Abstract
Objective: to investigate the prevalence and antimicrobial resistance of Enterococcus species isolated from a university hospital, and explore the mechanisms underlying the antimicrobial resistance, so as to provide clinical evidence for the inappropriate clinical use of antimicrobial agents and the control and prevention of enterococcal infections. Methods: a total of 1,157 enterococcal strains isolated from various clinical specimens from January 2010 to December 2012 in the General Hospital of Ningxia Medical University were identified to species level with a VITEK-2 COMPACT fully automated microbiological system, and the antimicrobial susceptibility of Enterococcus species was determined using the Kirby-Bauer disc diffusion method. The multiple-drug resistant enterococcal isolates were screened from the clinical isolates of Enterococcus species from the burns department. The minimal inhibitory concentration (MIC) of Enterococcus species to the three fluoroquinolones, including ciprofloxacin, gatifloxacin and levofloxacin was determined with the agar dilution method, and the changes in the MIC of Enterococcus species to the three fluoroquinolones following reserpine treatment were evaluated. The β-lactam, aminoglycoside, tetracycline, macrolide, glycopeptide resistance genes and the efflux pump emeA genes were detected in the enterococcal isolates using a polymerase chain reaction (PCR) assay. Results: the 1,157 clinical isolates of Enterococcus species included 679 E. faecium isolates (58.7%), 382 E. faecalis isolates (33%), 26 E. casseliflavus isolates (2.2%), 24 E. avium isolates (2.1%), and 46 isolates of other Enterococcus species (4%). The prevalence of antimicrobial resistance varied significantly between E. faecium and E. faecalis, and ≤1.1% of these two Enterococcus species were found to be resistant to vancomycin, teicoplanin or linezolid. In addition, the Enterococcus species isolated from different departments of the hospital exhibited various resistances to the same antimicrobial agent, while reserpine treatment reduced the resistance of Enterococcus species to ciprofloxacin, gatifloxacin and levofloxacin. The β-lactamase gene TEM, aminoglycoside-modifying-enzyme genes aac(6')-aph(2"), aph(3')-III, ant(6)-I and ant(2")-I, tetracycline resistance gene tetM, erythromycin resistance gene ermB, vancomycin resistance gene vanA and the enterococcal multidrug resistance efflux emeA gene were detected in 77%, 62%, 26%, 13%, 36%, 31%, 66%, 5% and 55% of the 100 multiple-drug resistant enterococcal isolates. Conclusions: similar to previous findings, E. faecium and E. faecalis are predominant conditionally pathogenic bacteria that cause hospital-acquired infections that can cause urinary and respiratory system infections. Multiple and high-level antimicrobial resistance is highly prevalent in the hospital isolates of Enterococcus species. Reserpine treatment inhibits the active efflux of Enterococcus species to ciprofloxacin, gatifloxacin and levofloxacin in vitro and reduces the MIC of Enterococcus species to these three fluoroquinolones. The presence of the enterococcal multidrug resistance efflux emeA gene is associated with the resistance to antibiotics in Enterococcus species. The monitoring of the prevalence and antimicrobial resistance of Enterococcus species is of great significance to guide the control and prevention of enterococcal infections.
Keywords: Enterococcus spp., antimicrobial resistance, active efflux mechanism, reserpine, fluoroquinolones
1. Introduction
Enterococci are commensal bacteria inhabiting the intestines of both humans and animals, which are the major conditionally pathogenic bacteria that cause hospital-acquired infections [1]. Recently, frequent inappropriate use of antimicrobial agents, increase in invasive therapy, and wide use of immunosuppressants has resulted in a growing rise in the number of clinical infections caused by Enterococcus spp., notably Enterococcus faecium [2]. In addition, the emergence of high-level aminoglycoside-resistant (HLAR) enterococci and vancomycin-resistant enterococci (VRE) causes great difficulties in clinical anti-infective therapy [3,4,5]. In this hospital-based study, a total of 1,157 Enterococcus strains isolated from a university hospital during the period from January 2010 through December 2012 were detected and identified to investigate the prevalence and antimicrobial resistance of Enterococcus species. In addition the mechanisms underlying the antimicrobial resistance were explored so as to provide clinical evidence for the inappropriate clinical use of antimicrobial agents and the control and prevention of enterococcal infections.
2. Materials and Methods
2.1. Enterococcus Strains
A total of 1,157 enterococcal strains were isolated from 1,157 diverse clinical specimens obtained from January 2010 to December 2012 at the General Hospital of Ningxia Medical University (Yinchuan, China). All strains were identified to the species level with a VITEK-2 COMPACT fully automated microbiological system (bioMérieux, Inc.; Durham, NC, USA). The quality control strain Enterococcus faecalis ATCC 29212 was purchased from Shanghai Harmony Biotechnology Co., Ltd. (Shanghai, China).
2.2. Antibiotic Susceptibility Testing
The susceptibility of Enterococcus species to 16 antibiotics was determined using the Clinical Laboratory Standard Institute (CLSI) recommended, WHO modified Kirby-Bauer disc diffusion method [6].
2.3. Screening of Multiple-drug Resistant Enterococcal Isolates
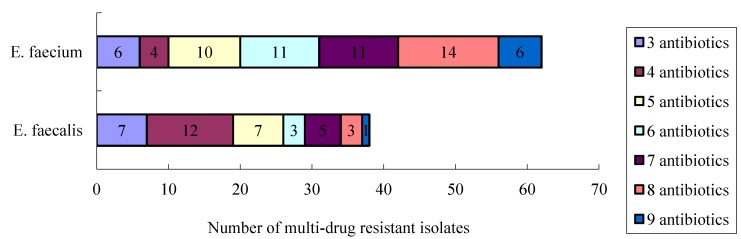
A total of 100 multiple-drug resistant enterococcal isolates (resistant to at least three antibiotics) were screened from the 182 isolates of Enterococcus species from the burns department during the period between January 2010 and December 2012, and the antimicrobial resistance in these 100 multiple-drug resistant enterococcal strains is described in Figure 1 and Table 1.
Figure 1.
Antimicrobial resistance in 100 multiple-drug resistant enterococcal isolates.
Table 1.
