six-year-old fresh Korean ginseng roots (P. ginseng) were purchased from 10 different ginseng stores in Geumsan (Fig. 1A), which is the most famous ginseng-distributing market town in Korea. Various ginseng products such as dried root slices and flower teas of P. ginseng and P. quinquefolius were purchased at Changchun and Fusong in Jilin province, China. Standard control DNA for P. ginseng and P. quinquefolius was obtained from leaves of plants growing at the farm of Seoul National University, Suwon. All DNAs from the commercial products were prepared based on the method of Allen [25]. The concentration of the DNA was checked by UV spectrophotometer (NanoDrop ND-1000; Thermo Scientific, Nanodrop Technologies, Wilmington, DE) and agarose gel electrophoresis (AGE).
Fig. 1.
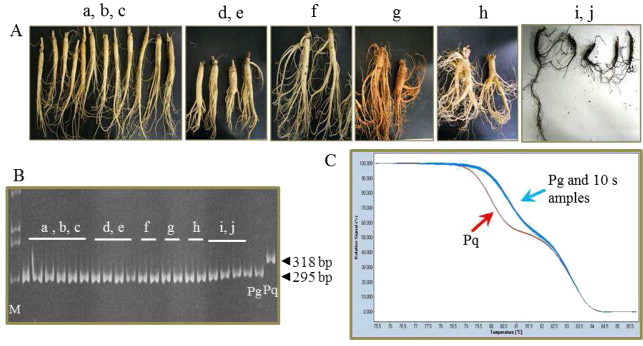
Authentication of Panax species using the pgcpir 035 marker in fresh ginseng roots purchased from markets. (A) Ten fresh ginseng root lots were purchased from different markets in Korea: a-c, 3-year-old roots; d & e, 4-year-old roots; f & g, 5-year-old roots; h, 6-year-old roots cultivated in ginseng fields; i & j, 10-year-old mountain-grown ginseng roots. (B) Gel electrophoresis for 10 root lots. Two roots from each lot were analyzed. Pg and Pq indicate P. ginseng and P. quinquefolius standard DNA, respectively. M indicates 100 bp DNA ladder. (C) HRM analysis of the same 20 root samples and standard DNAs, Pg and Pq.
2.2. Purchase of processed ginseng and red ginseng products, and DNA extraction
Ten kinds of processed ginseng or red ginseng products including powder, pellets, extract, dried roots, ginseng preserved in sugar or honey, drinks, shredded slices, and tea powder were purchased from the Korea ginseng market and used for preparation of DNA using different protocols [26]. We modified or added additional steps for different products. The ginseng extracts were in a concentrated form of red ginseng and thus were sticky. Accordingly, the ginseng extracts were diluted with water. After centrifuging the samples, pellets were visible in the tubes. This step was repeated three times. Discarding supernatants, the pellet was washed twice, and then DNA extraction was begun using the pellet. The same protocols were used for DNA extraction from liquid extracts and drinks. Products preserved in honey or sugar required additional washing with water to remove sugar and other components. Then, materials were ground with liquid nitrogen. Subsequent steps were the same as the previous method [25].
2.3. PCR and electrophoresis
PCR was carried out in a total volume of 25 μL containing 20 ng DNA, 2.5 mM each dNTP, 10 pmol each primer (Macrogen, Seoul, Korea) and 0.4 U Taq polymerase (Vivagen, Seongnam, Korea). The PCR cycling conditions were as follows: 4 min at 94°C for initial DNA denaturation; 35 cycles of 20 s at 94°C, 20 s at 60°C, and 30 s at 72°C; followed by 10 min at 72°C. PCR products were separated by 1.0–2.0% AGE or 5–9% mini polyacrylamide gel electrophoresis (PAGE; CBS Scientific, San Diego, CA, USA).
