Abstract
The plasma membrane quality control system of eukaryotic cells is able to recognize and degrade damaged cell surface proteins. Recent studies have identified two mechanisms involved in the recognition of unfolded transmembrane proteins. One system uses chaperones to detect unfolded cytoplasmic domains of transmembrane proteins, whereas the second mechanism relies on an internal quality control system of the protein, which can trigger degradation when the protein deviates from the folded state. Both quality control mechanisms are key to prevent proteotoxic effects at the cell surface and to ensure cell integrity.
Protein degradation is crucial for proper cellular function. It can fine-tune cellular pathways by reducing or eliminating the activity of a particular protein or clear away nonfunctional or dysfunctional proteins that can arise upon damage or unfolding. For the latter case, cells use numerous monitoring systems to identify unfolded proteins and trigger their rapid degradation before cell damage can occur. The best-studied systems of cellular protein quality control (QC) are those of soluble, cytoplasmic proteins. In addition, highly efficient QC systems act at the plasma membrane where the accumulation of even a few damaged transmembrane proteins could cause loss of cell integrity and death. As a consequence, cells appear to have evolved multilayered and efficient QC systems that function at different locations and use different mechanisms to ensure fidelity in the recognition of damaged proteins. This review focuses on the recent advances in the understanding how eukaryotic cells detect unfolded cell surface proteins. For more information on the pathways that function in the degradation of plasma membrane protein we refer to previously published reviews (Okiyoneda et al., 2011; MacGurn et al., 2012).
A key feature of QC pathways that target soluble proteins is the recognition of unfolded proteins by molecular chaperones that bind to exposed hydrophobic regions of unfolded proteins, assist in refolding, and if refolding fails, assist in the degradation of the damaged protein (Chen et al., 2011; Kästle and Grune, 2012; Doyle et al., 2013). Guided by these insights from cytoplasmic protein QC, researchers have for many years focused on the identification of chaperone-like factors that might act similarly as sensors for unfolded transmembrane proteins at the plasma membrane. Several studies have used the model organism Saccharomyces cerevisiae due to its genetic tractability to identify quality control factors that are responsible for the rapid degradation of unfolded cell surface proteins (Li et al., 1999; Liu and Chang, 2006; Lin et al., 2008; Wang et al., 2011). Depending on the model protein and the method used for the analysis, these studies identified many factors involved in endocytosis, endosomal trafficking, and vacuolar/lysosomal degradation of cell surface proteins. In fact, these studies confirmed or identified the function of many proteins that play a role in the general degradation route for cell surface proteins, referred to as the multivesicular body (MVB) pathway (Henne et al., 2011; Hurley and Stenmark, 2011; Babst and Odorizzi, 2013), but did not identify specific quality control proteins, which shall be discussed later.
Degradation of cell surface proteins is mediated by the MVB pathway
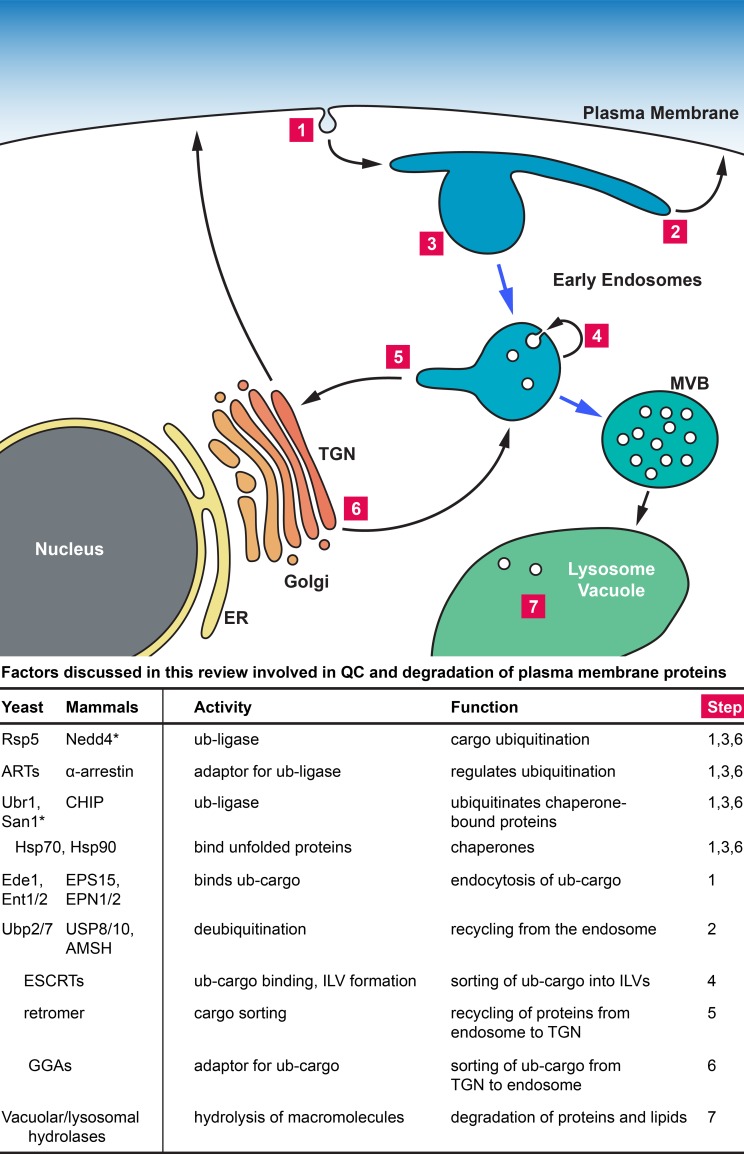
In yeast, degradation of most plasma membrane proteins requires the ubiquitin ligase Rsp5, a HECT-type E3 ligase that tags proteins destined for degradation with ubiquitin (Hein et al., 1995; Wang et al., 1999; Belgareh-Touzé et al., 2008). Rsp5 seems to perform all the ubiquitination reactions associated with the endocytic pathway. Although a single ubiquitin has been shown to be sufficient to trigger degradation of the tagged protein (Terrell et al., 1998), polyubiquitination of cell surface proteins seems to be common (Lauwers et al., 2010). The ubiquitinated transmembrane cargo is then recognized by factors of the endocytosis machinery, such as Ede1, and clathrin-mediated endocytosis delivers the ubiquitinated protein to an early endosome (Fig. 1, step 1; Haglund and Dikic, 2012). At the endosome, the cargo (proteins sorted through the endosomal system) has two options: recycling to the cell surface either directly or indirectly via the trans-Golgi (Fig. 1, step 2 and 5, respectively) or continuing to the vacuole for degradation. Although the precise mechanism is not known, it is thought that competing de-ubiquitination and re-ubiquitination reactions at the endosome are key to deciding the fate of the cargoes (Piper et al., 2014). This important regulatory step of the endocytic pathway is discussed in more detail later. If ubiquitination prevails, the cargo will be captured by the ESCRT (endosomal sorting complex required for transport) machinery, a complex of proteins that sort ubiquitin-tagged transmembrane proteins into vesicles that bud into the lumen of the endosome (Fig. 1, step 4; Hurley, 2010). The presence of intraluminal vesicles (ILVs) is a hallmark of late endosomes, which are thus also called multivesicular bodies (MVBs).