Antimicrobial resistance in 100 isolates of E. faecium and E. faecalis.
| Antibiotics | E. faecium (n = 62) | E. faecalis (n = 38) | ||
|---|---|---|---|---|
| Antibiotics-resistant isolate | Prevalence (%) | Antibiotics-resistant isolate | Prevalence (%) | |
| Penicillin | 55 | 88.7 | 2 | 5.3 |
| Ampicillin | 51 | 82.3 | 2 | 5.3 |
| High-level gentamicin | 1 | 1.6 | 1 | 2.6 |
| Rifampicin | 49 | 79.0 | 17 | 44.7 |
| Ciprofloxacin | 36 | 58.1 | 6 | 15.8 |
| Levofloxacin | 28 | 45.2 | 5 | 13.2 |
| Fosfomycin | 15 | 24.2 | 3 | 7.9 |
| Erythromycin | 56 | 90.3 | 20 | 52.6 |
| Furadantin | 7 | 11.3 | 1 | 2.6 |
| Linezolid | 0 | 0.0 | 0 | 0.0 |
| Vancomycin | 0 | 0.0 | 0 | 0.0 |
| Teicoplanin | 0 | 0.0 | 0 | 0.0 |
| Chloramphenicol | 3 | 4.8 | 10 | 26.3 |
| Quinupristin/dalfopristin | 0 | 0.0 | 25 | 65.8 |
| Minocycline | 20 | 32.3 | 18 | 47.4 |
| Tetracycline | 30 | 48.4 | 26 | 68.4 |
2.3.1. Efflux Pump Inhibition Assay
The minimal inhibitory concentration (MIC) of Enterococcus species to the three fluoroquinolones, including ciprofloxacin, gatifloxacin and levofloxacin (Dalian Meilun Biology Technology Co., Ltd.; Dalian, China) at final concentrations of 0.25–512 mg/L, was determined with the agar dilution method [7], while E. faecalis ATCC 29212 served as a control isolate. In addition, the MIC of Enterococcus species to ciprofloxacin, gatifloxacin and levofloxacin following treatment with an efflux pump inhibitor reserpine (Dalian Meilun Biology Technology Co., Ltd.; (Dalian, China) at a concentration of 20 mg/L, while the antibiotics-free Mueller-Hinton (M-H) agar (Oxoid, Basingstoke, UK) medium containing 20 mg/L reserpine served as controls.
2.3.2. Detection of Antimicrobial Resistance Genes
The 100 multiple-drug resistant enterococcal strains were isolated in pure cultures. Then, 6–8 enterococcal colonies were randomly selected, diluted with 200 µL of ddH2O, centrifuged, boiled at 95 °C for 10 min, followed by centrifugation at 12,000 r/min for 5 min, and the supernatant was the DNA of the enterococcal strains. The β-lactam, aminoglycoside, tetracycline, macrolide, glycopeptide resistance genes and the efflux pump genes were detected in the 100 multiple-drug resistant enterococcal isolates using a polymerase chain reaction (PCR) assay with primers (Table 2) synthesized by the Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China). PCR was performed with a 25 μL system containing 12.5 μL Premix Taq (BioTeke Biotech Co., Ltd.; Beijing, China), 1 μL DNA template, 1 μL of the forward and reverse primers, and 9.5 μL ddH2O under the following conditions: pre-degeneration at 94 °C for 3 min (at 93 °C for 2 min for the ant(6)-I and tetM genes and at 94 °C for 5 min for the emeA gene), followed by 35 cycles of degeneration at 94 °C for 40 s, annealing at 55 °C for 40 s, and extension at 72 °C for 40 s (35 cycles of degeneration at 93 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 60 s for the ant(6)-I gene; 35 cycles of degeneration at 93 °C for 60 s, annealing at 55 °C for 60 s, and extension at 72 °C for 60 s for the tetM gene; 30 cycles of degeneration at 94 °C for 45 s, annealing at 57 °C for 60 s, and extension at 72 °C for 90 s for the emeA gene), and final extension at 72 °C for 2 min (for 5 min for the ant(6)-I and tetM genes and for 10 min for the emeA gene). The amplification products were electrophoresed on 1.5% agarose gels (TAKARA Biotechnology (Dalian) Co., Ltd.; Dalian, China). Following electrophoresis, the agarose gels were stained with ethidium bromide for 15 min, and then visualized with a gel imaging analysis system with Quantity One software (Bio Rad; Hercules, CA, USA).
Table 2.
Sequences of the primers for amplification of antibiotics-resistant genes in Enterococcus spp.
| Antibiotic-resistant Enterococcus spp. | Representative Gene | Sequence (5'-3') | Amplification Product Size (bp) |
|---|---|---|---|
| β-lactam-resistant Enterococcus spp. | TEM | P1:AGGAAGAGTATGATTCAACA | 535 |
| P2:CTCGTCGTTTGGTATGGC | |||
| Aminoglycoside-resistant Enterococcus spp. | aac(6')/aph(2') | P1:CCAAGAGCAATAAGGGCATA | 220 |
| P2:CACTATCATAACCACTACCG | |||
| aph(3')-II | P1:GCCGATGTGGATTGCGAAAA | 292 | |
| P2:GCTTGATCCCCAGTAAGTCA | |||
| ant(6)-I | P1:ACTGGCTTAATCAATTTGGG | 597 | |
| P2:GCCTTTCCGCCACCTCACC | |||
| ant(2'')-I | P1:GAGCGAAATCTGCCGCTCTGG | 320 | |
| P2:CTGTTACAACGGACTGGCCGC | |||
| ant(4', 4”) | P1:GCAAGGACCGACAACATTTC | 165 | |
| P2:TGGCACAGATGGTCATAACC | |||
| Tetracycline-resistant Enterococcus spp. | tetM | P1:GTGTGACGAACTTTACCGAA | 501 |
| P2:GCTTTGTATCTCCAAGAACAC | |||
| Macrolide-resistant Enterococcus spp. | ermB | P1:GAAAAGGTACTAAACCAAATA | 616 |
| P2:AGTAACGGTACTTAAATTGTTTAC | |||
| mefA | P1:ACTATCATTAATCACTAGTGC | 346 | |
| P2:TTCTTCTGGTACTAAAAGTGG | |||
| Glycopeptide-resistant Enterococcus spp. | vanA | P1: GGGAAAACGACAATTGC | 732 |
| P2:GTACAATGCGGCCGTTA | |||
| vanB | P1:CATCGCCGTCCCCGAATTTCAAA | 297 | |
| P2:GATGCGGAAGATACCGTGGCT | |||
| vanC1 | P1:GGTATCAAGGAAACCTC | 822 | |
| P2:CTTCCGCCATCATAGCT | |||
| vanC2/3 | P1:CTCCTACGATTCTCTTG | 439 | |
| P2:CGAGCAAGACCTTTAAG | |||
| Multidrug resistance efflux pump | emeA | P1:GTGACAGCCTTTGTGGCAGAT | 687 |
| P2:TAGTCCGTTGATGGTTCCTTG |
2.4. Statistical Analysis
All data were managed using the software WHONET version 5.6, and all statistical analyses were performed with the statistical software SPSS version 17.0 (SPSS Inc.; Chicago, IL, USA). The difference of antimicrobial sensitivity in Enterococcus species was compared with chi-square test, with a p-value < 0.05 indicative of statistical significance.
3. Results
3.1. Distribution of Enterococcus Species in Various Clinical Specimens
The 1,157 Enterococcus species were isolated from 1,157 clinical specimens collected between January 2010 to December 2012 in the hospital, including 679 E. faecium isolates (58.7%, 679/1,157), 382 E. faecalis isolates (33%, 382/1,157), 26 E. casseliflavus isolates (2.2%, 26/1,157), 24 E. avium isolates (2.1%, 24/1,157), and 46 isolates of other Enterococcus species (4%, 46/1,157). The MIC50 and MIC90 values of the 16 antibiotics against the four major enterococcal strains are shown in Table 3. The top five departments from which Enterococcus species were isolated (Table 4) included the burns department (15.7%), intensive care unit (ICU; 14.4%), pediatrics department (13.5%), urology department (5.8%) and respiratory medicine department (4.1%), and the highest prevalence of Enterococcus species was detected in urine specimens (31.4%), followed by pus specimens (24.4%) and secretion specimens (16%).
