Summary
Ectopic expression of reprogramming factors has been widely adopted to reprogram somatic nucleus into a pluripotent state (induced pluripotent stem cells [iPSCs]). However, genetic aberrations such as somatic gene mutation in the resulting iPSCs have raised concerns regarding their clinical utility. To test whether the increased somatic mutations are primarily the by-products of current reprogramming methods, we reprogrammed embryonic fibroblasts of inbred C57BL/6 mice into either iPSCs (8 lines, 4 previously published) or embryonic stem cells (ESCs) with somatic cell nuclear transfer (SCNT ESCs; 11 lines). Exome sequencing of these lines indicates a significantly lower mutation load in SCNT ESCs than iPSCs of syngeneic background. In addition, one SCNT-ESC line has no detectable exome mutation, and two pairs of SCNT-ESC lines only have shared preexisting mutations. In contrast, every iPSC line carries unique mutations. Our study highlights the need for improving reprogramming methods in more physiologically relevant conditions.
Graphical Abstract
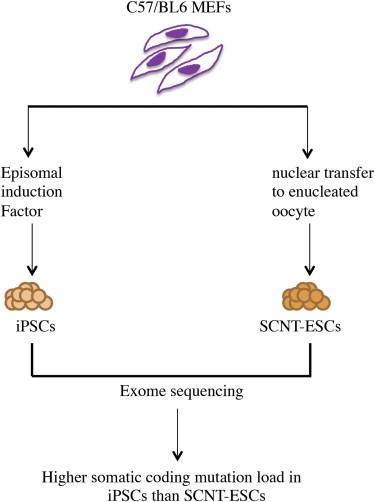
Highlights
-
•
Comparison of somatic-coding mutation load in SCNT ESCs and iPSCs in mice
-
•
Lower mutational load in SCNT ESCs than iPSCs of syngeneic background
By exome sequencing of pluripotent stem cell lines reprogrammed from embryonic fibroblasts of inbred C57BL/6 mice with somatic cell nuclear transfer-embryonic stem cell (SCNT-ESC) or ectopic expression of reprogramming factors (induced pluripotent stem cells [iPSC]), Zhang, Deng, Xu, and colleagues show that the SCNT-ESC lines harbor a significantly lower somatic-coding mutation load than iPSC lines.
Introduction
Reprogramming of somatic nuclei into a pluripotent state can be achieved through either somatic cell nuclear transfer (SCNT) (Campbell et al., 1996) or ectopic expression of reprogramming factors in somatic cells to generate induced pluripotent stem cells (iPSCs) (Takahashi et al., 2007, Yu et al., 2007). The latter approach has become widely adopted because it is ethically more acceptable and technically more feasible to many organisms such as humans. The iPSCs are functionally indistinguishable from embryonic stem cells (ESCs). However, recent studies have revealed genetic and epigenetic aberrations in the resulting iPSCs (Gore et al., 2011, Cheng et al., 2012, Ji et al., 2012, Lister et al., 2011, Ruiz et al., 2012, Ruiz et al., 2013, Young et al., 2012). For example, it has been shown that iPSCs always possess somatic-coding mutations (Gore et al., 2011, Ji et al., 2012, Ruiz et al., 2013, Young et al., 2012), leading to the concern for the safety of such cells in clinical application.
SCNT has been widely used to reprogram the somatic nucleus into a pluripotent state by the injection of a donor nucleus into an enucleated oocyte. The technique mimics the process of early embryonic development except that the blastocyst formed contains identical genomic DNA as the donor (Figure 1). Because SCNT provides a physiologically relevant condition for nuclear reprogramming and allows development, it has been widely adopted to produce viable cloned mammals such as sheep (Campbell et al., 1996), mice (Wakayama et al., 1998), and rabbit (Chesné et al., 2002) from primary culture. There is also an ongoing effort to derive human ESC-like lines using SCNT (SCNT ESCs) for patient-specific therapy (Egli et al., 2011, Tachibana et al., 2013). However, the genetic integrity of these SCNT ESCs has yet been reported.
Figure 1.
