Abstract
Background
Intestinal ischemia-reperfusion is a frequent clinical event associated to injury in distant organs, especially the heart.
Objective
To investigate the gene expression of oxidative stress and antioxidant defense in the heart of inbred mice subjected to intestinal ischemia and reperfusion (IR).
Methods
Twelve mice (C57BL / 6) were assigned to: IR Group (GIR) with 60 minutes of superior mesenteric artery occlusion followed by 60 minutes of reperfusion; Control Group (CG) which underwent anesthesia and laparotomy without IR procedure and was observed for 120 minutes. Intestine and heart samples were processed using the RT-qPCR / Reverse transcriptase-quantitative Polymerase Chain Reaction method for the gene expression of 84 genes related to oxidative stress and oxidative defense (Student's "t" test, p < 0.05).
Results
The intestinal tissue (GIR) was noted to have an up-regulation of 65 genes (74.71%) in comparison to normal tissue (CG), and 37 genes (44.04%) were hyper-expressed (greater than three times the threshold allowed by the algorithm). Regarding the remote effects of intestinal I/R in cardiac tissue an up-regulation of 28 genes (33.33%) was seen, but only eight genes (9.52%) were hyper-expressed three times above threshold. Four (7.14%) of these eight genes were expressed in both intestinal and cardiac tissues. Cardiomyocytes with smaller and pyknotic nuclei, rich in heterochromatin with rare nucleoli, indicating cardiac distress, were observed in the GIR.
Conclusion
Intestinal I/R caused a statistically significant over expression of 8 genes associated with oxidative stress in remote myocardial tissue.
Keywords: Gene Expression, Mice, Oxidative Stress, Intestines / pathology
Introduction
Tissue ischemia and reperfusion (IR), and the cell and humoral changes that follow have a great impact in human homeostasis1. Although reactive oxygen species (ROS) are involved in homeostasis, if a number greater than the natural limits of cell defense of antioxidants and sweepers is present, an unbalance will be seen, known as oxidative stress2. Although the oxygen is a critical substrate for relieving ischemia, it paradoxically acts as a harmful metabolite during the reperfusion period2-5, creating a predisposition for injury in distant organs, in addition to local lesions6.
IR, and the subsequent oxidative stress, is associated to clinical and surgical conditions, such as: sepsis, artery occlusion, cardiac arrest, thrombosis, embolism, vasospasm, tumor, organ transplantation, organ excision, cerebrovascular accident and myocardial infarction2,6-8.
Oxidative stress in the cardiovascular system predisposes to severe cardiomyocyte and cardiac vessel injury, changing their contraction and relaxation mechanisms, having as consequence their disadaptation and impaired function6.
Gene expression assessment associated with oxidative stress in distant organs is a new tool for clarifying different aspects of the phenomenon, both related to diagnosis and monitoring, and therapeutic procedure results9,10. A recent technological resource makes it possible to assess a large number of genes in a safe and relatively simple way, using RT-qPCR (Reverse transcriptase-quantitative Polymerase Chain Reaction) method11-14. Thus, the genomic response of the heart to the oxidative stress caused by renal, hepatic, and mesenteric ischemia, infrarenal aortic occlusion or lower limb ischemia has been described in animal models13-14.
Considering splanchnic ischemia, especially of the small intestine, is a severe event, and that in addition to its local manifestations it has a systemic repercussion, this paper proposes to assess the expression of 84 genes related to the oxidative stress and the antioxidant defense in the cardiac tissue after intestinal ischemia and reperfusion in an inbred mice model.
Methods
The research was approved by the Research Ethics Committee of Hospital São Paulo, bonded to Universidade Federal de São Paulo-UNIFESP under number 1379-1308. The project was ratified by the Research Ethics Committee of Universidade Federal da Grande Dourados-UFGD (protocol 306/08). Experimental procedures followed international norms about animal research and the guidelines of the Brazilian Society of Lab Animal Science (SBCAL). The animals were kept according to the Guide for Caring and Using Lab Animals (Institute of Lab Animal Research, 1996).