Figure 1.
Trafficking pathways of cell surface proteins in eukaryotic cells. Numbers refer to trafficking steps that are involved in QC. *, function in QC of transmembrane proteins has not been shown; ub, ubiquitin.
Several of the ESCRT proteins contain ubiquitin-binding domains that interact with tagged cargo and initiate their sorting into the ILVs (Shields and Piper, 2011). The ESCRTs are also responsible for the formation of the ILVs, a membrane deformation event with a topology reversed to that of all other vesicle formation events of the cell. During the packaging of the cargo into the ILVs, ubiquitin is removed from the cargo and recycled for further use (Amerik et al., 2000; Luhtala and Odorizzi, 2004). At this stage, the transmembrane cargo is isolated from the cytoplasm and therefore no longer able to affect cellular function. After the MVB is fully matured (all the protein sorting has occurred), the limiting membrane of the MVB fuses with the vacuolar membrane and releases the ILVs into the lumen of the vacuole. The lumen of the vacuole contains a large number of hydrolases that degrade the lipids and proteins contained within the vesicles (Fig. 1, step 7).
Protein sorting at the TGN also feeds into the MVB pathway. The TGN receives both newly synthesized transmembrane proteins that passed the ER QC system and proteins that have been retrieved from the endosome with the help of the retromer protein complex (Fig. 1, step 5; Seaman, 2012). The TGN QC system seems to be able to recognize unfolded proteins and tag these proteins with ubiquitin (MacGurn et al., 2012). Ubiquitinated proteins are then bound by sorting receptors such as the GGA proteins that concentrate the ubiquitinated cargoes into vesicles destined for the endosome (Fig. 1, step 6; Puertollano and Bonifacino, 2004; Scott et al., 2004; Shiba et al., 2004). At the endosome the ubiquitinated proteins are sorted into the MVB pathway and delivered to the lysosome for degradation. Examples of TGN-based QC in yeast are the rapid degradation of mutant forms of Pma1 (plasma membrane ATPase; Pizzirusso and Chang, 2004) and Wsc1 (cell wall integrity sensor; Wang et al., 2011). It should be noted that the TGN not only sorts unfolded proteins to the endosome for degradation. For example, at high nutrient concentrations, newly synthesized nutrient transporters are redirected at the TGN away from the secretory pathway toward the endosome by a ubiquitin-dependent sorting mechanism (Blondel et al., 2004). Therefore, similar to endocytosis at the plasma membrane, the TGN-to-endosome trafficking pathway serves not only protein QC but functions in the general protein turnover.
The studies in yeast identified many genes that not only function as protein QC factors but were also involved in the general degradation of transmembrane proteins (see Box 1 with regard to the definition of QC factors). However, the question remained, which factors are specifically required for the quality control of plasma membrane proteins? A first answer to this question was obtained by QC studies in mammalian cells.
Box 1. What is a QC factor?
The term “QC factor” is not clearly defined and is used differently by researchers in the field. A broad definition of a QC factor would be: A factor that plays a role in the turnover of an unfolded protein. This definition includes any factors that are involved in the degradation of the unfolded protein, such as ubiquitin ligases, vacuolar peptidases, the endocytic machinery, and proteins involved in fusion of endosomal membranes. A much more narrow definition for QC factor is: A factor that is only involved in the degradation of unfolded proteins. Based on this definition, none of the above-mentioned proteins and protein complexes would qualify as QC factors. Both of these definitions are problematic because of the overlap between systems involved in protein QC and other degradation pathways. Therefore, terms such as “folding sensors” (systems that recognize unfolding), “pro-folding factors” (proteins that support folding), and “degradation machinery” (general protein degradation system) might be better suited to define the function of a particular protein involved in QC.
Chaperone-dependent QC of plasma membrane proteins
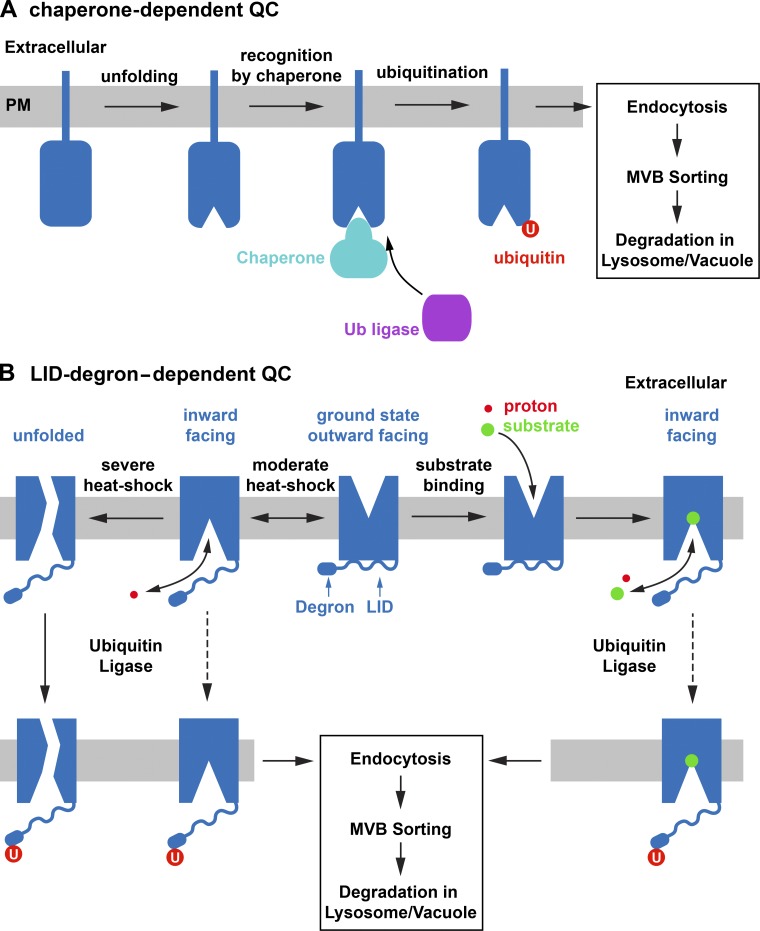
Two recent studies using mammalian cell lines identified QC genes important for the rapid turnover of unfolded model plasma membrane proteins. One study used an artificial fusion protein composed of a plasma membrane anchor and a protein domain that unfolds at elevated temperature (mutant bacteriophage lambda repressor) to identify QC factors (Apaja et al., 2010). The findings based on this fusion protein were then confirmed using mutant forms of G protein–coupled receptors (GPCRs). The second study was based on the unstable CFTR mutant ΔF508, a mutated ABC transporter that is the most common cause of the genetic disorder cystic fibrosis (Okiyoneda et al., 2010). Interestingly, the molecular chaperones Hsp70 and Hsp90 and the ubiquitin ligase CHIP were found by both studies to be important for plasma membrane QC (Fig. 1). In both cases, the data suggested that chaperones are recognizing unfolded cytoplasmic regions of the cell surface proteins and that these chaperones recruit the ubiquitin ligase CHIP. The ubiquitinated proteins were then rapidly endocytosed and delivered in an ESCRT-dependent manner to the lysosome for degradation (Fig. 2 A).