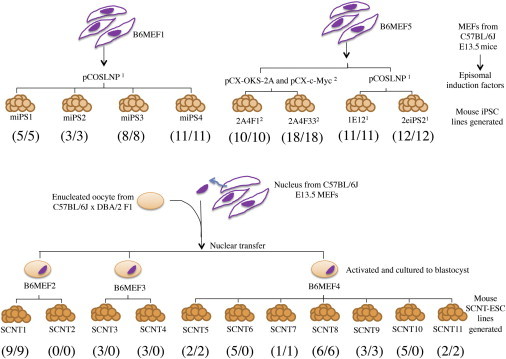
Summary of Pairwise Comparison between Mouse Fibroblast and Pluripotent Stem Cell Lines
The bold numbers in parentheses indicate validated/unique somatic-coding mutations. Footnote 1 is previously described in Zhao et al., 2011. Footnote 2 is previously described in Araki et al., 2013. E13.5, embryonic day 13.5.
In this study, we characterized the protein-coding region of 8 iPSC lines and 11 SCNT-ESC lines derived from a syngeneic inbred mouse background at single-nucleotide resolution. We chose to focus on exome not only due to cost considerations but also because exome mutations are more interpretable, and the iPSC mutation load in exome has been well characterized (Gore et al., 2011, Ji et al., 2012, Ruiz et al., 2013, Young et al., 2012). We observed significantly lower somatic-coding mutation load in SCNT ESCs than iPSCs. These findings suggest that current reprogramming methods to generate iPSCs could be improved in more physiologically relevant conditions to optimize nuclear reprogramming for clinical application.
Results
Previous studies have reported that human iPSCs derived from diverse somatic origins and reprogramming methods all carried between 2 and 14 point mutations in protein-coding regions (Gore et al., 2011, Ji et al., 2012, Ruiz et al., 2013). In this study, we sought to determine whether acquisition of protein-coding mutations must occur to allow successful nuclear reprogramming. To avoid the influence of distinct genetic backgrounds on reprogramming, we derived 11 mouse SCNT-ESC lines and 4 mouse iPSC lines from the syngeneic mouse embryonic fibroblasts (MEFs) of the inbred C57BL/6 (B6) mice (Figure 1). The B6 iPSCs were reprogrammed with the integration-free approaches by Abe’s and our group (Araki et al., 2013, Zhao et al., 2011), and the SCNT ESCs were generated as we previously described (Bai et al., 2007, Jiang et al., 2008, Yu et al., 2005). The pluripotency of these cell lines was extensively characterized by the surface expression of ESC-specific markers, their capability to differentiate into each of the three germ layers, or in most cases, by their capability to contribute to adult chimeric mice and germline transmission (Figures 2A–2D; Figures S1 and S2 available online). We next performed exome sequencing and pairwise comparison of the 19 pluripotent stem cell lines and progenitor MEF cells (Figure 1). After mapping sequenced reads, 90% or more of protein-coding regions had sufficiently high sequence coverage (>10×) and consensus quality (>30) to identify somatic-coding mutations in each cell line (Table 1). We identified and validated a total of 78 unique somatic-coding mutations within 8 iPSC lines (Tables 1, 2, and S1), or an average of 9.8 mutations per line, consistent with previously observed mutational load in human iPSC lines (Gore et al., 2011). In contrast, 31 unique mutations were identified and validated in the 11 SCNT-ESC lines, leading to a projection of 2.8 mutations per line in protein-coding regions (Tables 1, 2, and S1). The mutational load of iPSC lines was significantly higher than that of SCNT-ESC lines (p < 0.004, Mann-Whitney U test).
Figure 2.
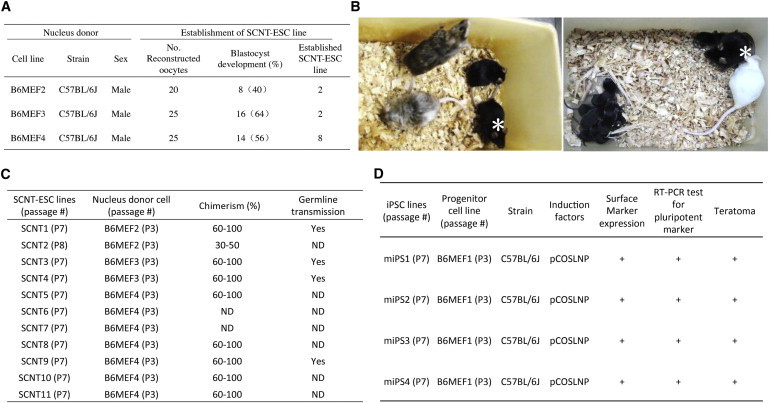
Summary of Mouse SCNT-ESC and iPSC Line Characterization
(A) Establishment of SCNT-ESC lines from MEFs.