Table 3.
MIC50 and MIC90 scales of 16 antibiotics against Enterococcus species (µg/mL).
| Antibiotics | E. faecium (n = 679) | E. faecalis (n = 382) | E. casseliflavus (n = 26) | E. avium (n = 24) | ||||
|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | |
| Penicillin | 64 | 64 | 2 | 8 | 0.5 | 2 | 1 | 64 |
| Ampicillin | 32 | 32 | 2 | 16 | 2 | 2 | 2 | 16 |
| High-level gentamicin * | - | - | - | - | - | - | - | - |
| Rifampicin | 16 | 16 | 8 | 32 | 1 | 2 | 2 | 2 |
| Ciprofloxacin | 64 | 128 | 1 | 16 | 0.5 | 2 | 0.5 | 1 |
| Levofloxacin | 8 | 128 | 1 | 8 | 2 | 4 | 1 | 2 |
| Fosfomycin | 64 | 128 | 32 | 64 | 64 | 256 | 32 | 32 |
| Erythromycin | 64 | 256 | 16 | 256 | 1 | 8 | 8 | 8 |
| Furadantin | 64 | 256 | 16 | 16 | 16 | 32 | 32 | 128 |
| Linezolid | 2 | 2 | 2 | 2 | 2 | 4 | 1 | 2 |
| Vancomycin | 1 | 1 | 1 | 2 | 2 | 4 | 0.5 | 1 |
| Teicoplanin | 2 | 4 | 2 | 2 | 4 | 8 | 2 | 2 |
| Chloramphenicol | 8 | 16 | 8 | 32 | 2 | 4 | 2 | 4 |
| Quinupristin/dalfopristin | 0.5 | 1 | 4 | 8 | 1 | 2 | 2 | 4 |
| Minocycline | 8 | 32 | 16 | 64 | 2 | 8 | 4 | 8 |
| Tetracycline | 8 | 16 | 16 | 16 | 1 | 16 | 16 | 16 |
Note: * Only resistance found was against high-level gentamicin.
Table 4.
Distribution of 1157 Enterococcus species isolated from various clinical departments.
| Clinical department | E. faecium (n = 679) | E. faecalis (n = 382) | E. casseliflavus (n = 26) | E. avium (n = 24) | E. raffinosus (n = 18) | E. gallinarum (n = 7) | Other Enterococcus species (n = 21) |
|---|---|---|---|---|---|---|---|
| Department of burns | 73 | 93 | 7 | 2 | 2 | 2 | 3 |
| ICU | 116 | 37 | 4 | 3 | 1 | 1 | 5 |
| Department of pediatrics | 99 | 44 | 5 | 1 | 3 | 2 | 2 |
| Department of urology | 27 | 38 | 0 | 1 | 1 | 0 | 0 |
| Department of respiratory medicine | 42 | 5 | 1 | 0 | 0 | 0 | 0 |
| Department of: hepatobiliary surgery | 30 | 8 | 5 | 0 | 3 | 0 | 1 |
| Department of orthopedics | 16 | 18 | 1 | 2 | 0 | 1 | 3 |
| Department of endocrinology | 8 | 9 | 0 | 1 | 1 | 0 | 1 |
| Department of neurology | 13 | 4 | 1 | 0 | 0 | 0 | 0 |
| Department of gynecology | 6 | 8 | 0 | 0 | 2 | 0 | 0 |
| Other department | 249 | 118 | 2 | 14 | 5 | 1 | 6 |
3.2. Sensitivity of Enterococcus Species to Antibiotics
Of the 1,157 Enterococcus isolates, a low prevalence of resistance to linezolid, vancomycin and teicoplanin was detected, while over 40% prevalence of resistance to most antibiotics tested in this study was found. The prevalence of antimicrobial resistance in isolates of E. faecium, E. faecalis, E. casseliflavus and E. avium is presented in Table 5.
Table 5.
Antimicrobial resistance in Enterococcus species.
| Antibiotics | E. faecium (n = 679) | E. faecalis (n = 382) | E. casseliflavus (n = 26) | E. avium (n = 24) | ||||
|---|---|---|---|---|---|---|---|---|
| Antibiotics-Resistant Isolate | Prevalence (%) | Antibiotics-resistant Isolate | Prevalence (%) | Antibiotics-Resistant Isolate | Prevalence (%) | Antibiotics-Resistant Isolate | Prevalence (%) | |
| Penicillin | 621 | 91.4 | 22 | 5.8 | 1 | 3.8 | 8 | 33.3 |
| Ampicillin | 610 | 89.8 | 9 | 2.4 | 0 | 0.0 | 6 | 25.0 |
| High-level gentamicin | 22 | 3.2 | 8 | 2.1 | 0 | 0.0 | 1 | 4.5 |
| Rifampicin | 566 | 83.3 | 191 | 50.0 | 0 | 0.0 | 0 | 0.0 |
| Ciprofloxacin | 585 | 86.1 | 66 | 17.4 | 1 | 3.8 | 1 | 4.5 |
| Levofloxacin | 552 | 81.3 | 65 | 17.1 | 1 | 3.8 | 0 | 0.0 |
| Fosfomycin | 170 | 25.0 | 35 | 9.1 | 9 | 33.3 | 0 | 0.0 |
| Erythromycin | 615 | 90.6 | 235 | 61.5 | 8 | 32.0 | 22 | 91.7 |
| Furadantin | 238 | 35.0 | 10 | 2.6 | 0 | 0.0 | 5 | 20.0 |
| Linezolid | 6 | 0.9 | 4 | 1.1 | 0 | 0.0 | 0 | 0.0 |
| Vancomycin | 5 | 0.7 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Teicoplanin | 4 | 0.6 | 0 | 0.0 | 2 | 7.1 | 0 | 0.0 |
| Chloramphenicol | 65 | 9.5 | 149 | 39.1 | 0 | 0.0 | 0 | 0.0 |
| Quinupristin/dalfopristin | 12 | 1.8 | 310 | 81.2 | 1 | 3.8 | 4 | 17.4 |
| Minocycline | 272 | 40.0 | 200 | 52.4 | 2 | 7.1 | 4 | 17.4 |
| Tetracycline | 360 | 53.0 | 277 | 72.5 | 6 | 23.1 | 18 | 73.9 |
3.3. Comparison of Antimicrobial Resistance between E. faecium and E. faecalis
E. faecium and E. faecalis comprised 91.7% of the 1,157 Enterococcus species isolates collected from the hospital from January 2010 to December 2012. A significantly higher prevalence of resistance to penicillin, ampicillin, rifampicin, ciprofloxacin, levofloxacin, fosfomycin, erythromycin and furadantin was detected in E. faecium than that in E. faecalis (p < 0.05), while a greater prevalence of resistance to chloramphenicol, quinupristin/dalfopristin, minocycline and tetracycline was found in E. faecalis than that in E. faecium (p < 0.05). In addition, a low prevalence of resistance to linezolid, vancomycin and teicoplanin was detected in both E. faecium and E. faecalis.
