Abstract
Background
A mutant screening was carried out previously to look for new genes related to the Cucumber mosaic virus infection response in Arabidopsis. A Pumilio RNA binding protein-coding gene, Arabidopsis Pumilio RNA binding protein 5 (APUM5), was obtained from this screening.
Results
APUM5 transcriptional profiling was carried out using a bioinformatics tool. We found that APUM5 was associated with both biotic and abiotic stress responses. However, bacterial and fungal pathogen infection susceptibility was not changed in APUM5 transgenic plants compared to that in wild type plants although APUM5 expression was induced upon pathogen infection. In contrast, APUM5 was involved in the abiotic stress response. 35S-APUM5 transgenic plants showed hypersensitive phenotypes under salt and drought stresses during germination, primary root elongation at the seedling stage, and at the vegetative stage in soil. We also showed that some abiotic stress-responsive genes were negatively regulated in 35S-APUM5 transgenic plants. The APUM5-Pumilio homology domain (PHD) protein bound to the 3′ untranslated region (UTR) of the abiotic stress-responsive genes which contained putative Pumilio RNA binding motifs at the 3′ UTR.
Conclusions
These results suggest that APUM5 may be a new post-transcriptional regulator of the abiotic stress response by direct binding of target genes 3′ UTRs.
Background
Post-transcriptional/translational control of gene expression is a powerful strategy for the eukaryotes to adapt to biotic and abiotic stresses. In particular, this process is regulated by various RNA-binding proteins (RBPs) that regulate several processes involving RNA processing, mRNA transport, mRNA stability, and mRNA translation through direct or indirect interactions with target mRNAs [1-5]. Many RBPs have several conserved RNA binding domains (RBDs) to interact with target RNAs; these are the RNA-recognition motif (RRM), the K Homology domain, the Pumilio homology domain (PHD), and the double-stranded RNA binding domain [6-8].
The Pumilio RNA binding family proteins termed Puf proteins have repeats of a conserved PHD, which recognizes a highly conserved 8–10 nucleotide core motif including the ‘UGUA’ tetranucleotide in the 3′ mRNA untranslated region (UTR) to specifically regulate target mRNA stability and translation. Mutated key amino acids of this PHD alter the sequence specificity of Pumilio 1, a human Pumilio protein 1 [8,9]. Most of the identified Puf proteins are conserved throughout evolution in mammals, fungi, protozoa, and plants [10-13]. Many Puf proteins have been identified as essential factors for several aspects of development such as germline stem cell maintenance, synaptic plasticity, embryonic axis patterning, and mating type switching [13-17]. Many approaches such as DNA microarrays, bioinformatics approaches, and RNA immunoprecipitation have been performed in Saccharomyces cerevisiae and Drosophila melanogaster to identify specific target RNAs associated with Puf proteins [15,18,19]. Many RNAs with binding motifs interact with Puf proteins at different developmental stages and in different tissues. Puf proteins may be involved in much larger and richer post-transcriptional regulation in mammals. However, it is largely unknown how plant Pufs control post-transcriptional/translational processing by binding to their target 3′ UTR transcripts, and their functions have been poorly analyzed.
Recent work has identified putative mRNA targets of Arabidopsis Pumilio (APUM) proteins, and a comparative analysis of plant Puf proteins was performed [11,20]. These results demonstrate that plant Puf proteins may also act as post-transcriptional/translational repressors through an evolutionarily conserved mechanism, and their recombinant PHD protein binds to the nanos response element (NRE) sequence within the 3′ UTR of hunchback (hb) mRNA of the Drosophila Pumilio target. Additionally, their putative target RNA candidates are associated with plant growth and development similar to mammalian Puf proteins [11,20]. Some Puf proteins affect rRNA processing apparently without target sequence specificity. For example, analysis of APUM23 knock-out plant phenotypes revealed that APUM23 function is involved in rRNA processing in the Arabidopsis nucleolar region [21].
We previously isolated an Arabidopsis Pumilio RNA binding protein 5 (APUM5) mutant as a susceptibility-reduced mutant to Cucumber mosaic virus (CMV) infection and this mutant exhibits up-regulated APUM5 gene expression [22]. APUM5 has a conserved eight tandem repeats of PHD but their function in Arabidopsis is still largely unknown. Thus, we analyzed APUM5 gene expression patterns using a bioinformatics tool and found it could be up-regulated by biotic and abiotic stresses. In this study, we conducted loss-of-function and gain-of-function studies using APUM5 transgenic plants to further elucidate the specific roles and regulation of APUM5 under biotic and abiotic stress conditions. The results showed that APUM5 functions as a negative regulator under salt, osmotic, and drought stress conditions although APUM5 transgenic plants did not show any phenotypic change upon bacterial and fungal pathogen infections.
Results
APUM5 is a pathogen-responsive gene but does not affect susceptibility or resistance to Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) and Alternaria brassicicola infections
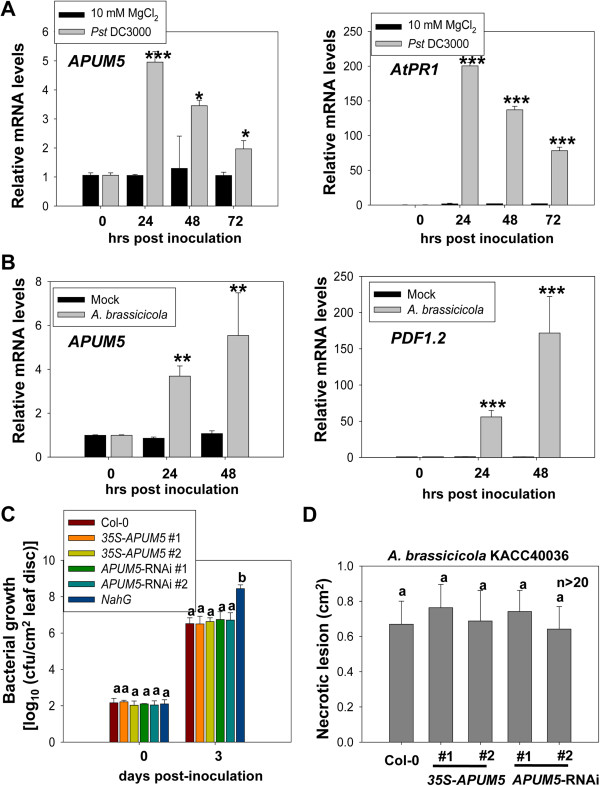
APUM5 is involved in susceptibility to CMV [22,23]. An APUM5 gene transcript analysis was performed via the Arabidopsis eFP Browser to further elucidate the functional role of APUM5 [24]. Interestingly, APUM5 was induced by biotic and abiotic stresses in the bioinformatics analysis. Furthermore, APUM5 gene expression was enhanced in the mpk4-1 mutant background compared with accession Landsberg erecta (Ler) control plants (Additional file 1). Arabidopsis MAP kinase 4 (AtMPK4) is a negative regulator involved in salicylic acid-dependent disease resistance. mpk4-1 exhibits enhanced resistance to biotrophs and increased susceptibility to necrotrophs [25]. Based on these results, we postulated that APUM5 might be associated with the biotic/abiotic defense response pathway. First, we confirmed whether APUM5 expression is responsive to infection by bacterial pathogens such as Pst DC3000. As a result, APUM5 mRNA expression increased upon Pst DC3000 infection at 24 h post infection (hpi) and started to diminish at 48 and 72 hpi (Figure 1A). AtPR1 was used as a positive control and was successfully induced upon Pst DC3000 infection (Figure 1A). Thus, we checked the possibility that APUM5 might be involved in the pathogen defense response and could affect bacterial growth. However, APUM5 transgenic plants (both 35S-APUM5 and APUM5-RNAi plants) did not show changes in susceptibility or resistance in the Pst DC3000 growth assay (Figure 1C). These data indicate that APUM5 is not required for bacterial pathogen resistance although APUM5 was induced upon bacterial infection.
Figure 1.
APUM5 was induced in response to Pst DC3000 and A. brassicicola infection but did not affect susceptibility. (A)APUM5 gene expression pattern upon Pst DC3000 inoculation. AtPR1 was used as a positive control for Pst DC3000 inoculation. Error bars indicate standard deviations (n = 3). (Student’s t-test; *P < 0.01, ***P < 0.0001). (B)APUM5 was induced by A. brassicicola infection. PDF1.2 was used as a positive control for fungal pathogen infection. Error bars indicate standard deviations (n = 3). (Student’s t-test; **P < 0.001, ***P < 0.0001). (C) Bacterial growth analysis in APUM5 transgenic plants. NahG plant was used as a positive control for Pst DC3000 inoculation. Error bars indicate standard deviations (n = 3). (D) Quantitative analysis of necrotic lesion size upon A. brassicicola infection at 5 dpi.