(B) Pluripotency of SCNT ESCs in vivo. Left panel shows chimeric mice derived from SCNT-ESC lines. SCNT1 cells were injected into eight-cell embryos of ICR mice, and shown are 8-week-old offspring, in which black coat color is derived from the SCNT-ESC contribution. Right panel shows the male that was crossed with a white ICR female, producing a litter containing nine black offspring, confirming the contribution of SCNT1 to the germline. Asterisks in the left and right panels indicate the same male.
(C) Summary of mouse SCNT-ESC line characterization. The percentage of chimerism was established based on coat color. ND, not determined.
(D) Summary of mouse iPSC line characterization.
Table 1.
Summary of Sequencing Statistics for Mouse SCNT-ESC and iPSC Lines
| Cell Line | Quality-Filtered Sequences (bp) | 10× Exome Coverage | 20× Exome Coverage | dbSNP Percentages | Shared High-Quality Coding Regions (bp) | No. of Validated/Projected Coding Mutations | No. of Unique Coding Mutationsa |
|---|---|---|---|---|---|---|---|
| SCNT1 | 13,147,531,800 | 95% | 88% | 30 | 31,194,049 | 9/9 | 9 |
| SCNT2 | 10,427,221,300 | 94% | 85% | 29 | 31,222,097 | 0/0 | 0 |
| SCNT3 | 4,996,365,000 | 90% | 75% | 33 | 30,799,305 | 3/3 | 0 |
| SCNT4 | 4,744,153,500 | 90% | 74% | 33 | 30,790,251 | 3/3 | 0 |
| SCNT5 | 6,213,421,800 | 92% | 80% | 33 | 31,200,714 | 2/2 | 2 |
| SCNT6 | 5,168,988,000 | 89% | 74% | 33 | 31,155,933 | 5/5 | 0 |
| SCNT7 | 5,686,592,400 | 90% | 76% | 33 | 31,171,629 | 1/5 | 1 |
| SCNT8 | 5,525,811,400 | 90% | 77% | 32 | 31,159,710 | 6/6 | 6 |
| SCNT9 | 6,177,436,900 | 91% | 80% | 33 | 31,176,729 | 3/3 | 3 |
| SCNT10 | 5,527,450,200 | 90% | 76% | 34 | 31,140,431 | 5/5 | 0 |
| SCNT11 | 5,127,529,500 | 89% | 71% | 33 | 31,093,228 | 2/2 | 2 |
| miPS1 | 5,118,424,100 | 90% | 76% | 31 | 31,067,998 | 5/5 | 5 |
| miPS2 | 5,463,084,500 | 91% | 79% | 30 | 31,098,810 | 3/3 | 3 |
| miPS3 | 4,959,169,500 | 90% | 75% | 30 | 31,048,768 | 8/8 | 8 |
| miPS4 | 4,730,980,100 | 90% | 75% | 32 | 31,048,646 | 11/11 | 11 |
| 2A4F1 | 5,450,265,400 | 90% | 75% | 30 | 30,823,695 | 10/10 | 10 |
| 2A4F33 | 5,277,562,300 | 89% | 73% | 31 | 30,805,337 | 18/19 | 18 |
| 1E12 | 4,291,623,180 | 84% | 60% | 30 | 30,766,265 | 11/11 | 11 |
| 2eiPS2 | 3,850,799,960 | 80% | 50% | 31 | 30,721,856 | 12/13 | 12 |
Quality-filtered sequence represents the amount of sequence data generated that passed the Illumina quality filter with a sequencing depth of at least 8 and a consensus quality score of at least 30 (bp). The dbSNP percentage is the percentage of identified variants that are in the Single Nucleotide Polymorphism Database. The shared coding region is the portion of the genome that was sequenced at high depth and quality in both the pluripotent stem cell line and matched fibroblast (as shown in Figure 1). The projected number of somatic-coding mutations is calculated by the fraction of consensus coding sequence identified in both pluripotent stem cells and fibroblasts.
