Abstract
OBJECTIVE
The biobreeding diabetes-prone (BBDP) rat spontaneously develops type 1 diabetes. Two of the genetic factors contributing to this syndrome are the major histocompatibility complex (Iddm1) and a Gimap5 mutation (Iddm2) responsible for a T-lymphopenia. Susceptibility to experimentally induced type 1 diabetes is widespread among nonlymphopenic (wild-type Iddm2) rat strains provided they share the BBDP Iddm1 allele. The question follows as to whether spontaneous and experimentally induced type 1 diabetes share susceptibility loci besides Iddm1. Our objectives were to map a novel, serendipitously discovered Iddm locus, confirm its effects by developing congenic sublines, and assess its differential contribution to spontaneous and experimentally induced type 1 diabetes.
RESEARCH DESIGN AND METHODS
An unexpected reduction in spontaneous type 1 diabetes incidence (86 to 31%, P < 0.0001) was observed in a BBDP line congenic for a Wistar Furth–derived allotypic marker, RT7 (chromosome 13). Genome-wide analysis revealed that, besides the RT7 locus, a Wistar Furth chromosome 8 fragment had also been introduced. The contribution of these intervals to diabetes resistance was assessed through linkage analysis using 134 F2 (BBDP × double congenic line) animals and a panel of congenic sublines. One of these sublines, resistant to spontaneous type 1 diabetes, was tested for susceptibility to experimentally induced type 1 diabetes.
RESULTS
Both linkage analysis and congenic sublines mapped a novel locus (Iddm24) to the telomeric 10.34 Mb of chromosome 8, influencing cumulative incidence and age of onset of spontaneous type 1 diabetes but not insulitis nor experimentally induced type 1 diabetes.
CONCLUSIONS
This study has identified a type 1 diabetes susceptibility locus that appears to act after the development of insulitis and that regulates spontaneous type 1 diabetes exclusively.
The biobreeding diabetes-prone (BBDP) rat spontaneously develops type 1 diabetes, with a polygenic mode of inheritance (1). Two type 1 diabetes susceptibility loci, Iddm1 and Iddm2, have been identified. Iddm1 maps to the u haplotype of the class II major histocompatibility complex (RT1 in the rat) on chromosome 20 (2–6). The Iddm2 susceptibility gene on chromosome 4 is Gimap5 (7,8). The BBDP Gimap5 allele results in the truncation of two-thirds of the predicted gene product (7,8). This recessive mutation induces a peripheral T-cell lymphopenia and most likely contributes to type 1 diabetes through an altered development of regulatory T-cells (1,9).
Attempts to map other spontaneous type 1 diabetes loci by linkage have identified multiple loci that, with the exception of Iddm3 on chromosome 2, only show weak linkage (10,11). Furthermore, only Iddm8 on chromosome 6 has been mapped using multiple type 1 diabetes–resistant strains and subsequently confirmed using congenic lines (11,12). These mapping results suggest that BBDP type 1 diabetes is complex, requiring multiple predisposing gene alleles, most of which have minor contributions to the overall autoimmune process, thus complicating the positional cloning of type 1 diabetes genes (13).
To overcome these difficulties in identifying susceptibility genes for spontaneous type 1 diabetes, investigators have taken advantage of the striking observation that many strains of rats are highly susceptible to experimentally induced type 1 diabetes (6). These experimentally induced type 1 diabetes syndromes develop rapidly and with high penetrance following either Kilham’s virus infection or toll-like receptor ligation by poly I:C, with or without simultaneous depletion of regulatory T-cells by treatment with depleting anti-ART2 monoclonal antibody (mAb) (1,14). Importantly, virus-induced, experimentally induced type 1 diabetes requires both the BBDP Iddm1 allele and the wild-type Iddm2 allele (1,6,14,15). Using crosses between parental strains satisfying both these requirements, loci conferring susceptibility to experimentally induced type 1 diabetes, has been successfully mapped. However, notable observations have emerged from these studies. Thus far, there has been no overlap between the loci conferring susceptibility to experimentally induced and spontaneous type 1 diabetes, despite the fact that in all of these studies one of the parental strains was either the BBDP or the genetically related biobreeding diabetes-resistant (BBDR) strain (16,17). One possible explanation for this is that although both syndromes share susceptibility alleles, experimentally induced type 1 diabetes bypasses several steps critical to the pathogenesis of spontaneous type 1 diabetes, therefore requiring fewer susceptibility alleles, which facilitates their detection. Alternatively, some of the susceptibility alleles implicated in spontaneous and experimentally induced type 1 diabetes are distinct, and, among these, some could be specific for the procedure used to experimentally induce type 1 diabetes. For example, Iddm14 on chromosome 4 is implicated in the pathogenesis of type 1 diabetes induced by either virus infection or treatment with poly I:C and anti-ART2 mAb, while Iddm20 on chromosome 17 is specific for virus-induced type 1 diabetes (18).
