Abstract
Collagen is the most prominent protein of human tissues. Its content and organization define to a large extent the mechanical properties of tissue as well as its function. Methods that have been used traditionally to visualize and analyze collagen are invasive, provide only qualitative or indirect information, and have limited use in studies that aim to understand the dynamic nature of collagen remodeling and its interactions with the surrounding cells and other matrix components. Second harmonic generation (SHG) imaging emerged as a promising noninvasive modality for providing high-resolution images of collagen fibers within thick specimens, such as tissues. In this article, we present a fully automated procedure to acquire quantitative information on the content, orientation, and organization of collagen fibers. We use this procedure to monitor the dynamic remodeling of collagen gels in the absence or presence of fibroblasts over periods of 12 or 14 days. We find that an adaptive thresholding and stretching approach provides great insight to the content of collagen fibers within SHG images without the need for user input. An additional feature-erosion and feature-dilation step is useful for preserving structure and noise removal in images with low signal. To quantitatively assess the orientation of collagen fibers, we extract the orientation index (OI), a parameter based on the power distribution of the spatial-frequency-averaged, two-dimensional Fourier transform of the SHG images. To measure the local organization of the collagen fibers, we access the Hough transform of small tiles of the image and compute the entropy distribution, which represents the probability of finding the direction of fibers along a dominant direction. Using these methods we observed that the presence and number of fibroblasts within the collagen gel significantly affects the remodeling of the collagen matrix. In the absence of fibroblasts, gels contract, especially during the first few days, in a manner that allows the fibers to remain mostly disoriented, as indicated by small OI values. Subtle changes in the local organization of fibers may be taking place as the corresponding entropy values of these gels show a small decrease. The presence of fibroblasts affects the collagen matrix in a manner that is highly dependent on their number. A low density of fibroblasts enhances the rate of initial gel contraction, but ultimately leads to degradation of collagen fibers, which start to organize in localized clumps. This degradation and reorganization is seen within the first days of incubation with fibroblasts at a high density and is followed by de novo collagen fiber deposition by the fibroblasts. These collagen fibers are more highly oriented and organized than the fibers of the original collagen gel. These initial studies demonstrate that SHG imaging in combination with automated image analysis approaches offer a noninvasive and easily implementable method for characterizing important features of the content and organization of collagen in tissuelike specimens. Therefore, these studies could offer important insights for improving tissue engineering and disease diagnostic efforts.
INTRODUCTION
Collagen is the most prominent protein in the human body, making up 30% of its total protein content. As a biomolecule, collagen has a unique role in the extracellular matrix (ECM), providing both structural support and protection of living tissue. Variations in the type, content, and organization of collagen result in tissues with distinctly different mechanical properties such as bone, cartilage, and connective tissue. Monitoring and detection of the collagen characteristics within tissue has important implications in disease detection, wound healing, angiogenesis, and regenerative medicine applications.1, 2, 3, 4, 5, 6, 7, 8 For example, changes in the organization and crosslinking or content of collagen are characteristic of cancerous transformations.9, 10, 11, 12 Therefore, it is important to develop quantitative methods that can provide detail with regards to the content, organization, and dynamic remodeling of collagen fibers.
In this study, we present the use of second harmonic generation (SHG) imaging in combination with analytical approaches as a means to characterize collagen fiber content, organization, and orientation in a noninvasive, fully automated and quantitative way. When an electric field is incident upon an object, it induces a polarization that is described by
where is a constant (permittivity of free space) and is the -order susceptibility of the material. SHG is related to the second term of this equation and, thus, depends on the second-order susceptibility of the object. SHG requires phase matching of the electric field radiated from all the molecules within the focal volume of nonlinear excitation. Thus, no SHG can be detected when the material possesses centrosymmetry or when the molecules that make up a material are randomly oriented because of destructive interference. The fibrillar organization of collagen makes it an ideal natural SHG source.13 In SHG, two low energy photons interact simultaneously with a molecule to yield coherent scattering of a single photon that has the same energy as the two incident photons. In SHG microscopy, a focused beam of light is scanned across the specimen to create a two-dimensional (2D) or three-dimensional high-resolution image. The two-photon requirement confines the volume of SHG to the focal region of the light beam where the density of photons is highest.
The morphology of collagen fibrils has been imaged with SHG in engineered, ex vivo and in vivo tissues.10, 13, 14, 15, 16 It has been shown that SHG signal from collagen fibers is generated at the surface layers of the fibrils in a manner that is highly dependent on the ionic strength of the surrounding solution.14 In addition, characterization of the directionality of the SHG signal, even in a simple form such as the ratio of the SHG signal detected in the forward and backward directions, can provide information about the size of the fibrils, with the backward scattered signal conferring higher sensitivity to smaller, immature fibers.14 Furthermore, fiber orientation affects the intensity and directionality of SHG with fibers lying along the plane that is perpendicular to the plane of propagation of the excitation beam yielding the strongest signal.17, 18 Finally, the degree of organization and alignment within fibrillar bundles has been found to significantly influence the detected SHG signal.19
The analytical tools that have been presented so far for characterizing collagen organization relied at least to a certain extent either on user input,8, 12, 17, 19, 20, 21, 22 or on computationally intense calculations that confine the region of interest to a limited region.9 Here, we present the use of an adaptive thresholding approach in combination with Fourier- and a Hough-transform based analysis as a relatively simple and efficient means of acquiring quantitative information on the content, orientation, and local organization of collagen fibrils. The Fourier-transform-based approach is an established approach that has been used previously to characterize collagen organization, which relies on the autocorrelation of intensity fluctuations within an image and reports indirectly on the level of fiber alignment along a dominant direction within an image. The Hough-transform approach is examined in this study as a complementary approach to characterize directly the local orientation of collagen fibers throughout an entire image. We use this information to describe dynamic remodeling that occurs within collagen type I gels in the absence or presence of two concentrations of fibroblasts, the type of cells that deposit collagen in tissues. These studies illustrate the potential use of these approaches to detect and characterize collagen organization for disease diagnostic and tissue engineering applications.
METHODS
Cell culture
Mouse J2 fibroblasts were cultured in Dulbecco's modified eagle medium (DMEM) (Gibco) and supplemented with 10% fetal bovine serum (FBS) with 1% Gentamicin (Gibco). As the fibroblast cells reached 90% confluence, they were treated with 0.25% trypsin and 0.05% EDTA, centrifuged, and subsequently passaged onto a tissue culture plastic plate at a concentration of . Cell media was changed every two days until seeding into the collagen gel.
Collagen gels
In order to create a simplified ECM model, we used type I rat tail collagen gels. Briefly, a volume of rat tail collagen (BD Bioscience) with an initial concentration of 4 mg/ml was mixed on ice with the appropriate volume of 10× Dubelco’s minimal essential medium (DMEM, Gibco) supplemented with FBS (Gibco) and L-glutamine (Gibco) to yield final collagen concentrations of 1.0, 2.0, and 3.0 mg/ml. The pH of the mix was adjusted to 7.4 with a sodium bicarbonate solution. Finally of the acellular collagen solution was pipetted into a 24 well plate, covering the bottom of the well, and incubated at for 1 h to allow polymerization before adding fibroblast media consisting of high glucose DMEM supplemented with 10% FBS and 0.05% gentamicin (Gibco). After 24 h the gels were removed from the 24 well plates using a scalpel and transferred to a glass bottom petri dish (MatTek Corporation) so that they can be imaged over the course of two weeks. Immediately upon the transfer, 1 ml of media was added to the dish and changed every other day for the duration of the experiment.