2.4. High-resolution melting analysis
High-resolution melting (HRM) analysis was carried out as previously described [24]. The melting analysis was performed by raising the temperature to 95°C for 1 min, lowering the temperature to 40°C for 1 min, raising the temperature to 70°C for 5 s, and finally increasing the temperature to 90°C, with continuous fluorescence acquisition followed by a cool down to 40°C using a LightCycler 480 (Roche Applied Science, Indianapolis, IN, USA). The fluorescence signal was plotted in real time versus temperature to produce melting curves for each sample. The melting curves were then converted into negative derivative curves of fluorescence with respect to temperature, and the results were analyzed using the Roche LightCycler 480 Data Analysis software (Roche Applied Science).
3. Results and Discussion
3.1. Evaluation of fresh roots in Korean ginseng markets using a codominant rps2–rpoC2 marker
From among the 24 InDel markers derived from CIS regions of P. ginseng and P. quinquefolius [24], we initially utilized the pgcpir 035 marker showing the largest InDel between both species for analysis of fresh ginseng root products from Korean ginseng markets. The pgcpir 035 marker produced 295-bp and 318-bp bands for P. ginseng and P. quinquefolius, respectively, and the products were clearly distinguishable by AGE (Fig. 1B) and HRM (Fig. 1C).
We purchased fresh ginseng roots from 10 different ginseng stores in the Geumsan ginseng market in Korea (Fig. 1A). Root ages varied from 3 yr to 6 yr for regularly cultivated ginseng (Fig. 1A a–h) and up to 10 yr for mountain-grown ginseng (Fig. 1A i,j). All of the ginseng roots purchased from the 10 different ginseng stores were revealed to be P. ginseng. It is not unexpected that we did not find any American ginseng roots among the tested fresh ginseng roots, because American ginseng is not officially allowed to be imported into Korea at present.
The pgcpir 035 marker is based on a 23-bp InDel that is derived from copy number variation of a 23-bp tandem repeat, with two and three copies present in the intergenic spacers of rps2–rpoC2 genes of P. ginseng and P. quinquefolius, respectively [24]. The CIS of the rps2–rpoC2 genes has previously been used for genetic diversity analysis of a grass subfamily and Apocynaceae plants [27,28]. Here, we found that the rps2–rpoC2 CIS also provided a reproducible and credible marker to identify Korean ginseng and American ginseng. We inspected many Korean ginseng samples including all 10 registered cultivars, various landraces, and various products in addition to the 10 fresh ginseng root samples described above, and all gave rise to results identical to that of P. ginseng standard DNA [14,15]. We did not inspect various P. quinquefolius accessions, but the 23 bp InDel in the CIS of rps2–rpoC2 pgcpir 035 should allow reproducible and credible discrimination between most Korean and American ginseng products, even though the roots of P. ginseng and P. quinquefolius can barely be distinguished from one another.
3.2. Application of the pgcpir 035 marker to commercial processed ginseng and red ginseng products
Authentication of commercial processed ginseng products is more difficult than that of fresh roots because products such as powder, shredded slices, pellets, liquid extracts, and tea look identical, even when they are made from different species (Fig. 2A). This facilitates the illegal practice of disguising American ginseng (P. quinquefolius) as P. ginseng in ginseng trade markets. To optimize the method for authentication of ginseng species in commercial products, we tested the ability of the pgcpir 035 marker to detect the original species used to make the processed products. First, we optimized the DNA extraction methods for various processed ginseng products based on the previous report [26]. PCR usually requires 10–50 ng/μL DNA, but only low amounts of DNA were extracted from the commercial ginseng products using conventional DNA isolation protocols or even commercial DNA extraction kits. However, we could amplify the pgcpir 035 marker using the trace amounts of DNA extracted from various processed ginseng products including red ginseng products because the marker that is targeted to cp genome DNA is over several hundred times greater than the number of nuclear genome copies in plant tissues [17].
Fig. 2.
