Abstract
The membrane anchors of viral envelope proteins play essential roles in cell entry. Recent crystal structures of the ectodomain of envelope protein E2 from a pestivirus suggest that E2 belongs to a novel structural class of membrane fusion machinery. Based on geometric constraints from the E2 structures, we generated atomic models of the E1 and E2 membrane anchors using computational approaches. The E1 anchor contains two amphipathic perimembrane helices and one transmembrane helix; the E2 anchor contains a short helical hairpin stabilized in the membrane by an arginine residue, similar to flaviviruses. A pair of histidine residues in the E2 ectodomain may participate in pH sensing. The proposed atomic models point to Cys987 in E2 as the site of disulfide bond linkage with E1 to form E1–E2 heterodimers. The membrane anchor models provide structural constraints for the disulfide bonding pattern and overall backbone conformation of the E1 ectodomain.
Keywords: Membrane anchor, Transmembrane helix, Amphipathic helix, Arginine snorkel, Histidine pH sensor, Type I membrane protein, Membrane fusion protein, Secondary structure prediction
Graphical abstract
Highlights
-
•
Structures of pestivirus E2 proteins impose constraints on E1, E2 membrane anchors.
-
•
Atomic models of the E1 and E2 membrane anchors were generated in silico.
-
•
A “snorkeling” arginine completes the short helical hairpin in the E2 membrane anchor.
-
•
Roles in pH sensing and E1–E2 disulfide bond formation are proposed for E1 residues.
-
•
Implications for E1 ectodomain structure and disulfide bonding pattern are discussed.
Introduction
Viruses from the pestivirus genus in the Flaviviridae family are economically important pathogens that infect livestock. Within this family, pestiviruses are the closest relatives to Hepatitis C virus (HCV), which remains an important human pathogen (Lindenbach et al., 2013, Shepard et al., 2005). The pestivirus BVDV (bovine viral diarrhea virus) is often used as a model system for studying HCV for which there is currently no effective vaccine (De Francesco and Migliaccio, 2005). A mechanistic understanding at the molecular level of the lifecycles of pesti- and hepaciviruses could open new avenues for developing antiviral vaccines and therapeutics.
To deliver their genome into the cytoplasm, enveloped viruses must fuse their lipid envelope with a cellular membrane. This critical membrane fusion step is catalyzed by the viral envelope proteins, which are anchored in the viral membrane by helical transmembrane (TM) anchors. These membrane fusion proteins respond to the reduced pH of endocytic compartments, or to other environmental cues, with conformational changes that expose a hydrophobic fusion motif allowing it to insert into the endosomal membrane. These proteins then fold back on themselves, forcing the cell membrane (held by the fusion motif) and the viral membrane (held by the transmembrane anchor) against each other, resulting in fusion of the viral and endosomal membranes (Harrison, 2008). Viral fusion proteins fall into at least three distinct structural classes. “Class I” fusion proteins are found in ortho- and paramyxoviruses, retroviruses, filoviruses, and coronaviruses (Lamb and Jardetzky, 2007). The unifying structural feature of class I fusion proteins is a core consisting of three bundled α-helices (Kielian and Rey, 2006, Schibli and Weissenhorn, 2004). Class II fusion proteins are found in flaviviruses, alphaviruses, phleboviruses, and rubella virus (Dessau and Modis, 2013, DuBois et al., 2013, Lescar et al., 2001, Rey et al., 1995). Class II proteins have a three-domain architecture, with tightly folded “fusion loops” in the central domain serving as the fusion motif (Kielian and Rey, 2006, Modis, 2014). The class II fusion protein in flaviviruses contains an unusual, membrane curvature-inducing transmembrane anchor consisting of a short helical hairpin stabilized in the bilayer by an arginine residue with its guanidinium moiety “snorkeling” to the phosphate layer of the membrane׳s inner leaflet (Zhang et al., 2013). Class III fusion proteins, found in herpesviruses, rhabdoviruses and baculoviruses, possess structural features from both class I proteins (a core three-helix bundle) and from class II proteins (a central β-stranded fusion domain) (Backovic and Jardetzky, 2011, Heldwein et al., 2006, Kadlec et al., 2008, Roche et al., 2006, Roche et al., 2007). Interestingly, all viral fusion proteins are homotrimers in their postfusion conformation (Harrison, 2008, Kielian and Rey, 2006, Modis, 2014).
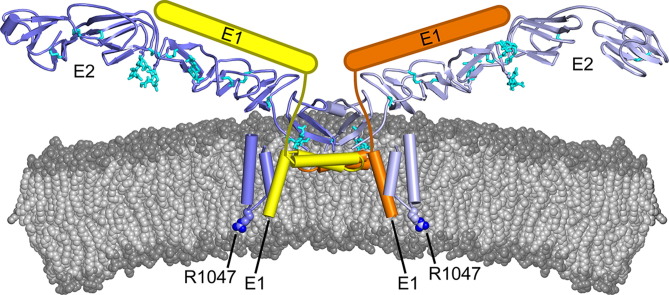
In contrast to other enveloped viruses, which contain a single fusion protein, two glycoproteins, E1 and E2, are necessary and sufficient for membrane fusion in pestiviruses and hepaciviruses. E1 and E2 are both type I TM proteins with membrane-anchored C-terminal tails (MATs) (Ronecker et al., 2008, Wang et al., 2004). Class II folds had been proposed for both HCV E1 and pestivirus E2 (Garry and Dash, 2003). However, two recent crystal structures of the BVDV1 E2 ectodomain showed that BVDV E2 has a novel fold (El Omari et al., 2013, Li et al., 2013). Similarly, a recent structure of the HCV E2 ectodomain core fragment showed that HCV E2 has a novel fold distinct from that of BVDV E2 (Kong et al., 2013). Neither BVDV E2 nor HCV E2 contains an internal or terminal fusion motif with any clear resemblance to those of other viral fusion proteins. The reduced pH of endocytic compartments is not sufficient to initiate membrane fusion of pesti- and hepaciviruses (Krey et al., 2005, Mathapati et al., 2010, Meertens et al., 2006, Tscherne et al., 2006, Tscherne et al., 2008). Viruses from both families are acid-resistant prior to endocytosis, and require a poorly understood activation step to become fusogenic at endosomal pH (Krey et al., 2005, Meertens et al., 2006, Tscherne et al., 2006). Pestiviruses are also unique among enveloped viruses in that their glycoproteins are extensively crosslinked by intermolecular disulfide bonds. The predominant disulfide linkage is between E1 and E2. E2–E2 disulfide linkages have also been observed, albeit at lower abundance (Thiel et al., 1991, Weiland et al., 1990). Notably, the BVDV E2 ectodomain forms disulfide-linked homodimers in the absence of E1 (Li et al., 2013). E1–E2 dimers are required for virus infectivity. In contrast E2–E2 disulfide linkages are associated with reduced infectivity (Branza-Nichita et al., 2001, Durantel et al., 2001, Ronecker et al., 2008). Moreover, three positively charged residues, two in the E1 MAT and one in the E2 MAT, are essential for cell entry of non-infectious pseudoviruses that contain only the E1 and E2 proteins (Ronecker et al., 2008). Similarly, in HCV the TM segment of E2 is required for correct folding and assembly of E1–E2 heterodimers (Patel et al., 2001). Together, these findings suggest that interactions within the E1/E2 MAT assembly are essential for activation of pestiviruses for cell entry.