The cellular matrices were created following the same protocol as the 3 mg/ml acellular gels. However, upon pH adjustment, a suspension of of fibroblast media containing either 50 000 (low density) or 150 000 (high density) fibroblasts was added to the collagen solution and placed in the incubator.
Second harmonic generation image acquisition
SHG images of the collagen fibers were captured using a Leica TCS SP2 laser scanning confocal microscope with an inverted DM IRE2 microscope. Excitation was achieved at 800 nm using a tunable Ti:sapphire laser (Mai-Tai, Spectra Physics) emitting 100 fs pulses at 80 MHz. The SHG signal scattered in the forward direction was detected using a high transmittance, 20 nm bandpass filter centered at 400 nm designed for two-photon imaging measurements (Chroma, HQ400\20 m-2P).
We used a 63×, 1.2 numerical aperture water immersion objective (Leica) yielding a image of a field in approximately 1 s. We collected depth stacks of 50 images at axial steps, with the most superficial optical section acquired within from the gel surface adjacent to the coverslip (the entire gel was approximately thick). Each image within the stack was frame averaged four times. Imaging data were acquired from two sets of gels from each group and from three distinct locations within each gel. Each gel was approximately 10 mm in diameter and all images were recorded from central regions of the gel that were at least 3 mm away from the edges to avoid artifacts.
Image Analysis: Thresholding and collagen fiber density calculations
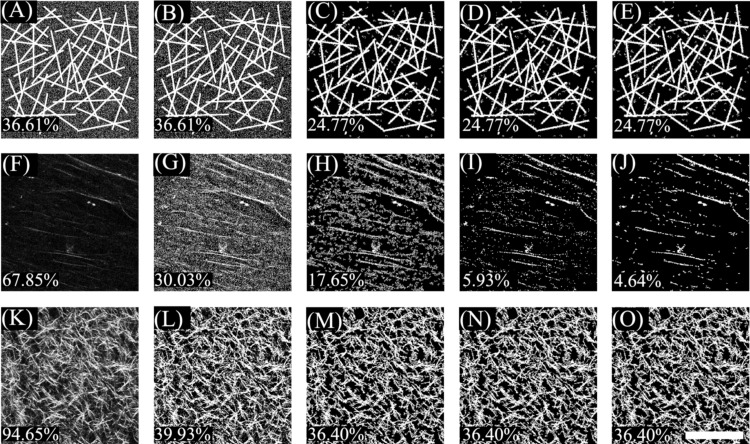
Image analysis was performed using the MATLAB software platform on 2D optical SHG sections. An automated thresholding procedure was developed to enhance the signal to noise ratio of the 8 bit images in an objective manner, so as to extract quantitative information about the content, orientation, and organization of the collagen fibers. The effects of this procedure for a model image (consisting of thin rectangles and 20% Gaussian noise) and two recorded collagen images representative of the signal to noise ratio achieved from the different gels over time, are shown in Fig. 1 (panels A, F, and K). Initially all images were processed using an adaptive threshold algorithm with a kernel size of . Briefly, to calculate the intensity of a pixel with coordinates , we determined the average intensity of all pixels within the square bound by pixels ,, , and and subtracted it from the intensity at pixel . If the resulting intensity was negative, it was reset to zero. While the model image is not affected by this procedure (panel B), the percentage of “on” pixels is reduced significantly for the two acquired collagen images and results in enhanced visualization of the fibers (panels G and L). However, in the case of the fibers whose intensity is only slightly higher above the background (panels F and G), adaptive thresholding results in enhancement of the background noise as well. Following this adaptive thresholding procedure, we also set to zero all pixels that did not belong to a group of 15 interconnected pixels. This procedure results in elimination of most of the Gaussian noise added to the model image (panel C) and the background for the collagen fiber image with a low signal to noise ratio (panel H). An additional noise reduction step was implemented for some of the images that consisted of a few weak SHG producing fibers, as is the case for panels F–H. To automatically classify the images that needed additional processing, the mean and standard deviation of the intensity of the original image was calculated. If the sum of the mean and standard deviation was less than 50 (approximately 20% of the dynamic range of the images), then a element erosion and dilation procedure was implemented using standard MATLAB functions. This yielded a significant reduction in the background noise (panel I), which was improved further by a final step that involved stretching the image so that the intensity of 5% of the pixels with the lowest signal was set to zero and that of 10% of the pixels with the highest intensity was set to 255. All pixels with resulting values less than 55 were set to 0 and a final erosion and dilation step was implemented (panel J). The threshold value of 55 was chosen as it represents the lower 20% values of the dynamic range of the image intensities and it resulted consistently in reasonable levels of noise reduction. The number of pixels that had nonzero intensity was calculated and reported as a percentage of the total number of pixels within the field of view. This number is referred to as the fiber density. The final fiber density number of the model collagen fiber image (24.77%) is very close to the actual fiber density in the image (21.15%).
Figure 1.