Next, we checked whether APUM5 affected fungal pathogen infection. When Col-0 plants were inoculated with A. brassicicola, APUM5 mRNA expression levels increased at the 24 and 48 h time points (Figure 1B). PDF1.2, which is a positive control for fungal pathogen infection, was induced by the A. brassicicola infection (Figure 1B). We evaluated the necrotic lesion size in Col-0 and APUM5 transgenic plants upon A. brassicicola infection. The APUM5 transgenic plants did not exhibit any significant increase or decrease in necrotic lesion sizes compared with those in Col-0 plants (Figure 1D). Thus, APUM5 overexpression or knockdown did not change the fungal pathogen growth phenotype, although APUM5 is a fungal pathogen-responsive gene. These results suggest that APUM5 is not associated with defense against Pst DC3000 and A. brassicicola infections even though APUM5 expression response to bacterial and fungal pathogens.
Expression of APUM5 increases strongly following mannitol, salt, and ABA treatments
We found that APUM5 is a pathogen-responsive gene following bacterial and fungal pathogen infections. However, the function of APUM5 was not associated with defense against these pathogens, although APUM5 inhibits CMV infection by binding to the CMV 3′ UTR [22,23]. Furthermore, APUM5 transgenic plants did not show enhanced or repressed PR gene expression [22]. These results might explain that APUM5 did not directly regulate defense-related genes.
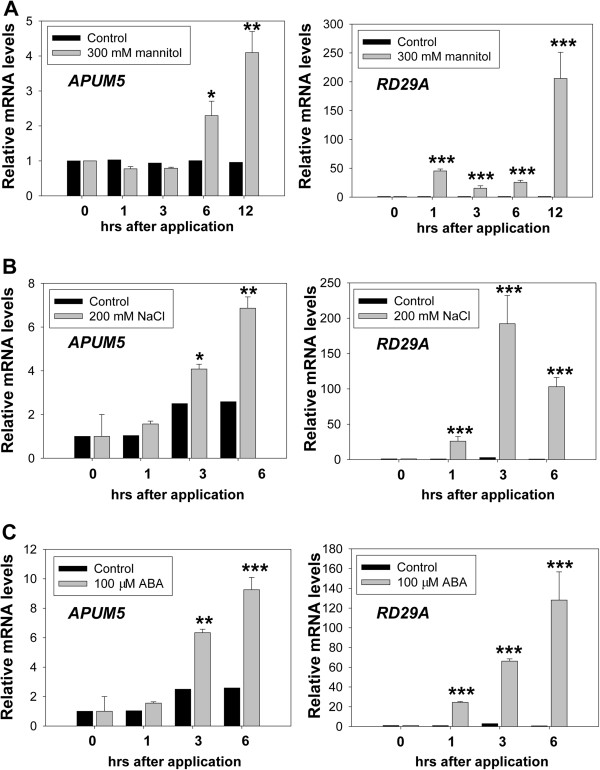
APUM5 is significantly induced by abiotic stressors such as mannitol, salt, ABA treatments [24]. We verified that APUM5 expression increased rapidly in response to mannitol treatment in 10-day-old seedlings (Figure 2A). The RD29A promoter contains both a dehydration-responsive element (DRE) and an ABA-responsive element (ABRE), which are two major cis-acting elements involved in ABA-independent and -dependent gene expression, respectively [26]. RD29A was used as a positive control and was successfully induced by mannitol stress (Figure 2A). To further investigate the effects of other osmotic stressors and ABA treatment on APUM5 expression, the expression levels of APUM5 in 10-day-old seedlings treated with NaCl and ABA were measured by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). APUM5 expression increased gradually and strongly following NaCl treatment (Figure 2B). We also evaluated the effect of ABA. As expected, APUM5 gene expression was enhanced by ABA treatment (Figure 2C). RD29A expression was also increased by the NaCl and ABA treatments (Figure 2). These observations suggest that APUM5 may play a role during the osmotic and ABA stress response.
Figure 2.
Expression analysis of APUM5 by qRT-PCR following osmotic stress, salt, and ABA treatments. (A) Expression patterns of APUM5 and RD29A in response to mannitol treatment. (B) Expression patterns of APUM5 and RD29A in response to salt treatment. (C) Transcription levels of APUM5 and RD29A following ABA treatment. Expression patterns of RD29A were used as a positive control for the mannitol, NaCl, and ABA treatments. APUM5 and RD29A transcription levels in 10-day-old wild-type Arabidopsis seedlings grown on 1/2 MS medium containing 1.5% sucrose and treated without (control) or with 300 mM mannitol, 200 mM NaCl, or 100 μM ABA were analyzed by qRT-PCR. Error bars indicate standard deviations (n = 3). (Student’s t-test; *P < 0.01, **P < 0.001, ***P < 0.0001).
Tissue-specific expression of GUS in APUM5pro-GUS transgenic plants
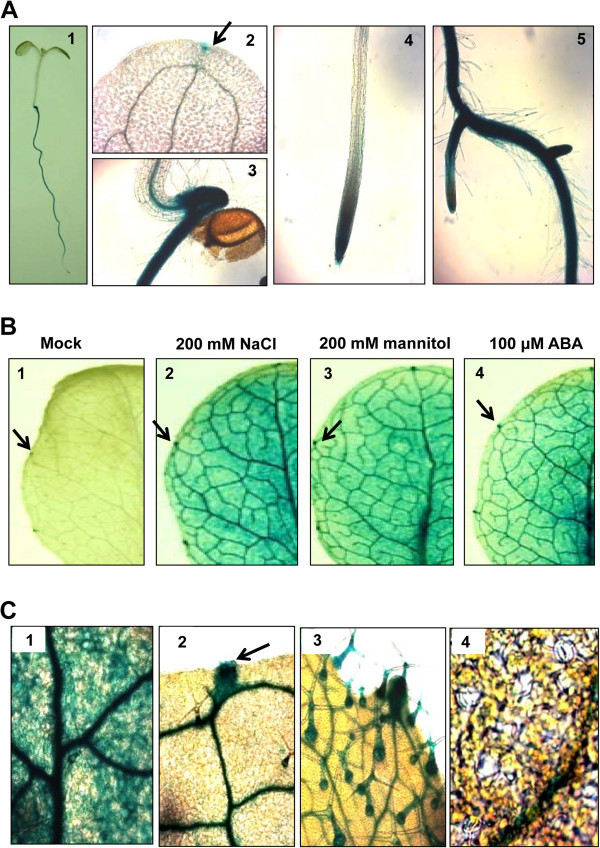
A 1.3-kb fragment of the APUM5 promoter region was fused to the GUS reporter gene, and this construct was introduced into Arabidopsis to analyze the spatial expression of APUM5. APUM5pro-GUS expression was analyzed in the 10-day-old seedling stage of T3 transgenic plants, with strong GUS activity in the root tip, primary root, lateral root, and shoot apical meristem region (Figure 3A, Sections 1, 3, 4, and 5). Although low GUS activity was detected in leaf tissue, much higher activity was observed in the hydathodal cells (Figure 3A, Section 2). High levels of GUS activity were also detected in cauline leaves, flowers, and silique ends (Additional file 2). These observations confirmed results described previously [27].
Figure 3.
Tissue-specific expression of GUS in APUM5pro-GUS transgenic plants. (A)APUM5 promoter activity was determined by GUS histochemical staining. Section 1, 4-day-old seedlings. Section 2, hydathode on the cotyledon indicated by an arrow. Section 3, shoot apical meristem region. Section 4, primary root. Section 5, lateral roots. (B) The effect of salt, osmotic, and ABA treatments on APUM5pro-GUS activity. Three-week-old seedlings were treated with 200 mM NaCl, 200 mM mannitol, or 100 μM ABA for 6 h before GUS staining. Enhanced APUM5 promoter activity on the rosette leaf following abiotic stress. (C) Enlarged image from 3B. Section 1, APUM5 promoter activity expressed in leaf vasculature and mesophyll cell regions. Section 2, hydathode and leaf vascular bundle. Section 3, trichomes on a rosette leaf. Section 4, guard cells.
