Abstract
An increasing number of population studies are assessing epigenetic variation in relation to early-life outcomes in tissues accessible to epidemiologic researchers. Epigenetic mechanisms are highly tissue specific, however, and it is unclear whether the variation observed in one of the tissue types is representative of other sources or whether the variation in DNA methylation is distinct, reflecting potential functional differences across tissues. To assess relations between DNA methylation in various samples from newborns and children in early infancy, we measured promoter or gene-body DNA methylation in matched term placenta, cord blood, and 3–6 mo saliva samples from 27 unrelated infants enrolled in the Rhode Island Child Health Study. We investigated 7 gene loci (KLF15, NR3C1, LEP, DEPTOR, DDIT4, HSD11B2, and CEBPB) and global methylation, using repetitive region LINE-1 and ALUYb8 sequences. We observed a great degree of interlocus, intertissue, and interindividual epigenetic variation in most of the analyzed loci. In correlation analyses, only cord blood NR3C1 promoter methylation correlated negatively with methylation in saliva. We conclude that placenta, cord blood, and saliva cannot be used as a substitute for one another to evaluate DNA methylation at these loci during infancy. Each tissue has a unique epigenetic signature that likely reflects their differential functions. Future studies should consider the uniqueness of these features, to improve epigenetic biomarker discovery and translation.—Armstrong, D. A., Lesseur, C., Conradt, E., Lester, B. M., Marsit, C. J. Global and gene-specific DNA methylation across multiple tissues in early infancy: implications for children's health research.
Keywords: cord blood, epigenetic epidemiology, placenta, saliva
The term epigenetics refers to heritable changes in gene expression without alterations in the underlying DNA sequence (1). DNA methylation of cytosine residues within CpG dinucleotides is the most commonly studied epigenetic mark (2). It is relevant during development and is thought to mediate, at least in part, epigenetic programming (3, 4). Epigenetics could provide a molecular basis for the epidemiologically derived theory of developmental origin of health and disease (DOHaD; ref. 5). In addition, recent emphasis has been placed on the role of environmental and lifestyle exposures in epigenetic programming and development (6), including prenatal socioeconomic adversity (7), nutrition (8), maternal depression (9), air and water quality (10), smoking (11), and exercise (12).
Epigenetic epidemiology is an emerging field that attempts to elucidate associations of human diseases, disorders, and conditions with epigenetic variation (4, 13, 14). For this purpose, multiple tissue sources have been examined. Peripheral blood is a common source of DNA in adults (15), due to its relative accessibility and because it offers the possibility of prospective sampling, especially attractive in the context of dynamic epigenetic changes (16). Birth cohort studies, on the other hand, frequently collect cord blood and placenta to assess the newborn's epigenome (16–19), and evidence from these studies has strengthened the relation between epigenetics and the DOHaD theory. However, cord blood and placenta can be sampled at only one time point, their predictive capability beyond newborn and early-life outcomes has yet to be shown, and their collection requires somewhat complicated and time-dependent protocols that may be available only in large studies at academic medical centers. Similarly, blood sampling through venipuncture is not ideal during early infancy.
Alternatively, saliva is a more practical DNA source, because it can be collected through noninvasive protocols and can be collected more readily at repeated time points throughout childhood and beyond. The use of saliva for clinical assays dates back to 1928 (20), and today it is commonly used for various molecular assessments (21). Recently, an increasing number of epigenetic epidemiologic studies have used saliva as a DNA source. For example, in a study performed in adult twins, saliva DNA methylation at 3 CpG sites in different genes predicted age in a population ranging from 18 to 70 yr (22). Moreover, a recent study (19) found a positive correlation in TACSTD2 promoter methylation between saliva and peripheral blood in 11-yr-old children. This evidence suggests that, at least for some loci, saliva DNA methylation can act as a surrogate for leukocyte methylation, but in general, few data are available on the relationship during early infancy between DNA methylation in saliva samples and other tissue sources, such as placenta and cord blood. Given the high tissue specificity of epigenetic mechanisms, understanding the relationship of epigenetic marks, such as DNA methylation between different tissues, can have important consequences on study design and data interpretation arising from their assessment.
Therefore, we assessed global and gene-specific DNA methylation in paired tissues from newborns: umbilical cord blood and placental tissue and saliva collected in early infancy. Our goal was to investigate the extent to which these tissues could serve as surrogates for one another, particularly for genes relevant to the developmental origins of health and disease during infancy.
MATERIALS AND METHODS
Study population
Infants included in this study are part of the ongoing Rhode Island Child Health Study (RICHS; ref. 23), which enrolls mother–infant dyads after delivery at Women and Infants Hospital (Providence, RI,. USA). Percentiles based on birth weight and gestational age are calculated from the Fenton growth chart (24) and used to enroll term infants born small for gestational age (SGA; <10th percentile) or large for gestational age (LGA; >90th percentile), along with an appropriate for gestational age (AGA) infant matched on sex, gestational age (±3 d), and maternal age (±2 yr). Only singleton, viable infants are included in the study. Other exclusion criteria include maternal age < 18 yr, maternal life-threatening medical complications, and infant congenital or chromosomal abnormalities. Information from the delivery medical record was collected with a structured chart review form, followed by an interviewer-administered structured questionnaire to obtain demographic, lifestyle, and exposure histories. All participants provided written, informed consent for involvement in this study. The protocols used were approved by the Institutional Review Boards of Women and Infants Hospital and Dartmouth College and were performed in accordance with the Declaration of Helsinki.
Biological sample collection and nucleic acid extraction
Placental biopsies (n=27) free of maternal decidua were collected within 4 h of delivery. In total, 12 fragments (∼1 g), 3 from each quadrant, were excised from the maternal side of the placenta 2 cm from the umbilical cord insertion site. Immediately after collection, the samples were placed in RNAlater (Life Technologies, Carlsbad, CA, USA) and stored at 4°C. After ≥72 h, tissue segments from each placental region were blotted dry, snap-frozen in liquid nitrogen, homogenized by pulverization in a stainless steel cup and piston unit (Cellcrusher, Cork, Ireland), and stored at −80°C until needed. When possible, infant cord blood (n=16) was obtained from residual samples collected for clinical purposes with a syringe from the umbilical cord after delivery and stored in blood collection tubes (4°C) until DNA extraction. From 26 of the infants, saliva samples were collected with the Oragene Saliva Collection Kit (DNA Genotek Inc., Ottawa, ON, Canada) at 1 follow-up visit between 3 and 6 mo of age. DNA was extracted from homogenized placental samples and an aliquot of whole umbilical cord blood with the DNeasy Blood and Tissue Kit (Qiagen, Inc, Valencia, CA, USA). DNA isolation from saliva samples was performed with the Oragene extraction kit (DNA Genotek). The DNA was quantified with the ND2000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). DNA from each sample was sodium bisulfite modified with the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA). All tissue samples from the same individual were modified in the same plate. The procedures were performed according to the manufacturer's instructions.