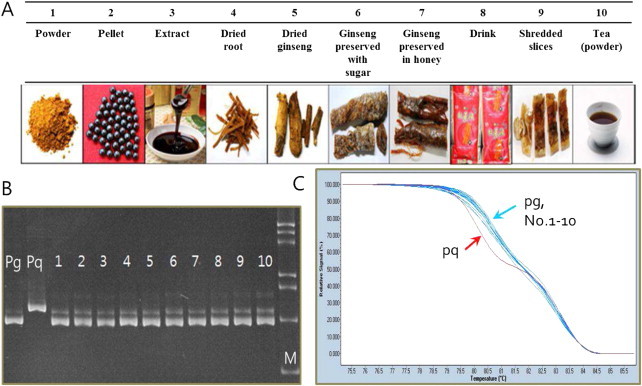
Authentication of processed ginseng products using the pgcpir 035 marker. (A) Various processed ginseng products purchased from Korean ginseng markets. (B) PAGE of PCR products using template DNA extracted from commercial products shown in (A). 1-10 indicate products in (A) and Pg and Pq indicate P. ginseng and P. quinquefolius standard DNA, respectively. M indicates 100 bp DNA ladder. (C) HRM analysis using the same samples as in (B).
We inspected 10 different ginseng or red ginseng products purchased from Korean ginseng markets (Fig. 2A). Although an additional nonspecific band was sometimes detected, all of the products were found to be made from P. ginseng (Fig. 2B). HRM analysis was also performed to confirm the PCR results, and again, different patterns were observed for the P. quinquefolius control DNA (Fig. 2C). HRM analysis can be utilized to detect not only small InDels, but also SNPs from PCR amplicons in several plant species [24,29–31]. Our HRM results were consistent with those of the AGE that all of the processed ginseng products were composed of P. ginseng.
3.3. Development of a species-specific dominant marker set for identification of P. ginseng and P. quinquefolius
Codominant markers such as pgcpir 035 are useful at the experimental level because they distinguish both genotypes at once. However, detection of codominant markers is dependent on high-resolution gel electrophoresis. Other markers derived from small InDel regions might be more difficult to detect than the large pgcpir 035 InDel. By contrast, species-specific dominant markers amplify only one species-unique band and can be detected by simple gel electrophoresis or by other DNA diagnostic kits. In addition, species-specific dominant markers can be useful for detection of intentional mixing between two species.
The pgcpir 030 CIS marker derived from the CIS between rbcL and accD shows an 8-bp InDel between P. ginseng and P. quinquefolius [24]. The 8-bp InDel is not easily distinguished by AGE. Therefore, we developed species-specific dominant markers using the sequences unique to either P. ginseng or P. quinquefolius (Fig. 3). The dominant markers include the SSP-PG-030-F2 and pgcpir 030R primer combination for amplifying a P. ginseng-unique dominant band (Pg-specific marker) and the SSP-PQ-030-F2 and pgcpir 030 R primer pair for amplifying a P. quinquefolius-unique dominant band (Pq-specific marker; Fig. 3B,C). These two primer sets reproducibly produced species-specific unique bands.
Fig. 3.
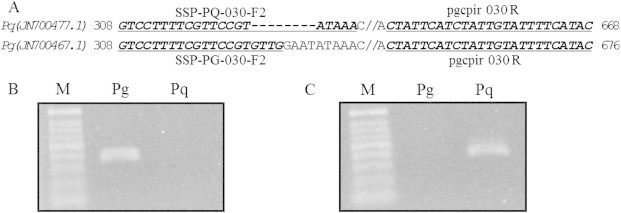
Development of species-specific dominant marker sets for the CIS of rbcL-accD. (A) Nucleotide sequence comparison of InDel regions between P. ginseng (Pg) and P. quinquefolius (Pq) and development of species-unique primers. In parentheses are the NCBI accession numbers. Underlined bold italic characters indicate primers. (B) Pg-unique 370-bp PCR band amplified using SSP-PG-030-F2 and pgcpir 030R. (C) Pq-unique 362-bp PCR product amplified using SSP-PQ-030-F2 and pgcpir 030R. M indicates 100 bp DNA ladder.