Sample
The animals were kept under controlled conditions of temperature and noise, and had a 12-hour cycle of light and darkness. They had free access to water and food adequate to the species. Surgical procedures were performed in the Experimental Surgery labs of the Surgical Department of Universidade Federal da Grande Dourados (UFGD). Twelve adult male mice were selected (C57BL/6 strain), with an average weight of 35 grams. They were supplied by the Center for the Development of Experimental Models for Medicine and Biology UNIFESP (CEDEME) and randomly assigned to either one of these two groups: Ischemia and Reperfusion Group (GIR), where six animals were subjected to superior mesenteric artery occlusion for 60 minutes followed by 60 minutes of reperfusion; and Control Group (GC), where six animals were subjected to median laparotomy under anesthesia, with no vascular clamping of the intestine, and observed for 120 minutes.
Anesthetic and surgical procedures
The mice were administered the following anesthetics intramuscularly: ketamine hydrochloride (44 mg.Kg-1, Ketamina Agener - União Química, São Paulo), xylazine hydrochloride (2.5 mg.Kg-1, Calmium - União Química, São Paulo) and acepromazine (0.75 mg.Kg-1, Acepran - Rhobifarma). Room temperature was kept at 38ºC. The surgical procedure included median laparotomy, and after opening of the peritoneal cavity, abdominal viscera were kept away and identified, and the superior mesenteric artery and vein were isolated and occluded by a microsurgery clamp (metal vascular clip). Artery occlusion was confirmed by pale coloration of the intestine and no pulsing of the artery. After 60 minutes of vascular occlusion, the metal clamp was removed, and reperfusion was initiated, characterized by the return of artery pulsing and different coloration.
Material collection procedure
After the reperfusion period in the GIR group or the observation period in the GC group was finished, samples of the small intestine of the mice were collected for the assessment of ischemia effects and gene expression. Sample collection was performed under anesthesia. Subsequently, the heart was removed, still beating, and sectioned lengthwise at the larger axis, by the ventricles. Heart fragments were imbibed into 10% formaldehyde individually and subjected to standard optical microscope histological processing for hematoxiline and eosin (HE). Another lengthwise fragment of the heart and a 30 mm segment of the intestine were carefully washed with saline solution, wrapped in aluminium foil, labeled and placed in cryogenic vials containing liquid nitrogen (-196ºC). These samples were taken to the Molecular Biology Lab of the Gynecology Department - UNIFESP for gene expression processing.
Gene expression procedures
The RNA of the 84 genes related to the effect of the oxidative stress and antioxidant defense (Table 1) was extracted from the small intestine and heart samples of mice in the GC and GIR using TRIzol(r) (Life Technologies, Grand Island, NY, USA) reagent, and purified by inverse measures (Rneasy MiniKit Qiagen, Co - USA). The concentration of total RNA samples was determined by spectrophotometry, and result quality was assured by the same 2% agarose gel analysis. The first complementary DNA chain (cDNA) was synthesized using 1 µg of total RNA and RT2 First Strand kit (SABiosciences). Identical amounts of cDNA and Master Mix SYBR(r) Green qPCR Mastermix (SABiosciences, QIAGEN Company) were distributed to each PCR array well containing portions of specific genes, which had been previously selected. PCR was performed according to manufacturer's instructions in 96 wells for the expression of 84 genes related to oxidative stress, five endogenous control genes used for measuring PCR array information (ACTB, Gapdh, Hsp90ab1, Hprt1, Gusb), and one negative control for checking for potential contamination with genomic DNA. The negative control primary sample detected non-transcribed and repetitive genomic DNA with a high level of sensitivity. Three reverse transcription control (RTC) wells were used to check for RT reaction efficiency with qPCR test, which aims at detecting patterns synthesized by the control RNA of the first strand synthesis kit. Replication of positive PCR controls (PPCs) was used to determine the efficiency of the polimerase chain reaction. These controls use an artificial DNA sequence predefined in the detection process. Replicated control wells (RTC and PPC) also assess the consistency among wells and plates. The equipment software (MxPro Equipment Real Time Systems, Stratagene, GE, Co) calculated the values of the beginning of the cycle (Ct) for all genes under analysis. At last, the software compared the pair by calculating gene expression change from the pure initial cycle, using [2-Δ ΔC] method. The method used for determining the relative expression of interest genes in this study was shown in the data analysis table of PCR Array v3.3 (PCR Array Data Analysis v3.3-SABiosciences - Qiagen, Company)15.