Validated unique mutations after removing shared mutations.
Table 2.
Summary of Somatic Mutations Validated in Mouse SCNT-ESC and iPSC Lines
| Cell Line | Mutated Genes |
|---|---|
| SCNT1 | Tchhl1, Neil1, Spag9, Irf9, Opcm1, Ubr3, Lrrtm4, Susd1, and Speg |
| SCNT2 | n/a |
| SCNT3 | Spata7, Col24a1, and Aff3 |
| SCNT4 | Spata7, Col24a1, and Aff3 |
| SCNT5 | Mast1 and Ccy2a4 |
| SCNT6 | Olfr963, Vmn1r197, Nckipsd, Olfr330, and Klhl22 |
| SCNT7 | Tanc1 |
| SCNT8 | Bbs1, Cyp2c39, Ndufaf3, Chd6, Cacnali, and Aldh8a1 |
| SCNT9 | Fat1, Efhd1, and Olfr118 |
| SCNT10 | Olfr963, Vmn1r197, Nckipsd, Olfr330, and Klhl22 |
| SCNT11 | Kdm3b and Il17rb |
| miPS1 | Srsf11, Odam, Greb1, Card6, and 4930443G12Rik |
| miPS2 | Siglec5, Tenc1, and Dusp10 |
| miPS3 | Adam5, Muc5b, Crbn, Disp2, Mtmr10, Moap1, Entpd8, and Cntn5 |
| miPS4 | Eif3b, Acox2, Olfr1349, Ensa, Ankrd13b, Inad1, Slc39a6, March1, 4933422H20Rik, 1700011F14Rik, and Slc12a2 |
| 2A4F1 | Irx1, Gjb6, Akap6, Atp6v1e2, Abcg2, Atp9b, 2810021J22Rik, Scaf1a, and Brsk1 |
| 2A4F33 | Gm16432, Zfp777, Pdzrn3, Bc016423, Rabgap11, D16Ertd472e, Smarcc2, Arhgap17, Catsperg2, Akr7a5, Prune2, Dip2a, Tt1, Opn5, P1xna1, Prcp, Olfr1382, and Zfp316 |
| 1E12 | Klk8, Serpinb9b, Pick1, Vps33ba, Myo5b, Grm1, Hivep1, Ank3, Adamts12, and Cebpe |
| 2eiPS2 | Paqr6, Pold1, BC030867, Adss, Tcf4, Oas1g, M112, Ube4a, Zfp827, Hmgc1, Dnahc2, and Tex 19.2 |
n/a, not applicable.
Two unique somatic-coding mutations were detected in this gene.
In contrast to the findings that every iPSC line examined in this and previous studies harbored protein-coding mutations, we did not detect any protein-coding mutation in one SCNT-ESC line (Table 1, SCNT2). Moreover, there were two pairs of SCNT-ESC lines (SCNT3 and SCNT4 and SCNT6 and SCNT10) that only had shared somatic mutations (Table 2), suggesting that these mutations were not introduced during reprogramming but, instead, were present in the fibroblast progenitors. In SCNT-ESC lines, the blastocyst formed contains identical genomic information as the donor (Figure 1). Given the nature of this technique, any shared mutations in SCNT ESCs derived from the same donor cells were possibly inherited from a rare parental fibroblast carrying these mutations. None of these mutations located in any of the known mutation hot spots, so the possibility of seeing two mutations occurring at the same position due to “mutation hot spots” was too low even for one line. After removing these potentially preexisting shared mutations, four SCNT-ESC lines carried no detectable coding mutations introduced during reprogramming. In contrast, all mutations identified in the iPSC lines were unique. We observed great variability in somatic-coding mutational load across SCNT-ESC and iPSC lines that contributed to chimeric mice. Therefore, these detectable mutations appear to have no apparent functional consequence in development. Furthermore, none of the mutated genes clusters in a specific functional pathway.