Figure 2.
Plasma membrane protein QC mechanisms. (A) Chaperones recognize unfolded cytoplasmic domains and recruit a ubiquitin ligase. Ubiquitinated proteins are rapidly endocytosed and degraded via the MVB pathway. PM, plasma membrane. (B) Nutrient transporters use the same intrinsic mechanism, the LID-degron system, for both substrate-dependent degradation and QC. Deviations from the ground state of the transporter, caused either by substrate binding or by stress (e.g., heat shock), result in ubiquitination of the degron. A moderate heat shock might trigger the conformational changes of the import cycle even in absence of substrate. Ubiquitinated transporters are rapidly endocytosed and degraded. Ubiquitination efficiency of substrate-bound transporters depends on the cytoplasmic substrate concentration (high concentrations stabilize the substrate-bound state).
The mammalian studies concluded that the plasma membrane QC depends on the same players that are responsible for recognizing damaged cytoplasmic proteins. The main difference is the degradation pathway the proteins take after ubiquitination: degradation by the proteasome in the case of soluble proteins and the MVB pathway in the case of cell surface proteins. It is interesting to note that CHIP modifies plasma membrane proteins preferentially with K63-linked polyubiquitin chains (Apaja et al., 2010, 2013), the preferred ubiquitin tag for the MVB pathway (Lauwers et al., 2009; Erpapazoglou et al., 2012), whereas in the cytoplasm CHIP mainly adds K48-type ubiquitin chains to its substrates (Xu et al., 2009).
It is surprising that the various genetic screens in yeast did not identify these components of the chaperone system as being involved in the QC of peripheral transmembrane proteins. One obvious explanation might be functional redundancy among the chaperones, which would make the identification by a genetic screen very unlikely. However, a study in yeast using a similar temperature-sensitive protein domain that was used in the mammalian experiments (a mutant lambda repressor) found that anchoring the domain to the plasma membrane prevented the rapid degradation of the unfolded domain (Lewis and Pelham, 2009). This observation might indicate that, in contrast to the mammalian system, QC at the yeast plasma membrane and cytoplasm differs, either in the recognition of unfolded regions by chaperones or in the recruitment of the ubiquitin ligase. For one, yeast does not express a ubiquitin ligase homologous to mammalian CHIP. Two yeast ligases, Ubr1 and San1, have been identified that seem to function similarly to CHIP in cytoplasmic QC (Heck et al., 2010). However, all studies of peripheral QC in yeast identified Rsp5 as the major ubiquitin ligase responsible for the degradation of damaged plasma membrane proteins. Rsp5 is not known to interact with chaperones, and thus it is possible that yeast does not rely on a chaperone-based system for its plasma membrane QC. That said, QC of only a small set of yeast surface proteins has been studied and therefore it is possible that future studies might find cases where plasma membrane QC is chaperone dependent.
The ART proteins
If chaperones are not essential for plasma membrane QC in yeast, what mechanism recognizes the unfolded state of cell surface proteins? This is a difficult question to answer because the reason why a particular protein is degraded is often not known. In the case of nutrient transporters, at least three reasons have been described that can trigger the rapid degradation of these proteins. Many transporters are rapidly degraded in the presence of high concentrations of the nutrient they import, referred to as substrate-dependent down-regulation (Blondel et al., 2004; Felice et al., 2005). This negative feedback system ensures that the cytoplasmic concentration of the nutrient does not exceed a level that could negatively affect the metabolism of the cell or even become toxic. The rapid degradation of transporters is also caused by cellular responses to stress such as amino acid starvation (Jones et al., 2012) and exposure to the translation inhibitor cycloheximide (MacGurn et al., 2011). Finally, nutrient transporters have been shown to be endocytosed and degraded in the presence of cellular insults such as heat shock and exposure to toxic chemicals, which are thought to cause protein unfolding (Volland et al., 1994; Keener and Babst, 2013). Of these, only the latter reason for protein degradation would be considered quality control. However, factors identified in yeast to be important for heat shock–induced degradation of nutrient transporters also play a role in the degradation of the transporters under all other conditions. A good example is the class of proteins called the arrestin-related transport receptors (ARTs; Becuwe et al., 2012a). The first member of this group of proteins was found in a genetic screen that aimed to identify genes involved in the quality control and degradation of the yeast arginine transporter Can1 (Lin et al., 2008). The ARTs function as Rsp5 adaptors for nutrient transporters in that they bind to transporters at the plasma membrane, TGN, and possibly endosome and aid in the recruitment of Rsp5 (Fig. 1). As a consequence, the ARTs increase the efficiency of Rsp5-dependent ubiquitination of nutrient transporters (Nikko et al., 2008; Nikko and Pelham, 2009). However, the ARTs do not function analogous to chaperones because the binding of ARTs to transporters does not seem to require unfolding of these transmembrane proteins. Furthermore, there is no data that would suggest that ARTs act as pro-folding factors. Finally, the ARTs are not folding sensors that recognize general features of unfolded proteins (e.g., exposed hydrophobic protein region), but instead function together with Rsp5 in the degradation of a specific set of cell surface transporters under all conditions. Nevertheless, the ART-Rsp5 system plays an essential role in the efficient QC of many cell surface transporters (Zhao et al., 2013).
A key aspect of the ARTs is the fact that these proteins can be regulated by phosphorylation and ubiquitination (Lin et al., 2008; MacGurn et al., 2011; Becuwe et al., 2012b; Merhi and André, 2012). Regulation of ART proteins has been observed as consequence of changes in the environment and the cell’s metabolism. Each ART protein targets a specific set of nutrient transporters, allowing the cell to fine-tune the turnover rate of these transporters dependent on the metabolic state. Increasing the degradation rate of a particular set of nutrient transporters is expected to cause a rapid drop in the cellular concentration of the corresponding nutrient. This regulatory mechanism is able to react faster to metabolic changes than transcriptional control alone could achieve. In addition, by targeting all ARTs, the cell is able to trigger down-regulation of most transporters in unison, a phenomenon that is observed during amino acid starvation and cycloheximide poisoning (MacGurn et al., 2011; Jones et al., 2012).
The LID-degron system, a protein-intrinsic QC mechanism
Studies of yeast Fur4 degradation have given important insights into the mechanism of nutrient transporter QC. Fur4, the uracil importer of yeast, belongs to the family of APC transporters, a large protein family conserved from bacteria to humans. These transporters contain 12 transmembrane domains and operate based on an alternative-access model, meaning that the transporter switches between a conformation that allows binding of extracellular substrate at a central binding pocket to a conformation in which substrate is released from the binding site into the cytoplasm (Boudker and Verdon, 2010; Forrest et al., 2011). Fur4 uses the proton gradient across the plasma membrane of yeast to import uracil from the extracellular space.