3.4. Inspection of primary processed ginseng products in Chinese ginseng markets
Many different products made from P. ginseng and P. quinquefolius are sold in Chinese ginseng markets (Fig. 4A). We purchased various forms of primary processed ginseng, such as dried root slices, dried flowers, flakes, dried ginseng, and powder, in which the original species was labeled as American ginseng (P. quinquefolius) or Korean ginseng (P. ginseng). Results using the codominant marker pgcpir 035 and the species-specific dominant marker sets were in agreement with regard to genotype and also coincided with the species names denoted on the product labels, suggesting that both markers are credible for evaluation of species (Fig. 4B,C). However, some products gave rise to bands for both species-specific markers, suggesting that Korean and American ginseng might be mixed during manufacturing or harvesting in some products (data not shown). Polymorphism of CIS is rarely identified among accessions in the same species [20,24,32,33], although a few CIS markers polymorphic in the same species were reported for Allium cepa, such as markers for identification of cytoplasmic male sterile genotypes among various onion accessions [34,35]. Therefore, although it is unlikely, we cannot preclude the possibility that an unrecognized variation among American ginseng accessions in the target regions might coincide with the region in Korean ginseng by chance. Inspection of more large collections and regular monitoring will be necessary to address this possibility.
Fig. 4.
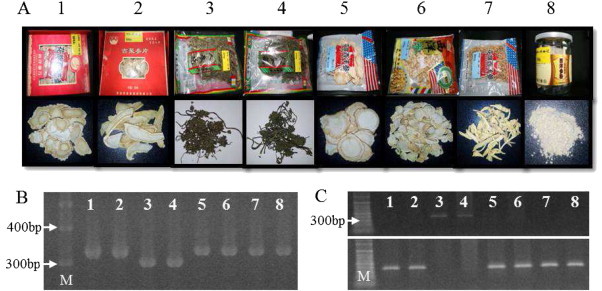
Authentication of Panax species in primary processed commercial ginseng products purchased from the Chinese market. (A) Various kinds of processed ginseng products: dried root slices (1, 2, 5, 6), dried roots (7), root powder (8), and dried flowers (3, 4). Each product denotes the original species on its label: P. ginseng (3 and 4) and P. quinquefolius (1, 2, 5, 6, 7, and 8). (B) Agarose gel electrophoresis of PCR products amplified using pgcpir 035 for each sample. (C) Agarose gel electrophoresis of PCR products amplified using Pg-specific primers (upper panel) and Pq-specific primers (lower panel) for each sample. M indicates 100 bp DNA ladder.
3.5. Detection of intentional mixing between Korean and American ginseng products
The above results show that the codominant pgcpir 035 DNA marker and species-specific dominant marker set can be successfully applied to identify the original species from fresh roots and various processed ginseng products. Codominant markers have been utilized to identify heterozygosity in individuals and mixing of samples in other species. We tested our markers for the detection of mixtures of the two ginseng species because intentional or unintentional mixing of the species could be common in the ginseng market, as our preliminary results suggested for the Chinese market. Therefore, we used both markers on samples of mixed DNA or tissues that included P. ginseng and P. quinquefolius in various ratios (Fig. 5). As expected, the codominant pgcpir 035 marker gave rise to various intensities of both bands that coincided with the mixing ratio. Mixtures of dried root slices containing <10% of the second species could be clearly identified using the codominant pgcpir 035 DNA marker (Fig. 5).
Fig. 5.