Previous results showed that APUM5 expression was highly enhanced under the osmotic stress condition (Figure 2), indicating that APUM5 expression could be observed in the leaf tissue in the GUS activity assay following osmotic stress treatment. Thus, the APUM5 promoter cis-elements were examined using Athena, the Arabidopsis promoter analysis tool [28]. The promoter analysis revealed that the APUM5 promoter contains ABA response elements (ABFs binding motif, ABREATRD22) and MYB recognition elements (MYB4 binding motif, MYB1AT), all of which are cis-acting elements often found in ABA- or environmental stress-related genes (Additional file 3) [26]. APUM5pro-GUS transgenic plants were treated with mannitol, NaCl, and exogenous ABA in soil to further investigate APUM5 expression. After applying of NaCl, mannitol, and ABA, most of the leaf tissues showed strong GUS activity and were activated in the entire plant vasculature (Figure 3B, C). This was consistent with qRT-PCR analyses of the APUM5 gene expression pattern during osmotic stress and exogenous ABA applications (Figure 2). Interestingly, GUS activity was not detected in guard cells (Figure 3C, Section 4), whereas strong GUS activity was detected in the hydathodes, trichomes, mesophyll cells, main veins, and vascular tissues (Figure 3C, Sections 1–3). These results indicate that APUM5 may not directly affect stomatal regulation through an ABA-dependent pathway of stomatal closure and opening. In contrast, APUM5 may affect the physical endurance of the plants under osmotic or drought stress via hydathodes and trichomes.
Overexpression of APUM5 leads to hypersensitivity and down-regulation to reduced susceptibility to salt and mannitol in Arabidopsis
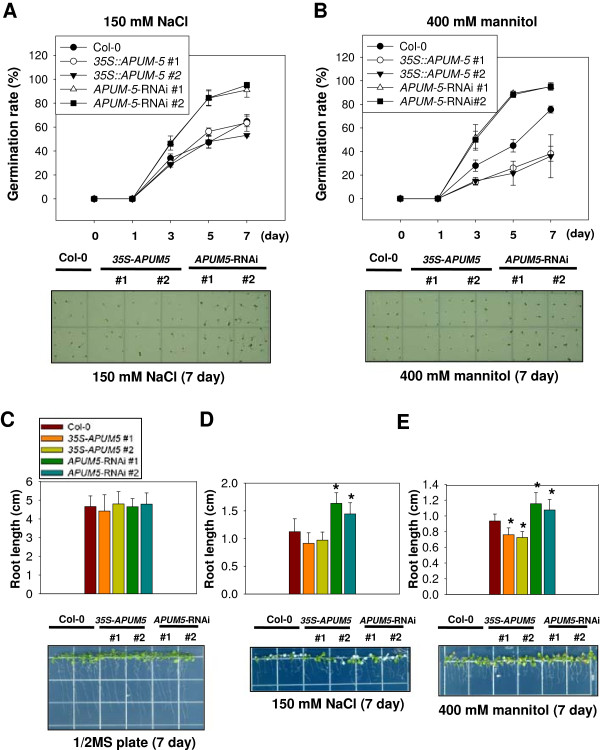
The above results demonstrated that osmotic stress and exogenous ABA treatment up-regulated APUM5 expression (Figure 2) and this result was consistent with APUM5pro-GUS expression under similar stress conditions (Figure 3B and C). Additionally, these results are supported by the APUM5 promoter cis-acting elements analysis (Additional file 3). These data suggest that APUM5 may be involved in osmotic, drought, and ABA sensitivity. Germination and post-germination growth efficiency of APUM5 transgenic and Col-0 wild-type plants were examined when the plants were treated with various concentrations of salt or mannitol to determine whether the physiological role of APUM5 in Arabidopsis is associated with salt or dehydration stress. The germination rate of Col-0, 35S-APUM5, and APUM5-RNAi plants was similar on the control 1/2 MS medium plate (Additional file 4). However, the germination rate of APUM5-RNAi line #1 and #2 plants was about 20% higher than that of Col-0 on the 1/2 MS medium supplemented with 150 mM NaCl, whereas no obvious difference in germination rate was observed between the Col-0 and 35S-APUM5 transgenic plants (Figure 4A). However, primary root elongation of 35S-APUM5 transgenic plants was hypersensitive in the 1/2 MS plate containing 100 mM NaCl (Additional file 5A). On the other hand, the germination rate of APUM5-RNAi line #1 and #2 plants increased about 18% compared with that of the Col-0 plants on 1/2 MS medium supplemented with 400 mM mannitol, whereas the germination rate of 35S-APUM5 transgenic line #1 and #2 plants decreased significantly (about 35%) by 7 days (Figure 4B). Primary root elongation of 35S-APUM5 was also hypersensitive on the 1/2 MS plate containing 300 mM mannitol (Additional file 5B). These results suggest that APUM5 may negatively contribute to salt and dehydration stress tolerance.
Figure 4.
Effect of salt and dehydration stress on growth of wild-type and APUM5 transgenic seedlings. (A) The germination ability of wild-type, 35S-APUM5, and APUM5-RNAi plants was measured on 1/2 MS medium containing 150 mM NaCl, and germination was scored at the indicated days. (B) The germination test of wild-type, 35S-APUM5, and APUM5-RNAi transgenic plants upon 1/2 MS medium containing 400 mM mannitol. These experiments were performed for three independent sets (60–70 seeds per experiment). (C) Normal root growth of Col-0 and APUM5 transgenic plants. (D) The effect of 150 mM NaCl on primary root elongation. (E) The effect of 400 mM mannitol on primary root elongation. Seeds were germinated for 3 days on 1/2 MS medium and the seedlings were transferred (n = 30, triplicates) to 1/2 MS containing 150 mM NaCl or 400 mM mannitol. Primary root length was measured 7 days after transfer. Error bars indicate standard deviations (n = 3). (Student’s t-test; *P < 0.01).
Fully germinated 3-day-old seedlings in normal 1/2 MS medium were transferred to plates supplemented with 150 mM NaCl or 400 mM mannitol to further assess the effect of salt or dehydration on growth of APUM5 transgenic seedlings, and primary root elongation was monitored after 7 days. Primary root elongation of the APUM5 transgenic plants was similar to that of the Col-0 plants under normal conditions (Figure 4C). However, primary root length of APUM5-RNAi plants was longer than that of Col-0 plants under the 150 mM NaCl supplement condition, whereas that of 35S-APUM5 transgenic plants was shorter (Figure 4D). This phenotype is thus concentration-dependent effect in root elongation of 35S-APUM5 plants. In contrast, APUM5-RNAi transgenic plants showed enhanced primary root growth under the salinity stress condition (Figure 4D). The primary root length of APUM5-RNAi transgenic plants was 14–15% longer compared with that of Col-0 plants when the plants were grown on plates supplemented with 400 mM mannitol, whereas 35S-APUM5 plants showed reduced primary root growth of 16–17% compared to that of Col-0 plants (Figure 4E).
We assessed whether germination and primary root growth were affected by an exogenous ABA application. The germination rate of 35S-APUM5 transgenic plants decreased about 39% and 49% compared with that of wild-type and APUM5-RNAi plants following 0.5 μM and 0.7 μM ABA treatments, respectively (Additional file 6A). Primary root length of 35S-APUM5 plants was shorter than that of the wild-type and APUM5-RNAi plants following ABA treatment (Additional file 6B). These results show that APUM5-overexpressing plants are more hypersensitive to dehydration or salt stress, suggesting that APUM5 might regulate the abiotic stress response.
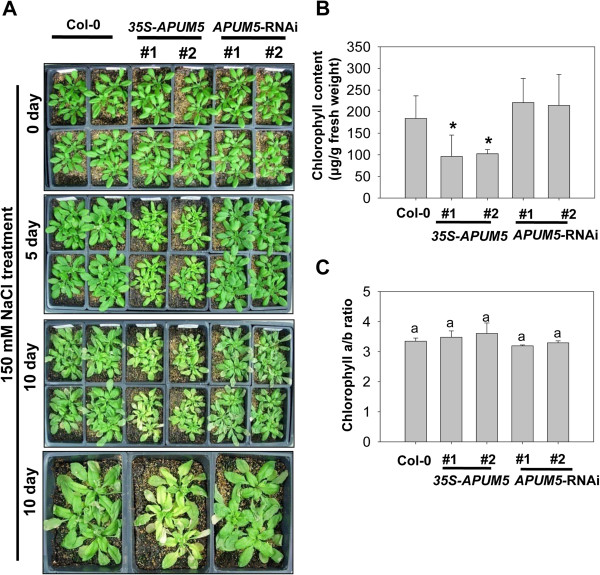
APUM5 seems to be directly involved in the osmotic stress response. Phenotypes of wild type and APUM5 transgenic plants treated with salt were evaluated in soil to investigate the possible role of APUM5 in the salt stress response. A high-salinity treatment resulted in symptoms on Col-0 leaves such as chlorosis, leaf burn, and senescence as well as a decrease in leaf area compared with those in non-stressed plants [29]. Wild-type and APUM5 transgenic plants showed similar normal growth at the vegetative stage. However, 35S-APUM5 transgenic plants exhibited a slightly more shrinking phenotype compared with Col-0 and APUM5-RNAi plants when irrigated with 150 mM NaCl for 5 days (Figure 5A). 35S-APUM5 transgenic plants showed significantly enhanced chlorosis, leaf burn, and reduced leaf area at 10 days, compared with that in Col-0 and APUM5-RNAi plants (Figure 5A). To further analyze the effect of salt stress, chlorophyll contents and chlorophyll a/b ratio were measured because chlorosis was enhanced in APUM5-overexressing plants. Chlorophyll content decreased but the chlorophyll a/b ratio remained unchanged in 35S-APUM5 transgenic plants compared to those in Col-0 and APUM5-RNAi plants (Figure 5B and C). Taken together, these results indicate that APUM5-overexpressing plants exhibit hypersensitivity to salt stress at the vegetative and primary root elongation stage, suggesting that APUM5 may act as a negative regulator when plants are subjected to salt stress.
