Summary
The adult newt brain has a marked neurogenic potential and is highly regenerative. Ventricular, radial glia-like ependymoglia cells give rise to neurons both during normal homeostasis and after injury, but subpopulations among ependymoglia cells have not been defined. We show here that a substantial portion of GFAP+ ependymoglia cells in the proliferative hot spots of the telencephalon has transit-amplifying characteristics. In contrast, proliferating ependymoglia cells, which are scattered along the ventricular wall, have stem cell features in terms of label retention and insensitivity to AraC treatment. Ablation of neurons remodels the proliferation dynamics and leads to de novo formation of regions displaying features of neurogenic niches, such as the appearance of cells with transit-amplifying features and proliferating neuroblasts. The results have implication both for our understanding of the evolutionary diversification of radial glia cells as well as the processes regulating neurogenesis and regeneration in the adult vertebrate brain.
Highlights
-
•
Most ventricular ependymoglia cells have stem cell properties
-
•
Some ependymoglia in neurogenic hot spots have transit-amplifying characteristics
-
•
Injury transforms non-hot spots to regions with features of neurogenic niches
-
•
Injury leads to changes in the cellular response to inhibition of Notch signaling
Kirkham, Simon, and colleagues report that in the newt brain, most ependymoglia cells have stem cell properties. Ablation of neurons remodels the proliferation dynamics in the telencephalon and leads to de novo formation of regions displaying features of neurogenic niches. Neuronal ablation also leads to changes in the cellular response to inhibition of Notch signaling.
Introduction
Adult neurogenesis is a distinctive feature of the telencephalon in the mammalian brain. Neurogenesis proceeds by neural stem cells (NSCs), giving rise to transit-amplifying cells, which subsequently differentiate into neuroblasts and mature neurons (Bonaguidi et al., 2012, Malatesta et al., 2000, Noctor et al., 2001, Seri et al., 2004). Despite the presence of NSCs and the apparent constitutive neurogenesis in the subventricular zone of the lateral ventricles and in the hippocampus, the ability of mammals to replace neurons that are lost due to injury or during the course of progressive neurodegenerative diseases are modest at best (Arias-Carrión et al., 2007, Arias-Carrión et al., 2009, Kernie and Parent, 2010).
In contrast to mammals, several nonmammalian vertebrate species, such as teleost fishes and salamanders, display a remarkable ability to regenerate brain tissue by processes that involve extensive neurogenic events (for a recent review, see Grandel and Brand, 2013). Studies over the past years have substantially increased our understanding of adult neurogenesis in these species (e.g., Chapouton et al., 2007). Both nongenetic and genetic cell-tracking studies revealed that cells with radial glia features act as neuronal progenitors in fishes and salamanders. These cells line the ventricular system, express GFAP, and have long processes reaching to the pial surface (Berg et al., 2010, Kroehne et al., 2011, Maden et al., 2013, Pérez-Cañellas and García-Verdugo, 1996). The zebrafish telencephalon has been shown to have a distinctive heterogeneity among ventricular cells, in terms of anatomical localization and protein-expression profiles (Chapouton et al., 2010, Ganz et al., 2010, März et al., 2010).
Neurogenic regions have been mapped and revealed an uneven distribution of actively dividing cells with progenitor potential along the ventricular system in anamniotes (Adolf et al., 2006, Berg et al., 2010, Kaslin et al., 2009). Some of these studies indicated that a correlation between the distribution of active neurogenic niches and regions with neuroregenerative capacity exists (Zupanc and Zupanc, 2006); however, the two are not necessarily linked to each other. For example, studies in the aquatic salamander Notophthalmus viridescens (red-spotted newt) showed extensive regeneration following ablation of neurons in regions that are essentially devoid of neurogenesis under normal conditions (Berg et al., 2010, Parish et al., 2007). Nevertheless, the newt telencephalon harbors several proliferative hot spots, such as the lateral wall of the ventricle adjacent to the dorsal pallium (Dp) and the lateral wall of the ventricle adjacent to the bed nucleus of the stria terminalis (Bst) (Berg et al., 2010). Hence, the telencephalon is an ideal model for studying the cellular composition and regulatory mechanisms of neuronal regeneration in an environment, which is permissive for constitutive neurogenesis.
Here, we started to address to what extent GFAP+ ventricular cells, denoted as ependymoglia cells (Parish et al., 2007), within and outside of the constitutively active niches are different from each other in the newt telencephalon. We define two different types of ependymoglia cells, which display uneven distribution along the ventricle. Unexpectedly, we find that the majority of ependymoglia cells display stem cell features in terms of label retention and insensitivity to treatment that eliminates rapidly dividing cells. However, these cells are not restricted to the proliferation hot spots but are dispersed along the ventricular wall and create de novo neurogenic regions after ablation of neurons. The proliferation hot spots on the other hand are largely composed of cells with characteristics of transit-amplifying populations. We also characterize dynamical changes in the cellular composition of both hot spots and non-hot spots after neuronal ablation and how Notch signaling relates to these changes.
Results
GFAP+ Ependymoglia Cells Form Neurospheres
Neurosphere formation is a hallmark of NSCs (Reynolds and Weiss, 1992). We first tested whether the newt brain harbors cells that can form neurospheres from different parts of the brain. We observed that culturing cells in media containing epidermal growth factor (EGF) and fibroblast growth factor 2 (FGF-2) led to the formation of sphere-like structures from both the telencephalon (Figures 1A and 1B) and from dien- and mesencephalon (Figure S1 available online). Over 14 days, these spheres increased in size and we frequently observed large numbers of GFAP-expressing cells located in the core of the spheres (Figure 1C). Double labeling with proliferating cell nuclear antigen (PCNA) indicated that GFAP+ cells were proliferating (Figure 1D). Furthermore, after pulsing with the nucleotide analog, 5-ethynyl-2′-deoxyuridine (EdU), we could find EdU/GFAP double-labeled cells in the center of the spheres (Figure 1E). Upon changing to media without growth factors and allowing the spheres to attach, we found cells expressing the neuronal marker, Tuj1, and we also observed the appearance of GFAP cells in the periphery of the spheroid structures, along with cells migrating away (Figures 1F–1H). These data indicated that GFAP+ cells have stem cell properties.
Figure 1.
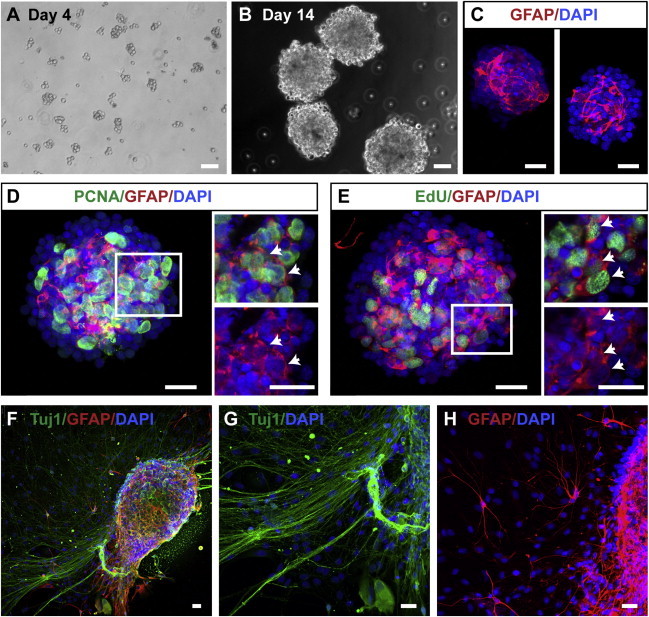
Neurosphere-Forming Cells in the Newt Brain
(A and B) Isolated cells from newt brain cultured in DMEM/F12, FGF-2, and EGF formed small spheres after 4 days (A). These spheres increased in size over time (B).