Similar to other nutrient transporters, high substrate concentrations in the medium trigger the rapid down-regulation of Fur4 (Séron et al., 1999). This degradation requires ubiquitination by Rsp5, which targets two lysine residues in the cytoplasmic N-terminal domain of Fur4 (Marchal et al., 2000). Ubiquitinated Fur4 is endocytosed and degraded via the MVB pathway. Surprisingly, the same two lysines ubiquitinated during substrate-dependent down-regulation are also required for the degradation of Fur4 under stress conditions such as heat shock or peroxide treatment, even though 15 other lysines are predicted to be exposed to the cytoplasm. Additional analysis of Fur4 turnover supported the idea that both substrate-dependent down-regulation and QC-dependent degradation rely on the same intrinsic, chaperone-independent mechanism, the so-called LID-degron system (Fig. 2 B; Keener and Babst, 2013). A degron refers to a domain containing lysines that are targeted for ubiquitination in a regulated manner. Degrons are often found in proteins where degradation is used to regulate the function of the protein (e.g., degradation of cyclins during cell cycle; Ravid and Hochstrasser, 2008). The LID (loop interaction domain) refers to the cytoplasmic ∼20 amino acids before the first transmembrane domain of Fur4. Based on the crystal structure analysis of a bacterial Fur4 homologue (Weyand et al., 2008), the LID is predicted to contact all cytoplasmic loops and the C-terminal region of Fur4 via hydrogen bonding (hence the name “loop interaction domain”). However, many of these interactions are only present when substrate is not bound to the transporter (referred to as ground state). Uracil binding induces large conformational changes in the transmembrane regions of Fur4 that cause disruption of LID–loop interactions. As a consequence, the LID is predicted to have increased flexibility when Fur4 is actively transporting substrate.
One functional consequence of this flexibility may be to expose the degron region of the protein, which allows the ubiquitin ligase Rsp5 access to the lysines of the degron. Therefore, the LID functions as a conformation sensor that detects deviations from the ground state and exposes a nearby degron. The degron becomes ubiquitinated, which triggers degradation of the transporter via endocytosis and the MVB pathway (Fig. 2 B). This system explains why degradation by high substrate concentrations or unfolding is mediated by the same mechanism: both conditions cause Fur4 to be in the nonground state.
Chaperone-mediated QC and the LID-degron system are similar in that both mechanisms rely on conformational changes that cause the exposure of a peptide sequence that can be recognized by the degradation machinery. However, an important difference between these two QC mechanisms is the fact that the LID-degron system does not require protein unfolding to cause ubiquitination. Protein conformations that are part of the normal substrate import mechanism can trigger the ubiquitination of Fur4’s degron. Chaperones bind to hydrophobic peptide sequences that, in a functional protein, are not exposed to the cytoplasm. An important function of this chaperone interaction is to prevent client protein aggregation and allow for refolding to occur. In contrast, the LID-degron system seems to specifically trigger degradation of the transporter. The LID shares hydrogen bonding with all cytoplasmic loops and thus is able to sense conformational changes that occur throughout the protein, in the transmembrane regions or the extracellular domains of the transporter. As a consequence, under various conditions, such as substrate binding, heat shock, or oxidation, the same Fur4 degron (lysine residues) is targeted for ubiquitination independent of the cause or type of conformational change. The LID functions as an internal sensor that directs the information of any larger conformational change of the transporter to a dedicated ubiquitination site, the degron. In contrast, chaperone-based QC targets the particular domain or region of the protein that unfolds, thereby directing the ubiquitin ligase to a lysine in proximity of the unfolded region. Therefore, depending on the insult, the lysine targeted for ubiquitination might differ.
Rsp5 recruitment by the LID-degron.
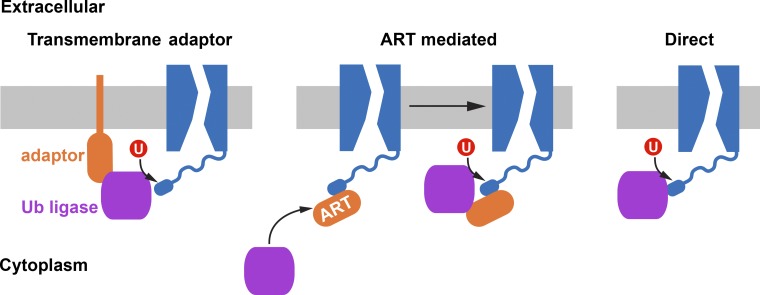
Currently it is not understood how cytoplasmic Rsp5 is recruited to the degron of Fur4. Experiments with modified versions of the degron suggested that the predicted increase in degron accessibility caused by the loss of LID–loop interactions is key for the ubiquitination reaction (Keener and Babst, 2013). Although no specific ART protein has been implicated in the recruitment of Rsp5 to Fur4, data suggest that the Rsp5 adaptors Bul1 and Bul2 might be involved in Fur4 ubiquitination (Nikko and Pelham, 2009). Alternatively, Rsp5 might be recruited to the plasma membrane by transmembrane adaptors such as Bsd2, Tre1, and Ear1, which have been shown to function in recruiting Rsp5 to Golgi and endosomes (Hettema et al., 2004; Sullivan et al., 2007; Léon et al., 2008). Membrane-associated Rsp5 might be able to directly recognize the exposed degron of Fur4 without the need of additional adaptors (Fig. 3). A third possibility is the direct recruitment of cytoplasmic Rsp5 to the exposed degron (Fig. 3). Although Fur4 does not contain PPXY (Pro-Pro-X-Tyr) motifs, amino acid sequences that have been shown to mediate Rsp5 recruiting (Lin et al., 2008), direct Rsp5 recruitment to Fur4 via a different type of interaction cannot be excluded. For example, direct substrate recognition without chaperones has been observed for the ubiquitin ligases Ubr1/2 and San1 (Nillegoda et al., 2010; Rosenbaum et al., 2011), which seem to be able to bind directly to unfolded protein regions. Similarly, Rsp5 might be able to recognize the exposed degron of Fur4 as an unfolded protein sequence and target it for ubiquitination.
Figure 3.
Possible mechanisms of ubiquitin ligase recruitment by the degron. The ubiquitin ligase might be recruited to transporters either by transmembrane adaptors, via arrestin-related trafficking adaptors (ARTs), or by direct binding to the degron region of the transporter.
Because Fur4 belongs to the conserved APC superfamily of transporters it is likely that the LID-degron system is found in many other family members. Several of these members (e.g., Can1, Mup1, Tat2, Lyp1) have been shown to require the ARTs for efficient degradation (Lin et al., 2008; Nikko and Pelham, 2009), suggesting that in these cases the ARTs might functionally interact with the LID-degron system. In fact, the LID-degron mechanism could explain how the ARTs are able to specifically interact with transporters that are either unfolded or substrate bound. In these cases the degron might function as the binding site for the ART proteins, which when bound to the transporter recruit Rsp5 (Fig. 3). This model is supported by data that localized the ART-binding site to the N-terminal, cytoplasmic tail of the transporters Lyp1 and Can1 (Lin et al., 2008), the same region that occupies the LID-degron in Fur4. Therefore, it seems likely that QC of most of the nutrient transporters may depend on a similar intrinsic conformation sensor, the LID, which regulates Rsp5 recruitment by regulating an ART-binding site or degron (Fig. 3). A similar mechanism is likely present in mammalian cells, where studies have shown cooperation between Nedd4-type ubiquitin ligases and ART homologues (referred to as α-arrestin) in the degradation of plasma membrane proteins (Foot et al., 2008; Patwari et al., 2009; Nabhan et al., 2010).
