Abstract
Today it is generally accepted that B cells require cognate interactions with CD4+ T cells to develop high-affinity antibodies against proteins. CD4+ T cells recognize peptides (epitopes) presented by MHC class II molecules that are expressed on antigen-presenting cells. Structural features of both the MHC class II molecule and the peptide determine the specificity of CD4+ T cells that can bind to the MHC class II-peptide complex. We used a new humanized hemophilic mouse model to identify FVIII peptides presented by HLA-DRB1*1501. This model carries a knock-out of all murine MHC class II molecules and expresses a chimeric murine-human MHC class II complex that contains the peptide-binding sites of the human HLA-DRB1*1501. When mice were treated with human FVIII, the proportion of mice that developed antibodies depended on the application route of FVIII and the activation state of the innate immune system. We identified 8 FVIII peptide regions that contained CD4+ T-cell epitopes presented by HLA-DRB1*1501 to CD4+ T cells during immune responses against FVIII. CD4+ T-cell responses after intravenous and subcutaneous application of FVIII involved the same immunodominant FVIII epitopes. Interestingly, most of the 8 peptide regions contained promiscuous epitopes that bound to several different HLA-DR proteins in in vitro binding assays.
Introduction
The most serious complication in replacement therapy with FVIII products is the development of neutralizing antibodies against FVIII (FVIII inhibitors), which is observed in approximately 25% to 30% of patients with severe hemophiliaA.1Although several genetic2 and nongenetic3 factors that contribute to the risk for patients to develop these antibodies have been identified, why some patients develop antibodies while others do not remains largely unknown.
Today it is generally accepted that B cells require cognate interactions with CD4+ T cells to develop high-affinity antibodies against protein antigens.4,5 In line with this perception, several lines of evidence have supported the involvement of CD4+ T cells in the generation of antibody responses against FVIII in patients with hemophilia A and in murine hemophilia models.6,7 CD4+ T cells express T-cell receptors that recognize antigen-derived peptides (CD4+ T-cell epitopes) presented by MHC class II molecules, which are expressed on specialized antigen-presenting cells.8 Structural features of both the MHC class II molecule and the peptide determine the specificity of CD4+ T cells that can bind to the MHC class II-peptide complex.8,9 The conditions under which CD4+ T cells interact with this complex determine whether the immune system reacts with nonresponsiveness, is activated to develop specific antibodies, or is tolerized to suppress antibody responses.9,10 Therefore, it is crucial to understand which FVIII peptides are presented by MHC class II complexes under conditions of FVIII replacement therapy and how CD4+ T cells interact with MHC class II-FVIII peptide complexes expressed by antigen-presenting cells. The available information on FVIII peptides presented in the context of specific human MHC class II molecules is limited. Several studies used peripheral blood cells of patients and healthy controls11 to identify CD4+ T-cell epitopes in the A2 domain,12 A3 domain,13 and C2 domain of FVIII.14 However, these studies lack information on the specific MHC class II molecules associated with the FVIII peptides identified. Jacquemin et al identified T-cell epitopes of FVIII using CD4+ T-cell clones isolated from a mild hemophilia A patient carrying an Arg2150His mutation in the C1 domain of FVIII.15 All clones recognized FVIII peptides encompassing residue Arg2150. Peptides were presented by HLA-DRB1*0401/HLA-DRB4*01 or HLA-DRDRB1*1501/HLA-DRB5*0101. One of the peptides identified was a promiscuous epitope that bound to several different HLA-DR proteins. James et al used MHC class II tetramers to analyze FVIII-specific CD4+ T cells obtained from a mild hemophilia A patient carrying an Ala2201Pro mutation in the C2 domain of FVIII.16 Responses of CD4+ T cells to sequences containing Ala2201 (wild-type), Pro2201 (hemophilic), and other predicted T-cell epitopes were evaluated and resulted in the identification of an HLA-DRB1*0101 restricted T-cell epitope containing the wild-type Ala2201 sequence of FVIII. In another study, James et al investigated 2 patients carrying an Arg593Cys mutation in the A2 domain of FVIII.17 The authors found a promiscuous CD4+ T-cell epitope that contained the wild-type Arg593 sequence of FVIII and bound to 3 different HLA-DR proteins (HLA-DRB1*0101, HLA-DRB1*1101, and HLA-DRB1*1501). Recently, van Haren et al identified FVIII-specific CD4+ T-cell epitopes by analyzing FVIII peptides eluted from human monocyte-derived dendritic cells that were obtained from healthy blood donors expressing different MHC class II haplotypes (DRB1*0101/DRB1*1301; DRB1*0701/DRB1*1501; DRB1*0401/DRB1*0404; DRB1*0101/DRB1*1302).18 Several of the FVIII epitopes identified were promiscuous and bound to different HLA-DR proteins.
We created a humanized hemophilic mouse model19 to identify FVIII peptides presented by HLA-DRB1*1501 and to study the regulation of antibody responses against FVIII by the interaction of CD4+ T-cell subsets with FVIII peptides presented by HLA-DRB1*1501. We were particularly interested in HLA-DRB1*1501 because this MHC class II protein was shown to be associated with an increased risk for patients to develop FVIII inhibitors.20-22 The new humanized hemophilic mouse model (E17 HLA-DRB1*1501 mice) carries a knockout of the entire murine MHC class II complex and expresses a chimeric murine-human MHC class II complex, which contains the sequences of the human HLA-DRA*0101 and HLA-DRB1*1501 proteins responsible for peptide binding, and the murine binding sides for the murine CD4 protein. This chimeric murine-human MHC class II complex was shown to facilitate an optimal human MHC class II-restricted CD4+ T-cell response.23
Early studies indicated that only a fraction of E17 HLA-DRB1*1501 mice developed antibodies when intravenously treated with human FVIII.19 These findings evoked the question of whether the chimeric murine-human MHC class II complex is able to select a polyclonal repertoire of naive CD4+ T cells. We present evidence that the repertoire of naive CD4+ T cells selected by the chimeric murine-human MHC class II complex expresses the same degree of polyclonality as the repertoire of naive CD4+ T cells selected by the murine MHC class II complex in conventional E17 hemophilic mice. Next, we wondered whether the lack of antibody development in nonresponder mice is associated with the specific nature of the intravenous application route. We compared intravenous application with subcutaneous application of FVIII and combined intravenous application with a concomitant activation of the innate immune system. Our data indicate that both subcutaneous application of FVIII and a combination of intravenous FVIII together with an activation of the innate immune system result in the development of antibodies in all mice included in the study. Based on these data, we asked whether differences in responder rates after intravenous and subcutaneous application of FVIII might be associated with different immunodominant CD4+ T-cell epitopes involved in the immune response to intravenous and subcutaneous FVIII. We identified a set of 8 FVIII peptide regions containing epitopes that were presented by HLA-DRB1*1501 during in vivo immune responses against FVIII. Importantly, there was no major difference in the FVIII peptide regions identified between intravenous and subcutaneous application of FVIII. Most of the 8 FVIII peptide regions contained promiscuous epitopes that bound to several different HLA-DR proteins in in vitro binding assays.
We think that the new E17 HLA-DRB1*1501 hemophilic mouse model represents an interesting opportunity to uncover mechanisms that drive the immune system’s decision whether or not to develop antibodies against FVIII.
Methods
Mouse studies
All mouse studies were done in accordance with Austrian federal law (Act BG 501/1989) regulating animal experimentation and were approved by the Baxter Bioscience Institutional Animal Care and Use Committee. Mice were male and 8 to 12 weeks old at the beginning of the experiments.
Humanized E17 HLA-DRB1*1501 mice
Humanized E17 HLA-DRB1*1501 mice carry a knockout of all mouse MHC class II molecules and express a chimeric human-mouse MHC class II protein, which contains the human sequences responsible for binding peptides and the murine sequence responsible for binding murine CD4.19 Furthermore, these mice express the hemophilic E17 FVIII knockout as described by Bi et al.24 Humanized E17 HLA-DRB1*1501 mice had a mixed C57BL6/S129 background.
Conventional E17 mice
Our colony of conventional E17 hemophilic mice was established with a breeding pair from the original colony24 and crossed into the C57BL/6J background.25
Treatment of mice with recombinant human FVIII
Mice received a minimum of 4 intravenous or subcutaneous doses of 1 μg recombinant human FVIII (full-length FVIII; Baxter BioScience) in 200 μL Advate formulation buffer (Baxter BioScience) at weekly intervals as indicated.
In some experiments, 2 μg lipopolysaccaride (LPS, ligand for toll-like receptor 4; InvivoGen) was administered together with FVIII. LPS was reconstituted according to the manufacturer’s instructions and subsequently mixed with FVIII.