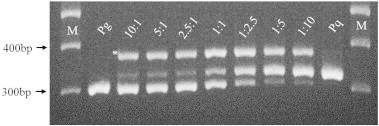
PCR products from proportional mixtures of P. ginseng (Pg) and P. quinquefolius (Pq) root slices. Dried root samples of P. ginseng and P. quinquefolius were mixed in the ratios of 10:1, 5:1, 2.5:1, 1:1, 1:2.5, 1:5, and 1:10, and DNA extracted from the mixed samples was used for PCR reactions using the co-dominant pgcpir035 marker. M indicates 100 bp DNA ladder. * The additional heteroduplex band that appeared in the mixed samples.
In addition to the species-unique bands, an additional band (* in Fig. 5) was always observed for the mixed samples. Multiple artifact bands can be produced during the amplification of simple sequence repeat sequences from heterozygous plants [36–38] and have been found in our previous studies of ginseng [14,15,39]. Based on specific reamplification of this band from multiple tested bands, we concluded that the artifact bands could be derived from the formation of heteroduplexes during PAGE analysis when more than two similar bands coexisted in the same PCR product [39]. Therefore, we consider the appearance of the heteroduplex artifact bands as a signature for the mixture of the two species that can be beneficial for authentication or identification of mixtures in large volumes of processed ginseng samples [40].
The InDel-based codominant marker has limitations in high-throughput analysis to detect mixtures of the species because genotyping with the marker depends on high-resolution gel electrophoresis. Even though HRM can detect both individual genotypes without gel electrophoresis, the method has limited application to mixed samples [24,29–31]. To address this, we tested the ability of the species-specific markers to identify mixtures. The Pg-specific marker could reveal the presence of P. ginseng at a 1% level in the American ginseng products (Fig. 6A). Conversely, the Pq-specific marker could identify down to 1% P. quinquefolius in P. ginseng products (Fig. 6B).
Fig. 6.
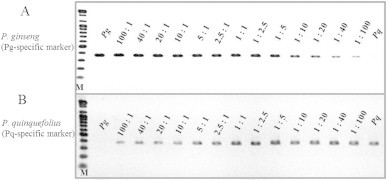
PCR results of using the species-specific dominant marker set on proportionally mixed DNA of P. ginseng and P. quinquefolius. Gel electrophoresis of PCR products amplified using Pg-specific primers (A) and Pq-specific primers (B) for proportionally mixed DNA of P. ginseng and P. quinquefolius. The same concentrations of both DNAs were mixed in 100:1 ∼ 1:100 ratios and the same amount of mixed DNA was used for PCR. The mixing ratio is denoted on each lane. M indicates 100 bp ladder.
Quantitative PCR with the same primer set was consistent with the AGE results, and revealed quantitative mixing ratios down to 1% (Fig. 7). The quantitative PCR method reports the quantitative mixing ratio without requiring gel electrophoresis, which is an advantage for mass and high-throughput analysis for monitoring mislabeling or false trading in commercial ginseng products [41]. These markers will be useful to prevent the illegal distribution or intentional mixing of American and Korean ginseng in the ginseng market.
Fig. 7.
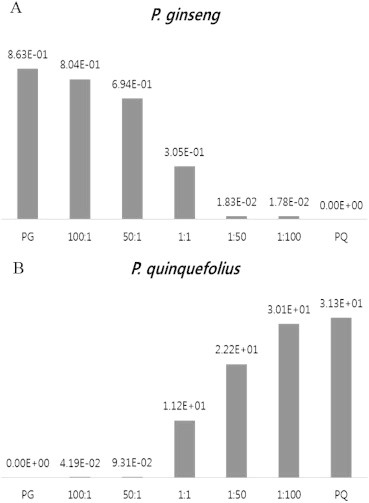
Quantification of intentional mixing of DNA from P. ginseng and P. quinquefolius by qPCR using species-specific markers. Five mixtures were quantified by qPCR using Pg-specific marker (A) and Pq-specific marker (B). The ratio of mixed DNA is indicated and the quantification of each qPCR is denoted on the bars.