In this study, we applied computational modeling tools to elucidate the role of the BVDV1 E1 and E2 MAT domains in activation of viral membrane fusion. Using the geometric constraints imposed by the known structure and disulfide bonding pattern of the E2 ectodomain dimer (El Omari et al., 2013, Li et al., 2013), and by the requirement for a disulfide linkage between E1 and E2, we constructed atomic models of the E1 and E2 MATs. Our models identify two amphipathic perimembrane helices in the E1 MAT, one of which contains a potential disulfide linkage site, and a helical hairpin in the E2 MAT. These models also impose geometric constraints on the conformation of the E1 ectodomain for which no structural information is available.
Results and discussion
Possible role in pH sensing of two juxtamembrane histidine residues in BVDV E2
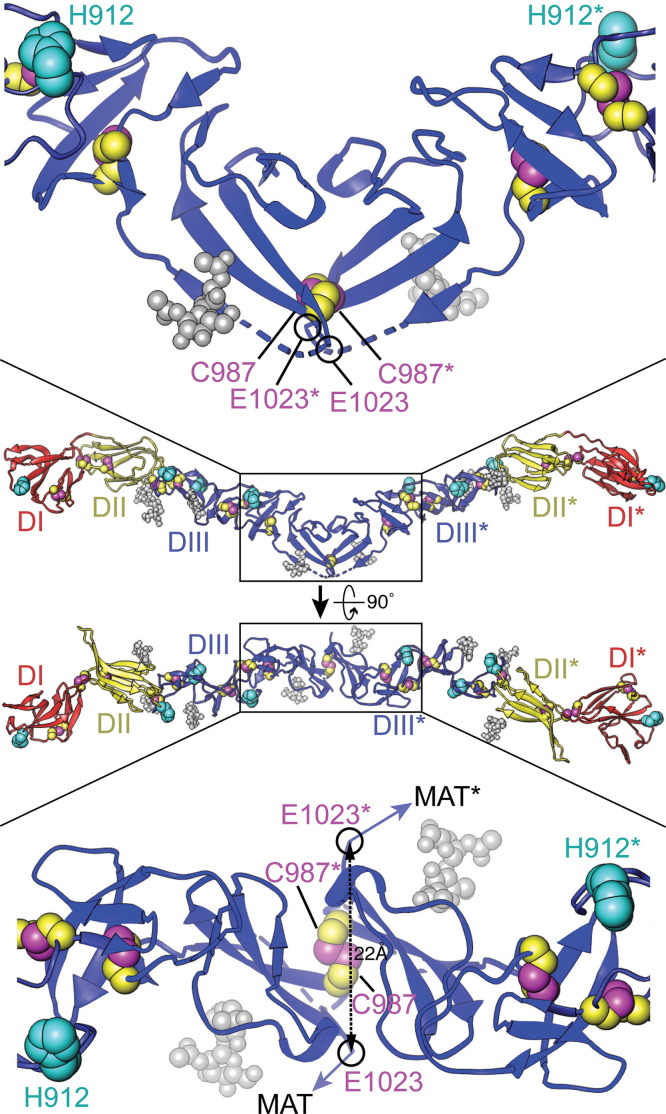
The dimeric BVDV E2 ectodomain contains an intersubunit disulfide bond across the dyad between C987 of each subunit, within the hydrophobic dimer interface enriched with aromatic residues (El Omari et al., 2013, Li et al., 2013). The last modeled residue in the E2 crystal structure is E1023 (in polyprotein numbering). The distance between the Cα atoms of E1023 of the two subunits is 22 Å ( Fig. 1). The remaining seven residues present in the crystallized construct are disordered in the crystals, including two histidine residues (H1027 and H1028) conserved in BVDV. These histidines could conceivably extend toward the dyad to form an intersubunit tetrahedral 4-His metal chelator across the dyad. The coordination of a metal (e.g. Zn2+) at the dimer interface provide additional stability to the E2 homodimer. Histidine protonation during endosomal acidification is a common mechanism for pH sensing in viral fusion proteins (de Boer et al., 2012, Dessau and Modis, 2013, Fritz et al., 2008, Nayak et al., 2009, Qin et al., 2009, Zheng et al., 2011). Protonation of the H1027 and H1028 side chains would reduce their metal binding affinity and could hence contribute to low pH-dependent destabilization of the E2 dimer. Assuming a pH-dependent conformational change is required for fusion activity, H1027 and H1028 may contribute to the pH-sensing mechanism of BVDV.
Fig. 1.
Two orthogonal views of the dimeric BVDV E2 ectodomains with close-up views near the dyad. Numbering refers to the polyprotein of BVDV-1 strain NADL. N-linked glycans are shown in gray space-filling representation, His residues (H762, H912, H846, and H871) in cyan, and Cys side chains in magenta (Sγ atoms) and yellow (carbon atoms). The locations of proposed intermolecular disulfide bonds between C987 of E2 and C668 of E1 are indicated by magenta arrows. E2 is colored by domain: I, red; II, yellow; III blue. The most C-terminal residue in the E2 crystal structure, E1023 (circled in black), connects to the membrane anchor and C-terminal tail (MAT). Seven additional C-terminal residues in the crystallized E2 construct were disordered in the crystal, including two conserved His residues (H1027 and H1028) between the membrane anchor and the ectodomain. The distance between the Cα atoms of E1023 from the two subunits is 22 Å. The Cα–Cα distance for C987 from the two subunits is 6.5 Å.