Detection of antibodies against human FVIII and against the C2 domain of human FVIII
Blood samples were collected 7 days after the last dose of FVIII. Whole blood was added to sodium citrate at a 4:1 ratio, and plasma was separated by centrifugation. Antibodies against human FVIII in plasma samples were measured using ELISAs as described.26 ELISA plates were coated with either 1 μg/mL recombinant human FVIII or 1 μg/mL recombinant C2 domain of human FVIII (ProBioGen). Inhibitory antibodies were analyzed with a modified Bethesda assay (Technoclone).
Detection of antigen-specific CD4+ T cells
Antigen-specific CD4+ T cells were detected by expression of CD154 after short-term in vitro restimulation with the antigen of interest as described by Frentsch et al.27 Mice were treated with 8 weekly doses of human FVIII. Three days after the last dose, spleens were obtained and spleen cells were prepared as described.28 Spleen cells were in vitro restimulated for 16 hours at 37°C and 5% CO2 with either human FVIII (20 μg/mL) or human FVIII peptides (1 μg/mL per peptide) as indicated. After 1 hour of restimulation, 2.5 μg brefeldin A (BD Biosciences PharMingen) was added to the cultures to inhibit intracellular protein transport. After incubation, cells were washed and nonspecific binding sites were blocked by a mixture of anti-CD16 and anti-CD32 antibodies (Fc Block; BD Biosciences PharMingen). Afterward, cells were stained with a peridinin-chlorophyll protein complex-labeled antibody against extracellular CD4 (clone RM4-5, BD Biosciences PharMingen) and subsequently permeabilized for intracellular staining using Cytofix/Cytoperm (BD Biosciences PharMingen) according to the manufacturer’s instructions. Finally, cells were stained with an allophycocyanin-labeled antibody against intracellular CD154 (clone MR1; eBioscience) as described.27 Frequencies of CD4+CD154+ double-positive cells were analyzed using a BD LSR Fortessa flow cytometer (BD Biosciences) and FlowJo software (Version 7.6.3; TreeStar).
A live/dead cell marker (LIVE/DEAD Fixable Violet Dead Cell Stain Kit; Invitrogen) was included in all staining procedures and used according to the manufacturer’s protocol.
Generation of FVIII-specific T-cell hybridoma clones and assessment of peptide specificity
Mice were treated with 4 to 8 weekly doses of human FVIII. Three days after the last dose, spleens were obtained and spleen cells were prepared as described.28 Spleen cells were restimulated with 20 μg/mL recombinant human FVIII and kept for 3 or 10 days at 37°C and 5% CO2 to generate CD4+ T-cell blasts. Cells were then washed, fused with BW5147 cells (BW5147.G.1.4; ATCC #TIB 48) using polyethylene glycol fusion as described,29 and plated into 96 well microplates (TPP Techno Plastic Products). Microcultures were kept at 37°C and 5% CO2 until growing clones could be detected. The culture medium was changed to hypoxanthine-aminopterin-thymidine selection medium (hypoxanthine-aminopterin-thymidine medium supplement; Sigma-Aldrich) after 48 hours and maintained for 2 weeks. T-cell hybridoma clones were picked and screened for specificity to human FVIII.
Screening for FVIII-specific hybridoma clones
A total of 1 × 105 hybridoma cells were cocultured with antigen-presenting cells in the presence of 10 μg/mL human FVIII for 24 hours at 37°C and 5% CO2. Culture supernatants were collected and analyzed for the release of IL-2 using either an IL-2 ELISA Kit (BioLegend) or an IL-2 Bio-Plex assay (Bio-Rad Laboratories). Hybridoma clones with an at least 5-fold increase in IL-2 release (ratio between FVIII-stimulated cultures and medium controls) were considered to be FVIII-specific and were subsequently subcloned by limiting dilution (0.3 cells/well).
Assessment of peptide specificity
A total of 1 × 105 hybridoma cells were cocultured with antigen-presenting cells in the presence of human FVIII peptide pools containing up to 33 FVIII peptides (1 μg/mL for each peptide). Peptide specificities of individual hybridomas were identified using a cross matrix scheme as described by Ay et al.30 In brief, 2 identical peptide membranes were synthesized and cut into vertical or horizontal strips. Each strip was processed into one pool of peptides. Each T-cell hybridoma was tested against all peptide pools that were processed from the vertical and horizontal strips. The crossing point of positive hits for the vertical and the horizontal pools revealed the peptide that was recognized by a particular T-cell hybridoma clone.30 Peptide specificities were confirmed by testing the peptides identified individually against scrambled versions of the same peptides.
Spleen cells obtained from either naive humanized E17 HLA-DRB1*1501 mice or naive conventional hemophilic E17 mice were used as antigen-presenting cells. For comparison, MGAR cells (ECACC #88022014), obtained from a human antigen-presenting cell line expressing HLA-DRB1*1501, were used as antigen-presenting cells for the human HLA-DRB1*1501 complex.
Human dendritic cells
In some experiments, human monocyte-derived dendritic cells were used as human antigen-presenting cells. Human monocyte-derived dendritic cells were generated from CD14+ peripheral monocytes obtained from healthy blood donors homozygously expressing the MHC class II protein HLA-DRB1*1501, as described by Pickl et al.31
Human FVIII peptides
Human FVIII peptide libraries (15 mers, shifted by 3 amino acids) covering the whole sequence of full-length human FVIII were synthesized using SPOT synthesis as described by Ay et al30 (Rudolf Volkmer; Charité Berlin, Humboldt-University). Peptide pools with up to 33 peptides per pool were dissolved in DMSO (DMSO Hybrimax; Sigma-Aldrich) and further diluted in Dulbecco PBS (Invitrogen).
Single peptides were synthesized using solid-phase peptide synthesis (Rudolf Volkmer), dissolved in DMSO and further diluted in Dulbecco PBS (Invitrogen).
Amino acid numbering in the human FVIII peptides is based on the full-length human FVIII protein, without the 19 amino acid signal peptide.
Binding of peptides to HLA-DRB1* haplotypes
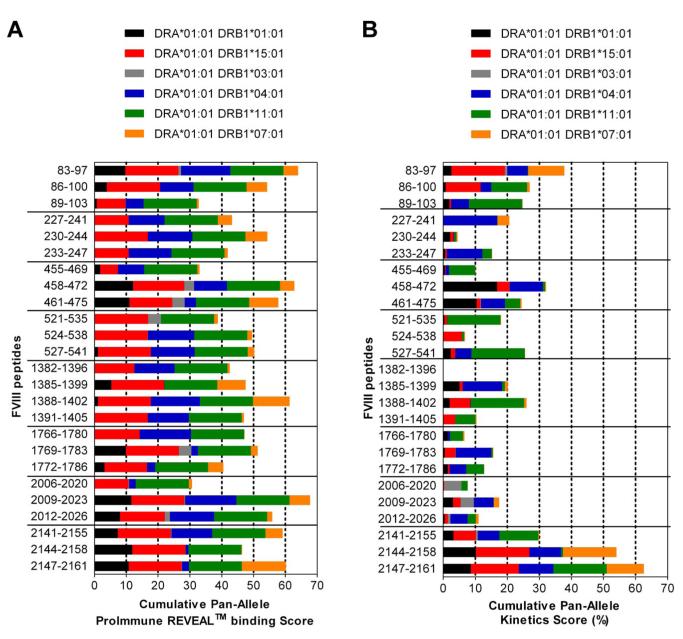
Binding of peptides to HLA-DRB1* haplotypes was analyzed by ProImmune using their cell-free REVEAL class II binding technology (ProImmune; www.proimmune.com). The Cumulative Pan-Allele ProImmune REVEAL score for each peptide was calculated by adding up the binding scores calculated for each peptide and each of the 6 HLA-DRB1* haplotypes analyzed and dividing the sum by 6 (number of HLA-DRB1* haplotypes analyzed). Different colors corresponding to the contribution of each of the different HLA-DRB1* haplotypes tested were used for illustration (see Figure 6A).