(C) GFAP+ cells are found in the center of the 14-day-old neurospheres.
(D and E) Neurospheres cultured for 14 days in DMEM/F12, FGF-2, and EGF contain proliferating GFAP+ cells as indicated by PCNA labeling (D, arrows) and incorporate EdU (E, arrows).
(F–H) Neurospheres plated in poly-D-lysine plates and cultured in differentiation medium for 14 days produce Tuj1+ cells with long extensions (F and G). GFAP+ cells were also observed away from the neurosphere (F and H).
The scale bars represent 50 μm.
Heterogeneity among GFAP+ Ependymoglia Cells
We previously determined that proliferating ependymoglia cells are predominantly found in hot spots, which were previously defined by Berg et al. (2010) and are illustrated in Figure 2A. However, heterogeneity among ependymoglia cells was not further studied. In order to test whether we could define subpopulations among the seemingly homogenous GFAP+ ependymoglia cells, we performed immunohistochemical analyses using antibodies against known glial markers in combination with proliferation markers. These efforts resulted in the distinction of two subpopulations, which we subsequently denoted as type-1 and type-2 cells, respectively.
Figure 2.
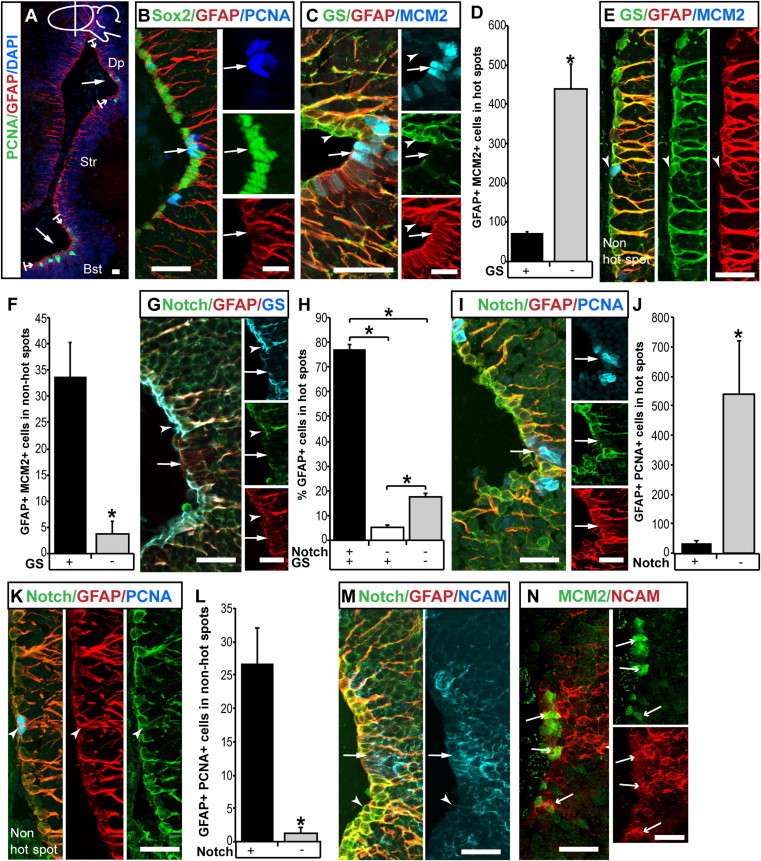
Immunohistochemical Characterization of Ventricular Ependymoglia Cells
(A) All ependymoglia cells are GFAP+; proliferation hot spots such as the dorsal pallium (Dp) and the bed nucleus of the stria terminalis (Bst) are marked as well as the striatum (Str). The drawing of the newt brain indicates the plane of section. Lines with arrows demarcate the borders of the hot spots. Arrows highlight even expression of GFAP in the hot spots.
(B) Sox2 and GFAP are expressed in all ependymoglia cells. Arrow highlights even expression of Sox2 in proliferating PCNA+ cells.
(C) GS/GFAP/MCM2 staining reveals type-1 and type-2 cells. Arrow points to MCM2+/GFAP+/GS− (type-2) ependymoglia cells in hot spots. Arrowhead points to the surrounding nonproliferating GFAP+/GS+ (type-1) ependymoglia cells.
(D) Quantification of ependymoglia cells in hot spots coexpressing GFAP, GS, and MCM2. Note that most MCM2+ ependymoglia cells are type-2 cells (GFAP+/GS−). n = 5; p < 0.05.
(E) GS/GFAP/MCM2 expression in non-hot spot. Arrowhead points to proliferating type-1 cell (GFAP+/GS+).
(F) Quantification of ependymoglia cells in non-hot spots coexpressing GFAP, GS, and MCM2. Note that most MCM2+ ependymoglia cells are type-1 cells (GFAP+/GS+). n = 5; p < 0.05.
(G) Notch1−, type-2 ependymoglia cells (arrow) surrounded by Notch1+, type-1 ependymoglia cells (arrowhead). Note the colocalization of GS and Notch1 immunoreactivity.
(H) Quantification of GFAP-, Notch1-, and GS-expressing ependymoglia cells shows that, in hot spots, the vast majority of GFAP+ cells were type-1 (Notch1+/GS+). n = 4; p < 0.05.
(I) The majority of PCNA+ cells in hot spots are type-2 ependymoglia cells (GFAP+/Notch1−; arrow).
(J) Quantification of ependymoglia cells in hot spot regions coexpressing GFAP, Notch1, and PCNA. Note that almost all PCNA+ ependymoglia cells are type-2 cells (GFAP+/Notch1−). n = 5; p < 0.05.
(K and L) Few PCNA+ type-1 (GFAP+/Notch1+) ependymoglia cells were observed outside of hot spots. Quantified in (L). n = 5; p < 0.05.
(M) PSA-NCAM (NCAM) and type-2 (GFAP+/Notch1−) ependymoglia cells in hot spots (arrow). The surrounding region contains type-1 (GFAP+/Notch1+) ependymoglia cells and is devoid of PSA-NCAM expression (arrowhead).
(N) MCM2-expressing PSA-NCAM+ cells in hot spots (arrow).
Data represented as mean ± SEM. The scale bars represent 50 μm.