Korean and American ginseng are important herbal medicines and each species has some unique medicinal functions [42,43]. Applying the evaluation system we have developed here will promote and increase the value of Korean ginseng as well as American ginseng in Korea and worldwide, by allowing consumers to be confident in the contents of commercial ginseng products.
Conflicts of interest
All authors declare no conflicts of interest.
Acknowledgments
This study was supported by the Next-Generation BioGreen21 Program (No. PJ008202), Rural Development Administration, Korea.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Park M.J., Kim M.K., In J.G., Yang D.C. Molecular identification of Korean ginseng by amplification refractory mutation system-PCR. Food Res Int. 2006;39:568–574. [Google Scholar]
- 2.Yun T.K. Brief introduction of Panax ginseng CA Meyer. J Korean Med Sci. 2001;16:S3–S5. doi: 10.3346/jkms.2001.16.S.S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., Show P., Sze C., Wang Z., Tong Y. Molecular authentication of Chinese herbal materials. J Food Drug Analysis. 2007;15:1–9. [Google Scholar]
- 4.Ho I., Leung F. Isolation and characterization of repetitive DNA sequences from Panax ginseng. Mol Genet Genomics. 2002;266:951–961. doi: 10.1007/s00438-001-0617-6. [DOI] [PubMed] [Google Scholar]
- 5.Liu J., Wang S., Liu H., Yang L., Nan G. Stimulatory effect of saponin from Panax ginseng on immune function of lymphocytes in the elderly. Mech Ageing Dev. 1995;83:43–53. doi: 10.1016/0047-6374(95)01618-a. [DOI] [PubMed] [Google Scholar]
- 6.Shin H.R., Kim J.Y., Yun T.K., Morgan G., Vainio H. The cancer-preventive potential of Panax ginseng: a review of human and experimental evidence. Cancer Causes Control. 2000;11:565–576. doi: 10.1023/a:1008980200583. [DOI] [PubMed] [Google Scholar]
- 7.Kitts D.D., Hu C. Efficacy and safety of ginseng. Public Health Nutr. 2000;3:473–485. doi: 10.1017/s1368980000000550. [DOI] [PubMed] [Google Scholar]
- 8.Nam M.H., Kim S.I., Liu J.R., Yang D.C., Lim Y.P., Kwon K.H., Yoo J.S., Park Y.M. Proteomic analysis of Korean ginseng (Panax ginseng CA Meyer) J Chromatogr B. 2005;815:147–155. doi: 10.1016/j.jchromb.2004.10.063. [DOI] [PubMed] [Google Scholar]
- 9.Jung J.D., Park H.W., Hahn Y., Hur C.G., In D.S., Chung H.J., Liu J.R., Choi D.W. Discovery of genes for ginsenoside biosynthesis by analysis of ginseng expressed sequence tags. Plant Cell Rep. 2003;22:224–230. doi: 10.1007/s00299-003-0678-6. [DOI] [PubMed] [Google Scholar]
- 10.Chan T.W.D., But P.P.H., Cheng S.W., Kwok I.M.Y., Lau F.W., Xu H.X. Differentiation and authentication of Panax ginseng, Panax quinquefolius, and ginseng products by using HPLC/MS. Anal Chem. 2000;72:1281–1287. doi: 10.1021/ac990819z. [DOI] [PubMed] [Google Scholar]
- 11.Wang X., Sakuma T., Asafu-Adjaye E., Shiu G.K. Determination of ginsenosides in plant extracts from Panax ginseng and Panax quinquefolius L. by LC/MS/MS. Anal Chem. 1999;71:1579–1584. doi: 10.1021/ac980890p. [DOI] [PubMed] [Google Scholar]
- 12.Gostimsky S.A., Kokaeva Z.G., Konovalov F.A. Studying plant genome variation using molecular markers. Russ J Genet. 2005;41:378–388. doi: 10.1007/s11177-005-0101-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma K.H., Dixit A., Kim Y.C., Lee D.Y., Kim T.S., Cho E.G., Park Y.J. Development and characterization of new microsatellite markers for ginseng (Panax ginseng CA Meyer) Conserv Genet. 2007;8:1507–1509. [Google Scholar]
