A contractile actomyosin meshwork at the top of a cell is mechanically coupled to dorsal actin fibers that are anchored via focal adhesions to the cell surface, generating a counterbalanced adhesion/contraction system that drives cell shape changes.
Abstract
How adherent and contractile systems coordinate to promote cell shape changes is unclear. Here, we define a counterbalanced adhesion/contraction model for cell shape control. Live-cell microscopy data showed a crucial role for a contractile meshwork at the top of the cell, which is composed of actin arcs and myosin IIA filaments. The contractile actin meshwork is organized like muscle sarcomeres, with repeating myosin II filaments separated by the actin bundling protein α-actinin, and is mechanically coupled to noncontractile dorsal actin fibers that run from top to bottom in the cell. When the meshwork contracts, it pulls the dorsal fibers away from the substrate. This pulling force is counterbalanced by the dorsal fibers’ attachment to focal adhesions, causing the fibers to bend downward and flattening the cell. This model is likely to be relevant for understanding how cells configure themselves to complex surfaces, protrude into tight spaces, and generate three-dimensional forces on the growth substrate under both healthy and diseased conditions.
Introduction
Cells modulate their shape to crawl through different substrates, extend out from cell masses, and adapt to different tissue-specific environments, processes that are critical for the morphogenetic pathways underlying tissue regeneration and remodeling, as well as in disease progression in cancer (Aman and Piotrowski, 2010; Watanabe and Takahashi, 2010; Levin, 2012; Riahi et al., 2012). Cell shape changes rely upon spatial and temporal coordination of biochemical and physical processes at the molecular, cellular, and tissue scale (Keren et al., 2008; Mogilner and Keren, 2009; Gardel et al., 2010; Zhang et al., 2010; DuFort et al., 2011; Farge, 2011). Yet, progress in understanding how these processes interact to control 3D cell shape has proved challenging. Limitations in image resolution, as well as a lack of 3D models of the cytoskeleton, have made it difficult to understand, for example, what contractile elements drive particular cell 3D shape changes and how they are spatio-dynamically regulated. Whether the subcellular systems controlling 3D cell shape have interdependence with other systems involved in cell morphodynamics, such as adhesion and migration, is also not clear.
Upon crawling across a surface, motile cells extend a flat leading edge, called the lamella (Ponti et al., 2004). The emergence of this flat structure provides a testable model system for cell shape morphogenesis in vertebrates. The lamella is enriched in actin, myosin II, and substrate adhesion components, and plays important roles in generating traction forces on the growth substrate for cell movement and mechanotransduction (Ponti et al., 2004; Hotulainen and Lappalainen, 2006; Hu et al., 2007; Gardel et al., 2008). There are three classes of actin filament–based stress fibers participating in these functions that reside in the lamella: transverse actin arcs, dorsal stress fibers (DSFs), and ventral stress fibers (Hotulainen and Lappalainen, 2006). The actin arcs run parallel to the leading edge and are enriched in myosin II (Heath, 1981; Hotulainen and Lappalainen, 2006; Medeiros et al., 2006). DSFs extend vertically upwards from focal adhesions to the dorsal side of the cell and largely lack myosin II (Small et al., 1998; Hotulainen and Lappalainen, 2006). Ventral stress fibers, however, reside at the cell bottom and connect to the substrate at both ends by focal adhesions (Hotulainen and Lappalainen, 2006). Previous studies have suggested how the different actin stress fibers generate force on the growth substrate and help drive cell movement (Gardel et al., 2010). But no model has yet explained how these filaments help generate the lamella’s flat shape.
In this study, we combined 3D superresolution analyses of crawling cells with the development of a biophysical modeling scheme to show that the seemingly complex process of lamella flattening in the crawling cell can be explained based on mechanical principles and cytoskeletal reorganization. Structured illumination microscopy (SIM; Shao et al., 2011) helped clarify the fine 3D contractile organization of actin filaments in the lamella, revealing that the primary actin filaments undergoing myosin II–based contraction were transverse actin arcs running parallel to the top of the cell. As the arcs contracted, they pulled on DSFs, which resisted by pivoting on their attached focal adhesions at the cell bottom, generating 3D forces on the growth substrate. This caused the dorsal membrane of the cell to tilt downward and the lamella to flatten. Removing myosin IIA contractility caused the lamella to lose its flatness, whereas adding myosin IIA to nonmotile cells, which lack a flat lamella, caused cells to create one. Together, our results suggest that myosin II contractile machinery mediates lamella flattening in a process involving counterbalanced contractile and adhesive forces.
Results
Resolving actin filament–based structures in 3D
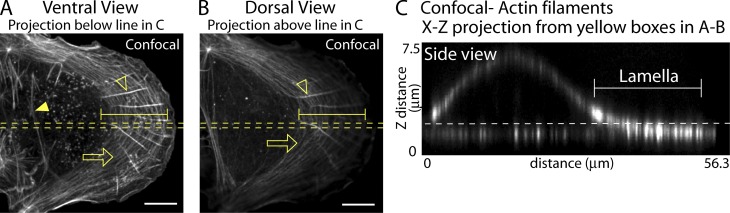
To analyze the distribution of actin fibers within crawling cells, we examined actin organization in U2OS cells, a well-studied cell type known for its directed motility (Hotulainen and Lappalainen, 2006). Conventional confocal microscopy imaging of these cells after labeling with fluorescent phalloidin to visualize actin structures confirmed that most actin fibers are in the flat front surface of the cell known as the lamella (Fig. 1, A–C; Gardel et al., 2010). Because of the extreme flatness of the lamella, the different actin fiber types associated with it could be visualized in a single confocal slice (∼500-nm thickness) sectioned nearest the cell bottom (Fig. 1 A, ventral view), with actin fibers largely absent in the next confocal slice upward (Fig. 1 B, dorsal view). The lateral orientations of different stress fibers seen in the ventral view (Fig. 1 A) were readily distinguishable, and included: ventral stress fibers (closed arrowhead) and DSFs (open arrowhead) that ran perpendicular to the leading edge; and, transverse actin arcs (open arrow) running parallel to the front edge, as described previously (Small et al., 1998; Hotulainen and Lappalainen, 2006). Notably, when the cell was viewed sidewise in z, little vertical organization of these actin fibers could be distinguished because all the fibers resided in the same diffraction-limited z slice (Fig. 1 C). The vertical arrangements of the actin filaments are likely to be important for understanding how these filaments function to flatten the lamella and control cell shape. We therefore sought higher resolution imaging approaches to clarify the functional 3D organization of actin and myosin II filaments in the lamella.
Figure 1.
Actin organization in the lamella is axially diffraction-limited. (A–C) Actin filaments localized in a U2OS cell by confocal imaging. (A and B) x-y views of projections from below (A) and above (B) the dotted white line in C. Open arrows, open arrowheads, and closed arrowheads denote actin arcs, DSF, and ventral stress fibers, respectively. Brackets denote lamella. (C) Side view of a cell from the region of interest denoted by the broken yellow lines in A and B. The broken white line shows the axially diffraction-limited layer at the bottom of the cell. Bracket denotes the lamella. Bars, 10 µm.
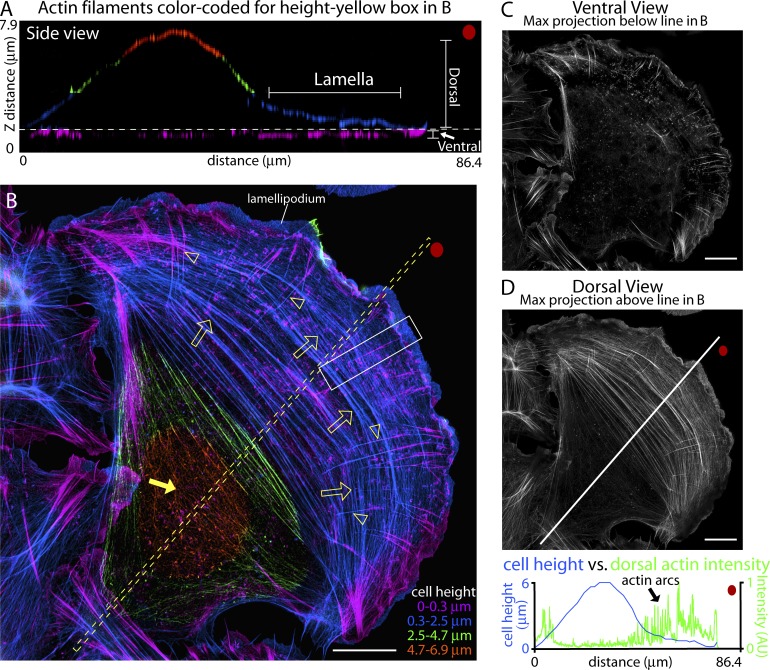
SIM yields a twofold improvement in x-y and z resolution relative to diffraction-limited microscopy, and can be used to section through an entire cell to generate high-resolution multispectral 3D images (Gustafsson et al., 2008; Shao et al., 2011; Fiolka et al., 2012). We thus used SIM to examine the vertical organization of actin filaments in the flat lamellae of U2OS cells. Cells were plated on fibronectin-coated coverslips, then fixed with paraformaldehyde, and their actin filaments were labeled with fluorescent phalloidin. This protocol was used to preserve the shape of the lamella since we found that common cell preparation techniques involving extraction of membranes before fixation, used previously to view the cell edge either by EM or 3D stochastic optical reconstruction microscopy (STORM; Verkhovsky et al., 1995; Cramer et al., 1997; Svitkina et al., 1997; Burnette et al., 2011; Xu et al., 2012), artificially flatten the lamella (Fig. S1). A side view of actin structures associated with the cell fixed using our protocol shows a typical profile in which the leading edge is comprised of a thin lamella extending ∼20–30 µm from the cell body, which is 6–8 µm high (Fig. 2 A). Note that 3D SIM has the needed resolution to resolve the vertical organization of the actin filament layers in the lamella (Fig. 2 A, lamella bracket). Looking top-down at the cell revealed distinct actin filaments differentially associated with different z heights (Fig. 2 B, see color coding by height; and Video 1). Actin arcs, running parallel to the leading edge, were the prominent actin filament–based structure 0.3 µm above the ventral surface, close to the upper dorsal cell membrane. The arcs remained in close association with the dorsal cell membrane throughout the lamella (Fig. 2 B, arrows).