3.4. Antimicrobial Resistance in Enterococcus Species Isolated from Various Departments of the Hospital
The prevalence of antimicrobial resistance varied in the Enterococcus species isolated from different departments of the hospital. A lower prevalence was detected in the Enterococcus species isolated from the department of pediatrics, where a high prevalence of penicillin resistance was found, while the highest prevalence was found in the burns department (Table 6).
Table 6.
Prevalence of antimicrobial resistance in Enterococcus species isolated from different departments of the hospital (%).
| Antibiotics | Department of burns (n = 182), ICU (n = 171) | Department of pediatrics (n = 164) | ||||
|---|---|---|---|---|---|---|
| E. faecium | E. faecalis | E. faecium | E. faecalis | E. faecium | E. faecalis | |
| Penicillin | 81.2 | 4.5 | 88.0 | 11.4 | 93.6 | 2.5 |
| Ampicillin | 77.9 | 2.2 | 89.0 | 2.7 | 92.2 | 0.0 |
| Gentamicin | 5.5 | 0.0 | 4.3 | 0.0 | 1.5 | 0.0 |
| Ciprofloxacin | 85.8 | 8.9 | 83.2 | 18.9 | 77.1 | 4.7 |
| Levofloxacin | 86.2 | 7.9 | 84.7 | 17.1 | 60.5 | 2.5 |
| Erythromycin | 83.8 | 47.3 | 89.9 | 59.5 | 92.9 | 38.6 |
| Furadantin | 20.0 | 3.4 | 40.7 | 2.9 | 10.0 | 0.0 |
| Quinupristin/dalfopristin | 1.5 | 77.2 | 0.0 | 86.5 | 1.0 | 66.7 |
| Tetracycline | 62.7 | 67.4 | 50.0 | 67.6 | 70.0 | 69.0 |
3.5. Effect of Reserpine Treatment on Antimicrobial Sensitivity in Enterococcus Species
All 100 of the clinical isolates of enterococci grew well on the M-H agar plates with or without reserpine, indicating that reserpine had no inhibitory effects on the growth of Enterococcus species. The number of enterococcal isolates resistant to ciprofloxacin, gatifloxacin and levofloxacin was reduced from 42 to 30 following treatment with 20 mg/L reserpine, with a corresponding reduction in the prevalence of resistance from 42% to 30%, while the number of enterococcal isolates resistant to the all three fluoroquinolones was reduced from 30 to 15, with a significant reduction also observed. The MIC alteration of three fluoroquinolones for 100 multiple-drug enterococcal strains before and after reserpine treatment is shown in Table 7. Reduced MIC was observed in 84 clinical isolates of Enterococcus species following reserpine treatment, including 72 isolates with increased sensitivity to ciprofloxacin, 55 isolates with increased sensitivity to gatifloxacin and 39 isolates with increased sensitivity to levofloxacin. Following reserpine treatment, 36 isolates had an increased sensitivity to all the three fluoroquinolones, 18 isolated showed an increased sensitivity to two fluoroquinolones, while 30 isolates exhibited an increased sensitivity to a fluoroquinolone (Table 8).
Table 7.
Changes in MIC50 and MIC90 of three fluoroquinolones for 100 multiple-drug enterococcal strains before and after reserpine treatment.
| Time | Ciprofloxacin | Gatifloxacin | Levofloxacin | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Prevalence of Drug Resistance (%) | MIC50 (mg/L) | MIC90 (mg/L) | Prevalence of Drug Resistance (%) | MIC50 (mg/L) | MIC90 (mg/L) | Prevalence of Drug Resistance (%) | MIC50 (mg/L) | MIC90 (mg/L) | |
| Before reserpine treatment | 42.0 | 2 | 256 | 30.0 | 1 | 32 | 33.0 | 2 | 64 |
| After reserpine treatment | 28.0 | 0.25 | 128 | 17.0 | 0.5 | 8 | 23.0 | 2 | 32 |
Table 8.
Changes of antimicrobial sensitivity in 100 enterococcal isolates following treatment with 20 mg/L reserpine.
| Antibiotics | Drug Sensitivity Test | No. Isolates | No. of enterococcal Isolates with Reduced MIC following Treatment with 20 mg/L Reserpine | ||||
|---|---|---|---|---|---|---|---|
| MIC Reduction by 1/2 | MIC Reduction by 1/4 | MIC Reduction by 1/8 | MIC Reduction by >1/8 | No Reduction | |||
| Ciprofloxacin | Resistant | 42 | 10 | 6 | 3 | 21 | 2 |
| Sensitive | 58 | 4 | 9 | 19 | 0 | 26 | |
| Gatifloxacin | Resistant | 30 | 7 | 8 | 1 | 13 | 1 |
| Sensitive | 70 | 16 | 4 | 3 | 3 | 44 | |
| Levofloxacin | Resistant | 33 | 11 | 8 | 5 | 3 | 6 |
| Sensitive | 67 | 11 | 1 | 0 | 0 | 55 | |
3.6. Prevalence of Antimicrobial Resistance Genes
Of the 100 multiple-drug resistant enterococcal isolates, there were 38 isolates of E. faecalis and 62 isolates of E. faecium, while the TEM, aac(6')/aph(2''), aph(3')-III, ant(6)-I, ant(2'')-I, tetM, ermB, vanA and emeA genes were detected in 77, 62, 26, 13, 36, 31, 66, 5 and 55 multiple-drug resistant enterococcal isolates, respectively. The detection of these antimicrobial resistance genes in 38 isolates of E. faecalis and 62 isolates of E. faecium is shown in Table 9. The emeA gene was detected in 73.8% of the ciprofloxacin-resistant enterococcal isolates, 76.7% of the gatifloxacin-resistant enterococcal isolates, and 75.8% of the levofloxacin-resistant enterococcal isolates, while the prevalence of the emeA gene was 41.4%, 45.7% and 44.8% in the ciprofloxacin-, gatifloxacin- and levofloxacin- sensitive enterococcal isolates, respectively (Table 10).
Table 9.
Detection of antibiotic resistance genes in multiple-drug resistant E. faecalis and E. faecium.
| Antibiotic Resistance Gene | E. faecalis Isolate (n = 38) | E. faecium Isolate (n = 62) | ||
|---|---|---|---|---|
| No. of Isolates with Resistance Gene Detected | Prevalence (%) | No. of Isolates with Resistance Gene Detected | Prevalence (%) | |
| TEM | 18 | 47.4 | 59 | 95.1 |
| aac(6’)/aph(2”) | 30 | 78.9 | 32 | 52.4 |
| Aph(3’)-III | 12 | 31.6 | 14 | 23.3 |
| Ant(6)-I | 4 | 10.5 | 9 | 14.3 |
| Ant(2”)-I | 11 | 28.9 | 25 | 39.8 |
| tetM | 12 | 31.6 | 19 | 30.1 |
| ermB | 27 | 71.1 | 39 | 62.1 |
| vanA | 0 | 0.0 | 5 | 8.1 |
| emeA | 10 | 26.3 | 45 | 72.6 |
Table 10.