Gene selection and DNA methylation analysis
We selected genes for methylation analysis by using a multifaceted approach. First, we focused on several genes that are currently under study in epidemiologic investigations of DOHaD. Group 1: hydroxysteroid dehydrogenase 11 β2 (HSD11B2; 3 CpGs), nuclear receptor subfamily 3, group C, member 1 (NR3C1; 13CpGs), and leptin (LEP; 23 CpGs) (23, 25, 26). Second, we included genes that are downstream targets, have been noted to interact with the genes in group 1, or have been reported to be highly expressed in placental tissue. Group 2: DNA-damage-inducible transcript 4 (DDIT4; 4 CpGs), Krüppel-like factor 15 (KLF15; 3 CpGs), CCAAT/enhancer-binding protein β (CEBPB (7 CpGs), and DEP domain-containing, mTOR-interacting protein (DEPTOR; 4 CpGs) (27–29). Most of the CpGs localize to gene promoter regions for HSD11B2, NR3C1, LEP, CEBPB, and KLF15. However, we also assessed gene body CpG methylation for DEPTOR and DDIT4. Table 1 displays the sequence, chromosomal position, and number of CpG sites for each gene locus. Finally, we examined 2 repetitive-element regions that have been used in epidemiologic and exposure assessment studies as surrogate measures of global DNA methylation: Arthrobacter luteus subfamily Yb8 (AluYb8), a short stretch of DNA originally characterized by the action of the A. luteus (Alu) restriction endonuclease, and long interspersed nuclear element 1 (LINE-1).
Table 1.
Gene loci–chromosomal position: sequence and CpGs for pyrosequencing analysis
| Gene locus | Sequence analyzed |
CpG sites |
||
|---|---|---|---|---|
| Sequence, nonconverted | Chromosomal position | CpGs in assay | CpG chromosomal position | |
| KLF15 | TCTGCGTGATTGAGCGGGAAGCG | 3:126076655–126076677 | 3 | 126076659, −669, −676 |
| NR3C1(1) |
CGCGGAGCTGGGCGGGGGCGGGAAGGAGGTAG CGAGAAAAGAAACTGGAGAAA |
5:142783588–142783640 | 5 | 142783592, −599, −602, −608, −611 |
| NR3C1(2) |
CGGTGGCCCTCTTAACGCCGCCCCAGAGAGAC CAGGTCGGCCCCCGCCGCTGCCGCCGCCACC CTTTTTCCTGGGGAGTTGGGGG |
5:142783501–142783585 | 8 | 142783501, −503, −513, −519, −533, −555, −570, −573 |
| LEP(1) |
CGCGCGTGGCTCCTGGCGCGCCGAGGCCCTC CCTCGAGGCCCCGCGAGGTGCACACTGCGGG CCCAGGGCTAGCAGCCGCCCGGCACGTCGCT ACCCTGAGGGGCGGGGCGGGAGCTG GCGCTAG |
7:127881127–127881204 | 11 | 127881127, −129, −131, −143, −145, −148, −161, −169, −171, −185, −204 |
| LEP(2) |
AAATGCGCCGGGGCCTGCGGGGCAGTTGCGCA AGTTGTGATCGGGCCGCTATAAGAGGGGCGG GCAGGCATGGAGCCCCGTAGGAATCGCAGC GCCAGCGGTT |
7:127881246–127881350 | 12 | −246, −257, −260, −269, −280, −293, −298, −312, −330, −339, −344, −350 |
| DEPTOR |
GCGGGAGTGGCGGGGCGCAGCAAAGGGAGCTG GAGCG |
8:120886136–120886172 | 4 | 120886137, −146, −151, −171 |
| DDIT4 | CCCCCGCCTGTGCGTTTCGTTTTGAAGCCG | 10:74034388–74034417 | 4 | 74034392, −400, −405, −416 |
| HSD11B2 | CCGGGAGCGGTCGTCCTGTTCCCCCGCCAG | 16:67464387–67464417 | 4 | 67464389, −395, −399, −412 |
| CEBPB | CGGGGCGCGATCCTGCCCGGTCGCGCCG | 20:48806715–48806742 | 7 | 48806715, −720, −722, −732, −736, 738, −741 |
Bisulfite pyrosequencing was performed to detect DNA methylation, as described elsewhere for the genes HSD11B2 (23, 30), NR3C1 (31), and LEP (26) and the repetitive sequences LINE-1 (32) and AluYb8 (33). PCR products were amplified from bisulfite-modified DNA with the Pyromark PCR Kit (Qiagen). PCR and sequencing primers (Integrated DNA Technologies, Coralville, IA, USA) for HSD11B2, LEP, NR3C1, AluYb8, and LINE-1 are presented in Supplemental Table S1. Predesigned Pyromark CpG assays (Qiagen) were used to analyze DNA methylation within the following genes: CEBPB (PM00197596), DDIT4 (PM00044275), DEPTOR (PM00036400), and KLF15 (PM00015855). Cycling conditions for predesigned assays were 94°C for 15 min, followed by 45 cycles at 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min, with a final extension of 7 min at 72°C. PCR products were sequenced with the PyroMark MD instrument (Qiagen), and the percentage of methylation at each CpG site was quantified with Pyro Q-CpG 1.0.11 software (Qiagen). All samples were run in triplicate, and average values were used in subsequent analyses. Non-CpG cytosines in each read served as internal controls to verify bisulfite DNA modification efficiency (≥95% in all samples from 3 tissues). All procedures were performed according to the manufacturer's protocols.
Statistical analyses
Spearman rank coefficients were calculated to evaluate correlations between CpG units within the loci. The mean methylation across the CpGs analyzed within the loci was used to obtain an overall measurement for each of the examined regions, which was used in subsequent analyses. Summary statistics, including variances, were obtained for each tissue across all loci studied. Mean methylation between tissues within a given locus was compared by using linear mixed-effects models that accounted for the presence of paired samples. Spearman rank coefficients were calculated to evaluate correlations between tissues for each locus. Statistical significance was set at P < 0.05; all tests were 2-sided. All analyses were conducted in RStudio 0.97.314 (RStudio, Boston, MA, USA) using R 3.0.1 (34).