Discussion
The major advantage of using cells from an inbred mouse strain for the comparison of various reprogramming technologies was that the MEFs isolated from these mice were genetically identical, in contrast to the human cell lines that have much more genetic variability. It also enables more comprehensive pluripotent functionality testing such as the contribution to chimeric mice. In addition, genetic difference between inbred mice is usually present as homozygous variants, whereas somatic mutations would always appear as heterozygous. By exome sequencing of SCNT ESCs and iPSCs reprogrammed from syngeneic mouse B6 fibroblasts, we were able to perform a direct comparison of the somatic mutation load between the two nuclear-reprogramming methods.
Previous studies reported an average of 6–12 somatic-coding mutations in human iPSC lines when compared against their corresponding somatic cell of origin (Gore et al., 2011, Ji et al., 2012, Ruiz et al., 2013). We discovered that, at least in mouse cells, the somatic mutation load in SCNT-ESC lines was significantly lower than that of iPSC lines. Furthermore, one of the SCNT-ESC lines has no detectable coding mutation. Studies have suggested that some but not all identified somatic variants in iPSCs, such as point mutations and copy number variations, were present in their progenitor cells, whereas others were introduced during reprogramming (Abyzov et al., 2012, Gore et al., 2011, Young et al., 2012). Therefore, our findings of two pairs of SCNT-ESC lines that only harbor shared mutations suggest that genetic variants most likely preexisted in the somatic population of origin, without acquiring any additional coding somatic mutation during reprogramming. The differential somatic mutation load in pluripotent stem cells reprogrammed with the two methods could be due to the difference in their derivation time. In this context, the iPSCs are established 2–4 weeks after ectopic expression of transcription factors, whereas SCNT ESCs are established 4 days after oocyte activation. Therefore, it is very likely that formation of iPSCs has to go through additional rounds of cell division and a potentially more stressful condition when compared to SCNT ESCs. Taking into consideration the differential reprogramming time, the iPSCs spent an average of 50–77 days in culture, whereas SCNT ESCs spent an average of 25–32 days in culture. By dividing the average number of mutations observed with the number of days in culture, iPSCs give rise to at least twice the number of mutations per day compared to SCNT ESCs. Although it has been reported that reprogramming associated somatic-coding mutations individually does not provide a selective advantage to facilitate the acquisition of pluripotency during reprogramming, it remains to be determined whether a combination of mutations could have a role in reprogramming (Ruiz et al., 2013). However, we could not rule out the possibility that, during an extended period of induced pluripotency, somatic mutations might selectively accumulate and/or enrich over time.
Our data suggest that, when compared to induced pluripotency, SCNT might be a safer way to reprogram somatic cells into a pluripotent state with a lower mutation load. Therefore, it is important to optimize the condition of induced pluripotency into a more physiologically relevant context to minimize genetic aberrations and improve the feasibility for clinical application.
Experimental Procedures
Mice and Cell Culture
All mouse work was approved by the Institutional Animal Care and Use Committee of Peking University and UCSD. B6D2F1 mice (8–10 weeks old) were superovulated with 7 U of pregnant mare’s serum gonadotropin and 9 U of human chorionic gonadotropin (HCG) 48 hr later. Metaphase II (MII) oocytes were then collected 14 hr after HCG injection for nuclear transplantation experiments. MEF cells were derived from 13.5 days postcoitum (dpc) embryos collected from B6 mice. Primary MEFs were cultured in Dulbecco’s modified Eagle’s media (DMEMs) supplemented with 10% fetal bovine serum (FBS).
Nuclear Transfer
The spindle-chromosome complexes (SCCs) of MII oocytes were enucleated using a blunt Piezo-driven pipette in a droplet of HEPES-CZB medium containing 5 μg/ml cytochalasin B (CB). After enucleating, oocytes were maintained in CZB medium until injection. The fibroblast nucleus was separated and injected into the enucleated oocytes using a Piezo drill micromanipulator. The reconstructed oocytes were cultured in CZB medium for approximately 1–3 hr before activation. Then activation was achieved for 6 hr in calcium-free CZB medium containing 10 mM of strontium chloride and 5 μg/ml of CB supplemented with 250 nM scriptaid. Following activation, the reconstructed embryos were cultured in G1 with scriptaid at the same concentrations for the next 4 hr, then subsequently cultured in G1 and G2 medium (Vitrolife) at 37°C under 5% CO2 for 3.5 days.