Table 1.
Distribution of investigated genes according to family groups with similar functions and their respective identification acronyms in the gene bank.
| # | Families | Number of genes | Genes investigated |
| 1 | Glutathione Peroxidases (GPX): | 10 | Gpx1, Gpx2, Gpx3, Gpx4, Gpx5, Gpx6, Gpx7, Gpx8, Gstk1, Gsr. |
| 2 | Peroxiredoxins (TPX): | 8 | Ehd2, Prdx1, Prdx2, Prdx3, Prdx4, Prdx5, Prdx6, Prdx1rs1. |
| 3 | Peroxidases | 16 | Aass, Apc, Cat, Ctsb, Duox1, Epx, Lpo, Mpo, Ptgs1, Ptgs2, Rag2, RGD1560658 (Serpinb1b), RGD1565187 (Kif9), Slc41a3, Tmod1, Tpo. |
| 4 | Reactive Oxygen Species | 16 | Ccs, Cyba, Fmo2, Il19, Il22, Ncf2, Nos2, Nox1, Nox4, Noxa1, Noxo1, Recql4, Scd1, Sod1, Sod2, Sod3. |
| 5 | Oxidative Stress | 22 | Als2, Apoe, Ercc2, Ercc6 Gab1, Idh1, Mpp4, Nqo1, Nudt15, Nxn, Park7, Ppp1r15b, Prnp, Psmb5, Srxn1, Txnip, Txnrd1, Txnrd2, Txnrd3, Ucp3, Xpa, Zmynd17. |
| 6 | Oxygen Carriers | 12 | Aqr, LOC367198 (Atr), Cygb, Dnm2, Fancc, Hbq1, Slb (Ift172), Mb, Ngb, Slc38a1, Vim , Xirp1. |
Statistical Analysis
Each sample was assessed in triplicate for gene expression data. Student's t-test (p < 0.05) was used to validate the homogeneity of each gene expression reaction. For comparison between the two groups, the computer program calculated the quantification cycle variation (Ct) of the study group in relation to the quantification cycle (Ct) of the control group expressed in the logarithm basis (2) by the 2^(-Delta Delta Ct) formula. The gene expression results are shown as positive expression (GIR higher than GC) or negative expression (GIR lower than GC). The numbers represent how many times each gene was expressed, with the positive sign (+, higher) or the negative sign (-, lower). The software calculated results three times higher (hyper-expression) or three times lower (hypo-expression) than the threshold allowed by the algorithm for statistical significance (p < 0.05).
Results
Gene Expression Assessment
From the 84 genes assessed in the intestine, 65 genes (74.71%) had an up-regulation, and out of these, 37 genes (44.04%) were hyper-expressed, that is, the expression was three times higher than the threshold established by the algorithm in comparison to the control group (Chart 1).
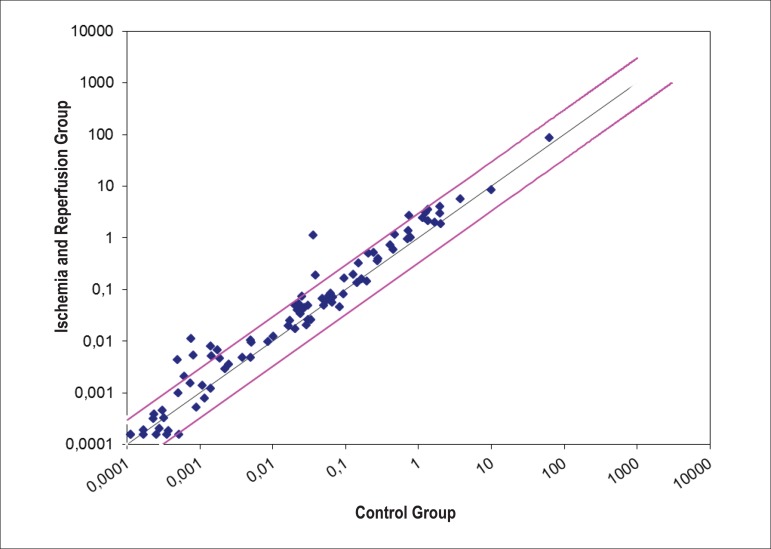
Chart 1.