RESULTS
Study population characteristics
Table 2 displays the sociodemographic and clinical characteristics of the study population. The sample of infants enrolled in this study was evenly distributed between males and females and, in accordance with the RICHS cohort design, was oversampled for SGA (11%) and LGA (33%) infants. All infants were considered term (mean gestational age, 39 wk). The majority of the infants (63%) were born by Cesarean section of Caucasian mothers whose age ranged between 18 and 39 yr. Only 1 mother reported smoking during pregnancy. The average age of infant saliva sample collection was 18 wk (range, 13–24 wk).
Table 2.
Population characteristics
| Characteristic | Obs | % | Mean ± sd | Min | Max |
|---|---|---|---|---|---|
| Infant | |||||
| Birth weight (g) | 27 | 3692.8 ± 573.4 | 2225 | 4640 | |
| Birth weight group | |||||
| AGA | 15 | 55.6 | |||
| LGA | 9 | 33.3 | |||
| SGA | 3 | 11.1 | |||
| Sex | |||||
| Female | 14 | 51.9 | |||
| Male | 13 | 48.1 | |||
| Gestational age (wk) | 27 | 39 ± 0.82 | 37 | 41 | |
| Delivery method [n (%)] | |||||
| C-section | 17 | 63 | |||
| Vaginal | 10 | 37 | |||
| Age at saliva sample collection (wk) | 26 | 18.16 ± 2.81 | 13 | 24 | |
| Maternal | |||||
| Age (yr) | 27 | 31 ± 6.5 | 18 | 39 | |
| Ethnicity | |||||
| Other | 4 | 14.8 | |||
| White | 23 | 85.2 | |||
| Tobacco use during pregnancy | |||||
| No | 26 | 96.3 | |||
| Yes | 1 | 3.7 |
obs, observed; min, minimum; max, maximum.
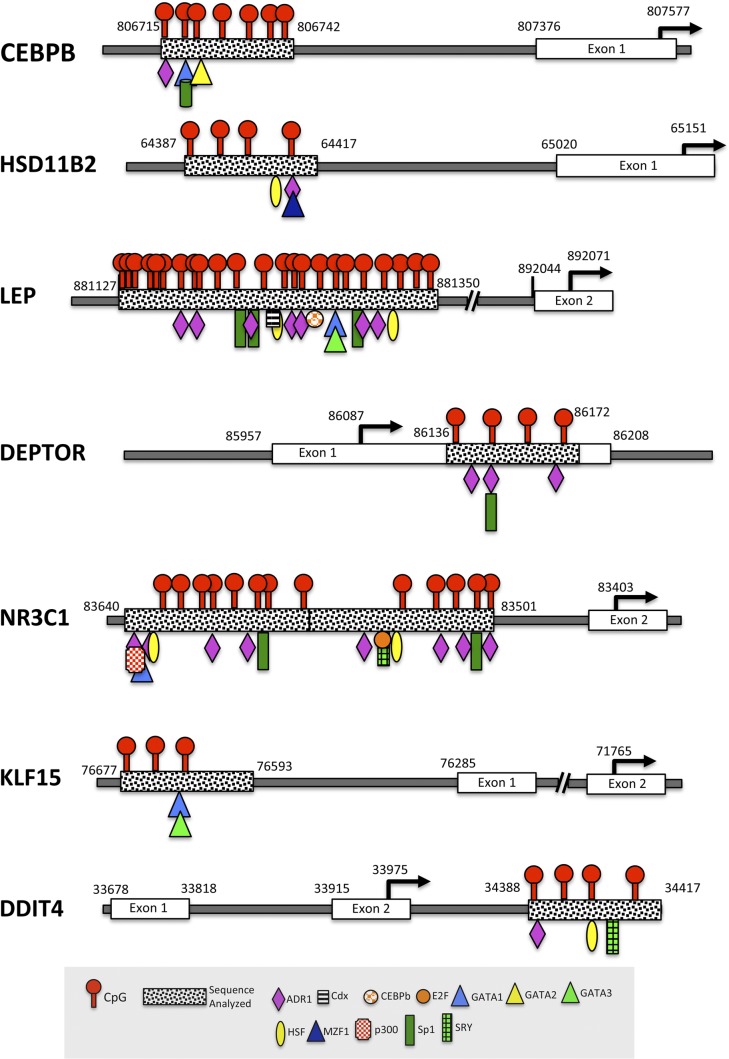
Putative transcription factor (TF) binding site maps
We used the TF binding site online database TFSEARCH 1.3 (http://www.cbrc.jp/research/db/TFSEARCH.html) (35) to create an overlapping map of potential TF binding sites associated with the DNA sequences and CpG sites analyzed. Figure 1 shows diagrams of each gene with sequence position relative to the nearest exons, ATG start site, CpG sites, and potential TFs associated with each sequence. Some common TFs that overlapped CpG sites within the gene groups include alcohol dehydrogenase regulator 1 (ADR1), GATA binding protein 1 (GATA1), heat shock factor (HSF), and specificity protein 1 (Sp1).
Figure 1.
CpG assay/TF binding site maps. Diagram of chromosomal position for each gene, including the pyrosequencing sequence, the intra-assay CpG locations with overlapping putative TF binding sites, the nearest exon, and the gene ATG start site. CEBPB, HSD11B2, LEP, NR3C1, and KLF15 CpG assays are located in the putative gene promoter region, upstream of the ATG start site for each gene. DEPTOR and DDIT4 are examples of CpG assays located downstream of the ATG start site in the gene body.
DNA methylation correlations between CpGs
We examined DNA methylation across 68 CpG units distributed between different genes and repetitive-elements loci. With the exception of NR3C1 and LEP, the loci examined contained <10 CpG units and spanned short sequences. We calculated Spearman rank coefficients between CpGs of each locus across each of the examined tissues. The average coefficients and ranges are presented in Supplemental Table S2. These correlations were locus and tissue dependent, with the placenta exhibiting higher coefficients than the cord blood or saliva. Of note, the saliva samples exhibited the least amount of correlation between the examined CpG units. Given this tissue heterogeneity and according to the generally accepted method in most of the literature, we calculated mean methylation across CpG units of each of the examined regions.