Prevalence of the emeA gene in multiple-drug resistant enterococcal isolates.
| Antibiotics | Antibiotic-resistant enterococcal Isolate | Antibiotic-sensitive enterococcal Isolate | χ2 | p | ||||
|---|---|---|---|---|---|---|---|---|
| Total Isolates | No. Isolate with emeA Gene Detected | Prevalence (%) | Total Isolates | No. Isolate with emeA Gene Detected | Prevalence (%) | |||
| Ciprofloxacin | 42 | 31 | 73.8% | 58 | 24 | 41.4 | 13.02 | <0.005 |
| Gatifloxacin | 30 | 23 | 76.7% | 70 | 32 | 45.7 | 8.13 | <0.005 |
| Levofloxacin | 33 | 25 | 75.8% | 67 | 30 | 44.8 | 8.57 | <0.005 |
4. Discussion
Due to the spread of enterococcal antimicrobial resistance [8,9], the tracing of the infectious sources is of great significance for the control of enterococcal infections and its spreading. Among the 289 enterococcal strainss isolated from a tertiary-care pediatric hospital in Mexico City during an 18-month period, E. faecalis and E. faecium comprised 81.2% of the total isolates, and antimicrobial resistance in Enterococcus spp. was found to be common [10]. Of the 415 enterococcal isolates obtained from clinical samples between January 1999 and 31 December 2001 in the Mubarak Al-Kabeer, Amiri, Adan, Ibn Sina and Maternity hospitals in Kuwait, E. faecalis (85.3%) and E. faecium (7.7%) accounted for 93% of the samples [11]. Salem-Bekhit and colleagues identified 69.2% E. faecalis and 11.3% E. faecium in 206 enterococcal species obtained from the clinical samples in Riyadh hospitals of King Saud University, Saudi Arabia [12].
Maschieto et al. reported that the distribution of Enterococcus spp. isolated from the intestinal tracts of patients from a university hospital in Brazil was E. faecium (34%), followed by E. faecalis (33%), E. gallinarum (23.7%), E. casseliflavus (5.2%), E. avium (1%), and E. hirae (1%) [13]. In China, E. faecium and E. faecalis were also found to be predominant in the enterococci isolated from clinical specimens [14,15,16,17]. Similar to these findings, the current study showed that E. faecium (58.7%) and E. faecalis (33%) were predominant in the 1157 clinical isolates of Enterococcus species isolated from our hospital. However, the present study involved a large sample size, compared the antimicrobial resistance in enterococcal strains isolated from different departments of the hospital, and investigated the efflux mechanism of resistance in enterococci, which is rarely reported previously. The Enterococcus species were mainly isolated from the urinary system clinical specimens, which was in agreement with the detection of Enterococcus species isolated from the First Affiliated Hospital of Chongqing Medical University [18]. In addition, Enterococcus species were found to be predominantly isolated from the burns department, ICU and pediatrics department, which was associated with the patients’ critical illness, long-term antibiotic use and decline in immune function [19].
Enterococcus species are found to be intrinsically resistant to cephalosporins and aminoglycosides. Even though bacteria were found to be sensitive to these drugs in in-vitro experiments, unsatisfactory efficacy was found in clinical practice [2,20,21]. Multiple-antimicrobial resistance has been widely reported in Enterococcus species [22,23,24,25].
In the current study, a significantly higher prevalence of resistance to penicillin, ampicillin, rifampicin, ciprofloxacin, levofloxacin, fosfomycin, erythromycin and furadantin was detected in E. faecium than in E. faecalis (p < 0.05), while a greater prevalence of resistance to chloramphenicol, quinupristin/dalfopristin, minocycline and tetracycline was found in E. faecalis than in E. faecium (p < 0.05). In addition, a low prevalence of resistance to linezolid, vancomycin and teicoplanin was detected in both E. faecium and E. faecalis. Therefore, linezolid, vancomycin and teicoplanin are currently widely used drugs for the effective treatment of enterococcal infections [22,23,26]. Quinupristin/dalfopristin, a novel streptogramin antibiotic agent, has been widely used for the treatment of vancomycin-resistant enterococcal infections in USA and Europe, and a high therapeutic efficacy has been achieved [27,28,29]. The mechanism of action of the agent is found to involve early and late stage inhibition of bacterial protein synthesis [30,31], however, the drug shows poor efficacy against E. faecalis [32,33]. High rates of resistance to quinupristin-dalfopristin have been detected in enterococci isolated from poultry production environments [34], chickens [35], and clinical specimens [36,37,38]. In the present study, the prevalence of quinupristin-dalfopristin resistance was 81.2% in E. faecalis, which was significantly higher than that the 1.8% in E. faecium (p < 0.05). In addition, quinupristin/dalfopristin has been recommended by CLSI for the treatment of vancomycin-resistant enterococcal infections. Since antimicrobial resistance varies in Enterococcus species, there is a great need to identify enterococcal strains to the species level, which would facilitate the appropriate selection of antibiotics.
Like previous reports [16,17], our findings also found that the prevalence of antimicrobial resistance varied in the enterococci isolated from different departments of the hospital. A lower prevalence of antibiotic resistance was detected in the enterococci isolated from the department of pediatrics as compared to those from other departments of the hospital, while a high prevalence of penicillin resistance was found, which may be associated with the frequent application of penicillin, a commonly used drug in pediatrics. A high prevalence of antimicrobial resistance was found in the enterococci isolated from the burns department and ICU of the hospital, which may be attributed to the patients’ critical illness, poor immunity and long-term antibiotic use, or the habit of the antibiotic use [19].
To understand the shift of antimicrobial resistance in enterococci in our hospital, we compared the results from this study to the distribution of antimicrobial resistance in enterococci isolated from clinical specimens during the period from January 2007 through December 2009 [39], and found a great rise in the number of enterococcal isolates, in which E. faecium was still predominant, but its constituent ratio increased. In addition, the enterococcal isolates were still resistant to more than 40% of the commonly used antibiotics; however, no significant rise was found in the prevalence of antimicrobial resistance. In the current study, we identified 10 linezolid-resistant enterococcal strains, which were not detected in the enterococci isolated between 2007 and 2009. It is considered that the continuous antibiotic pressure causes the secondary resistance to linezolid in enterococci [40].
Reserpine has been proved to reduce the MIC of fluoroquinolones against antimicrobial-resistant bacteria [41,42,43]. It is found that the combination of the multidrug efflux inhibitor reserpine and fluoroquinolone enhances the sensitivity of fluoroquinolone-resistant Streptococcus pneumonia and Staphylococcus aureus to fluoroquinolones [44]. Our findings showed that reserpine treatment caused a significant reduction in the resistance to the three fluoroquinolones ciprofloxacin, gatifloxacin and levofloxacin in Enterococcus species, and the MIC of fluoroquinolones was reduced by over 2-fold in 72% of the enterococcal isolates.