Here, we report on a novel type 1 diabetes susceptibility locus in the BBDP rat, Iddm24, which has not been identified through any of the previous studies summarized above but has a powerful effect on spontaneous type 1 diabetes incidence. We have fine mapped this locus to the telomeric 10.34 Mb of rat chromosome 8. Moreover, we show that this locus is specific for spontaneous type 1 diabetes as it has no effect on susceptibility to type 1 diabetes induced by poly I:C and anti-ART2 mAb. This locus has been designated Iddm24 by the Rat Genome Nomenclature Committee and given the RGD_ID no. 1599689. Information on this and each of the rat Iddm loci can be found on the rat genome database (http://rgd.mcw.edu/) and T1dbase (http://www.t1dbase.org/page/Welcome/display/?species=Rat) (accessed 7 February 2007).
RESEARCH DESIGN AND METHODS
BBDP and Wistar Furth rats were purchased from BRM (Worcester, MA) and Charles River (Frederick, MD), respectively. CD45 congenic inbred BBDP rats were developed through introgression of the Wistar Furth CD45 (RT7.2) allele onto BBDP rats by phenotypic selection of backcross breeders for >10 generations, followed by intercrossing. F2 rats were derived from a cross-intercross of the congenic and BBDP strains. A cohort of these F2 rats (n = 134) was used for linkage analysis. Others exhibiting potentially informative recombinations were bred with BBDP rats. Resulting littermates retaining the parental recombination were intercrossed to generate congenic sublines homozygous for these regions. BBnon-lyp rats are an Iddm2 congenic inbred line derived by the introgression of the wild-type Iddm2 locus from the BBDR strain into the BBDP strain. Backcross breeders were selected for a normal proportion of peripheral blood T-lymphocytes by flowcytometry for >10 generations, then intercrossed (19). The resulting BBnon-lyp rats are nonlymphopenic, hence, spontaneous type 1 diabetes resistant. Animals were maintained under specific pathogen- and virus antibody–free conditions, and, starting at 2 months, were screened for type 1 diabetes development as previously described (19). Nondiabetic littermates were killed at 4 months, after which the pancreas was fixed in 10% formalin and assessed histologically after hematoxylin and eosin staining.
Type 1 diabetes was triggered through treatment of animals with poly I:C (1/g body wt intraperitoneally three times per week) and DS4.23, an anti-ART2.1 mAb (50 μg intraperitoneally five times per week) starting at 30 days of age (16). Animals were tested for type 1 diabetes development after 2 weeks of treatment. All animal protocols were approved by the Sunnybrook Animal Care Committee.
Genetic analysis
Genotyping was performed as previously described (19,20). All microsatellite primers were purchased from Sigma-Genosys (Oakville, Ontario, Canada) and their physical location obtained from the Rat Genome Database (http://rgd.mcw.edu/). Genome coordinates indicated in Figs. 2 and 4A are derived from the Rat Genome Sequencing Consortium (RGSC) 3.4 assembly of December 2004. Additional primer information is available at www.well.ox.ac.uk/rat_mapping_resources. Novel microsatellite markers were identified through tandem repeat finder (21) analysis. Before linkage analysis, R/QTL (22) was used to identify genotyping errors and check the marker order by reestimating the genetic map for each order by calculating logarithm of odds (LOD) scores (log10 likelihood ratios) relative to the initial order. MAPMAKER/EXP, version 3.0 (23,24), was used to construct the genetic linkage map and imported into Windows QTL Cartographer, version 2.5 (25), for interval mapping (26) and permutation tests to determine experimental-wide significance levels for each trait. LOD score thresholds were determined by permutation testing (n = 1,000 permutations) (27). Significant loci were defined as those that exceeded the 95th percentile (P < 0.05) of permutation distribution, while highly significant loci exceeded the 99th percentile (P < 0.01). Raw data used for this linkage analysis is available at http://t1dbase.org/downloads/Iddm24/.