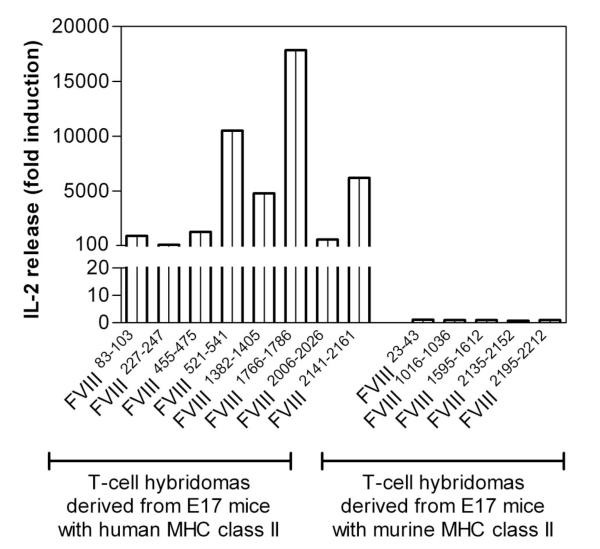
Figure 6. Human monocyte-derived dendritic cells expressing HLA-DRB1*1501 process and present immunodominant epitopes of human FVIII that were identified in E17 HLA-DRB1*1501 mice.
T-cell hybridomas derived from E17 HLA-DRB1*1501 mice (E17 mice with human MHC class II) or from conventional E17 mice (E17 mice with murine MHC class II) were cocultured with human monocyte-derived dendritic cells, which express HLA-DRB1*1501, in the presence of human FVIII for 24 hours. Culture supernatants were collected and analyzed for the release of IL-2. Presented is the fold induction of IL-2, reflecting the IL-2 levels found in culture supernatants of cultures restimulated with FVIII compared with culture supernatants of cultures restimulated in the absence of FVIII with culture medium only.
The kinetics score for binding of each peptide to HLA-DRB1* haplotypes was calculated based on the on-rates and off-rates of peptide binding using the full rate assay technology from ProImmmune. The cumulative pan-allele kinetics score for each peptide was calculated by adding up the kinetics scores calculated for each peptide and each of the 6 HLA-DRB1* haplotypes analyzed and dividing the sum by 6 (number of HLA-DRB1* haplotypes analyzed). Different colors corresponding to the contribution of each of the different HLA-DRB1* haplotypes tested were used for illustration (see Figure 6B).
Details of all the technologies used by Proimmune are provided in Supplemental Methods.
Analysis of the combinatorial diversity of the T-cell receptor-β chain
The diversity of the T-cell receptor-β chain was analyzed in CD4+ T cells purified from spleen cells of naive mice. Spleens were obtained from naive humanized E17 HLA-DRB1*1501 mice and from naive conventional E17 mice, and spleen cells were prepared as described.28 CD4+ T cells were purified from spleen cells by magnetic cell sorting using mouse CD4 Dynabeads (Invitrogen). The diversity of the T-cell receptor-β chain repertoire was analyzed by ImmunID Technologies using an Immun-TraCkeR test as described.32 Details of this technology are provided in the Supplemental Methods.
Statistical analysis
The unpaired 2-tailed Student t test was used for comparison of means between groups. P < .05 was considered statistically significant.
Results
The human HLA-DRB1*1501 selects a polyclonal repertoire of naive CD4+ T cells in humanized E17 HLA-DRB1*1501 mice
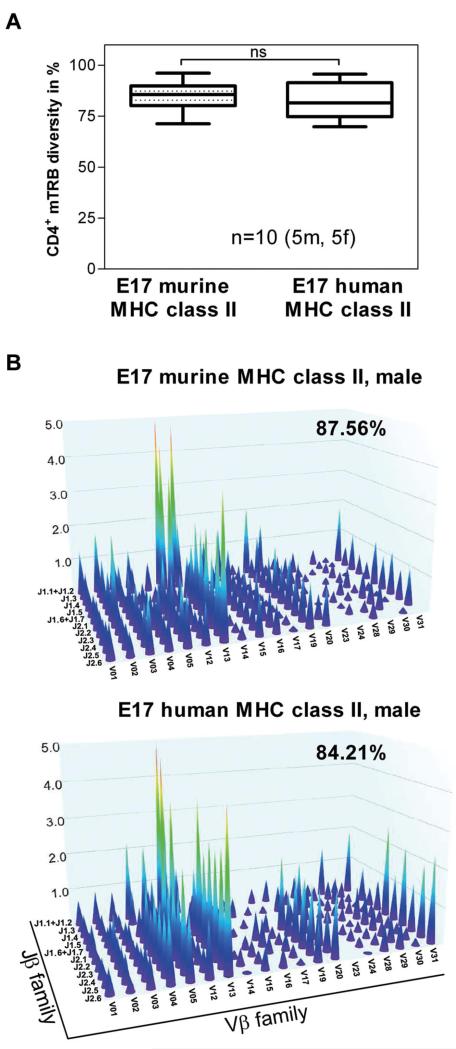
MHC class II molecules expressed by antigen-presenting cells are essential for selecting the CD4+ T-cell repertoire in the thymus.33,34 Therefore, we asked whether the chimeric human MHC class II molecule expressed in humanized E17 HLA-DRB1*1501 mice is able to select a polyclonal repertoire of naive CD4+ T cells. The results of our studies indicate that splenic naive CD4+ T cells obtained from humanized E17 HLA-DRB1*1501 mice express a degree of polyclonality similar to that of naive CD4+ T cells found in conventional E17 mice expressing a murine MHC class II complex (Figure 1A-B).
Figure 1. The degree of polyclonality of the naive CD4+ T-cell repertoire in E17 HLA-DRB1*1501 mice is similar to the degree of polyclonality in E17 mice expressing a murine MHC class II complex.
Genomic DNA was isolated from splenic CD4+ T cells obtained from naive conventional E17 mice (E17 murine MHC class II) and naive E17 HLA-DRB1*1501 mice (E17 human MHC class II) and analyzed for the combinational diversity of V-J gene rearrangements of the T-cell receptor-β chain using multiplex PCR. (A) Box plots demonstrating the diversity of V-J gene rearrangements of the T-cell receptor-β chain (TRB) in naive conventional E17 mice (E17 murine MHC class II) and naive E17 HLA-DRB1*1501 mice (E17 human MHC class II) in percent (CD4+ mTRB diversity). Ten animals, 5 male (5m) and 5 female (5f), from each mouse strain were included in the analysis. ns indicates not significant. (B) Three-dimensional representation of the repertoire of the CD4+ T-cell receptor-β chain for one representative naive conventional E17 mouse (E17 murine MHC class II) and one representative naive E17 HLA-DRB1*1501 mouse (E17 human MHC-class II). Each peak represents the rearrangement of a family of V genes, relative to the rearrangement of the J genes. The intensity of these rearrangements is represented on the z-axis.
Based on these data, we conclude that the chimeric human MHC class II molecule expressed in humanized E17 HLA-DRB1*1501 mice is functionally active and able to select a polyclonal naive CD4+ T-cell repertoire.
Application route of FVIII determines the incidence of antibody responses but not the selection of immunodominant CD4+ T-cell epitopes
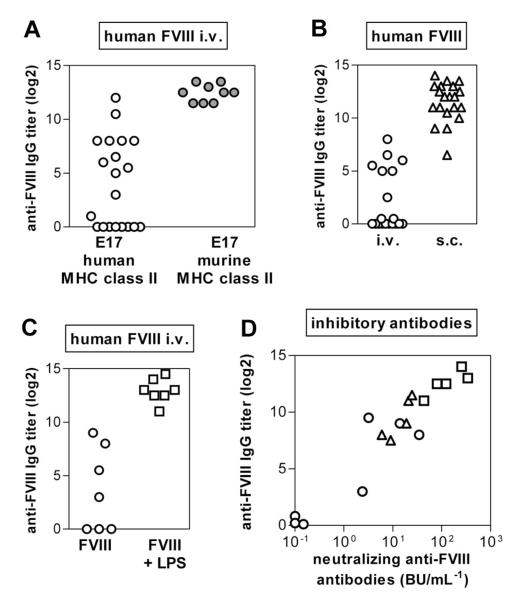
Previously, we reported that intravenous application of human FVIII induced antibodies in only a proportion of humanized E17 HLA-DRB1*1501 mice.19 Up to now, we have analyzed antibody responses of 228 mice included in 12 studies. The responder rates have varied between 30% and 80%. The differences in responder rates in different studies might be the result of the mixed background of the mice. Based on these data, we now asked whether the nonresponsiveness of a proportion of humanized E17 HLA-DRB1*1501 mice reflects the inability of these mice to respond to human FVIII or is associated with the intravenous application route. We treated one group of mice with intravenous FVIII and another group of mice with subcutaneous FVIII. Our results confirmed that only a fraction of mice developed antibodies after intravenous application of human FVIII (Figure 2A), in contrast to conventional E17 mice that we included as a control group (Figure 2A). However, all humanized E17 HLA-DRB1*1501 mice developed antibodies (Figure 2B) when treated with subcutaneous FVIII. Based on these results, we wondered whether the proportion of mice responding to treatment with intravenous FVIII would increase by concurrent activation of the innate immune system. Our results show that all mice responded with antibodies to FVIII when treated intravenously with a mixture of FVIII and LPS, a well-known stimulator of the innate immune system35 (Figure 2C).