Activity-dependent down-regulation as a QC mechanism.
A unique aspect of the LID-degron system is the fact that it performs both the substrate-dependent down-regulation as well as QC. As a consequence, high substrate concentrations and insults on the protein (e.g., higher temperature) act synergistically in the turnover of the transporter. Therefore, in most cases it is impossible to distinguish between protein degradation caused by high import activity or by a temporary unfolding event. In particular, increasing temperatures are expected to cause first an increase in import activity because the energy barriers associated with the conformational changes required for nutrient import are easier to overcome at higher temperature. As a consequence, a moderate temperature increase is expected to cause a higher transporter turnover rate, even though substrate concentration is unchanged.
Increasing the temperature further might overcome the energy barriers of the import cycle even in the absence of substrate (Fig. 2 B). At this point the import mechanism is uncoupled from substrate binding and occurs randomly. This idea is supported by the observation that a temperature shift from 30°C to 37°C causes rapid substrate-independent degradation of Fur4, even though the same temperature shift does not render Fur4 unfolded and nonfunctional (Keener and Babst, 2013). Finally, at high temperatures we would expect to see transporter conformations that are not part of the normal import cycle, which would be referred to as “unfolding.” In any case, the LID-degron system would be able to recognize the conformational change and trigger degradation of the transporter (Fig. 2 B).
The model described above has important consequences for the QC not only of transporters such as Fur4, but any plasma membrane protein that (1) undergoes conformational changes as part of the protein’s function and (2) is down-regulated when highly active. There are numerous yeast and mammalian proteins that fit these two conditions, including ion channels and signaling receptors (Nabhan et al., 2010; Hyun et al., 2013). For example, the binding of an extracellular signaling molecule to a GPCR induces conformational changes in the receptor that activate signaling cascades on the cytoplasmic side of the membrane (Oldham and Hamm, 2008). Furthermore, for many GPCRs it has been shown that high signaling activity causes endocytosis and degradation of the receptor, a negative feedback system that plays an important regulatory role in many signaling pathways (Hanyaloglu and von Zastrow, 2008). It is conceivable that increased temperatures might cause GPCRs to randomly switch to the activated state, even in the absence of the signaling molecule. The result would be the rapid degradation of the GPCR because the high activity of the receptor would trigger the down-regulation mechanism of the signaling receptor. Therefore, analogous to the LID-degron system in Fur4, activity-dependent down-regulation of GPCRs might also function as a QC system for these receptors, a model that will have to be tested experimentally.
Activity-dependent down-regulation can explain the heat shock–induced turnover of many cell surface proteins. However, severe disruption of the protein conformation caused by chemicals, mutations, or very high temperatures might unfold the protein to an extent in which it is no longer able to trigger activation-dependent down-regulation. Under these conditions the chaperone-CHIP system might play the key role in QC of these plasma membrane proteins, as shown for mammalian GPCRs (Apaja et al., 2010).
The endosome reevaluates QC decisions
Endocytosis delivers ubiquitinated proteins to an early endosome, where the competition between de-ubiquitination and re-ubiquitination decides the fate of many of the endocytosed transmembrane proteins (Hurley and Stenmark, 2011; Piper et al., 2014). Proteins that have lost their ubiquitin tags are recycled back to the plasma membrane, whereas ubiquitinated proteins remain in the endosomal system and are ultimately degraded. Recycling to the plasma membrane occurs either via a fast, direct pathway or an indirect pathway via retromer-mediated recycling to the TGN (Fig. 1, step 2 and 5; Hsu and Prekeris, 2010; Seaman, 2012; Taguchi, 2013). The factors involved in regulating these recycling pathways are not well understood. In yeast, the major ligase involved in the ubiquitination of plasma membrane proteins, Rsp5, forms a complex with two de-ubiquitinating enzymes, Ubp2 and Ubp7 (Kee et al., 2005; Ren et al., 2007). This protein complex contains both ubiquitination and de-ubiquitination activities, and thus might be involved in the decision between degradation and recycling. However, Ubp2 seems also to positively regulate Rsp5 activity, which complicates the study of the role of endosomal de-ubiquitination in cargo recycling (Lam and Emili, 2013). In mammalian cells, several de-ubiquitinating enzymes have been identified that play an important role in the recycling of endosomal cargoes (Fig. 1; Mizuno et al., 2005; Bomberger et al., 2009; Hasdemir et al., 2009; Berlin et al., 2010; Meenhuis et al., 2011).
A surprising aspect of the endosomal sorting system is the observation that a large proportion of endocytosed proteins are ultimately returned to the cell surface (e.g., ∼75% recycling of CFTR; Swiatecka-Urban et al., 2005). This recycling step seems to be counterproductive and poses the question of why the proteins were endocytosed in the first place. In the case of nutrient transporters, a possible explanation for a high endocytosis and recycling rate might be found in the structural flexibility of these proteins. To facilitate nutrient import, transporters undergo large conformational changes that seem to require only thermal energy (Forrest et al., 2011). This observation suggests that even under normal growth conditions, and in the absence of substrate, these cell surface proteins have a high probability to flex and temporarily appear unfolded, thereby triggering ubiquitination and endocytosis of the protein. However, at the endosome, proteins seem to be de-ubiquitinated and “re-checked” (Fig. 1, step 3). One explanation may be that if by the time the transporter reaches the endosome it has regained the proper structure, re-ubiquitination does not occur and the protein recycles back to the plasma membrane. If the transporter remains at the endosome in an unfolded state, re-ubiquitination is efficient and the protein traffics via the MVB pathway to the vacuole for degradation. This second QC step at the endosome is likely mediated by the same mechanisms that are involved in plasma membrane QC (chaperones or LID system).
The combination of plasma membrane and endosomal QC is the solution to two competing goals of the cell. On the one hand, unfolded cell surface proteins are a serious danger to cellular integrity, and thus the plasma membrane QC system is tuned to endocytose proteins even when they are only transiently unfolded. On the other hand, the consequence of high protein turnover is a large energy cost to the cell because the degraded proteins have to be replaced. The endosomal QC system allows the cell to recheck the endocytosed protein at a safe place to ensure that mainly permanently damaged proteins are degraded.