Intertissue and interindividual variation in DNA methylation
The distribution of mean DNA methylation in each region is shown in Table 3. Overall, mean DNA methylation varied widely, ranging from 0 to 89%. As expected, global DNA methylation measured at the LINE-1 and Aluy8b repetitive elements was higher (>40%) than the methylation observed at the gene-specific loci surveyed in this study. Although higher, mean methylation levels differed significantly between tissues for both the repetitive sequences LINE-1 and Aluy8b (P<0.001, all comparisons; Table 3). Mean DNA methylation at the CEBPB, HSD11B2, and LEP promoters was low to moderate, with values ranging from 1.7 to 36.6% and means that differed significantly between tissues (P<0.001, all comparisons; Table 3;). In contrast, DEPTOR, DDIT4, KLF15, and NR3C1 mean methylation was <5% in the tissues examined, but similar to our previous results, these 4 low-methylation regions exhibited different mean methylation values between tissues (P<0.001, all comparisons; Table 3).
Table 3.
Global and gene-specific summary statistics of percentage of mean methylation in placenta, cord blood, and saliva
| Gene | Placenta |
Cord blood |
Saliva |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | M | Var | Min | Max | n | M | Var | Min | Max | n | M | Var | Min | Max | |
| AluYb8 | 27 | 69.6 | 5.06 | 65.3 | 73.6 | 15 | 86.4 | 1.35 | 83.5 | 87.9 | 26 | 87.9 | 0.74 | 85.4 | 89.3 |
| LINE-1 | 27 | 45.2 | 4.80 | 41.8 | 49.1 | 16 | 76.6 | 0.64 | 74.8 | 77.8 | 26 | 58.2 | 3.80 | 52.9 | 61.2 |
| CEBPB | 27 | 4.1 | 0.25 | 3.3 | 5.4 | 16 | 13.7 | 2.76 | 10.9 | 16.4 | 26 | 1.2 | 0.62 | 0.6 | 4.7 |
| HSD11B2 | 27 | 15.7 | 4.93 | 12.0 | 20.2 | 16 | 4.9 | 0.08 | 4.3 | 5.4 | 26 | 1.5 | 1.14 | 0.9 | 5.2 |
| LEP | 27 | 25.5 | 32.04 | 11.8 | 36.6 | 16 | 15.7 | 3.17 | 12.8 | 19.2 | 25 | 5.3 | 4.62 | 2.1 | 10.4 |
| DEPTOR | 27 | 0.8 | 0.07 | 0.6 | 2.1 | 16 | 0.6 | 0.02 | 0.4 | 0.9 | 16 | 0.6 | 0.02 | 0.4 | 0.9 |
| DDIT4 | 27 | 4.2 | 0.29 | 2.6 | 5.5 | 16 | 2.6 | 0.04 | 2.2 | 2.9 | 25 | 2.0 | 0.12 | 1.7 | 3.1 |
| KLF15 | 27 | 0.7 | 0.01 | 0.4 | 0.8 | 16 | 0.9 | 0.01 | 0.8 | 1.2 | 26 | 0.7 | 0.04 | 0.4 | 1.2 |
| NR3C1 | 19 | 1.9 | 0.03 | 1.5 | 2.2 | 11 | 2.4 | 0.12 | 1.7 | 2.9 | 19 | 4.0 | 3.76 | 2.2 | 8.9 |
M, mean; var, variance; min, minimum; max, maximum.
Table 3 also describes the variance of the mean methylation in each region across tissues. Of the tissues analyzed in this study, the placenta showed the greatest interindividual variability when measured with a dispersion statistic, such as the variance. Of the gene loci examined, placental LEP exhibited considerably more interindividual variability than any of the other regions. Across 7 gene loci, only LEP and CEBPB displayed variability within the 3 tissues analyzed. For HSD11B2 and NR3C1, interindividual variability in DNA methylation was high only in placenta and saliva, respectively. Global DNA methylation at the LINE-1 and ALUYB8 repetitive elements displayed ample variance, especially in placental tissue.
To inquire about interindividual variation within each CpG locus in the regions examined, we estimated variances of the CpG sites of each locus. For global DNA methylation, placental samples had the greatest degree of variability in both elements, but CpG4 in Aluy8b in saliva had the highest variance. Interestingly, these elements showed greater variation in saliva, followed by placental tissue and then cord blood (Supplemental Table S3). The largest area analyzed, LEP, displayed higher variance values for most of the 23 CpGs in placenta and saliva, but not in cord blood (Supplemental Table S4). The 13 CpGs of NR3C1 exhibited very low variation in all placental and cord blood samples, but at the same loci, methylation variance at saliva CpGs was much higher. The CEBPB region showed low variability in placental tissue, intermediate in saliva, and higher in cord blood. HSD11B2 methylation variability was low in cord blood and high across the loci in placenta and saliva (Supplemental Table S4). Overall, DNA methylation at the examined loci displayed high interindividual variability that was both locus and tissue dependent, and we did not observe any conserved pattern of variation across tissues.
DNA methylation correlations between tissues
Using Spearman's rank correlations between paired infant tissues, we observed a significant correlation in DNA methylation between cord blood and placental tissue for Aluy8b (ρ=0.57; P=0.03) and a positive correlation for LINE-1 methylation (ρ=0.39; P=0.14; Table 4). Between the placental DNA methylation and saliva methylation data collected in infants between 3 and 6 mo of age, we did not observe correlations of any of the investigated regions. Correspondingly, in comparisons of saliva and cord blood methylation, we did not find significant correlations, with the exception of a significant inverse correlation in the NR3C1 promoter (ρ=−0.58; P=0.02). Hence, in this study, we found scarce intraindividual correlations between tissues in DNA methylation for most of the loci.
Table 4.