Effects of the different steps involved in image thresholding for (a) model image consisting of bright sticks that model collagen fibers and 20% Gaussian noise, (f) an acquired collagen fiber image with low signal to noise ratio, and (k) an acquired collagen fiber image with high signal to noise ratio. Adaptive thresholding is performed on those images to achieve the corresponding fields shown on panels (b), (g), and (l). The results of eliminating any pixels that do not belong to a cluster of at least 15 interconnected “on” pixels is shown on panels (c), (h), and (m). Only panel (h) meets the requirements for further processing, which involves erosion and dilation (i) stretching, thresholding, and a final erosion and dilation step (j). The corresponding fiber densities are displayed at the bottom left corner of each panel. Scale
Image analysis: Fiber alignment assessment
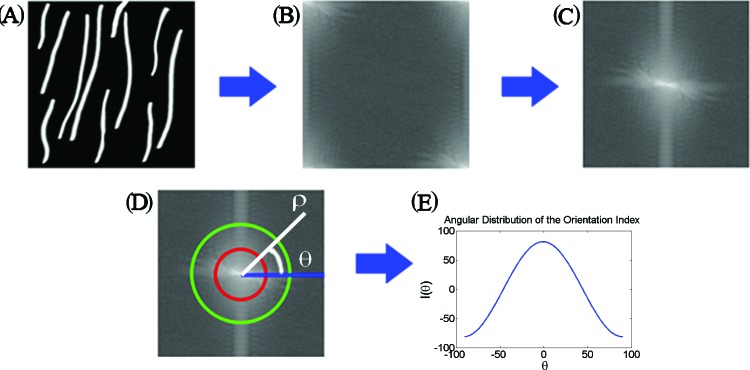
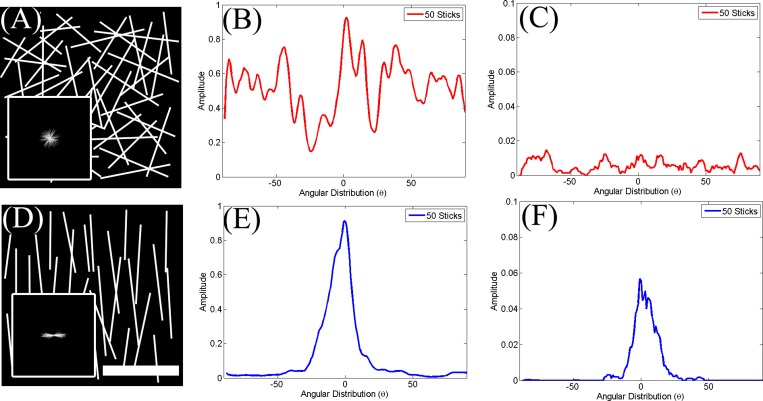
The level of collagen fiber alignment was quantitatively assessed using the orientation index (OI) as described previously.23 The main steps of this approach are depicted schematically in Fig. 2. Initially, we decomposed the image [Fig. 2a] into its spatial frequency components by performing a 2D discrete Fourier transform (DFT). When using the DFT routine supplied by MATLAB, the resulting image [Fig. 2b] contained the amplitude of the lower frequency components at the four corners of the image and that of the highest frequency components at the center. This image was remapped into a polar coordinate representation, with the distance from the origin representing the amplitude of increasing spatial frequency components and the angle from the horizontal representing the direction on the original image along which the corresponding spatial frequency decomposition was calculated. The power spectral density (PSD) was then determined as the squared amplitude or autocorrelation of the 2D DFT, as depicted in Fig. 2c. The PSD of collagen fibers that are arranged randomly within a field [Fig. 3a] appears round and uniform [Fig. 3a, inset], while the PSD of mostly aligned fibers [Fig. 3d] is highly elongated and directional [Fig. 3d, inset]. The information in this 2D image can be compressed into a line plot of the spatial frequency averaged intensity as a function of angle, [Fig. 2e]. This function fluctuates randomly for collagen fibers arranged randomly over the range of angles [Fig. 3b] and exhibits a well defined peak when the fibers are highly aligned [Fig. 3e]. From we identified the angle at which the intensity was maximum and we defined that as the dominant direction , which was set to a value of 0°. Then the OI, representing the percentage of fibrils within the image parallel to , was defined as
Thus, an OI of 100% represents an image with all fibers aligned along this direction, while an OI of 0% is representative of random alignment. The OI for the randomly and highly aligned model collagen fiber images shown in Fig. 3 were 2.3% and 70.4%, respectively.
Figure 2.
Schematic of the steps followed for the extraction of the OI for a model SHG image. Initially, the 2D Fourier transform of the image shown in panel (a) is calculated, as shown in (b). The coordinates of the Fourier image are transformed so that the spatial frequency is lowest at the origin and increases as we move toward the image edges (c). The frequency components along different orientations are represented by the amplitude along a specific angle θ from the horizontal (d). The information in each map can be compacted into a line plot of the spatial frequency averaged intensity as a function of the angle θ (e). From such plots the OI of each image is calculated.
Figure 3.
OI and entropy distributions for model images consisting of sticks (collagen fibers) arranged in a (a) highly random or (d) aligned orientation. The corresponding spatially frequency averaged angular power spectral density distributions are shown in (b) and (e), while the probability distributions from the Hough-transform-based analysis are shown in (c) and (f). The distributions are broad and exhibit strong fluctuations for the randomly oriented sticks, while they consist of a well-defined peak in the highly aligned configuration. Scale represents .
Image analysis: Fiber organization
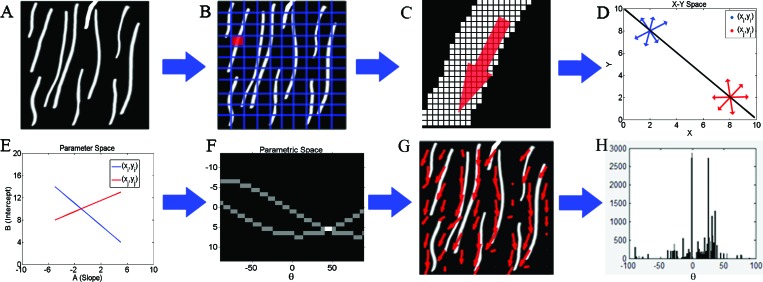
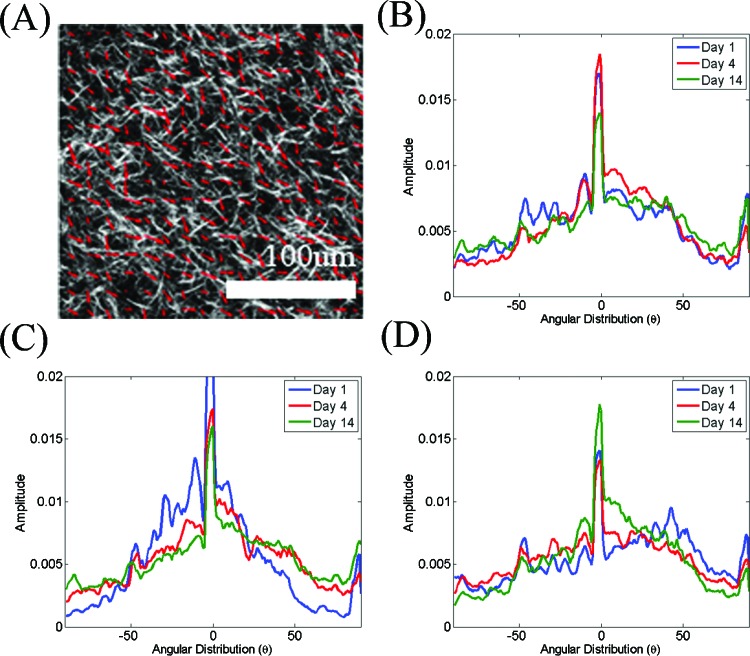
To acquire an additional quantitative measure of the level of organization of collagen fibers, we used the entropy distribution of the image based on the Hough transform.24 Unlike the Fourier-based method, which relies on analysis of the intensity fluctuations within an image from a statistical perspective, the Hough transform is a more direct measure of the local orientation of collagen fibers. The approach is depicted schematically in Fig. 4. Specifically, we divided each frame [Fig. 4a] into tiles [Fig. 4b]. The Hough transform was used to identify the direction and relative magnitude of the dominant line in each tile. A magnified picture of one of the tiles is shown in Fig. 4c, with the presence of multiple points along a fibril segment. The infinite number of lines crossing a point with slopes and intercepts and such that , is represented by a line in space, i.e., . Now if we consider a second point in space, there would be another line , in space representing all the lines that intersect this point in space. If and are points of a line in space, this line is represented by the point , where the two lines in space intersect [Figs. 4d, 4e]. Therefore, the point where most of the lines in space intersect represents the dominant direction of the lines within a tile. The number of lines that intersect at that point represents the level of dominance of that direction. Therefore, for each tile, the Hough transform provides the direction and magnitude of the dominant lines. In order to avoid problems arising from representing vertical lines with infinite slope values, the Hough transform employs a parametric description of a line through a point such that , where θ is the angle from the horizontal direction and ρ represents the distance from the origin. So, the set of lines going through a point is represented by a curved line in space. The points where these curved lines cross represent the coordinates of lines within the original image as in the representation [Fig. 4f]. Using this representation the direction and amplitude of the dominant lines with each tile were calculated, as depicted in Fig. 4g. The probability of encountering a dominant line along a particular direction was then estimated as [Fig. 4h] and the entropy of this distribution was defined as . The distributions for the model images of Fig. 3 for the random and highly aligned collagen fiber cases are shown in Figs. 3c, 3f, respectively, demonstrating that a with a well-defined peak corresponds to a field with high levels of organization. The entropy is a measure of order of this distribution and in our study it represents a quantitative measure of the local fibril organization, with lower ε values corresponding to more organized fibers. For example, the entropy was 6.58 and 4.9 for the images in Figs. 3a, 3c, respectively.