Figure 2.
SIM can resolve actin arcs on the dorsal surface of the lamella. (A–E) 3D SIM of actin filaments in a U2OS cell. (A) Side view from the region of interest denoted by the box in B. Red dots show orientation. The bottom focal plane (purple) is similar to a total internal reflection fluorescence (TIRF) microscopy image. Subsequent 2.2-µm sections above the bottom focal plane are labeled blue, green, and orange, respectively. (B) x-y view of colored layers in A. Arrowheads denote DSFs and open arrows denote actin arcs. The closed arrow denotes actin on top of the cell body. Intensity levels of each section in A and B were normalized so that structure throughout the cell could be displayed in a single image irrespective of relative intensity. (C) Maximum projection of the ventral actin filaments from below the yellow broken line in B. (D) Maximum projection of dorsal actin filaments from above the dotted line in B. The graph plots the intensity of dorsal actin filaments (green line) against cell height (blue line) along the region from the line in B. Bars, 10 µm.
Actin arcs and DSFs were enriched in the lamella zone relative to the rest of the cell, whereas ventral stress fibers were not. This could be seen by comparing the cell height versus the intensity of the actin filament signal on the ventral side of the cell (where ventral stress fibers reside) and the dorsal side of the cell (where actin arcs reside; Fig. 2, C and D). Actin arcs together with DSFs were found where the lamella was flat, whereas no consistent correlation was observed between the position of ventral stress fibers and lamellar height (Fig. 2 D). Indeed, the region where arcs and DSFs disappeared (∼20–30 µm from the edge) was where the cell body began to bulge upward (see the height vs. actin intensity graph in Fig. 2 D).
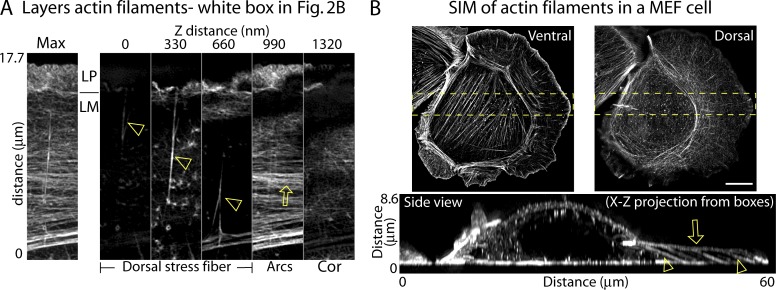
Investigating the orientation of DSFs in the lamella (Fig. 3 A), we found that a single DSF (open arrowhead) appeared at the ventral surface and could be seen extending up through subsequent z sections to the dorsal surface, where they met actin arcs (open arrow) enriched near the top of the lamella. Only a thin, isotropic meshwork of actin was found above the actin arcs and DSFs. In addition to U2OS cells, we also examined the orientation of DSFs in spreading primary mouse embryonic fibroblast (MEF) cells using SIM. A similar 3D organization of actin structures was found, with DSFs arising at the ventral surface of the cell near focal adhesions and reaching upward to intersect with actin arcs at the dorsal surface (Fig. 3 B, Fig. S2, and Video 2).
Figure 3.
DSFs connect ventral and dorsal sides of the lamella. (A) Maximum projection and z sections every 330 nm from the box in Fig. 2 B showing the layers of actin structures at the edge. Open arrowheads and open arrows in A denote DSF and actin arcs, respectively. (B) Ventral, dorsal, and side views (taken from the yellow boxed region) of the actin filaments in a spreading primary MEF cell. Open arrowheads and open arrows in side view denote DSF and actin arcs, respectively. Bar, 10 µm.
Based on these results, we concluded that actin fibers in crawling cells show a specific 3D organization in which DSFs and arcs are preferentially enriched in the flat lamellar region. The arcs run along the top of the cell, whereas the DSFs run vertically (bottom-to-top of cell). Importantly, the DSFs connect the actin arcs at the top of the cell with focal adhesions at the cell bottom. We next sought to understand how this organization couples to contractile forces necessary to flatten the lamella.
Determining which actin filaments are contractile and how they contract
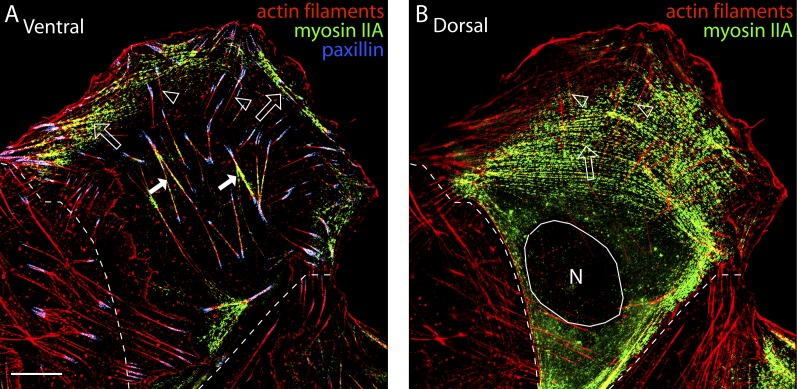
Either actin arcs or DSFs could form the contractile system responsible for lamella flattening. To distinguish between these possibilities, we used 3D SIM to examine the localization of myosin IIA, a motor protein that causes actin contraction in the lamella (Vicente-Manzanares et al., 2009; Vallenius, 2013). The other myosin II isoform expressed by U2OS cells, myosin IIB, is found at the back of the cell (Vallenius, 2013). At the cell bottom, seen in the ventral-most view of the cell (Fig. 4 A), a significant fraction of myosin IIA filaments were localized along actin arcs at the front of cells (Fig. 4, A and B, open arrows; and Fig. S3), with virtually no myosin IIA localized on DSFs (Fig. 4, A and B, open arrowheads) and only some localized on ventral stress fibers (Fig. 4 A, solid arrows). This was consistent with previous observations (Hotulainen and Lappalainen, 2006). Examining the top of the cell in the dorsal view (Fig. 4 B), we found that myosin IIA was enriched on actin arcs, and did not localize on DSFs intersecting with actin arcs (Fig. 4 B, open arrowheads). Given that the myosin II contractile machinery in the lamella is restricted to actin arcs, we concluded that these arcs form the contractile system in the lamella.
Figure 4.
Myosin II localizes to the dorsal surface of the lamella. (A and B) Myosin IIA localizes within actin arcs on the cell’s dorsal surface. (A) Ventral view of actin filaments (red), myosin IIA–GFP, and paxillin in a U2OS cell. Open arrowheads denote DSF, open arrows denote newly formed actin arcs, and closed arrows denote ventral stress fibers. (B) Dorsal view showing mature actin arcs (open arrow) and DSF (open arrowheads). Broken lines denote the sides of the cell and the circle denotes the position of the nucleus (N). Bar, 10 µm.
To understand how actomyosin contractile machinery is assembled and contractile force is generated in the lamella, we examined the orientation of myosin II filaments along actin arcs. Myosin II filaments form by tail–tail association of two or more hexamers consisting of two heavy chains, two regulatory light chains, and two essential light chains (Vicente-Manzanares et al., 2009). The resulting filament contains groups of motor domains on each end (Fig. 5 A, schematic), which can pull on actin filaments (Vicente-Manzanares et al., 2009). We found that although the substructure of a myosin IIA filament is diffraction-limited in crawling cells (Fig. 5 A, widefield), the two groups of motor domains and tail domain can be resolved using SIM (Fig. 5, A–C).
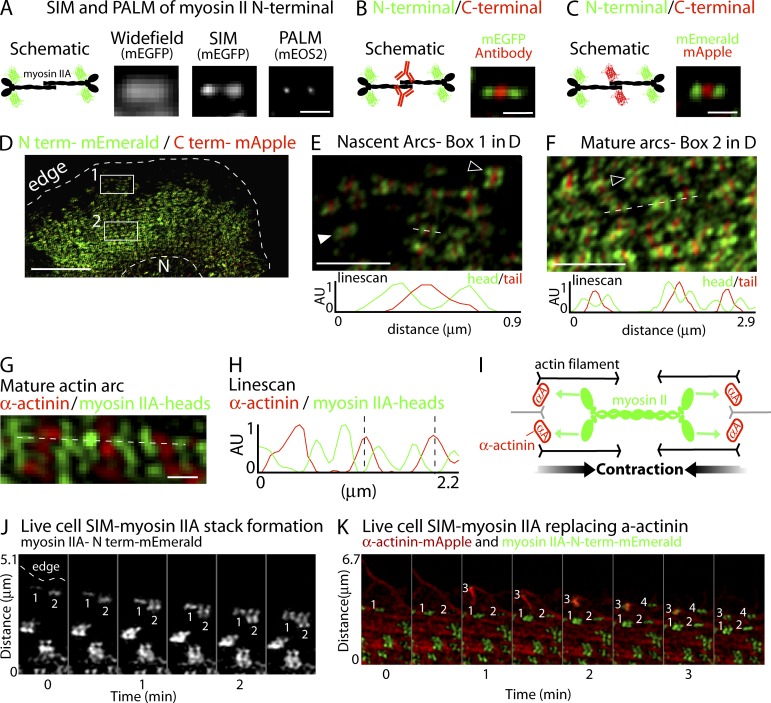
Figure 5.