Figure 5.
Analysis of salt sensitivity in wild type and APUM5 transgenic plants in soil. (A) Photographs show plants treated with 150 mM NaCl at the indicated time points. Four-week-old Col-0, 35S-APUM5, and APUM5-RNAi plants were treated with 150 mM NaCl. (B) Chlorophyll contents of salt-treated plants on day 10. Pigments were extracted from the salt-treated plant leaves. Data are mean values of six independent experiments. (C) Chlorophyll a/b ratio of salt-treated plant leaves. Error bars represent ± SD (Student’s t-test; *P < 0.01).
APUM5-overexpressing plants are more hypersensitive whereas APUM5-knockdown plants are more tolerant to drought stress compared to wild-type plants in soil
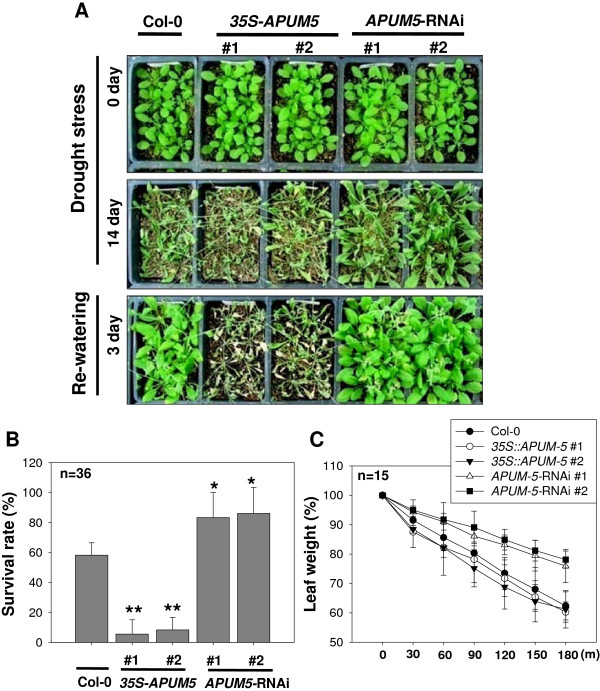
We further investigated whether APUM5 transgenic plants showed an altered phenotype to drought tolerance. Both Col-0 and 35S-APUM5 plants became severely wilted when water was withheld from soil for 14 days (Figure 6A). However, down-regulation of APUM5 by APUM5-RNAi resulted in the enhanced tolerant phenotype at the same stage compared with that of Col-0 (Figure 6A). Survival rate was examined after re-watering the 14 day-water-withheld plants. Approximately 77% of the APUM5-RNAi plants survived, whereas 11–13% of the 35S-APUM5 and 58% of the Col-0 plants survived (Figure 6B). Additionally, water loss rate of the wild-type and APUM5 transgenic plants was measured in detached leaves. Wild-type plants exhibited similar weight loss of detached leaves as APUM5-RNAi plants, whereas 35S-APUM5 transgenic plants had highly enhanced water loss rates (Figure 6C). Thus, similar to the salt stress results, APUM5-overexpressing plants exhibited more drought sensitivity, whereas repressing of APUM5 expression led to more drought tolerance compared with that of wild-type plants in soil.
Figure 6.
Drought sensitivity and water loss analysis of wild type and APUM5 transgenic plants. (A) The photographs show increased drought sensitivity of 35S-APUM5 transgenic plants and decreased sensitivity of APUM5-RNAi plants. Irrigation was stopped in 4-week-old plants for 14 days and then they were re-watered for 3 days afterwards. (B) Percentage survival of wild type and APUM5 transgenic (35S-APUM5 and APUM5-RNAi) plants. Each group of 30–40 plants was averaged to measure survival rate (Student’s t-test; *P < 0.01, **P < 0.001). (C) Water loss measurements in detached rosette leaves of wild-type and APUM5 transgenic plants after the drought stress treatment. Detached rosette leaves were measured and monitored for loss of fresh weight at the indicated time points. Data are mean values of three independent experiments (n = 15).
Altered transcription levels of abiotic stress-responsive genes in APUM5-overexpressing plants under drought stress
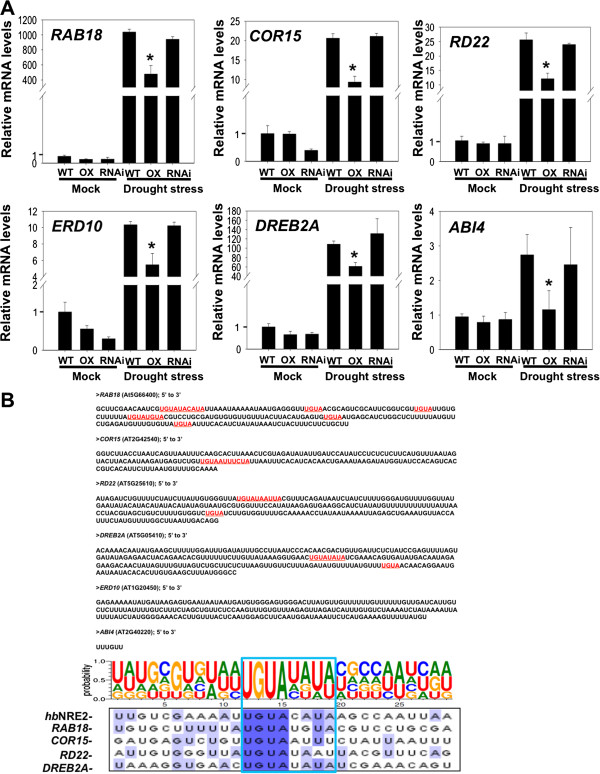
A gene expression analysis of various genes was performed by qRT-PCR to examine whether the enhanced sensitivity of 35S-APUM5 transgenic plants to salt and drought stresses was accompanied by changed transcription levels of abiotic stress-responsive genes. The transcription levels of some abiotic stress-responsive genes decreased in 35S-APUM5 transgenic plants under the drought stress condition compared to those in wild type plants (Figure 7A). The putative APUM5 binding site was searched in the 3′ UTR region of abiotic stress-responsive genes to investigate the possible role of APUM5 as a post-transcriptional repressor. We conducted this search because mammalian Pumilio proteins directly interact with target transcripts containing the conserved ‘UGUA’ tetranucleotide motif [9,15,17]. Some of 3′ UTRs of abiotic stress-responsive genes contained the conserved ‘UGUA’ tetranucleotide motif (Figure 7B and Additional file 7B). The 3′ UTRs of significantly downregulated genes in 35S-APUM5 transgenic plants by drought stress were re-analyzed and compared with the Drosophila hbNRE2 sequence. The ‘UGUA’ tetranucleotide motif of the 3′ UTRs was highly conserved among the most abiotic stress-responsive genes and hbNRE2 (Figure 7B). ERD10 and ABI4 transcript levels also decreased about 52% and 42% in APUM5-overexpressing plants compared with wild-type plants upon drought stress, respectively (Figure 7A). However, ERD10 and ABI4 did not have the ‘UGUA’ motif in the 3′ UTR region (Figure 7B). In contrast, KIN1, AtMYB6, AAO3, and RD29B did not contain the ‘UGUA’ tetranucleotide motif and their transcript levels did not change significantly in 35S-APUM5 plants upon drought stress (Additional file 7). APUM5-RNAi plants did not exhibit a dramatic alteration in ABA-response genes expression partly because APUM5-RNAi transgenic plants showed only 50% silencing levels of the APUM5 gene and the effect may be similar to that of the heterozygote mutant (Huh et al., 2013). These results suggest that transcripts of some abiotic stress-responsive genes could be negatively regulated by binding of the APUM5 protein in their 3′ UTR regions.
Figure 7.