Lipids and the QC of transmembrane proteins
The lipid environment plays a key role in the function and stability of many transmembrane proteins. A set of plasma membrane proteins localize to subdomains of the membrane, also referred to as rafts, that contain a unique lipid composition (Simons and Sampaio, 2011). Interfering with the lipid raft composition or the packaging of the transmembrane proteins into rafts can lead to loss of protein function and rapid degradation (Lauwers et al., 2007; Pineau et al., 2008; Payet et al., 2013). In yeast, this rapid protein turnover has been shown to require the ubiquitin ligase Rsp5. Therefore, one of the functions of the TGN and plasma membrane QC systems is to ensure that transmembrane proteins are surrounded by the proper lipid environment. Particularly, nutrient transporters would be expected to be sensitive to changes in fluidity of the surrounding membrane. These multi-spanning transmembrane proteins undergo large conformational changes as part of their transport activity and thus the fluidity of the surrounding membrane might affect the transport activity of these proteins. A rigid membrane interferes with the conformational changes necessary for transport and thus will inhibit normal function. In contrast, a fluid membrane will allow for too much flexibility of the transporter, and even in the absence of substrate, cause fast turnover of the protein via QC pathways. This model highlights how important a tight regulation of membrane fluidity is in order to adapt to changing environmental conditions such as temperature and salt concentration.
Concluding remarks
Recent studies have identified two different conformation-sensing mechanisms that function in the QC of membrane proteins beyond the ER. One mechanism uses a chaperone-based system to monitor membrane proteins and recognize unfolding events in cytoplasmic domains. The other mechanism, the LID system, is a sensor that is intrinsic to the proteins themselves. In the ground state of the protein, this intrinsic mechanism hides a ubiquitination site from ubiquitin ligases, but allows access to the site at any other conformational state. This system triggers rapid protein degradation even if the unfolded protein region is not accessible from the cytoplasm. It is possible that many multi-spanning transmembrane proteins use this or a similar system to ensure that unfolding events occurring within the transmembrane region or in extracellular domains are detected. In fact, the activity-dependent degradation mechanisms found for many channels, transporters, and signaling receptors is expected to function as a QC system that triggers degradation of these proteins during heat shock. Some proteins might combine several QC mechanisms to deal with unfolding events in different parts of the protein. Inefficient degradation of unfolded proteins can lead to toxic accumulation of these proteins and the subsequent death of the cell (Wang et al., 2011; Keener and Babst, 2013; Zhao et al., 2013). Therefore, amino acid sequences are under strong selection not only to produce functional proteins, but also proteins that if damaged are easily recognized and removed. As a consequence proteins have evolved to efficiently communicate with the QC systems of a particular cell and organelle. One important conclusion from this evolutionary view of protein unfolding is that proteins foreign to the organism or the organelle are not good tools for the study of QC systems. Future studies analyzing QC of endogenous cell surface proteins are likely to identify additional QC mechanisms unique for a subset of proteins or a particular stress situation.
Acknowledgments
I thank Matt Curtiss and Tamara Darsow for critical reading of the manuscript.
The research conducted in my laboratory on the ESCRT proteins is supported by National Institutes of Health grant R01 GM074171.
The author declares no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ART
- arrestin-related trafficking adaptor
- ESCRT
- endosomal sorting complex required for transport
- GPCR
- G protein–coupled receptor
- ILV
- intraluminal vesicle
- LID
- loop interaction domain
- MVB
- multivesicular body
- QC
- quality control
References
- Amerik A.Y., Nowak J., Swaminathan S., Hochstrasser M. 2000. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell. 11:3365–3380 10.1091/mbc.11.10.3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apaja P.M., Xu H., Lukacs G.L. 2010. Quality control for unfolded proteins at the plasma membrane. J. Cell Biol. 191:553–570 10.1083/jcb.201006012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apaja P.M., Foo B., Okiyoneda T., Valinsky W.C., Barriere H., Atanasiu R., Ficker E., Lukacs G.L., Shrier A. 2013. Ubiquitination-dependent quality control of hERG K+ channel with acquired and inherited conformational defect at the plasma membrane. Mol. Biol. Cell. 24:3787–3804 10.1091/mbc.E13-07-0417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Odorizzi G. 2013. The balance of protein expression and degradation: an ESCRTs point of view. Curr. Opin. Cell Biol. 25:489–494 10.1016/j.ceb.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becuwe M., Herrador A., Haguenauer-Tsapis R., Vincent O., Léon S. 2012a. Ubiquitin-mediated regulation of endocytosis by proteins of the arrestin family. Biochem. Res. Int. 2012:242764 10.1155/2012/242764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becuwe M., Vieira N., Lara D., Gomes-Rezende J., Soares-Cunha C., Casal M., Haguenauer-Tsapis R., Vincent O., Paiva S., Léon S. 2012b. A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. J. Cell Biol. 196:247–259 10.1083/jcb.201109113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh-Touzé N., Léon S., Erpapazoglou Z., Stawiecka-Mirota M., Urban-Grimal D., Haguenauer-Tsapis R. 2008. Versatile role of the yeast ubiquitin ligase Rsp5p in intracellular trafficking. Biochem. Soc. Trans. 36:791–796 10.1042/BST0360791 [DOI] [PubMed] [Google Scholar]
- Berlin I., Higginbotham K.M., Dise R.S., Sierra M.I., Nash P.D. 2010. The deubiquitinating enzyme USP8 promotes trafficking and degradation of the chemokine receptor 4 at the sorting endosome. J. Biol. Chem. 285:37895–37908 10.1074/jbc.M110.129411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel M.O., Morvan J., Dupré S., Urban-Grimal D., Haguenauer-Tsapis R., Volland C. 2004. Direct sorting of the yeast uracil permease to the endosomal system is controlled by uracil binding and Rsp5p-dependent ubiquitylation. Mol. Biol. Cell. 15:883–895 10.1091/mbc.E03-04-0202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomberger J.M., Barnaby R.L., Stanton B.A. 2009. The deubiquitinating enzyme USP10 regulates the post-endocytic sorting of cystic fibrosis transmembrane conductance regulator in airway epithelial cells. J. Biol. Chem. 284:18778–18789 10.1074/jbc.M109.001685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudker O., Verdon G. 2010. Structural perspectives on secondary active transporters. Trends Pharmacol. Sci. 31:418–426 10.1016/j.tips.2010.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Retzlaff M., Roos T., Frydman J. 2011. Cellular strategies of protein quality control. Cold Spring Harb. Perspect. Biol. 3:a004374 10.1101/cshperspect.a004374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S.M., Genest O., Wickner S. 2013. Protein rescue from aggregates by powerful molecular chaperone machines. Nat. Rev. Mol. Cell Biol. 14:617–629 10.1038/nrm3660 [DOI] [PubMed] [Google Scholar]