Figure 4.
Schematic of the steps followed for the extraction of the entropy for a model SHG image, included in (a). Initially, the image is segmented into tiles (b), so that each tile is likely to consist of a fairly linear segment of a collagen fiber (C is a magnified version of a single tile). The Hough transform is performed on each tile to determine the dominant direction of the collagen fiber segments present. If we consider two points in space, there is an infinite number of lines that go through these points represented by red and blue linear segments (panel D). All the lines intersecting point shown in blue are represented by a line (also shown in blue) in the space representing the slopes and intercepts of these lines (panel E). The lines intersecting the second point are represented by the red line. The point where the red and blue lines cross in panel E represents the intercept and slope of the line that connects the two points in space (shown in black in panel D). To avoid the need to include infinite slope numbers to represent vertical lines, we use a parametric description of a line through a point such that , where θ is the angle from the horizontal direction and ρ represents the distance from the origin. So, the set of lines going through a point is represented by a curved line in space (panel F). The point where the two lines in F intersect represents the line connecting the two points in panel D. Thus, for each tile the points where most of the lines intersect represents the dominant direction of the lines present in the tile. This is represented by the red arrows in panel G. The probability of finding the dominant direction along a particular angle for a given tile of the image is shown in H. The entropy of this distribution is used a measure of local organization of the fibers.
Statistical analysis
The two-tailed student t-test was used to assess statistical difference with between the mean values of parameters representing two groups of gels. The tests were performed using the MATLAB statistics toolbox.
RESULTS
Dynamic changes in acellular collagen matrices
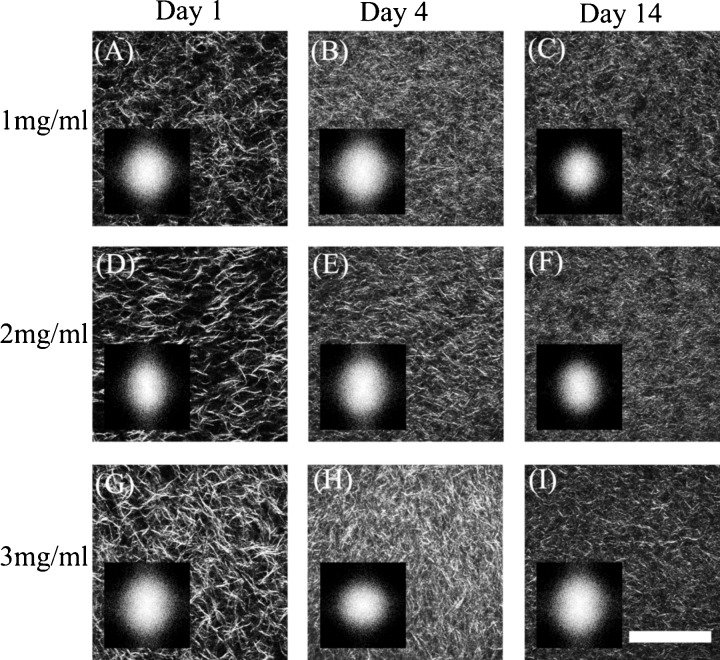
The density and organization of collagen fibrils were monitored within the same set of gels over the course of two weeks. Representative optical sections from collagen gels with an initial concentration of 1, 2, and 3 mg/ml are shown in Fig. 5 for days 1, 4, and 14. Simple visual inspection of these images reveals initial differences in the density of collagen fibrils present in the gels during the onset of the experiment. These differences become less obvious by day 4.
Figure 5.
Representative SHG images from gels with original collagen concentration of 1.0 (A–C), 2.0 (D–F), and 3.0 (G–I) mg/ml following 1 (A, D, G), 3 (B, E, H), and 14 (C, F, I) days upon the onset of gelation. The 2D PSD for each SHG image is shown as an inset. .
We employed three different metrics to quantify the dynamic changes within a particular concentration of collagen gels and to identify differences in the morphological properties and remodeling between gels of different initial collagen concentrations. To assess the overall density of collagen fibers detectable within an image, we thresholded the images following an adaptive threshold and stretching approach and calculated the percentage of pixels that were on, relative to the total number of pixels within a frame, as described in detail in the methods section. As shown in Table 1, the mean percentage of the positive pixels on the first day of each experiment increased linearly with the initial concentration of collagen within the gels. The results from day 4 and 14 indicate that these initial differences do not persist as a function of time. While all types of gels contract leading to a significant increase in the percentage of on pixels and thus in collagen density , the relative level of contraction is more significant for the lowest concentration gels during the first few days. In fact, by day 4 the correlation between the initial collagen concentration and the collagen fiber density in our SHG images is eliminated.
Table 1.
Quantitative assessment of collagen fiber content and organization in acellular collagen gels.
| Fiber density(%) | OI | Entropy | |
|---|---|---|---|
| Day 1 | |||
| 1 mg/ml | |||
| 2 mg/ml | |||
| 3 mg/ml | |||
| Day 4 | |||
| 1 mg/ml | |||
| 2 mg/ml | |||
| 3 mg/ml | |||
| Day 14 | |||
| 1 mg/ml | |||
| 2 mg/ml | |||
| 3 mg/ml | |||
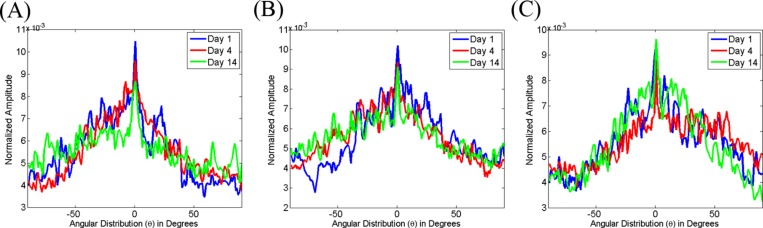
The 2D PSDs for each image included in Fig. 5 are shown in the corresponding insets. The shapes of these PSDs are very similar, suggesting that the relative changes in organization of the collagen fibers within these images are subtle. These subtle changes can be quantified by examining more carefully the form of the spatial frequency-averaged PSD distributions as a function of angle, i.e., . Representative area normalized distributions from each set of collagen gels on days 1, 4, and 14 are shown in Fig. 6. The shape of these distributions is fairly broad and remains rather consistent for the duration of the experiment. A more pronounced change in the shape of the distribution can be observed for day 14 of the 1 mg/ml collagen gel. The OI represents a quantitative measure that can characterize these distributions, as it is the percentage of collagen fibers aligned along the dominant direction (set to in Fig. 6). The mean OI values for each type of gel are included in Table 1 and reveal the mostly disoriented appearance of the collagen fibers within these gels. In most cases the OI has values that are lower than 15%, with the 1 mg/ml gels on day 14 exhibiting the lowest OI of approximately 7%.