Real-time qRT-PCR analysis of ABA-responsive genes in wild type and APUM5 transgenic plants under drought stress. (A) Expression of the ABA-regulated genes in wild-type and APUM5 transgenic plants under drought stress. mRNA levels were determined by qRT-PCR using total RNAs isolated from control and 6 h drought-stressed detached rosette leaves of wild-type and APUM5 transgenic plants. Data are mean values of three independent experiments (Student’s t-test; *P < 0.01). (B) Sequence analysis of the putative APUM5 binding sites in the 3′ UTRs of abiotic stress-related genes. The putative binding sequence including ‘UGUA’ is highlighted. hbNRE2 was used as a conserved Pumilio RNA binding motif. The ABA-responsive genes down-regulated in the 35S-APUM5 transgenic plants under drought stress were analyzed at the 3′ UTR sequence data base of The Arabidopsis Information Resource (TAIR) and UTRdb (http://utrdb.ba.itb.cnr.it/). The motif was used to search for Drosophila hbNRE2-related sequences and reconstructed using sequence alignment (CLUSTALW) and WebLogo software.
APUM5-PHD binds to the 3′ UTR motifs of abiotic stress-responsive putative target genes
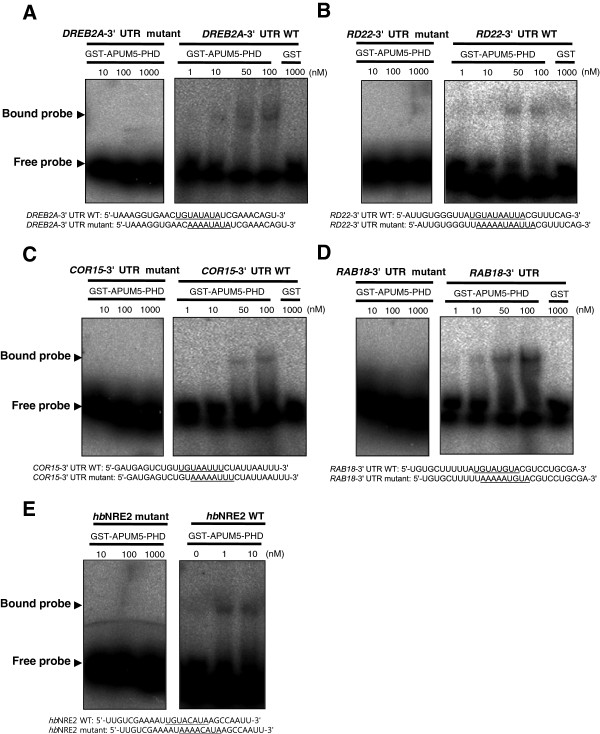
An electrophoretic mobility shift assay (EMSA) was performed to determine if the APUM5-PHD binds to the putative target RNAs. Genes that were highly down-regulated by APUM5 and had a ‘UGUA’ binding motif were selected. 32P-labeled synthetic 30 nucleotide RNAs along with ‘UGUA’ core sequence mutants were incubated with recombinant GST-APUM5-PHD protein. GST was used as a negative control. The EMSA results revealed that GST-APUM5-PHD bound effectively to DREB2A, RD22, COR15, and RAB18 but not to mutant RNAs, whereas the GST protein did not interact with these RNAs (Figure 8A–D). Furthermore, APUM5-PHD also showed strong binding affinity for hbNRE2 RNA (Figure 8E). APUM5 recognized the 8–10 nucleotide ‘UGUA’ core motifs. These results indicate that APUM5 binding affinity might be flexible for target binding motif recognition and this flexibility could contribute to multi-regulation of abiotic stress-responsive genes by destabilizing target mRNAs. This result confirmed that the Arabidopsis APUM5 protein has RNA binding activity and that the binding is important for regulating putative target 3′ UTRs.
Figure 8.
Gel mobility shift assay of the recombinant containing APUM5-PHD for 3′ UTR motifs of abiotic stress-responsive genes. (A-D). Indicated 32P-labeled RNA probes were incubated with recombinant GST-APUM5-PHD (1, 10, 50, and 100 nM) for 30 min in 20 μl RNA binding buffer at room temperature. RNA-protein complexes were separated on a 5% native gel and analyzed by autoradiography. GST protein (1000 nM) was used as a negative control. (E) 32P-labeled hbNRE2 RNA probes were incubated with recombinant GST-APUM5-PHD (1 and 10 nM) and used as a positive binding control.
APUM5 negatively regulates the RD22 and RAB18 3′ UTR reporters
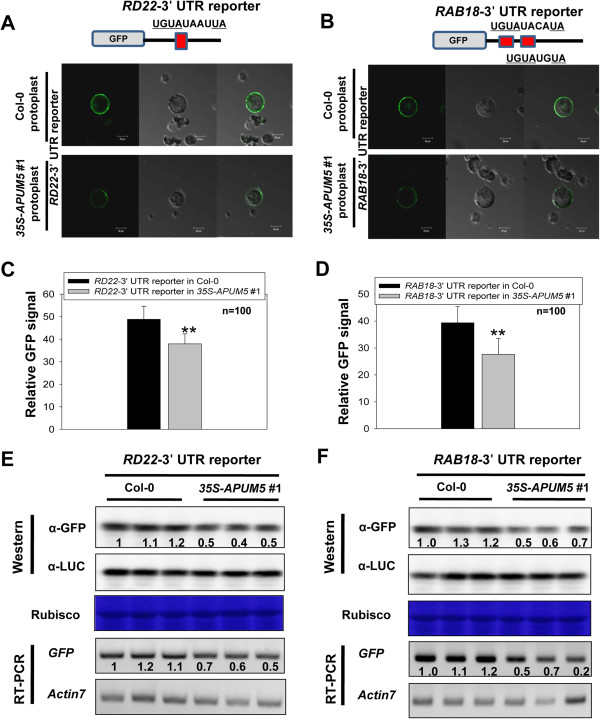
We found that the APUM5-PHD protein directly bound to 3′ UTRs of ABA-responsive genes in vitro. This phenomenon might explain that APUM5 negatively regulates ABA-responsive genes via binding to target 3′ UTRs. We made reporter constructs with 3′ UTRs of ABA-responsive genes and expressed reporter constructs in Col-0 and 35S-APUM5 transgenic protoplasts to identify the function of this binding. The RD22-3′ UTR reporter normally expressed GFP signals in Col-0 protoplasts, whereas the RD22-3′ UTR reporter showed reduced GFP signals in 35S-APUM5 transgenic protoplasts (Figure 9A). Next, these GFP signals were quantified by confocal LSM700 microscopy and ImageJ software. The signal intensity of the RD22-3′ UTR reporter in 35S-APUM5 transgenic protoplasts decreased approximately 20% compared with that in Col-0 protoplasts (Figure 9C). Western blot with a GFP antibody and RT-PCR analyses were performed to further confirm these data. Both the GFP protein and RNA levels of the RD22-3′ UTR reporter in 35S-APUM5 transgenic protoplasts decreased compared with the levels in Col-0 protoplasts (Figure 9E). We also performed the reporter assay with the RAB18-3′ UTR reporter using a similar procedure. The RAB18-3′ UTR reporter in 35S-APUM5 transgenic protoplasts showed about a 45% reduction in GFP signal intensity compared with that in Col-0 protoplasts (Figure 9B and D). In the Western blot and RT-PCR analyses, GFP protein and RNA levels decreased slightly in 35S-APUM5 transgenic protoplasts compared with those in Col-0 protoplasts (Figure 9F).
Figure 9.
APUM5 negatively regulates the 3′ UTRs of ABA-responsive genes in the reporter assay. (A and B) Protoplasts of Col-0 and 35S-APUM5 transgenic plants were transformed with the RD22-3′ UTR or RAB18-3′ UTR reporter construct by PEG-mediated transformation. GFP signals were detected by LSM 700 confocal microscopy under identical conditions. (C, D) GFP signal intensity of the RD22-3′ UTR and RAB18-3′ UTR reporter were analyzed with Zen software of the LSM 700 confocal microscope and ImageJ software (rsbweb.nih.gov/ij/). Error bars represent ± SD (Student’s t-test; **P < 0.001). (E, F) Western blot and RT-PCR analysis were performed with protoplasts of Col-0 and 35S-APUM5 transgenic plants transformed with the RD22-3′ UTR or RAB18-3′ UTR reporter construct. Protein and RNA samples were obtained from four-independent experiments and extracted. Rubisco was used as the protein loading control and AtActin7 was used as the internal control. The numbers designate relative intensity.
Previous results showed that APUM5-PHD also bound to the DREB2A and COR15 3′ UTRs (Figure 8A and C), and these putative target RNAs also contained the PHD binding RNA motif. Thus, we expected that these reporters would also exhibit a decrease of the GFP signal in 35S-APUM5 transgenic protoplasts. However, the GFP protein and RNA levels of the DREB2A and COR15 3′ UTR reporters were not affected in 35S-APUM5 transgenic protoplasts compared with Col-0 protoplasts (Additional file 8). These data indicate that APUM5 negatively regulates some ABA-responsive genes by binding to 3′ UTR post-transcriptionally. Furthermore, RNA binding motifs may be very flexible and other RNA residues in addition to the ‘UGUA’ core motif or RNA structure could be important for the in vivo APUM5 binding system.