- 14.Choi H.I., Kim N.H., Kim J.H., Choi B.S., Ahn I.O., Lee J.S., Yang T.J. Development of reproducible EST-derived SSR markers and assessment of genetic diversity in Panax ginseng cultivars and related species. J Ginseng Res. 2011;35:399–412. doi: 10.5142/jgr.2011.35.4.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim N.H., Choi H.I., Ahn I.O., Yang T.J. EST-SSR marker sets for practical authentication of all nine registered ginseng cultivars in Korea. J Ginseng Res. 2012;36:298–307. doi: 10.5142/jgr.2012.36.3.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reboud X., Zeyl C. Organelle inheritance in plants. Heredity. 1994;72:132–140. [Google Scholar]
- 17.Pyke K.A. Plastid division and development. Plant Cell. 1999;11:549–556. doi: 10.1105/tpc.11.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall H.D., Newton C., Ritland K. Sequence-repeat polymorphisms exhibit the signature of recombination in lodgepole pine chloroplast DNA. Mol Biol Evol. 2001;18:2136–2138. doi: 10.1093/oxfordjournals.molbev.a003757. [DOI] [PubMed] [Google Scholar]
- 19.Harris S.A., Ingram R. Chloroplast DNA and biosystematics: the effects of intraspecific diversity and plastid transmission. Taxon. 1991;40:393–412. [Google Scholar]
- 20.Wolfe A.D., Randle C.P. Recombination, heteroplasmy, haplotype polymorphism, and paralogy in plastid genes: implications for plant molecular systematics. Syst Bot. 2004;29:1011–1020. [Google Scholar]
- 21.McCauley D.E. The use of chloroplast DNA polymorphism in studies of gene flow in plants. Trends Ecol Evol. 1995;10:198–202. doi: 10.1016/s0169-5347(00)89052-7. [DOI] [PubMed] [Google Scholar]
- 22.Britten R.J., Rowen L., Williams J., Cameron R.A. Majority of divergence between closely related DNA samples is due to indels. Proc Natl Acad Sci USA. 2003;100:4661–4665. doi: 10.1073/pnas.0330964100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K., Lee H. Complete chloroplast genome sequences from Korean ginseng (Panaxs chinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004;11:247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- 24.Kim J.H., Jung J.Y., Choi H.I., Kim N.H., Park J.Y., Lee Y., Yang T.J. Diversity and evolution of major Panax species revealed by scanning the entire chloroplast intergenic spacer sequences. Genet Resour Crop Evol. 2013;60:413–425. [Google Scholar]
- 25.Allen G.C., Flores-Vergara M.A., Krasynanski S., Kumar S., Thompson W.F. A modified protocol for rapid DNA isolation from plant tissues using acetyltrimethylammonium bromide. Nat Protoc. 2006;1:2320–2325. doi: 10.1038/nprot.2006.384. [DOI] [PubMed] [Google Scholar]
- 26.Mihalov J.J., Marderosian A.D., Pierce J.C. DNA Identification of commercial ginseng samples. J Agric Food Chem. 2000;48:3744–3752. doi: 10.1021/jf000011b. [DOI] [PubMed] [Google Scholar]
- 27.Barker N.P., Galley C., Verboom G.A., Mafa P., Gilbert M., Linder H.P. The phylogeny of the austral grass subfamily Danthonioideae: evidence from multiple data sets. Pl Syst Evol. 2007;264:135–156. [Google Scholar]
- 28.Ku C., Chung W.C., Chen L.L., Kuo C.H. The complete plastid genome sequence of Madagascar periwinkle Catharanthus roseus (L.) G. Don: plastid genome evolution, molecular marker identification, and phylogenetic implications in asterids. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068518. e68518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arthofer W., Steiner F.M., Schlick-Steiner B.C. Rapid and cost-effective screening of newly identified microsatellite loci by high-resolution melting analysis. Mol Genet Genomics. 2011;286:225–235. doi: 10.1007/s00438-011-0641-0. [DOI] [PubMed] [Google Scholar]