First, we looked at Sox2 and GLAST expressions, but both of these proteins were ubiquitously expressed in ependymoglia cells (Figures 2B, S2A, and S2B). Next, we determined the expression pattern of glutamine synthetase (GS). We saw that GS staining clearly marked two distinct populations of ependymoglia cells. One of the populations expressed GS in addition to GFAP (type-1), whereas the other population of cells did not express GS (type-2; Figure 2C). The GS− cells are found in clusters in hot spots and make up 32% ± 4% of the ependymoglia cells in hot spots (Figures 2C and S2C–S2E) and most (85% ± 2%) of the proliferating ependymoglia cells in this region (Figure 2D). The remaining proliferating ependymoglia cells (15% ± 2%) in hot spots are GS+ type-1 cells (Figure 2D). In contrast, in non-hot spots, GS− cells make up 0.3% ± 0.2% of the ependymoglia cells, whereas most (90% ± 6%) of the proliferating ependymoglia cells in the non-hot spots are GS+ (Figures S2C–S2E, 2E, and 2F). From these data, we concluded that hot spots contain both proliferating type-1 and type-2 cells, whereas non-hot spots essentially lack type-2 cells.
Given the documented role of Notch signaling in the regulation of neural stem and progenitor cell fate, we next determined the expression pattern of Notch1 receptor among the ependymoglia subpopulations defined by GS expression in hot spots and non-hot spots. We found active Notch signaling occurring in Notch1+ ependymoglia cells as assayed by electroporation of a reporter driven by the 12xCSL promoter (Hansson et al., 2006) and by in situ hybridization (Figures S2F–S2H). The antibody recognized a band of expected size in western blots (Figure S2I). Consistently, the epitope region against which the antibody was raised is highly conserved across species (Figures S2J and S2K). We found a marked unequal distribution of Notch1 expression among the GFAP+ ependymoglia cells in the hot spots (Figure 2G). Of GFAP+ cells, 77% ± 2% were Notch1+, whereas 23% ± 2% were Notch1− (Figure 2H). In addition, the majority of GFAP+ cells were either Notch1+/GS+ or Notch1−/GS− (Figure 2H). This is consistent with type-1 cells being GFAP+/GS+/Notch1+ and type-2 cells being GFAP+/GS−/Notch1−.
We next analyzed the proliferation pattern among the Notch1+ ependymoglia cells. We found that the vast majority (94% ± 1%) of proliferating ependymoglia cells in the hot spots were devoid of Notch1 expression (Figures 2I and 2J), whereas most of the proliferating ependymoglia cells in the non-hot spots were Notch1+ (Figures 2K and 2L). This is in accordance with the observation that most proliferating ependymoglia cells in the hot spots were devoid of GS expression, whereas proliferating ependymoglia cells in the non-hot spots expressed GS (Figures 2D and 2F).
In close proximity of the proliferation hot spots, we found GFAP− cells away from the ventricle, which expressed the cell-adhesion molecule polysialylated neuronal cell adhesion molecule (PSA-NCAM), a marker for immature neurons (Rousselot et al., 1995), and which were proliferating as assayed by minichromosome maintenance protein 2 (MCM2) expression (Figures 2M and 2N). On average, we found 211 ± 44 MCM2+/ PSA-NCAM+ cells in hot spots. In contrast, regions around the dispersed proliferating type-1 cells were devoid of proliferating type-2 and PSA-NCAM-expressing cells (data not shown). These observations further corroborated the view that the proliferation hot spots are constitutively active neurogenic regions, composed by type-1 and type-2 cells and by neuroblasts, whereas the non-hot spots are essentially devoid of type-2 cells and neuroblasts.
Type-1 Ependymoglia Cells Have Stem Cell Properties
The selective localization of type-2 cells to the proliferating hot spots led us to hypothesize that type-2 cells have stem cell properties. Unexpectedly, our data showed the opposite, indicating that a majority of stem cells are type-1 cells. Long-term label retention in pulse-chase experiments using nucleotide analogs is a distinctive feature of stem cells. We pulsed animals for 3 days with the nucleotide analog bromodeoxyuridine (BrdU) and subsequently chased the analog before sacrificing the animals at various time points. Following a chase period of 3 days, we found 22% ± 3% of BrdU being in type-1 cells. This percentage increased gradually and reached 63% ± 13% after 90 days (Figures 3A–3C and S3A).The remaining BrdU label was found in GFAP− nonventricular cells after 90 days (data not shown). In addition, type-2 cells in hot spots showed the opposite trend of label retention, with hardly any BrdU found after 90 days (Figure 3B).
Figure 3.
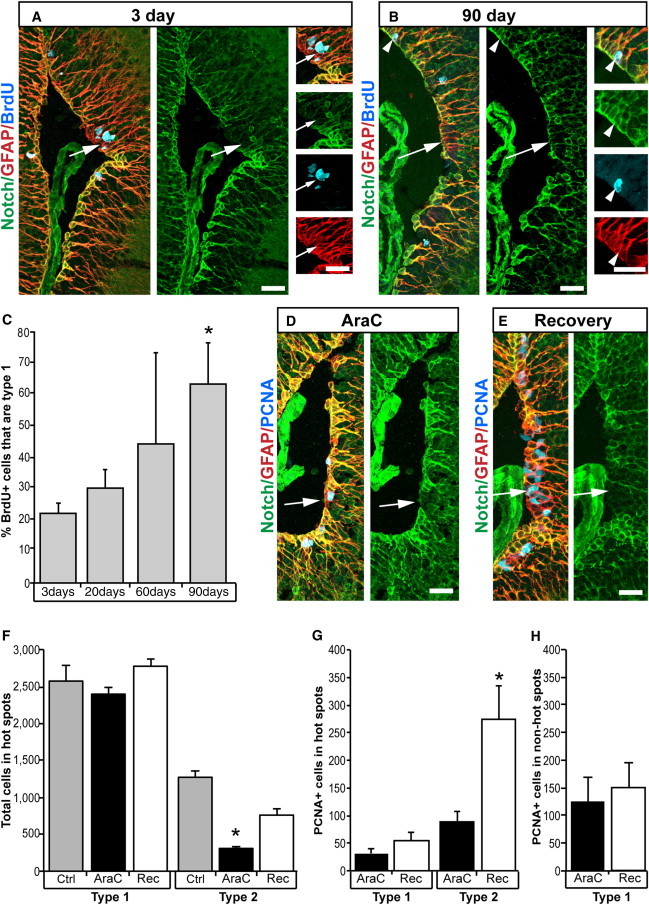
Type-1 Cells Are Resistant to AraC and Retain BrdU Labeling after Extended Chase
(A–C) A pulse of BrdU was chased for 3 days (A). BrdU-labeled type-2 ependymoglia cells in hot spots (arrow). BrdU-labeled type-1 ependymoglia cells after a 90-day chase (arrowhead; B). Type-2 ependymoglia cells were identified by lack of Notch1 expression (arrow). Percentage of GFAP+/BrdU+ cells that were type-1 ependymoglia cells (C). p < 0.05 between 3 days chase and 90 days chase. n = 4.
(D–H) AraC treatment (D) reduces the number of type-2 ependymoglia cells (arrow) but had no effect on type-1 ependymoglia cells in hot spots. After 14 days of recovery (E), the number of type-2 ependymoglia cells increased (F). p < 0.05 between control and AraC treatment. n = 5. Recovery for 14 days after AraC treatment also caused an increase in PCNA+ type-2 ependymoglia cells but had no effect on the proliferation of type-1 ependymoglia cells (G). p < 0.05; n = 5. The number of PCNA+ type-1 cells was also unaffected in non-hot spots (H). n = 5.