Discussion
Arabidopsis and rice have multiple Puf members [11]. However, the function of plant Pufs is poorly understood. We performed an APUM5 gene expression analysis using a bioinformatics tool to functionally characterize APUM5, which is related to the biotic and abiotic stress responses. APUM5 showed increased expression patterns following exposure to biotic and abiotic stressors. We showed here that the Arabidopsis Puf protein, APUM5, negatively regulated some abiotic stress responsive genes and could be involved in the osmotic and drought stress response, although APUM5 was not involved in susceptibility or resistance to bacterial and fungal pathogen infection (Figure 1C and D). However, APUM5 is a pathogen responsive gene upon bacterial and fungal pathogen infection (Figure 1A and B). APUM5 function may still be related to defense response regulation, although we did not show any definitive results for specific bacterial or fungal infections. Similar to animal Puf proteins, APUM5 may regulate diverse target genes and be regulated by defense-related factors such as MPK4 (Additional file 1).
ABA-mediated plant immunity has been highlighted in terms of virus-host interaction. When Tobacco mosaic virus (TMV-cg) infects ABA mutant plants, TMV-cg accumulation increases in the systemic region and TMV-cg could repress ABA signaling by repressing WRKY8 [30]. Furthermore, CMV 2b, a silencing suppressor, interferes with ABA-mediated signaling, and transgenic plants that overexpress CMV 2b exhibit enhanced tolerance to drought stress [31]. In contrast, overexpression of APUM5 led to more resistance to CMV infection and slightly diminished Turnip mosaic virus accumulation, although APUM5 may negatively regulate some of the ABA responsive genes [22]. This phenotype demonstrates that some plant viruses could negatively regulate ABA signaling by regulating host proteins. However, APUM5 defense function against CMV infection might not be directly connected with regulation of some ABA responsive genes.
The APUM5-overexpressing transgenic plants exhibited reduced expression of some abiotic-stress response genes (Figure 7A). Furthermore, the core Puf binding motif in the putative target genes is found at the 3′ UTR (Figure 7B), indicating that APUM5 is likely to control these putative target mRNAs by binding to the 3′ UTR. Puf3, a yeast Puf homolog protein, binds to the 3′ UTR of COX17 mRNA and regulates COX17 mRNA by enhancing RNA degradation [32]. Interestingly, the CMV 3′ UTR is not polyadenylated but contains the tRNA-like structure (TLS). APUM5 does not affect the RNA level of the CMV 3′ UTR reporter [22]. Thus, the function of APUM5 during abiotic stress could be different from the plant defense mechanism against CMV infection because APUM5 slightly affected reporter mRNA level (Figure 9E and F). This result indicates that APUM5 function could possibly be associated with a putative deadenylase complex such as the mammalian Puf-deadenylase complex [17], although we showed that the putative Ccr4-Pop2p-NOT mRNA deadenylase complex is not a APUM5 binding partner [23].
The putative APUM5 targets, DREB2A, RD22, COR15, and RAB18 contained ‘UGUA’ core binding motifs in the 3′ UTRs when compared with the hbNRE2 motif. One of them, RAB18 3′ UTR motif showed stronger binding affinity to APUM5-PHD protein compared to other motifs by binding even at 1 nM concentration (Figure 8). This that APUM5-PHD directly bound to these motifs in vitro, suggesting that the ‘UGUA’ core motif confers RNA-binding activity of Puf proteins but other RNA residues or secondary structures of the core motif are also important for Puf protein binding affinity in vivo[33]. In our results, ABI4 gene expression level in 35S-APUM5 transgenic plants decreased upon drought stress (Figure 7A). However, it did not contain the ‘UGUA’ core motif in its 3′ UTR sequence (Figure 7B), indicating that APUM5 might directly and indirectly regulate targets negatively.
ABA plays a key role in osmotic tolerance by regulating ABA-responsive transcription factors such as ABI3 and ABI5 [34]. ABA also affects seed germination [35]. APUM5 was highly expressed under the mannitol, NaCl, and exogenous ABA treatments (Figure 2). The seed germination and root growth experiments exhibited 35S-APUM5 transgenic plants were hypersensitive while APUM5-RNAi plants were insensitive compared to Col-0 wild-type plants under osmotic stress and exogenous ABA treatment of various concentrations (Figure 4 and Additional file 6). Interestingly, 35S-APUM5 transgenic plants were hypersensitive to salinity and drought stress in soil (Figures 5 and 6) although most ABA-sensitive mutants show salinity or drought tolerance phenotypes in soil [36,37]. Furthermore, some mutants exhibit development-dependent phenotypes to abiotic stress treatments. For example, AREB1, AREB2, and ABF3 mutants do not exhibit ABA sensitive phenotypes in single, double, or triple mutants during germination. However, these mutants show the ABA-sensitive phenotype during primary root growth [38]. These data indicate that some mutants exhibit development-dependent phenotypes to exogenous ABA application. In contrast, some Arabidopsis proteins control the ABA-dependent and ABA-independent pathways to the abiotic stress response [37,39]. For example, OSM1, a SNARE superfamily protein, is important to osmotic stress tolerance and the osm1 mutant exhibits a hypersensitive phenotype during seed germination under an osmotic stress condition. The osm1 mutant plants also show increased sensitivity to salt and soil desiccation [39].
An examination of tissue-specific activities of APUM5pro-GUS transgenic plants revealed high-level expression in roots under normal condition (Figure 3A). Furthermore, the APUM5pro-GUS activity assay showed enhanced levels under diverse abiotic stressors in hydathodal cells and trichomes (Figure 3B and C). Hydathodal cells are involved in secreting water [40,41], and trichomes generally protect leaves by insulating them from heat, salt, insects, and water loss [42]. These expression patterns indicate that APUM5 may be involved in this tissue-specific regulation and affect the salinity and drought stress condition, although APUM5 was not expressed in guard cells (Figure 3C).
Conclusions
Our results provide evidence that Arabidopsis Pumilio proteins have functionally conserved RNA binding domain and activity. We found that APUM5 is a defense-responsive gene and negatively regulated the RD22-3′ UTR and RAB18-3′ UTR reporters at the mRNA and protein levels. This negative regulation of APUM5 could be connected to a more sensitive 35S-APUM5 transgenic plant phenotype under abiotic stress. These data suggest that the APUM5 protein could have dual functions in both transcriptional regulation and translational control by binding with target 3′ UTR motifs and may have diverse functions during biotic stress, abiotic stress, and development. But still we do not know the real targets of APUM5 in vivo. To understand the Puf protein functions further, the real targets of APUM5 should be identified by immunoprecipitation of mRNA-APUM5 complexes.
Methods
Plant material, growth conditions, and transgenic plants
APUM5 transgenic plants were prepared as described previously [22]. A. thaliana Col-0 and APUM5 transgenic plants were grown in a 16 h light/8 h dark photoperiod at 23°C in soil.
Pathogen inoculation
Pst DC3000 was cultured in King’s B medium and resuspended in 10 mM MgCl2 for bacterial pathogen inoculation. Leaves of 4-week-old Col-0, APUM5 transgenic, and NahG plants were syringe infiltrated with Pst DC3000 suspensions (OD600 = 0.0001 in 10 mM MgCl2). Leaf discs were collected from Col-0 and other mutant leaves at 0 and 3 days post-inoculation (dpi) to detect bacterial growth. Bacterial counts on leaf discs were measured after 28°C incubation for 2 days as described previously [43]. A. brassicicola strain KACC40036 was grown on potato dextrose agar for the fungal pathogen disease assay (Difco Scientific, Detroit, MI, USA). A spore suspension (5 × 104 spores/mL in potato dextrose media) was dropped on the detached leaves of Col-0 and APUM5 transgenic plants and was kept under high humidity. The spore population was counted at 5 dpi [43].
Germination rate and root growth assays
Surface-sterilized seeds of Arabidopsis Col-0, APUM5-RNAi, and 35S-APUM5 plants were grown on plates containing 1/2 Murashige and Skoog Medium Basal Salt Mixture (Duchefa, Haarlem, The Netherlands), 1.5% sucrose, and 0.8% phytoagar with or without various concentrations of NaCl, mannitol, and ABA at 22°C under a long day condition to determine the effect of osmotic stress and ABA on germination. The percentage of germinated seeds was scored daily in three independent experiments (60–70 seeds per experiment).
A root growth assay was carried out by transferring 3-day-old seedlings to the minimal medium supplemented with different concentrations of NaCl, mannitol, and ABA to determine the effect of osmotic stress and ABA on root growth. After 7 or 10 days, root growth was measured in three independent experiments (30–40 seedlings per experiment).