- Erpapazoglou Z., Dhaoui M., Pantazopoulou M., Giordano F., Mari M., Léon S., Raposo G., Reggiori F., Haguenauer-Tsapis R. 2012. A dual role for K63-linked ubiquitin chains in multivesicular body biogenesis and cargo sorting. Mol. Biol. Cell. 23:2170–2183 10.1091/mbc.E11-10-0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felice M.R., De Domenico I., Li L., Ward D.M., Bartok B., Musci G., Kaplan J. 2005. Post-transcriptional regulation of the yeast high affinity iron transport system. J. Biol. Chem. 280:22181–22190 10.1074/jbc.M414663200 [DOI] [PubMed] [Google Scholar]
- Foot N.J., Dalton H.E., Shearwin-Whyatt L.M., Dorstyn L., Tan S.S., Yang B., Kumar S. 2008. Regulation of the divalent metal ion transporter DMT1 and iron homeostasis by a ubiquitin-dependent mechanism involving Ndfips and WWP2. Blood. 112:4268–4275 10.1182/blood-2008-04-150953 [DOI] [PubMed] [Google Scholar]
- Forrest L.R., Krämer R., Ziegler C. 2011. The structural basis of secondary active transport mechanisms. Biochim. Biophys. Acta. 1807:167–188 10.1016/j.bbabio.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Haglund K., Dikic I. 2012. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J. Cell Sci. 125:265–275 10.1242/jcs.091280 [DOI] [PubMed] [Google Scholar]
- Hanyaloglu A.C., von Zastrow M. 2008. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu. Rev. Pharmacol. Toxicol. 48:537–568 10.1146/annurev.pharmtox.48.113006.094830 [DOI] [PubMed] [Google Scholar]
- Hasdemir B., Murphy J.E., Cottrell G.S., Bunnett N.W. 2009. Endosomal deubiquitinating enzymes control ubiquitination and down-regulation of protease-activated receptor 2. J. Biol. Chem. 284:28453–28466 10.1074/jbc.M109.025692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck J.W., Cheung S.K., Hampton R.Y. 2010. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. USA. 107:1106–1111 10.1073/pnas.0910591107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein C., Springael J.Y., Volland C., Haguenauer-Tsapis R., André B. 1995. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol. Microbiol. 18:77–87 10.1111/j.1365-2958.1995.mmi_18010077.x [DOI] [PubMed] [Google Scholar]
- Henne W.M., Buchkovich N.J., Emr S.D. 2011. The ESCRT pathway. Dev. Cell. 21:77–91 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Hettema E.H., Valdez-Taubas J., Pelham H.R. 2004. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 23:1279–1288 10.1038/sj.emboj.7600137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu V.W., Prekeris R. 2010. Transport at the recycling endosome. Curr. Opin. Cell Biol. 22:528–534 10.1016/j.ceb.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H. 2010. The ESCRT complexes. Crit. Rev. Biochem. Mol. Biol. 45:463–487 10.3109/10409238.2010.502516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H., Stenmark H. 2011. Molecular mechanisms of ubiquitin-dependent membrane traffic. Annu Rev Biophys. 40:119–142 10.1146/annurev-biophys-042910-155404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun J.H., Eom K., Lee K.H., Ho W.K., Lee S.H. 2013. Activity-dependent downregulation of D-type K+ channel subunit Kv1.2 in rat hippocampal CA3 pyramidal neurons. J. Physiol. 591:5525–5540 10.1113/jphysiol.2013.259002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C.B., Ott E.M., Keener J.M., Curtiss M., Sandrin V., Babst M. 2012. Regulation of membrane protein degradation by starvation-response pathways. Traffic. 13:468–482 10.1111/j.1600-0854.2011.01314.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kästle M., Grune T. 2012. Interactions of the proteasomal system with chaperones: protein triage and protein quality control. Prog. Mol. Biol. Transl. Sci. 109:113–160 10.1016/B978-0-12-397863-9.00004-3 [DOI] [PubMed] [Google Scholar]
- Kee Y., Lyon N., Huibregtse J.M. 2005. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 24:2414–2424 10.1038/sj.emboj.7600710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J.M., Babst M. 2013. Quality control and substrate-dependent downregulation of the nutrient transporter Fur4. Traffic. 14:412–427 10.1111/tra.12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M.H., Emili A. 2013. Ubp2 regulates Rsp5 ubiquitination activity in vivo and in vitro. PLoS ONE. 8:e75372 10.1371/journal.pone.0075372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E., Grossmann G., André B. 2007. Evidence for coupled biogenesis of yeast Gap1 permease and sphingolipids: essential role in transport activity and normal control by ubiquitination. Mol. Biol. Cell. 18:3068–3080 10.1091/mbc.E07-03-0196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E., Jacob C., André B. 2009. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185:493–502 10.1083/jcb.200810114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E., Erpapazoglou Z., Haguenauer-Tsapis R., André B. 2010. The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 20:196–204 10.1016/j.tcb.2010.01.004 [DOI] [PubMed] [Google Scholar]
- Léon S., Erpapazoglou Z., Haguenauer-Tsapis R. 2008. Ear1p and Ssh4p are new adaptors of the ubiquitin ligase Rsp5p for cargo ubiquitylation and sorting at multivesicular bodies. Mol. Biol. Cell. 19:2379–2388 10.1091/mbc.E08-01-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M.J., Pelham H.R. 2009. Inefficient quality control of thermosensitive proteins on the plasma membrane. PLoS ONE. 4:e5038 10.1371/journal.pone.0005038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Kane T., Tipper C., Spatrick P., Jenness D.D. 1999. Yeast mutants affecting possible quality control of plasma membrane proteins. Mol. Cell. Biol. 19:3588–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.H., MacGurn J.A., Chu T., Stefan C.J., Emr S.D. 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 135:714–725 10.1016/j.cell.2008.09.025 [DOI] [PubMed] [Google Scholar]
- Liu Y., Chang A. 2006. Quality control of a mutant plasma membrane ATPase: ubiquitylation prevents cell-surface stability. J. Cell Sci. 119:360–369 10.1242/jcs.02749 [DOI] [PubMed] [Google Scholar]
- Luhtala N., Odorizzi G. 2004. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166:717–729 10.1083/jcb.200403139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn J.A., Hsu P.C., Smolka M.B., Emr S.D. 2011. TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell. 147:1104–1117 10.1016/j.cell.2011.09.054 [DOI] [PubMed] [Google Scholar]
- MacGurn J.A., Hsu P.C., Emr S.D. 2012. Ubiquitin and membrane protein turnover: from cradle to grave. Annu. Rev. Biochem. 81:231–259 10.1146/annurev-biochem-060210-093619 [DOI] [PubMed] [Google Scholar]
- Marchal C., Haguenauer-Tsapis R., Urban-Grimal D. 2000. Casein kinase I-dependent phosphorylation within a PEST sequence and ubiquitination at nearby lysines signal endocytosis of yeast uracil permease. J. Biol. Chem. 275:23608–23614 10.1074/jbc.M001735200 [DOI] [PubMed] [Google Scholar]
- Meenhuis A., Verwijmeren C., Roovers O., Touw I.P. 2011. The deubiquitinating enzyme DUB2A enhances CSF3 signalling by attenuating lysosomal routing of the CSF3 receptor. Biochem. J. 434:343–351 10.1042/BJ20101628 [DOI] [PubMed] [Google Scholar]