Disposition of the expression of 84 genes in the cardiac tissue related to intestine ischemia and reperfusion in the animals of GC and GIR groups. The black line indicates normal expression; the pink lines indicate three times higher than the gene expression allowed by the algorithm.
The cardiac tissue showed an up-regulation of 28 genes (33.33%) after intestinal I/R (Table 2). Only 8 genes (9.52%) were three times above the algorithm threshold (Table 3 and chart 2). Four genes (4.76%) were positively and simultaneously expressed in both intestinal and cardiac tissues (Table 4).
Table 2.
Distribution of 28 genes out of the 84 genes investigated in the heart with positive expression (+) or negative expression (-) 2^(- Delta Delta Ct) in the animals subjected to a 60-minute ischemia in the small intestine followed by a 60-minute reperfusion in comparison to the control group that was not subjected to ischemia/reperfusion; (* = significant p-value < 0.05)
| # | Gene Basis | Gene Symbol | Gene Name | GIR Intestine | P-value | GIR Heart | P-value |
| 1 | NM_013930 | Aass | Aminoadipate-semialdehyde synthase | +19.63* | 0.000001 | +6.65* | 0.01209 |
| 2 | NM_009696 | Apoe | Apolipoprotein E | +1.76* | 0.002533 | +3.60* | 0.01737 |
| 3 | NM_007985 | Fancc | Fanconi anemia. complementation group C | +7.42* | 0.000015 | +1.87* | 0.04313 |
| 4 | NM_018881 | Fmo2 | Flavin containing monooxygenase 2 | +1.52* | 0.019633 | +4.93* | 0.00772 |
| 5 | NM_010343 | Gpx5 | Glutathione peroxidase 5 | +5.10* | 0.001774 | +0.50* | 0.03717 |
| 6 | NM_027127 | Gpx8 | Glutathione peroxidase 8 (putative) | +1.43 | 0.072334 | +2.19* | 0.01271 |
| 7 | NM_010344 | Gsr | Glutathione reductase | +1.64 | 0.149896 | +2.34* | 0.02997 |
| 8 | NM_080420 | Lpo | Lactoperoxidase | +23.88* | 0.00002 | -2.28* | 0.00326 |
| 9 | NM_010877 | Ncf2 | Neutrophil cytosolic factor 2 | +4.31* | 0.000018 | +2.43* | 0.02876 |
| 10 | NM_010927 | Nos2 | Nitric oxide synthase 2. inducible | +4.71* | 0.000117 | +1.31* | 0.00129 |
| 11 | NM_015760 | Nox4 | NADPH oxidase 4 | +12.47* | 0.000005 | +5.73* | 0.02561 |
| 12 | NM_172204 | Noxa1 | NADPH oxidase activator 1 | +3.06* | 0.000083 | +1.49* | 0.00072 |
| 13 | NM_027988 | Noxo1 | NADPH oxidase organizer 1 | +1.33* | 0.000177 | +2.10* | 0.03623 |
| 14 | NM_008750 | Nxn | Nucleoredoxin | +1.98* | 0.002268 | +2.53* | 0.036 |
| 15 | NM_133819 | Ppp1r15b | Protein phosphatase 1. regulatory (inhibitor) subunit 15b | +1.06 | 0.962143 | +1.99* | 0.04908 |
| 16 | NM_012021 | Prdx5 | Peroxiredoxin 5 | -1.54* | 0.008534 | +2.14* | 0.01926 |