Data represented as mean ± SEM. The scale bars represent 50 μm.
As an additional tool, we treated animals with AraC, which kills proliferating cells. AraC treatment is an established method to selectively eliminate transit-amplifying cells, which divide more frequently than slowly dividing stem cells (Doetsch et al., 1999). Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labelling (TUNEL) staining shows that AraC causes the death of ventricular GFAP+ cells (Figure S3B). In accordance with the label-retention experiment, administration of AraC led to the elimination of type-2 cells, as assayed by Notch1/GFAP/PCNA triple labeling (Figures 3D and 3F). Twelve hours after AraC injection, the number of type-2 cells was reduced by 86% ± 2% and their number increased by 2.5-fold after a 14 days recovery period following the AraC treatment (Figures 3E and 3F). Consistently, the number of proliferating type-2 cells increased 3-fold during recovery (Figures 3E and 3G), indicating the compensatory proliferative burst after the elimination of the type-2 cells. In contrast, neither the total number nor the number of proliferating type-1 cells changed significantly after AraC treatment and during the recovery period (Figures 3D–3H).
These data show that type-1 ependymoglia cells that make up 92% ± 4% of the ventricle wall of the telencephalon (Figures S2C–S2E) have stem cell properties and suggest that type-2 cells in the hot spots have properties of transit-amplifying cells.
Ependymoglia Cell Dynamics during Neuronal Regeneration
Next, we tested how ependymoglia cells respond to the ablation of neurons. We tested two possibilities. The first possibility was that only the hot spots respond by increased proliferation of type-1 and type-2 cells and neuroblasts. The second possibility was that both the hot spots and the non-hot spots respond to loss of neurons. To distinguish between these two possibilities, we ablated cholinergic neurons and analyzed hot spots and non-hot spots, which are localized in proximity to the ablated neuronal population (Figure S4). Injection of the selective neurotoxin AF64A led to the ablation and subsequent regeneration of choline acetyltransferase (ChAT)-expressing neurons (Figures 4A and 4B). During the course of regeneration of cholinergic neurons, we observed an increase of both type-1 and type-2 cells (Figures 4C–4H). The most striking change that we found was the appearance of type-2 and proliferating PSA-NCAM+ cells in and around the non-hot spots (Figures 4F–4H and 4L–4N). These data indicated the de novo generation of a neurogenic niche as a response to neuronal ablation.
Figure 4.
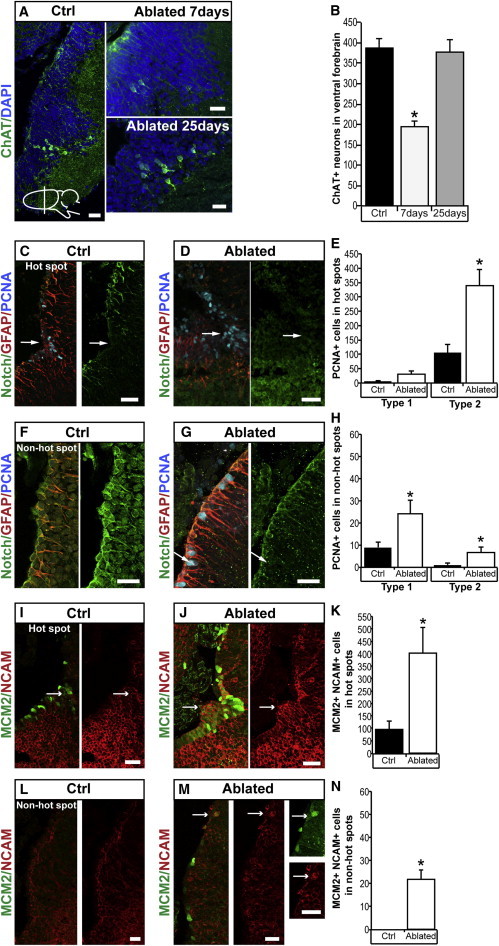
Ablation of ChAT+ Neurons Leads to De Novo Generation of Type-2 and Proliferating PSA-NCAM Cells in Non-Hot Spots.
(A and B) ChAT+ neurons are present in the parenchyma of the bed nucleus of the stria terminalis (A). The drawing of the newt brain indicates the plane of section. ChAT+ neurons are lost 7 days after injection of AF64A (A) and subsequently regenerate after 25 days (A). Quantified in (B). p < 0.05; n = 5.
(C–E) Compared to control (C), there is an increase of PCNA+ type-2 ependymoglia cells (arrow) in the hot spot adjacent to the ablated ChAT+ neurons (D). Quantified in (E). p < 0.05; n = 5.
(F–H) Compared to control (F), ablation of ChAT+ neurons caused an increase of PCNA+ type-1 and the appearance of PCNA+ type-2 ependymoglia cells (arrow) in the ventral non-hot spot. Quantified in (H). p < 0.05; n = 5.
(I–K) Compared to control (I), ablation of ChAT+ neurons caused an increase of PSA-NCAM (NCAM) MCM2+ cells in the hot spot (arrow). Quantified in (K). p < 0.05; n = 5.
(L–N) Lack of PSA-NCAM+/MCM2+ cells in the ventral non-hot spot (L). Ablation of ChAT+ neurons caused the appearance of PSA-NCAM+/MCM2+ cells in non-hot spots (arrow; M). Quantified in (N). p < 0.05; n = 5.
Data represented as mean ± SEM. The scale bars represent 50 μm.
Ependymoglia Cell Dynamics and Notch Signaling during Homeostasis and Neuronal Regeneration
Next, we tested how manipulation of Notch signaling impinges on ependymoglia cell dynamics during normal homeostasis and after ablation of neurons. To interfere with Notch signaling, we used the small molecule, DAPT, which is a γ-secretase inhibitor (Geling et al., 2002). In the absence of neuronal ablation, DAPT treatment led to increased proliferation of type-2 cells in the hot spots (Figures 5A–5C). On the other hand, we did not observe any statistically significant change in the proliferation of type-1 cells—neither in the hot spots nor in the non-hot spots (Figures 5B–5D). We also determined how DAPT treatment influenced the number of proliferating PSA-NCAM+ cells and found that it was increased by 45% ± 7% (Figures 5E–5G).
Figure 5.
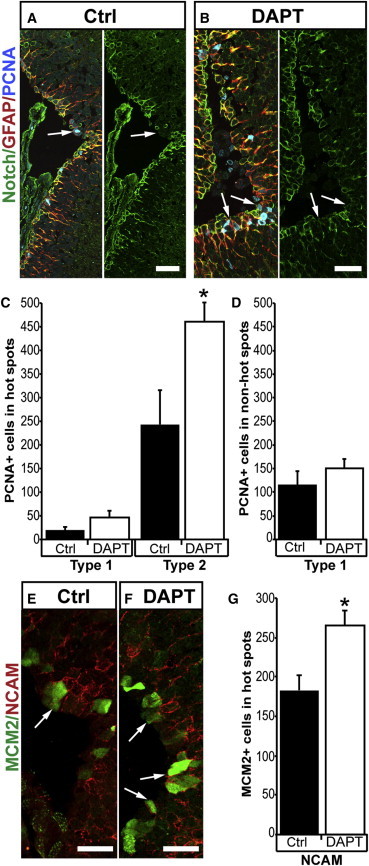
Inhibition of Notch Signaling Causes an Increase in Cell Proliferation during Homeostatic Conditions
(A–D) Control (A) versus DAPT-treated (B) brains shows an increase in PCNA+ type-2 ependymoglia cells in hot spots (arrows). Quantified in (C). p < 0.05; n = 5. No significant effect of DAPT treatment on non-hot spot cells (D). n = 5.