Manipulation of RNA and quantitative gene expression analysis under the various stress conditions
Ten-day-old seedlings were submerged in ddH2O supplemented with the indicated concentrations of mannitol, NaCl, or ABA for the gene expression analysis. Rosette leaves of 4-week-old plants were dehydrated on Whatman paper at 25°C under light for the indicated time period to test the effect of desiccation. Four-week-old plants were syringe infiltrated with a bacterial suspension (OD600 = 0.001 in 10 mM MgCl2) of virulent Pst DC3000 for the gene expression analysis under the biotic stressors. Arabidopsis plants were inoculated with A. brassicicola by spraying a spore suspension (5 × 105 spores/mL) in potato dextrose media for the gene expression pattern analyses. These samples were harvested at the indicated time points, and total RNA was isolated by the modified hot-phenol method [44] and reverse-transcribed with MMLV Reverse Transcriptase (Promega, Madison, WI, USA). Real-time PCR was performed according to the instructions provided by the LightCycler Real-Time PCR Systems (Roche, Mannheim, Germany). The primers used for real-time PCR reactions are listed in Additional file 9.
Histological assays
For histochemical analysis of the GUS activity, seedlings or plants from the T3 populations under the normal or stressed conditions were vacuum-infiltrated with Agrobacterium carrying the constructs in X-Gluc staining solution (0.5 M sodium phosphate buffer, pH7.0, 10% Triton X-100, 0.1 M K3Fe(CN)6, 0.1 M K4Fe(CN)6, 2 mM X-GlcA), incubated at 37°C overnight, and transferred into 70% (v/v) ethanol to remove chlorophyll [45]. GUS activity was observed under an Axioplan 2 imaging microscope (Carl Zeiss, Jena, Germany) and COOLPIX 5200 digital camera (Nikon, Osaka, Japan).
High-salinity and drought stress treatment in soil
Four-week-old Col-0, APUM5-RNAi, and 35S-APUM5 plants were irrigated with 150 mM NaCl every 3 days and allowed to grow for 3 weeks to test the effect of high-salinity stress. For the drought stress treatment, 4-week-old stage plants grown in soil were deprived of water for 14 days. Then, these plants were provided water. Surviving plants were counted at 3 days after re-watering. Three independent measurements of 30–40 plants were averaged.
Gel mobility shift assay
Synthetic RNA oligonucleotides were obtained from Bio Basic Inc. (Markham, ONT, Canada). An RNA probe was radiolabeled with 32P using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA, USA) and incubated with recombinant GST-APUM5-PHD for 30 min in a binding solution (20 mM Tris at pH 7.4, 50 mM KCl, 3 mM MgCl2, 2 mM dithiothreitol, 5% glycerol) at room temperature. Samples were separated on a 4% native gel containing 5% glycerol in 0.5 × TBE buffer, and subjected to autoradiography.
Reporter assay
To generate reporter constructs, the full RD22, RAB18, DREB2A, and COR15-3′ UTR sequences were isolated and ligated into the N-terminus of the modified 326-GFP3G vector [22]. Protoplasts of Col-0 and APUM5 transgenic plants were isolated, and reporter constructs were introduced into the protoplasts by PEG-mediated transformation [46], with several modifications. GFP signal intensity was quantified by Zen software for the LSM 700 confocal microscope and ImageJ software (rsbweb.nih.gov/ij/).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SUH and KHP conceived the study, designed the experiments and drafted the manuscript. SUH performed the experiments. SUH and KHP read and approved the final manuscript.
Supplementary Material
Expression analysis of APUM5 in mpk4-1 compared to Ler control plants. Total RNAs were extracted from 3-week old plants and mRNA levels were determined by qRT-PCR analysis. Error bars represent ± SD (n = 3). (Student’s t-test; ***P < 0.0001).
GUS activity analysis of APUM5pro-GUS transgenic plants at the flowering and silique development stages.APUM5 promoter activity was determined by histochemical GUS staining at the flower stage. (A) Cauline leaves and flowers. (B) Flowers. (C) Silique. Arrows indicate the end regions of the silique.
Cis-element analysis of the APUM5 promoter. A 1.3-kb promoter region was isolated using Athena analysis tools (http://www.bioinformatics2.wsu.edu/Athena) to analyze APUM5promoter sequences.
Germination rate analysis of APUM5 transgenic plants in 1/2 MS medium plates (n = 150).
Salt and mannitol sensitivity of wild-type and APUM5 transgenic plants. (A) The effect of 100 mM NaCl on primary root elongation. (B) The effect of 300 mM mannitol on primary root elongation. Seeds were germinated for 3 days on 1/2 MS medium, and the seedlings were transferred (n = 30, triplicates) to 1/2 MS containing 100 mM NaCl or 300 mM mannitol. Primary root length was measured at 10 days after transfer.
ABA effect on seed germination and primary root growth in wild-type and APUM5 transgenic plants. (A) The effect of ABA on seed germination. Seeds were germinated and grown on 1/2 MS medium containing 0.5 and 0.7 μM ABA for 7 days, and seedlings with green cotyledons were counted (n > 60, triplicates). (B) The effect of ABA on primary root elongation. Seeds were germinated for 3 days on 1/2 MS medium, and the seedlings were transferred (n = 30, triplicates) to 1/2 MS medium containing 2 and 5 μM ABA, respectively. Primary root length was measured at 10 days after transfer.
Expression analysis of some abiotic stress-responsive genes that do not have a putative APUM5 target site in APUM5 transgenic plants upon drought stress. (A) mRNA levels were determined by qRT-PCR analysis using total RNAs isolated from control and 6 h drought-stressed detached leaves. Error bars represent ± SD (n = 3). (B) The 3′ UTRs of abiotic stress-responsive genes were analyzed using the Arabidopsis Information Resource (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/) and the 3′ UTR sequence database UTRdb (http://utrdb.ba.itb.cnr.it/). Red indicates ‘UGUA’ tetranucleotide Puf target motif.
Reporter assay. (A, B) Protoplasts of Col-0 and 35S-APUM5 transgenic plants were transformed with the DREB2A-3′ UTR reporter or the COR15-3′ UTR construct by PEG-mediated transformation. GFP signals were detected by LSM 700 confocal microscopy under identical conditions. (C, D) GFP signal intensity of the DREB2A-3′ UTR and COR15-3′ UTR reporters was quantified with Zen software of the LSM 700 confocal microscope and ImageJ software (rsbweb.nih.gov/ij/). Error bars represent ± SD. (E, F) Western blot and RT-PCR analyses were performed with protoplasts of Col-0 and 35S-APUM5 transgenic plants transformed with the DREB2A-3′ UTR reporter or the COR15-3′ UTR construct. Protein and RNA samples were extracted from four-independent experiments. Rubisco was used as a protein loading control and AtActin7 was used as the internal control.
Coupled RT and qRT-PCR primers used for expression analysis of abiotic stress-related genes.
Contributor Information
Sung Un Huh, Email: sung.huh@tsl.ac.uk.
Kyung-Hee Paek, Email: khpaek95@korea.ac.kr.
Acknowledgements
This study was supported by the Wujangchoon Project (PJ007850) from the Rural Development Administration and the Mid-career Researcher Program (2012R1A2A2A02014387) through NRF grant funded by the MEST, Republic of Korea.