Figure 2. The incidence of antibody responses to FVIII in E17 HLA-DRB1*1501 mice depends on the application route of FVIII and on the activation state of the innate immune system.
Mice were treated with human FVIII at weekly intervals for 7 (A) or 8 (B-C) consecutive weeks. One week after the last dose, plasma samples were collected and analyzed by ELISA for the presence of anti-FVIII antibodies. (A) Comparison of the incidence of antibody responses after intravenous FVIII treatment of E17 HLA-DRB1*1501 mice (○) and conventional E17 mice expressing murine MHC class II ( ). Each data point represents an individual mouse. (B) Comparison of the incidence of antibody responses after intravenous (○) and subcutaneous (△) FVIII treatment of E17 HLA-DRB1*1501 mice. Each data point represents an individual mouse. (C) Comparison of the incidence of antibody responses after intravenous FVIII treatment of E17 HLA-DRB1*1501 mice in the absence (○; FVIII) or presence (□; FVIII + LPS) of a concomitant activation of the innate immune system. Each data point represents an individual mouse. (D) Correlation of neutralizing and total binding antibodies after intravenous (○) or subcutaneous (△) FVIII or intravenous FVIII + LPS (□) of HLA-DRB1*1501 mice. Each data point represents an individual mouse.
). Each data point represents an individual mouse. (B) Comparison of the incidence of antibody responses after intravenous (○) and subcutaneous (△) FVIII treatment of E17 HLA-DRB1*1501 mice. Each data point represents an individual mouse. (C) Comparison of the incidence of antibody responses after intravenous FVIII treatment of E17 HLA-DRB1*1501 mice in the absence (○; FVIII) or presence (□; FVIII + LPS) of a concomitant activation of the innate immune system. Each data point represents an individual mouse. (D) Correlation of neutralizing and total binding antibodies after intravenous (○) or subcutaneous (△) FVIII or intravenous FVIII + LPS (□) of HLA-DRB1*1501 mice. Each data point represents an individual mouse.
Previously, we demonstrated a correlation between binding antibodies as detected in ELISA assays and neutralizing antibodies as detected in Bethesda assays when mice were treated with intravenous FVIII.19 We now asked whether antibodies that develop after subcutaneous FVIII and after intravenous FVIII + LPS also express neutralizing activity against FVIII. Data presented in Figure 2D show that plasma samples containing antibodies against FVIII express neutralizing activity after all 3 treatment regimens (intravenous FVIII, intravenous FVIII + LPS, subcutaneous FVIII).
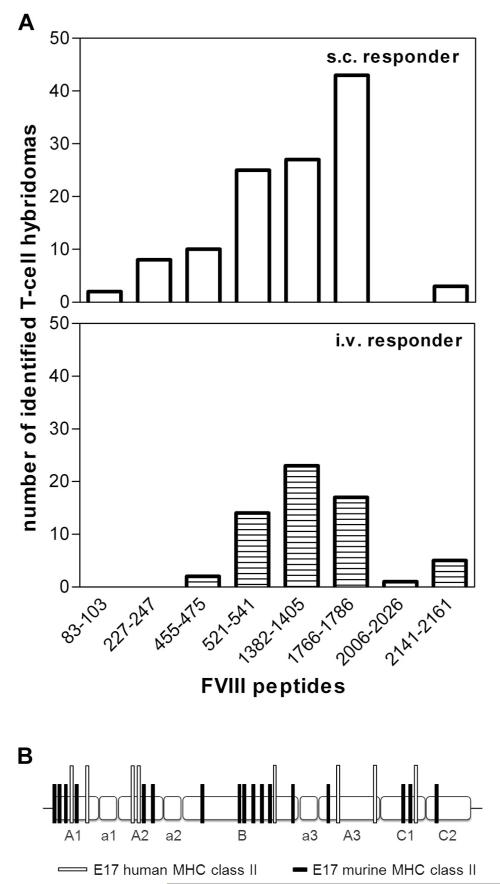
Next, we asked whether differences in the incidence of antibody responses after intravenous and subcutaneous application of FVIII could be caused by differences in CD4+ T-cell epitopes involved in the immune response to intravenous and subcutaneous FVIII. To address this question, we established and characterized FVIII-specific T-cell hybridoma libraries using splenic CD4+ T cells obtained from humanized E17 HLA-DRB1*1501 mice treated with up to 8 doses of human FVIII, given either intravenously or subcutaneously. We used a peptide library of human FVIII that contained 15-mer peptides shifted by 3 amino acids to characterize the peptide specificity of each individual T-cell hybridoma clone. In total, we analyzed 63 hybridoma clones obtained from mice treated with intravenous FVIII and 118 hybridoma clones obtained from mice treated with subcutaneous FVIII. We identified 8 FVIII peptide regions containing T-cell epitopes (Figure 3A; supplemental Table 1). Five of these 8 peptide regions were identical, including the 3 dominant peptide regions, after intravenous and subcutaneous application of FVIII (Figure 3A). Three minor peptide regions were only found for 1 of the 2 application routes. However, these minor differences might disappear when analyzing a larger number of hybridoma clones.
Figure 3. Immune response after intravenous and subcutaneous FVIII treatment of E17 HLA-DRB1*1501 mice is driven by the same immunodominant CD4+ T-cell epitopes, which are completely different from CD4+ T-cell epitopes found in E17 mice expressing a murine MHC class II complex.
E17 HLA-DRB1*1501 mice (E17 human MHC class II) and conventional E17 mice (E17 murine MHC class II) were immunized 4 to 8 times by intravenous or subcutaneous FVIII at weekly intervals. Spleen cells were obtained at 3 to 7 days after the last immunization and restimulated with FVIII in vitro. T-cell hybridomas were generated and characterized for their peptide specificity using a 15-mer peptide library (12 amino acids overlap) covering the sequence of full-length FVIII. Peptide specificities for all T-cell hybridoma clones could be identified, indicating that the large number of peptides per pool did not interfere with peptide recognition. (A) The number of T-cell hybridomas obtained from E17 HLA-DRB1*1501 mice that were identified for each of the indicated FVIII epitopes. A total of 118 hybridomas obtained after subcutaneous FVIII and 63 hybridomas obtained after intravenous FVIII treatment were analyzed. (B) The distribution of CD4+ T-cell epitopes identified in E17 HLA-DRB1*1501 mice (E17 human MHC class II) and in conventional E17 mice (E17 murine MHC class II) over the different domains (A1, a1, A2, a2, B, a3, A3, C1, and C2) of FVIII.
For comparison, we identified FVIII peptide regions containing T-cell epitopes involved in the immune response to intravenous FVIII in conventional hemophilic E17 mice. We analyzed 50 hybridoma clones and demonstrated that FVIII peptide regions containing T-cell epitopes are completely different in conventional hemophilic E17 mice and in humanized hemophilic E17 HLA-DRB1*1501 mice (Figure 3B; supplemental Table 1).
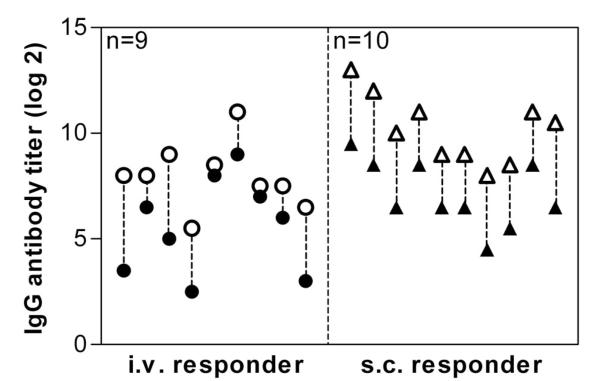
The C2 domain of human FVIII is generally considered to be very immunogenic. However, this assumption is mainly based on epitope data for antibodies against FVIII. Interestingly, we did not find a major CD4+ T-cell epitope in the C2 domain in humanized E17 HLA-DRB1*1501 mice. Therefore, we were wondering whether these mice would still develop antibodies against the C2 domain. We treated mice either intravenously or subcutaneously with FVIII and analyzed antibodies against full-length human FVIII and against the C2 domain of human FVIII. Our data indicate that all mice that developed antibodies against full-length FVIII also developed antibodies against the C2 domain of human FVIII (Figure 4).
Figure 4. Antibody response to full-length FVIII and to the C2 domain of FVIII.