| 17 | NM_011186 | Psmb5 | Proteasome (prosome. macropain) subunit. beta type 5 | +1.21 | 0.078442 | +1.32* | 0.00488 |
| 18 | NM_008969 | Ptgs1 | Prostaglandin-endoperoxide synthase 1 | +1.56* | 0.012124 | +2.94* | 0.00004 |
| 19 | NM_011198 | Ptgs2 | Prostaglandin-endoperoxide synthase 2 | +10.07* | 0.000003 | +3.94* | 0.00691 |
| 20 | NM_009020 | Rag2 | Recombination activating gene 2 | +15.79* | 0.000293 | +3.42* | 0.04444 |
| 21 | NM_009127 | Scd1 | Stearoyl-Coenzyme A desaturase 1 | +6.85* | 0.00042 | +31.73* | 0.000099 |
| 22 | NM_134086 | Slc38a1 | Solute carrier family 38. member 1 | +4.59* | 0.000106 | +8.86* | 0.01516 |
| 23 | NM_013671 | Sod2 | Superoxide dismutase 2. mitochondrial | +1.47* | 0.009249 | -1.80* | 0.02156 |
| 24 | NM_011435 | Sod3 | Superoxide dismutase 3. extracellular | +2.51* | 0.002347 | +1.28* | 0.00081 |
| 25 | NM_021883 | Tmod1 | Tropomodulin 1 | +15.01* | 0.000161 | -1.05* | 0.00619 |
| 26 | NM_023719 | Txnip | Thioredoxin interacting protein | +1.87* | 0.00026 | +2.02* | 0.00127 |
| 27 | NM_009464 | Ucp3 | Uncoupling protein 3 (mitochondrial. proton carrier) | +15.85* | 0.000102 | +1.62* | 0.01441 |
| 28 | NM_011701 | Vim | Vimentin | +2.20* | 0.000155 | +2.59* | 0.01509 |
Table 3.
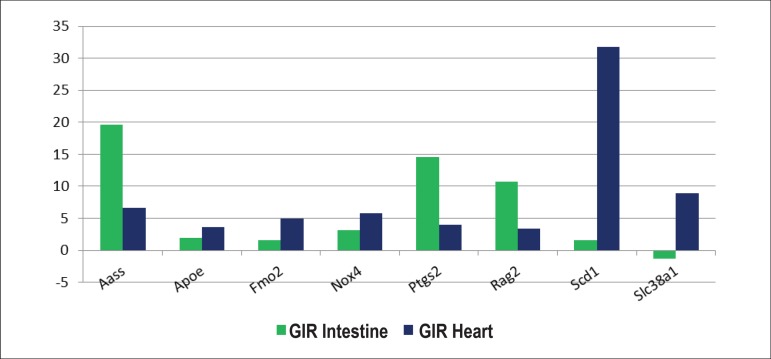
Distribution of 8 hyper-expressed genes 2^(- Delta Delta Ct) out of the 84 genes investigated in the heart compared to the genes expressed in the intestine of the animals subjected to a 60-minute ischemia in the small intestine followed by a 60-minute reperfusion in comparison to the control group that was not subjected to ischemia/reperfusion; (* = p-value < 0.05)
| # | Gene Basis | Gene Symbol | Gene Name | GIR Intestine | P-value | GIR Heart | P-value |
| 1 | NM_013930 | Aass | Aminoadipate-semialdehyde synthase | +19.64 | 0.000001* | +6.65 | 0.012092* |
| 2 | NM_009696 | Apoe | Apolipoprotein E | +1.94 | 0.002533* | +3.6 | 0.017365* |
| 3 | NM_018881 | Fmo2 | Flavin containing monooxygenase 2 | +1.6 | 0.019633* | +4.93 | 0.007719* |
| 4 | NM_015760 | Nox4 | NADPH oxidase 4 | +3.13 | 0.000005* | +5.73 | 0.025614* |
| 5 | NM_011198 | Ptgs2 | Prostaglandin-endoperoxide synthase 2 | +14.6 | 0.000003* | +3.94 | 0.006908* |
| 6 | NM_009020 | Rag2 | Recombination activating gene 2 | +10.76 | 0.000293* | +3.42 | 0.044442* |
| 7 | NM_009127 | Scd1 | Stearoyl-Coenzyme A desaturase 1 | +1.62 | 0.000420* | +31.73 | 0.000099* |
| 8 | NM_134086 | Slc38a1 | Solute carrier family 38. member 1 | -1.29 | 0.000106* | +8.86 | 0.015159* |
Chart 2.