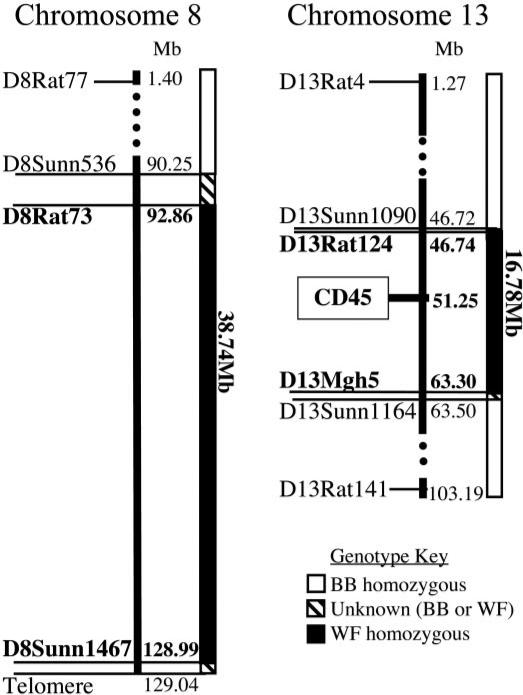
FIG. 2.
Physical mapping of congenic intervals on chromosomes 8 and 13 of the BB.WF chromosome 8/13 strain. Physical location of relevant microsatellite markers is indicated in megabases (RGSC 3.4 assembly, available at http://www.ensembl.org/Rattus_norvegicus/index.html, accessed 8 February 2007). The introgressed Wistar Furth regions are shown in black, while the BBDP background is shown in white. Marker names are indicated in bold text when the allele is of Wistar Furth origin and in plain text when of BB origin. The Wistar Furth introgressed region on chromosome 8 is ≤38.74 Mb in length, and the introgressed region on chromosome 13 is ≤16.78 Mb.
FIG. 4.
A: Six BB.WF chromosome 8 congenic sublines, each harboring different and/or overlapping sections of Wistar Furth alleles on chromosome 8, were developed. The position of relevant microsatellite markers are shown on the map of chromosome 8 on the left in megabases. Regions that are Wistar Furth in origin are shown in black and regions of BBDP origin shown in white. Areas bordering each congenic interval that have no marker coverage are shown in diagonal stripes. The size of each introgressed interval is indicated in megabases. Above each line, the cumulative incidence of type 1 diabetes (T1D) is shown with highly susceptible and resistant lines designated + and –, respectively. Congenic sublines BB.WF chromosome 8a, 8b, and 8c are highly susceptible to type 1 diabetes with ≥74% cumulative incidence, whereas BB.WF chromosome 8d and 8e, as well as the congenic retaining the whole fragment (BB.WF chromosome 8), are resistant to type 1 diabetes with cumulative incidences ≤41% (P < 0.0001 between type 1 diabetes + and – sublines). B: The rates of type 1 diabetes–free survival in BB.WF congenic sublines, which carry different Wistar Furth intervals of the chromosome 8 region. The congenic sublines BB.WF 8a, 8b, and 8c, which retain proximal portions of chromosome 8 Wistar Furth alleles, and the BB.WF chromosome 13 line are all highly susceptible to type 1 diabetes with no significant differences between their four survival curves. In contrast, the BB.WF chromosome 8 line and 8d and 8e congenic sublines, which retain distal portions of chromosome 8 Wistar Furth alleles, are all highly resistant to type 1 diabetes, again with no significant differences between their three survival curves. However, highly significant differences (P < 0.0001 by Kaplan-Meier analysis log-rank statistic) were found between the survival curves of any of the susceptible and any of the resistant lines.