In the current study, the emeA gene was detected in 73.8% of the ciprofloxacin-resistant enterococci, 76.7% of the gatifloxacin-resistant enterococci, and 75.8% of the levofloxacin-resistant enterococci, respectively, suggesting the presence of other mechanisms involved in the resistance of enterococci to the three fluoroquinolones in addition to drug efflux, and such a gene was present in 41.4% of the ciprofloxacin-sensitive enterococci, 45.7% of the gatifloxacin-sensitive enterococci, and 44.8% of the levofloxacin-sensitive enterococci, respectively, indicating no expression of the emeA gene in some enterococcal isolates. In addition, the occurrence of the emeA gene was significantly greater in the fluoroquinolone-resistant enterococci than that in the fluoroquinolone-sensitive enterococci (p < 0.05), indicating that the distribution of the emeA gene was associated with the resistance to the three fluoroquinolones in the Enterococcus species.
It is indicated that the resistance of enterococci to β-lactam is caused by the production of β-lactamase, which is encoded by the TEM gene, or modification in the penicillin-binding proteins (PBPs) [45,46]. In the current study, a high prevalence of penicillin resistance was detected in E. faecium, while a low prevalence was found in E. faecalis, and the occurrence of the TEM gene was 95.1% and 47.4% in E. faecium and E. faecalis, respectively. The aminoglycosides resistance in enterococci is mainly attributable to the production of aminoglycoside-modifying enzymes [47]. Currently, over 30 aminoglycoside modifying enzymes have been identified, in which bifunctional 6'-aminoglycoside acetyltransferase (AAC(6')) 2"-aminoglycoside phosphotransferase (APH(2")) enzyme encoded by the aac(6')/aph(2") gene is the most common one, which eliminates the synergistic effect between penicillin or glycopeptide antibiotics and aminoglycosides [48]. Our findings showed that the occurrence of the aac(6')/aph(2") gene, the aph(3')-III gene that encodes aminoglycoside 3'-type III phosphotransferase (APH(3')-III), the ant(6)-I gene that encodes 6-nucleotidyltransferase I (ANT(6)-I) and the ant(2")-I gene that encodes aminoglycoside- 2"-O-nucleotidyltransferase I (ANT(2")-I) was 62%, 26%, 13% and 36% in the 100 multiple-drug resistant enterococcal isolates, respectively. The resistance of enterococci to tetracyclines is mainly caused by the binding of the tetM gene-encoded ribosomal protection proteins to the ribosome, thereby avoiding the effect of tetracyclines [49]. In the current study, the prevalence of tetracycline resistance gene was 31.6% and 30.1% in E. faecalis and E. faecium, respectively. It is therefore considered that the resistance of Enterococcus species to β-lactam, aminoglycosides and tetracyclines is attributable to the presence of the gene that encodes the corresponding enzymes. Two mechanisms are considered to be responsible for macrolides resistance in enterococci, including the change in the target site of erythromycin mediated by the erm gene, and mef gene-mediated antimicrobial efflux [50,51]. ermB gene is the predominant type of erm gene in enterococci [50]. Our findings showed that the occurrence of the ermB gene was 71.1% and 62.1% in E. faecalis and E. faecium, respectively, indicating that the macrolides resistance in the Enterococcus species isolated from Ningxia region is mainly associated with the presence of the ermB gene. Like previous studies [52,53], the mefA gene was detected in enterococci in the current study; however, Liang et al. [54] detected the mefA gene in 9 of 53 clinical isolates of Enterococcus species, which may be due to the regional variation in the occurrence of the mefA gene in enterococci.
It is indicated that the resistance to glycopeptides in enterococci is mainly caused by the alteration of peptidoglycan precursors on the cell wall of enterococci, which leads to the failure of the glycopeptides to inhibiting the synthesis of the cell walls of enterococci, thereby resulting in the emergence of glycopeptide resistance [55,56]. In the present study, the vanA gene was detected in all of the 5 vancomycin-resistant isolates of enterococci. Vancomycin-resistant enterococci may transfer the vanA gene to S. aureus, which leads to the emergence of vancomycin-resistant S. aureus, thereby resulting in more difficulty in the clinical treatment of enterococcal infections [5]. Therefore, vancomycin should be used cautiously in the clinical therapy of enterococcal infections, and the management of vancomycin-resistant enterococci should be improved [22,57].
5. Conclusions
In summary, enterococci have become the major pathogenic bacteria that cause hospital-acquired infections due to multiple-antimicrobial resistance, and the clinical enterococcal infections predominantly occur in the urinary system. Antimicrobial sensitivity varies in different Enterococcus species, and the resistance of enterococci to antimicrobial agents is mainly attributable to the emergence of antimicrobial resistance genes. Reserpine, as an active efflux inhibitor, inhibits the active efflux of Enterococcus species, and reduces the MIC of antimicrobial-resistant Enterococcus species. The occurrence of the enterococcal multidrug resistance efflux emeA gene is associated with the resistance of enterococci to antimicrobial agents. The monitoring of the prevalence and antimicrobial resistance of Enterococcus species would provide a guide for the appropriate selection of antibiotics and prevent the occurrence of more antimicrobial-resistant enterococcal isolates.
Acknowledgments
This study was supported by the grant from Ningxia Science and Technology Project.
Author Contributions
Jia Wei conceived and designed the study; Gang Li and Wen Wang conducted the study, collected the data and performed analysis of data. Gang Li prepared the first draft of the manuscript; Wei Jia provided strategic advice and assisted with editing of the manuscript. All authors read and approved the final version of the manuscript.
Conflicts of Interests
The authors declare no conflict of interest.