Nanoscale organization and dynamics of myosin II filaments in the lamella. (A–C) Motor domains and tails of myosin II filaments resolved with SIM. (A) Schematic depicting the myosin II filament with mEGFP fused to the N terminus of the heavy chain. Examples of myosin II filaments imaged with wide-field, SIM, and PALM are shown. (B) Schematic and SIM imaging of a myosin II filament labeled with mEGFP on the N terminus of the heavy chain (green) and an antibody labeling the tail domains (red). (C) Schematic and SIM imaging of myosin II filaments with mEmerald fused to the N terminus (green) and mApple fused to the C terminus (red). (D) Low magnification of a U2OS cell expressing the mEmerald/mApple construct. Bar, 10 µm. (E and F) High-magnification views of boxes 1 and 2 from D. (E) mEmerald/mApple construct incorporated into myosin II filaments in newly formed actin arcs in box 1 from D showing single filaments (black arrowhead) and stacks of filaments (white arrowhead) with their long axis parallel to the edge. (F) Mature actin arcs in box 2 from D showing that myosin II filaments exist in stacks (arrowhead) with their long axes parallel to the edge. (G) High-magnification view of an actin arc from a cell expressing α-actinin–mApple (red) and myosin IIA–mEGFP–N-terminal (green). Fig. S4 D shows the entire cell. (H) Line scan from the broken line in G showing two myosin IIA head groups (green line) alternating with α-actinin (red line). Broken lines denote one sarcomere-like unit. (I) Schematic showing sarcomere-like contraction of actin arcs. Green arrows denote myosin II walking and black arrows denote α-actinin zones coming together (i.e., contraction). (J) Montage showing the expansion of two myosin II filaments into filament stacks at the edge of a cell. (K) Montage showing that α-actinin surrounds, but does not colocalize with, expanding stacks of myosin II filaments. Bars: (B and C) 400 nm; (D–F) 2 µm; (G) 400 nm.
To investigate the head-to-tail orientation of myosin IIA filaments along actin arcs by SIM, we expressed myosin IIA constructs tagged with mEmerald in the motor domain and mApple in the tail domain (Fig. 5, D–F; and Fig. S4, A–C). SIM analysis revealed that arc-associated myosin filaments are always aligned parallel to the arc, irrespective of the age of the arcs (Fig. 5, E and F). Moreover, in mature arcs (located further from the edge), myosin IIA filaments packed more closely, and became arranged in regular intervals (Fig. 5 F), which is reminiscent of myosin IIA filaments in muscle sarcomeres (Sanger et al., 2010). The appearance of a sarcomeric-like contractile unit in actin arcs is consistent with EM studies of myosin II structure on the dorsal cellular surface (Heath and Holifield, 1993; Verkhovsky et al., 1995; Cramer et al., 1997) and supports a model of myosin IIA–mediated actin arc constriction based on sarcomeric-like contraction.
Previous studies using diffraction-limited imaging have shown an alternating pattern of myosin II and α-actinin similar to what occurs in sarcomeres (Hotulainen and Lappalainen, 2006; Sanger et al., 2010; Tojkander et al., 2011). Imaging the motor domains of myosin IIA filaments along with α-actinin, we also found that myosin II structures in actin arcs were flanked by α-actinin labeling, which is consistent with a sarcomeric-like arrangement (Fig. 5, G–I; and Fig. S4 D). Interestingly the sarcomeric-like organization was unique to actin arcs; myosin II filament organization in ventral stress fibers had overlapping myosin II and α-actinin structures (Fig. S4 E). The sarcomeric-like pattern of myosin IIA filaments on arcs, and the increasingly tight packing of myosin IIA filaments as arcs mature, suggested that myosin II filament activity drives actin arc contraction and shortening.
To examine how myosin IIA structures assembled onto actin arcs, we performed SIM on live cells. We found that growth of myosin IIA–containing structures preferentially occurred by addition of new filaments onto single myosin IIA filaments, which formed near the cell edge (Fig. 5 J and Video 3). As the myosin IIA filaments grew, they underwent side-by-side association into a ribbon-like, stacked morphology (Fig. 5 J). The resultant stacks of myosin IIA were similar to those reported previously using EM (Verkhovsky et al., 1995). Interestingly, expanding stacks of myosin IIA filaments displaced α-actinin as they grew (Fig. 5 K). The growing myosin IIA filament structures appeared to gradually displace α-actinin in actin arc filaments, converting them into sarcomeric-like forms capable of large-scale contraction (Fig. 5 K and Video 3). In this way, myosin IIA–driven contraction of the arcs would help translocate the arc away from the leading edge, with arcs shortening as they moved inward from the edge (Video 3). The gradual replacement of α-actinin by expanding stacks of myosin II filaments represents a new model of how actin arcs form, as previous models have proposed that discrete units of α-actinin–containing actin filaments and myosin II–containing filaments are preformed and then stitched together (Hotulainen and Lappalainen, 2006; Tojkander et al., 2011). Further exploration of these different views of actin arc formation will be make for interesting future investigations.
Developing a model of how arc contraction causes the cell to flatten
Having clarified the type of actin filaments in migrating cells that are contractile (i.e., transverse actin arcs) and how they contract (through sarcomeric-like contraction), we next asked whether this contractile system could mediate flattening of the cell. The key clue, suggested by our SIM data and previous EM studies (Small et al., 1998), was that actin arcs and DSFs are interconnected. That is, DSFs running vertically from cell bottom to cell top bind focal adhesions at the cell bottom and interact with actin arcs at the cell top (see Figs. 2 and 3). This raised the possibility that the two filament systems are mechanically coupled in a way that when the arcs contract they force the DSFs to bend downward and cause the cell to flatten.
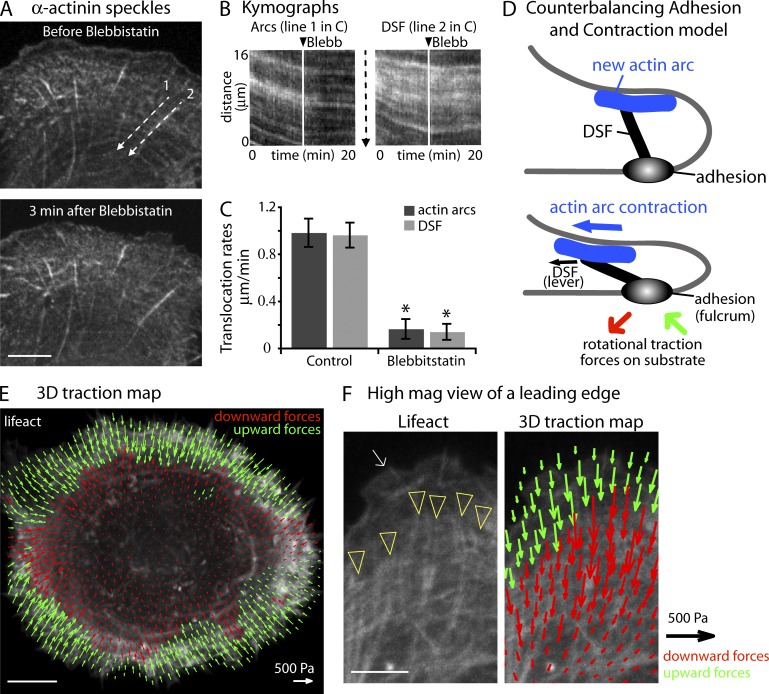
To test whether arcs and DSFs are mechanically coupled, we monitored actin arc and DSF movements with α-actinin speckles created by expressing a low amount of α-actinin–mApple (Fig. 6 A). α-Actinin localizes to both actin arcs and DSFs (Fig. S4 D), so it can be used to simultaneously monitor the motion of both structures. We found that individual α-actinin molecules incorporated into DSF bundles close to a focal adhesion and then moved away from this adhesion along the length of the actin bundle toward the top of the cell. Quantitative time-lapse imaging of α-actinin speckles revealed that actin arcs and DSFs moved away from the leading edge at the same rate in control cells (Fig. 6, A–C). Given the lack of myosin IIA on DSFs (Fig. 6, A and B), these data suggested that actin arc contraction generates forces that pull on DSFs. Consistent with this, when we inhibited myosin II contractility with 50 µM of the myosin II ATPase inhibitor, blebbistatin (Straight et al., 2003), translocation of both actin arcs and DSF was found to dramatically slow down despite DSFs having no myosin II associated with them (Fig. 6, B and C). Based on these data, we concluded that actin arcs are mechanically coupled to DSFs, with arc contraction exerting a force on DSFs.
Figure 6.
Actin arc contraction is coupled to DSFs. (A) α-Actinin–mApple speckles in a U2OS cell before and 3 min after 50 µM blebbistatin. Line 1 (arcs) and line 2 (DSF) were drawn parallel to the direction of speckle translocation. (B) Kymographs show the actin arc and an adjacent DSF flow before and after blebbistatin. (C) Actin arc and DSF translocation rates showing similar speeds before and after blebbistatin treatment (n = 3 experiments). *, P < 0.001. (D) Counterbalance model shows the arrangement of an actin arc (blue), DSF (black), and focal adhesion (gray) when a new actin arc is formed (top) and after the arc moves away from the edge (bottom). As the actin arc moves away from the edge it pulls on the dorsal side of the DSF. Because the DSF is attached to the focal adhesion, the force from actin arcs causes the DSF to bend downward and the edge of the cell to flatten. (E) 3D traction force map showing upward forces (green arrows) and downward forces (red arrows) exerted by a single U2OS cell spread out on a 2D gel (3 kPa). (F) High-magnification view of the edge of another U2OS cell with accompanying traction map. The white arrow denotes lamellipodium and yellow arrowheads denote the beginning of DSF. Bars: (A and E) 10 µm; (F) 5 µm.