Eight genes related to oxidative stress and expressed three times above the threshold allowed by the algorithm in intestine and cardiac tissues.
Table 4.
Amount and percentage of genes hyper-expressed with statistically significant values and simultaneous occurrence in the intestine and heart tissues
| GIR Intestine | GIR Heart | Simultaneous occurrence | |||
| Genes considered to be significant (p < 0.05) | 65 (74.71%) | 28 (33.33%) | 24 (28.57%) | ||
| Genes considered to be hyper-expressed | 37 (44.04%) | 8 (9.52%) | 4 (4.76%) | ||
Histological Assessment
In the qualitative histological analysis, heart tissue samples of the animals from both groups were formed by strips of cardiac muscle fibers separated from one another by type I collagen fibers. These strips contained elongated cardiac muscle cells with one or two nuclei at the center, and transversal grooves in the cytoplasm. Around the cardiomyocytes, there are connective tissue cells, type III collagen fibers and blood capillary.
The GC was noted to have cardiomyocytes with one or two large nuclei and very marked nucleoli (Figure 1). The GIR (one hour of ischemia and one hour of reperfusion) was noted to have smaller cardiomyocytes with pyknotic and darker nuclei, rich in heterochromatin with rare nucleoli, indicating cardiac distress (Figure 2).
Figure 1.
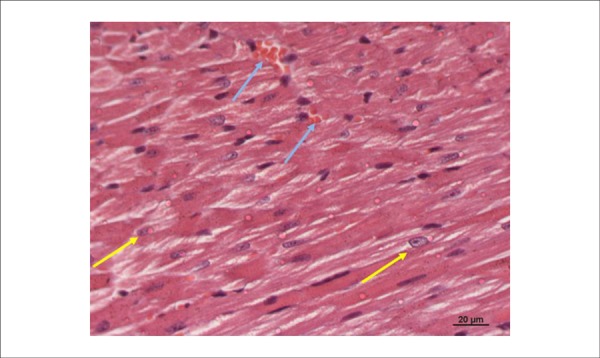
Photomicrography of the left ventricle of a mouse in the Control Group (GC). Please note large volume and euchromatic nuclei of cardiomyocytes centralized in the cell (yellow arrows). Preserved vessels with no red blood cell extravasation (blue arrows). Cardiac fibers correctly positioned. (HE 400x).
Figure 2.
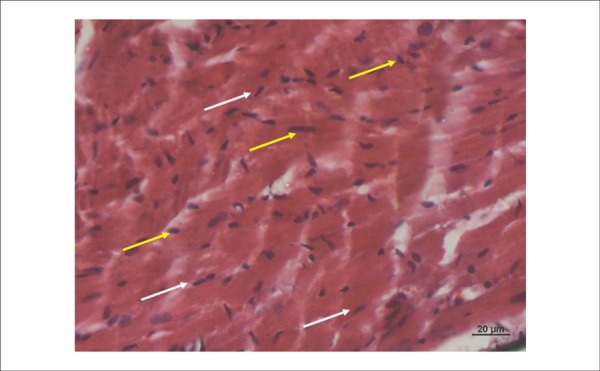
Photomicrography of the left ventricle of a mouse in the Ischemia and Reperfusion Group (GIR). Please note smaller volume and pyknotic nuclei (yellow cell) of cardiomyocytes in the edge of the cell (white cell). Swollen cells and cardiac fibers in a disorganized direction. (HE 400x).
Discussion
Data obtained from the research on oxidative stress and cell antioxidant response have showed that, in a short term model of intestinal ischemia (one hour of ischemia followed by one hour of reperfusion) in inbred mice, certain genes were positively expressed in the cardiac tissue (Table 2 and chart 1).