Spearman rank correlations of mean DNA methylation between tissues
| Gene | n | Cord blood vs. placenta |
n | Cord blood vs. saliva |
n | Saliva vs. placenta |
|||
|---|---|---|---|---|---|---|---|---|---|
| ρ | P | ρ | P | ρ | P | ||||
| ALUYB8 | 15 | 0.57 | 0.03 | 15 | −0.09 | 0.74 | 26 | 0.22 | 0.27 |
| LINE-1 | 16 | 0.39 | 0.14 | 15 | 0.15 | 0.60 | 26 | 0.13 | 0.51 |
| CEBPB | 16 | 0.14 | 0.59 | 15 | 0.44 | 0.10 | 26 | 0.27 | 0.19 |
| HSD11B2 | 16 | −0.05 | 0.86 | 15 | −0.39 | 0.15 | 26 | 0.06 | 0.78 |
| LEP | 16 | −0.12 | 0.66 | 14 | −0.02 | 0.93 | 25 | 0.26 | 0.22 |
| DEPTOR | 16 | 0.19 | 0.48 | 14 | 0.10 | 0.75 | 25 | −0.05 | 0.80 |
| DDIT4 | 16 | 0.21 | 0.43 | 15 | −0.42 | 0.12 | 25 | −0.26 | 0.21 |
| KLF15 | 16 | 0.08 | 0.77 | 15 | −0.07 | 0.79 | 26 | 0.20 | 0.32 |
| NR3C1 | 16 | 0.07 | 0.78 | 11 | −0.58 | 0.02 | 19 | −0.27 | 0.20 |
DISCUSSION
Several groups have investigated DNA methylation in paired samples from tissues of adult humans (36–39) or related maternal and fetal DNA methylation patterns (40), but only 2 have investigated DNA methylation in different fetal tissues (41, 42). We are the first to assess methylation signatures from healthy, unrelated infants in 3 paired tissues commonly collected in birth cohort studies. We focused on 7 genes that have been analyzed in the DOHaD epidemiologic literature (23, 25, 26, 43) and their potential downstream targets (27) or on genes reported to be highly expressed during fetal development (28, 29).
DNA methylation inhibits gene expression, mainly by 2 mechanisms: preventing the binding of TFs with their cognate DNA recognition sequence (44) or recruiting methyl-CpG-binding proteins (MBPs) that can repress transcription (45, 46). Therefore, it is important to understand the genomic context of the regions being examined for DNA methylation by quantitative methylation assessment. In examining putative TF binding sites within the sequences examined, we commonly identified, most notably, HSFs and Sp1 binding sites in most of the genes. HSFs are transcriptional regulators that are inducible during stress responses, but also during normal development and physiology (47). Similarly, Sp1 regulates transcription of a large number of genes and is involved in a variety of processes such as growth, differentiation, immune responses, and stem cell regulation (48). The Sp1 binding region contains GC-rich motifs and has been identified in the 5′ region of several genes (49), which explains the high frequency of Sp1 binding sites within the investigated loci that are generally CpG-rich promoter regions. Although, the TF binding site analysis performed is based on putative sites, some have been confirmed. For example, Sp1 and CEBP have been confirmed to interact with the LEP promoter (50). Given the technical limitations of sequence-based TF queries, it remains to be determined whether these or other TFs are actively involved in regulation of the genes analyzed in this study, but this could be an intriguing endeavor, especially in light of the tissue-specific expression of TFs (51). The tissue-specific nature of the TFs binding to these sites may also be critical in driving the differences in tissue-specific DNA methylation observed in this study.
Within the loci examined, correlations between DNA methylation of CpG units varied across tissues. Placenta exhibited the highest degree of correlation and saliva the lowest degree. Correspondingly, the loci LINE-1, HSD11B2, CEBPB, NR3C1, and LEP displayed average coefficients >0.2 in 3 of the tissues, whereas other loci, such as DEPTOR, had low correlation between adjacent CpGs. The length of the sequence does not appear to play a role in the degree of correlation. These results highlight the dynamic nature and tissue specificity of methylation marks and suggest that correlation should be assessed and appropriate statistical modeling of the data should be used. This finding, along with the data regarding TF binding, supports, at least in some cases, examining CpG loci individually.
As expected, we observed a higher degree of methylation at the LINE-1 and Aluy8b elements, but, consistent with our findings in gene-specific regions, methylation of these sequences differed between tissues. This intertissue variation may be explained by the polymorphic and ubiquitous location of the elements across the genome (52) or tissue-specific differences in the need to control their expression. Given their abundance, these sequences have been the subject of a considerable amount of study, especially in the cancer biomarker field (53). In addition, placental global DNA methylation has been shown to be associated with in utero exposures (54), highlighting the need for early-life epigenetic profiling of these elements (55–57).
Among the gene-specific loci, we observed a great degree of intertissue and interindividual variation. Most notably, placental LEP and HSD1B2 exhibited abundant variation when compared to the other loci examined. These results are consistent with studies by our group (23, 26) and others (58, 59) that have profiled placental methylation and found a negative correlation with gene expression in this tissue (23, 60). This is the only study that has assessed saliva LEP and HSD11B2 methylation in infants; we found lower methylation of LEP in saliva than in the other profiled tissues. The low level may be explained by previous observations of leptin in microglobules in whole saliva and salivary glands (61). Similarly, the lower HSD11B2 methylation in saliva could be related to the expression of this glucocorticoid antagonist in sweat and salivary glands (62). Notably, although there is likely to be greater cellular heterogeneity in blood than in the cells found in saliva, LEP had higher interindividual variation in saliva than in cord blood.
This is the first report of DNA methylation patterns in these newborn/infant tissues within CEBPB, a TF implicated in numerous cellular processes, including myeloid cell differentiation (63). We observed higher promoter methylation in cord blood, a somewhat unexpected finding, as these samples are likely to contain hematopoietic progenitors including myeloid cells (64), although these are probably a minor cell type in the samples. We did not quantify gene expression; hence, we cannot state the functional relevance of this finding in cord blood. More research is needed to explore CEBPB epigenetic variation and potential relation to gene expression, but our preliminary results showed an interesting pattern of intertissue and interindividual variation at this locus.
The promoter of KLF15, a downstream target of NR3C1 (27), and gene body of DEPTOR were also examined. Our results indicate very low methylation levels and low variability in all tissues, demonstrating that at least at the examined CpGs, methylation profiling may not be useful in larger studies. Methylation at the gene body of DDIT4 was higher, and interindividual variation was found at this locus in placental tissue in comparison to that in cord blood or saliva. Although, DDIT4 interacts with NR3C1 in skeletal muscle (27), it is unknown whether this gene is epigenetically regulated or is expressed in the tissues examined.
The glucocorticoid receptor NR3C1 is a known mediator of glucocorticoid signaling. Methylation in the examined area (exon 1F) correlates inversely with expression in placental tissue (25). Although we observed generally low levels of methylation at NR3C1, this promoter further exemplifies the tissue specificity of this epigenetic mark, and even these low levels of methylation have been linked to variability in expression. Saliva methylation was the highest and most variable, and, in this region, it was negatively associated with total salivary cortisol in female adults who participated in a social stress test (65). Such findings demonstrate the potential utility of these biomarkers, even though the intertissue correlation may be low, as these markers can still be representative of key phenotypes.