Mice were treated with human FVIII, intravenously (○ ), or subcutaneously (△▲), at weekly intervals for 8 consecutive weeks. One week after the last dose, plasma samples were collected and analyzed by ELISA for the presence of antibodies against FVIII (○△) and against the C2 domain of human FVIII (
), or subcutaneously (△▲), at weekly intervals for 8 consecutive weeks. One week after the last dose, plasma samples were collected and analyzed by ELISA for the presence of antibodies against FVIII (○△) and against the C2 domain of human FVIII ( ▲). Presented are IgG antibody titers of individual mice.
▲). Presented are IgG antibody titers of individual mice.
FVIII-specific T cells that recognize major epitopes identified by hybridoma technology are detectable in the spleen of mice treated with FVIII
After identifying the major FVIII peptide regions containing T-cell epitopes involved in the immune response against FVIII in humanized E17 HLA-DRB1*1501 mice, we asked whether FVIII-specific T cells that recognize these major T-cell epitopes can be detected in the spleen of mice treated with FVIII.
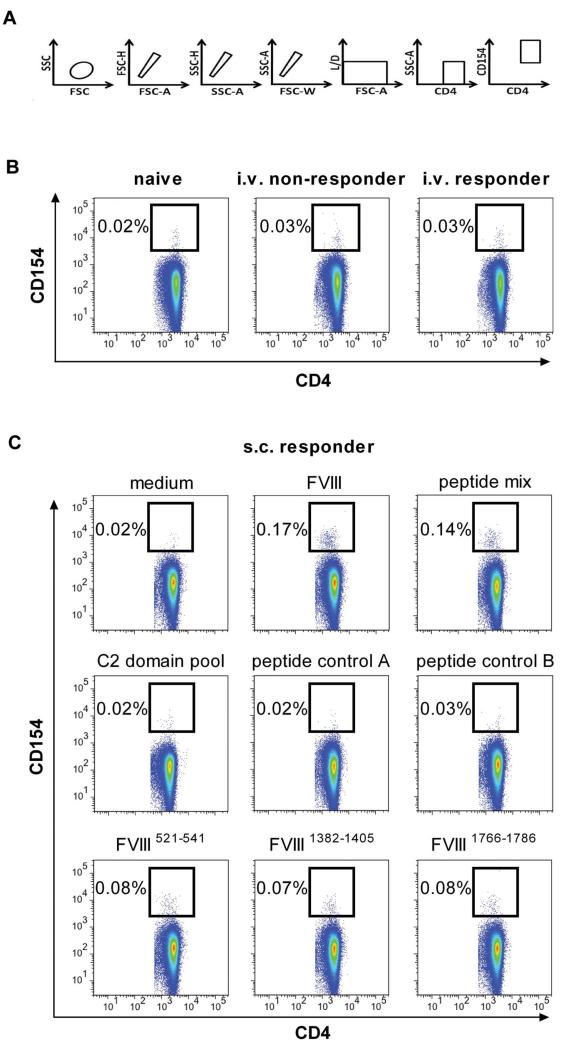
At first, we studied the frequency of FVIII-specific CD4+ T cells in the spleen after treatment of mice with 8 doses of human FVIII, given either intravenously or subcutaneously. For this purpose, we used multicolor flow cytometry involving staining of intracellular CD154 as a marker for activated CD4+ T cells (Figure 5A). The frequency of FVIII-specific CD4+ T cells after intravenous treatment with FVIII was 0.03% in both responder and nonresponder mice and, therefore, indistinguishable from background staining (0.02%-0.03%; Figure 5B). However, when we analyzed spleen cells obtained from mice treated with subcutaneous FVIII, we found a distinct population of FVIII-specific CD4+ T cells that accounted for approximately 0.17% of all CD4+ T cells in the spleen (Figure 5C). These results indicate that subcutaneous application of FVIII induced a higher frequency of FVIII-specific CD4+ T cells than intravenous application.
Figure 5. Different frequencies of FVIII-specific CD4+ T cells in the spleen after intravenous and subcutaneous application of FVIII in E17 HLA-DRB1*1501 mice.
Spleen cells were prepared from mice treated with 8 weekly doses of intravenous (B) or subcutaneous (C) FVIII and in vitro restimulated with either full-length human FVIII (B-C; 20 μg/mL) or FVIII peptides (C; 1 μg/mL per peptide) for 16 hours. The frequency of FVIII-specific or peptide-specific CD4+ T cells expressed as the percentage of CD4+ CD154+ T cells in relation to total CD4+ T cells is presented. Data are the results of a representative experiment. A total of 3 experiments with identical settings were done; all generated similar data. (A) The gating strategy for the quantification of CD4+ T cells that express intracellular CD154. (B) Frequencies of FVIII-specific CD4+ T cells (CD4+ T cells expressing intracellular CD154) after intravenous FVIII treatment. The results presented were obtained from mice with (intravenous responder) and without (intravenous nonresponder) anti-FVIII antibodies. Naive mice (naive) were included as a negative control. (C) Frequencies of FVIII-specific CD4+ T cells (CD4+ T cells expressing intracellular CD154) after subcutaneous FVIII treatment. Spleen cells were restimulated with 1 of the 6 following items: FVIII (FVIII), a mix of the 3 FVIII epitopes identified as dominant epitopes in E17 HLA-DRB1*1501 mice as shown in Figure 3A (peptide mix), a mix of scrambled versions of the 3 dominant epitopes (peptide control A), a mix of 3 FVIII epitopes identified as dominant epitopes in conventional E17 mice, which express the murine MHC class II molecule (peptide control B), a peptide pool spanning the C2 domain of FVIII, and one of the individual FVIII peptide regions containing dominant epitopes in E17 HLA-DRB1*1501 mice as indicated. Spleen cells restimulated with medium only (medium) were included as a negative control.
Next, we asked whether FVIII-specific CD4+ T cells that recognize the major FVIII peptide regions containing T-cell epitopes identified by hybridoma technology can be detected in spleen cells of mice treated with subcutaneous FVIII. We treated E17 HLA-DRB1*1501 mice with 8 weekly doses of subcutaneous FVIII, isolated spleen cells, and restimulated them in vitro with 1 of the 6 following items: human FVIII protein, a mix of the 3 immunodominant FVIII peptide regions containing T-cell epitopes, a mix of scrambled immunodominant FVIII peptide regions containing T-cell epitopes (peptide control A), a mix of FVIII peptide regions containing T-cell epitopes identified in conventional E17 mice expressing the murine MHC class II molecule (peptide control B), a peptide pool spanning the C2 domain of human FVIII (C2 domain pool), and 1 of the 3 individual immunodominant FVIII peptide regions containing T-cell epitopes. We found distinct populations of activated CD4+ T cells after stimulation with the mix of the 3 immunodominant FVIII peptide regions and after stimulation with each of the 3 dominant FVIII peptide regions identified by hybridoma technology. All negative controls showed negative results that were indistinguishable from the medium controls (Figure 5C).
Based on these data, we conclude that FVIII-specific T cells that recognize major epitopes as identified by hybridoma technology are detectable in the spleen of mice treated with FVIII.
FVIII T-cell epitopes identified in humanized E17 HLA-DRB1*1501 mice are generated and presented by human antigen-presenting cells
After identifying and confirming dominant FVIII peptide regions containing T-cell epitopes in humanized E17 HLA-DRB1*1501 mice, we asked whether these FVIII peptides are generated and presented by human antigen-presenting cells derived from healthy blood donors expressing the MHC class II protein HLA-DRB1*1501. Vidovic et al demonstrated that MHC class II-matched human antigen-presenting cells can stimulate T-cell hybridomas generated from human MHC class II transgenic mice if they present the correct peptide.36 We isolated monocytes from healthy blood donors and differentiated them into dendritic cells in vitro. Subsequently, we incubated the human dendritic cells with human FVIII and asked whether these cells would be able to stimulate FVIII-specific T-cell hybridoma clones of known specificity obtained from humanized E17 HLA-DRB1*1501 mice treated with FVIII. The stimulation of T-cell hybridoma clones would indicate that human dendritic cells are able to generate and present the same FVIII peptides as the ones identified in humanized E17 HLA-DRB1*1501 mice treated with human FVIII. Our results show that human dendritic cells obtained from HLA-DRB1*1501 positive healthy blood donors are indeed able to stimulate all T-cell hybridoma clones generated from humanized E17 HLA-DRB1*1501 mice treated with human FVIII (Figure 6). By contrast, human dendritic cells are unable to stimulate T-cell hybridoma clones generated from conventional E17 mice (Figure 6), which further confirms that the stimulation of T-cell clones obtained from humanized E17 HLA-DRB1*1501 mice depends on peptides presented by the human HLA-DRB1*1501.
