Abstract
Background
The hard clam or northern quahog, Mercenaria mercenaria, is one of the most valuable seafood products in the United States representing the first marine resource in some Northeastern states. Severe episodes of hard clam mortality have been consistently associated with infections caused by a thraustochytrid parasite called Quahog Parasite Unknown (QPX). QPX is considered as a cold/temperate water organism since the disease occurs only in the coastal waters of the northwestern Atlantic Ocean from Maritime Canada to Virginia. High disease development at cold temperatures was also confirmed in laboratory studies and is thought to be caused predominantly by immunosuppression of the clam host even though the effect of temperature on QPX virulence has not been fully investigated. In this study, the QPX transcriptome was sequenced using Roche 454 technology to better characterize this microbe and initiate research on the molecular basis of QPX virulence towards hard clams.
Results
Close to 18,000 transcriptomic sequences were generated and functionally annotated. Results revealed a wide array of QPX putative virulence factors including a variety of peptidases, antioxidant enzymes, and proteins involved in extracellular mucus production and other secretory proteins potentially involved in interactions with the clam host. Furthermore, a 15 K oligonucleotide array was constructed and used to investigate the effect of temperature on QPX fitness and virulence factors. Results identified a set of QPX molecular chaperones that could explain its adaptation to cold temperatures. Finally, several virulence-related factors were up-regulated at low temperature providing molecular targets for further investigations of increased QPX pathogenicity in cold water conditions.
Conclusions
This is one of the first studies to characterize the transcriptome of a parasitic labyrinthulid, offering new insights into the molecular bases of the pathogenicity of members of this group. Results from the oligoarray study demonstrated the ability of QPX to cope with a wide range of environmental temperatures, including those considered to be suboptimal for clam immunity (low temperature) providing a mechanistic scenario for disease distribution in the field and for high disease prevalence and intensity at low temperature. These results will serve as basis for studies aimed at a better characterization of specific putative virulence factors.
Keywords: Quahog Parasite Unknown (QPX), Virulence factors, Oligoarrays, Gene expression
Background
QPX (Quahog Parasite Unknown) is a unicellular eukaryote that infects hard clam (Mercenaria mercenaria) populations along the northeastern coast of North America [1-4]. QPX is thought to be an opportunistic pathogen occurring at low abundance in coastal waters, sediments and organic debris [5-7], but occasionally becoming invasive to clams exhibiting lower resistance [8-10], suppressed immunity and/or during unfavorable environmental conditions [11-13]. QPX is an undescribed species belonging to thraustochytrids in the phylogenetic group Labyrinthulomycetes[14-16]. Labyrinthulids (slime nets) are composed of ecologically important but greatly understudied marine protists closely related to water molds (oomycetes), diatoms, golden and brown algae, all of which collectively form a monophyletic cluster named stramenopiles [17].
Labyrinthulids are marine saprophytes associated with marine algal and animal detritus, but some are found as parasitic, commensal and mutualistic species [18]. Labyrinthula zostera is the most known labyrinthulid parasite which has caused extensive devastation of marine seagrasses worldwide [19]. In the past, several labyrinthulids were reported to cause disease in mollusks including abalones, squids and nudibranchs [20-22]. In recent years, new labyrinthulid species have been emerging as parasites in other marine invertebrates [23-26]. All of these parasitic labyrinthulids lack any significant genomic or transcriptomic sequence information, making it difficult to determine how they evolved their parasitic abilities, and what the virulence mechanisms are which enable them to invade and survive in host tissues.
Even though significant clam mortality has been associated with QPX disease since the early 1950’s [3], many aspects of the disease remain unexplored and understudied. Little progress has been made to understand QPX basic cell biology, including its life stages, nutritional requirements, and factors mediating its virulence toward clams. As is often the case for opportunistic infections, QPX disease occurrence and development is significantly dependent upon external environmental conditions affecting both the parasite and the host. QPX’s natural geographic range of distribution shows that this parasite can be considered as a temperate/cold species, as it has not been described in the relatively warm waters south of Virginia. Interestingly, QPX growth in vitro is optimal between 20°C and 23°C, and growth decreases at both colder (below 8°C) and warmer (above 27°C) temperatures [27]. The major impact of temperature on disease development is reflected in the seasonal fluctuations of disease prevalence in clams, with peaks ranging from fall in MA to early summer and spring in NY and VA, respectively [1-4]. On the other hand, experimental transmission studies showed maximal disease development at low water temperatures, which has been attributed to the suppression of clam immunity [11,12] and it is hypothesized that immunosuppression is a major factor controlling QPX disease development. In fact, the effect of temperature on clam immunity was demonstrated [12] providing a possible mechanism for increased disease development at cold temperature. On the contrary, very little is known about the effect of temperature on QPX physiology, fitness or its virulence characteristics and ability to cause disease. Very recently, the partial sequencing of the QPX genome and two transcriptomes of the parasite provided some sequence resources for QPX investigations [28]. The present study was also conducted to gather transcriptomic sequence information for a better understanding of QPX biology and exploration of its virulence factors. For that purpose, transcriptome sequences of QPX were generated, assembled and annotated and then used to build a custom designed 15 K oligonucleotide array that was subsequently used to investigate QPX molecular response to temperature conditions known to regulate disease development [11,12]. Findings were discussed with a focus on the characterization of possible links between temperature and the expression profiles of virulence-related factors.
Results and discussion
Characterization of QPX transcriptome
In the present study, we sequenced, assembled and annotated the transcriptome of quahog parasite unknown, the thraustochytrid protist that causes disease and mortality in an important bivalve species, the hard clam Mercenaria mercenaria. The cDNA sequence library was generated from QPX cells (isolated from Raritan Bay, New York) grown in a variety of culture media to obtain satisfactory coverage for transcripts that may have different expression profiles under various culture conditions. A total of 490,726 sequence reads (average length of 215 bp) were generated by the 454 GS-FLX Titanium platform. Quality and length filtering yielded 223,652 high-quality reads that were assembled using MIRA. The resulting QPX transcriptome library consists of 17,934 transcript sequences including contigs (continuous sequences, 14,636), singletons (single copy sequences, 1,636), and long repetitive contigs (lrc, 1,662) with sequence length ranging from 40 bp to over 2,700 bp and a total of 7,111,495 bp in assembled library. Sequences longer than 200 bp have been deposited into DDBJ/EMBL/GenBank under the TSA (Transcript Shotgun Assembly) accession number: GALJ00000000, BioProject accession number: PRJNA207819, BioSample accession number: SAMN02194536, and Sequence Read Archive accession number: SRR900258. The number of assembled bases produced in this study represent about 20% of the genomic sequence assembly (34,620,595 bp assembled into 21,280 contigs) recently produced for QPX [28] which represents a smaller transcriptome coverage than that of other stramenopiles with fully sequenced genomes such as Thalassiosira pseudonana for which about 32% of the genome is represented by coding sequences [29].
Blast2GO server was used to find homologous sequences to QPX transcripts in publicly available databases and to assign functional information to these transcripts. A total of 7,093 transcripts were found to have positive blastx homology matches (E < 10-3) to other sequences in NCBI database (Additional file 1). The top four species showing the most similarity to QPX sequences were the oomycete Phytophthora infestans, the brown alga Ectocarpus siliculosus, and the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana, all of which are stramenopilan species, thus confirming QPX affiliation to this phylogenetic group [17]. A total of 4,644 sequences were identified to have a match to at least one conserved protein domain in the InterPro database (Additional file 1) and 5,764 sequences were assigned at least one functional annotation term based on the Gene Ontology vocabulary database with a cut-off E-value of 10-3 (Additional file 1). Summary of the GO annotation is present in Figure 1. The level-2 functional annotation of QPX transcriptome returned 5,768 and 5,459 gene ontology phrases associated with the biological process (Figure 1A) and molecular function (Figure 1B) categories, respectively. The combined level 2 and 3 functional annotation of QPX transcripts resulted in 3,081 cellular component gene ontologies (Figure 1C).
Figure 1.
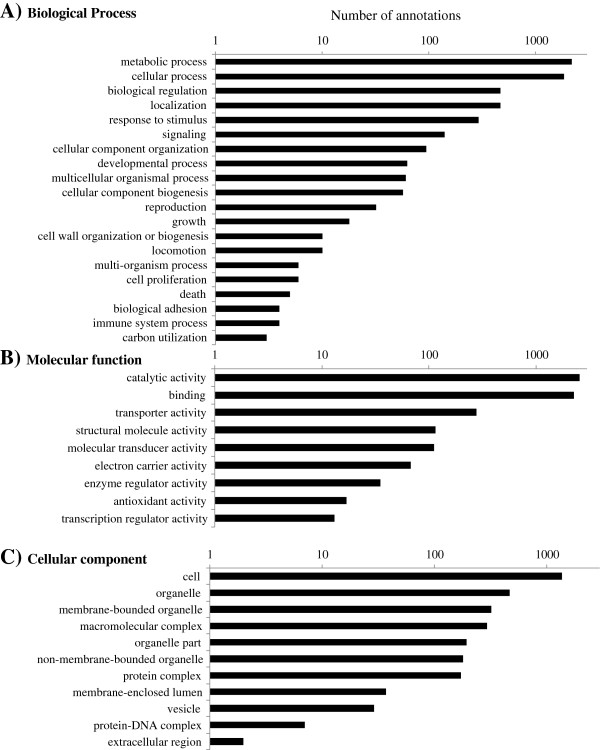
Gene Ontology (GO) annotations of the QPX transcriptome. GO phrases identified by the Blast2GO search tool in QPX transcriptome sequence library A) a total of 5,768 level-2 annotations within the biological process ontology; B) 5,459 molecular function level-2 annotations C) combined level-2 and level-3 annotations (3,081) within the cellular component ontology.
Putative virulence factors in QPX transcriptome
The primary goal of generating a transcriptome library for QPX was to provide resources for future studies on QPX virulence factors. The GO terms are commonly used as clues in the discovery of molecules involved in the infection process, pathogenicity and host-pathogen interactions [30]. Likely the most destructive virulence molecules are hydrolytic enzymes produced by pathogenic organisms to destroy host-derived biopolymers (proteins, polysaccharides and lipids) and to invade and colonize their hosts [31]. The annotation carried out in this study revealed 777 QPX transcripts encoding hydrolytic enzymes including glycosyl hydrolases (pectinase, amylases, cellulases, xylanases, chitnases, glucanases), peptidases, lipases and phospholipases (Table 1, Additional file 1), all of which have been found to be involved in pathogenicity of many microorganisms [32,33]. For example, chitinases from the oomycete pathogen Aphanomyces astaci of freshwater crayfish have been investigated as potential virulence factors [34]. Since the molluscan shell is partially composed of chitin [35,36] and QPX cells have been previously found associated with empty molluskan shells [5], QPX’s ability to digest shell material represents a venue for future investigations. Similarly, phospholipases are important enzymes utilized by many pathogenic organisms, including bacteria, fungi and protozoans, to hydrolyze cell membranes of host cells [37-41].
Table 1.