Figure 6.
Representative area normalized distributions of determined from the 2D Fourier transform of the images are shown for day 1 (blue), 3 (red), and 14 (green) of observation from collagen gels with initial collagen concentration of (a) 1, (b) 2, and (c) 3 mg/ml. Note that the shape of these distributions does not change significantly over time.
A more direct measure of alignment is represented by the probability distribution of finding the dominant direction of collagen fibers within a particular image section along a particular direction θ, , and is shown in Fig. 7. The shape of these distributions remains fairly consistent for all types of gels. The entropy values of these distributions are included in Table 1 and confirm the presence of consistently disorganized fibers. Nevertheless, a trend toward slightly smaller entropy values for day 14 as compared to day 1 is observed and may reveal very subtle changes in organization. Interestingly, the OI and entropy values do not always exhibit the same trends, suggesting that these two parameters report on somewhat different aspects of organization of the image elements.
Figure 7.
Representative area normalized distributions of the probability that the dominant direction of the linear segments within each image tile is along a particular direction as assessed by the Hough transform. Data is shown for day 1 (blue), 3 (red), and 14 (green) of observation from collagen gels with initial collagen concentration of (a) 1, (b) 2, and (c) 3 mg/ml.
In summary, dynamic monitoring of acellular collagen gels allows us to observe significant levels of contraction over the course of two weeks, which occurs at different rates depending on the initial concentration of collagen. This contraction is accompanied by a small decrease in the OI and entropy values of the gels. Generally, both the OI and entropy values reveal collagen fibers that are not well organized or oriented along a dominant direction.
Dynamic changes in collagen gels with a low concentration of fibroblasts
When fibroblasts are present in the collagen gels, the dynamic changes that occur over the same period of time are significantly different from the acellular gels. Representative SHG images of a gel seeded initially with 50 000 fibroblasts are shown for a period of two weeks in Fig. 8, with the corresponding PSD distributions as insets. We observe an increase in the density of collagen fibers during the first seven days, followed by the appearance of significant gaps by day 12. The collagen fiber density values reported in Table 2 indicate that the presence of fibroblasts affects significantly the observed fiber density achieved following 1 day of incubation. The level of contraction that occurs during the first 3–7 days is higher in these 3 mg/ml gels than in the corresponding acellular gels. However, the fiber density decreases by day 12, probably due to enzymes secreted by the fibroblasts that degrade the collagen.
Figure 8.
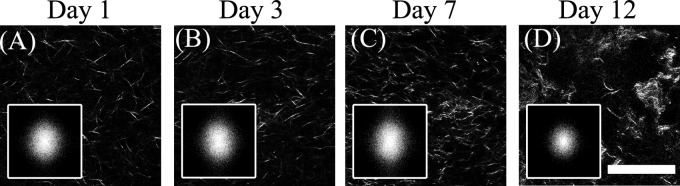
Representative SHG images from gels with 3 mg/ml original collagen concentration seeded with 70 000 fibroblasts following (a) 1, (b) 3, (c) 7, and (d) 12 days from the onset of gelation. A significant change in the appearance of the gels is seen on day 12. The corresponding 2D PSDs are included in the insets, demonstrating a noticeably more uniform distribution for the last day of measurements. Scale .
Table 2.
Quantitative assessment of collagen fiber content and organization in collagen gels containing a low concentration of fibroblasts.
| Fiber density | OI | Entropy | |
|---|---|---|---|
| Day 1 | |||
| Day 3 | |||
| Day 7 | |||
| Day 12 |
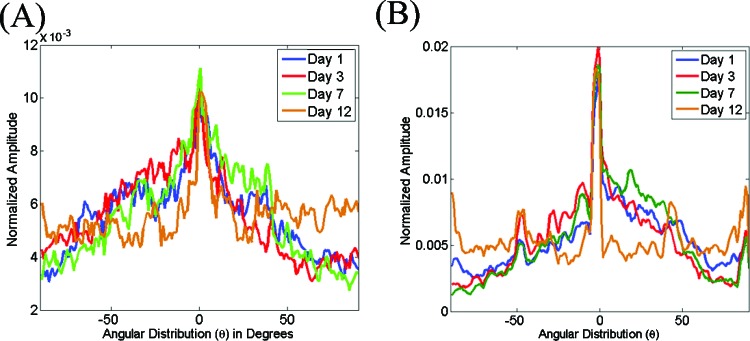
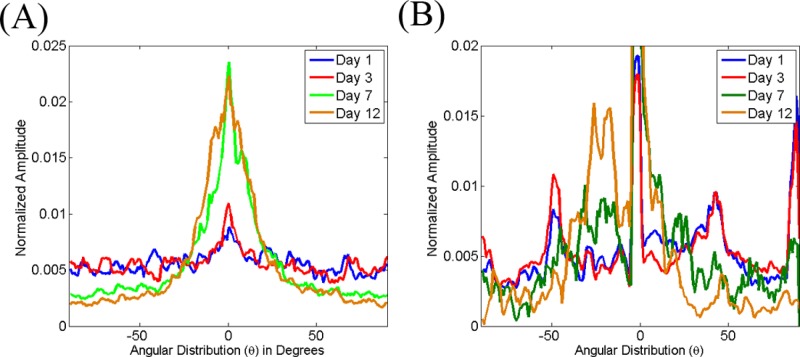
From the insets in Fig. 8, we can see that the shape of the PSD on day 12 appears rounder than the ones for days 1–7. This is consistent with a significant change in the shape of the distribution, which is flat over almost the entire angular range [Fig. 9a]. The corresponding mean OI values for these gels provide a quantitative measure for these changes, indicating a significant decrease in the percentage of fibers oriented along a dominant direction to only 5% for day 12 (Table 2). Interestingly the mean entropy values (Table 2) characterizing the probability distribution of fibers along a particular direction within each tile of the images decreases consistently as a function of time, indicating gradual enhancements in the overall level of organization of the gels. Representative distributions from a gel are included in Fig. 9b, demonstrating explicitly the significant changes that are observed during the last time point. This may suggest that the organization of the collagen fiber clumps at the later time point remain organized and this organization is perceived more accurately by the entropy values, which result from the direct localized characterization of fibers, rather than by the OI values, which are derived from a statistical evaluation of the entire field.
Figure 9.
Representative area normalized distributions extracted from the 2D Fourier transform (a) and Hough transform (b) of SHG images acquired from gels seeded with 70 000 fibroblasts following 1 (blue), 3 (red), 7 (green), and 12 (brown) days from the onset of gelation. Note the change in distribution observed for the day 12 measurements.