RESULTS AND DISCUSSION
CD45, a tyrosine protein phosphatase expressed on the surface of all hemopoietic cells, exists in two allelic forms in the rat that can be distinguished by specific mAbs. To facilitate lymphocyte transfer experiments, we developed CD45 congenic BBDP rats through introgression of the Wistar Furth RT7.2 allele onto the BBDP (normally RT7.1) genetic background as described above. Unexpectedly, we observed a markedly reduced incidence of type 1 diabetes in this congenic line (Fig. 1). Specifically, 31% of the RT7.2 congenic rats (n = 36) vs. 86% of BBDP rats (n = 14) developed type 1 diabetes by 120 days (P < 0.0001; Kaplan-Meier log-rank statistic). Importantly, prospective analysis of pancreatic histology did not reveal significant differences in the kinetics or severity of islet inflammation between BBDP and RT7.2 congenic strains (data not shown). The approximate threefold reduction in type 1 diabetes incidence suggests that Wistar Furth alleles harbored within the congenic line confer diabetes resistance on a BBDP genetic background. The type 1 diabetes incidence in F1(BBDP × RT7.2 congenics), 37%, was similar to that of the RT7.2 congenic line but significantly (P = 0.0006; Kaplan-Meier log-rank statistic) reduced compared with BBDP rats, indicating a dominant mode of inheritance of the Wistar Furth–derived type 1 diabetes resistance (Fig. 1).
FIG. 1.
Analysis of type 1 diabetes–free survival in BBDP and congenic strains. The number of days to onset of diabetes is labeled on the horizontal axis. Data from the parental BBDP strain are compared with both the original double-congenic line and F1 offspring of the two. Both the congenic line and heterozygous animals have significantly decreased disease incidence compared with BBDP (P < 0.0001 and <0.0006 respectively, by Kaplan-Meier analysis log-rank statistic).
To find out whether Wistar Furth–derived chromosomal regions other than the CD45 locus had also been introgressed into the congenic line, we used 208 genetic markers covering the genome at an average interval of <10 Mb (maximum intermarker distance 19 Mb; maximum marker-to-telomere/centromere distance 23 Mb). This analysis revealed that over 38 Mb of the telomeric end of chromosome 8 had also been captured from the Wistar Furth strain (Fig. 2). No other chromosomal regions of Wistar Furth origin were found, and, importantly, Iddm12 previously linked to type 1 diabetes induced by poly I:C + anti-ART2.1.1 mAb (16) was excluded from the chromosome 13 congenic interval.
The apparent resistance to recombination in this large Wistar Furth–derived chromosome 8 interval during >10 backcrosses may have been due to a chromosomal rearrangement at this location. To determine whether this was the case and to map the type 1 diabetes resistance locus, we performed an F2 intercross between the RT7.2 congenic line and BBDP rats. F2 animals (n = 134) exhibited normal recombination rates in both congenic regions. Furthermore, linkage analysis of this cohort revealed highly significant linkage on chromosome 8 (>99.9% confidence), with age of type 1 diabetes onset (peak LOD 4.72) between D8Rat119 and D8Rat118 and with significant linkage (>95% confidence) with type 1 diabetes (peak LOD 2.92) at D8Rat119 (Fig. 3). No linkage of either trait to the chromosome 13 interval was found. As expected, given the similarity in pancreatic pathology between the two parental lines, insulitis and its severity failed to reach significant linkage to either interval. However, the strong linkage of disease incidence and age of onset to a region that does not seem to influence insulitis suggests that the chromo-some 8 locus may control either the cellular composition of insulitis and/or a step of the diabetogenic process occurring after the development of insulitis.