Examples of gene ontology terms commonly associated with virulence identified in QPX transcriptome; F: molecular function, P: biological process
| Gene Ontology term | GO category | GO level | GO ID | Seqs no. |
|---|---|---|---|---|
|
Degradation of biopolymers hydrolase activity |
|
|
||
|
Hydrolase activity |
F |
3 |
GO:0016787 |
777 |
| Nuclease activity |
F |
5 |
GO:0004518 |
29 |
| Phosphatase activity |
F |
6 |
GO:0016791 |
27 |
| Lipase activity |
F |
5 |
GO:0016298 |
9 |
| Phospholipase activity |
F |
6 |
GO:0004620 |
4 |
| Chitinase activity |
F |
6 |
GO:0004568 |
3 |
| Glucosidase activity |
F |
6 |
GO:0015926 |
2 |
| Amylase activity |
F |
6 |
GO:0016160 |
1 |
| Galactosidase activity |
F |
6 |
GO:0015925 |
1 |
|
Peptidase activity |
F |
4 |
GO:0008233 |
203 |
| Metallopeptidase activity |
F |
6 |
GO:0008237 |
47 |
| Serine-type peptidase activity |
F |
5 |
GO:0008236 |
45 |
| Cysteine-type peptidase activity |
F |
6 |
GO:0008234 |
35 |
| Metalloendopeptidase activity |
F |
7 |
GO:0004222 |
30 |
| Aspartic-type endopeptidase activity |
F |
7 |
GO:0004190 |
10 |
| Threonine-type peptidase activity |
F |
6 |
GO:0070003 |
7 |
| Peptidase inhibitor activity |
F |
4 |
GO:0030414 |
9 |
|
Cell homeostasis and protection against oxidative stress |
|
|
||
|
Antioxidant activity |
F |
2 |
GO:0016209 |
17 |
| Peroxidase activity |
F |
3 |
GO:0004601 |
10 |
| Superoxide metabolic process |
P |
5 |
GO:0006801 |
5 |
| Catalase activity |
F |
4 |
GO:0004096 |
4 |
| Thioredoxin-disulfide reductase activity |
F |
3 |
GO:0004791 |
2 |
| Peroxiredoxin activity |
F |
5 |
GO:0051920 |
2 |
| Thioredoxin peroxidase activity |
F |
4 |
GO:0008379 |
1 |
| Glutathione peroxidase activity |
F |
4 |
GO:0004602 |
1 |
|
Secretion and polysaccharide production |
|
|
||
| Polysaccharide metabolic process |
P |
4 |
GO:0005976 |
21 |
| Polysaccharide biosynthetic process |
P |
6 |
GO:0000271 |
13 |
| Lipopolysaccharide biosynthetic process |
P |
6 |
GO:0009103 |
3 |
| Extracellular polysaccharide metabolic process |
P |
6 |
GO:0046379 |
2 |
| Secretion |
P |
5 |
GO:0046903 |
8 |
| Exocytosis |
P |
6 |
GO:0006887 |
4 |
| Vesicle docking involved in exocytosis |
P |
5 |
GO:0006904 |
3 |
| Vesicle-mediated transport |
P |
5 |
GO:0016192 |
38 |
|
Adhesion and recognition |
|
|
||
| Cell adhesion |
P |
3 |
GO:0007155 |
4 |
| Receptor activity |
F |
4 |
GO:0004872 |
34 |
| G-protein coupled receptor protein signaling pathway |
P |
5 |
GO:0007186 |
18 |
| Cell communication | P | 3 | GO:0007154 | 13 |
Peptidases can degrade host protein material such as collagen, fibrin and actin in the connective tissue of the extracellular matrix, but are also cable of destroying immunoglobulins [42-45]. In this study, we found over 200 transcripts annotated to encode peptidases belonging to five major peptidase clans: serine (45), cysteine (35), metallo (47), aspartate (10), and threonine peptidases (7) (Table 1). Within all peptidases, QPX was found to possess several peptidases commonly associated with pathogenicity. These include papain-type cysteine peptidases (IPR000169; IPR000668, Additional file 1) also known as cathepsins, which are the dominant peptidases in pathogenic protozoans [46]. Further, QPX possesses serine peptidases in the S8 family known as subtilisins or subtilases (IPR000209; IPR015500, Additional file 1), which are universal virulence factors found in pathogenic bacteria [47,48], protozoa [49,50], and fungi. QPX also possesses sequences encoding peptidases found to be virulence factors in other pathogens [44,51-54] including aspartic peptidases (family A1), serine peptidases (families S1, IPR001254; S9, IPR001375; and S10, IPR001563) and metallopeptidases (families M16, IPR011237; M28 and M35) (Additional file 1). Based on the sequence information, QPX is expected to use hydrolytic enzymes, including peptidases, to break down extracellular material of clam tissue to be able to propagate throughout the host body. In situ observation of degraded clam tissues surrounding QPX cells [8,9,48] indirectly supports the involvement of peptidases in the pathogenesis of this parasite.
QPX infection inside the clam tissue leads to an inflammatory response of clam hemocytes aggregating around QPX cells. It is also known that the hemocytes of bivalves, including M. mercenaria, produce reactive oxygen species (ROS) to combat invading organisms [55]. To survive the clam defense, QPX would be expected to produce antioxidant molecules which can neutralize the toxic effect of ROS. In the generated QPX transcriptome library, seventeen sequences were identified to be involved in antioxidant activity (GO:0016209, Table 1) including catalases (IPR002226), superoxide dismutases (IPR001424, IPR001189), thioredoxin peroxidases (IPR012336, IPR005746), glutathione peroxidases (IPR000889, IPR002109), and ascorbate peroxidases (IPR010255 IPR002207) (Additional file 1). All of these antioxidant molecules have been shown to be effective against endogenous (metabolism related) and exogenous (from host defense) ROS species and are essential for resistance to oxidative stress in many pathogenic protozoans during disease development [56,57]. QPX antioxidant armor suggests that the pathogen is prepared to overcome M. mercenaria hemocyte-derived ROS, and facilitate the establishment of an infection.
During infection, molecules that play the most important roles are the ones on the interface between the host cell or its extracellular matrix and the pathogen cell. These molecules are involved in cell recognition, cell adhesion and cell communication. The QPX library allowed the identification of 34 sequences annotated to molecular function term receptor activity (GO:0004872), 18 linked to G-protein coupled receptor protein signaling pathways (GO:0007186), 13 transcripts involved in cell communication (GO:0007154), and 4 transcripts related to cell adhesion (GO:0007155) (Table 1). These molecules are excellent targets for future research studies on the capabilities of QPX to anchor itself to the extracellular matrix within clam tissue. For example, some adhesive and ligand binding molecules of plant and animal pathogens are related to thrombospondin [58,59], integrin [59,60] and lectins [61-66]. Our results show that the QPX transcriptome contains transcripts with homology to lectins (qpx_c8760, IPR008985; qpx_c13004, qpx_c14444), fibronectin-related proteins (qpx_c14678, IPR003961 and qpx_c7827, IPR008957), integrins (qpx_lrc16604, qpx_lrc4391, qpx_lrc9075), and thrombospodin-related molecules (qpx_c12015) (Additional file 1).
QPX oligoarray gene expression profiles in response to temperature and QPX adaptation to temperature changes
The gene expression profiles of QPX cultivated at four different temperatures (27°C, 23°C, 13°C, 10°C) were investigated using 15 K 60-mer oligonucleotide arrays. A total of 1,580 transcripts were differentially expressed (DE, at least 1.5 fold in conjunction to ANOVA p < 0.01) in response to temperature changes (Figure 2, Additional file 2). To confirm gene expression patterns obtained by the oligoarray analysis, transcription levels of seven different QPX peptidases (four serine and three cysteine peptidases: S8-1, qpx_c765; S8-2, qpx_c1822; S8-3, qpx_c2550; S01B, qpx_c674; C1A-1, qpx_c534; C1A-2, qpx_c5487; C1A-3, qpx_c3110; Table 2) at four different temperatures were examined using quantitative real time PCR. Using qPCR method, the mRNA expression patterns of four of the seven examined peptidases altered by the temperature conditions were statistically significant (ANOVA, p < 0.001) and the data was significant for six out of the seven peptidases based on oligoarrays results (Figure 3). The comparison of the relative transcript levels determined by the two methods, oligoarray and qPCR resulted in a strong statistically significant correlation (Pearson, r = 0.86).
Figure 2.
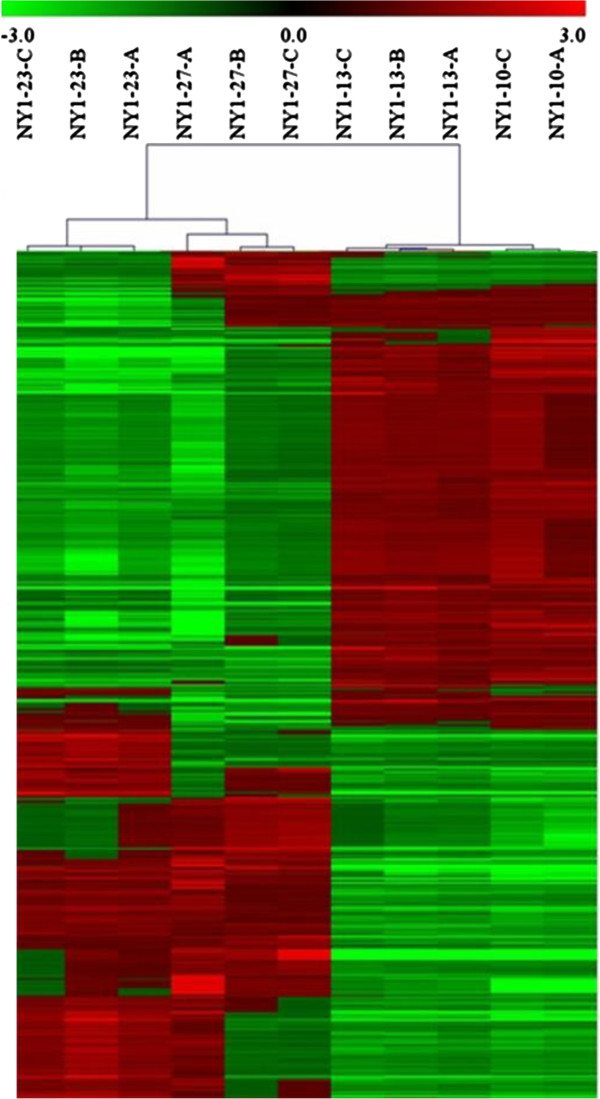
Heat map of differentially expressed QPX transcripts. Hierarchical clustering of 1,580 QPX transcripts (Pearson’s correlation centered) identified to be differentially expressed at four temperature treatments by oligoarrays. The clustering of samples within each treatment (27°C, 23°C, 13°C, n = 3 and 10°C, n = 2) highlights the reproducibility of the assay.
Table 2.