- 30.Botticella E., Sestili F., Hernandez-Lopez A., Phillips A., Lafiandra D. High resolution melting analysis for the detection of EMS induced mutations in wheat Sbella genes. BMC Plant Biol. 2011;11:156. doi: 10.1186/1471-2229-11-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montgomery J., Wittwer C.T., Palais R., Zhou L. Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis. Nat Protoc. 2007;2:59–66. doi: 10.1038/nprot.2007.10. [DOI] [PubMed] [Google Scholar]
- 32.Gao L., Zhou Y., Wang Z.W., Su Y.J., Wang T. Evolution of the rpoB-psbZ region in fern plastid genomes: notable structural rearrangements and highly variable intergenic spacers. BMC Plant Biol. 2011;11:64. doi: 10.1186/1471-2229-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y.J., Ma P.F., Li D.Z. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae) PLoS One. 2011;6 doi: 10.1371/journal.pone.0020596. e20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho K.S., Yang T.J., Hong S.Y., Kwon Y.S., Woo J.G., Park H.G. Determination of cytoplasmic male sterile factors in onion plants (Allium cepa L.) using PCR-RFLP and SNP markers. Mol Cells. 2006;21:411–417. [PubMed] [Google Scholar]
- 35.Kim S., Lee Y.P., Lim H., Ahn Y.S., Sung S.K. Identification of highly variable chloroplast sequences and development of cpDNA based molecular markers that distinguish four cytoplasm types in radish (Raphanus sativus L.) Theor Appl Genet. 2009;119:189–198. doi: 10.1007/s00122-009-1028-z. [DOI] [PubMed] [Google Scholar]
- 36.Walsh P.S., Fildes N.J., Reynolds R. Sequence analysis and characterization of stutter products at the tetranucleotide repeat locus vWA. Nucleic Acids Res. 1996;24:2807–2812. doi: 10.1093/nar/24.14.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernando P., Evans B.J., Morales J.C., Melnick D.J. Electrophoresis artefacts - a previously unrecognized cause of error in microsatellite analysis. Mol Ecol Notes. 2001;1:325–328. [Google Scholar]
- 38.Bovo D., Rugge M., Shiao Y.H. Origin of spurious multiple bands in the amplification of microsatellite sequences. Mol Path. 1999;52:50–51. doi: 10.1136/mp.52.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim N.H., Choi H.I., Kim K.H., Jang W., Yang T.J. Evidence of genome duplication revealed by sequence analysis of multi loci EST-SSR bands in Panax ginseng CA Meyer. J Ginseng Res. 2014;38:130–135. doi: 10.1016/j.jgr.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J., Zhang X., Wang T., Li Z., Guan G., Hong Y. Efficient detection, quantification and enrichment of subtle allelic alterations. DNA Res. 2012;19:423–433. doi: 10.1093/dnares/dss023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovatt A. Applications of quantitative PCR in the biosafety and genetic stability assessment of biotechnology products. J Biotechnol. 2002;82:279–300. doi: 10.1016/S1389-0352(01)00043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baeg I.H., So S.H. The world ginseng market and the ginseng (Korea) J Ginseng Res. 2013;37:1–7. doi: 10.5142/jgr.2013.37.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Punja Z.K. American ginseng: research developments, opportunities, and challenges. J Ginseng Res. 2011;35:368–374. doi: 10.5142/jgr.2011.35.3.368. [DOI] [PMC free article] [PubMed] [Google Scholar]