References
- 1.Jarvis W.R., Martone W.J. Predominant pathogens in hospital infections. J. Antimicrob. Chemother. 1992;29:19–24. doi: 10.1093/jac/29.suppl_A.19. [DOI] [PubMed] [Google Scholar]
- 2.Sood S., Malhotra M., Das B.K., Kapil A. Enterococcal infections & antimicrobial resistance. Indian J. Med. Res. 2008;128:111–121. [PubMed] [Google Scholar]
- 3.Bonten M.J., Willems R., Weinstein R.A. Vancomycin-resistant enterococci: Why are they here, and where do they come from? Lancet Infect. Dis. 2001;1:314–325. doi: 10.1016/S1473-3099(01)00145-1. [DOI] [PubMed] [Google Scholar]
- 4.Adhikari L. High-level aminoglycoside resistance and reduced susceptibility to vancomycin in nosocomial enterococci. J. Glob. Infect. Dis. 2010;2:231–235. doi: 10.4103/0974-777X.68534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cetinkaya Y., Falk P., Mayhall C.G. Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 2000;13:686–707. doi: 10.1128/CMR.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biemer J.J. Antimicrobial susceptibility testing by the Kirby-Bauer disc diffusion method. Ann. Clin. Lab. Sci. 1973;3:135–140. [PubMed] [Google Scholar]
- 7.Tsaur S.M., Chang S.C., Luh K.T., Hsieh W.C. Antimicrobial susceptibility of enterococci in vitro. J. Formos. Med. Assoc. 1993;92:547–552. [PubMed] [Google Scholar]
- 8.Shay D.K., Goldmann D.A., Jarvis W.R. Reducing the spread of antimicrobial-resistant microorganisms. Control of vancomycin-resistant enterococci. Pediatr. Clin. North. Am. 1995;42:703–716. doi: 10.1016/s0031-3955(16)38986-6. [DOI] [PubMed] [Google Scholar]
- 9.Klare I., Konstabel C., Badstübner D., Werner G., Witte W. Occurrence and spread of antibiotic resistances in Enterococcus faecium. Int. J. Food. Microbiol. 2003;88:269–290. doi: 10.1016/S0168-1605(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 10.Miranda G., Lee L., Kelly C., Solórzano F., Leaños B., Muñoz O., Patterson J.E. Antimicrobial resistance from enterococci in a pediatric hospital. Plasmids in Enterococcus faecalis isolates with high-level gentamicin and streptomycin resistance. Arch. Med. Res. 2001;32:159–163. doi: 10.1016/S0188-4409(01)00265-X. [DOI] [PubMed] [Google Scholar]
- 11.Udo E.E., Al-Sweih N., Phillips O.A., Chugh T.D. Species prevalence and antibacterial resistance of enterococci isolated in Kuwait hospitals. J. Med. Microbiol. 2003;52:163–168. doi: 10.1099/jmm.0.04949-0. [DOI] [PubMed] [Google Scholar]
- 12.Salem-Bekhit M.M., Moussa I.M., Muharram M.M., Alanazy F.K., Hefni H.M. Prevalence and antimicrobial resistance pattern of multidrug-resistant enterococci isolated from clinical specimens. Indian J. Med. Microbiol. 2012;30:44–51. doi: 10.4103/0255-0857.93032. [DOI] [PubMed] [Google Scholar]
- 13.Maschieto A., Martinez R., Palazzo I.C., Darini A.L. Antimicrobial resistance of Enterococcus sp. isolated from the intestinal tract of patients from a university hospital in Brazil. Mem. Inst. Oswaldo Cruz. 2004;99:763–767. doi: 10.1590/S0074-02762004000700018. [DOI] [PubMed] [Google Scholar]
- 14.Ling J.M., Char T.S., Cheng A.F. Distribution of enterococci in Hong Kong. J. Infect. 2002;45:257–262. doi: 10.1053/jinf.2002.1054. [DOI] [PubMed] [Google Scholar]
- 15.Lu J.F., Qian L., Huo J.F., Shao K.Y. Clinical distribution and drug sensitivity analysis of 302 strains of Enterococcus. Med. Pharm. J. Chin. PLA. 2011;23:42–43. [Google Scholar]
- 16.Guo Z.S., Zhang L.H., Lin S.S. Analysis of the isolation and variance of resistance to antibiotics of Enterococcus in blood specimens. Lab. Med. Clin. 2012;9:537–539. [Google Scholar]
- 17.Qian Y.H., Guo J.W., Ma C., Hao X.H., Liu L.J., Li Y.J., Ma X.B. Distribution and antibiotics sensitivity status of 243 strains of enterococcus from clinical samples. Chin. J. Nosocomiol. 2011;21:1892–1894. [Google Scholar]
- 18.Liu Y., Huang W.X., Dong Y.S., Jia B., Zheng X.P., Liu C.W., Yu D.G., Li C.Z. Antimicrobial susceptibility of Enterococci isolated from the first affiliated hospital of Chongqing Medical University during 2006–2009. Chin. J. Antibiot. 2010;35:703–706. [Google Scholar]
- 19.Xiao Y.H., Wang J., Li Y., MOH National Antimicrobial Resistance Investigation Net Bacterial resistance surveillance in China: A report from Mohnarin 2004–2005. Eur. J. Clin. Microbiol. Infect. Dis. 2008;27:697–708. doi: 10.1007/s10096-008-0494-6. [DOI] [PubMed] [Google Scholar]
- 20.Hollenbeck B.L., Rice L.B. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence. 2012;3:421–433. doi: 10.4161/viru.21282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arias C.A., Murray B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arias C.A., Contreras G.A., Murray B.E. Management of multidrug-resistant enterococcal infections. Clin. Microbiol. Infect. 2010;16:555–562. doi: 10.1111/j.1469-0691.2010.03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arias C.A., Murray B.E. Emergence and management of drug-resistant enterococcal infections. Expert. Rev. Anti. Infect. Ther. 2008;6:637–655. doi: 10.1586/14787210.6.5.637. [DOI] [PubMed] [Google Scholar]
- 24.Huycke M.M., Sahm D.F., Gilmore M.S. Multiple-drug resistant enterococci: The nature of the problem and an agenda for the future. Emerg. Infect. Dis. 1998;4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammerum A.M., Lester C.H., Heuer O.E. Antimicrobial-resistant enterococci in animals and meat: A human health hazard? Foodborne. Pathog. Dis. 2010;7:1137–1146. doi: 10.1089/fpd.2010.0552. [DOI] [PubMed] [Google Scholar]
- 26.El-Khoury J., Fishman J.A. Linezolid in the treatment of vancomycin-resistant Enterococcus faecium in solid organ transplant recipients: Report of a multicenter compassionate-use trial. Transpl. Infect. Dis. 2003;5:121–125. doi: 10.1034/j.1399-3062.2003.00024.x. [DOI] [PubMed] [Google Scholar]
- 27.Moellering R.C. Quinupristin/dalfopristin: Therapeutic potential for vancomycin-resistant enterococcal infections. J. Antimicrob. Chemother. 1999;44:25–30. doi: 10.1093/jac/44.suppl_1.25. [DOI] [PubMed] [Google Scholar]
- 28.Moellering R.C., Linden P.K., Reinhardt J., Blumberg E.A., Bompart F., Talbot G.H. The efficacy and safety of quinupristin/dalfopristin for the treatment of infections caused by vancomycin-resistant Enterococcus faecium. Synercid Emergency-Use Study Group. J. Antimicrob. Chemother. 1999;44:251–261. doi: 10.1093/jac/44.2.251. [DOI] [PubMed] [Google Scholar]
- 29.Winston D.J., Emmanouilides C., Kroeber A., Hindler J., Bruckner D.A., Territo M.C., Busuttil R.W. Quinupristin/Dalfopristin therapy for infections due to vancomycin-resistant Enterococcus faecium. Clin. Infect. Dis. 2000;30:790–797. doi: 10.1086/313766. [DOI] [PubMed] [Google Scholar]
- 30.Linden P. Quinupristin-dalfopristin. Curr. Infect. Dis. Rep. 1999;1:480–487. doi: 10.1007/s11908-999-0062-y. [DOI] [PubMed] [Google Scholar]