Sequence information for transcripts selected for the quantitative PCR validation of oligoarray data
| Sequence name | Sequence ID | Sequence description | Primer sequences |
Top blast match |
||
|---|---|---|---|---|---|---|
| Acc. no. | E-value | |||||
| C1A-1 |
qpx_c534 |
Cysteine peptidase |
F: |
ACGGCAATGTTACCGAAGAGGCTA |
XP_002507788 |
1.6E-48 |
| |
|
R: |
TAGTTATCCAATGGGCCCGCGTTA |
|
|
|
| C1A-2 |
qpx_c5487 |
Cysteine peptidase |
F: |
ACTGGAGCAAGAAGGGAGCAGTAA |
CBJ28832 |
1.9E-29 |
| |
|
R: |
AAGACCACCTGTGGTGGAGAAACT |
|
|
|
| C1A-3 |
qpx_c3110 |
Cysteine peptidase |
F: |
TGCAGGTCGTCGTTGCTTTAGTCT |
EAY93080 |
3.8E-14 |
| |
|
R: |
TAGCCAACGATTGAAACTGCGTGG |
|
|
|
| S8-1 |
qpx_c765 |
Serine peptidase |
F: |
TCGTGCTGGACACATAGTTGTCGT |
ABI79453 |
6.3E-30 |
| |
|
R: |
TATCGGTGGCTCCAACGCTTATCA |
|
|
|
| S8-2 |
qpx_c1822 |
Serine peptidase |
F: |
TATGGCTACTCCATTTGTCGCTGG |
ABI79453 |
1.9E-29 |
| |
|
R: |
ACGAGCAAATTGGGAGATTCGTGC |
|
|
|
| S8-3 |
qpx_c2550 |
Serine peptidase |
F: |
AAGAGCGGTTGGGAATATGGGAGT |
ABI79453 |
7.5E-51 |
| |
|
R: |
AACAACAAGCATGCCCTCTTCTGC |
|
|
|
| S01B |
qpx_c674 |
Serine peptidase |
F: |
AGGTCTACGTATGGCTCCACTTCA |
XP_002906640 |
1.7E-22 |
| |
|
R: |
TTGAGCCCACATTTCTCAGCAGGA |
|
|
|
| Actin |
qpx_c116 |
Actin |
F: |
TGAAGATCTTGACCGAGCGTGGTT |
ABC85743 |
1.0E-173 |
| R: | AGCGGTCTTCATCTCCTGGTCAAA | |||||
Figure 3.
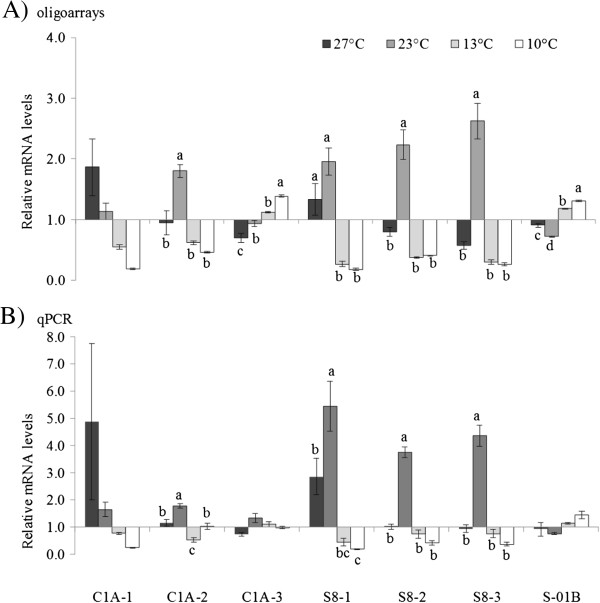
Validation of oligoarray results. Relative mRNA expression levels (mean ± std error, n = 3) of six peptidases in QPX grown at four different temperatures as determined by oligoarrays (A) and quantitative RT-PCR (B). Significant differences (ANOVA, p < 0.01, Tukey post-hoc test) between treatments for each gene are indicated by letter labels.
Among the 1,580 DE transcripts, only 617 had known predicted biological or molecular function (Additional file 2) and a subset of these is presented in Table 3. The GO annotations, as well as the information on the best blast hit result for the remaining sequences are given in Additional file 2. These 617 sequences were grouped into 17 functional categories (Figure 4) to identify biological processes differentially expressed under different temperature treatments. In general, the highest number of differentially expressed genes included those involved in cell metabolic processes such as amino acid, carbohydrate, protein, fatty acid and lipid metabolism (241 transcripts in total, Figure 4). This pattern, however is biased by the fact that public databases are skewed toward most conserved genes which tend to be genes belonging to the metabolic pathways. More than half of the QPX genes regulated in response to temperature have unknown molecular functions (61%, Additional file 2).
Table 3.
Selected differentially expressed genes in QPX exposed to four temperature treatments
|
Sequence description |
Mean fold changed |
Seq. ID number |
Top BLAST Hit |
||||
|---|---|---|---|---|---|---|---|
| 27°C | 23°C | 13°C | 10°C | Accession no. | E-value | ||
|
Protein stability |
|
|
|
|
|
|
|
| dnaj protein 1 |
-1.5 |
-1.6 |
1.4 |
1.2 |
qpx_c101 |
XP_002287676 |
4.13E-06 |
| dnaj protein 2 |
-1.3 |
-2.3 |
1.4 |
1.5 |
qpx_c9231 |
XP_002505603 |
7.23E-23 |
| dnaj protein 3 |
-2.1 |
-1.9 |
1.5 |
1.6 |
qpx_c11973 |
EGZ29544 |
1.71E-06 |
| Heat shock protein 40 kDa |
-2.2 |
-1.7 |
1.4 |
1.6 |
qpx_c37 |
XP_003496481 |
1.35E-41 |
| Heat shock protein 70 kDa 1 |
-2.7 |
-4.1 |
1.6 |
1.9 |
qpx_c366 |
XP_002788182 |
1.24E-33 |
| Heat shock protein 70 kDa 2 |
-2.2 |
-3.1 |
1.6 |
1.6 |
qpx_c2869 |
EGZ24428 |
4.92E-79 |
| Heat shock protein 70 kDa 3 |
-1.7 |
-1.8 |
1.4 |
1.4 |
qpx_c1020 |
XP_001694468 |
9.67E-58 |
| Heat shock protein 90 kDa |
-0.9 |
-4.0 |
1.5 |
1.6 |
qpx_c16851 |
XP_002291118 |
6.50E-49 |
| Heat shock protein 100 kDa |
0.2 |
-3.8 |
1.7 |
0.1 |
qpx_c3466 |
XP_635137 |
5.11E-27 |
| Heat shock protein 18 kDa |
2.2 |
0.5 |
-5.5 |
-6.0 |
qpx_c56 |
NP_662846 |
1.95E-16 |
| Heat shock protein 16 kDa |
0.3 |
1.8 |
-1.9 |
-2.2 |
qpx_c2700 |
ADR66511 |
4.49E-15 |
| Heat shock protein 15 kDa |
0.4 |
1.5 |
-1.6 |
-1.6 |
qpx_c5637 |
YP_969657 |
1.64E-13 |
|
Proteolysis |
|
|
|
|
|
|
|
| Cysteine peptidase C1-1 |
1.9 |
0.5 |
-1.9 |
-5.3 |
qpx_c534 |
XP_002507788 |
1.07E-56 |
| Cysteine peptidase C1-2 |
0.2 |
1.8 |
-1.6 |
-2.2 |
qpx_c5487 |
XP_002178071 |
6.01E-34 |
| Cysteine peptidase C1-3 |
-2.1 |
-1.3 |
0.4 |
2.0 |
qpx_c4154 |
CAB43538 |
1.44E-09 |
| Cysteine peptidase C1-4 |
-3.9 |
4.6 |
-5.5 |
-2.7 |
qpx_c1221 |
EGD76061 |
1.37E-06 |
| Cysteine peptidase C1-5 |
-2.2 |
-2.3 |
1.5 |
1.6 |
qpx_c8710 |
XP_003212177 |
2.04E-13 |
| Metallopeptidase M12B |
-5.9 |
-5.9 |
1.8 |
2.3 |
qpx_c6353 |
ADW54356 |
3.64E-06 |
| Metallopeptidase M32 |
-1.7 |
-1.4 |
1.6 |
1.1 |
qpx_c5681 |
ZP_09027479 |
5.64E-21 |
| Serine peptidase S8-1 |
0.7 |
2.0 |
-3.9 |
-5.7 |
qpx_c765 |
ABI79453 |
1.82E-34 |
| Serine peptidase S8-2 |
-1.3 |
2.2 |
-2.7 |
-2.5 |
qpx_c1822 |
ABI79453 |
3.26E-34 |
| Serine peptidase S8-3 |
-1.8 |
2.6 |
-3.5 |
-3.8 |
qpx_c2550 |
ABI79453 |
2.46E-53 |
| Serine carboxypeptidase S10-1 |
0.3 |
1.7 |
-2.0 |
-1.7 |
qpx_c2447 |
EGB09909 |
1.21E-47 |
| Serine carboxypeptidase S10-2 |
0.7 |
2.0 |
-2.8 |
-2.7 |
qpx_c8079 |
XP_001747631 |
8.39E-08 |
| Serine carboxypeptidase S28-1 |
0.3 |
1.5 |
-1.4 |
-1.4 |
qpx_c13341 |
CCA21035 |
1.27E-13 |
| Serine carboxypeptidase S28-2 |
-0.2 |
1.8 |
-2.2 |
-2.8 |
qpx_c2535 |
EGD81876 |
6.83E-53 |
|
Oxidative stress |
|
|
|
|
|
|
|
| Catalase |
-1.6 |
-2.1 |
1.2 |
1.9 |
qpx_c2181 |
AEX91749 |
1.50E-33 |
| Cu/Zn superoxide dismutase 1 |
-0.6 |
1.9 |
-1.7 |
-2.0 |
qpx_c1801 |
AAN85727 |
6.97E-28 |
| Cu/Zn superoxide dismutase 2 |
-0.3 |
1.7 |
-1.7 |
-1.8 |
qpx_c3152 |
ADN04915 |
9.10E-40 |
| Mg/Fe superoxide dismutase |
-0.4 |
1.9 |
-2.2 |
-1.9 |
qpx_c6686 |
XP_002958078 |
2.13E-27 |
| Superoxide NADPH oxidase |
-1.8 |
-1.8 |
1.5 |
1.5 |
qpx_c1054 |
EGB04413 |
5.01E-24 |
| Thioredoxin 1 |
-0.4 |
1.9 |
-1.9 |
-2.2 |
qpx_c1718 |
XP_765168 |
4.11E-25 |
| Thioredoxin 2 |
-1.2 |
1.6 |
-1.4 |
-1.3 |
qpx_c2033 |
XP_002402396 |
3.08E-27 |
| Thioredoxin 3 |
-0.2 |
1.9 |
-2.2 |
-2.5 |
qpx_c4189 |
XP_001800478 |
8.60E-19 |
| Thioredoxin reductase 1 |
0.4 |
-1.2 |
-1.2 |
1.5 |
qpx_c1123 |
EEE25525 |
2.45E-38 |
| Thioredoxin reductase 2 |
-1.8 |
-2.0 |
1.5 |
1.4 |
qpx_c2283 |
EGB06390 |
7.90E-46 |
| Thioredoxin reductase 3 |
1.1 |
1.3 |
-1.2 |
-1.6 |
qpx_c3882 |
XP_002181543 |
4.13E-33 |
|
Polysaccharide synthesis and secretion |
|
|
|
|
|
|
|
| Glycosyltransferase |
-0.8 |
-2.4 |
1.4 |
1.4 |
qpx_c2871 |
XP_002296954 |
3.04E-05 |
| Bligosaccharyl transferase |
-1.4 |
-2.2 |
1.4 |
1.5 |
qpx_c5264 |
XP_002185367 |
2.06E-22 |
| Beta-amylase/glucosidase |
-4.2 |
-1.2 |
1.3 |
1.8 |
qpx_c1194 |
ZP_08507128 |
2.84E-13 |
| Alpha-amylase/glucosidase |
-4.9 |
-4.8 |
1.7 |
2.2 |
qpx_c114 |
YP_750476 |
1.26E-32 |
| Sugar transporter 1 |
-1.5 |
-1.4 |
1.3 |
1.5 |
qpx_c6741 |
XP_003452761 |
1.54E-13 |
| Sugar transporter 2 |
-3.4 |
-2.1 |
1.5 |
2.0 |
qpx_c730 |
XP_642887 |
2.37E-12 |
| gdp-mannose-3-epimerase |
-1.5 |
-1.1 |
1.2 |
1.3 |
qpx_c4458 |
XP_002956437 |
8.07E-94 |
| udp-galactose-4-epimerase |
0.1 |
-3.1 |
1.4 |
1.5 |
qpx_c5964 |
ZP_08151628 |
5.96E-34 |
| udp-glucose gdp-mannose dehydrogenase |
-7.9 |
-5.1 |
2.5 |
1.2 |
qpx_c15700 |
ZP_06587676 |
1.90E-28 |
| udp-glucose 6-dehydrogenase |
-2.6 |
-3.7 |
1.7 |
1.9 |
qpx_c7532 |
CBJ28343 |
1.02E-25 |
| Integrin-related protein 1 |
-2.0 |
-1.4 |
1.5 |
1.5 |
qpx_c5688 |
EGB09976 |
3.30E-07 |
| Integrin-related protein 2 |
-2.2 |
-2.2 |
1.6 |
1.5 |
qpx_lrc9075 |
ZP_08823332 |
9.08E-08 |
| Syntaxin | -2.5 | -2.6 | 1.6 | 1.8 | qpx_c324 | EGB05290 | 1.40E-15 |
Mean fold changes are shown (3 biological replicates).