(E–G) Control (E) versus DAPT-treated (F) brains shows an increase in MCM2+ PSA NCAM+ (NCAM) cells (arrows) in hot spots. Quantified in (G). p < 0.05; n = 4.
Data represented as mean ± SEM. The scale bars represent 50 μm.
After ablation of cholinergic neurons, the injury-responsive increased proliferation of type-1, type-2, and PSA-NCAM+ cells in the hot spots was not significantly altered by DAPT treatment (Figures 6A and 6B). Conversely, whereas normal proliferation was not influenced in the non-hot spots by DAPT treatment, the ablation-responsive increase in type-1 cells was inhibited by DAPT (Figure 6C). Counting the PSA-NCAM+ cells in the non-hot spots showed increased proliferation after DAPT treatment (Figure 6D).
Figure 6.
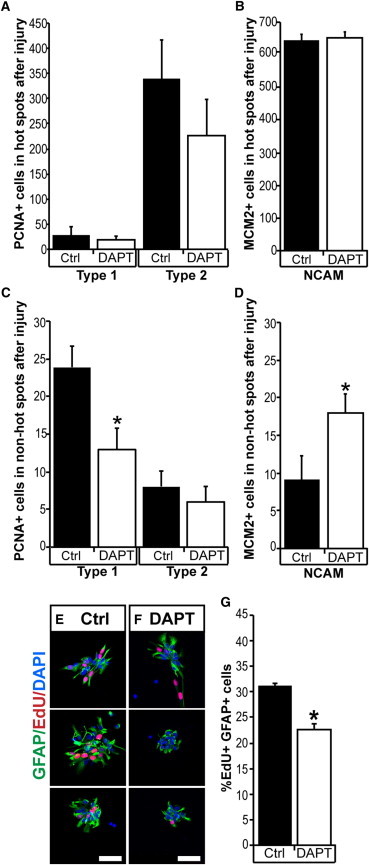
Injury Leads to Changes in the Cellular Response to Inhibition of Notch Signaling
(A and B) After ablation of cholinergic neurons, DAPT treatment does not significantly affect the injury-induced proliferation neither of type-1 and type-2 ependymoglia cells (A) nor the PSA NCAM+ (NCAM) cells (B) in hot spots. n = 5.
(C and D) After ablation of cholinergic neurons, DAPT treatment leads to a decrease in injury-induced proliferation of type-1 ependymoglia (C) and an increase in PSA-NCAM+ cells in non-hot spots. p < 0.05; n = 5.
(E–G) DAPT treatment caused a decrease in the incorporation of EdU by GFAP+ neurosphere cells (E and F). Quantified in (G). p < 0.05; n = 4.
Data represented as mean ± SEM. The scale bars represent 50 μm.
In order to corroborate this interpretation, we analyzed neurosphere cultures. We reasoned that the neurosphere cultures mimic an injury response due to the dissociation process and would primarily be composed by type-1 cells derived from non-hot spots, as these are the quantitatively dominating population (81% ± 3%) in the forebrain. In agreement with the in vivo results, we found that DAPT treatment decreased the proliferation of the neurosphere-forming ependymoglia cells as assayed by EdU incorporation (Figures 6E–6G).
These results indicated that, during normal homeostasis, stem cell (type-1) proliferation is not Notch-signaling sensitive and proliferation of transit-amplifying cells (type-2) and neuroblasts (PSA-NCAM+) is Notch-signaling sensitive. The insensitivity of stem cells to Notch signaling remains in the hot spots also during regeneration; however, the injury-responsive increased stem cell proliferation in the non-hot spots is dependent on Notch signaling.
Discussion
In this paper, we characterized cellular heterogeneity among ventricular radial glia-like ependymoglia cells in the newt telencephalon. Our findings reveal two principal subpopulations among these cells: one having stem cell features (type-1) and the other with features of transit-amplifying cells (type-2; summarized in Figure 7). We also define differential sensitivity among ependymoglia cells to treatment with the γ-secretase inhibitor DAPT during homeostatic and regenerative conditions, suggesting that Notch signaling has a context-dependent and cell-type-specific role in regulating precursor cell fate in the newt telencephalon (summarized in Figure 7).
Figure 7.
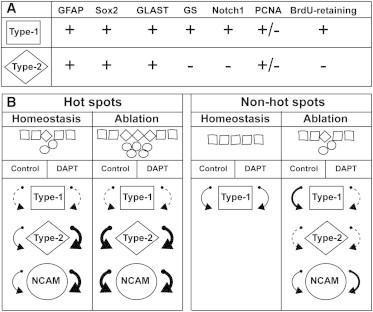
Summary of Proliferation Dynamics during Homeostasis and Regeneration in the Presence or Absence of Notch-Signaling Inhibitor
(A) Expression patterns of various glial and proliferation markers expressed by type-1 and type-2 ependymoglia cells.
(B) In the graphic representation of the cells lining the ventricle, squares represent type-1 cells (GFAP+, Notch1+, and GS+), diamonds depict type-2 cells (GFAP+, Notch1−, and GS−), and circles denote PSA-NCAM+ neuroblasts. The thickness of the arrows indicates the relative number of cycling cells, with dashed line being the lowest and thick bold being the highest.
The newt telencephalon harbors distinct proliferation hot spots (Berg et al., 2010). An unexpected observation presented in this paper is that the seemingly randomly proliferating type-1 ependymoglia cells are label-retaining and insensitive to AraC treatment. Hence, the ventricular system in the newt telencephalon appears to be lined by rarely self-renewing stem-like ependymoglia cells, and this may be one factor behind these animals’ marked neuroregenerative capacity.
Indeed, upon ablation of cholinergic neurons, we found several indications for the formation of neurogenic niches in non-hot spots. Although, due to the lack of selective tracing methods, the fate of non-hot spot cells in relation to cells generated in the hot spots can at present not be determined, our observations suggest a de novo generation of a lineage in non-hot spots as a response to injury. First, we observed the increased number of proliferating type-1 cells. Second, we observed the appearance of proliferating type-2 cells in vicinity of the type-1 cells. Third, in proximity to the type-1 and newly formed type-2 cells, we also noted an injury-specific appearance of proliferating PSA-NCAM-expressing cells.
The newt brain lacks GFAP-expressing parenchymal astrocytes (Berg et al., 2011, Parish et al., 2007). It is likely that ependymoglia cells are responsible for functions that are carried out by astrocytes in mammals, such as protecting neurons from excitotoxicity by metabolic regulation of glutamate involving GS (Norenberg and Martinez-Hernandez, 1979). Whereas many glial markers, for example GFAP and GLAST, are common for all ependymoglia cells in the newt brain, GS selectively labels the type-1 cells. This finding is noteworthy in an evolutionary comparative context. Because GS does not mark cell populations with transit-amplifying characters, this observation may support the view of astrocytes being NSCs in vertebrates (Kriegstein and Alvarez-Buylla, 2009).