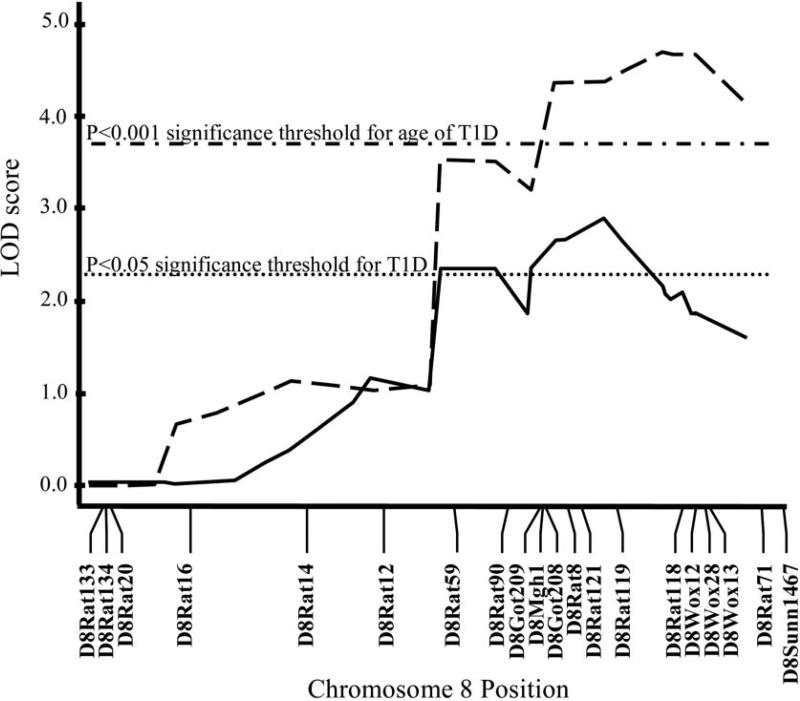
FIG. 3.
Linkage analysis using an F2 (BBDP × BB.WF chromosome 8/13) intercross (n = 134), showing significant linkage with both type 1 diabetes (T1D, solid line) and age of type 1 diabetes onset (dashed line). LOD scores were calculated using Windows QTL Cartographer, version 2.5 (25). MAPMAKER/EXP, version 3.0 (23,24), was used to construct the genetic linkage map using the 18 microsatellite markers shown. LOD score thresholds were determined by permutation testing (n = 1,000 permutations) (27). Significant loci were defined as those that exceeded the 95th percentile (P < 0.05, dotted line) of permutation distribution, while highly significant loci exceeded the 99.9th percentile (P < 0.001, dash-dotted line). Highly significant linkage (>99.9% confidence) was reached with age of type 1 diabetes onset (peak LOD 4.72) between D8Rat119 (120.53 Mb) and D8Rat118 (123.89 Mb), and significant linkage (>95% confidence) was reached with type 1 diabetes (peak LOD 2.92) at D8Rat119.
To confirm the results of the linkage analysis, congenic sublines isolating the entire chromosome 8 and 13 intervals in homozygous form were developed as described in RESEARCH DESIGN AND METHODS and their type 1 diabetes susceptibility assessed (Fig. 4B). The incidence of type 1 diabetes in BBDP Wistar Furth (BB.WF) chromosome 13 rats (82%, n = 40) was similar to that of BBDP rats but significantly higher (P < 0.0001) than that of BB.WF chromosome 8 rats (41%, n = 61). Importantly, the incidence of type 1 diabetes in BB.WF chromosome 8 rats was similar to that of the original double-congenic line. Furthermore, type 1 diabetes onset in BB.WF chromosome 8 rats was significantly delayed (P < 0.0001) compared with both BBDP and BB.WF chromosome 13 strains (Fig. 4B).