Secondary structure analysis and modeling of the BVDV E2 membrane anchor
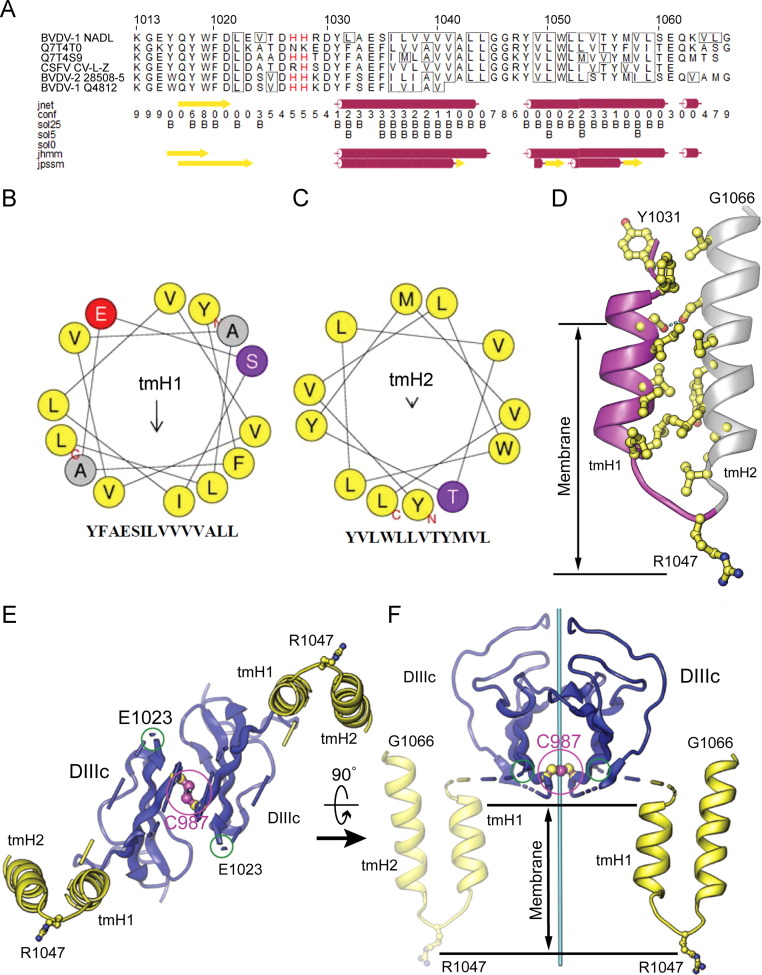
Residues 1036–1054 in E2 are hydrophobic except for R1047 and were predicted by several algorithms (Bernsel et al., 2009) to form a TM segment. The presence of the sequence Gly–Gly–Arg within this segment suggests that the anchor may have a similar topology as the unusual membrane anchor of dengue E protein (Zhang et al., 2013). In this topology, a short helical hairpin spans across part of the bilayer and the guanidinium moiety of the arginine “snorkels” to the hydrophilic layer of the inner monolayer, forming a salt bridge with the phosphate of a phospholipid head group ( Fig. 2). In this model, R1047 is essential for stabilizing the two hairpin helices (hpH1 and hpH2) in the viral membrane. Consistent with this model, substitution of the arginine with alanine prevents formation of E1–E2 heterodimers and inactivates the virus (Ronecker et al., 2008). An interhelical hydrogen bond between two serine side chains (Ser1035–Ser1060) stabilizes the otherwise hydrophobic packing between the two helices of the hairpin (Fig. 2D). Such interhelical hydrogen bonds are known to drive the assembly of TM segments (Zhou et al., 2000). The atomic model of the E2 MAT based on these constraints and on secondary structure predictions spans residues 1031–1066 (Fig. 2). The seven residues linking the ectodomain to the MAT (1024–1030) were not modeled. Based on the observed E1023–E1023 Cα distance of 22 Å (Fig. 1), we cannot rule out the possibility the E2 TM hairpins form homodimers as the unmodeled linker may span up to 28 Å.
Fig. 2.
Sequence analysis and atomic modeling of the BVDV E2 membrane anchor. (A) Two transmembrane helices (tmH1 and tmH2) were predicted with JPRED-3. The reference sequence and numbering refer to the BVDV-1 NADL polyprotein (Xu et al., 1997). This sequence was used to query the UniRef90 database. The following pestivirus E1 UniProt sequences returned from the search are shown: Q7T4T0, pestivirus PG-1; Q7T4S9, border disease virus strain V2536/2; Q9WPE6, classical swine fever virus strain C-V-LZ; Q9WP29, BVDV-2 strain 28508-5; Q65793, BVDV-1 strain Q4812. (B and C) Helical wheel representations of the core portions of tmH1 (B), and tmH2 (C). The arrows represent the direction and magnitude of the amphipathic moment of each helix. (D) Computationally generated model of the E2 membrane anchor. An interhelical hydrogen bond (dashed line) stabilizes the otherwise hydrophobic packing between the two helices of the hairpin. (E and F) Two orthogonal views of a computational model for dimeric E2 domain IIIc (DIIIc) followed by the membrane anchors. The positions and orientations of the membrane anchors relative to the DIIIc domains may vary depending on the structure of the linker between DIIIc and the anchor (blue/yellow dashed lines). C987 and the last residue in the crystal structure, E1023 are circled in magenta and green, respectively. R1047, which stabilizes the short helical hairpin by “snorkeling” to the inner surface of the viral membrane, is in ball-and-stick representation. The E2–E2 dyad is shown in cyan.