Establishment of SCNT-ESC Lines
The SCNT-ESC lines were established as we previously described (Bai et al., 2007, Jiang et al., 2008, Yu et al., 2005), using knockout DMEM (Gibco) medium supplemented with 15% FBS, 1 μM PD0325901, and 3 μM CHIR99021, and 1,000 U/ml leukemia inhibitory factor (LIF; Chemicon).
In Vitro Differentiation
SCNT ESCs were dissociated into single cells with 0.25% Trypsin/EDTA (Gibco). After MEF feeder cells were depleted by incubating cell suspension for 10 min at 37°C, SCNT ESCs were transferred onto a low-adherence plate containing the differentiation medium that includes Iscove’s modified Eagle medium, 15% FBS, 2 mM GlutaMAX (Invitrogen), 1% nonessential amino acids (Invitrogen), 0.1 mM 2-mercaptoethanol (Invitrogen), and 1% penicillin-streptomycin (Invitrogen). After embryoid bodies were cultured in suspension for 4 days with a daily medium change, they were transferred onto a 24-well plate coated with 0.1% gelatin for an additional 5 days of spontaneous differentiation.
Immunohistochemistry
Differentiated cells were washed with PBS, fixed in 4% paraformaldehyde at room temperature for 20 min, and washed and blocked with PBS containing 0.1% Triton X-100 (Sigma-Aldrich) and 10% normal donkey serum (Jackson ImmunoResearch) at room temperature for 1 hr. Samples were incubated with primary antibodies at 4°C overnight, washed with PBS, and incubated with Alexa 488-conjugated and/or Alexa 549-conjugated secondary antibodies (Jackson ImmunoResearch) for 1 hr, then rinsed in PBS and counterstained with 1 μg/ml DAPI (Roche). The following primary antibodies and dilutions were used: β III-TUBULIN (1:100; Santa Cruz Biotechnology); NEUROD2 (1:200; Santa Cruz); α-SMA (1:200; Millipore); VIMENTIN (1:1:100; Santa Cruz); SOX17 (1:200; Millipore); and AFP (1:200; Abcam). Images were visualized by laser-scanning confocal microscopy (PerkinElmer; UltraView VoX).
Chimera Construction
Host eight-cell embryos were collected from imprinting control region (ICR) female mice at 2.5 dpc, and seven to ten SCNT ESCs were injected into them by a XYClone Laser System (Hamilton Thorne Biosciences). Chimeric mice were identified by coat color, and the male chimera was assessed for germline transmission by mating with ICR female mice.
iPSC Generation and Characterization
Mouse iPSCs were generated as previously described by Zhao et al. (2011). Briefly, MEFs were transfected with pCOSLNP vector using the Basic Nucleofector Kit for Primary Mammalian Fibroblasts (Lonza) followed by puromycin selection for 3 days and then were plated on irradiated B6 MEF feeders, cultured in mES medium, and treated with 5 μM PS48, 0.25 mM NaB (Stemgent), 0.5 μM A-83-01 (Stemgent), and 0.5 μM PD0325901 (Stemgent) for 4 weeks. After iPSC colonies were picked, the cell lines were established and expanded at roughly 3–4 days per passage, and the lack of random integration of the episomal vector was confirmed by Southern blotting analysis with a combination of probes that cover the entire episomal vector.
Whole-Genome Library Construction
Genomic DNA was collected and then extracted using the DNeasy Blood & Tissue Kit (QIAGEN). Next, the DNA (3 μg in 50 μl volumes) was fragmented with Covaris AFA and then processed following the manufacturer’s protocol (NEBNext DNA Library Prep Master Mix Set for Illumina). Briefly, fragmented DNA was end polished, A tailed, and then ligated to adaptors compatible with Illumina sequencing primers. The purified and ligated products were amplified by PCR to generate whole-genome libraries.
Quantitative Real-Time PCR
Total RNA was extracted and reverse transcribed into cDNA according to the manufacturer’s protocols. Briefly, total RNA from fibroblasts, ESCs, iPSCs, or SCNT ESCs was purified using the QIAshredder and RNeasy Mini Kit (QIAGEN) and then reserved transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR was performed using the SYBR Green PCR Master Mix (Applied Biosystems) and previously described primers for Oct3/4 (endogenous specific), Sox2 (endogenous specific), and Nanog (Zhao et al., 2011). Using Microsoft Excel, the values were averaged and normalized to GAPDH, then calculated relative to wild-type MEFs.