Assuming that actin arc contraction generates forces to pull DSFs away from focal adhesions, than resistance by the focal adhesion attachment to the substrate should counterbalance this force. In this scenario, cell shape control would involve a contractile machine at the top of the cell (composed of myosin IIA and actin arcs) counterbalanced by an adherence machine at the cell bottom (composed of focal adhesions bound to the substrate; Fig. 6 D). In this model, the DSF and focal adhesion would behave like a lever (i.e., rigid rod pivoting at a fixed hinge or fulcrum). The force of actin arc contraction pulling the DSF away from the edge of the cell would cause the DSF (i.e., rod) to pivot on the focal adhesion (i.e., fulcrum), causing the dorsal side of the lamella to move downward, thus flattening the cell (Fig. 6 D).
Envisioning DSFs pivoting on focal adhesions predicts not only that actin arcs pulling DSFs away from the edge will flatten the cell, but that, in response, focal adhesions will generate rotational 3D traction forces on the growth substrate, specifically an upward force in front of focal adhesions and a downward force behind adhesions (Fig. 6 D). Indeed, a recent report has shown that 3D traction forces from the plane of the 2D growth substrate are exerted by focal adhesions (Legant et al., 2013). To test if our U2OS cells generated such predicted 3D traction forces, we placed cells expressing the actin filament probe, Lifeact-mEGFP, on polyacrylamide gels imbedded with fluorescent beads. We then calculated the 3D forces from the confocal image z series. The results revealed that U2OS cells do indeed pull up on the substrate at the very leading edge and push down further away from the edge (Fig. 6 E). Furthermore, the transition from upward forces to downward forces occurred at the front of the DSF (Fig. 6 F).
Together, these results supported the idea of a counterbalanced contraction–adhesion system regulating lamellar cell shape. In this model, a contractile system on the dorsal surface flattens the lamella by coupling to adhesions on the ventral surface through noncontractile elements. During this process, cells exert a pulling force on the substrate in response to changes in contraction forces arising on the top of the cell.
Testing the counterbalanced contraction–adhesion model
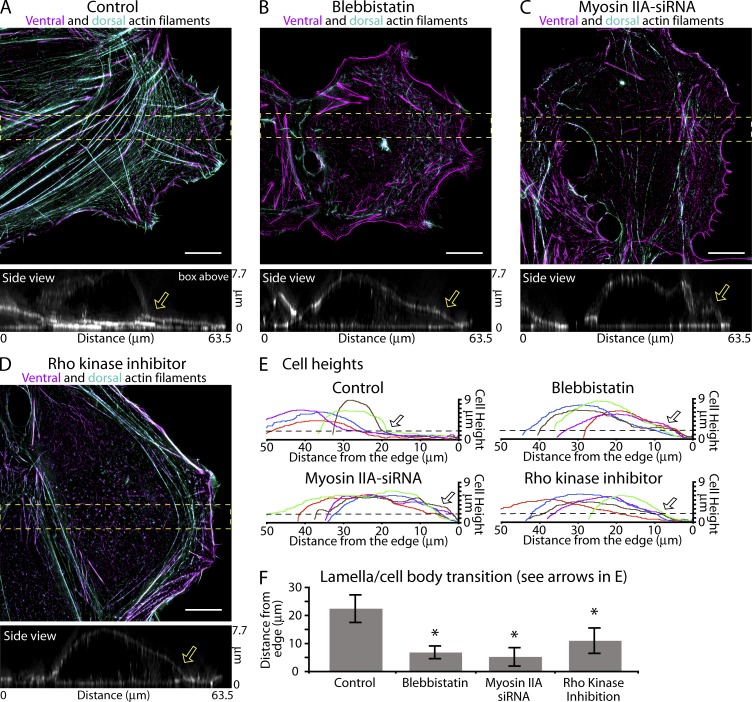
The contractile and counterbalancing adhesion system model described in the previous section predicts that actin arcs and DSFs are required to shape the lamella. We therefore examined the effect of abolishing the actin arcs/DSFs on lamella height. We compared cells before and after treatments reducing actin arc and DSF networks (Fig. 7, A–F). We found that treatment of U2OS cells either with blebbistatin (Fig. 7 B) or siRNA of myosin IIA (Fig. 7 C and Fig. S5, A–D) reduced both actin arcs and DSFs compared with control cells (Fig. 7 A). We also disrupted actin arcs by inhibiting Rho-associated kinase with the drug Y-27632 (Fig. 7 D). Y-27632 does not interfere with actin arc formation but does reduce actin arc lifetime (Zhang et al., 2003), thus the actin arc network does not extend as far into the cell as in control cells (Fig. 7 D, closed arrow). Inspecting the side views of cells (Fig. 7, A–D) and quantifying the heights of cells from each of these treatments revealed a clear pattern (Fig. 7 E). Control cells had a flat lamella, which is typically <2 µm in height and extends a mean 22.4 ± 4.9 µm from the edge before the cell body rises up (Fig. 7, E and F, open arrows). In cells treated with blebbistatin, myosin IIA siRNA, or Y-27632, we found that the distance the flat portion of the lamella extended from the edge was reduced to 6.8 ± 2.3 µm, 5.2 ± 3.3 µm, and 11.0 ± 4.5 µm, respectively (Fig. 7 E, open arrows; and Fig. 7 F). These results support the hypothesis that myosin IIA–mediated arc contraction flattens the lamella. To further support a specific role for actin arcs in flattening the lamella, we also found no correlation between changes in focal adhesion morphology and cell shape during myosin II or Rho-kinase perturbations (Fig. S5 F).
Figure 7.
Actin arcs/DSF play a role in flattening U2OS cells. (A–F) Removing actin arcs/DSFs abolishes the lamella’s flatness. (A–D) Ventral (purple) and dorsal (blue-green) actin filament organization in a control U2OS cell (A), a cell treated with 20 µM blebbistatin for 2 h (B), a cell treated with myosin IIA–siRNA (C), and a cell treated with 10 µM Y-27632 for 2 h (D). Side views are x-z maximum projections from the yellow boxes. Arrows denote the transition between the flat portion of the cell (lamella) and the cell body. (E) Cell heights from five cells from the treatments in A–D. Arrows show the mean distance cell height rises above 2 µm (dotted lines). (F) Quantification of the distance from the leading edge at which cells become higher than 2 µm (n = 6 cells per condition from three separate experiments). Error bars show standard deviation. *, P < 0.001. Bars, 10 µm.
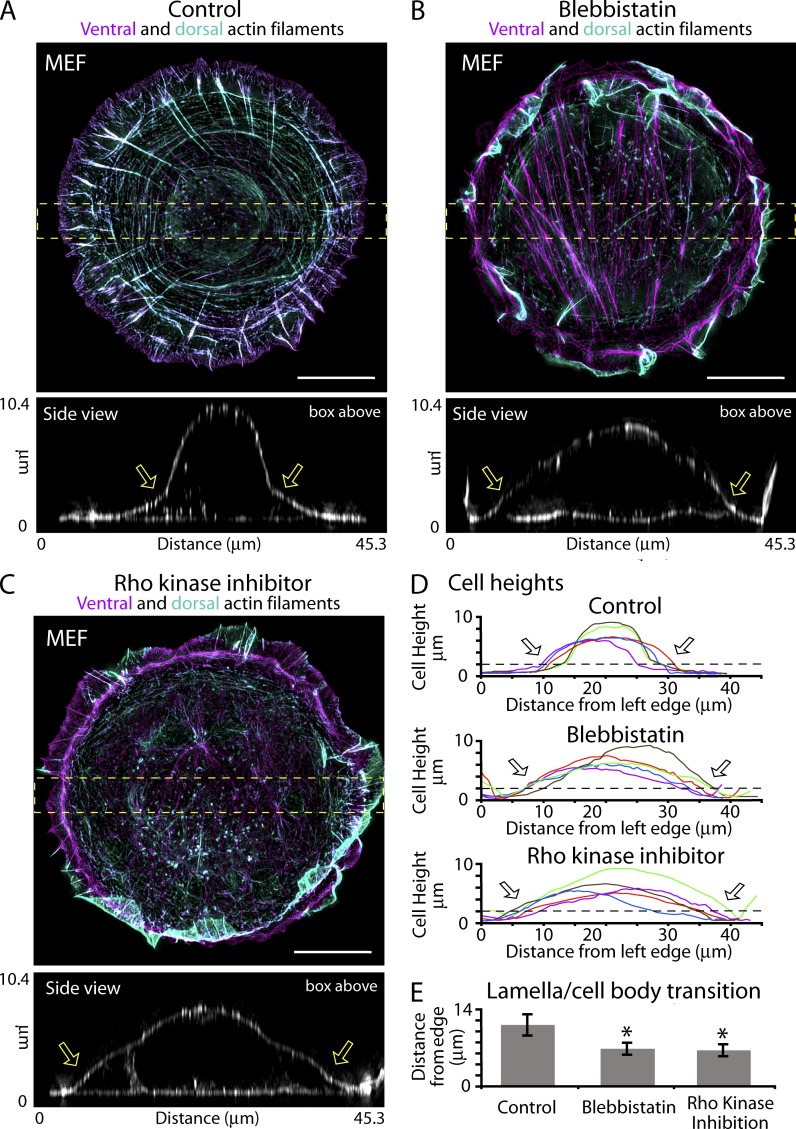
Next, we tested if removing actin arcs/DSFs resulted in the loss of a flat lamella in nontransformed cells. We used primary MEFs spread out on adhesive micropatterned substrates (Fig. 8 A). The micropatterns constrain the area on which cells can spread and thus create a population of cells with identical 2D areas. This is particularly useful when using fibroblasts because they tend to assume a wide variety of 2D shapes after their initial spreading phase (Döbereiner et al., 2004), which complicates 3D shape comparisons from cell to cell. MEFs were allowed to spread out on 1,100-µm2 circles coated with fibronectin for 1 h. After 1 h, the media was replaced with fresh control media, blebbistatin-containing media, or Y-27632–containing media. We found that the control media population contained a radial array of DSF and connecting actin arcs on the dorsal side of the cell (Fig. 8 A). However, the blebbistatin- and Y-27632–treated cells displayed a marked reduction in actin arcs/DSF (Fig. 8, B and C). Additionally, we also found that the length the flat lamella extended from the edge was reduced from 11.0 ± 1.9 mm in control cells to 6.8 ± 1.1 and 6.5 ± 1.0 mm in blebbistatin- or Y-27632–treated cells, respectively (Fig. 8, A–C, side views; and Fig. 8, D and E, quantification). This data were consistent with a role of actin arcs/DSF in flattening the edge of MEF cells.