Secondary structure analysis and modeling of the BVDV E1 membrane anchor
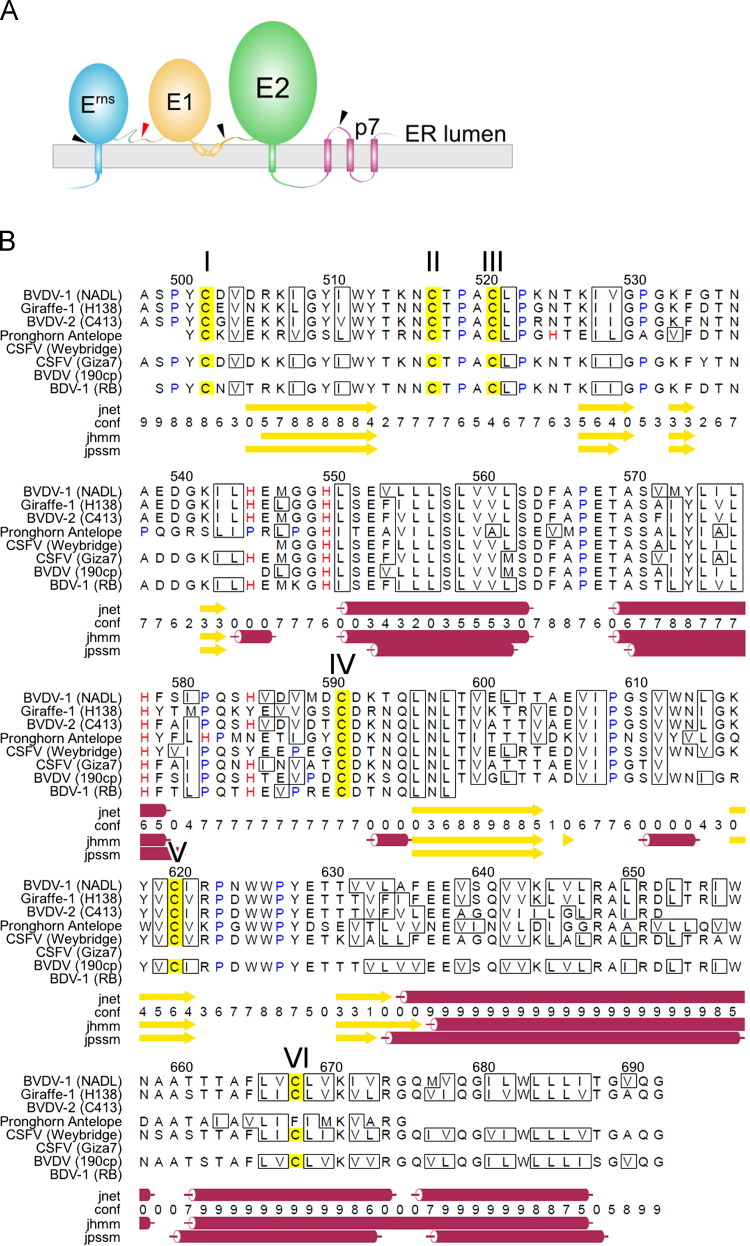
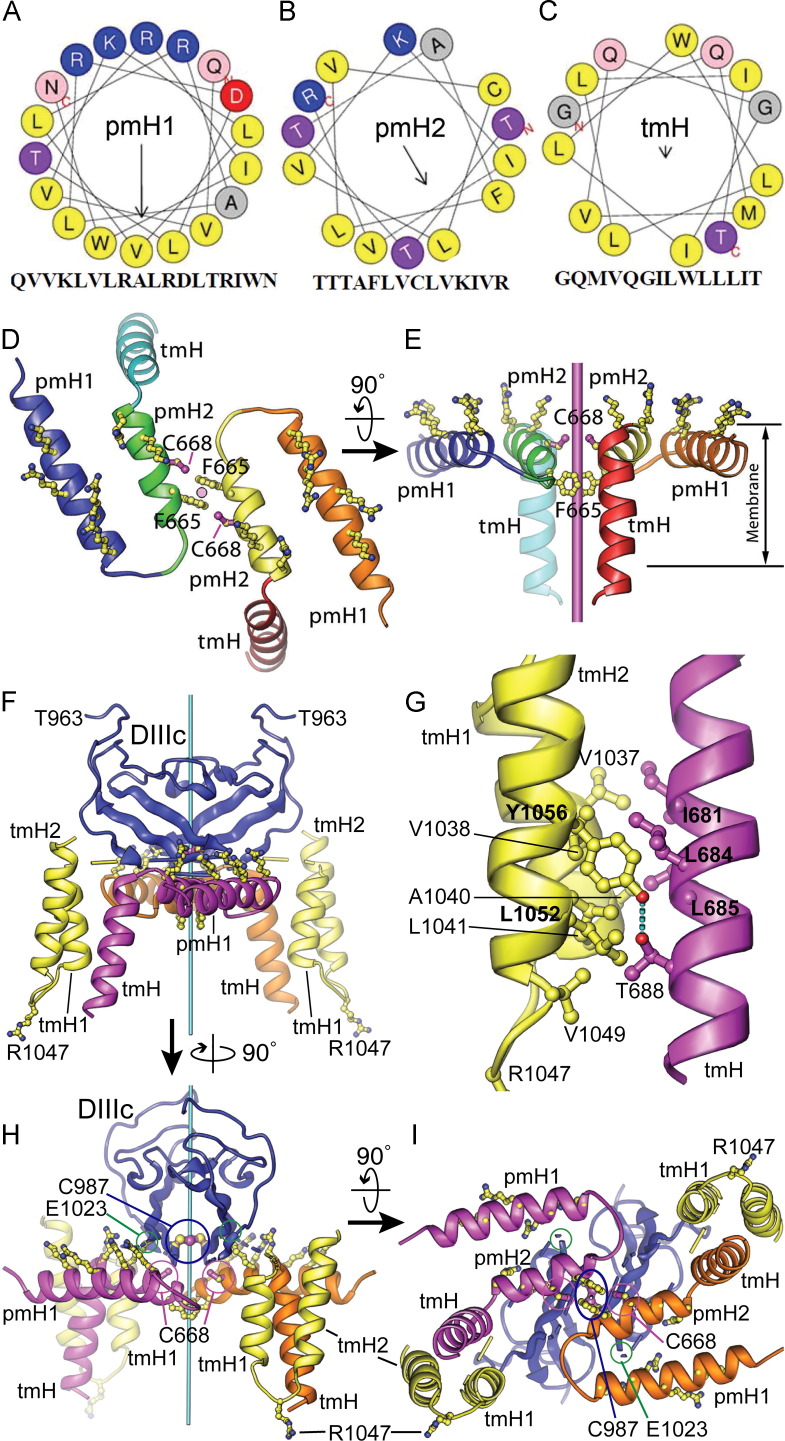
Secondary structure predictions based on multiple sequence alignments (Cole et al., 2008) suggest that the E1 membrane anchor contains two perimembrane helices (pmH) and one transmembrane helix (tmH): pmH1, A635 to N658; pmH2, T661 to R674; and tmH, Q676 to I687 ( Fig. 3). In a helical wheel representation (Mount, 2004), pmH1 and pmH2 are strongly amphipathic. A KxxxRxxRxxxR motif in pmH1 and a CxxKxxR motif in pmH2 create a positively charged face on each helix in the otherwise hydrophobic outer surfaces ( Fig. 4). These positively charged residues are proposed to interact with negatively charged phospholipid head groups, whereas the remaining hydrophobic residues are presumably buried inside the bilayer. The pmH1 and pmH2 helices of E1 interact via their LTVL and TVLV sequences, respectively (Fig. 4). Notably, the dengue virus M and E envelope proteins each contain a pair of amphipathic perimembrane helices (Zhang et al., 2013). The third predicted helix in E1 has no charged residues and is predicted to be a transmembrane helix (tmH). Our atomic model of the E1 MAT based on these constraints and on secondary structure predictions spans residues 636–692 (Fig. 4).
Fig. 3.