Most FVIII peptides containing T-cell epitopes identified in humanized E17 HLA-DRB1*1501 mice are promiscuous and bind to several different human MHC class II proteins
Finally, we asked whether FVIII peptides containing T-cell epitopes identified in humanized E17 HLA-DRB1*1501 mice only bind to HLA-DRB*1501 or also bind to other human MHC class II haplotypes. We did binding studies that included the 6 most common HLA-DRB1* haplotypes (Figure 7A). The results of these binding studies indicate that all FVIII peptides identified in humanized E17 HLA-DRB1*1501 mice bind to 3 or more of the HLA-DRB1* haplotypes tested (Figure 7A).
Figure 7. Immunodominant T-cell epitopes of human FVIII identified in E17 HLA-DRB1*1501 mice bind to a variety of different HLA-DRB1* haplotypes.
(A) The binding of dominant T-cell epitopes of FVIII, as identified in E17 HLA-DRB1*1501 mice to the 6 most common HLA-DRB1* haplotypes, was analyzed using the REVEAL class II technology of ProImmune, as described in “Binding of peptides to HLA-DRB1* haplotypes.” The Cumulative Pan-Allele ProImmune REVEAL score for each peptide across all of the 6 HLA-DRB1* haplotypes was calculated by adding up the ProImmune REVEAL scores of each of the 6 HLA-DRB1* haplotypes analyzed divided by 6. For illustration, different colors corresponding to the contribution of each of the different HLA-DRB1* haplotypes tested were used. (B) The kinetics score for binding of the dominant T-cell epitopes of FVIII to the 6 most common HLA-DRB1* haplotypes was determined using the full rate assay technology of ProImmune, as described in “Binding of peptides to HLA-DRB1* haplotypes.” The Cumulative Pan-Allele kinetics score for each peptide across all of the 6 HLA-DRB1* haplotypes was calculated by adding up the kinetics scores of each of the 6 HLA-DRB1* haplotypes analyzed divided by 6. Different colors corresponding to the contribution of each of the different HLA-DRB1* haplotypes tested were used for illustration.
Once we had discovered that all FVIII peptides containing T-cell epitopes identified in humanized E17 HLA-DRB1*1501 mice bound to several different human HLA-DRB1* haplotypes, we wanted to obtain more insight into the binding kinetics of the peptides to the different HLA-DRB1* haplotypes. We measured the on-rates and off-rates for binding of each peptide with each of the 6 HLA-DRB1* haplotypes and calculated kinetics scores (Figure 7B; supplemental Tables 2 and 3). The results of these kinetics studies indicate that FVIII peptides that bind to different HLA-DRB1* haplotypes express different binding kinetics for each of the HLA-DRB1* haplotypes.
Discussion
MHC class II proteins expressed on antigen-presenting cells are essential for shaping the repertoire of CD4+ T cells in the thymus. Moreover, they determine the peptides that are presented to CD4+ T cells in peripheral lymphoid organs during adaptive immune responses, such as the initiation of antibody responses against FVIII.37 Previously, we reported the development of a new hemophilic E17 mouse model that expresses the peptide binding parts of the human MHC class II protein HLA-DRB1*1501 on the background of a complete knockout of all murine MHC class II genes.19 In this study, we show that hemophilic E17 HLA-DRB1*1501 mice express a fully diverse repertoire of naive CD4+ T cells with a polyclonality that is similar to the polyclonality of naive CD4+ T cells found in hemophilic E17 mice, confirming the functional competence of the chimeric human MHC class II complex. The differences in the actual pattern of V-J gene rearrangements that we found in naive CD4+ T cells of the 2 mouse strains are probably the result of the different MHC class II molecules that are responsible for the selection of the CD4+ T-cell repertoire.38
When we treated mice with intravenous human FVIII, only a fraction of them developed antibodies against FVIII, and the responder rate varied between 30% and 80%. Similar to patients, antibodies against FVIII expressed neutralizing activity. We used 1000 ng FVIII per dose, which was required to induce antibodies after 8 weekly intravenous doses. The 1000 ng FVIII in our studies corresponded to approximately 280 IU FVIII/kg. We also tested a clinically more relevant dose of 200 ng FVIII, corresponding to approximately 56 IU FVIII/kg but found very low responder rates after 8 weekly doses. Patients develop neutralizing antibodies against FVIII after an average of 10 to 50 exposure days.39 Future studies with the new hemophilic E17 HLA-DRB1*1501 mouse model will show whether 56 IU/kg and an extended treatment regimen of 10 to 50 weekly doses will increase the responder rates to the low dose.
The fact that only a fraction of hemophilic E17 HLA-DRB1*1501 mice developed antibodies after intravenous FVIII indicated that the new mouse model might be suitable to unravel the complex mechanisms responsible for the immune system’s decision whether or not to develop antibodies against FVIII. However, a couple of questions arose that needed to be addressed to evaluate the quality of this new model. First, we had to demonstrate that the lack of antibody development in a proportion of mice after intravenous FVIII treatment is associated with the intravenous application route and does not reflect the inability of these mice to respond to FVIII. Several reports have indicated that the intravenous application route for proteins is associated with a less immunogenic response than the subcutaneous or intramuscular application routes.40,41 Therefore, we expected to see a higher incidence of antibodies against FVIII after subcutaneous application if mice were able to respond to FVIII. Our data confirmed this assumption. All mice developed antibodies after subcutaneous application of FVIII. The increased incidence of antibody development after intravenous FVIII application in the presence of a concomitant activation of the innate immune system further supports the conclusion that all mice are able to respond to FVIII. We speculate that intravenous application of FVIII induces peripheral immune tolerance in nonresponder mice and that the activation of the innate immune system by LPS creates pro-inflammatory conditions that counter the potential tolerogenic immune response after intravenous FVIII. We are currently testing this hypothesis in follow-up studies.
Several studies indicate that the induction of antibody responses against FVIII in hemophilia A is T-cell dependent.6,7 The identification of CD4+ T-cell epitopes of FVIII has therefore been the subject of previous studies in mice and humans.11-18 We used humanized E17 HLA-DRB1*1501 mice to identify CD4+ T-cell epitopes of FVIII in the context of HLA-DRB1*1501. We identified 8 major FVIII peptide regions that contain CD4+ T-cell epitopes. A comparison of these peptide regions with the FVIII peptides regions identified in conventional E17 mice carrying the murine MHC class II complex revealed major differences, which highlight the importance of the nature of the MHC class II for the selection of peptides to be presented to CD4+ T cells. The question arose whether FVIII peptide regions containing CD4+ T-cell epitopes identified in humanized E17 HLA-DRB1*1501 mice by hybridoma technology could be confirmed in bulk spleen cell cultures obtained from mice treated with FVIII. Our results show that peptide-specific CD4+ T cells are detectable in bulk spleen cells, which confirms the relevance of peptides identified by hybridoma technology. Currently, we are in the process of identifying the minimum FVIII peptide sequences required to stimulate FVIII-specific T-cell hybridomas that we generated. These studies will reveal the exact FVIII sequences that constitute the actual T-cell epitopes. So far we have used only 15-mer and 21-mer peptides.
One of the FVIII peptide regions that we identified is located in the B domain. This finding raises the question of whether CD4+ T-cell epitopes in the B domain of FVIII could contribute to potential differences in immunogenicity between full-length FVIII and B-domain-deleted FVIII. CD4+ T-cell epitopes in the B domain of FVIII could contribute to either antibody responses or to the induction of immune tolerance. Aledort recently indicated that B domain-deleted FVIII and full-length FVIIII might express different immunogenicities.42 Based on these data, it will be interesting to determine whether CD4+ T-cell epitopes in the B domain of FVIII contribute to immunogenicity of FVIII or behave like Tregitopes that favor the induction of immune tolerance, as recently described by de Groot et al.43
Several studies have demonstrated that peptides presented to CD4+ T cells in human MHC class II transgenic mice were identical to peptides presented to CD4+ T cells in patients who carry the same MHC class II haplotype.44-46 We generated human monocyte-derived dendritic cells expressing HLA-DRB1*1501 and asked whether these cells would be able to generate the CD4+ T-cell epitopes of FVIII that we identified in humanized E17 HLA-DRB1*1501 mice. Our results indicate that this is indeed the case, which supports the potential relevance for humans of the FVIII peptide regions identified in humanized mice. Three of the 8 HLA-DRB1*1501-restricted FVIII peptide regions that we identified had already been described by Jacquemin et al15 and van Haren et al.18 These authors used in vitro binding assays15 or analyzed FVIII peptides eluted from human monocyte-derived dendritic cells.18 These data further support the relevance for the situation in humans of the FVIII peptides containing T-cell epitopes that we identified in humanized E17 HLA-DRB1*1501 mice.