Figure 4.
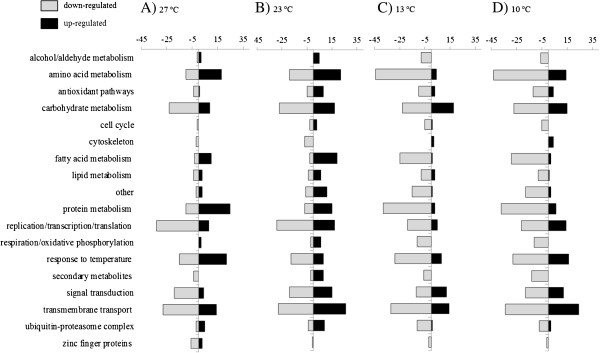
Molecular processes affected by temperature treatments. Number of annotated QPX transcripts differentially expressed (DE) in response to four temperature treatments (A: 27 °C, B: 23 °C, C: 13 °C, D: 10 °C). Transcripts are grouped by their putative biological process and molecular function into 17 operational groups.
The patterns of DE metabolic process vary between the temperature treatments with the most noticeable difference showing suppression of metabolic processes at cold temperatures: 178 transcripts down regulated at 13°C and 184 down regulated at 10°C in comparison to 79 transcripts down regulated at 27°C and 88 at 23°C (Figure 4). The slowing down of metabolism is a common response of ectothermic organisms to cold temperatures [67]. In addition, results suggest that the cold temperature conditions lowered the respiration rate of QPX (Figure 4) supporting a lower metabolic rate for the parasite. In contrast, seven transcripts within the respiration and oxidative phosphorylation process were induced at 23°C, suggesting an increased respiration rate at this temperature. The enrichment analysis revealed just a few biological and molecular processes being affected by the temperature changes but not in all temperature treatments (Table 4). For instance, the histidine catabolic processes were enriched by 151 fold among genes up-regulated at 27°C, while cellular aldehyde metabolic processes were enriched by 49 fold among genes up-regulated at 23°C and by 69 fold among genes down-regulated at 10°C (Table 4). These changes reflect the adjustment of metabolic rate of the parasite at different temperatures as shown by the overall patterns presented in Figure 4.
Table 4.
Gene Ontology terms enriched in response to temperature changes (Fisher’s exact test with multiple testing correction of FDR < 0.05, Benjamini and Hochberg)
| GO term ID | Gene ontology term description | GO type | FDR | p-value | % in DE group a | % on array b | Fold enrich c |
|---|---|---|---|---|---|---|---|
|
Up-regulated at 27°C |
|
|
|
|
|
|
|
| GO:0006548 |
Histidine catabolic process |
P |
2.9E-02 |
2.8E-05 |
5.4 |
0.0 |
151 |
| GO:0016829 |
Lyase activity |
F |
2.5E-02 |
8.0E-06 |
19.6 |
3.7 |
5 |
|
Down-regulated at 27°C |
|
|
|
|
|
|
|
| GO:0019915 |
Lipid storage |
P |
1.8E-02 |
2.2E-05 |
3.7 |
0.0 |
104 |
| GO:0005840 |
Ribosome |
C |
2.9E-02 |
5.7E-05 |
11.1 |
2.0 |
6 |
| GO:0003735 |
Structural constituent of ribosome |
F |
1.8E-02 |
2.9E-05 |
11.1 |
1.8 |
6 |
|
Up-regulated at 23°C |
|
|
|
|
|
|
|
| GO:0006081 |
Cellular aldehyde metabolic process |
P |
2.5E-02 |
3.2E-05 |
3.5 |
0.1 |
49 |
| GO:0003755 |
Peptidyl-prolyl cis-trans isomerase activity |
F |
1.3E-02 |
9.7E-06 |
4.4 |
0.1 |
30 |
| GO:0000502 |
proteasome complex |
C |
1.3E-02 |
1.2E-05 |
5.3 |
0.3 |
16 |
|
Down-regulated at 10°C |
|
|
|
|
|
|
|
| GO:0006548 |
Histidine catabolic process |
P |
8.1E-03 |
1.0E-05 |
2.4 |
0.0 |
69 |
| GO:0008233 |
Peptidase activity |
F |
4.9E-02 |
1.7E-04 |
11.0 |
3.9 |
3 |
| GO:0016829 |
Lyase activity |
F |
4.9E-02 |
1.9E-04 |
10.4 |
3.6 |
3 |
| GO:0006081 |
Cellular aldehyde metabolic process |
P |
4.9E-02 |
1.4E-04 |
2.4 |
0.1 |
33 |
| GO:0016616 | Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | F | 2.7E-02 | 4.3E-05 | 6.1 | 1.1 | 6 |
aPercent of sequences with that gene ontology term in the differentially expressed gene group.
bPercent of sequences with that gene ontology term on the whole array.
cFold enrichment: Fold increase of the GO term in the differentially expressed gene group.
The 23°C is in a range of QPX optimal growth conditions, however this temperature condition was also associated with an up-regulation of genes related to oxidative stress (eight up regulated transcripts, Figure 4). Slight oxidative stress, characterized as an accumulation of reactive oxygen species (ROS), happens in every cell as a result of imbalance between oxygen availability and oxygen consumption from respiration [67]. If a small amount of ROS, specifically superoxide anion, is produced during the normal respiration process as a simple by-product, an increased respiration rate would cause increased production of ROS. QPX possesses six different sequences encoding superoxide dismutase enzymes (SODs, data not shown), including four copper/zinc related SODs which are usually found in the cytosol of a cell, and two iron/manganese SODs which are usually found in mitochondria. The expression of only three QPX SODs (two Cu/Zn SOD and one Fe/Mn enzymes) were up-regulated at 23°C, suggesting that these three SODs are the main enzymes expressed in QPX cells to cope with the accumulation of endogenous ROS during the normal respiration process.
A well-documented response of organisms to temperature change is the regulation of expression of molecular chaperones known as heat shock proteins which are responsible for maintaining protein stability in situ. Temperature stress can lead to changes in proteins configuration and their potential malfunction but heat shock proteins control aggregation of denatured proteins in the cell, and are responsible for their reconfiguration, translocation across membranes and degradation. Overall 49 QPX transcripts coding for heat shock proteins were found to be regulated with temperature changes (Figure 4, Additional file 2). The amino acid sequence alignments of theses transcripts show that QPX has three different heat shock proteins which can be classified into a group of molecular chaperones of low monomeric molecular mass, ranging from 12 to 43 kDa [68]. These low molecular weight chaperones were up-regulated at the elevated temperature of 27°C (heat shock proteins 15, 16 and 18 kDa, Table 3). The transcription levels of these small molecular weight chaperones have been commonly investigated in pathogenic protozoans affecting humans and their expression is usually regulated during heat stress related to fever (e.g. Trypanosoma cruzi[68], Toxoplasma gondii[69]). Nine molecular weight chaperons: three dnaj-like proteins, three heat-shock protein 70, one heat-shock proteins 40, one 90 and one 101 kDa (Table 3) were up-regulated in response to cold temperature conditions revealing that they fulfill a role of cold shock chaperons. Heat shock proteins 70 and 90 have been previously documented to act as cold temperature chaperons in bacteria [70], nematode larvae [71], and oomycetes [72]. The dnaj proteins are co-chaperones of hsp70 and contain a conserved amino acid J domain which binds to hsp70 and stabilizes the interaction between the substrate and hsp70 [73]. Their up-regulation in QPX at low temperature highlights a similar response to cold stress and underlines the ability of the parasite to cope with suboptimal temperatures.