References
- Kim JY, Kim WY, Kwak KJ, Oh SH, Han YS, Kang H. Zinc finger-containing glycine-rich RNA-binding protein in Oryza sativa has an RNA chaperone activity under cold stress conditions. Plant Cell Environ. 2010;33(5):759–768. doi: 10.1111/j.1365-3040.2009.02101.x. [DOI] [PubMed] [Google Scholar]
- Lorkovic ZJ. Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 2009;14(4):229–236. doi: 10.1016/j.tplants.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Mata J, Marguerat S, Bahler J. Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem Sci. 2005;30(9):506–514. doi: 10.1016/j.tibs.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309(5740):1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Huh SU, Paek K-H. Plant RNA binding proteins for control of RNA virus infection. Front in Physiol. 2013;4(397) doi: 10.3389/fphys.2013.00397. doi:10.3389/fphys 00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic ZJ, Barta A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 2002;30(3):623–635. doi: 10.1093/nar/30.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat Rev Mol Cell Biol. 2004;5(12):1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110(4):501–512. doi: 10.1016/S0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- Cheong CG, Hall TM. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci U S A. 2006;103(37):13635–13639. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassov DS, Jurecic R. The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life. 2003;55(7):359–366. doi: 10.1080/15216540310001603093. [DOI] [PubMed] [Google Scholar]
- Tam PP, Barrette-Ng IH, Simon DM, Tam MW, Ang AL, Muench DG. The Puf family of RNA-binding proteins in plants: phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 2010;10:44. doi: 10.1186/1471-2229-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7(4):855–865. doi: 10.1016/S1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′ UTR regulation as a way of life. Trends Genet. 2002;18(3):150–157. doi: 10.1016/S0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Deng Y, Singer RH, Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 2008;22(8):1037–1050. doi: 10.1101/gad.1611308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci USA. 2006;103(12):4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JP, Schoderboeck L, Gingl E, Luzi E, Riefler J, Di Leva F, Karra D, Thomas S, Kiebler MA, Macchi P. Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc Natl Acad Sci USA. 2010;107(7):3222–3227. doi: 10.1073/pnas.0907128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RP, Aggarwal AK. mRNA regulation by Puf domain proteins. Sci STKE. 2006;2006(354):e37. doi: 10.1126/stke.3542006pe37. [DOI] [PubMed] [Google Scholar]
- Chen G, Li W, Zhang QS, Regulski M, Sinha N, Barditch J, Tully T, Krainer AR, Zhang MQ, Dubnau J. Identification of synaptic targets of Drosophila pumilio. PLoS Comput Biol. 2008;4(2):e1000026. doi: 10.1371/journal.pcbi.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6(10):e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francischini CW, Quaggio RB. Molecular characterization of Arabidopsis thaliana PUF proteins–binding specificity and target candidates. FEBS J. 2009;276(19):5456–5470. doi: 10.1111/j.1742-4658.2009.07230.x. [DOI] [PubMed] [Google Scholar]
- Abbasi N, Kim HB, Park NI, Kim HS, Kim YK, Park YI, Choi SB. APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010;64(6):960–976. doi: 10.1111/j.1365-313X.2010.04393.x. [DOI] [PubMed] [Google Scholar]
- Huh SU, Kim MJ, Paek KH. Arabidopsis Pumilio protein APUM5 suppresses Cucumber mosaic virus infection via direct binding of viral RNAs. Proc Natl Acad Sci USA. 2013;110(2):779–784. doi: 10.1073/pnas.1214287110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SU, Paek KH. Role of Arabidopsis Pumilio RNA binding protein 5 in virus infection. Plant Signal Behav. 2013;8(5):e23975. doi: 10.4161/psb.23975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2(1):e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Bjorn Nielsen H, Zhu S, Newman MA, Shokat KM, Rietz S, Parker J, Mundy J. Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 2006;47(4):532–546. doi: 10.1111/j.1365-313X.2006.02806.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10(2):88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Abbasi N, Park YI, Choi SB. Pumilio Puf domain RNA-binding proteins in Arabidopsis. Plant Signal Behav. 2011;6(3):364–368. doi: 10.4161/psb.6.3.14380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor TR, Dyreson C, Wyrick JJ. Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics. 2005;21(24):4411–4413. doi: 10.1093/bioinformatics/bti714. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Plant salt tolerance. Trends Plant Sci. 2001;6(2):66–71. doi: 10.1016/S1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang L, Li D, Wang F, Yu D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA. 2013;110(21):E1963–1971. doi: 10.1073/pnas.1221347110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood JH, McCann L, Naish M, Dixon H, Murphy AM, Stancombe MA, Bennett MH, Powell G, Webb AA, Carr JP. A viral RNA silencing suppressor interferes with abscisic acid-mediated signalling and induces drought tolerance in Arabidopsis thaliana. Mol Plant Pathol. 2013;14(2):158–170. doi: 10.1111/j.1364-3703.2012.00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas W, Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 2000;19(23):6602–6611. doi: 10.1093/emboj/19.23.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Dolgner SJ, Hall TM. Understanding and engineering RNA sequence specificity of PUF proteins. Curr Opin Struct Biol. 2009;19(1):110–115. doi: 10.1016/j.sbi.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH. AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 2003;17(3):410–418. doi: 10.1101/gad.1055803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Chen Z, Zhang H, Jablonowski D, Zhou X, Ren X, Hong X, Schaffrath R, Zhu JK, Gong Z. Mutations in ABO1/ELO2, a subunit of holo-Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol Cell Biol. 2006;26(18):6902–6912. doi: 10.1128/MCB.00433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61(4):672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- Zhu J, Gong Z, Zhang C, Song CP, Damsz B, Inan G, Koiwa H, Zhu JK, Hasegawa PM, Bressan RA. OSM1/SYP61: a syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell. 2002;14(12):3009–3028. doi: 10.1105/tpc.006981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Bogenschutz NL, Torii KU. The bHLH protein, MUTE, controls differentiation of stomata and the hydathode pore in Arabidopsis. Plant Cell Physiol. 2008;49(6):934–943. doi: 10.1093/pcp/pcn067. [DOI] [PubMed] [Google Scholar]
- Pilot G, Stransky H, Bushey DF, Pratelli R, Ludewig U, Wingate VP, Frommer WB. Overexpression of GLUTAMINE DUMPER1 leads to hypersecretion of glutamine from Hydathodes of Arabidopsis leaves. Plant Cell. 2004;16(7):1827–1840. doi: 10.1105/tpc.021642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada E, Kim JA, Meyer AJ, Hell R, Clemens S, Choi YE. Expression profiling of tobacco leaf trichomes identifies genes for biotic and abiotic stresses. Plant Cell Physiol. 2010;51(10):1627–1637. doi: 10.1093/pcp/pcq118. [DOI] [PubMed] [Google Scholar]
- Choi J, Huh SU, Kojima M, Sakakibara H, Paek KH, Hwang I. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev Cell. 2010;19(2):284–295. doi: 10.1016/j.devcel.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17(6):2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression analysis of APUM5 in mpk4-1 compared to Ler control plants. Total RNAs were extracted from 3-week old plants and mRNA levels were determined by qRT-PCR analysis. Error bars represent ± SD (n = 3). (Student’s t-test; ***P < 0.0001).
GUS activity analysis of APUM5pro-GUS transgenic plants at the flowering and silique development stages.APUM5 promoter activity was determined by histochemical GUS staining at the flower stage. (A) Cauline leaves and flowers. (B) Flowers. (C) Silique. Arrows indicate the end regions of the silique.
Cis-element analysis of the APUM5 promoter. A 1.3-kb promoter region was isolated using Athena analysis tools (http://www.bioinformatics2.wsu.edu/Athena) to analyze APUM5promoter sequences.
Germination rate analysis of APUM5 transgenic plants in 1/2 MS medium plates (n = 150).
Salt and mannitol sensitivity of wild-type and APUM5 transgenic plants. (A) The effect of 100 mM NaCl on primary root elongation. (B) The effect of 300 mM mannitol on primary root elongation. Seeds were germinated for 3 days on 1/2 MS medium, and the seedlings were transferred (n = 30, triplicates) to 1/2 MS containing 100 mM NaCl or 300 mM mannitol. Primary root length was measured at 10 days after transfer.
ABA effect on seed germination and primary root growth in wild-type and APUM5 transgenic plants. (A) The effect of ABA on seed germination. Seeds were germinated and grown on 1/2 MS medium containing 0.5 and 0.7 μM ABA for 7 days, and seedlings with green cotyledons were counted (n > 60, triplicates). (B) The effect of ABA on primary root elongation. Seeds were germinated for 3 days on 1/2 MS medium, and the seedlings were transferred (n = 30, triplicates) to 1/2 MS medium containing 2 and 5 μM ABA, respectively. Primary root length was measured at 10 days after transfer.
Expression analysis of some abiotic stress-responsive genes that do not have a putative APUM5 target site in APUM5 transgenic plants upon drought stress. (A) mRNA levels were determined by qRT-PCR analysis using total RNAs isolated from control and 6 h drought-stressed detached leaves. Error bars represent ± SD (n = 3). (B) The 3′ UTRs of abiotic stress-responsive genes were analyzed using the Arabidopsis Information Resource (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/) and the 3′ UTR sequence database UTRdb (http://utrdb.ba.itb.cnr.it/). Red indicates ‘UGUA’ tetranucleotide Puf target motif.
Reporter assay. (A, B) Protoplasts of Col-0 and 35S-APUM5 transgenic plants were transformed with the DREB2A-3′ UTR reporter or the COR15-3′ UTR construct by PEG-mediated transformation. GFP signals were detected by LSM 700 confocal microscopy under identical conditions. (C, D) GFP signal intensity of the DREB2A-3′ UTR and COR15-3′ UTR reporters was quantified with Zen software of the LSM 700 confocal microscope and ImageJ software (rsbweb.nih.gov/ij/). Error bars represent ± SD. (E, F) Western blot and RT-PCR analyses were performed with protoplasts of Col-0 and 35S-APUM5 transgenic plants transformed with the DREB2A-3′ UTR reporter or the COR15-3′ UTR construct. Protein and RNA samples were extracted from four-independent experiments. Rubisco was used as a protein loading control and AtActin7 was used as the internal control.
Coupled RT and qRT-PCR primers used for expression analysis of abiotic stress-related genes.