- Merhi A., André B. 2012. Internal amino acids promote Gap1 permease ubiquitylation via TORC1/Npr1/14-3-3-dependent control of the Bul arrestin-like adaptors. Mol. Cell. Biol. 32:4510–4522 10.1128/MCB.00463-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno E., Iura T., Mukai A., Yoshimori T., Kitamura N., Komada M. 2005. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol. Biol. Cell. 16:5163–5174 10.1091/mbc.E05-06-0560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan J.F., Pan H., Lu Q. 2010. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep. 11:605–611 10.1038/embor.2010.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E., Pelham H.R. 2009. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic. 10:1856–1867 10.1111/j.1600-0854.2009.00990.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E., Sullivan J.A., Pelham H.R. 2008. Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep. 9:1216–1221 10.1038/embor.2008.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda N.B., Theodoraki M.A., Mandal A.K., Mayo K.J., Ren H.Y., Sultana R., Wu K., Johnson J., Cyr D.M., Caplan A.J. 2010. Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol. Biol. Cell. 21:2102–2116 10.1091/mbc.E10-02-0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T., Barrière H., Bagdány M., Rabeh W.M., Du K., Höhfeld J., Young J.C., Lukacs G.L. 2010. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 329:805–810 10.1126/science.1191542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T., Apaja P.M., Lukacs G.L. 2011. Protein quality control at the plasma membrane. Curr. Opin. Cell Biol. 23:483–491 10.1016/j.ceb.2011.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham W.M., Hamm H.E. 2008. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 9:60–71 10.1038/nrm2299 [DOI] [PubMed] [Google Scholar]
- Patwari P., Chutkow W.A., Cummings K., Verstraeten V.L., Lammerding J., Schreiter E.R., Lee R.T. 2009. Thioredoxin-independent regulation of metabolism by the alpha-arrestin proteins. J. Biol. Chem. 284:24996–25003 10.1074/jbc.M109.018093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payet L.A., Pineau L., Snyder E.C., Colas J., Moussa A., Vannier B., Bigay J., Clarhaut J., Becq F., Berjeaud J.M., et al. 2013. Saturated fatty acids alter the late secretory pathway by modulating membrane properties. Traffic. 14:1228–1241 10.1111/tra.12117 [DOI] [PubMed] [Google Scholar]
- Pineau L., Bonifait L., Berjeaud J.M., Alimardani-Theuil P., Bergès T., Ferreira T. 2008. A lipid-mediated quality control process in the Golgi apparatus in yeast. Mol. Biol. Cell. 19:807–821 10.1091/mbc.E07-06-0600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper R.C., Dikic I., Lukacs G.L. 2014. Ubiquitin-dependent sorting in endocytosis. Cold Spring Harb. Perspect. Biol. 6:a016808 10.1101/cshperspect.a016808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzirusso M., Chang A. 2004. Ubiquitin-mediated targeting of a mutant plasma membrane ATPase, Pma1-7, to the endosomal/vacuolar system in yeast. Mol. Biol. Cell. 15:2401–2409 10.1091/mbc.E03-10-0727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R., Bonifacino J.S. 2004. Interactions of GGA3 with the ubiquitin sorting machinery. Nat. Cell Biol. 6:244–251 10.1038/ncb1106 [DOI] [PubMed] [Google Scholar]
- Ravid T., Hochstrasser M. 2008. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9:679–690 10.1038/nrm2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Kee Y., Huibregtse J.M., Piper R.C. 2007. Hse1, a component of the yeast Hrs-STAM ubiquitin-sorting complex, associates with ubiquitin peptidases and a ligase to control sorting efficiency into multivesicular bodies. Mol. Biol. Cell. 18:324–335 10.1091/mbc.E06-06-0557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J.C., Fredrickson E.K., Oeser M.L., Garrett-Engele C.M., Locke M.N., Richardson L.A., Nelson Z.W., Hetrick E.D., Milac T.I., Gottschling D.E., Gardner R.G. 2011. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol. Cell. 41:93–106 10.1016/j.molcel.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott P.M., Bilodeau P.S., Zhdankina O., Winistorfer S.C., Hauglund M.J., Allaman M.M., Kearney W.R., Robertson A.D., Boman A.L., Piper R.C. 2004. GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat. Cell Biol. 6:252–259 10.1038/ncb1107 [DOI] [PubMed] [Google Scholar]
- Seaman M.N. 2012. The retromer complex - endosomal protein recycling and beyond. J. Cell Sci. 125:4693–4702 10.1242/jcs.103440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séron K., Blondel M.O., Haguenauer-Tsapis R., Volland C. 1999. Uracil-induced down-regulation of the yeast uracil permease. J. Bacteriol. 181:1793–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y., Katoh Y., Shiba T., Yoshino K., Takatsu H., Kobayashi H., Shin H.W., Wakatsuki S., Nakayama K. 2004. GAT (GGA and Tom1) domain responsible for ubiquitin binding and ubiquitination. J. Biol. Chem. 279:7105–7111 10.1074/jbc.M311702200 [DOI] [PubMed] [Google Scholar]
- Shields S.B., Piper R.C. 2011. How ubiquitin functions with ESCRTs. Traffic. 12:1306–1317 10.1111/j.1600-0854.2011.01242.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., Sampaio J.L. 2011. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 3:a004697 10.1101/cshperspect.a004697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J.A., Lewis M.J., Nikko E., Pelham H.R. 2007. Multiple interactions drive adaptor-mediated recruitment of the ubiquitin ligase rsp5 to membrane proteins in vivo and in vitro. Mol. Biol. Cell. 18:2429–2440 10.1091/mbc.E07-01-0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatecka-Urban A., Brown A., Moreau-Marquis S., Renuka J., Coutermarsh B., Barnaby R., Karlson K.H., Flotte T.R., Fukuda M., Langford G.M., Stanton B.A. 2005. The short apical membrane half-life of rescued DeltaF508-cystic fibrosis transmembrane conductance regulator (CFTR) results from accelerated endocytosis of DeltaF508-CFTR in polarized human airway epithelial cells. J. Biol. Chem. 280:36762–36772 10.1074/jbc.M508944200 [DOI] [PubMed] [Google Scholar]
- Taguchi T. 2013. Emerging roles of recycling endosomes. J. Biochem. 153:505–510 10.1093/jb/mvt034 [DOI] [PubMed] [Google Scholar]
- Terrell J., Shih S., Dunn R., Hicke L. 1998. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol. Cell. 1:193–202 10.1016/S1097-2765(00)80020-9 [DOI] [PubMed] [Google Scholar]
- Volland C., Urban-Grimal D., Géraud G., Haguenauer-Tsapis R. 1994. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J. Biol. Chem. 269:9833–9841 [PubMed] [Google Scholar]
- Wang G., Yang J., Huibregtse J.M. 1999. Functional domains of the Rsp5 ubiquitin-protein ligase. Mol. Cell. Biol. 19:342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Thibault G., Ng D.T. 2011. Routing misfolded proteins through the multivesicular body (MVB) pathway protects against proteotoxicity. J. Biol. Chem. 286:29376–29387 10.1074/jbc.M111.233346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyand S., Shimamura T., Yajima S., Suzuki S., Mirza O., Krusong K., Carpenter E.P., Rutherford N.G., Hadden J.M., O’Reilly J., et al. 2008. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 322:709–713 10.1126/science.1164440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Duong D.M., Seyfried N.T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. 2009. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 137:133–145 10.1016/j.cell.2009.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Macgurn J.A., Liu M., Emr S. 2013. The ART-Rsp5 ubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. Elife. 2:e00459 10.7554/eLife.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]