Figure 8.
Actin arcs/DSF play a role in flattening MEF cells. (A–C) MEF cells plated on 1,100-mm2 micropatterned circles. Ventral (purple) and dorsal (blue-green) actin filaments are shown for a control cell (A) and cells treated with 5 µM blebbistatin (B) or 10 µm Y-27632 (C) for 2 h. Side views are x-z maximum projections from the yellow boxes. Arrows denote the transition between the flat portion of the cell (lamella) and the cell body. (D) Cell heights from five cells from the treatments in A–C. Arrows show the mean distance cell height rises above 2 µm (broken lines). (E) Quantification of the distance from the leading edge at which cells become higher than 2 µm (n = 12 cells per condition from three separate experiments). Error bars show standard deviation. *, P < 0.001. Bars, 10 µm.
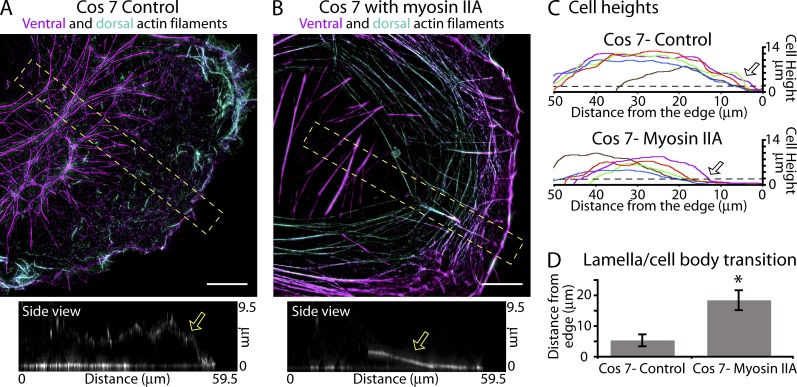
Because removing actin arcs/DSFs from cells abolished their flat lamellae, we wondered if adding actin arcs/DSFs to cells lacking them would create a flat lamella. To test this, we used COS-7 cells, which do not endogenously express myosin IIA (Even-Ram et al., 2007). We found that COS-7 cells contained ventral stress fibers but relatively little actin filament–based structures on their dorsal surface and only a short flat lamella-like region at the cell’s front (Fig. 9, A and C). However, when we exogenously expressed myosin IIA, not only did these cells acquire actin arcs/DSFs, but they also acquired flat lamellae (Fig. 9, B and C; and Fig. S5 E). The mean flat portion of the lamella of a myosin IIA–expressing COS-7 cell extended to a distance of 18.4 ± 3.3 µm compared with a control length of 5.3 ± 2.0 µm (Fig. 9, C and D). These results, along with the data from myosin II depletion and inhibition, demonstrate that myosin IIA is necessary and sufficient to create the dorsal contractile network that generates a flat lamella.
Figure 9.
Adding actin arcs/DSFs to cells creates a flat lamella. (A and B) Ventral (purple) and dorsal (blue-green) actin filament organization in a control Cos7 cell (A) and in a Cos7 cell expressing myosin II–mApple (B). Side views are x-y maximum projections from the yellow boxes. Arrows in side views denote the height of the leading edge. (C) Heights of control Cos7 cells and those expressing myosin IIA–mApple. Arrows show the mean distance cell height rises above 2 µm (broken lines). *, P < 0.001. (D) Quantification of the distance from the leading edge at which cells become higher than 2 µm (n = 6 cells). Error bars show standard deviation. Bars, 10 µm.
Discussion
Lamella flattening is a basic morphological feature of migrating cells, with its molecular and physical mechanisms likely crucial for events underlying cell motility and mechanotransduction. Our results addressing these mechanisms reveal a complex interdependency of actin, myosin IIA, and focal adhesions in generating lamella flattening through counterbalanced contraction and adhesion.
Using the improved resolution of SIM, we dissected actin fiber organization and cytoskeletal interactions underlying lamella shape, beginning by molecularly characterizing the vertical 3D layering and dynamics of cytoskeletal elements in the thin lamella, which were previously unresolvable by conventional light microscopy (Fig. 1; Hotulainen and Lappalainen, 2006) and only partially described by EM (Small et al., 1998). The vertical layering we observed included: myosin IIA–rich actin arcs, aligned along the dorsal cell surface tangentially to the leading edge; and noncontractile DSFs, aligned perpendicular to the cell edge and extending vertically (Figs. 2–4). Notably, DSFs were interconnected with both actin arcs at the top of the cell and focal adhesions at the cell bottom, as previously suggested (Small et al., 1998), with every focal adhesion at the edge of the cell having a DSF extending from it up to an actin arc on the dorsal surface (Fig. 4). The actin arcs underwent myosin-dependent contraction, which supports the phenomenon of myosin-dependent retrograde flow in the lamella (Ponti et al., 2004). This occurred through a mechanism involving sarcomeric-like contraction of the arcs, with adjacent head domains of myosin II filaments (visible by SIM) interspersed with the actin bundling protein α-actinin along contracting arcs (Fig. 5), akin to myosin filaments in muscle (Sanger et al., 2005). Myosin II filaments displaced α-actinin as they formed on the arc, setting up the sarcomere-like contractile organization of alternating myosin II and α-actinin zones along the arc (Fig. 5).
Intriguingly, contraction and movement of the arcs inward along the dorsal surface of the cell was coordinated with DSF dynamics (Fig. 6). Fluorescence speckle microscopy of fluorescently tagged α-actinin speckles revealed that speckles on contracting arcs and DSFs moved at identical rates, with this rate dependent on the contractibility of arcs (i.e., blebbistatin treatment immediately stopped the movement of both arcs and DSFs despite the DSFs having no myosin II associated with them). This suggested that actin arc contraction generates forces that can pull DSFs away from focal adhesions, and that arcs and DSFs are mechanically coupled. Given that forces driving arc movement impact DSF behavior, one interesting remaining question is whether the myosin II contractility of arcs helps generate or maintain DSFs, as is suggested by the observation that long-term blebbistatin treatment reduces the number of DSFs (Hotulainen and Lappalainen, 2006).
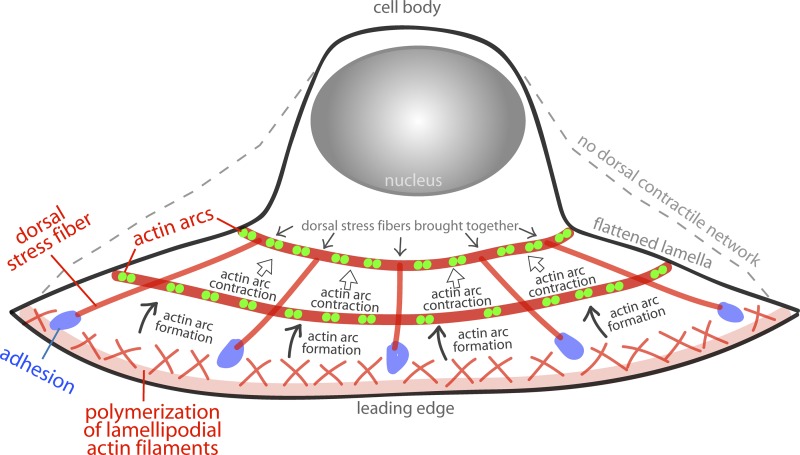
DSFs in the lamella were shown to tilt downward, which suggests a role in lamella flattening because of DSFs interconnecting the top and bottom surfaces of the cell (Figs. 2 and 3). We therefore asked what caused DSFs to tilt. Our data suggested two factors: the contractile activity of arcs moving along DSFs, and the anchoring of DSFs to focal adhesions. We found that as a newly formed actin arc moves away from the edge of the cell through myosin II contractility, it pulls the dorsal end of a DSF with it (Fig. 6). Because the ventral end of the DSF is attached to a relatively stable focal adhesion, the DSF acts as a lever and pivots on the adhesion in response to arc contraction (see cartoon in Fig. 6 D). The pivoting allows the DSF to tilt and move closer to the substrate. As actin arcs contract and shorten their lengths all along the leading edge, they would pull the ends of multiple DSFs attached to the arcs on the dorsal surface closer together (as shown in Fig. 10), resulting in all the DSFs tilting inward and the cell flattening.
Figure 10.
A role for actin arc contraction in cell flattening. A diagram depicting actin arc dynamics in a crawling cell. Actin filaments in the lamellipodium are created by polymerization at the leading edge of the cell (bottom of the image). A subset of these actin filaments are condensed into the actin filament bundles that make up a new actin arc (“actin arc formation”). Myosin II loads on to the newly forming actin arc, giving the actin arc the ability to contract. Dual green dots along the actin arc denote the two motor domains of a myosin II filament as revealed by SIM imaging. An actin arc contracts through myosin II activity and shortens along its length (“actin arc contraction”), thus leading to a decrease in the circumference of the actin arc as it moves away from the edge. As its shrinks, the actin arc pulls the non-adhesion-attached ends of DSFs closer together (“dorsal stress fibers brought together”). The force the actin arc exerts on DSFs is balanced by the attachment of the DSFs to the substrate. Thus, as the actin arc pulls the DSFs together, the dorsal contractile system moves closer to the ventral side of the cell, creating a flat lamella (“flattened lamella”). The broken line depicts the shape of the cell without actin arcs or DSFs (“no dorsal contractile network”).