We observed a positive correlation between cord blood and placental AluYb8 methylation, and similar results were observed for LINE-1, although with limited statistical power. However, saliva global DNA methylation did not correlate with placental or cord blood. We could hypothesize that this discrepancy is due to the extemporaneousness of saliva sample collection, but we did not have saliva collected at birth to test this hypothesis. In addition, we observed a negative correlation between cord blood and saliva NR3C1 methylation, but not for any of the other gene loci or repetitive elements. If the observed correlation is confirmed in future studies, it could offer the possibility of prospective saliva sampling and NR3C1 DNA methylation profiling in infants and eliminate the need of venipuncture in a susceptible population. Similar correlations between blood and buccal cell methylation have been observed in a study in adults (39), but the NRC31 locus was not investigated, and a correlation for LEP methylation was found that we did not observe in our study. These contrasting results highlight the need for future studies that assess DNA methylation in different tissues at different time points in life, especially with the recognized dynamic nature of the epigenome.
This study has several advantages. We profiled methylation in 3 paired tissues from infants, and we investigated several gene candidates and global DNA methylation by using a highly quantitative technique. However, our sample size was limited, which may have limited our power to detect associations, and we did not take into account the cellular heterogeneity of these tissues that, at least in cord blood, has been shown to influence DNA methylation (18). In addition, we did not have saliva DNA samples collected at birth; thus, we could not assess how temporal changes may affect the correlations between saliva and the other tissues studied. Overall, our findings, in line with the growing literature on epigenetic mechanisms, suggest highly tissue-specific DNA methylation that would not support using these tissues as surrogates for one another at the loci examined. Each tissue's unique epigenetic signature is likely to reflect its differential functions. At the same time, numerous studies have begun to link both environmental factors and phenotypes with DNA methylation in these various tissues. Such studies strongly support the utility of accessible epigenetic features as biomarkers of both exposure and effect in epidemiologic and clinical/translational studies. It is critical to carefully assess how various tissue epigenetic marks relate to these factors and outcomes and to interpret findings appropriately in light of this specificity (7, 25, 66–72).
Supplementary Material
Acknowledgments
The authors thank Joyce Lee (Department of Pediatrics, Women's and Infants Hospital) for her efforts in sample collection and Susan Simon (Media Learning Technologist, Jones Media Center, Dartmouth College Library) for assistance with figure preparation.
The study was funded by grants from the National Institute of Environmental Health Sciences (NIEHS; P01 ES022832/EPA RD83544201 and R01ES022223) and the National Institute of Mental Health (NIMH; R01MH094609), U.S. National Institutes of Health (Bethesda, MD, USA).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- AGA
- appropriate for gestational age
- AluYb8
- Arthrobacter luteus subfamily Yb8
- CEBPB
- CAAT-enhancing binding protein β
- DDIT4
- DNA-damage-inducible transcript 4 protein
- DEPTOR
- DEP domain-containing, mTOR-interacting protein
- DOHaD
- developmental origin of health and disease
- HSD11B2
- hydroxysteroid dehydrogenase 11 β2
- HSF
- heat shock factor
- KLF15
- Kruppel-like factor 15
- LEP
- leptin
- LGA
- large for gestational age
- LINE-1
- long interspersed nuclear element 1
- NR3C1
- nuclear receptor subfamily 3, group C, member 1
- RICHS
- Rhode Island Child Health Study
- SGA
- small for gestational age
- Sp1
- specificity protein 1
- TF
- transcription factor
REFERENCES
- 1. Bird A. (2007) Perceptions of epigenetics. Nature 447, 396–398 [DOI] [PubMed] [Google Scholar]
- 2. Jones P. A., Liang G. (2009) Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 10, 805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jones P. A. (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 [DOI] [PubMed] [Google Scholar]
- 4. Michels K. B., Waterland R. A. (2012) The role of epigenetics in the developmental origins of health and disease. In Epigenetic Epidemiology (Michels K. B, ed), pp, 105–116, Springer Verlag, New York [Google Scholar]
- 5. Gluckman P. D., Hanson M. A. (2004) Living with the past: evolution, development, and patterns of disease. Science 305, 1733–1736 [DOI] [PubMed] [Google Scholar]
- 6. Gluckman P. D., Hanson M. A., Cooper C., Thornburg K. L. (2008) Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med. 359, 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Appleton A. A., Armstrong D. A., Lesseur C., Lee J., Padbury J. F., Lester B. M., Marsit C. J. (2013) Patterning in placental 11-B hydroxysteroid dehydrogenase methylation according to prenatal socioeconomic adversity. PLoS One 8, e74691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parle-McDermott A., Ozaki M. (2011) The impact of nutrition on differential methylated regions of the genome. Adv. Nutr. 2, 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Conradt E., Lester B. M., Appleton A. A., Armstrong D. A., Marsit C. J. (2013) The role of DNA methylation of NR3C1 and 11β-HSD2 and exposure to maternal mood disorder in utero on newborn neurobehavior. Epigenetics 8, 1321–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baccarelli A., Wright R. O., Bollati V., Tarantini L., Litonjua A. A., Suh H. H., Zanobetti A., Sparrow D., Vokonas P. S., Schwartz J. (2009) Rapid DNA methylation changes after exposure to traffic particles. Am. J. Respir. Crit. Care Med. 179, 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Breton C. V., Byun H.-M., Wenten M., Pan F., Yang A., Gilliland F. D. (2009) Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am. J. Respir. Crit. Care Med. 180, 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rönn T., Volkov P., Davegårdh C., Dayeh T., Hall E., Olsson A. H., Nilsson E., Tornberg Å., Nitert M. D., Eriksson K.-F. (2013) A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 9, e1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Christensen B. C., Marsit C. J., Kelsey K. T. (2012) Influence of environmental factors on the epigenome. In Epigenetic Epidemiology (Michels K. B., ed) pp. 197–224, Springer, New York [Google Scholar]
- 14. Mill J., Heijmans B. T. (2013) From promises to practical strategies in epigenetic epidemiology. Nat. Rev. Genet. 14, 585–594 [DOI] [PubMed] [Google Scholar]