It is interesting to note that the zebrafish telencephalon, which is also highly regenerative (Baumgart et al., 2012, Kroehne et al., 2011), lacks a distinct GS− cell population that expresses other canonical glial genes, such as S100β and GFAP (Ganz et al., 2010, März et al., 2010). However, the fish telencephalon seems to harbor label-retaining cells with low or no glial marker expression (Ganz et al., 2010). Another interesting comparison with the zebrafish telencephalon is that Notch1 is mainly expressed in quiescent radial glia cells in the fish (Chapouton et al., 2010). In contrast, in the newt telencephalon, the Notch1 antibody equally labels proliferating and nonproliferating cells in the non-hot spots. In addition, no increase in proliferating PSA-NCAM+ cells was observed after stab injury to the zebrafish telencephalon (Baumgart et al., 2012). These observations reflect differences in the cellular organization and the injury response comparing newts and zebrafish. Nevertheless, DAPT treatment has been shown to reduce the injury-induced production of cells with active neurogenin reporter in the zebrafish telencephalon (Kishimoto et al., 2012), which is similar to our findings in the newt telencephalon showing that DAPT treatment reduces the injury-induced proliferation of type-1 cells in the non-hot spots. The altered sensitivity to DAPT treatment of the type-1 cells after injury comparing hot spots and non-hot spots further reinforces the differences between injury-induced and constitutive neurogenesis, as previously suggested by Berg et al. (2010) and Kizil et al. (2012).
The importance of Notch signaling in the regulation of neuronal precursor cell fate has been well documented also in mammals, and perturbation of Notch signaling has profound effects both during homeostatic conditions as well as in injury models (Ables et al., 2010, Basak et al., 2012, Carlén et al., 2009, Chapouton et al., 2010, Ehm et al., 2010, Imayoshi et al., 2010). In the mouse brain, quiescent NSCs are insensitive to the loss of Notch signaling. However, upon injury-induced activation of NSCs, Notch signaling is required to fully reinstate neurogenesis (Basak et al., 2012). Further cross-species comparisons, preferentially by involving targeted fate-mapping approaches, of how Notch signaling impinges on neurogenesis are likely to reveal novel ways of enhancing regeneration in the adult vertebrate brain.
Experimental Procedures
Animals
Adult red-spotted newt Notophthalmus viridescens (Charles Sullivan) were housed at 15°C–20°C. All experimental procedures were performed according to European and local ethical permits.
Treatments
BrdU (Sigma) 20 mg/kg was injected intraperitoneally twice daily for 3 days and chased for various amounts of time. Cholinergic ablations were carried out according to Berg et al. (2011) by intracranial injection into the lateral ventricle of 50 ng AF64A. ChAT+ cells were quantified in the bed nucleus of the stria terminalis. The Notch-signaling reporter 12xCLS-H2BYFP (Hansson et al., 2006) was electroporated into the lateral wall of the lateral ventricle as described previously (Berg et al., 2010). AraC (500 mg/kg; Sigma) was delivered via intraperitoneal injections twice daily for 5 days. Animals were either sacrificed 12 hr after the last injection or allowed to recover for 14 days before being scarified. DAPT (N-[N-(3,5-difluorophenacetyl-L-alanyl)]- S-phenylglycine t-butyl ester; Axxora) was administered as previously described (Chapouton et al., 2010). Briefly, a stock solution of 10 mM DAPT in DMSO was diluted in swimming water to a final concentration of 100 μM. Control animals were placed in equal concentration of DMSO. Newts were treated for 48 hr. Newts ablated with AF64A were treated with DAPT between 5 and 7 days after toxin injection.
Cell Culture
All media were diluted by 33% with water to match the osmolality of the newt cells. Newts were anesthetized with a solution of 0.1% MS-222 (Sigma). The isolated brain was digested in 30 U/ml Papain (Sigma), L15 (Gibco), 40 μg/ml DNase (Roche), and 2 mg/ml ovamucoid (Worthington Biochemical) for 1 hr at room temperature. Next, L15 containing 2 mg/ml ovamucoid and 40 μg/ml DNases was added at a ratio 1:1 and left at room temperature for 5 min. Cells were spun down and resuspended in L15 and triturated with a p1000. Cell suspension was filter through 30 μm cell strainer (Gibco) before being spun down and resuspended in Dulbecco’s modified Eagles medium (DMEM)/F12/Glutamax (Gibco), 2% B27 (Gibco), 100 U/ml penicillin-streptomycin (Gibco), 20 ng/ml EGF (Gibco) and 20 ng FGF-2 (Gibco). Cells were plated in uncoated 24-well plates and cultured at 25°C at 2% CO2. Every fourth day, cells were supplemented with fresh growth media. After 14 days, neurospheres were plated out on dish coated with poly-D-lysine (Sigma) and allowed to differentiate in DMEM/F12/Glutamax media with no growth factors. Neurospheres were generated and cultured as described above either in the presence of 10 μM DAPT (Axxora) or vehicle from days 7 to 9. Twenty micromolar EdU (Invitrogen) was added to the medium for 8 hr at day 9; spheres were dissociated and plated on poly-D-lysine-coated plates (Sigma) for immunohistochemistry.
Immunohistochemistry
Immunohistochemical staining of brain tissue was performed as described previously (Berg et al., 2011, Kirkham et al., 2011). Neurospheres were fixed in 4% formaldehyde for 15 min at room temperature and treated with 0.1% Triton X-100 in PBS (Sigma). Cells and 20 μm newt brain coronal sections were incubated at 4°C for 15–18 hr in blocking solution containing either PBS, 4% goat serum (Invitrogen), and 0.1% Triton X-100 or PBS, 0.2% fish skin gelatin (Sigma), 0.2% BSA (Sigma), and 0.2% Triton X-100 with the following primary antibodies: mouse anti-GFAP-CY3 (1:500; Sigma), goat anti-GFP (1:500; Abcam), guinea pig anti-GLAST (1:1,000; Frontier Institute), rabbit anti-Sox2 (1:500; Abcam), rabbit anti-glutamine synthetase (1:500; Millipore), rabbit anti-MCM2 antibody (1:200; Abcam), PSA-NCAM (1:8,000; Millipore), rabbit anti-Notch1 (1:100; Santa Cruz Biotechnology), mouse anti-PCNA (1:500; Chemicon), rat anti-BrdU (1:500; Accurate Chemical and Scientific), and anti-ChAT (1:250; Millipore). For BrdU and PCNA staining, sections and cells were treated with PBS containing 2 M HCL and 0.5% Triton X-100 for 20 min at 37°C. For PSA-NCAM and MCM2 staining, sections were incubated in citrate buffer (10 mM sodium citrate, 0.05% Tween 20 [pH 6]) at 37°C for 30 min. After labeling with the primary antibody, sections and cells were washed 3 × 5 min in PBS before the appropriate secondary antibodies (Molecular Probes) were applied in blocking solution for 2 hr at room temperature. Triple labeling of GFAP with two other antibodies required additional 5 min incubation in 4% formaldehyde at room temperature after incubation with the secondary antibodies. This was followed by 1 hr incubation at room temperature with GFAP-CY3 (1:1,000; Sigma) in blocking solution. Sections were mounted in mounting medium containing 1 mg/ml DAPI (Dako). TUNEL was performed post-antibody incubation following the manufacturer’s instructions for cryosections (In Situ Cell Death Detection Kits for detection by fluorescent microscopy; Roche). Images for cell counting of tissue sections were captured with LSM-700 using ZEN software (both Zeiss), Angstrom Grid confocal microscope (Leica Microsystems) using velocity software (PerkinElmer), or Axioplan2 using Axiovision software (both Zeiss). In areas of high cell density, confocal z stacks were captured, and for large areas, confocal tile montages were created. Images were manipulated with Volocity (PerkinElmer) or Photoshop (Adobe) using linear adjustments. Cell counts were performed on a region starting with the proliferation hot spot adjacent to the bed nucleus of the stria terminalis to the most caudal part of the telencephalon.