Topology and secondary structure predictions of BVDV E1. (A) Topology of the pestivirus E1 and E2 proteins. Wedges indicate the sites at which proteases cleave during polyprotein maturation. (B) Secondary structure prediction of BVDV E1 based on a pestivirus multiple sequence alignment. Secondary structure elements were predicted with Jpred 3. Sequence numbering refers to the BVDV-1 NADL polyprotein. The sequence contains six cysteines, labeled I–VI and referred to in the text as such. Conserved His residues flank the first two predicted α-helices. The following pestivirus E2 UniProt sequences were selected for the sequence alignment based on search results using the BVDV-1 NADL sequence as the query in search against the UniRef90 database: Q9PYB2, pestivirus giraffe-1 strain H138; UPI0000167D3B, BVDV-2 strain C413; Q533M7, pronghorn antelope pestivirus; Q68954, classical swine fever virus strain Weybridge; O90243, classical swine fever virus strain Giza7; Q76B25, bovine viral diarrhea virus strain 190cp; P89049, pestivirus type 3 (border disease virus 1) strain RB.
Fig. 4.
Helical wheel modeling of the E1 membrane anchor, and atomic modeling of the E1–E2 oligomeric assembly. (A–C) Helical wheel representations of the core portions of the three membrane-anchored helices in E1: perimembrane helices 1 and 2 (pmH1 and pmH2), and the transmembrane helix (tmH). Interaction of the left side of pmH1 with the right side of pmH2 would position Cys668 (“C”) near the dyad and avoid burying the charged aspartate side chain (“D”) at the interface between the two helices. The arrows represent the direction and magnitude of the amphipathic moment of each helix. (D–E) Orthogonal views of a computational model of two E1 membrane anchors placed to dock optimally on the model of E2 shown in Fig. 2. The dyad is shown in cyan. (F) Computational model of an E1–E2 heterotetramer consistent with the crystallographic E2 homodimer and with the formation of a disulfide bond between E1 C668 and E2 C987. The two E2 fragments are shown in blue (domain IIIc, DIIIc) and yellow (TM helices tmH1, tmH2). The two E1 membrane anchors are in magenta and orange, respectively. The relative position and orientation of DIIIc relative to the membrane anchor assembly may vary depending on the structure of the linker between DIIIc and tmH1 (yellow/blue dashed lines). (G) Close-up of the interface between the E1 tmH and E2 tmH1 helices. A hydrogen bond between E2 Y1056 and E1 T688 stabilizes the otherwise hydrophobic interface. (H-I) Two alternative views of panel F. In the modeled prefusion conformation, the Cα atoms of E1 C668 (cysteine VI, circled in magenta) and E2 C987 (circled in blue) are 6.9 Å apart, within the range for disulfide bond formation.
Modeling of a heterotetrameric BVDV E1–E2 membrane anchor assembly
Having obtained models of the membrane anchors of E1 and E2 we sought to construct a model of the E1–E2 assembly, integrating all available biochemical and structural constraints. The crystal structures of BVDV E2 reveal an extensive network of non-covalent interactions at the E2 homodimer interface. The extent of this network indicates that the E2 homodimers represent a biologically relevant species and are unlikely to dissociate (El Omari et al., 2013, Li et al., 2013). Moreover, E2 ectodomain homodimers are remarkably stable in solution even in the presence of reducing agents (Li et al., 2013). Hence any structural model of the E1–E2 assembly would need to be consistent with a stable E2–E2 homodimer interface.
The requirement for disulfide-linked E1–E2 heterodimers for infectivity (Ronecker et al., 2008) imposes additional constraints. All of the cysteines in E2 form intramolecular disulfide bonds, except for C987, which forms a disulfide bond across the E2 dimer interface in the crystal structures and in solution in the absence of E1 (El Omari et al., 2013, Li et al., 2013). However, E1–E2 disulfide crosslinks are the dominant crosslinks in pestivirus particles (Rumenapf et al., 1993, Thiel et al., 1991, Weiland et al., 1990). Thus, the E2–E2 crosslink through Cys987 observed in crystal structures is either an artifact due to E1 being absent, or it may form after the fusogenic conformational change in E1/E2 has proceeded. Alternatively, E2–E2 crosslinks may be present in an immature form of the virus. Either way, since E2–E2 crosslinks are a minor species in fusogenic virions, E2 C987 is by elimination the best candidate for forming the observed and necessary disulfide link with E1. It is less clear which of the E1 cysteines is (or are) involved in E1–E2 crosslinking. Any one of the six cysteines in E1 from classical swine fever virus (CSFV, formerly hog cholera virus) can be mutated without disrupting E1–E2 heterodimers in SDS-PAGE under non-reducing conditions (Fernandez-Sainz et al., 2011). Conversely, mutation of either one of the charged residues in the CxxKxxR motif of the E1 MAT (in the pmH2 helix) leads to the loss of E1–E2 dimers in non-reducing SDS-PAGE despite the conservation of all cysteine residues (Ronecker et al., 2008).
To reconcile the available biochemical and structural data, we sought a structural model of the E1–E2 assembly that was consistent with both a stable E2–E2 homodimer interface and E1–E2 disulfide crosslinks. Based on the structure of E2, for simultaneous access to two C987 residues in an E2 dimer two E1 molecules must dimerize through pmH2–pmH2 interactions to form a heterotetramer consisting of two molecules of E1 and two molecules of E2 (Fig. 4D and E). We were able to construct a heterotetrameric model in this configuration, in which E2 C987 is near E1 C668. In this model, the Cα atoms of E1 C668 and E2 C987 are 6.9 Å apart, which is within the range for disulfide bond formation (Fig. 4F–I). The E1–E1 and E1–E2 interfaces have good shape complementarity. An interhelical hydrogen bond between E2 Y1056 and E1 T688 stabilizes the otherwise hydrophobic interface between the E1 tmH and E2 tmH1 helices (Fig. 4G). This model is consistent with the previously established importance of interhelical hydrogen bonding in driving strong and specific interactions in membrane proteins (Zhou et al., 2000). The C668 residues in the two E1 subunits are far enough from each other in this model (13 Å Cα–Cα distance) that they would not form an intermolecular E1–E1 disulfide bond, which has not been observed experimentally. Additionally, the side chain of F665, by forming a π-stacking interaction with its dyad-related mate in our model, could prevent the two C668 residues from forming a disulfide bond (Fig. 4). The position of E1 C668 in the CxxKxxR motif of pmH2, near the membrane anchor of E1, makes it a good candidate for disulfide bonding with E2 C987. The requirement of K671 and R674 for E1–E2 heterodimer formation suggests that these charged residues promote disulfide bonding between C668 and E2 C987, possibly by keeping E1 helix pmH2 positioned on the membrane surface rather than inside the membrane. However, since no single cysteine in E1 is indispensable, residues other than E1 C688 may participate in E1–E2 heterodimer formation (Fernandez-Sainz et al., 2011, Ronecker et al., 2008).