A key prediction of this model of lamella shaping is that inhibiting actin arc structure or contractility should create a thicker lamella by increasing the distance between the ventral and dorsal surfaces. Consistent with this prediction, when myosin IIA was inactivated using blebbistatin treatment or was removed using siRNA, the affected cell failed to flatten its lamella (Figs. 6 and 7). Moreover, adding myosin IIA to COS-7 cells, which lack a flat lamella, caused the cells to create one (Fig. 8). The results, therefore, support lamella shape being controlled by counterbalanced contraction/adhesion forces.
Several factors could impact this mechanism of lamella shape control. One factor is the contractile activity of myosin IIA, which controls the magnitude of the forces generated by actin arcs on DSFs, and therefore adhesions. Another factor is the length of the DSF. The cartoon in Fig. 6 D depicts the simplest scenario in which the DSF stays a constant length while it is being pulled by the actin arc. However, the DSF is not just attached to but also grows out of focal adhesions. Our α-actinin speckling data showed that actin bundles within the DSF flow away from their respective adhesions, with newly formed actin bundles incorporating into the DSF close to the adhesions (Fig. 6). Moreover, actin components within the DSF move at identical rates to actin components within contracting arcs. This implies that the DSF’s length is controlled by changes in rates of DSF assembly at the adhesion and arc contractility, which has consequences. For example, if DSF growth increases while myosin II contractility stays the same, the force transmitted to the adhesion would likely decrease. The converse would also be true. If the DSF slowed down its assembly rate, then there would be more force transmitted from the pulling actin arcs. Further studies are needed to explore these possibilities, including how the rate of DSF growth is regulated by myosin II contractility of arcs.
In conclusion, our data suggest that cytoskeletal reorganization events are the primary factor controlling cell shape in the lamella. These events are orchestrated by a contractile system on the dorsal surface coupled to adhesions on the ventral surface through noncontractile elements. The counterbalanced forces operating in this system explain how a cell can alter its pulling force on the substrate in response to changes in contraction forces arising on the top of the cell or in growth of noncontractile elements at adhesions. Even in a complex 3D environment, the same principles for cell shape control are predicted to operate as with cells in a 2D environment. In both environments, positions of adhesions would be coupled to a contractile machine at some distance, possibly through noncontractile actin filament–based elements. Cells may therefore use this counterbalanced contraction/adhesion system to control their shape/behavior in a variety of morphogenetic processes.
Materials and methods
Cell culture and chemicals
U2OS and Cos7 cells were purchased from American Type Culture Collection and cultured in DMEM F12/10% FBS (Corning). Primary MEFs were a generous gift from A. Rambold (National Institutes of Health, Bethesda, MD) and cultured in DMEM/10% FBS (Corning) as described previously (Rambold et al., 2011). Growth substrates were prepared by coating glass coverslips, ranging between 168 and 172 µm, with 10 µg/ml of fibronectin in PBS at 37°C for 2 h. For protein expression experiments, cells were transiently transfected with Fugene 6 (Promega) according to the manufacturer’s instructions overnight in a 25-cm flask before plating on a growth substrate. U20S and Cos7 cells were cultured overnight. Spreading MEF cells shown in Fig. 1 G and Fig. S2 were cultured for 30 min after plating on growth substrates at 37°C. MEFs were also plated on adhesive micropatterned substrates containing 1,100-µm2 circles coated with fibronectin purchased from CYTOO Inc. Cells were plated according to the manufacturer’s instructions and left to spread for 1 h before drug treatments. Blebbistatin, fibronectin, Y-27632, TRITC-phalloidin, mouse–α-paxillin antibody, and mouse–α-tubulin antibody were purchased from Sigma-Aldrich. Alexa Fluor 488–phalloidin and secondary antibodies, Alexa Fluor 488–goat–α-rabbit and Alexa Fluor 405–goat–α-mouse, were purchased from Invitrogen. Rabbit–α-Myosin IIA was purchased from Convance. Knockdown of nonmuscle myosin IIA was performed with the Accell SMARTpool siRNA to human MYH9 purchased from Thermo Fisher Scientific according to the manufacturer’s instructions.
Plasmids
The plasmid encoding Myosin IIA–mEGFP was used as described previously (Chua et al., 2009). In brief, nonmuscle myosin IIA was in an enhanced GFP-N3 vector (Takara Bio Inc.) and its expression driven by a CMV promoter. All other fusions were first constructed using an advanced EGFP variant with several mutations designed to enhance brightness and folding efficiency (mEmerald: wild-type GFP + F64L, S65T, S72A, N149K, M153T, I167T, and A206K). The resulting targeting vectors were used as pilots to demonstrate proper localization of the intended fusions. To construct mouse Myosin IIA (available from GenBank under accession no. NM_022410), we purchased a synthetic version of the cDNA (Blue Heron) that included several silent point mutations to remove restriction enzyme sites in the sequence, as well as to create an 18–amino acid linker that would separate the fluorescent protein from the targeting protein (SGLRSGSGGGSASGGSGS). This fragment, along with an mEmerald-C1 cloning vector backbone (Clontech-style), was then digested with XhoI and EcoRI, gel purified, and ligated to generate mEmerald-MyosinIIA-C-18. This plasmid, along with mEos2-C1 and mApple-C1 cloning vectors, was double digested with BglII and NheI, gel purified, and ligated to form mEos2-MyosinIIA-C-18 and mApple-MyosinIIA-C-18.
To create the N-terminal (with respect to the florescent protein) Myosin IIA fusions, the following primers were used to amplify the Myosin IIA and create the 14–amino acid linker (GGSGGSAGDPPVAT): XhoI forward, 5′-GTGAGGTCCCTCGAGGCCACCATGGCTCAGCAGGCTGCAGACAAGTACCTCTATGTGG-3′; and BamHI reverse, 5′-GGTGGCGACCGGTGGATCCCCTGCGCTCCCGCCGCTTCCGCCTTCAGCTGCCTTGGCATCGGCCC-3′.
After digestion and gel purification, the PCR product was ligated into a similarly digested mEmerald-N1 cloning vector to produce mEmerald-MyosinIIA-N-14. The resulting fusion, along with mApple-N1 cloning vector, was double digested with BamHI and NotI, gel purified, and ligated to produce mApple-MyosinIIA-N-14.
To construct the mEmerald-C-16-mApple-N-14-MyosinIIA plasmid, PCR was used to create a 16–amino acid linker (GGASAGGGASGGLEAT) and to amplify mEmerald using the following primers: XhoI forward, 5′-CAGATCCCTCGAGGCCACCATGGTGAGCAAGGGCGAGGAGCTGTTCAC-3′; and XhoI reverse, 5′-GCCTCACTCGAGCCCTCCGCTGGCACCTCCCCCTGCGGATGCCCCTCCCTTGTACAGCTCGTCCATGCCGAGAGTGATC-3′.
After the PCR product was digested and purified, it was ligated into a similarly digested mApple-MyosinIIA-N-14 vector to produce the mEmerald-C-16-mApple-N-14-MyosinIIA.
To construct human α-actinin (GenBank accession no. NM_001130005; a gift from T. Keller, Florida State University, Tallahassee, FL), the cDNA was amplified using PCR to create a 19–amino acid linker (KLRILQSTVPRARDPPVAT) with the following primers: XhoI forward, 5′-GTGCGCTCGAGCTCAAGCTTCGCACCATCATGGACCATTATGATTCTCAGCAAACCAACGATTACATGCAGCCAG-3′; and EcoRI reverse, 5′-ACCTCGAATTCGAAGCTTGAGGTCACTCTCGCCGTACAGCGCCGTGGAG-3′.
The resulting product was digested, gel purified, and ligated into a similarly digested mEmerald-N1 cloning vector, creating mEmerald-α-actinin-19. After sequence verification, this vector, along with an mApple-N1 cloning vector, was double digested with EcoRI and NotI, gel purified, and ligated to form mApple-α-actinin-19.
All DNA used for transfection was prepared using the Plasmid Maxi kit (QIAGEN). To ensure proper localization, the constructs were characterized by wide-field and confocal fluorescence microscopy after transfection of ∼1 µg of DNA in HeLa cells (CCL2 line; American Type Culture Collection) using Effectene (QIAGEN). Transfected cells were grown on coverslips in DMEM/F12, fixed after 48 h, and mounted using Gelvatol.
Fixation and immunocytochemistry
Cells were fixed with 4% paraformaldehyde in PBS at room temperature for 20 min and then extracted for 5 min with 1% Triton X-100 in 4% paraformaldehyde in PBS. Primary antibodies for myosin IIA or paxillin at a 1:500 concentration suspended in 5% BSA/PBS were applied to cells for 1 h at room temperature. After washing, secondary antibodies Alexa Fluor 488–goat α-rabbit and Alexa Fluor 405–goat α-mouse were added to myosin IIA– or paxillin-labeled cells, respectively, for 45 min. Staining of actin filaments with either TRITC-phalloidin or Alexa Fluor 488 phalloidin was performed as described previously.
We also tried SIM imaging of cells prepared by alternative fixation protocols commonly used for the preparation of cells for electron microscopy and recently for 3D STORM analysis of actin structures (Cramer et al., 1997; Svitkina et al., 1997; Burnette et al., 2008, 2011; Xu et al., 2012). These techniques involve extracting the membrane of live cells before fixation in the presence of cytoskeletal stabilizers. We found that extracting cells before fixation flattens the lamella of U2OS and Cos7 cells (Fig. S1). For these experiments we followed the detailed protocol in Xu et al. (2012). Given the cell flattening associated with the live-cell membrane extraction protocol, we chose to fix samples before membrane extraction for our SIM experiments.