In Situ Hybridization
A fragment of Notch1 was amplified from newt adult brain cDNA using the following primers: For: TCT CCG TTT CAA CAG TCT CC; Rev: AAG TTG GTG GCT GGG AGT GT. The fragment was inserted into pCR4 topo cloning vector using TOPO TA cloning kit (Invitrogen). Digoxigenin-labeled probes were synthesized using T7 and Sp6 RNA polymerase kit (Roche). Brains were dissected out and prepared in a similar manner to immunohistochemical procedure (see above), except that ribonuclease (RNase) free reagents were used. Ten micrometer coronal sections were fixed in 4% formaldehyde for 15 min, treated with 0.2 N HCL for 12 min, washed, and then incubated in acetylation buffer (0.1 M trietholamine and 0.25% acetic anhydride) for 10 min. Next, the slides were rinsed in RNase free water and permeabilized with a solution of 1 μg/ml Proteinase K (Roche) and 2 mM CaCl2 for 15 min at 37°C. The slides were incubated in prehybridization buffer (50% deionized form of amide, 5× saline sodium citrate [SSC], 5× Denhardt’s solution, 250 μg/ml yeast RNA, 500 μg/ml herring sperm single-stranded DNA [ssDNA]; Sigma) for 2 hr at room temperature before being incubated in hybridization solution (50 ng probe, 50% deionized form of amide, 5× SSC, 5× Denhardt’s solution, 250 μg/ml yeast RNA, 500 μg/ml herring sperm ssDNA) for 16 hr at 60°C. The slides were then washed with 0.2× SSC buffer containing 0.05% tween 20 at 70°C for 3 × 1 hr. Finally, the probes were detected by TSA Plus Fluorescence for in situ hybridization (PerkinElmer) following the manufacturer’s instructions.
Western Blot
Newt brains were homogenized using downs homogenizer in lysis buffer containing 50 mM Tris (pH 8; Sigma), 150 mM NaCl (Sigma), 10% glycerol (Sigma), 1% NP40 (Sigma), 0.5% deoxycholate (Sigma), and proteinase inhibitors mix (Roche). Protein extract was boiled in sample buffer (Invitrogen) and ran on SDS-PAGE gel (Invitrogen) before being transferred onto polyvinylidene fluoride membrane. Membranes were incubated overnight at 4°C with rabbit anti-Notch1 antibody (1:1,000; Santa Cruz Biotechnology) in blocking buffer containing Tris-buffered saline, 5% BSA (Sigma), and 0.1% Triton X-100. Anti-rabbit-horseradish peroxidase (1:10,000; GE Healthcare) in blocking buffer was incubated for 1 hr at room temperature before the membrane was treated with enhanced chemiluminescence following the manufacturer’s instructions (GE Healthcare) and imaged using Image Quant LAS 4000 (GE Healthcare).
Alignments
Notch1 sequences for Homo sapiens (ref: NP_060087.3), Mus musculus (ref: NP_032740.3), and Xenopus laevis (ref: NP_001081074.1) were obtained from the National Center for Biotechnology database. Notch1 sequence for Notophthalmus viridescens was obtained from an inferred proteome (Abdullayev et al., 2013). Alignment was performed using DNAstar mega align.
Statistical Analyses
Student’s t test was used where p values are indicated. In all figures, n represents the number of biological replicates.
Acknowledgments
We would like to thank E. Andersson and U. Lendahl for providing reagents and for helpful discussions. This work was supported by grants from AFA Insurances, Cancerfonden, Swedish Research Council, and European Research Council to A.S. M.K. was supported by a HFSPO postdoc fellowship.
Published: March 20, 2014
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information includes four figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2014.01.018.
Contributor Information
Matthew Kirkham, Email: matthew.kirkham@ki.se.
András Simon, Email: andras.simon@ki.se.
Supplemental Information
References
- Abdullayev I., Kirkham M., Björklund Å.K., Simon A., Sandberg R. A reference transcriptome and inferred proteome for the salamander Notophthalmus viridescens. Exp. Cell Res. 2013;319:1187–1197. doi: 10.1016/j.yexcr.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Ables J.L., Decarolis N.A., Johnson M.A., Rivera P.D., Gao Z., Cooper D.C., Radtke F., Hsieh J., Eisch A.J. Notch1 is required for maintenance of the reservoir of adult hippocampal stem cells. J. Neurosci. 2010;30:10484–10492. doi: 10.1523/JNEUROSCI.4721-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolf B., Chapouton P., Lam C.S., Topp S., Tannhäuser B., Strähle U., Götz M., Bally-Cuif L. Conserved and acquired features of adult neurogenesis in the zebrafish telencephalon. Dev. Biol. 2006;295:278–293. doi: 10.1016/j.ydbio.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Arias-Carrión O., Freundlieb N., Oertel W.H., Höglinger G.U. Adult neurogenesis and Parkinson’s disease. CNS Neurol. Disord. Drug Targets. 2007;6:326–335. doi: 10.2174/187152707783220875. [DOI] [PubMed] [Google Scholar]
- Arias-Carrión O., Yamada E., Freundlieb N., Djufri M., Maurer L., Hermanns G., Ipach B., Chiu W.-H., Steiner C., Oertel W.H., Höglinger G.U. Neurogenesis in substantia nigra of parkinsonian brains? J. Neural Transm. Suppl. 2009:279–285. doi: 10.1007/978-3-211-92660-4_23. [DOI] [PubMed] [Google Scholar]
- Basak O., Giachino C., Fiorini E., Macdonald H.R., Taylor V. Neurogenic subventricular zone stem/progenitor cells are Notch1-dependent in their active but not quiescent state. J. Neurosci. 2012;32:5654–5666. doi: 10.1523/JNEUROSCI.0455-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart E.V., Barbosa J.S., Bally-Cuif L., Götz M., Ninkovic J. Stab wound injury of the zebrafish telencephalon: a model for comparative analysis of reactive gliosis. Glia. 2012;60:343–357. doi: 10.1002/glia.22269. [DOI] [PubMed] [Google Scholar]
- Berg D.A., Kirkham M., Beljajeva A., Knapp D., Habermann B., Ryge J., Tanaka E.M., Simon A. Efficient regeneration by activation of neurogenesis in homeostatically quiescent regions of the adult vertebrate brain. Development. 2010;137:4127–4134. doi: 10.1242/dev.055541. [DOI] [PubMed] [Google Scholar]