- 15. Mehta D., Klengel T., Conneely K. N., Smith A. K., Altmann A., Pace T. W., Rex-Haffner M., Loeschner A., Gonik M., Mercer K. B. (2013) Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc. Natl. Acad. Sci. U. S. A. 110, 8302–8307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ng J. W., Barrett L. M., Wong A., Kuh D., Smith G. D., Relton C. L. (2012) The role of longitudinal cohort studies in epigenetic epidemiology: challenges and opportunities. Genome Biol. 13, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Filiberto A. C., Maccani M. A., Koestler D., Wilhelm-Benartzi C., Avissar-Whiting M., Banister C. E., Gagne L. A., Marsit C. J. (2011) Birthweight is associated with DNA promoter methylation of the glucocorticoid receptor in human placenta. Epigenetics 6, 566–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koestler D. C., Avissar-Whiting M., Houseman E. A., Karagas M. R., Marsit C. J. (2013) Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environ. Health Perspect. 121, 971–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Groom A., Potter C., Swan D. C., Fatemifar G., Evans D. M., Ring S. M., Turcot V., Pearce M. S., Embleton N. D., Smith G. D. (2012) Postnatal growth and DNA methylation are associated with differential gene expression of the TACSTD2 gene and childhood fat mass. Diabetes 61, 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mueller B. (1928) Über den Nachweis eingetrockneten Speichels in Tüchern. Int J. Legal Med. 11, 211–224 [Google Scholar]
- 21. Hales C. N., Barker D. J., Clark P. M., Cox L. J., Fall C., Osmond C., Winter P. D. (1991) Fetal and infant growth and impaired glucose tolerance at age 64. BMJ 303, 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bocklandt S., Lin W., Sehl M. E., Sánchez F. J., Sinsheimer J. S., Horvath S., Vilain E. (2011) Epigenetic predictor of age. PLoS One 6, e14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marsit C. J., Maccani M. A., Padbury J. F., Lester B. M. (2012) Placental 11-beta hydroxysteroid dehydrogenase methylation is associated with newborn growth and a measure of neurobehavioral outcome. PLoS One 7, e33794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fenton T. (2003) A new growth chart for preterm babies: Babson and Benda's chart updated with recent data and a new format. BMC Pediatr. 3, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bromer C., Marsit C. J., Armstrong D. A., Padbury J. F., Lester B. (2012) Genetic and epigenetic variation of the glucocorticoid receptor (NR3C1) in placenta and infant neurobehavior. Dev. Psychobiol. 55, 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lesseur C., Armstrong D. A., Paquette A. G., Koestler D. C., Padbury J. F., Marsit C. J. (2013) Tissue-specific Leptin promoter DNA methylation is associated with maternal and infant perinatal factors. Mol. Cell. Endocrinol. 381, 160–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shimizu N., Yoshikawa N., Ito N., Maruyama T., Suzuki Y., Takeda S., Nakae J., Tagata Y., Nishitani S., Takehana K., Sano M., Fukuda K., Suematsu M., Morimoto C., Tanaka H. (2011) Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab. 13, 170–182 [DOI] [PubMed] [Google Scholar]
- 28. Boruk M., Savory J. G., Hache R. J. (1998) AF-2-dependent potentiation of CCAAT enhancer binding protein beta-mediated transcriptional activation by glucocorticoid receptor. Mol. Endocrinol. 12, 1749–1763 [DOI] [PubMed] [Google Scholar]
- 29. Mparmpakas D., Zachariades E., Goumenou A., Gidron Y., Karteris E. (2012) Placental DEPTOR as a stress sensor during pregnancy. Clin. Sci. 122, 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alikhani-Koopaei R., Fouladkou F., Frey F. J., Frey B. M. (2004) Epigenetic regulation of 11 beta-hydroxysteroid dehydrogenase type 2 expression. J. Clin. Invest. 114, 1146–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oberlander T. F., Weinberg J., Papsdorf M., Grunau R., Misri S., Devlin A. M. (2008) Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics 3, 97–106 [DOI] [PubMed] [Google Scholar]
- 32. Bollati V., Baccarelli A., Hou L., Bonzini M., Fustinoni S., Cavallo D., Byun H. M., Jiang J., Marinelli B., Pesatori A. C., Bertazzi P. A., Yang A. S. (2007) Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 67, 876–880 [DOI] [PubMed] [Google Scholar]
- 33. Choi S. H., Worswick S., Byun H. M., Shear T., Soussa J. C., Wolff E. M., Douer D., Garcia-Manero G., Liang G., Yang A. S. (2009) Changes in DNA methylation of tandem DNA repeats are different from interspersed repeats in cancer. Int. J. Cancer 125, 723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. R Core Team (2013) A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 35. Heinemeyer T., Wingender E., Reuter I., Hermjakob H., Kel A. E., Kel O., Ignatieva E. V., Ananko E. A., Podkolodnaya O. A., Kolpakov F. (1998) Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 26, 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. De Bustos C., Ramos E., Young J. M., Tran R. K., Menzel U., Langford C. F., Eichler E. E., Hsu L., Henikoff S., Dumanski J. P., Trask B. J. (2009) Tissue-specific variation in DNA methylation levels along human chromosome 1. Epigenetics Chromatin 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Davies M. N., Volta M., Pidsley R., Lunnon K., Dixit A., Lovestone S., Coarfa C., Harris R. A., Milosavljevic A., Troakes C., Al-Sarraj S., Dobson R., Schalkwyk L. C., Mill J. (2012) Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 13, R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Byun H. M., Siegmund K. D., Pan F., Weisenberger D. J., Kanel G., Laird P. W., Yang A. S. (2009) Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum. Mol. Genet. 18, 4808–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Talens R. P., Boomsma D. I., Tobi E. W., Kremer D., Jukema J. W., Willemsen G., Putter H., Slagboom P. E., Heijmans B. T. (2010) Variation, patterns, and temporal stability of DNA methylation: considerations for epigenetic epidemiology. FASEB J. 24, 3135–3144 [DOI] [PubMed] [Google Scholar]
- 40. Papageorgiou E. A., Fiegler H., Rakyan V., Beck S., Hulten M., Lamnissou K., Carter N. P., Patsalis P. C. (2009) Sites of differential DNA methylation between placenta and peripheral blood: molecular markers for noninvasive prenatal diagnosis of aneuploidies. Am. J. Pathol. 174, 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eckmann-Scholz C., Bens S., Kolarova J., Schneppenheim S., Caliebe A., Heidemann S., von Kaisenberg C., Kautza M., Jonat W., Siebert R., Ammerpohl O. (2012) DNA-methylation profiling of fetal tissues reveals marked epigenetic differences between chorionic and amniotic samples. PLoS One 7, e39014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ollikainen M., Smith K. R., Joo E. J., Ng H. K., Andronikos R., Novakovic B., Abdul Aziz N. K., Carlin J. B., Morley R., Saffery R., Craig J. M. (2010) DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum. Mol. Genet. 19, 4176–4188 [DOI] [PubMed] [Google Scholar]