Flow Cytometric Analysis
Live cells from iPSCs, SCNT ESCs, or B6 ESCs were collected, washed with PBS, and incubated with fluorescein isothiocyanate (FITC)-conjugated secondary antibody such as SSEA-1-FITC (Stemgent) or IgM-FITC isotype control (BD Biosciences) prior to fluorescence-activated cell sorting analysis.
Teratoma Assay and Histology
Teratomas were formed by injecting one to three million iPSCs into severe combined immunodeficient mice as previously described by Zhao et al. (2011). The tumors were fixed in formalin or paraffin embedded, sectioned, hematoxylin and eosin stained, and imaged using an Olympus MVX10 MacroView Microscope for histology analysis.
In-Solution Hybridization Capture with DNA Baits
In-solution hybrid capture was performed according to the manufacturer’s protocol (Agilent SureSelect XT Mouse All Exon Kit), which the mouse whole-genome libraries were prepared for in-solution hybrid capture with SureSelect mouse exon RNA for 24–72 hr. The exome regions were recovered with streptavidin beads after incubation, then PCR amplified with 25 μl of template, 2 μM each of the primers Syb_FP5 and Syb_RP7, and 50 μl Phusion High-Fidelity Master Mix (New England Biolabs) at 98°C for 30 s, and 13 cycles of 98°C for 10 s, 60°C for 30 s, 72°C for 30 s, and 72°C for 5 min. The amplicons were purified with QIAGEN QIAquick columns, and the libraries were sequenced on an Illumina Genome Analyzer IIx or HiSeq.
Consensus Sequence Generation and Variant Calling
Variant calling was performed as previously described by Gore et al. (2011). Briefly, sequencing reads obtained from the Illumina Genome Analyzer or HiSeq were postprocessed and quality filtered using GERALD. The filtered reads were mapped to the mouse reference genome using BWA and down sampled using Picard. The consensus sequences generated by GATK in mouse iPSC or SCNT-ESC samples were then used to compare with progenitor samples to find candidate novel mutations. Each heterozygous SNP identified in iPSC or SCNT-ESC lines that was not observed in the progenitor line was considered candidate mutations.
Sanger Validation of Candidate Mutations
Genomic DNA (6 ng) extracted from mouse iPSC, SCNT ESC, and its somatic progenitor lines was amplified in 50 μl PCRs with 100 nM of specifically designed forward and reverse primers around the mutation site (primers available upon request), and 25 μl of Taq 2× master mix (NEB) at 94°C for 2 min, then 35 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 30 s, and final extension at 72°C for 3 min. Then the PCR product was purified with QIAquick columns. For Sanger sequencing validation, 10 ng of purified DNA was premixed with 25 pmol of the forward primer and submitted to Genewiz.
Statistical Analysis
The Mann-Whitney U test (unpaired, nonparametric) was performed to determine whether the mouse SCNT-ESC and iPSC lines have a significant amount of somatic-coding mutations. The result suggests that there is a statistically significant difference between the somatic-coding mutational load of the iPSC and SCNT-ESC lines (p < 0.004). We used DAVID (Dennis et al., 2003) to analyze the somatic-coding mutations identified in both iPSC and SCNT-ESC lines and check for commonly mutated pathways. The mutations do not seem to be mutated in any common pathways.
Acknowledgments
We thank Athurva Gore for bioinformatics support and UCSD pathology core for histologic analysis. This work was supported by a grant from National Basic Science Program of China, Chinese Ministry of Science and Technology (2013CB966900), and grants from California Institute for Regenerative Medicine (RB3-05083) to K.Z. and (TR1-01277 and RM-0173) to Y.X.
Published: March 27, 2014
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information includes two figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2014.02.005.
Contributor Information
Kun Zhang, Email: kzhang@bioeng.ucsd.edu.
Hongkui Deng, Email: hongkui_deng@pku.edu.cn.
Yang Xu, Email: yangxu@ucsd.edu.
Accession Numbers
The NCBI Sequence Read Archive accession number for the sequencing data reported in this paper is SRP038139.