QPX putative virulence factors regulated by temperature
Temperature is one of the environmental factors controlling QPX disease development in hard clams [11,12] and one of the goals in the present study was to investigate the response of QPX putative virulence factor to temperature changes. Overall 14 QPX peptidases were found to be regulated with temperature changes including seven serine peptidases, five cysteine peptidases, and two metallopeptidases (Table 3). One papain-like peptidase (qpx_c534) was up-regulated at 27°C, two (qpx_c5487, qpx_c1221) were up-regulated at 23°C, and two (qpx_c4154, qpx_c8710) were up-regulated at 13°C and 10°C (Table 3), suggesting different roles for these genes in basic metabolism and/or pathogenesis of the parasite. Papain-like cysteine peptidases are some of the most extensively studied virulence factors. They play a crucial role in parasite biology, including growth, development and replication. They are also implicated in host tissue degradation, including digestion of host extracellular matrix proteins and destruction of host immune-related proteins such as immunoglobulins [74]. Further specific experimental studies are needed to specifically characterize the biochemical function and role of each papain-like peptidase in QPX biology and virulence abilities. Other QPX peptidases for which expression was shown to be regulated at different temperatures were subtilases. The amino acid sequence alignments of QPX transcripts annotated as belonging to the S8 family revealed that QPX possesses at least 6 different subtilisin-like peptidases (data not shown). Only three subtilases (qpx_c765, qpx_c1822, qpx_c2550) were shown to be differentially expressed in response to temperature changes, with the highest levels measured at 23°C (Figure 3, Table 3) which corresponds to QPX’s optimal growth temperature. QPX is a saprophytic microorganism which needs to degrade nutrient proteins extra-cellularly and transport oligopeptides and amino acids into the cell via diffusion. QPX maximal growth at 23°C could then explain and justify the need for an efficient extracellular protein degradation and nutrient acquisition system associated with high expression of its digestive enzymes. Subtilases, however are also universally occurring secreted enzymes found in many medically important pathogenic microorganisms, including bacteria (e.g. Mycobacterium, Streptococcus[47,48,75-77]), protozoa (e.g. Plasmodium, Leshmania and Toxoplasma[50,78-80]), and fungi (e.g. Aspergillus spp. [81]). Therefore, the role of subtilisins as QPX virulence factors might also be a possibility, but these enzymes may be important for host tissue digestion and disease progression during advanced and late stages of infection when the overcoming of host immune system is less important but degradation of the tissue and pathogen proliferation takes over.
The M12B peptidase (qpx_c6353, Table 3) which was up-regulated at 13°C and 10°C also require special attention. It belongs to the ADAMs or A Dis-integrin And Metalloprotease family of zinc-dependent endopeptidases. The same QPX peptidase was also found to be regulated with temperature in another study [28]. In animals, ADAMs are membrane-anchored glycosylated enzymes capable of degrading proteins from the surface of cells, thus playing important roles in cell adhesion, signaling, cell-cell fusion, and cell-cell interactions [82,83]. They possess two different protein components in addition to the metalloprotease domain, including an integrin domain (with adhesion and receptor activities) and a cytosolic domain which provides an attachment for various signal transduction proteins [82]. Cell-surface glycol-metallo-proteases play an important role in the pathogenesis of trypanosomatids, as they allow these parasites to adhere and move through connective tissue of their hosts [84]. The specific role, if any, of adamysin-like protease in the virulence of the QPX requires additional studies.
The QPX M32 enzyme is a carboxy-Taq-metallopeptidase (qpx_c5681, Table 3). The first zinc-containing thermostable metallopeptidase was originally discovered and purified from Thermus aquaticus and had an optimal enzymatic activity at 80°C [85]. Most bacterial and archaebacterial species are known to possess the M32 carobxypeptidases, but they are considered absent in most eukaryotic genomes except for a few protozoan species [86]. Their biological function in these organisms remains unknown. However, the basic characteristics of two recombinant M32 peptidases of Trypanosoma cruzi have been investigated [86]. They were shown not to behave as themostable enzymes, with their activity significantly decreasing with increased temperature. The current study is the first report of M32 enzyme in a thraustochytrid species for which stability and activity increases at lower temperatures. This cold resistant QPX’s metallopeptidase is a very interesting target for investigation of its biological function and role in the infection process.
QPX cells release a gel-like muco-filamentous secretion which appears to protect QPX from clam hemocyte phagocytosis and encapsulation as seen during histo-pathologic observation of infected clam tissues [2,8,9,87]. In addition, QPX mucus causes necrosis of clam hemocytes in vitro [88] and provides protection against clam humoral defense [89]. Overall, the generated transcriptome library contains 21 sequences involved in polysaccharide metabolic processes (GO:0005976) including 13 sequences involved in the biosynthesis of polysaccharides (GO:0000271) but complete pathways of mucus production were not identified (Table 1). The consistency of QPX mucus secretion changes with different temperatures, from a gel with low viscosity at 27°C to a very rigid gel at 10°C, thus it is surmised that cold temperatures would benefit the parasite by providing stronger protection during the infection process. Thicker mucus secretion at lower temperature was associated with an up-regulation of GDP-mannose dehydrogenase (qpx_c15700, Table 3). This enzyme essentially catalyzes irreversible conversion of GDP-mannose to GDP-mannuronic acid and ultimately leads to the biosynthesis of alginates in Pseudomonas aeruginosa[90] and in the brown alga Ectocarpus siliculosus[91]. In addition, the expression of two enzymes involved in the production of mannose (gdp-mannose-3- epimerase) and galactose (udp-galactose-4-epimerase) were also up-regulated at the two cold water temperatures (qpx_c4458 and qpx_c5964, respectively, Table 3). Mannose is the most important monosaccharide required for the production of mannuronic acid, which is the major component of alginates [92]. Interestingly, the mRNA levels of several different transcripts coding for UDP-glucose/GDP-mannose dehydrogenase were up-regulated in QPX at the two cold temperatures (qpx_c7532 in Table 3, full list in Additional file 2). Further, there were also a few other molecules in the carbohydrate metabolism and vesicular secretion pathways that were induced at 10 and 13°C. These include two glycosyltransferases (qpx_c2871, qpx_c5264, Table 3) which transfer and add a range of different sugars to other sugars, phosphates and proteins, thus participating in glycosylation of proteins and in synthesis of polysaccharides and glycoconjugates [93]. If the up-regulated glycotransferases are involved in the glycosylation process of QPX extracellular mucopolysaccharides, it can provide additional support of the hypothesis of considerable changes to the QPX mucus structure at cold temperature. In addition, two sugar transporters (qpx_c730 and qpx_c6741, Table 3) which carry dissolved simple sugars across membranes in response to chemosmotic gradient [94], and a syntaxin-type protein (qpx_c324, Table 3) that is a membrane protein participating in the vesicular membrane fusion during exocytosis [95], were over-expressed at the colder temperatures. Again, it can be speculated that these molecules participate in mucus polymers production and/or rearrangements. Even though the composition and structure of QPX mucus is still to be determined, these findings identify specific candidates that may be involved in mucus biosynthesis and provide a plausible scenario for the regulation of mucus consistency at different temperatures. The present hypothesis that QPX mucus production is increased in cold conditions can also be supported by the up-regulation of several molecules in carbohydrate synthesis and secretion pathways.
Conclusions
In conclusion, this study characterized the transcriptome of a parasitic labyrinthulid, offering new insights into the molecular bases for the pathogenicity of these microorganisms. The gene ontology annotation completed in this study is an excellent source of information for future investigations of this parasite. Results from the oligoarray experiment demonstrated the ability of QPX to cope with a wide range of environmental temperatures, including those considered to be suboptimal for clam immunity (low temperature), providing a mechanistic scenario for disease distribution in the field and for high disease prevalence and intensity at low temperature. These results will serve as a basis for studies aimed at thorough characterization of specific putative virulence factors.
Methods
Library construction and sequencing
QPX isolate NY0313808BC7 [96] initially isolated from a diseased clam collected from Raritan Bay, New York, was used for RNA extraction and library construction. To enhance library coverage, QPX was separately grown in Minimum Essential Medium (MEM, Sigma) supplemented with one of the following protein sources: clam muscle homogenates (500, 1000 or 3000 ug ml-1 [97], fetal bovine serum (0%, 2.5%, 5% or 10%) or gelatin (0.1%, 0.2 or 0.3%). Additional cultures were grown in MEM supplemented with yeastolate (0.1%, 0.2% or 0.3%) or with clam muscle homogenates made in sterile seawater without MEM. Cultures were incubated in triplicate at 23°C and parasite cells were separately harvested on Day 8 and Day 14. Parasite biovolume in each sample was assessed using a fluorometric technique [98] before samples were pooled using the same parasite biovolume from each culture condition. Total RNA was extracted from pooled samples using Trizol (Molecular Research Center, Inc.). The quality of total RNA was verified using Agilent 2100 bioanalyser and quantified by assessing the A260/280 and A260/230 ratios using a Nanodrop (ND 1000) spectrophotometer. Poly(A+) RNA were isolated from 300 μg of total RNA using the PolyATract® mRNA Isolation system (Promega) following the manufacturer’s instructions and used for cDNA synthesis. Preparation and sequencing of the cDNA library were performed at the McGill University and Génome Québec Innovation Centre (Canada) following the manufacturer’s protocol (Roche-454 Life Sciences, Brandford, CT, USA). The SMART adaptor sequences were removed from reads using a Perl script. Reads less than 50 bp and low-quality (quality scores below 20) reads were filtered out using SeqClean and remaining high-quality reads were assembled using MIRA assembler (version 3.0.5) using the default parameters for de novo EST assembly of 454 reads [99].
Transcriptome annotation
The annotation of the QPX transcriptome was completed using the online bioinformatics tool Blast2GO [100]. First, all sequences were subjected to blastx (basic local alignment tool) searches against the National Center for Bioinformatics (NCBI) sequence database with E-value cut off of 10^-3. Next the sequences proceeded through mapping to Gene Ontology (GO) Consortium database of standardized phrases describing functional information of known gene products, and finally GO functional annotation was completed using cutoff value 10^-3. Using the same tool, the sequences in all six translation frames were subjected to the InterProScan to find conserved protein domain matches in the Integrated Protein database of the European Bioinformatics Institute [101].
In silico identification of QPX putative virulence factors
The selection of potential virulence factors was based on the above annotated QPX transcriptome library and homology of QPX transcripts and protein sequences to the virulence factors of other protistan parasites and pathogens. The QPX sequence library was screened for sequences encoding for a variety of peptidases, hydrolytic enzymes, antioxidants, cell surface receptor and adhesion molecules. Manual curation of automated annotation as well as alignments and translations into amino acid sequences was accomplished using Geneious software [102]. The translated amino acid sequences were manually checked for the correct protein signatures and conserved protein domain using MEROPS, the peptidase database at http://merops.sanger.ac.uk/[103] and in the InterPro database from the European Bioinformatics Institute, http://www.ebi.ac.uk/Tools/pfa/iprscan5/[101,104].
Transcriptomic changes in QPX in response to temperature
For the temperature treatment, 500 μl of an exponentially growing QPX (isolate NY0313808BC7) culture was inoculated into twelve 25-ml culture flasks (Falcon) filled with 5 ml of Minimum Essential Medium (MEM, Sigma) supplemented with 10% of fetal bovine serum (FBS, Sigma). The flasks were incubated at four different temperatures: 27°C, 23°C, 13°C and 10°C, in triplicates (except 10°C which was done in duplicate) for each treatment. After seven days of incubation, the cultures were diluted with equal volume of filter sterilized artificial seawater and passed several times through a syringe to facilitate liquefaction of QPX mucus secretion. The mixtures were then transferred into 15-ml conical tubes and centrifuged at 3000 g for 40 minutes at 4°C. The supernatant was discarded and cell pellets collected and kept on ice for immediate RNA extraction. Trizol reagent (Molecular Research Center, Inc.) was used to isolate RNA from all samples following manufacturer’s protocol. RNA quality and quantity were estimated spectophotometrically using a Nandrop spectrophotometer.