The presence of six positively charged E1 residues on the outer membrane leaflet and R1047 from E2 on the inner leaflet in our model could substantially reduce the thickness of the viral membrane, and increase its fusogenicity. A reduction of membrane thickness is consistent with the thickness of the hydrophobic portion of the E1–E2 membrane anchor assembly, which at approximately 26 Å is on the low end of the documented range for lipid bilayers (Mitra et al., 2004). Similar structural features in dengue virus were proposed to be responsible for distortions of the viral membrane (Zhang et al., 2013). Interestingly, the disordered C-terminus of the E2 ectodomain contains five conserved negatively charged residues (Fig. 2). Electrostatic interactions between these residues and the positively charged residues in the E1 MAT may promote association of E1 and E2 so that a disulfide bond can form between E2 C987 and E1 C668.
Constraints on the structure and disulfide bonding pattern of the E1 ectodomain
Despite the absence of structural information for the E1 ectodomain, recent studies identify specific epitopes in E1 and E2 as likely to participate in E1–E2 interactions. Antibody neutralization escape mutations in the conserved TAV epitope of CSFV E2 (residues 829–831 in domain II) were accompanied by two compensatory mutations in E1, Y575H and D583E, along with mutations in ERNS, suggesting that the TAV epitope of E2 interacts with residues 575 and 583 of E1 (Leifer et al., 2012). Similarly, CSFV strains with mutations in the E2 TAV epitope can replicate more efficiently if compensatory mutations occur in E1 (E634D) or ERNS (Kortekaas et al., 2010). More generally, the antigenic epitopes that have been mapped onto E2 from BVDV or CSFV are all located in the first two Ig-like domains of E2, domains I and II (Li et al., 2013). The lack of confirmed antigenic epitopes in domain III of E2 suggests that this domain may be shielded from the solvent by E1 on the viral surface. This would imply that the membrane distal face of E2 domain III forms contacts with E1. This would be consistent with the location of the TAV epitope on the membrane distal face of E2.
E1 contains six conserved cysteine residues, subsequently referred to as I–VI (Fig. 3). As is typical for viral glycoproteins, the cysteines are likely to form disulfide bonds. Our model of two E1 molecules docked onto the dimeric E2 crystal structure (Fig. 4), in combination with secondary structure prediction of the E1 ectodomain, imposes certain specific geometric constraints on the internal disulfide bonding pattern within E1. Secondary structure predictions place cysteines I and II on opposite ends of a β-strand (Fig. 3), suggesting that these residues are too far away from each other to form a disulfide bond. A IV–V disulfide bond is possible because cysteines IV and V are located on the opposite ends of two consecutive predicted β-strands (Fig. 3), which would place them in proximity to each other if the two strands contribute to the same β-sheet, or to two stacked sheets in the same domain. A II–III disulfide bond is also possible because the cysteines II and III are separated by only three residues, Thr–Pro–Ala, in which the proline may promote a non-linear or hairpin-like loop conformation. In summary, our analysis suggests that a I–II linkage is unlikely and that a possible disulfide bonding pattern in E1 is II–III and IV–V. This arrangement would leave cysteines I and IV unpaired and available for covalent or non-covalent interactions with E2, if they are not buried within E1. However, to definitively establish the disulfide bonding pattern in E1 it will be necessary to obtain structural information for the E1 ectodomain.
Geometric and sequence constraints on a putative fusion motif in E1
Since E2 does not contain any clearly recognizable fusion motifs to insert into the host cell membrane, E1 is thought to bear the fusion motif in pestiviruses (El Omari et al., 2013, Li et al., 2013). If so, E1 would have to at least transiently extend to span the distance between the cellular and viral membranes prior to membrane fusion, approximately 20 nm (Kim et al., 2011). With only 177 amino acids in its ectodomain, or 143 amino acids excluding the predicted perimembrane helices, E1 would have to adopt a highly elongated fold in order to span 20 nm. Since coiled-coils efficiently form rigid and highly elongated structures, the two predicted α-helices in the central region of the E1 ectodomain (Fig. 3) may form a helical coiled-coil.
Fusion motifs do not have any strict consensus sequences but their membrane anchoring activity stems from the intrinsic physical properties of aromatic side chains, usually tryptophan and phenylalanine, and hydrophobic side chains, most commonly leucine and isoleucine (Backovic and Jardetzky, 2009, Kielian and Rey, 2006, Skehel and Wiley, 2000). A candidate conserved fusion loop sequence with these properties is in residues 573–579, (I/L/M)YL(I/V/A)LH(F/Y) (Fig. 3). Alternatively, the GYIWY sequence near the N-terminus (residues 509–513) could serve as the fusion loop. Since E1 is unlikely to have a large hydrophobic core due to its elongated shape, the fusion motif would have to be sequestered at the E1–E2 interface or near the viral membrane in the prefusion state.
Conclusions
Using the geometric constraints imposed by the crystal structures of BVDV E2, along with secondary structure predictions and sequence alignments, we have generated theoretical models for the membrane anchors of BVDV E1 and E2. The model for the E2 anchor contains a truncated helical hairpin stabilized by an arginine residue, similar to the anchor reported recently for dengue E protein (Zhang et al., 2013). Based on this model we propose that that histidines 1027 and 1028 may participate in pH sensing by E2, possibly by forming a four-histidine metal-chelating cluster across the E2 dimer interface. The model for the E1 anchor contains two amphipathic perimembrane helices followed by a transmembrane helix. Our heterotetrameric model of the E1–E2 assembly suggests that Cys668 in E1 forms a disulfide bond with Cys987 in E2, thereby stabilizing the E1–E2 interaction that is required for virus infectivity. We extended our sequence and structural analysis to include the E1 ectodomain, for which we propose a partial disulfide bonding pattern and a possible α-helical coiled-coil configuration.
Our modeling studies allow us to propose the following tentative molecular mechanism for how E1–E2 heterodimers initiate membrane fusion: (i) the reduced endosomal pH protonates the side chains in the histidine cluster at the E2 dimer interface, destabilizing the cluster, exposing and possibly destabilizing the E2 homodimer interface, (ii) a second, previously postulated but poorly understood activation step (Krey et al., 2005) triggers a conformational rearrangement that exposes a fusion motif in E1, possibly in residues 509–513 or in residues 573–579, (iii) E1 extends to reach the target cellular membrane, with the fusion motif poised for membrane insertion. Future experiments to test or confirm this proposed mechanism of pestivirus fusion activation may be guided by the structural constraints and atomic models developed in this study.