Dynamic changes in collagen gels with a high concentration of fibroblasts
Remodeling of the collagen gels by fibroblasts is even more pronounced when they are embedded in the gels in higher numbers (150 000 fibroblasts per gel). Specifically, as shown in Fig. 10, the SHG images acquired during day 1 and 3 appear more like the collagen gels embedded with low concentration of fibroblasts for 12 days, with collagen fibers organized in clumps that are separated by significant gaps. On days 7 and 12 there is a significant decrease in the number of fibers that yield SHG signal. Those fibers appear long and straight, in contrast with the short, curved fibers that characterized the collagen fibers of the original gels. It is most likely that these long fibers are fibers deposited by the fibroblasts themselves.
Figure 10.
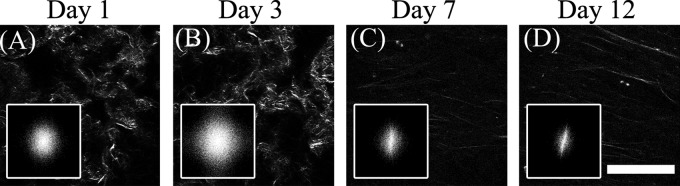
Representative SHG images from gels with 3 mg/ml original collagen concentration seeded with 150 000 fibroblasts following (a) 1, (b) 3, (c) 7, and (d) 12 days from the onset of gelation. On days 1 and 3 the gels appear similar to the gels seeded with a lower concentration of fibroblasts for 12 days. The collagen fibers detected on days 7 and 12 appear significantly longer and better aligned than the previous time points. These likely represent fibers deposited by the fibroblasts. The corresponding 2D PSDs are included in the insets, demonstrating highly nonuniform distributions for the measurements acquired on days 7 and 12. Scale .
The completely distinct morphology of the fibroblast deposited fibers results in very different distributions for the spatial frequency averaged PSD intensity [Fig. 11a] and the probability that the dominant direction of the fibers within each image tile will be along a particular angle [Fig. 11b]. The corresponding OI and entropy values report these changes quantitatively (Table 3). Specifically, we find that while the original fibroblast remodeled collagen gel fibers have very low OI and high entropy values (days 1 and 3), the fibroblast deposited collagen fibers are characterized by high OI (20%–30%) and low entropy (3.9–4.6) values. The decrease in the number of fibers observed within a given field on days 7 and 12 are also consistently reported by the collagen fiber density values, which decrease to only approximately 20% from original values that were twice as high.
Figure 11.
Representative area normalized distributions extracted from the 2D Fourier transform (a) and Hough transform (b) of SHG images acquired from gels seeded with 150 000 fibroblasts following 1 (blue), 3 (red), 7 (green), and 12 (brown) days from the onset of gelation. Note the substantial changes in the distributions observed for the measurements on days 7 and 12.
Table 3.
Quantitative assessment of collagen fiber content and organization in collagen gels containing a high concentration of fibroblasts.
| Fiber density | OI | Entropy |
|---|---|---|
DISCUSSION
Collagen content and organization determine to a large extent the tensile strength of human tissues and reveal important information with regards to the health status and normal development or regeneration of tissue. As such, they play a central role in tissue engineering, regenerative medicine, and disease diagnostic technology efforts. Therefore, their characterization and monitoring, especially in a dynamic, noninvasive manner could play a significant role in the advancement of these areas from the basic research to the clinical stages.
Standard methods for tissue staining, such as hematoxylin and eosin as well as antibody-based immunohistochemical methods, can be used to visualize collagen fibers and provide information with regards to the specific type of collagen fibers that are present.25 However, these approaches are invasive, require thin tissue sections, are time consuming, and yield rather qualitative results. Other methods such as real-time polymerase chain reaction, in situ hybridization and enzyme linked immunosorbent assays (ELISA) yield quantitative results on the expression of genes responsible for collagen synthesis; however, such approaches cannot assess the actual levels of collagen present or their organization.26, 27, 28 Radiolabeling-based approaches have been used to quantify collagen trafficking in a quantitative manner, but those are tedious and not suitable for in vivo applications.29 Scanning and transmission electron microscopy (SEM/TEM) can be used to probe the collagen microstructure, but only in thin samples and following extensive preprocessing.30 Other techniques such as polarization microscopy exploit the unique birefringence properties of collagen fibers, but yield quantitative results only in thin samples as well.31
Noninvasive imaging approaches to visualize the fibrillar types of collagen in thick specimens are being developed as more suitable alternatives for in vivo applications. For example, high resolution imaging of collagen fibers has been achieved using confocal reflectance and fluorescence imaging.9, 23, 32 Algorithms for three-dimensional reconstruction of collagen fibers have been developed.9, 32 This type of work is providing useful insights in the remodeling of the ECM during the development of early cancerous changes.9 However, so far these reconstructions have been limited to regions that extend only a few microns because they are computationally intensive. A 2D Fourier-based analysis approach was developed and used to describe collagen remodeling by fibroblasts in the absence or presence of Rho kinase.23 Results from this study suggest that coalignment of collagen fibrils and cells along the plane of greatest resistance depends on the generation of contractile forces that in turn depend on Rho kinase. This type of information may be ultimately useful in controlling the organization of collagen fibers within tissues. This is the approach that we also employed in our study for extraction of the OI values, since the method relies on direct image analysis (i.e., it does not require any model-based fitting of the data) to provide quantitative orientation information.
SHG imaging provides a number of advantages over confocal microscopy for in vivo imaging. For example, the use of longer wavelengths (typically around 800 nm) where tissue scattering and absorption are significantly lower than in the visible allows deeper penetration for SHG excitation. Also, the confinement of SHG excitation within a small volume eliminates the need for a confocal detection pinhole and allows for more efficient signal collection. However, the efficiency of SHG generation depends on the relative orientation of the collagen fibers with respect to the polarization of the incident beam,17, 18, 22, 33, 34 the arrangement of adjacent collagen fibrils34 and environmental factors such as ionic strength14 and this may introduce errors in the collagen content estimates performed by SHG image analysis. The use or circularly polarized light and three-dimensional image analysis approaches may offer options for overcoming some of these effects.
Despite such potential limitations, a number of studies have been recently performed that illustrate the potential of SHG imaging to provide nondestructively unique insights in ECM properties that play a critical role in the health status of a tissue. SHG images acquired in vivo have been analyzed to assess, for example, the effects of collagenases on the integrity of collagen fibers and their relationship to drug diffusivity.10 Furthermore, collagen content estimates extracted from analysis of SHG images were used to characterize lung fibrosis in ex vivo mouse tissues.35 Significant differences in the second order nonlinear susceptibility of collagen fibers have also been discovered in ex vivo biopsies of normal and malignant melanoma skin tissue.33 Quantitative characterization of the SHG intensity correlation as a function of pixel distance from ex vivo images of ovarian tissue revealed significant loss in the organization of fibers in the stroma of cancer epithelium when compared to normal ovarian tissue. Such organizational differences were also observed in the stroma of focal lesions and to a lesser extent in the stroma of normal-high risk patients.20 Finally, SHG imaging has been used to examine the collagen fiber morphology surrounding tumor cells in vivo and in tumor explants.12 The latter study clearly illustrated that the matrix reorganizes significantly in areas of cancer cell invasion, so that the collagen fibers align with the cancer cells and facilitate cell motility into the matrix. Fiber orientation was assessed using ImageJ functions and it was not clear what was the algorithm or the area over which quantitative calculations were performed.