Oligoarray design and hybridization
A subset of contigs produced by MIRA was used for the production of a 8 × 15 k 60-mer oligonucleotide array using the Agilent eArray application (https://earray.chem.agilent.com/earray/). These included 6,781 curated annotated sequences and 8,297 non-annotated sequences (minimal size = 215 b) to emphasize gene discovery. One probe was produced for each submitted sequence. Probes were synthesized in situ along with positive and negative controls using 8x15K-feature Agilent format slides. Labeled (Cy3 or Cy5) complementary RNA (cRNA) was synthesized from 150 ng of RNA purified from cultures submitted to different temperatures using Trizol (Invitrogen) and the Two-Color Microarray-Based Gene Expression Analysis Protocol (Quick Amp Labeling) following manufacturer’s protocol. Labeled cRNA was purified using Illustra CyScribe GFX Purification Kit (GE Healthcare). cRNA quantity and quality (including dye incorporation) were determined by spectrophotometry (Nanodrop). Samples were considered satisfactory if cRNA concentration and incorporation efficiency exceeded 300 ng/µl and 8 pmol Cy/µg cRNA, respectively. All arrays were hybridized with the same amount of cRNA (300 ng of each Cy3- and Cy5-labeled cRNA). Arrays hybridization and washes were conducted according to the kit protocol and the arrays were scanned with a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA, USA) using the suggested Agilent scan settings.
Oligoarray data normalization and analysis
Spot fluorescence intensities were extracted using GenePix software. LIMMA package in R software was used to normalize the intensities data and to remove within-array (method: global lowess) and between-array (method: quantile) non-biological variation [105]. After normalization, the intensities in separate color channels were exported into an excel spreadsheet for further data quality control and trimming. The intensities which were less than two-fold background intensities were eliminated from further analysis. The relative expression of each transcript at each temperature treatment was calculated as the ratio of the mean intensity (n = 3 for 27°C, 23°C, 13°C, and n = 2 for 10°C) for each temperature treatment and the mean intensity of that transcript in all temperature treatments (n = 11). Hierarchical clustering of all samples and genes (Pearson correlation) and the determination of statistically significant differentially expressed genes (one way ANOVA, p-value < 0.01) were completed in the TM4-suite using MeV program [106,107]. The final criteria for differential gene expression were the significance by ANOVA analysis and a one and half fold increase from the mean (up-regulation) or a one and half fold decrease from the mean (down-regulation) in at least one of the experimental treatments. The transcript sequences in the few categories of interest were investigated further by amino acid sequence alignment analysis to identify the number of unique QPX genes responsive to temperature. All translations and alignments were accomplished using Geneious software.
Real time PCR and oligo array validation
Real time PCR was performed on selected transcripts of interest which were shown to be differentially and no-differentially expressed by oligoarray methodology. Total RNA (2.5 µg) from each sample was used to synthesize cDNA using the MMLV reverse transcription kit (Promega) and oligo dT primers following manufactures protocol. Relative quantification was carried out in 10-µl reactions with Brilliant II SYBR green qPCR master mix (Agilent), 100 nM final primer concentration and 5 ng of RNA-equivalent cDNA. The PCR reactions were performed using Mastercycler ep realplex PCR machine (Eppendorf). Primers with a melting temperature of 60°C were designed using PrimerQuest program (Integrated DNA Technologies, IDT) within the open reading frames encoding for six different QPX peptidases and actin which was used as a reference gene for mRNA levels normalization (Table 2). The amplification products (135 – 148 bp) were confirmed using gel electrophoresis. The peptidases expression levels were normalized to the actin gene and relative transcript levels were calculated using the delta delta Ct method [108].
Availability of supporting data
This Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GALJ00000000.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BA, ER and AT designed the study. ER, MP, AT, EPE and BA carried out the experiments. ER and BA analyzed the data and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Annotation of QPX transcriptome. This is a Microsoft Excel worksheet that contains information on 17934 transcripts of quahog parasite unknown (QPX). The table contains the top blastx match accession number and E value, Gene Ontology terms and InterPro domain numbers associated with each QPX sequence.
A list of 1580 QPX transcripts differentially expressed (DE) in response to four temperature treatments. The file is a Microsoft Excel worksheet that contains a list of 1580 DE QPX transcripts and the mean fold change value for each transcript at each temperature treatment.
Contributor Information
Ewelina Rubin, Email: ewelinarubin@gmail.com.
Arnaud Tanguy, Email: atanguy@sb-roscoff.fr.
Mickael Perrigault, Email: mike.perrigault@gmail.com.
Emmanuelle Pales Espinosa, Email: emmanuelle.palesespinosa@stonybrook.edu.
Bassem Allam, Email: bassem.allam@stonybrook.edu.
Acknowledgments
This research was supported by projects R/XG-19 (to BA, AT and EPE) and R/FBM-34 (to BA and EPE), funded under awards NA07OAR4170010 from the National Sea Grant College Program of NOAA to the Research Foundation of State University of New York on behalf of New York Sea Grant. The study was also partially supported by project IOS 1050596 (to BA and EPE) supported by the National Science Foundation. The statements, findings, conclusions, views and recommendations are those of the authors and do not necessarily reflect the views of any of those organizations.
References
- Ragone Calvo LM, Walker JG, Burreson EM. Prevalence and distribution of QPX, Quahog Parasite Unknown, in hard clams Mercenaria mercenaria in Virginia, USA. Dis Aquat Organ. 1998;15:209–219. doi: 10.3354/dao033209. [DOI] [PubMed] [Google Scholar]
- Smolowitz R, Leavitt D, Perkins F. Observations of a Protistan Disease Similar to QPX in Mercenaria mercenaria (Hard Clams) from the Coast of Massachusetts. J Invertebr Pathol. 1998;15:9–25. doi: 10.1006/jipa.1997.4706. [DOI] [PubMed] [Google Scholar]
- Whyte SK, Cawthorn RJ, McGladdery SE. QPX (Quahog Parasite X), a pathogen of northern Quahaug Mercenaria mercenaria from the Gulf of St. Lawrence, Canada. Dis Aquat Organ. 1994;15:129–136. [Google Scholar]
- Dove ADM, Bowser PR, Cerrato RM. Histological analysis of an outbreak of QPX disease in wild hard ClamsMercenaria mercenariain New York. J Aquat Anim Health. 2004;15(4):246–250. [Google Scholar]
- Gast RJ, Moran DM, Audemard C, Lyons MM, DeFavari J, Reece KS, Leavitt D, Smolowitz R. Enviornmental distribution and persistence of Quahog Parasite Unknown (QPX) Dis Aquat Organ. 2008;15:219–229. doi: 10.3354/dao01948. [DOI] [PubMed] [Google Scholar]
- Liu Q, Allam B, Collier JL. Quantitative real-time PCR assay for QPX (Thraustochytriidae), a parasite of the hard clam (Mercenaria mercenaria) Appl Environ Microbiol. 2009;15(14):4913–4918. doi: 10.1128/AEM.00246-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons MM, Ward JE, Smolowitz R, Uhlinger KR, Gast RJ. Lethal marine snow: pathogen of bivalve mollusc concealed in marine aggregates. Limnol Oceanogr. 2005;15(6):1983–1988. [Google Scholar]
- Dahl SF, Perrigault M, Allam B. Laboratory transmission studies of QPX disease in the hard clam: Interactions between different host strains and pathogen isolates. Aquaculture. 2008;15(1–4):64–70. [Google Scholar]
- Dahl SF, Thiel J, Allam B. Field performance and QPX disease progress in culture and wild-type strains of Mercenaria mercenaria in New York waters. J Shellfish Res. 2010;15(1):83–90. [Google Scholar]
- Ragone Calvo LM, Ford SE, Kraeuter JN, Leavitt DF, Smolowitz R, Burreson EM. Influence of host genetic origin and geographic location on QPX disease in northern quahogs(=hard clams), Mercenaria mercenaria. J Shellfish Res. 2007;15(1):109–119. [Google Scholar]
- Dahl SF, Perrigault M, Liu Q, Collier JL, Barnes DA, Allam B. Effects of temperature on hard clam (Mercenaria mercenaria) immunity and QPX (Quahog Parasite Unknown) disease development: I. Dynamics of QPX disease. J Invertebr Pathol. 2011;15(2):314–321. doi: 10.1016/j.jip.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Perrigault M, Dahl SF, Espinosa EP, Gambino L, Allam B. Effects of temperature on hard clam (Mercenaria mercenaria) immunity and QPX (Quahog Parasite Unknown) disease development: II. Defense parameters. J Invertebr Pathol. 2011;15(2):322–332. doi: 10.1016/j.jip.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Ford SE, Kraeuter JN, Barber RD, Mathis G. Aquaculture-associated factors in QPX disease of hard clams: density and seed source. Aquaculture. 2002;15:23–38. [Google Scholar]
- Ragan MA, MacCallum GS, Murphy CA, Cannone JJ, Gutell RR, McGladdery SE. Protistan parasite QPX of hard-shell clam Mercenaria mercenaria is a member of Labyrinthulomycota. Dis Aquat Organ. 2000;15(3):185–190. doi: 10.3354/dao042185. [DOI] [PubMed] [Google Scholar]
- Maas PAY, Kleinschuster SJ, Dykstra MJ, Smolowitz R, Parent J. Molecular characterization of QPX (Quahog Parasite Unknown), a pathgogen of Mercenaria mercenaria. J Shellfish Res. 1999;15(2):561–567. [Google Scholar]
- Stokes NA, Ragone Calvo LM, Reece KS, Burreson EM. Molecular diagnostics, field validation, and phylogenetic analysis of Quahog Parasite Unknown (QPX), a pathogen of the hard clam Mercenaria mercenaria. Dis Aquat Organ. 2002;15:232–247. doi: 10.3354/dao052233. [DOI] [PubMed] [Google Scholar]
- Tsui CK, Marshall W, Yokoyama R, Honda D, Lippmeier JC, Craven KD, Peterson PD, Berbee ML. Labyrinthulomycetes phylogeny and its implications for the evolutionary loss of chloroplasts and gain of ectoplasmic gliding. Mol Phylogenet Evol. 2009;15(1):129–140. doi: 10.1016/j.ympev.2008.09.027. [DOI] [PubMed] [Google Scholar]
- Raghukumar S. Ecology of the marine protists, the Labyrinthulomycetes (Thraustochytrids and Labyrinthulids) Eur J Protistol. 2002;15(2):127–145. [Google Scholar]