Previous reports indicated that some CD4+ T-cell epitopes are promiscuous and bind to several different MHC class II molecules.47 Based on these reports, we asked whether the 8 FVIII peptide regions that we identified in humanized E17 HLA-DRB1*1501 mice contain such promiscuous epitopes. We tested the 6 most common HLA-DRB1* haplotypes and found that all 15mer peptides contained in the 8 FVIII peptide regions bind to 3 or more HLA-DR haplotypes. These findings indicate that the epitopes identified in E17 HLA-DRB1*1501 mice might be relevant not only for HLA-DRB1*1501 positive patients but also for a broader range of patients. However, the on-rates and off-rates of peptide binding were different for the different HLA-DR haplotypes. Future studies will show how different binding kinetics result in potential functional differences.
Interestingly, we did not find HLA-DRB1*1501-restricted T-cell epitopes in the C2 domain of FVIII although we were able to identify antibodies against the C2 domain. Reding et al described T-cell epitopes in the C2 domain of FVIII but did not specify the MHC class II haplotype of their study participants.14,48 Therefore, it is not possible to discuss their results in the context of our findings. However, it is to be expected that T-cell epitopes and B-cell epitopes of FVIII do not necessarily overlap. B-cell epitopes are often conformational epitopes composed of amino acids from different parts of a polypeptide chain, which are brought in close proximity by the 3-dimensional structure of the mature protein.49 By contrast, T-cell epitopes are short linear peptide sequences that are bound to the peptide-binding cleft of MHC class II molecules and are presented on the surface of antigen-presenting cells.50 Our studies emphasize the importance of both B-cell and T-cell epitopes in the immunogenicity assessment of FVIII proteins.
In conclusion, the humanized E17 HLA-DRB1*1501 hemophilic mouse model is a valuable tool to study the regulation of FVIII-specific CD4+ T-cell responses in the context of the human MHC class II molecule HLA-DRB1*1501. Furthermore, this mouse model represents an interesting opportunity to unravel the complex mechanisms responsible for the immune system’s decision of whether or not to develop antibodies against FVIII. The fact that E17 HLA-DRB1*1501 mice express only one MHC class II molecule is a clear limitation of this model. However, other hemophilic mouse models expressing alternative MHC class II molecules could complement this model in the future.
Supplementary Material
Acknowledgments
The authors thank Elisabeth Hopfner, Thomas Prenninger, Markus Pasztorek, Ginta Pordes, Lydia Suely, Nidha Abrar, and Monika Grewal for technical assistance and Elise Langdon-Neuner for editing the manuscript.
This work was supported by Baxter BioScience.
Footnotes
Contribution: K.N.S. designed research, performed most in vitro analysis, analyzed and interpreted data, and wrote the paper; P.M.v.H. designed research and analyzed and interpreted data; B.B. performed in vitro analysis and interpreted data; D.C.W. designed research and provided technology for the generation of T-cell hybridomas; S.U. performed molecular analysis for the characterization of the new mouse model and analyzed and interpreted data; C.H. analyzed and interpreted data; M.S. designed and supervised specific animal breeding schedules; R.U.A. and M.W. designed, supervised, and performed animal experiments; C.L. performed flow cytometric analysis of FVIII-specific CD4+ T cells; M.d.l.R. established methodology for the analysis of FVIII-specific CD4+ T cells and interpreted data; H.P.S. interpreted data; and B.M.R. designed research, analyzed and interpreted data, and wrote the paper.
Conflict-of-interest disclosure: K.N.S., B.B., S.U., C.H., M.S., R.U.A., M.W., C.L., M.d.l.R., H.P.S., and B.M.R. are employees of Baxter BioScience. P.M.v.H. was an employee of Baxter BioScience at the time she contributed to this study. The remaining author declares no competing financial interests.
References
- 1.Hoyer LW, Aledort LM, Lisher JM, Reisner HM, White GC. The incidence of factor VIII inhibitors in patients with severe hemophilia A. In: Aledort LM, Hoyer LW, Lusher JM, Reisner HM, White GC, editors. Inhibitors to Coagulation Factors. Plenum; New York, NY: 1995. pp. 35–45. [DOI] [PubMed] [Google Scholar]
- 2.Oldenburg J, Pavlova A. Genetic risk factors for inhibitors to factors VIII and IX. Haemophilia. 2006;12(suppl 6):15–22. doi: 10.1111/j.1365-2516.2006.01361.x. [DOI] [PubMed] [Google Scholar]
- 3.Gouw SC, van der Bom JG, van den Berg H. Marijke. Treatment-related risk factors of inhibitor development in previously untreated patients with hemophilia A: the CANAL cohort study. Blood. 2007;109(11):4648–4654. doi: 10.1182/blood-2006-11-056291. [DOI] [PubMed] [Google Scholar]
- 4.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381(6585):751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 5.Vinuesa CG, Tangye SG, Moser B, Mackay CR. Follicular B helper T cells in antibody responses and autoimmunity. Nat Rev Immunol. 2005;5(11):853–865. doi: 10.1038/nri1714. [DOI] [PubMed] [Google Scholar]
- 6.Ragni MV, Bontempo FA, Lewis JH. Disappearance of inhibitor to factor VIII in HIV-infected hemophiliacs with progression to AIDS or severe ARC. Transfusion. 1989;29(5):447–449. doi: 10.1046/j.1537-2995.1989.29589284147.x. [DOI] [PubMed] [Google Scholar]
- 7.Sasgary M, Ahmad RU, Schwarz HP, Turecek PL, Reipert BM. Single cell analysis of factor VIII-specific T cells in hemophilic mice after treatment with human factor VIII. Thromb Haemost. 2002;87(2):266–272. [PubMed] [Google Scholar]
- 8.Sinigaglia F, Hammer J. Defining rules for the peptide-MHC class II interaction. Curr Opin Immunol. 1994;6(1):52–56. doi: 10.1016/0952-7915(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 9.Suri A, Lovitch SB, Unanue ER. The wide diversity and complexity of peptides bound to class II MHC molecules. Curr Opin Immunol. 2006;18(1):70–77. doi: 10.1016/j.coi.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392(6673):245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 11.Pratt KP, Thompson AR. B-cell and T-cell epitopes in anti-factor VIII immune responses. Clin Rev Allergy Immunol. 2009;37(2):80–95. doi: 10.1007/s12016-009-8120-7. [DOI] [PubMed] [Google Scholar]
- 12.Hu GL, Okita DK, Conti-Fine BM. T cell recognition of the A2 domain of coagulation factor VIII in hemophilia patients and healthy subjects. J Thromb Haemost. 2004;2(11):1908–1917. doi: 10.1111/j.1538-7836.2004.00918.x. [DOI] [PubMed] [Google Scholar]
- 13.Reding MT, Okita DK, Diethelm-Okita BM, Anderson TA, Conti-Fine BM. Epitope repertoire of human CD4(+) T cells on the A3 domain of coagulation factor VIII. J Thromb Haemost. 2004;2(8):1385–1394. doi: 10.1111/j.1538-7836.2004.00850.x. [DOI] [PubMed] [Google Scholar]
- 14.Reding MT, Okita DK, Diethelm-Okita BM, Anderson TA, Conti-Fine BM. Human CD4+ T-cell epitope repertoire on the C2 domain of coagulation factor VIII. J Thromb Haemost. 2003;1(8):1777–1784. doi: 10.1046/j.1538-7836.2003.00251.x. [DOI] [PubMed] [Google Scholar]
- 15.Jacquemin M, Vantomme V, Buhot C, et al. CD4+ T-cell clones specific for wild-type factor VIII: a molecular mechanism responsible for a higher incidence of inhibitor formation in mild/moderate hemophilia A. Blood. 2003;101(4):1351–1358. doi: 10.1182/blood-2002-05-1369. [DOI] [PubMed] [Google Scholar]
- 16.James EA, Kwok WW, Ettinger RA, Thompson AR, Pratt KP. T-cell responses over time in a mild hemophilia A inhibitor subject: epitope identification and transient immunogenicity of the corresponding self-peptide. J Thromb Haemost. 2007;5(12):2399–2407. doi: 10.1111/j.1538-7836.2007.02762.x. [DOI] [PubMed] [Google Scholar]