- 31.Manzella J.P. Quinupristin-dalfopristin: A new antibiotic for severe gram-positive infections. Am. Fam. Physician. 2001;64:1863–1866. [PubMed] [Google Scholar]
- 32.Schouten M.A., Hoogkamp-Korstanje J.A. Comparative in vitro activities of quinupristin-dalfopristin against Gram-positive bloodstream isolates. J. Antimicrob. Chemother. 1997;40:213–219. doi: 10.1093/jac/40.2.213. [DOI] [PubMed] [Google Scholar]
- 33.Hershberger E., Donabedian S., Konstantinou K., Zervos M.J. Quinupristin-dalfopristin resistance in gram-positive bacteria: Mechanism of resistance and epidemiology. Clin. Infect. Dis. 2004;38:92–98. doi: 10.1086/380125. [DOI] [PubMed] [Google Scholar]
- 34.Hayes J.R., McIntosh A.C., Qaiyumi S., Johnson J.A., English L.L., Carr L.E., Wagner D.D., Joseph S.W. High-frequency recovery of quinupristin-dalfopristin-resistant Enterococcus faecium isolates from the poultry production environment. J. Clin. Microbiol. 2001;39:2298–2299. doi: 10.1128/JCM.39.6.2298-2299.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald L.C., Rossiter S., Mackinson C., Wang Y.Y., Johnson S., Sullivan M., Sokolow R., DeBess E., Gilbert L., Benson J.A., et al. Quinupristin-dalfopristin-resistant Enterococcus faecium on chicken and in human stool specimens. N. Engl. J. Med. 2001;345:1155–1160. doi: 10.1056/NEJMoa010805. [DOI] [PubMed] [Google Scholar]
- 36.Soltani M., Beighton D., Howard J.P., Woodford N. Mechanisms of resistance to quinupristin-dalfopristin among isolates of Enterococcus faecium from animals, raw meat, and hospital patients in western Europe. Antimicrob. Agents. Chemother. 2000;44:433–436. doi: 10.1128/AAC.44.2.433-436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luh K.T., Hsueh P.R., Teng L.J., Pan H.J., Chen Y.C., Lu J.J., Wu J.J., Ho S.W. Quinupristin-dalfopristin resistance among gram-positive bacteria in Taiwan. Antimicrob. Agents. Chemother. 2000;44:3374–3380. doi: 10.1128/AAC.44.12.3374-3380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh W.S., Ko K.S., Song J.H., Lee M.Y., Park S., Peck K.R., Lee N.Y., Kim C.K., Lee H., Kim S.W., et al. High rate of resistance to quinupristin-dalfopristin in Enterococcus faecium clinical isolates from Korea. Antimicrob. Agents. Chemother. 2005;49:5176–5178. doi: 10.1128/AAC.49.12.5176-5178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L.L., Yang X.Y., Li G., Li X.Y., Jia W. Clinical distribution of enterococci infection and drug resistance. Chin. J. Nasocomiol. 2011;21:1043–1045. [Google Scholar]
- 40.Xi R., Tian S.F., Chu Y.Z., Chen B.Y. Linezolid induces resistance in Enterococcus in vitro and the mechanism. Chin. J. Infect. Chemother. 2011;11:22–26. [Google Scholar]
- 41.Brenwald N.P., Gill M.J., Wise R. The effect of reserpine, an inhibitor of multi-drug efflux pumps, on the in vitro susceptibilities of fluoroquinolone-resistant strains of Streptococcus pneumoniae to norfloxacin. J. Antimicrob. Chemother. 1997;40:458–460. doi: 10.1093/jac/40.3.458. [DOI] [PubMed] [Google Scholar]
- 42.Beyer R.E., Pestova J.J., Millichap V., Stosor G., Noskin A., Peterson L.R. A convenient assay for estimating the possible involvement of efflux of fluoroquinolones by Streptococcus pneumoniae and Staphylococcus aureus: Evidence for diminished moxifloxacin, sparfloxacin, and trovafloxacin efflux. Antimicrob. Agents. Chemother. 2000;44:798–801. doi: 10.1128/AAC.44.3.798-801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brenwald N.P., Gill M.J., Wise R. Prevalence of a putative efflux mechanism among fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents. Chemother. 1998;42:2032–2035. doi: 10.1128/aac.42.8.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markham P.N. Inhibition of the emergence of ciprofloxacin resistance in Streptococcus pncumoiae by the multidrug efflux inhibitor reserpine. Antinicrob. Agents. Chemother. 1999;43:988–999. doi: 10.1128/aac.43.4.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilke M.S., Lovering A.L., Strynadka N.C.J. β-Lactam antibiotic resistance: A current structural perspective. Curr. Opin. Microbiol. 2005;8:525–533. doi: 10.1016/j.mib.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 46.Zapun A., Contreras-Martel C., Vernet T. Penicillin-binding proteins and β-lactam resistance. FEMS. Microbiol. Rev. 2008;32:361–385. doi: 10.1111/j.1574-6976.2007.00095.x. [DOI] [PubMed] [Google Scholar]
- 47.Chow J.W. Aminoglycoside resistance in enterococci. Clin. Infect. Dis. 2000;31:586–589. doi: 10.1086/313949. [DOI] [PubMed] [Google Scholar]
- 48.Daigle D.M., Hughes D.W., Wright G.D. Prodigious substrate specificity of AAC(6')-APH(2"), an aminoglycoside antibiotic resistance determinant in enterococci and staphylococci. Chem. Biol. 1999;6:99–110. doi: 10.1016/S1074-5521(99)80006-4. [DOI] [PubMed] [Google Scholar]
- 49.Roberts M.C. Tetracycline resistance determinants: Mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS. Microbiol. Rev. 1996;19:1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 50.Portillo A., Ruiz-Larrea F., Zarazaga M., Alonso A., Martinez J.L., Torres C. Macrolide resistance genes in Enterococcus spp. Antimicrob. Agents Chemother. 2000;44:967–971. doi: 10.1128/AAC.44.4.967-971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leclercq R. Mechanisms of resistance to macrolides and lincosamides: Nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 2002;34:482–492. doi: 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- 52.Reyes J., Hidalgo M., Díaz L., Rincón S., Moreno J., Vanegas N., Castañeda E., Arias C.A. Characterization of macrolide resistance in Gram-positive cocci from Colombian hospitals: A countrywide surveillance. Int. J. Infect. Dis. 2007;11:329–336. doi: 10.1016/j.ijid.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 53.Martel A., Devriese L.A., Decostere A., Haesebrouck F. Presence of macrolide resistance genes in streptococci and enterococci isolated from pigs and pork carcasses. Int. J. Food Microbiol. 2003;84:27–32. doi: 10.1016/S0168-1605(02)00390-2. [DOI] [PubMed] [Google Scholar]
- 54.Liang Q.H., Xu Y.J., Liu C.L., Zeng L.W. Analysis of antimicrobial resistance and antimicrobial resistance genes in 53 clinically isolated enterococcal isolates. J. Trop. Med. 2012;12:1458–1461. [Google Scholar]
- 55.Arthur M., Reynolds P., Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996;4:401–407. doi: 10.1016/0966-842X(96)10063-9. [DOI] [PubMed] [Google Scholar]
- 56.Arthur M., Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob. Agents. Chemother. 1993;37:1563–1571. doi: 10.1128/AAC.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kauffman C.A. Therapeutic and preventative options for the management of vancomycin-resistant enterococcal infections. J. Antimicrob. Chemother. 2003;51:23–30. doi: 10.1093/jac/dkg273. [DOI] [PubMed] [Google Scholar]