- Bergmann N, Fricke B, Schmidt MC, Tams V, Beining K, Schwitte H, Boettcher AA, Martin DL, Bockelmann AC, Reusch TB, Rauch G. A quantitative real-time polymerase chain reaction assay for the seagrass pathogen Labyrinthula zosterae. Mol Eco Resour. 2011;15(6):1076–1081. doi: 10.1111/j.1755-0998.2011.03051.x. [DOI] [PubMed] [Google Scholar]
- Bower SM. The life cycle and ultrastructure of a new species of thraustochytrid (Protozoa: Labyrinthomorpha) pathogenic to small abalone. Aquaculture. 1987;15(1–2):269–272. [Google Scholar]
- Jones GM, O’Dor RK. Ultrastructural observations on a thraustochytrid fungus parasitic in the gills of squid (illex illecebrosus LeSueur) J Parasitol. 1983;15(5):903–911. [Google Scholar]
- McLean N, Porter D. The yellow-spot disease of tritonia diomedea bergh, 1894 (mollusca: gastropoda: nudibranchia): encapsulation of the thraustochytriaceous parasite by host amoebocytes. J Parasitol. 1982;15(2):243–252. [Google Scholar]
- Kvingedal R, Owens L, Jerry DR. A new parasite that infects eggs of the mud crab, Scylla serrata, in Australia. J Invertebr Pathol. 2006;15(1):54–59. doi: 10.1016/j.jip.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Burge CA, Douglas N, Conti-Jerpe I, Weil E, Roberts S, Friedman CS, Harvell CD. Friend or foe: the association of Labyrinthulomycetes with the Caribbean sea fan Gorgonia ventalina. Dis Aquat Organ. 2012;15(1):1–12. doi: 10.3354/dao02487. [DOI] [PubMed] [Google Scholar]
- Schärer L, Knoflach D, Vizoso DB, Rieger G, Peintner U. Thraustochytrids as novel parasitic protists of marine free-living flatworms: thraustochytrium caudivorum sp. nov. parasitizes Macrostomum lignano. Mar Biol. 2007;15(5):1095–1104. [Google Scholar]
- Harel M, Ben-Dov E, Rasoulouniriana D, Siboni N, Kramarsky-Winter E, Loya Y, Barak Z, Wiesman Z, Kushmaro A. A new Thraustochytrid, strain Fng1, isolated from the surface mucus of the hermatypic coral Fungia granulosa. FEMS Micro Eco. 2008;15(3):378–387. doi: 10.1111/j.1574-6941.2008.00464.x. [DOI] [PubMed] [Google Scholar]
- Perrigault M, Bugge DM, Allam B. Effect of environmental factors on survival and growth of quahog parasite unknown (QPX) in vitro. J Invertebr Pathol. 2010;15(2):83–89. doi: 10.1016/j.jip.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Garcia-Vedrenne AE, Groner M, Page-Karjian A, Siegmund GF, Singh S, Sziklay J, Roberts S. Development of genomic resources for a thraustochytrid pathogen and investigation of temperature influences on gene expression. PLoS One. 2013;15(9):e74196. doi: 10.1371/journal.pone.0074196. doi:74110.71371/journal.pone.0074196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kroger N, Lau WW, Lane TW, Larimer FW, Lippmeier JC, Lucas S. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;15(5693):79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Chia-Ta T, Wen-Lin H, Shinn-Jang H, Li-Sun S, Shinn-Ying H. Virulent-GO: prediction of virulent proteins in bacterial pathogens utilizing gene ontology terms. World Acad Sci Eng Technol. 2009;15:80–87. [Google Scholar]
- Casadevall A, Pirofski L. Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect Immun. 1999;15(8):3703–3713. doi: 10.1128/iai.67.8.3703-3713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SJ, Fernandez-Robledo JA, Gardner MJ, El-Sayed NM, Kuo CH, Schott EJ, Wang H, Kissinger JC, Vasta GR. The Alveolate Perkinsus marinus: biological insights from EST gene discovery. BMC Genomics. 2010;15:228. doi: 10.1186/1471-2164-11-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto-Alalibo T, Tian M, Gajendran K, Waugh ME, van West P, Kamoun S. Expressed sequence tags from the oomycete fish pathogen Saprolegnia parasitica reveal putative virulence factors. BMC Microbiol. 2005;15:46. doi: 10.1186/1471-2180-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwimmer G, Tober R, Bibars-Reiter R, Licek E, Steinborn R. Identification of two GH18 chitinase family genes and their use as targets for detection of the crayfish-plague oomycete Aphanomyces astaci. BMC Microbiol. 2009;15:184. doi: 10.1186/1471-2180-9-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addadi L, Joester D, Nudelman F, Weiner S. Mollusk shell formation: a source of new concepts for understanding biomineralization processes. Chemistry. 2006;15(4):980–987. doi: 10.1002/chem.200500980. [DOI] [PubMed] [Google Scholar]
- Furuhashi T, Schwarzinger C, Miksik I, Smrz M, Beran A. Molluscan shell evolution with review of shell calcification hypothesis. Comp Biochem Physiol B Biochem Mol Biol. 2009;15(3):351–371. doi: 10.1016/j.cbpb.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Belaunzaran ML, Wilkowsky SE, Lammel EM, Gimenez G, Bott E, Barbieri MA, de Isola EL. Phospholipase A1: a novel virulence factor in Trypanosoma cruzi. Mol Biochem Parasitol. 2013;15(2):77–86. doi: 10.1016/j.molbiopara.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Calderone RA, Fonzi WA. Virulence factors of Candida albicans. Trends Microbiol. 2001;15(7):327–335. doi: 10.1016/s0966-842x(01)02094-7. [DOI] [PubMed] [Google Scholar]
- Cox GM, McDade HC, Chen SC, Tucker SC, Gottfredsson M, Wright LC, Sorrell TC, Leidich SD, Casadevall A, Ghannoum MA, Perfect JR. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol Microbiol. 2001;15(1):166–175. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- Ghannoum MA. Potential role of phospholipases in virulence and fungal pathogenesis. Clin Microbiol Rev. 2000;15(1):122–143. doi: 10.1128/cmr.13.1.122-143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehr F, Kretschmar M, Kröger C, Hube B, Schäfer W. Microbial lipases as virulence factors. J Mol Catal B Enzym. 2003;15(5–6):347–355. [Google Scholar]
- McKerrow JHSE, Rosenthal PJ, Bouvier J. The proteases and pathogenicity of parasitic protozoa. Annu Rev Microbiol. 1993;15:821–853. doi: 10.1146/annurev.mi.47.100193.004133. [DOI] [PubMed] [Google Scholar]
- Sajid M, McKerrow JH. Cysteine proteases of parasitic organisms. Mol Biochem Parasitol. 2002;15:1–21. doi: 10.1016/s0166-6851(01)00438-8. [DOI] [PubMed] [Google Scholar]
- Klemba M, Goldberg DE. Biological roles of proteases in parasitic protozoa. Annu Rev Biochem. 2002;15:275–305. doi: 10.1146/annurev.biochem.71.090501.145453. [DOI] [PubMed] [Google Scholar]
- Olivos-Garcia A, Saavedra E, Ramos-Martinez E, Nequiz M, Perez-Tamayo R. Molecular nature of virulence in Entamoeba histolytica. Infect Genet Evol. 2009;15(6):1033–1037. doi: 10.1016/j.meegid.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Atkinson HJ, Babbitt PC, Sajid M. The global cysteine peptidase landscape in parasites. Trends Parasitol. 2009;15(12):573–581. doi: 10.1016/j.pt.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifait L, Vaillancourt K, Gottschalk M, Frenette M, Grenier D. Purification and characterization of the subtilisin-like protease of Streptococcus suis that contributes to its virulence. Vet Microbiol. 2011;15(2–4):333–340. doi: 10.1016/j.vetmic.2010.09.024. [DOI] [PubMed] [Google Scholar]
- Hu Q, Liu P, Yu Z, Zhao G, Li J, Teng L, Zhou M, Bei W, Chen H, Jin M. Identification of a cell wall-associated subtilisin-like serine protease involved in the pathogenesis of Streptococcus suis serotype 2. Microb Pathog. 2010;15(3–4):103–109. doi: 10.1016/j.micpath.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Hernandez-Romano P, Hernandez R, Arroyo R, Alderete JF, Lopez-Villasenor I. Identification and characterization of a surface-associated, subtilisin-like serine protease in Trichomonas vaginalis. Parasitology. 2010;15(11):1621–1635. doi: 10.1017/S003118201000051X. [DOI] [PubMed] [Google Scholar]
- Yeoh S, O’Donnell RA, Koussis K, Dluzewski AR, Ansell KH, Osborne SA, Hackett F, Withers-Martinez C, Mitchell GH, Bannister LH, Bryans JS, Kettleborough CA, Blackman MJ. Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes. Cell. 2007;15(6):1072–1083. doi: 10.1016/j.cell.2007.10.049. [DOI] [PubMed] [Google Scholar]
- Dubovenko AG, Dunaevsky YE, Belozersky MA, Oppert B, Lord JC, Elpidina EN. Trypsin-like proteins of the fungi as possible markers of pathogenicity. Fungal Biol. 2010;15(2–3):151–159. doi: 10.1016/j.funbio.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Gvozdeva EL, Ievleva EV, Gerasimova NG, Ozeretskovskaia OL, Valueva TA. Exoproteinases of the Oomecete Phytophthora infestans. Appl Biochem Microbiol. 2004;15(2):165–169. [PubMed] [Google Scholar]
- Coetzer TH, Goldring JP, Huson LE. Oligopeptidase B: a processing peptidase involved in pathogenesis. Biochimie. 2008;15(2):336–344. doi: 10.1016/j.biochi.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Monod M, Capoccia S, Lechenne B, Zaugg C, Holdom M, Jousson O. Secreted proteases from pathogenic fungi. Int J Med Microbiol. 2002;15(5–6):405–419. doi: 10.1078/1438-4221-00223. [DOI] [PubMed] [Google Scholar]
- Bugge DM, Hegaret H, Wikfors GH, Allam B. Oxidative burst in hard clam (Mercenaria mercenaria) haemocytes. Fish Shellfish Immunol. 2007;15(1):188–196. doi: 10.1016/j.fsi.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Osorio L, Rios I, Gutierrez B, Gonzalez J. Virulence factors of Trypanosoma cruzi: who is who? Microbes Infect. 2012;15(15):1390–1402. doi: 10.1016/j.micinf.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Cheng XJ, Yoshihara E, Takeuchi T, Tachibana H. Molecular characterization of peroxiredoxin from Entamoeba moshkovskii and a comparison with Entamoeba histolytica. Mol Biochem Parasitol. 2004;15(2):195–203. doi: 10.1016/j.molbiopara.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Robold AV, Hardham AR. During attachment Phytophthora spores secrete proteins containing thrombospondin type 1 repeats. Curr Genet. 2005;15(5):307–315. doi: 10.1007/s00294-004-0559-8. [DOI] [PubMed] [Google Scholar]
- Yuda M, Sawai T, Chinzei Y. Structure and expression of an adhesive protein-like molecule of mosquito invasive-stage malarial parasite. J Exp Med. 1999;15(12):1947–1952. doi: 10.1084/jem.189.12.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JM, Hoff EF, Carruthers VB. Multimerization of the Toxoplasma gondii MIC2 integrin-like A-domain is required for binding to heparin and human cells. Mol Biochem Parasitol. 2004;15(2):201–212. doi: 10.1016/j.molbiopara.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Vines RR, Ramakrishnan G, Rogers JB, Lockhart LA, Mann BJ, Petri WA Jr. Regulation of adherence and virulence by the Entamoeba histolytica lectin cytoplasmic domain, which contains a beta2 integrin motif. Mol Biol Cell. 1998;15(8):2069–2079. doi: 10.1091/mbc.9.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist CA, Petri WA. Virulence factors of Entamoeba histolytica. Curr Opin Microbiol. 1999;15(4):433–437. doi: 10.1016/S1369-5274(99)80076-9. [DOI] [PubMed] [Google Scholar]