To refine the location of the chromosome 8 locus, five additional sublines dissecting this locus were developed and their type 1 diabetes susceptibility assessed (Fig. 4A). Sublines BB.WF chromosome 8a, 8b, and 8c rats are still highly susceptible to type 1 diabetes, with type 1 diabetes incidences >74%, while sublines BB.WF chromosome 8d and 8e rats are highly resistant to type 1 diabetes (33% type 1 diabetes incidence; P < 0.0001 vs. sublines BB.WF chromosome 8a, 8b, and 8c) (Fig. 4B). There were no significant differences in age of type 1 diabetes onset between sublines BB.WF chromosome 8d and 8e nor between sublines BB.WF chromosome 8a, 8b, and 8c. However, type 1 diabetes onset was significantly (P < 0.0001) delayed in BB.WF chromosome 8d rats when compared with BB.WF 8a, 8b, and 8c animals. The differential type 1 diabetes susceptibility of BB.WF chromosome 8c and 8e sublines shows that one or more factors necessary for type 1 diabetes resistance is present in the telomeric 10.34 Mb of chromosome 8. Using the December 2004 RGSC 3.4 assembly, the coordinates for this region, from D8Rat121 to the telomere of chromosome 8, are 118,705,294 to 129,041,809. The congenic line BB.WF chromosome 8e, harboring Iddm24, has been registered on the Rat Genome Database as RGD_ID no. 1599674. It is possible that type 1 diabetes resistance requires an interaction between this factor and other loci and that their separation during the development of congenic sublines disrupts this resistance effect. The lack of significant difference in age of onset between the type 1 diabetes–resistant BB.WF chromosome 8e line and each of the susceptible lines may reflect such a complex gene interaction within Iddm24. However, since both BB.WF chromosome 8d and 8e sublines are resistant to type 1 diabetes, all of the Wistar Furth elements necessary for resistance must be present within the telomeric 20.39 Mb of chromosome 8.
The identification of a novel Iddm locus on chromosome 8 was unexpected because no linkage to this region has previously been reported. Even more surprising was the identification of this locus in a cross between BBDP and Wistar Furth rats, which have previously been used to map Iddm loci (16). There are several possible but not exclusive explanations for this apparent discrepancy. First, the size of the cohort used in this previous mapping study was relatively small and hence could have missed an effect from chromosome 8. More importantly, this genetic analysis was applied to a cohort of animals in which type 1 diabetes did not develop spontaneously but was induced by treatment with poly I:C + anti-ART2.1 mAb (16). The differential contribution of some Iddm loci to spontaneous and experimentally induced type 1 diabetes has been well established. For example, the Iddm1 u haplotype is required for both syndromes. In contrast, the BBDP Iddm2 allele is necessary for spontaneous type 1 diabetes, while wild-type Gimap5 is required for virus-induced, experimentally induced type 1 diabetes (16). This suggests that the experimental procedures used for type 1 diabetes induction can bypass some of the steps leading to the spontaneous development of type 1 diabetes.
This led us to determine whether the Wistar Furth–derived Iddm24 allele confers resistance not only to spontaneous type 1 diabetes but to experimentally induced type 1 diabetes as well. To this end, nonlymphopenic BBnon-lyp rats were crossed with the lymphopenic BB.WF chromosome 8e subline and the susceptibility of the nonlymphopenic F1 offspring to type 1 diabetes induced by poly I:C + anti-ART2.1 mAb was assessed. All F1 rats (n = 10) developed type 1 diabetes within 35 days of treatment. This 100% incidence is similar to that previously reported in (BBDP × Wistar Furth) F1 (16) and indicates that the Wistar Furth–derived Iddm24 allele, while conferring dominant resistance to spontaneous type 1 diabetes, either does not protect against this form of experimentally induced type 1 diabetes or only confers resistance in a recessive manner. To distinguish between these two possible explanations, F1 animals were intercrossed and susceptibility to type 1 diabetes induced by poly I:C + anti-ART2.1 was assessed in the nonlymphopenic F2 rats that were Wistar Furth or BB homozygous or Wistar Furth or BB heterozygous for the 20.39 Mb telomeric fragment of chromosome 8, because the animals were either WF or BB homozygous. All of the F2 animals developed type 1 diabetes independent of their genotype at Iddm24 (10/10 Wistar Furth/Wistar Furth, 7/7 BB/BB, and 10/10 Wistar Furth/BB rats), demonstrating that this locus only affects susceptibility to spontaneous type 1 diabetes, hence its lack of detection in previous studies. This locus contains a cluster of cytokine and chemokine receptors. It is not implausible that allelic variation in one or more of these genes could result in a distinct pattern of differentiation and/or tissue migration of T-cell subsets required for the destruction of β-cells.