Materials and methods
Multiple sequence alignments and helical wheel projections
To assess the surface electrostatic potential of the predicted membrane-proximal and TM helices of E1 and E2 and their possible modes of assembly, we generated multiple sequence alignments of pestiviral E1 and E2 sequences. To identify pestivirus sequences to use in the alignments, we used the BVDV-1 strain NADL (Xu et al., 1997) protein sequence as the query to search against the UniRef90 database (Mount, 2004). Secondary structure predictions were generated using Jpred 3 (Cole et al., 2008). Helical wheel projections were carried out using a web service interface created by Everett and colleagues (Kryshtafovych et al., 2011).
Atomic modeling
Atomic models of the E1 and E2 membrane anchors were generated manually in the model building program Coot (Emsley and Cowtan, 2004) starting with α-helical template coordinates with idealized geometry. All connecting loop structures were built manually with Coot and then subjected to geometry idealization with Coot. Although E1 pmH1 and pmH2 were modeled as helical hairpins, they could alternatively form more extended non-hairpin structures, particularly after the opening of a fusion pore in the postfusion conformation. To assist in the structural analysis of the E1 ectodomain, we used Coot to generate complete atomic models of the E1 that contained the secondary elements predicted by Jpred 3 (Cole et al., 2008, Mount, 2004). Multiple polyalanine models with alternative backbone configurations were generated. The geometry of each E1 ectodomain model was idealized with Coot. When compared with the experimentally determined structure of the E2 ectodomain (El Omari et al., 2013, Li et al., 2013), the E1 ectodomain models allowed us to generate geometric constraints on the backbone configuration and disulfide bonding pattern of E1. The atomic model of the E1 MAT spans residues 636–692; the atomic model of the E2 MAT based on these constraints and on secondary structure predictions spans residues 1031–1066. The atomic coordinates of the heterotetrameric E1–E2 membrane anchor assembly as modeled in this study are available in the Supplementary online information. Figures were prepared using the graphics display program Ribbons (Carson, 1997).
Acknowledgments
We thank Dr. Francisco N. Barrera for inspirational discussions. This work was supported by a Burroughs Welcome Investigator in the Pathogenesis of Infectious Disease Award and National Institutes of Health Grants P01 GM022778 and R01 GM102869 (to Y.M.). J.W. was supported by the Steitz Center for Structural Biology, Gwangju Institute of Science and Technology, Republic of Korea.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2014.02.015.
Appendix A. Supplementary materials
Supplementary data
Supplementary data
References
- Backovic M., Jardetzky T.S. Class III viral membrane fusion proteins. Curr. Opin. Struct. Biol. 2009;19:189–196. doi: 10.1016/j.sbi.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backovic M., Jardetzky T.S. Class III viral membrane fusion proteins. Adv. Exp. Med. Biol. 2011;714:91–101. doi: 10.1007/978-94-007-0782-5_3. [DOI] [PubMed] [Google Scholar]
- Bernsel A., Viklund H., Hennerdal A., Elofsson A. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37:W465–W468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branza-Nichita N., Durantel D., Carrouee-Durantel S., Dwek R.A., Zitzmann N. Antiviral effect of N-butyldeoxynojirimycin against bovine viral diarrhea virus correlates with misfolding of E2 envelope proteins and impairment of their association into E1-E2 heterodimers. J. Virol. 2001;75:3527–3536. doi: 10.1128/JVI.75.8.3527-3536.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M. Ribbons. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- Cole C., Barber J.D., Barton G.J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer S.M., Kortekaas J., Spel L., Rottier P.J., Moormann R.J., Bosch B.J. Acid-activated structural reorganization of the Rift Valley fever virus Gc fusion protein. J. Virol. 2012;86:13642–13652. doi: 10.1128/JVI.01973-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Francesco R., Migliaccio G. Challenges and successes in developing new therapies for hepatitis C. Nature. 2005;436:953–960. doi: 10.1038/nature04080. [DOI] [PubMed] [Google Scholar]
- Dessau M., Modis Y. Crystal structure of glycoprotein C from Rift Valley fever virus. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1696–1701. doi: 10.1073/pnas.1217780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBois R.M., Vaney M.C., Tortorici M.A., Kurdi R.A., Barba-Spaeth G., Krey T., Rey F.A. Functional and evolutionary insight from the crystal structure of rubella virus protein E1. Nature. 2013;493:552–556. doi: 10.1038/nature11741. [DOI] [PubMed] [Google Scholar]
- Durantel D., Branza-Nichita N., Carrouee-Durantel S., Butters T.D., Dwek R.A., Zitzmann N. Study of the mechanism of antiviral action of iminosugar derivatives against bovine viral diarrhea virus. J. Virol. 2001;75:8987–8998. doi: 10.1128/JVI.75.19.8987-8998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Omari K., Iourin O., Harlos K., Grimes J.M., Stuart D.I. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 2013;3:30–35. doi: 10.1016/j.celrep.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fernandez-Sainz I., Holinka L.G., Gladue D., O׳Donnell V., Lu Z., Gavrilov B.K., Risatti G.R., Borca M.V. Substitution of specific cysteine residues in the E1 glycoprotein of classical swine fever virus strain Brescia affects formation of E1–E2 heterodimers and alters virulence in swine. J. Virol. 2011;85:7264–7272. doi: 10.1128/JVI.00186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz R., Stiasny K., Heinz F.X. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J. Cell Biol. 2008;183:353–361. doi: 10.1083/jcb.200806081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garry R.F., Dash S. Proteomics computational analyses suggest that hepatitis C virus E1 and pestivirus E2 envelope glycoproteins are truncated class II fusion proteins. Virology. 2003;307:255–265. doi: 10.1016/s0042-6822(02)00065-x. [DOI] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein E.E., Lou H., Bender F.C., Cohen G.H., Eisenberg R.J., Harrison S.C. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- Kadlec J., Loureiro S., Abrescia N.G., Stuart D.I., Jones I.M. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 2008;15:1024–1030. doi: 10.1038/nsmb.1484. [DOI] [PubMed] [Google Scholar]
- Kielian M., Rey F.A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.H., Donald J.E., Grigoryan G., Leser G.P., Fadeev A.Y., Lamb R.A., DeGrado W.F. Capture and imaging of a prehairpin fusion intermediate of the paramyxovirus PIV5. Proc. Natl. Acad. Sci. U. S. A. 2011;108:20992–20997. doi: 10.1073/pnas.1116034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Giang E., Nieusma T., Kadam R.U., Cogburn K.E., Hua Y., Dai X., Stanfield R.L., Burton D.R., Ward A.B., Wilson I.A., Law M. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortekaas J., Vloet R.P., Weerdmeester K., Ketelaar J., van Eijk M., Loeffen W.L. Rational design of a classical swine fever C-strain vaccine virus that enables the differentiation between infected and vaccinated animals. J. Virol. Methods. 2010;163:175–185. doi: 10.1016/j.jviromet.2009.09.012. [DOI] [PubMed] [Google Scholar]