- Mann BJ. Structure and function of the Entamoeba histolytica Gal/GalNAc lectin. Int Rev Cytol. 2002;15:59–80. doi: 10.1016/s0074-7696(02)16003-7. [DOI] [PubMed] [Google Scholar]
- Stanley SL Jr, Reed SL. Microbes and microbial toxins: paradigms for microbial-mucosal interactions. VI. Entamoeba histolytica: parasite-host interactions. Am J Physiol Gastrointest Liver Physiol. 2001;15(6):G1049–G1054. doi: 10.1152/ajpgi.2001.280.6.G1049. [DOI] [PubMed] [Google Scholar]
- Jacobs T, Erdmann H, Fleischer B. Molecular interaction of Siglecs (sialic acid-binding Ig-like lectins) with sialylated ligands on Trypanosoma cruzi. Eur J Cell Biol. 2010;15(1):113–116. doi: 10.1016/j.ejcb.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Gaulin E, Jauneau A, Villalba F, Rickauer M, Esquerre-Tugaye MT, Bottin A. The CBEL glycoprotein of Phytophthora parasitica var-nicotianae is involved in cell wall deposition and adhesion to cellulosic substrates. J Cell Sci. 2002;15(Pt 23):4565–4575. doi: 10.1242/jcs.00138. [DOI] [PubMed] [Google Scholar]
- Blagojevic DP, Grubor-Lajsic GN, Spasic MB. Cold defence responses: the role of oxidative stress. Front Biosci (Schol Ed) 2011;15:416–427. doi: 10.2741/s161. [DOI] [PubMed] [Google Scholar]
- Perez-Morales D, Ostoa-Saloma P, Espinoza B. Trypanosoma cruzi SHSP16: characterization of an alpha-crystallin small heat shock protein. Exp Parasitol. 2009;15(2):182–189. doi: 10.1016/j.exppara.2009.06.019. [DOI] [PubMed] [Google Scholar]
- de Miguel N, Braun N, Bepperling A, Kriehuber T, Kastenmuller A, Buchner J, Angel SO, Haslbeck M. Structural and functional diversity in the family of small heat shock proteins from the parasite Toxoplasma gondii. Biochim Biophys Acta. 2009;15(11):1738–1748. doi: 10.1016/j.bbamcr.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Lelivelt MJ, Kawula TH. Hsc66, an Hsp70 homolog in Escherichia coli, is induced by cold shock but not by heat shock. J Bacteriol. 1995;15(17):4900–4907. doi: 10.1128/jb.177.17.4900-4907.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Perez-Serrano J, Bernadina WE, Rodriguez-Caabeiro F. Stress response to cold in Trichinella species. Cryobiology. 2001;15(4):293–302. doi: 10.1006/cryo.2001.2363. [DOI] [PubMed] [Google Scholar]
- Judelson HS, Michelmore RW. Structure and expression of a gene encoding heat-shock protein Hsp70 from the Oomycete fungus Bremia lactucae. Gene. 1989;15(2):207–217. doi: 10.1016/0378-1119(89)90203-5. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;15(22):2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecaille F, Kaleta J, Bromme D. Human and parasitic papain-like cysteine proteases: their role in physiology and pathology and recent developments in inhibitor design. Chem Rev. 2002;15(12):4459–4488. doi: 10.1021/cr0101656. [DOI] [PubMed] [Google Scholar]
- Brown GD, Dave JA, van Pittius NC G, Stevens L, Ehlers MR, Beyers AD. The mycosins of Mycobacterium tuberculosis H37Rv: a family of subtilisin-like serine proteases. Gene. 2000;15(1–2):147–155. doi: 10.1016/s0378-1119(00)00277-8. [DOI] [PubMed] [Google Scholar]
- Coutte L, Antoine R, Drobecq H, Locht C, Jacob-Dubuisson F. Subtilisin-like autotransporter serves as maturation protease in a bacterial secretion pathway. EMBO J. 2001;15(18):5040–5048. doi: 10.1093/emboj/20.18.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennan RM, Wong W, Dhungyel OP, Han X, Wong D, Parker D, Rosado CJ, Law RH, McGowan S, Reeve SB, Levina V, Powers GA, Pike RN, Bottomley SP, Smith AI, Marsh I, Whittington RJ, Whisstock JC, Porter CJ, Rood JI. The subtilisin-like protease AprV2 is required for virulence and uses a novel disulphide-tethered exosite to bind substrates. PLoS Pathog. 2010;15(11):e1001210. doi: 10.1371/journal.ppat.1001210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagal V, Binder EM, Huynh MH, Kafsack BF, Harris PK, Diez R, Chen D, Cole RN, Carruthers VB, Kim K. Toxoplasma gondii protease TgSUB1 is required for cell surface processing of micronemal adhesive complexes and efficient adhesion of tachyzoites. Cell Microbiol. 2010;15(12):1792–1808. doi: 10.1111/j.1462-5822.2010.01509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Liu Y, Jung SY, Cho PY, Zheng B, Park H. Transcriptional activity of Plasmodium subtilisin-like protease 2 (Pf-Sub2) 5′untranslated regions and its interaction with hepatocyte growth factor. Korean J Parasitol. 2010;15(4):291–295. doi: 10.3347/kjp.2010.48.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenerton RK, Knudsen GM, Sajid M, Kelly BL, McKerrow JH. Leishmania subtilisin is a maturase for the trypanothione reductase system and contributes to disease pathology. J Biol Chem. 2010;15(41):31120–31129. doi: 10.1074/jbc.M110.114462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszewska A, Taylor JW, Szczesny P, Grynberg M. Independent subtilases expansions in fungi associated with animals. Mol Biol Evol. 2011;15(12):3395–3404. doi: 10.1093/molbev/msr176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;15(1):7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- van Goor H, Melenhorst WB, Turner AJ, Holgate ST. Adamalysins in biology and disease. J Pathol. 2009;15(3):277–286. doi: 10.1002/path.2594. [DOI] [PubMed] [Google Scholar]
- Masini d’Avila-Levy C, de Almeida Dias F, de Melo AC N, Martins JL, De Carvalho Santos Lopes AH, Souza Dos Santos AL, Vermelho AB, Branquinha MH. Insights into the role of gp63-like proteins in lower trypanosomatids. FEMS Microbiol Lett. 2006;15(1):149–156. doi: 10.1111/j.1574-6968.2005.00022.x. [DOI] [PubMed] [Google Scholar]
- Lee SH, Minagawa E, Taguchi H, Matsuzawa H, Ohta T, Kaminogawa S, Yamauchi K. Purification and characterization of a thermostable carboxypeptidase (carboxypeptidase Taq) from Thermus aquaticus YT-1. Biosci Biotechnol Biochem. 1995;15(11):1839–1844. doi: 10.1271/bbb.56.1839. [DOI] [PubMed] [Google Scholar]
- Niemirowicz G, Parussini F, Aguero F, Cazzulo JJ. Two metallocarboxypeptidases from the protozoan Trypanosoma cruzi belong to the M32 family, found so far only in prokaryotes. Biochem J. 2007;15(2):399–410. doi: 10.1042/BJ20060973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl SF, Allam B. Laboratory transmission studies of QPX disease in the northern quahog (=hard clam): Development of an infection procedure. J Shellfish Res. 2007;15(2):383–389. [Google Scholar]
- Perrigault M, Allam B. Cytotoxicity of quahog parasite unknown (QPX) toward hard clam (Mercenaria mercenaria) haemocytes and interactions between different pathogen isolates and host strains. Parasitology. 2009;15(11):1281–1289. doi: 10.1017/S0031182009990606. [DOI] [PubMed] [Google Scholar]
- Anderson RS, Kraus BS, McGladdery S, Smolowitz R. QPX, a pathogen of quahogs (hard clams), employs mucoid secretions to resist host antimicrobial agents. J Shellfish Res. 2003;15(1):205–208. [Google Scholar]
- Snook FC, Tipton PA, Beamer LJ. Crystal structure of GDP-mannose dehydrogenase: a key enzyme of alginate biosynthesis in P. Aeruginosa. Biochemistry. 2003;15:4658–4668. doi: 10.1021/bi027328k. [DOI] [PubMed] [Google Scholar]
- Tenhaken R, Voglas E, Cock JM, Neu V, Huber CG. Characterization of GDP-mannose dehydrogenase from the brown alga Ectocarpus siliculosus providing the precursor for the alginate polymer. J Biol Chem. 2011;15(19):16707–16715. doi: 10.1074/jbc.M111.230979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm BHA. In: Alginates: Biology and Applications, Microbiology Monographs 13. Rehm BHA, editor. Vol. 15. Berlin: Springer-Verlag Berlin Heidelberg; 2009. Alginate Production: Precursor Biosynthesis, Polymerization and Secretion; pp. 55–71. [Google Scholar]
- Breton C, Imberty A. Structure/function studies of glycosyltransferases. Curr Opin Struct Biol. 1999;15(5):563–571. doi: 10.1016/s0959-440x(99)00006-8. [DOI] [PubMed] [Google Scholar]
- S PS, Paulsen I, Saier MH. Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;15(1):1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacks JB, Doolittle WF. Molecular and phylogenetic characterization of syntaxin genes from parasitic protozoa. Mol Biochem Parasitol. 2004;15(2):123–136. doi: 10.1016/j.molbiopara.2004.02.014. [DOI] [PubMed] [Google Scholar]
- Qian H, Liu Q, Allam B, Collier JL. Molecular genetic variation within and among isolates of QPX (Thraustochytridae), a parasite of the hard clam Mercenaria mercenaria. Dis Aquat Organ. 2007;15(2):159–168. doi: 10.3354/dao01848. [DOI] [PubMed] [Google Scholar]
- Perrigault M, Bugge DM, Hao CC, Allam B. Modulatory effects of hard clam (Mercenaria mercenaria) tissue extracts on the in vitro growth of its pathogen QPX. J Invertebr Pathol. 2009;15(1):1–8. doi: 10.1016/j.jip.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Bugge DM, Allam B. A fluorometric technique for the in vitro measurment of growth and viability in quahog parasite unknown (QPX) J Shellfish Res. 2005;15(4):1013–1018. [Google Scholar]
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Muller WE, Wetter T, Suhai S. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;15(6):1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;15(10):3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, Finn RD, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Laugraud A, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Mulder N, Natale D, Orengo C, Quinn AF. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;15(Database issue):D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geneious v5.4. http://www.geneious.com/
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;15(Database issue):D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;15(9):847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;15(4):265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;15(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;15:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;15(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Annotation of QPX transcriptome. This is a Microsoft Excel worksheet that contains information on 17934 transcripts of quahog parasite unknown (QPX). The table contains the top blastx match accession number and E value, Gene Ontology terms and InterPro domain numbers associated with each QPX sequence.
A list of 1580 QPX transcripts differentially expressed (DE) in response to four temperature treatments. The file is a Microsoft Excel worksheet that contains a list of 1580 DE QPX transcripts and the mean fold change value for each transcript at each temperature treatment.