- 17.James EA, van Haren SD, Ettinger RA, et al. T-cell responses in two unrelated hemophilia A inhibitor subjects include an epitope at the factor VIII R593C missense site. J Thromb Haemost. 2011;9(4):689–699. doi: 10.1111/j.1538-7836.2011.04202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Haren SD, Herczenik E, Ten Brinke A, Mertens K, Voorberg J, Meijer AB. HLA-DR-presented peptide-repertoires derived from human monocyte-derived dendritic cells pulsed with blood coagulation factor VIII. Mol Cell Proteomics. 2011;10(6):M110.002246. doi: 10.1074/mcp.M110.002246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reipert BM, Steinitz KN, van Helden PM, et al. Opportunities and limitations of mouse models humanized for HLA class II antigens. J Thromb Haemost. 2009;7(suppl 1):92–97. doi: 10.1111/j.1538-7836.2009.03403.x. [DOI] [PubMed] [Google Scholar]
- 20.Hay CR, Ollier W, Pepper L, et al. HLA class II profile: a weak determinant of factor VIII inhibitor development in severe haemophilia A: UKHCDO Inhibitor Working Party. Thromb Haemost. 1997;77(2):234–237. [PubMed] [Google Scholar]
- 21.Oldenburg J, Picard JK, Schwaab R, Brackmann HH, Tuddenham EG, Simpson E. HLA genotype of patients with severe haemophilia A due to intron 22 inversion with and without inhibitors of factor VIII. Thromb Haemost. 1997;77(2):238–242. [PubMed] [Google Scholar]
- 22.Pavlova A, Delev D, Lacroix-Desmazes S, et al. Impact of polymorphisms of the major histocom-patibility complex class II, interleukin-10, tumor necrosis factor-alpha and cytotoxic T-lymphocyte antigen-4 genes on inhibitor development in severe hemophilia A. J Thromb Haemost. 2009;7(12):2006–2015. doi: 10.1111/j.1538-7836.2009.03636.x. [DOI] [PubMed] [Google Scholar]
- 23.Woods A, Chen HY, Trumbauer ME, Sirotina A, Cummings R, Zaller DM. Human major histocom-patibility complex class II-restricted T cell responses in transgenic mice. J Exp Med. 1994;180(1):173–181. doi: 10.1084/jem.180.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bi L, Lawler AM, Antonarakis SE, High KA, Gearhart JD, Kazazian HH., Jr. Targeted disruption of the mouse factor VIII gene produces a model of haemophilia A. Nat Genet. 1995;10(1):119–121. doi: 10.1038/ng0595-119. [DOI] [PubMed] [Google Scholar]
- 25.Muchitsch EM, Turecek PL, Zimmermann K, et al. Phenotypic expression of murine hemophilia. Thromb Haemost. 1999;82(4):1371–1373. [PubMed] [Google Scholar]
- 26.Hausl C, Maier E, Schwarz HP, et al. Long-term persistence of anti-factor VIII antibody-secreting cells in hemophilic mice after treatment with human factor VIII. Thromb Haemost. 2002;87(5):840–845. [PubMed] [Google Scholar]
- 27.Frentsch M, Arbach O, Kirchhoff D, et al. Direct access to CD4+ T cells specific for defined antigens according to CD154 expression. Nat Med. 2005;11(10):1118–1124. doi: 10.1038/nm1292. [DOI] [PubMed] [Google Scholar]
- 28.Hausl C, Ahmad RU, Schwarz HP, et al. Preventing restimulation of memory B cells in hemophilia A: a potential new strategy for the treatment of antibody-dependent immune disorders. Blood. 2004;104(1):115–122. doi: 10.1182/blood-2003-07-2456. [DOI] [PubMed] [Google Scholar]
- 29.Kruisbeek AM. Production of mouse T cell hybridomas. Curr Protoc Immunol. 2001;Chapter 3 doi: 10.1002/0471142735.im0314s24. Unit 3.14. [DOI] [PubMed] [Google Scholar]
- 30.Ay B, Streitz M, Boisguerin P, et al. Sorting and pooling strategy: a novel tool to map a virus proteome for CD8 T-cell epitopes. Biopolymers. 2007;88(1):64–75. doi: 10.1002/bip.20637. [DOI] [PubMed] [Google Scholar]
- 31.Pickl WF, Majdic O, Kohl P, et al. Molecular and functional characteristics of dendritic cells generated from highly purified CD14+ peripheral blood monocytes. J Immunol. 1996;157(9):3850–3859. [PubMed] [Google Scholar]
- 32.Pasqual N, Gallagher M, Aude-Garcia C, et al. Quantitative and qualitative changes in V-J alpha rearrangements during mouse thymocytes differentiation: implication for a limited T cell receptor alpha chain repertoire. J Exp Med. 2002;196(9):1163–1173. doi: 10.1084/jem.20021074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. 2009;9(12):833–844. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 34.von Boehmer H. Selection of the T-cell repertoire: receptor-controlled checkpoints in T-cell development. Adv Immunol. 2004;84:201–238. doi: 10.1016/S0065-2776(04)84006-9. [DOI] [PubMed] [Google Scholar]
- 35.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun. 2009;388(4):621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 36.Vidovic D, Graddis TJ, Stepan LP, Zaller DM, Laus R. Specific stimulation of MHC-transgenic mouse T-cell hybridomas with xenogeneic APC. Hum Immunol. 2003;64(2):238–244. doi: 10.1016/s0198-8859(02)00780-2. [DOI] [PubMed] [Google Scholar]
- 37.Germain RN. MHC-dependent antigen processing and peptide presentation: providing ligands for T lymphocyte activation. Cell. 1994;76(2):287–299. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- 38.Houston EG, Jr, Fink PJ. MHC drives TCR repertoire shaping, but not maturation, in recent thymic emigrants. J Immunol. 2009;183(11):7244–7249. doi: 10.4049/jimmunol.0902313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wight J, Paisley S. The epidemiology of inhibitors in haemophilia A: a systematic review. Haemophilia. 2003;9(4):418–435. doi: 10.1046/j.1365-2516.2003.00780.x. [DOI] [PubMed] [Google Scholar]
- 40.Peng A, Gaitonde P, Kosloski MP, Miclea RD, Varma P, Balu-Iyer SV. Effect of route of administration of human recombinant factor VIII on its immunogenicity in hemophilia A mice. J Pharm Sci. 2009;98(12):4480–4484. doi: 10.1002/jps.21765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schellekens H. Factors influencing the immuno-genicity of therapeutic proteins. Nephrol Dial Transplant. 2005;20(suppl 6):vi3–vi9. doi: 10.1093/ndt/gfh1092. [DOI] [PubMed] [Google Scholar]
- 42.Aledort LM, Navickis RJ, Wilkes MM. Can B-domain deletion alter the immunogenicity of recombinant factor VIII? A meta-analysis of prospective clinical studies. J Thromb Haemost. 2011;9(11):2180–2192. doi: 10.1111/j.1538-7836.2011.04472.x. [DOI] [PubMed] [Google Scholar]
- 43.de Groot AS, Moise L, McMurry JA, et al. Activation of natural regulatory T cells by IgG Fc-derived peptide ‘Tregitopes.’. Blood. 2008;112(8):3303–3311. doi: 10.1182/blood-2008-02-138073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gregersen JW, Holmes S, Fugger L. Humanized animal models for autoimmune diseases. Tissue Antigens. 2004;63(5):383–394. doi: 10.1111/j.0001-2815.2004.00243.x. [DOI] [PubMed] [Google Scholar]
- 45.Das P, Abraham R, David C. HLA transgenic mice as models of human autoimmune diseases. Rev Immunogenet. 2000;2(1):105–114. [PubMed] [Google Scholar]
- 46.Forsthuber TG, Shive CL, Wienhold W, et al. T cell epitopes of human myelin oligodendrocyte glycoprotein identified in HLA-DR4 (DRB1*0401) transgenic mice are encephalitogenic and are presented by human B cells. J Immunol. 2001;167(12):7119–7125. doi: 10.4049/jimmunol.167.12.7119. [DOI] [PubMed] [Google Scholar]
- 47.Hammer J, Valsasnini P, Tolba K, et al. Promiscuous and allele-specific anchors in HLA-DR-binding peptides. Cell. 1993;74(1):197–203. doi: 10.1016/0092-8674(93)90306-b. [DOI] [PubMed] [Google Scholar]
- 48.Reding MT, Wu H, Krampf M, et al. Sensitization of CD4+ T cells to coagulation factor VIII: response in congenital and acquired hemophilia patients and in healthy subjects. Thromb Haemost. 2000;84(4):643–652. [PubMed] [Google Scholar]
- 49.Mariuzza RA, Phillips SE, Poljak RJ. The structural basis of antigen-antibody recognition. Annu Rev Biophys Biophys Chem. 1987;16:139–159. doi: 10.1146/annurev.bb.16.060187.001035. [DOI] [PubMed] [Google Scholar]
- 50.Germain RN, Margulies DH. The biochemistry and cell biology of antigen processing and presentation. Annu Rev Immunol. 1993;11:403–450. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.