- Krey T., Thiel H.J., Rumenapf T. Acid-resistant bovine pestivirus requires activation for pH-triggered fusion during entry. J. Virol. 2005;79:4191–4200. doi: 10.1128/JVI.79.7.4191-4200.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryshtafovych A., Moult J., Bartual S.G., Bazan J.F., Berman H., Casteel D.E., Christodoulou E., Everett J.K., Hausmann J., Heidebrecht T., Hills T., Hui R., Hunt J.F., Seetharaman J., Joachimiak A., Kennedy M.A., Kim C., Lingel A., Michalska K., Montelione G.T., Otero J.M., Perrakis A., Pizarro J.C., van Raaij M.J., Ramelot T.A., Rousseau F., Tong L., Wernimont A.K., Young J., Schwede T. Target highlights in CASP9: experimental target structures for the critical assessment of techniques for protein structure prediction. Proteins. 2011;79(Suppl. 10):S6–S20. doi: 10.1002/prot.23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R.A., Jardetzky T.S. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifer I., Blome S., Blohm U., Konig P., Kuster H., Lange B., Beer M. Characterization of C-strain “Riems” TAV-epitope escape variants obtained through selective antibody pressure in cell culture. Vet. Res. 2012;43:33. doi: 10.1186/1297-9716-43-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescar J., Roussel A., Wien M.W., Navaza J., Fuller S.D., Wengler G., Rey F.A. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- Li Y., Wang J., Kanai R., Modis Y. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc. Natl. Acad. Sci. U. S. A. 2013;110:6805–6810. doi: 10.1073/pnas.1300524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D., Murray C.L., Thiel H.J., Rice C.M. Flaviviridae. In: Knipe D.M., Howley P.M., editors. Fields Virology. Sixth ed. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 712–746. [Google Scholar]
- Mathapati B.S., Mishra N., Rajukumar K., Nema R.K., Behera S.P., Dubey S.C. Entry of bovine viral diarrhea virus into ovine cells occurs through clathrin-dependent endocytosis and low pH-dependent fusion. In Vitro Cell Dev. Biol. 2010;46:403–407. doi: 10.1007/s11626-009-9263-9. [DOI] [PubMed] [Google Scholar]
- Meertens L., Bertaux C., Dragic T. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J. Virol. 2006;80:11571–11578. doi: 10.1128/JVI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K., Ubarretxena-Belandia I., Taguchi T., Warren G., Engelman D.M. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y. Relating structure to evolution in Class II viral membrane fusion proteins. Curr. Opin. Virol. 2014;5C:34–41. doi: 10.1016/j.coviro.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount D.M. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2004. Bioinformatics: Sequence and Genome Analysis. [Google Scholar]
- Nayak V., Dessau M., Kucera K., Anthony K., Ledizet M., Modis Y. Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion. J. Virol. 2009;83:4338–4344. doi: 10.1128/JVI.02574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J., Patel A.H., McLauchlan J. The transmembrane domain of the hepatitis C virus E2 glycoprotein is required for correct folding of the E1 glycoprotein and native complex formation. Virology. 2001;279:58–68. doi: 10.1006/viro.2000.0693. [DOI] [PubMed] [Google Scholar]
- Qin Z.L., Zheng Y., Kielian M. Role of conserved histidine residues in the low-pH dependence of the Semliki Forest virus fusion protein. J. Virol. 2009;83:4670–4677. doi: 10.1128/JVI.02646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey F.A., Heinz F.X., Mandl C., Kunz C., Harrison S.C. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Roche S., Bressanelli S., Rey F.A., Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- Roche S., Rey F.A., Gaudin Y., Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- Ronecker S., Zimmer G., Herrler G., Greiser-Wilke I., Grummer B. Formation of bovine viral diarrhea virus E1-E2 heterodimers is essential for virus entry and depends on charged residues in the transmembrane domains. J. Gen. Virol. 2008;89:2114–2121. doi: 10.1099/vir.0.2008/001792-0. [DOI] [PubMed] [Google Scholar]
- Rumenapf T., Unger G., Strauss J.H., Thiel H.J. Processing of the envelope glycoproteins of pestiviruses. J. Virol. 1993;67:3288–3294. doi: 10.1128/jvi.67.6.3288-3294.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibli D.J., Weissenhorn W. Class I and class II viral fusion protein structures reveal similar principles in membrane fusion. Mol. Membr. Biol. 2004;21:361–371. doi: 10.1080/09687860400017784. [DOI] [PubMed] [Google Scholar]
- Shepard C.W., Finelli L., Alter M.J. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Thiel H.J., Stark R., Weiland E., Rumenapf T., Meyers G. Hog cholera virus: molecular composition of virions from a pestivirus. J. Virol. 1991;65:4705–4712. doi: 10.1128/jvi.65.9.4705-4712.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherne D.M., Evans M.J., Macdonald M.R., Rice C.M. Transdominant inhibition of bovine viral diarrhea virus entry. J. Virol. 2008;82:2427–2436. doi: 10.1128/JVI.02158-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherne D.M., Jones C.T., Evans M.J., Lindenbach B.D., McKeating J.A., Rice C.M. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 2006;80:1734–1741. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Nie Y., Wang P., Ding M., Deng H. Characterization of classical swine fever virus entry by using pseudotyped viruses: E1 and E2 are sufficient to mediate viral entry. Virology. 2004;330:332–341. doi: 10.1016/j.virol.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Weiland E., Stark R., Haas B., Rumenapf T., Meyers G., Thiel H.J. Pestivirus glycoprotein which induces neutralizing antibodies forms part of a disulfide-linked heterodimer. J. Virol. 1990;64:3563–3569. doi: 10.1128/jvi.64.8.3563-3569.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Mendez E., Caron P.R., Lin C., Murcko M.A., Collett M.S., Rice C.M. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J. Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ge P., Yu X., Brannan J.M., Bi G., Zhang Q., Schein S., Zhou Z.H. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat. Struct. Mol. Biol. 2013;20:105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Sanchez-San Martin C., Qin Z.L., Kielian M. The domain I-domain III linker plays an important role in the fusogenic conformational change of the alphavirus membrane fusion protein. J. Virol. 2011;85:6334–6342. doi: 10.1128/JVI.00596-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F.X., Cocco M.J., Russ W.P., Brunger A.T., Engelman D.M. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat. Struct. Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data