- 43. Tobi E. W., Lumey L. H., Talens R. P., Kremer D., Putter H., Stein A. D., Slagboom P. E., Heijmans B. T. (2009) DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum. Mol. Genet. 18, 4046–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Watt F., Molloy P. L. (1988) Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 2, 1136–1143 [DOI] [PubMed] [Google Scholar]
- 45. Boyes J., Bird A. (1991) DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64, 1123–1134 [DOI] [PubMed] [Google Scholar]
- 46. Klose R. J., Bird A. P. (2006) Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, 89–97 [DOI] [PubMed] [Google Scholar]
- 47. Akerfelt M., Morimoto R. I., Sistonen L. (2010) Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell. Biol. 11, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Solomon S. S., Majumdar G., Martinez-Hernandez A., Raghow R. (2008) A critical role of Sp1 transcription factor in regulating gene expression in response to insulin and other hormones. Life Sci. 83, 305–312 [DOI] [PubMed] [Google Scholar]
- 49. Harrington M. A., Jones P. A., Imagawa M., Karin M. (1988) Cytosine methylation does not affect binding of transcription factor Sp1. Proc. Natl. Acad. Sci. U. S. A. 85, 2066–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mason M. M., He Y., Chen H., Quon M. J., Reitman M. (1998) Regulation of leptin promoter function by Sp1, C/EBP, and a novel factor. Endocrinology 139, 1013. [DOI] [PubMed] [Google Scholar]
- 51. Vaquerizas J. M., Kummerfeld S. K., Teichmann S. A., Luscombe N. M. (2009) A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 10, 252–263 [DOI] [PubMed] [Google Scholar]
- 52. Slotkin R. K., Martienssen R. (2007) Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285 [DOI] [PubMed] [Google Scholar]
- 53. Nelson H. H., Marsit C. J., Kelsey K. T. (2011) Global methylation in exposure biology and translational medical science. Environ. Health Perspect. 119, 1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wilhelm-Benartzi C. S., Houseman E. A., Maccani M. A., Poage G. M., Koestler D. C., Langevin S. M., Gagne L. A., Banister C. E., Padbury J. F., Marsit C. J. (2012) In utero exposures, infant growth, and DNA methylation of repetitive elements and developmentally related genes in human placenta. Environ. Health Perspect. 120, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pilsner J. R., Hall M. N., Liu X., Ilievski V., Slavkovich V., Levy D., Factor-Litvak P., Yunus M., Rahman M., Graziano J. H. (2012) Influence of prenatal arsenic exposure and newborn sex on global methylation of cord blood DNA. PLoS One 7, e37147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Boeke C. E., Baccarelli A., Kleinman K. P., Burris H. H., Litonjua A. A., Rifas-Shiman S. L., Tarantini L., Gillman M. (2012) Gestational intake of methyl donors and global LINE-1 DNA methylation in maternal and cord blood: prospective results from a folate-replete population. Epigenetics 7, 253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Haggarty P., Hoad G., Horgan G. W., Campbell D. M. (2013) DNA methyltransferase candidate polymorphisms, imprinting methylation, and birth outcome. PLoS One 8, e68896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hogg K., Blair J. D., von Dadelszen P., Robinson W. P. (2013) Hypomethylation of the LEP gene in placenta and elevated maternal leptin concentration in early onset pre-eclampsia. Mol. Cell. Endocrinol. 367, 64–73 [DOI] [PubMed] [Google Scholar]
- 59. Hogg K., Blair J. D., McFadden D. E., von Dadelszen P., Robinson W. P. (2013) Early onset pre-eclampsia is associated with altered DNA methylation of cortisol-signalling and steroidogenic genes in the placenta. PLoS One 8, e62969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bouchard L., Thibault S., Guay S. P., Santure M., Monpetit A., St-Pierre J., Perron P., Brisson D. (2010) Leptin gene epigenetic adaptation to impaired glucose metabolism during pregnancy. Diabetes Care 33, 2436–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. De Matteis R., Puxeddu R., Riva A., Cinti S. (2002) Intralobular ducts of human major salivary glands contain leptin and its receptor. J. Anat. 201, 363–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chapman K., Holmes M., Seckl J. (2013) 11β-Hydroxysteroid dehydrogenases: intracellular gate-keepers of tissue glucocorticoid action. Physiol. Rev. 93, 1139–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ramji D., Foka P. (2002) CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365, 561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Salahuddin S. Z., Markham P. D., Ruscetti F. W., Gallo R. C. (1981) Long-term suspension cultures of human cord blood myeloid cells. Blood 58, 931–938 [PubMed] [Google Scholar]
- 65. Edelman S., Shalev I., Uzefovsky F., Israel S., Knafo A., Kremer I., Mankuta D., Kaitz M., Ebstein R. P. (2012) Epigenetic and genetic factors predict women's salivary cortisol following a threat to the social self. PLoS One 7, e48597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Marsit C., Christensen B. (2013) Blood-derived DNA methylation markers of cancer risk. Adv. Exp. Med. Biol. 754, 233–252 [DOI] [PubMed] [Google Scholar]
- 67. Burris H. H., Baccarelli A. A. (2014) Environmental epigenetics: from novelty to scientific discipline. J. Appl. Toxicol. 34, 113–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kim J. H., Rozek L. S., Soliman A. S., Sartor M. A., Hablas A., Seifeldin I. A., Colacino J. A., Weinhouse C., Nahar M. S., Dolinoy D. C. (2013) Bisphenol A-associated epigenomic changes in prepubescent girls: a cross-sectional study in Gharbiah, Egypt. Environ. Health 12, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tyrka A. R., Price L. H., Marsit C., Walters O. C., Carpenter L. L. (2012) Childhood adversity and epigenetic modulation of the leukocyte glucocorticoid receptor: preliminary findings in healthy adults. PLoS One 7, e30148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Herbstman J. B., Wang S., Perera F. P., Lederman S. A., Vishnevetsky J., Rundle A. G., Hoepner L. A., Qu L., Tang D. (2013) Predictors and consequences of global DNA methylation in cord blood and at three years. PLoS One 8, e72824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Perera F., Herbstman J. (2011) Prenatal environmental exposures, epigenetics, and disease. Reprod. Toxicol. 31, 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ikegame T., Bundo M., Sunaga F., Asai T., Nishimura F., Yoshikawa A., Kawamura Y., Hibino H., Tochigi M., Kakiuchi C., Sasaki T., Kato T., Kasai K., Iwamoto K. (2013) DNA methylation analysis of BDNF gene promoters in peripheral blood cells of schizophrenia patients. Neurosci. Res. 77, 208–214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.