- Berg D.A., Kirkham M., Wang H., Frisén J., Simon A. Dopamine controls neurogenesis in the adult salamander midbrain in homeostasis and during regeneration of dopamine neurons. Cell Stem Cell. 2011;8:426–433. doi: 10.1016/j.stem.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Bonaguidi M.A., Song J., Ming G.-L., Song H. A unifying hypothesis on mammalian neural stem cell properties in the adult hippocampus. Curr. Opin. Neurobiol. 2012;22:754–761. doi: 10.1016/j.conb.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlén M., Meletis K., Göritz C., Darsalia V., Evergren E., Tanigaki K., Amendola M., Barnabé-Heider F., Yeung M.S.Y., Naldini L. Forebrain ependymal cells are Notch-dependent and generate neuroblasts and astrocytes after stroke. Nat. Neurosci. 2009;12:259–267. doi: 10.1038/nn.2268. [DOI] [PubMed] [Google Scholar]
- Chapouton P., Jagasia R., Bally-Cuif L. Adult neurogenesis in non-mammalian vertebrates. Bioessays. 2007;29:745–757. doi: 10.1002/bies.20615. [DOI] [PubMed] [Google Scholar]
- Chapouton P., Skupien P., Hesl B., Coolen M., Moore J.C., Madelaine R., Kremmer E., Faus-Kessler T., Blader P., Lawson N.D., Bally-Cuif L. Notch activity levels control the balance between quiescence and recruitment of adult neural stem cells. J. Neurosci. 2010;30:7961–7974. doi: 10.1523/JNEUROSCI.6170-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F., Caillé I., Lim D.A., García-Verdugo J.M., Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Ehm O., Göritz C., Covic M., Schäffner I., Schwarz T.J., Karaca E., Kempkes B., Kremmer E., Pfrieger F.W., Espinosa L. RBPJkappa-dependent signaling is essential for long-term maintenance of neural stem cells in the adult hippocampus. J. Neurosci. 2010;30:13794–13807. doi: 10.1523/JNEUROSCI.1567-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz J., Kaslin J., Hochmann S., Freudenreich D., Brand M. Heterogeneity and Fgf dependence of adult neural progenitors in the zebrafish telencephalon. Glia. 2010;58:1345–1363. doi: 10.1002/glia.21012. [DOI] [PubMed] [Google Scholar]
- Geling A., Steiner H., Willem M., Bally-Cuif L., Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3:688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandel H., Brand M. Comparative aspects of adult neural stem cell activity in vertebrates. Dev. Genes Evol. 2013;223:131–147. doi: 10.1007/s00427-012-0425-5. [DOI] [PubMed] [Google Scholar]
- Hansson E.M., Teixeira A.I., Gustafsson M.V., Dohda T., Chapman G., Meletis K., Muhr J., Lendahl U. Recording Notch signaling in real time. Dev. Neurosci. 2006;28:118–127. doi: 10.1159/000090758. [DOI] [PubMed] [Google Scholar]
- Imayoshi I., Sakamoto M., Yamaguchi M., Mori K., Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J. Neurosci. 2010;30:3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaslin J., Ganz J., Geffarth M., Grandel H., Hans S., Brand M. Stem cells in the adult zebrafish cerebellum: initiation and maintenance of a novel stem cell niche. J. Neurosci. 2009;29:6142–6153. doi: 10.1523/JNEUROSCI.0072-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernie S.G., Parent J.M. Forebrain neurogenesis after focal Ischemic and traumatic brain injury. Neurobiol. Dis. 2010;37:267–274. doi: 10.1016/j.nbd.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M., Berg D.A., Simon A. Microglia activation during neuroregeneration in the adult vertebrate brain. Neurosci. Lett. 2011;497:11–16. doi: 10.1016/j.neulet.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Kishimoto N., Shimizu K., Sawamoto K. Neuronal regeneration in a zebrafish model of adult brain injury. Dis. Model. Mech. 2012;5:200–209. doi: 10.1242/dmm.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizil C., Kyritsis N., Dudczig S., Kroehne V., Freudenreich D., Kaslin J., Brand M. Regenerative neurogenesis from neural progenitor cells requires injury-induced expression of Gata3. Dev. Cell. 2012;23:1230–1237. doi: 10.1016/j.devcel.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Kriegstein A., Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroehne V., Freudenreich D., Hans S., Kaslin J., Brand M. Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development. 2011;138:4831–4841. doi: 10.1242/dev.072587. [DOI] [PubMed] [Google Scholar]
- Maden M., Manwell L.A., Ormerod B.K. Proliferation zones in the axolotl brain and regeneration of the telencephalon. Neural Dev. 2013;8:1. doi: 10.1186/1749-8104-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta P., Hartfuss E., Götz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- März M., Chapouton P., Diotel N., Vaillant C., Hesl B., Takamiya M., Lam C.S., Kah O., Bally-Cuif L., Strähle U. Heterogeneity in progenitor cell subtypes in the ventricular zone of the zebrafish adult telencephalon. Glia. 2010;58:870–888. doi: 10.1002/glia.20971. [DOI] [PubMed] [Google Scholar]
- Noctor S.C., Flint A.C., Weissman T.A., Dammerman R.S., Kriegstein A.R. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Norenberg M.D., Martinez-Hernandez A. Fine structural localization of glutamine synthetase in astrocytes of rat brain. Brain Res. 1979;161:303–310. doi: 10.1016/0006-8993(79)90071-4. [DOI] [PubMed] [Google Scholar]
- Parish C.L., Beljajeva A., Arenas E., Simon A. Midbrain dopaminergic neurogenesis and behavioural recovery in a salamander lesion-induced regeneration model. Development. 2007;134:2881–2887. doi: 10.1242/dev.002329. [DOI] [PubMed] [Google Scholar]
- Pérez-Cañellas M.M., García-Verdugo J.M. Adult neurogenesis in the telencephalon of a lizard: a [3H]thymidine autoradiographic and bromodeoxyuridine immunocytochemical study. Brain Res. Dev. Brain Res. 1996;93:49–61. doi: 10.1016/0165-3806(96)00014-4. [DOI] [PubMed] [Google Scholar]
- Reynolds B.A., Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Rousselot P., Lois C., Alvarez-Buylla A. Embryonic (PSA) N-CAM reveals chains of migrating neuroblasts between the lateral ventricle and the olfactory bulb of adult mice. J. Comp. Neurol. 1995;351:51–61. doi: 10.1002/cne.903510106. [DOI] [PubMed] [Google Scholar]
- Seri B., García-Verdugo J.-M., Collado-Morente L., McEwen B.S., Alvarez-Buylla A. Cell types, lineage, and architecture of the germinal zone in the adult dentate gyrus. J. Comp. Neurol. 2004;478:359–378. doi: 10.1002/cne.20288. [DOI] [PubMed] [Google Scholar]
- Zupanc G.K., Zupanc M.M. New neurons for the injured brain: mechanisms of neuronal regeneration in adult teleost fish. Regen. Med. 2006;1:207–216. doi: 10.2217/17460751.1.2.207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