ACKNOWLEDGMENTS
This work was supported by grants from Genome Canada through the Ontario Genomics Institute and the Canadian Institutes of Health Research. R.H.W. is supported by a postdoctoral fellowship award from the Canadian Diabetes Association.
We thank Dominique Gauguier for generously providing microsatellite markers and polymorphism information and Trista Murphy for technical assistance.
Glossary
- BBDP
biobreeding diabetes prone
- BBDR
biobreeding diabetes resistant
- LOD
logarithm of odds
- mAb
monoclonal antibody
- RGSC
Rat Genome Sequencing Consortium
REFERENCES
- 1.Mordes JP, Poussier P, Rossini AR, Blankenhorn EP, Greiner DL. Rat Models of Type 1 Diabetes: Genetics, Environment and Autoimmunity. CRC Press; Boca Raton, FL: 2007. pp. 1–39. [DOI] [PubMed] [Google Scholar]
- 2.Colle E, Fuks A, Poussier P, Edouard P, Guttmann RD. Polygenic nature of spontaneous diabetes in the rat: permissive MHC haplotype and presence of the lymphopenic trait of the BB rat are not sufficient to produce susceptibility. Diabetes. 1992;41:1617–1623. doi: 10.2337/diab.41.12.1617. [DOI] [PubMed] [Google Scholar]
- 3.Colle E, Guttmann RD, Fuks A. Insulin-dependent diabetes mellitus is associated with genes that map to the right of the class I RT1: a locus of the major histocompatibility complex of the rat. Diabetes. 1986;35:454–458. doi: 10.2337/diab.35.4.454. [DOI] [PubMed] [Google Scholar]
- 4.Colle E, Guttmann RD, Seemayer T. Spontaneous diabetes mellitus syndrome in the rat. I. Association with the major histocompatibility complex. J Exp Med. 1981;154:1237–1242. doi: 10.1084/jem.154.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colle E, Ono SJ, Fuks A, Guttmann RD, Seemayer TA. Association of susceptibility to spontaneous diabetes in rat with genes of major histo-compatibility complex. Diabetes. 1988;37:1438–1443. doi: 10.2337/diab.37.10.1438. [DOI] [PubMed] [Google Scholar]
- 6.Ellerman KE, Like AA. Susceptibility to diabetes is widely distributed in normal class IIu haplotype rats. Diabetologia. 2000;43:890–898. doi: 10.1007/s001250051466. [DOI] [PubMed] [Google Scholar]
- 7.Hornum L, Romer J, Markholst H. The diabetes-prone BB rat carries a frameshift mutation in Ian4, a positional candidate of Iddm1. Diabetes. 2002;51:1972–1979. doi: 10.2337/diabetes.51.6.1972. [DOI] [PubMed] [Google Scholar]
- 8.MacMurray AJ, Moralejo DH, Kwitek AE, Rutledge EA, Van Yserloo B, Gohlke P, Speros SJ, Snyder B, Schaefer J, Bieg S, Jiang J, Ettinger RA, Fuller J, Daniels TL, Pettersson A, Orlebeke K, Birren B, Jacob HJ, Lander ES, Lernmark A. Lymphopenia in the BB rat model of type 1 diabetes is due to a mutation in a novel immune-associated nucleotide (Ian)-related gene. Genome Res. 2002;12:1029–1039. doi: 10.1101/gr.412702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramanathan S, Poussier P. BB rat lyp mutation and type 1 diabetes. Immunol Rev. 2001;184:161–171. doi: 10.1034/j.1600-065x.2001.1840115.x. [DOI] [PubMed] [Google Scholar]
- 10.Klaff LS, Koike G, Jiang J, Wang Y, Bieg S, Pettersson A, Lander E, Jacob H, Lernmark A. BB rat diabetes susceptibility and body weight regulation genes colocalize on chromosome 2. Mamm Genome. 1999;10:883–887. doi: 10.1007/s003359901108. [DOI] [PubMed] [Google Scholar]