Confocal imaging
Z-stacks of TRITC-phalloidin and Alexa Fluor 488–immunolabeled myosin IIA were acquired with an Apochromat 100× 1.4 NA objective lens (Carl Zeiss) on a Marinas spinning disk confocal imaging system (Intelligent Imaging Innovations) using an EM charge-coupled device camera (Evolve; Photometrics). Cells were imaged in PBS. To facilitate an approximation of the axial dimension, the sample was oversampled by taking 100-nm z steps between image acquisitions.
SIM imaging
SIM imaging of fixed cells was performed on a microscope (ELYRA SIM; Carl Zeiss) with an Apochromat 63× 1.4 NA oil objective lens, as described previously (Elia et al., 2011), at room temperature. Samples were mounted in Vectashield H-1000 (Vector Laboratories). Five orientation angles of the excitation grid with five phases each were acquired for each z plane. Raw images were acquired with a total magnification of 79 nm per cameral pixel with z spacing of 110 nm between planes. SIM processing was performed with the SIM module of the Zen software package (Carl Zeiss), then TIF stacks of processed SIM data were exported. SIM datasets were then imported into ImageJ, which was subsequently used to quantify data and create projection images.
Live-cell SIM data were acquired mostly as described previously (Shao et al., 2011; Fiolka et al., 2012). The main improvement is that for two-color image acquisition, the wavelength was switched at every camera exposure instead of after a whole 3D stack of one channel was acquired. Two cameras were used, one dedicated for each channel, to reduce fluorescence bleed-through while maintaining the highest acquisition speed. Live-cell samples were placed in an environment chamber containing Leibovitz’s L-15 medium maintained at 37°C temperature. A water immersion objective lens (C-Apochromat 63×/1.2 W Corr; Carl Zeiss) was used. We were able to acquire 15–25 two-color 3D SIM stacks of 12–15 planes, with 20–30 s between two time points. The excitation power used was 5–10 W/cm2 at the sample plane for both 488-nm and 560-nm lasers.
Selection of cells for this study
Polarized migrating cells were chosen for this study by finding cells that had two basic morphological features: (1) they were in contact with another cell at their rear and (2) they had a curved leading edge. By satisfying both of these criteria, the direction of movement could be determined in both live and fixed cell populations. The curved leading edge also made comparisons between different cells easier, as they had similar stress fiber organization (e.g., stress fibers on the surface of these cells were arc-shaped).
Creating ventral and dorsal views of the lamella
The diffraction-limited or SIM-limited layer at the bottom of cells was determined by creating a side projection of the actin filaments within the cell. The region of interest was a line drawn from the back to the front of the cell, which was positioned in a way that it crossed the top of the cell body and the middle of the curved leading edge. The bottom layer of actin filaments in the cell body of this side view was then inspected to determine the effective z resolution. Three to five of the smallest actin filament–based objects were used for this determination. The stack was then split into two sections: (1) Ventral containing the slices through the bottom layer of the cell and (2) dorsal containing the top slices. Maximum projection images for presentation or creation of overlays were then created in ImageJ. The vast majority of the signal from actin filament–based structures came from the populations associated with the membranes (aside from the DSFs, obviously). Therefore, we feel the maximum projections are accurate representations of the membrane-associated actin filament structures, as relatively little contributing structure can be found in the cell body in most cells.
Cell heights were measured from the side views (created as described in the previous paragraph) using ImageJ. The heights, in pixels, were exported to Excel (Microsoft) and converted into microns. P-values from Student’s t tests and standard deviations were calculated using Excel.
Measuring the translocation of α-actinin speckles in U2OS cells
Low levels of α-actinin–mApple were expressed in cells by titrating down the amount of plasmid DNA used for transient transfection until expressing cells could not be detected by eye. We empirically found that 50 ng of plasmid DNA per 1 ml of total media was the correct amount for our experiments. Expressing cells were then located by using an EM charge-coupled device camera (Evolve). Time-lapse recording of α-actinin speckles were then acquired with an Apochromat 100× 1.4 NA objective lens (Carl Zeiss) and an Evolve camera attached to a Marinas spinning disk confocal (Intelligent Imaging Innovations). Kymograph creation and flow rate quantification were performed as described previously (Burnette et al., 2008). In brief, pixel intensity values were averaged over a 3-pixel width along a region of interest line drawn through a DSF or actin arcs parallel to a DSF. Kymographs were created by stacking the intensities for every time point side by side to create a distance over time plot (i.e., kymograph). Velocities of α-actinin speckle movement were measured from their slope. P-values from Student’s t tests and standard deviations were calculated using Excel.
Gel fabrication
Polyacrylamide hydrogels were prepared with 5% acrylamide, 0.03% N, N-methylene-bis-acrylamide, ammonium persulfate (10% wt/vol solution, 1:125 volume), TEMED (1:1,250 volume; all from Bio-Rad Laboratories), and 0.22% vol/vol of 0.2 µm fluorescent beads (2% solid red beads; Molecular Probes). The unpolymerized hydrogel mixture was placed in a desiccator under vacuum for 30 min to remove excess gas. After 30 min of degasification, 30 µl of the mixture was poured on the glass slide. A hydrophobic cover slide equipped with 40-µm-thick wedges was deposited on the droplet. After polymerization occurred (15–30 min), the hydrophobic cover slide was gently removed, and the gel was left swelling in distilled water for 24 h. The Young modulus of the gel was measured as described in Wang et al. (2002). To calibrate the Young’s modulus of the gel, the mixture of bis-acrylamide and acrylamide was put in a glass cylinder obtained from a Pasteur pipette. After the mixture was polymerized, the gel was carefully removed from the piece of glass. Two plastic hooks were glued to each end of the gel. The elongation of the gel under the weight of different masses was measured and the force-displacement relationship of the gel was obtained. A linear relationship was found and the Young’s modulus was calculated as the slope of this relationship. We found a mean value of 5.8 ± 0.5 kPa. The composition of the gel was optimized to get displacement amplitudes that allow easy handling of particle tracking (fraction of microns averaged in plane displacement, as few as possible out of plane displacement).
Surface functionalization
Polyacrylamide gels were functionalized with a theoretical concentration of 5.4 µg/cm2 of fibronectin from human plasma (Roche). Fibronectin was covalently attached to the surface of the gel using a photoactivatable heterobifunctional reagent named Sulfo-LC-SDA (Thermo Fisher Scientific). This reagent differs from the commonly used Sulfo-SANPAH32 by its photosensitive group, a diazirine group, which is more stable and more reactive. A solution of 1.8 mg/ml of Sulfo-LC-SDA in PBS was prepared and kept protected from light. 1 ml of this solution was poured on the surface of the gel in a dark room. After 5 min, the solution was removed and the gels were exposed to a 365-nm UV light (18 mW/cm2) for 5 min. The gels were then washed with PBS. Next, the Sulfo-LC-SDA treatment was repeated (incubation, removal, and UV exposition). After washing the gels, the PBS solution was replaced by the fibronectin solution for 1 h. The gels were then washed with PBS and incubated with cell culture medium 1 h before cell seeding.
Detection of surface deformation and force calculation
Stacks of live cells in 37°C L-15 medium and beads were acquired before and after cells were removed with trypsin with our Marinas spinning disk. Stacks were then aligned at subpixel resolution in the x-y and z direction using image correlation with MATLAB software. We used the 2D particle image velocimetry (PIV) toolbox MatPIV written for MATLAB to calculate the bead displacements field. PIV was performed on a plane 1 µm below the cell surface with a window size of 64 × 64 pixels (i.e., 6.4 × 6.4 µm) and a 50% overlap between windows. We used a median filter to remove the very few aberrant vectors whose deviation from the neighboring ones were superior to twice the standard deviation in the neighboring area. The forces were calculated using this displacement field using regularization techniques as in Delanoë-Ayari et al. (2008) and using the L-curve criterion for the choice of the regularization parameter.
Online supplemental material
Fig. S1 shows examples of actin filament structures in U2OS and Cos 7 cells where the plasma membranes were extracted before fixation. Fig. S2 shows an additional example of actin filaments visualized by SIM in a spreading MEF cell. Fig. S3 shows the endogenous localization of myosin IIA in a U2OS cell. Fig. S4 shows additional examples of myosin IIA localizations as visualized by SIM. Fig. S5 shows the quantification of MYH9 knockdown in U2OS cells and the localization of exogenously expressed myosin IIA in a Cos 7 cell. Fig. S5 also shows several examples of focal adhesion localization in control cells and cells treated with blebbistatin or Y-27632. Videos 1 and 2 show z sections from the SIM of the actin filaments from a U2OS cell (Video 1) and a MEF cell (Video 2). Video 2 shows a SIM time-lapse recording of myosin IIA–mEmerald (green) and α-actinin–mApple (red) expressed in a U2OS cell. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201311104/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201311104.dv.
Supplementary Material
Acknowledgments
We thank Drs. Clare Waterman and John Hammer and the members of the Lippincott-Schwartz laboratory for critical comments and discussion.
D.T. Burnette was supported by a Pharmacology Research Associate Training (PRAT) Fellowship from National Institute of General Medical Sciences, National Institutes of Health, during part of this study.
The authors declare no competing financial interests.
Author contributions: D.T. Burnette and J. Lippincott-Schwartz conceived the project and designed the experiments. D.T. Burnette performed the experiments with contributions from L. Shao, C. Ott, A.M. Pasapera, R.S. Fischer, and M.J. Paszek. D.T. Burnette, C. Ott, H. Delanoe-Ayari, and C. Der Loughian performed data analysis. M.A. Baird and M.W. Davidson contributed novel fluorescence probes. D.T. Burnette, C. Ott, and J. Lippincott-Schwartz wrote the paper. All authors discussed the results and commented on the manuscript.