In addition to SHG measurements performed using animal or human tissues, a number of investigators employ collagen type I gels, similar to the ones described in this study, as a relatively simple in vitro model to assess the effects of specific factors on the detected SHG signal or to establish correlations between SHG image features and tissue properties of interest. For example, SHG images of 3 mg/ml collagen type I gels reported by Kirkpatrick et al. exhibit a very similar morphology to the images acquired from the acellular gels of our study, consisting of a highly disorganized mesh of collagen fibers.21 Analysis based on the correlation of SHG intensity as a function of pixel distance did not reveal any significant changes in the organization of collagen fibers during the first two days of observation following the onset of polymerization. However, by day 8 the gels exhibited lower correlation distances, consistent with the decreased levels of OI and entropy that we observe in the acellular gels of this study over 14 days of observation. Nevertheless, Kirkpatrick et al. did not observe any significant changes in the density of collagen fibers. The significant level of contraction that we observe in our gels is attributed to the presence of serum in the medium.
A series of elegant SHG studies has been performed by Raub et al. to characterize factors that affect the microstructure and mechanical properties of collagen gels. The investigators discovered that the polymerization temperature had a significant effect on the structure of collagen fibers, with colder temperatures resulting in more loosely packed, thicker collagen fibers with brighter SHG.17 Comparison between SHG image characteristics and gel mechanical properties revealed that the storage and elastic moduli of the gels correlated positively with SHG image fraction and negatively with the mean segmented SHG intensity.17 Therefore, the volume fraction of fibers (as reported by the SHG image fraction) rather than the fiber thickness (which correlates with SHG intensity) determines the bulk mechanical properties of the gels. Analysis of the acquired SHG images relied on segmentation that was based on the identification of regions that were devoid of collagen. Therefore, this analysis approach may introduce errors when the samples consist of densely packed collagen fibers, as in the case of their gels polymerized at higher temperatures and all our acellular gels.
More recently, Raub et al. exploited more sophisticated image correlation spectroscopy approaches to analyze SHG images of collagen type I gels and assess in greater detail the relationship between extracted SHG image features and the storage modulus of the gels.18 Specifically, SHG images were thresholded, based on the mean and standard deviation of the intensities of 12 sample regions, and filtered using a high and low pass frequency filter. Then, the 2D spatial autocorrelation function was calculated and fit to a 2D Gaussian function. The standard deviation of this Gaussian was used to define a characteristic fiber length scale, as , and a pore size scale, as . It was found that image thresholding was necessary to achieve a good correlation between the characteristic fiber size from SHG image analysis and the mean fiber diameter based on measurements of individual fibers identified manually. However, tended to be larger than the manual diameter estimates. When the same type of analysis was performed on SEM images, there was better agreement between the autocorrelation based and manually determined fiber diameters, suggesting that the accuracy of the SHG image analysis may be more susceptible to errors, possibly as a result of less optimal signal to noise ratio. In addition, it was found that the measured storage modulus of gels polymerized at different pH values correlated well with corresponding values extracted from SHG images analyzed using a particle analysis ImageJ function and manually calculated fiber diameters. This correlation was more modest when the investigators used the pore and fiber length scales extracted from their image correlation analysis approach. However, given that this analysis was fully automated and, thus, could be implemented to analyze images in real time, these initial results are highly promising.
Such studies demonstrate that SHG collagen imaging has the potential to characterize noninvasively important tissue mechanical properties that can in turn be used to determine the presence of diseases, the effects of drugs or the development of tissues with mechanical properties that would make them suitable for implantation. Our study offers an alternative approach to the automated characterization of SHG image analysis and also demonstrates the significant effects that the seeding density of fibroblasts can have on the remodeling process. We find that adaptive thresholding offers a robust initial processing step for images with varying degrees of signal to noise ratios, allowing us to extract reasonable estimates of fiber density within a given field of view. Of course, the dependence of SHG intensity on fiber orientation limits the sensitivity of our measurements to fibers that are not parallel to the image plane. Extension of our analysis to three-dimensional image stacks may alleviate some of these errors.
The OI and entropy calculations provide two alternative measures for assessing fiber organization at more global and local scales, respectively. Similar to the image correlation spectroscopy analysis approaches described by Raub et al.,18 the OI is based on assessment of the power spectral density features of the entire SHG image, but in the frequency domain not in the space domain. In addition, the OI relies on correlation of the image intensity fluctuations over a more extended frequency range. In fact, the combination of approaches may offer a more complete characterization of collagen fiber microstructure, which in turn could be more accurate in terms of sensing diagnostically useful changes or assessing bulk tissue mechanical properties. In contrast with such Fourier-based methods, which characterize intensity fluctuations within an image from a statistical perspective, the Hough-transform based approach provides a more direct means of assessing the local fiber orientation within an image in order to characterize global structure. It is likely to prove more useful when more localized changes in the collagen organization are of interest, as in the cases of angiogenesis8 and cancer cell invasion through the matrix,12 for example. Also, this approach may be more robust when significant gaps are present within a field. Polarization-dependent and polarization-modulated measurements could offer more accurate and detailed information on fiber organization, but they require more involved data acquisition procedures and may be more suitable for thin samples.22, 34
In summary, these studies illustrate that dynamic monitoring along with automated and easy to implement image analysis approaches could yield important information with regards to tissue status and function. These approaches will become increasingly more useful as SHG imaging finds broader use in in vivo studies for tissue engineering, wound healing, disease diagnostic, and drug assessment applications.
CONCLUSION
In this report, we present a fully automated and quantitative way to characterize the content and organization of collagen fibers visualized using SHG imaging. Adaptive thresholding and stretching is important for the initial image processing steps and the acquisition of meaningful collagen content information. Fourier- and Hough-transform based image analysis approaches can serve as useful methods for acquiring quantitative information with regards to the orientation and organization of collagen fibers. The OI, representing the percent of fibers aligned along a particular direction, reports the remodeling of collagen fibers within collagen gels in the presence or absence of fibroblasts. The entropy of Hough-transform-based distributions is sensitive to the local organization of collagen fibers and could serve as a second useful metric for characterizing the collagen matrix. Using these approaches, we are able to report significant differences in the morphology of collagen gels depending on the presence and absolute number of fibroblasts. These differences are reported in a highly quantitative manner as the collagen fiber density, OI and entropy of the gels. We anticipate that the approaches we present will be used to characterize collagen organization in engineered and human or animal tissues to assess their functional development or health status and gain improved understanding of the role and function of this important structural protein. The methods are easy and fast to implement and could be used for near real-time image analysis, which will be particularly important for clinical applications. It will be interesting to explore their expansion to the direct analysis of three-dimensional image cubes, potentially taking advantage of some of the methods that have already been developed.9, 32
ACKNOWLEDGMENTS
We wish to thank Olusola Akapo from the ECE department at Tufts University for development of the adaptive thresholding code. The study was supported by NSF (Grant No. BES0547292) and NIH (Grant No. P41EB002520).