- 11.Kloting II, van den Brandt J, Kovacs P. Quantitative trait loci for blood glucose confirm diabetes predisposing and protective genes, Iddm4 and Iddm5r, in the spontaneously diabetic BB/OK rat. Int J Mol Med. 1998;2:597–601. doi: 10.3892/ijmm.2.5.597. [DOI] [PubMed] [Google Scholar]
- 12.Kloting N, Kloting I. Congenic mapping of type 1 diabetes–protective gene(s) in an interval of 4 Mb on rat chromosome 6q32. Biochem Biophys Res Commun. 2004;323:388–394. doi: 10.1016/j.bbrc.2004.08.104. [DOI] [PubMed] [Google Scholar]
- 13.Jacob HJ, Pettersson A, Wilson D, Mao Y, Lernmark A, Lander ES. Genetic dissection of autoimmune type I diabetes in the BB rat. Nat Genet. 1992;2:56–60. doi: 10.1038/ng0992-56. [DOI] [PubMed] [Google Scholar]
- 14.Fowell D, Mason D. Evidence that the T-cell repertoire of normal rats contains cells with the potential to cause diabetes: characterization of the CD4+ T-cell subset that inhibits this autoimmune potential. J Exp Med. 1993;177:627–636. doi: 10.1084/jem.177.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guberski DL, Thomas VA, Shek WR, Like AA, Handler ES, Rossini AA, Wallace JE, Welsh RM. Induction of type I diabetes by Kilham's rat virus in diabetes-resistant BB/Wor rats. Science. 1991;254:1010–1013. doi: 10.1126/science.1658938. [DOI] [PubMed] [Google Scholar]
- 16.Martin AM, Blankenhorn EP, Maxson MN, Zhao M, Leif J, Mordes JP, Greiner DL. Non–major histocompatibility complex–linked diabetes susceptibility loci on chromosomes 4 and 13 in a backcross of the DP-BB/Wor rat to the WF rat. Diabetes. 1999;48:50–58. doi: 10.2337/diabetes.48.1.50. [DOI] [PubMed] [Google Scholar]
- 17.Martin AM, Maxson MN, Leif J, Mordes JP, Greiner DL, Blankenhorn EP. Diabetes-prone and diabetes-resistant BB rats share a common major diabetes susceptibility locus, iddm4: additional evidence for a “universal autoimmunity locus” on rat chromosome 4. Diabetes. 1999;48:2138–2144. doi: 10.2337/diabetes.48.11.2138. [DOI] [PubMed] [Google Scholar]
- 18.Blankenhorn EP, Rodemich L, Martin-Fernandez C, Leif J, Greiner DL, Mordes JP. The rat diabetes susceptibility locus Iddm4 and at least one additional gene are required for autoimmune diabetes induced by viral infection. Diabetes. 2005;54:1233–1237. doi: 10.2337/diabetes.54.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramanathan S, Bihoreau MT, Paterson AD, Marandi L, Gauguier D, Poussier P. Thymectomy and radiation-induced type 1 diabetes in nonlymphopenic BB rats. Diabetes. 2002;51:2975–2981. doi: 10.2337/diabetes.51.10.2975. [DOI] [PubMed] [Google Scholar]
- 20.Wallis RH, Wallace KJ, Collins SC, McAteer M, Argoud K, Bihoreau MT, Kaisaki PJ, Gauguier D. Enhanced insulin secretion and cholesterol metabolism in congenic strains of the spontaneously diabetic (type 2) Goto Kakizaki rat are controlled by independent genetic loci in rat chromosome 8. Diabetologia. 2004;47:1096–1106. doi: 10.1007/s00125-004-1416-5. [DOI] [PubMed] [Google Scholar]
- 21.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 23.Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 24.Lincoln S, Daly M, Lander E. Whitehead Institute Technical Report. Whitehead Institute; Cambridge, MA: 1992. Constructing Genetic Maps With MAP-MAKER/EXE 3.0. [Google Scholar]
- 25.Wang S, Basten CJ, Zeng Z-B. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University; Raleigh, NC: 2005. [Google Scholar]
- 26.Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]