Supplemental Information
Reference
- Abyzov A., Mariani J., Palejev D., Zhang Y., Haney M.S., Tomasini L., Ferrandino A.F., Rosenberg Belmaker L.A., Szekely A., Wilson M. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki R., Uda M., Hoki Y., Sunayama M., Nakamura M., Ando S., Sugiura M., Ideno H., Shimada A., Nifuji A., Abe M. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature. 2013;494:100–104. doi: 10.1038/nature11807. [DOI] [PubMed] [Google Scholar]
- Bai Z., Yong J., Qing T., Cheng J., Shen W., Ding M., Deng H. Serial nuclear transfer improves the developmental potential of mouse embryos cloned from oocytes matured in a protein-free medium. Mol. Reprod. Dev. 2007;74:560–567. doi: 10.1002/mrd.20614. [DOI] [PubMed] [Google Scholar]
- Campbell K.H., McWhir J., Ritchie W.A., Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- Cheng L., Hansen N.F., Zhao L., Du Y., Zou C., Donovan F.X., Chou B.K., Zhou G., Li S., Dowey S.N., NISC Comparative Sequencing Program Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell. 2012;10:337–344. doi: 10.1016/j.stem.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesné P., Adenot P.G., Viglietta C., Baratte M., Boulanger L., Renard J.P. Cloned rabbits produced by nuclear transfer from adult somatic cells. Nat. Biotechnol. 2002;20:366–369. doi: 10.1038/nbt0402-366. [DOI] [PubMed] [Google Scholar]
- Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Egli D., Chen A.E., Saphier G., Ichida J., Fitzgerald C., Go K.J., Acevedo N., Patel J., Baetscher M., Kearns W.G. Reprogramming within hours following nuclear transfer into mouse but not human zygotes. Nat. Commun. 2011;2:488. doi: 10.1038/ncomms1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A., Li Z., Fung H.L., Young J.E., Agarwal S., Antosiewicz-Bourget J., Canto I., Giorgetti A., Israel M.A., Kiskinis E. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Ng S.H., Sharma V., Neculai D., Hussein S., Sam M., Trinh Q., Church G.M., McPherson J.D., Nagy A., Batada N.N. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells. 2012;30:435–440. doi: 10.1002/stem.1011. [DOI] [PubMed] [Google Scholar]
- Jiang W., Bai Z., Zhang D., Shi Y., Yong J., Chen S., Ding M., Deng H. Differentiation of mouse nuclear transfer embryonic stem cells into functional pancreatic beta cells. Diabetologia. 2008;51:1671–1679. doi: 10.1007/s00125-008-1065-1. [DOI] [PubMed] [Google Scholar]
- Lister R., Pelizzola M., Kida Y.S., Hawkins R.D., Nery J.R., Hon G., Antosiewicz-Bourget J., O’Malley R., Castanon R., Klugman S. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S., Diep D., Gore A., Panopoulos A.D., Montserrat N., Plongthongkum N., Kumar S., Fung H.-L., Giorgetti A., Bilic J. Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA. 2012;109:16196–16201. doi: 10.1073/pnas.1202352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S., Gore A., Li Z., Panopoulos A.D., Montserrat N., Fung H.L., Giorgetti A., Bilic J., Batchelder E.M., Zaehres H. Analysis of protein-coding mutations in hiPSCs and their possible role during somatic cell reprogramming. Nat. Commun. 2013;4:1382. doi: 10.1038/ncomms2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Amato P., Sparman M., Gutierrez N.M., Tippner-Hedges R., Ma H., Kang E., Fulati A., Lee H.S., Sritanaudomchai H. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153:1228–1238. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Wakayama T., Perry A.C., Zuccotti M., Johnson K.R., Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- Young M.A., Larson D.E., Sun C.W., George D.R., Ding L., Miller C.A., Lin L., Pawlik K.M., Chen K., Fan X. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Yong J., Li X., Qing T., Qin H., Xiong X., You J., Ding M., Deng H. The proteasomal inhibitor MG132 increases the efficiency of mouse embryo production after cloning by electrofusion. Reproduction. 2005;130:553–558. doi: 10.1530/rep.1.00758. [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhao T., Zhang Z.N., Rong Z., Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–215. doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