Footnotes
Abbreviations used in this paper:
- DSF
- dorsal stress fiber
- MEF
- mouse embryonic fibroblast
- SIM
- structured illumination microscopy
References
- Aman A., Piotrowski T. 2010. Cell migration during morphogenesis. Dev. Biol. 341:20–33 10.1016/j.ydbio.2009.11.014 [DOI] [PubMed] [Google Scholar]
- Burnette D.T., Ji L., Schaefer A.W., Medeiros N.A., Danuser G., Forscher P. 2008. Myosin II activity facilitates microtubule bundling in the neuronal growth cone neck. Dev. Cell. 15:163–169 10.1016/j.devcel.2008.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette D.T., Manley S., Sengupta P., Sougrat R., Davidson M.W., Kachar B., Lippincott-Schwartz J. 2011. A role for actin arcs in the leading-edge advance of migrating cells. Nat. Cell Biol. 13:371–382 10.1038/ncb2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua J., Rikhy R., Lippincott-Schwartz J. 2009. Dynamin 2 orchestrates the global actomyosin cytoskeleton for epithelial maintenance and apical constriction. Proc. Natl. Acad. Sci. USA. 106:20770–20775 10.1073/pnas.0909812106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer L.P., Siebert M., Mitchison T.J. 1997. Identification of novel graded polarity actin filament bundles in locomoting heart fibroblasts: implications for the generation of motile force. J. Cell Biol. 136:1287–1305 10.1083/jcb.136.6.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoë-Ayari H., Iwaya S., Maeda Y.T., Inose J., Rivière C., Sano M., Rieu J.P. 2008. Changes in the magnitude and distribution of forces at different Dictyostelium developmental stages. Cell Motil. Cytoskeleton. 65:314–331 10.1002/cm.20262 [DOI] [PubMed] [Google Scholar]
- Döbereiner H.G., Dubin-Thaler B., Giannone G., Xenias H.S., Sheetz M.P. 2004. Dynamic phase transitions in cell spreading. Phys. Rev. Lett. 93:108105 10.1103/PhysRevLett.93.108105 [DOI] [PubMed] [Google Scholar]
- DuFort C.C., Paszek M.J., Weaver V.M. 2011. Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol. 12:308–319 10.1038/nrm3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N., Sougrat R., Spurlin T.A., Hurley J.H., Lippincott-Schwartz J. 2011. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. USA. 108:4846–4851 10.1073/pnas.1102714108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Ram S., Doyle A.D., Conti M.A., Matsumoto K., Adelstein R.S., Yamada K.M. 2007. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat. Cell Biol. 9:299–309 10.1038/ncb1540 [DOI] [PubMed] [Google Scholar]
- Farge E. 2011. Mechanotransduction in development. Curr. Top. Dev. Biol. 95:243–265 10.1016/B978-0-12-385065-2.00008-6 [DOI] [PubMed] [Google Scholar]
- Fiolka R., Shao L., Rego E.H., Davidson M.W., Gustafsson M.G. 2012. Time-lapse two-color 3D imaging of live cells with doubled resolution using structured illumination. Proc. Natl. Acad. Sci. USA. 109:5311–5315 10.1073/pnas.1119262109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel M.L., Sabass B., Ji L., Danuser G., Schwarz U.S., Waterman C.M. 2008. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J. Cell Biol. 183:999–1005 10.1083/jcb.200810060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel M.L., Schneider I.C., Aratyn-Schaus Y., Waterman C.M. 2010. Mechanical integration of actin and adhesion dynamics in cell migration. Annu. Rev. Cell Dev. Biol. 26:315–333 10.1146/annurev.cellbio.011209.122036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M.G., Shao L., Carlton P.M., Wang C.J., Golubovskaya I.N., Cande W.Z., Agard D.A., Sedat J.W. 2008. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94:4957–4970 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath J.P. 1981. Arcs: curved microfilament bundles beneath the dorsal surface of the leading lamellae of moving chick embryo fibroblasts. Cell Biol. Int. Rep. 5:975–980 10.1016/0309-1651(81)90214-9 [DOI] [PubMed] [Google Scholar]
- Heath J.P., Holifield B.F. 1993. On the mechanisms of cortical actin flow and its role in cytoskeletal organisation of fibroblasts. Symp. Soc. Exp. Biol. 47:35–56 [PubMed] [Google Scholar]
- Hotulainen P., Lappalainen P. 2006. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 173:383–394 10.1083/jcb.200511093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K., Ji L., Applegate K.T., Danuser G., Waterman-Storer C.M. 2007. Differential transmission of actin motion within focal adhesions. Science. 315:111–115 10.1126/science.1135085 [DOI] [PubMed] [Google Scholar]
- Keren K., Pincus Z., Allen G.M., Barnhart E.L., Marriott G., Mogilner A., Theriot J.A. 2008. Mechanism of shape determination in motile cells. Nature. 453:475–480 10.1038/nature06952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legant W.R., Choi C.K., Miller J.S., Shao L., Gao L., Betzig E., Chen C.S. 2013. Multidimensional traction force microscopy reveals out-of-plane rotational moments about focal adhesions. Proc. Natl. Acad. Sci. USA. 110:881–886 10.1073/pnas.1207997110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M. 2012. Morphogenetic fields in embryogenesis, regeneration, and cancer: non-local control of complex patterning. Biosystems. 109:243–261 10.1016/j.biosystems.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros N.A., Burnette D.T., Forscher P. 2006. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat. Cell Biol. 8:216–226 10.1038/ncb1367 [DOI] [PubMed] [Google Scholar]
- Mogilner A., Keren K. 2009. The shape of motile cells. Curr. Biol. 19:R762–R771 10.1016/j.cub.2009.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti A., Machacek M., Gupton S.L., Waterman-Storer C.M., Danuser G. 2004. Two distinct actin networks drive the protrusion of migrating cells. Science. 305:1782–1786 10.1126/science.1100533 [DOI] [PubMed] [Google Scholar]
- Rambold A.S., Kostelecky B., Elia N., Lippincott-Schwartz J. 2011. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA. 108:10190–10195 10.1073/pnas.1107402108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riahi R., Yang Y., Zhang D.D., Wong P.K. 2012. Advances in wound-healing assays for probing collective cell migration. J. Lab. Autom. 17:59–65 [DOI] [PubMed] [Google Scholar]
- Sanger J.W., Kang S., Siebrands C.C., Freeman N., Du A., Wang J., Stout A.L., Sanger J.M. 2005. How to build a myofibril. J. Muscle Res. Cell Motil. 26:343–354 10.1007/s10974-005-9016-7 [DOI] [PubMed] [Google Scholar]
- Sanger J.W., Wang J., Fan Y., White J., Sanger J.M. 2010. Assembly and dynamics of myofibrils. J. Biomed. Biotechnol. 2010:858606 10.1155/2010/858606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L., Kner P., Rego E.H., Gustafsson M.G. 2011. Super-resolution 3D microscopy of live whole cells using structured illumination. Nat. Methods. 8:1044–1046 10.1038/nmeth.1734 [DOI] [PubMed] [Google Scholar]
- Small J.V., Rottner K., Kaverina I., Anderson K.I. 1998. Assembling an actin cytoskeleton for cell attachment and movement. Biochim. Biophys. Acta. 1404:271–281 10.1016/S0167-4889(98)00080-9 [DOI] [PubMed] [Google Scholar]
- Straight A.F., Cheung A., Limouze J., Chen I., Westwood N.J., Sellers J.R., Mitchison T.J. 2003. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 299:1743–1747 10.1126/science.1081412 [DOI] [PubMed] [Google Scholar]
- Svitkina T.M., Verkhovsky A.B., McQuade K.M., Borisy G.G. 1997. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 139:397–415 10.1083/jcb.139.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojkander S., Gateva G., Schevzov G., Hotulainen P., Naumanen P., Martin C., Gunning P.W., Lappalainen P. 2011. A molecular pathway for myosin II recruitment to stress fibers. Curr. Biol. 21:539–550 10.1016/j.cub.2011.03.007 [DOI] [PubMed] [Google Scholar]
- Vallenius T. 2013. Actin stress fibre subtypes in mesenchymal-migrating cells. Open Biol. 3:130001 10.1098/rsob.130001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky A.B., Svitkina T.M., Borisy G.G. 1995. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J. Cell Biol. 131:989–1002 10.1083/jcb.131.4.989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Ma X., Adelstein R.S., Horwitz A.R. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10:778–790 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Tolić-Nørrelykke I.M., Chen J., Mijailovich S.M., Butler J.P., Fredberg J.J., Stamenović D. 2002. Cell prestress. I. Stiffness and prestress are closely associated in adherent contractile cells. Am. J. Physiol. Cell Physiol. 282:C606–C616 10.1152/ajpcell.00269.2001 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Takahashi Y. 2010. Tissue morphogenesis coupled with cell shape changes. Curr. Opin. Genet. Dev. 20:443–447 10.1016/j.gde.2010.05.004 [DOI] [PubMed] [Google Scholar]
- Xu K., Babcock H.P., Zhuang X. 2012. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat. Methods. 9:185–188 10.1038/nmeth.1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.F., Schaefer A.W., Burnette D.T., Schoonderwoert V.T., Forscher P. 2003. Rho-dependent contractile responses in the neuronal growth cone are independent of classical peripheral retrograde actin flow. Neuron. 40:931–944 10.1016/S0896-6273(03)00754-2 [DOI] [PubMed] [Google Scholar]
- Zhang H., Gally C., Labouesse M. 2010. Tissue morphogenesis: how multiple cells cooperate to generate a tissue. Curr. Opin. Cell Biol. 22:575–582 10.1016/j.ceb.2010.08.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.