References
- Bogenrieder T. and Herlyn M., Oncogene 22, 6524 (2003). 10.1038/sj.onc.1206757 [DOI] [PubMed] [Google Scholar]
- Connon C. J. and Meek K. M., Wound Repair Regen 11, 71 (2003). 10.1046/j.1524-475X.2003.11111.x [DOI] [PubMed] [Google Scholar]
- Friedl P., Histochem. Cell Biol. 122, 183 (2004). 10.1007/s00418-004-0682-0 [DOI] [PubMed] [Google Scholar]
- Friess W., Eur. J. Pharm. Biopharm. 45, 113 (1998). 10.1016/S0939-6411(98)00017-4 [DOI] [PubMed] [Google Scholar]
- Gelse K., Poschl E., and Aigner T., Adv. Drug Delivery Rev. 55, 1531 (2003). 10.1016/j.addr.2003.08.002 [DOI] [PubMed] [Google Scholar]
- Ingber D. E., Differentiation 70, 547 (2002). 10.1046/j.1432-0436.2002.700908.x [DOI] [PubMed] [Google Scholar]
- Vrana N. E., Elsheikh A., Builles N., Damour O., and Hasirci V., Biomaterials 28, 4303 (2007). 10.1016/j.biomaterials.2007.06.013 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick N. D., Andreou S., Hoying J. B., and Utzinger U., Am. J. Physiol. Heart Circ. Physiol. 292, H3198 (2007). 10.1152/ajpheart.01234.2006 [DOI] [PubMed] [Google Scholar]
- Arifler D., Pavlova I., Gillenwater A., and Richards-Kortum R., Biophys. J. 92, 3260 (2007). 10.1529/biophysj.106.089839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E., McKee T., diTomaso E., Pluen A., Seed B., Boucher Y., and Jain R. K., Nat. Med. 9, 796 (2003). 10.1038/nm879 [DOI] [PubMed] [Google Scholar]
- Friedl P., Maaser K., Klein C. E., Niggemann B., Krohne G., and Zanker K. S., Cancer Res. 57, 2061 (1997). [PubMed] [Google Scholar]
- Provenzano P. P., Eliceiri K. W., Campbell J. M., Inman D. R., White J. G., and Keely P. J., BMC Med. 4, 38 (2006). 10.1186/1741-7015-4-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagnola P. J. and Loew L. M., Nat. Biotechnol. 21, 1356 (2003). 10.1038/nbt894 [DOI] [PubMed] [Google Scholar]
- Williams R. M., Zipfel W. R., and Webb W. W., Biophys. J. 88, 1377 (2005). 10.1529/biophysj.104.047308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh A. T., Nassif N., Zoumi A., and Tromberg B. J., Opt. Lett. 27, 2082 (2002). 10.1364/OL.27.002082 [DOI] [PubMed] [Google Scholar]
- Zoumi A., Yeh A., and Tromberg B. J., Proc. Natl. Acad. Sci. U.S.A. 99, 11014 (2002). 10.1073/pnas.172368799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raub C. B., Suresh V., Krasieva T., Lyubovitsky J., Mih J. D., Putnam A. J., Tromberg B. J., and George S. C., Biophys. J. 92, 2212 (2007). 10.1529/biophysj.106.097998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raub C. B., Unruh J., Suresh V., Krasieva T., Lindmo T., Gratton E., Tromberg B. J., and George S. C., Biophys. J. 94, 2361 (2008). 10.1529/biophysj.107.120006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoller P., Reiser K. M., Celliers P. M., and Rubenchik A. M., Biophys. J. 82, 3330 (2002). 10.1016/S0006-3495(02)75673-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick N. D., Brewer M. A., and Utzinger U., Cancer Epidemiol. Biomarkers Prev. 16, 2048 (2007). 10.1158/1055-9965.EPI-07-0009 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick N. D., Hoying J. B., Botting S. K., Weiss J. A., and Utzinger U., J. Biomed. Opt. 11, 054021 (2006). 10.1117/1.2360516 [DOI] [PubMed] [Google Scholar]
- Stoller P., Kim B. M., Rubenchik A. M., Reiser K. M., and Da Silva L. B., J. Biomed. Opt. 7, 205 (2002). 10.1117/1.1431967 [DOI] [PubMed] [Google Scholar]
- Kim A., Lakshman N., and Petroll W. M., Exp. Cell Res. 312, 3683 (2006). 10.1016/j.yexcr.2006.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R. and Woods R., Digital Image Processing (Prentice-Hall, Upper Saddle River, NJ, 2002). [Google Scholar]
- Ponsioen T. L., van Luyn M. J., van der Worp R. J., van Meurs J. C., Hooymans J. M., and Los L. I., Invest. Ophthalmol. Visual Sci. 49, 4089 (2008). 10.1167/iovs.07-1456 [DOI] [PubMed] [Google Scholar]
- Damoulis P. D., Drakos D. E., Gagari E., and Kaplan D. L., Ann. N.Y. Acad. Sci. 1117, 367 (2007). 10.1196/annals.1402.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber H. E., Ingram J. A., Davis D. E., and Hanley E. N., Eur. Spine J. 9, 210 (2009). 10.1016/j.spinee.2008.01.012 [DOI] [PubMed] [Google Scholar]
- Quasnichka H. L., Tarlton J. F., Anderson-Mackenzie J. M., Billingham M. E., Bailey A. J., and Pickford A. R., J. Immunol. Methods 297, 133 (2005). 10.1016/j.jim.2004.12.016 [DOI] [PubMed] [Google Scholar]
- Abraham L. C., Dice J. F., Finn P. F., Mesires N. T., Lee K., and Kaplan D. L., Biomaterials 28, 151 (2007). 10.1016/j.biomaterials.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Gobeaux F., Mosser G., Anglo A., Panine P., Davidson P., Giraud-Guille M. M., and Belamie E., J. Mol. Biol. 376, 1509 (2008). 10.1016/j.jmb.2007.12.047 [DOI] [PubMed] [Google Scholar]
- Newton R., Haffegee J., and Ho M., J. Microsc. 180, 127 (1995). [Google Scholar]
- Wu J., Rajwa B., Filmer D. L., Hoffmann C. M., Yuan B., Chiang C., Sturgis J., and Robinson J. P., J. Microsc. 210, 158 (2003). 10.1046/j.1365-2818.2003.01191.x [DOI] [PubMed] [Google Scholar]
- Erikson A., Örtegren J., Hompland T., de Lange Davies C., and Lindgren M., J. Biomed. Opt. 12, 044002 (2007). 10.1117/1.2772311 [DOI] [PubMed] [Google Scholar]
- Stoller P., Celliers P. M., Reiser K. M., and Rubenchik A. M., Appl. Opt. 42, 5209 (2003). 10.1364/AO.42.005209 [DOI] [PubMed] [Google Scholar]
- Pena A. M., Fabre A., Debarre D., Marchal-Somme J., Crestani B., Martin J. L., Beaurepaire E., and Schanne-Klein M. C., Microsc. Res. Tech. 70, 162 (2007). 10.1002/jemt.20400 [DOI] [PubMed] [Google Scholar]