A global analysis confirms that intestinal ischemia followed by reperfusion is a phenomenon closely related to the generation and modulation of reactive oxygen species. Local changes in the intestine promoted humoral and cell responses which, after being disseminated through blood flow, affected the genomic apparatus of the cardiac tissue, inducing gene expression responses related to the generation and modulation of ROS.
The aminoadipate-semialdehyde synthase (Aass) gene encodes a catalyzing peroxidase protein whose substrates are hydrogen peroxide and organic hydroperoxide. It can be found in the majority of the tissues, functioning as a bifunctional enzyme which degradates lysine up to acetyl coenzyme A (acetyl-CoA) through two different metabolic pathways (saccharopine and pipecolic acid)16,17. Lysine causes lipid peroxidation, thereby reducing the concentration of glutathione peroxidases and harming cell antioxidant defenses. It acts by inhibiting Na+/K+-ATPase with changes in cell membrane ionic exchange mechanisms, inducing oxidative stress18,19. Lysine degradation by acetylation influences myocardial motility, increasing the actin sliding speed on myosin in comparison to their respective non-acetylated isoforms19. In the light of the known activities of the enzyme encoded by the Aass gene, it is possible to infer that its hyper-expression in the cardiac tissue is associated with protection of cardiac tissue against the harmful action of ROS.
Prostaglandin-endoperoxide synthase (Ptgs-2) is a peroxidase that converts the arachidonic acid intro prostaglandin and it is associated with injuries caused by the inflammatory process and cell proliferation20. It has two functions in the inflammatory process: initially, it contributes to the onset of the inflammatory response, and then it acts in process resolution21,22. Ptgs2 is not detected in normal cells, but it is present in responses to inflammatory cytokines in different types of cells and oxidative stress situations. Its presence has been reported in the myocardium in many different diseases which resulted in cardiac failure. In the context of this research paper, the hyper-expression of Ptgs2 gene has showed that myocardial tissue is sensitive to intestinal ischemia, reacting in a defensive way against oxidative stress22.
Xenobiotic neutralization processes, including amines, sulfides and some phosphorus and selenium compounds, are performed by a mono-oxygenase enzyme (Fmo2) containing flavin nucleotide. The reaction depends on the presence of NADPH (Nicotinamide adenine dinucleotide phosphate) hydrogenase. In the absence of NADP (nicotinamide adenine dinucleotide phosphate) Fmo2, it would work as NADPH oxidase consuming NADPH and causing oxidative stress upon the excessive production of hydrogen peroxide23. Mammals express five different mono oxygenases, and Fmo2 isoform is the most prevalent. A study in humans has associated Fmo2 with a higher resistance to the oxidative stress caused by environmental factors24,25. In this research paper, Fmo2 gene hyper expression suggests that intestinal ischemia induces protein encoding for antioxidant defense of the cardiac tissue.
NADPH oxidase (Nox) is a group of seven oxidase enzymes with intracellular and perinuclear manifestation, present in different tissues. Its presence is quite relevant in the cardiovascular tissue, although with no clear distinction between myocytes and other vascular components. Nox4 is the isoforms involved in ROS production, especially hydrogen peroxide and superoxide anion in a smaller scale26. It is involved in oxygen detection, vasomotor control, cell proliferation, differentiation, migration, apoptosis, senescence, fibrosis and angiogenesis27. Inflammatory cytokines, including TNF-alpha, function as Nox4 gene expression modulators28. Both play an important role in the pathogenesis of cardiovascular diseases, stimulating the extrinsic pathway in the formation of a death inducing signaling complex, resulting in the activation of the caspase cascade. The correlation of these in cardiovascular diseases is clear (atherosclerosis, blood hypertension, cardiac failure, and ischemic cerebrovascular accident)26,28,29. Its hyper-expression confirms intestinal ischemia/reperfusion had a harmful effect on the heart. This effect can also be morphologically confirmed by the changes identified in HE stained slides (Figure 2).
Apolipoprotein E (Apoe) is fundamental for the regular metabolism of triglycerides and cholesterol lipoprotein components. Primarily produced by the liver and macrophages, it's a cholesterol carrier, protects vessel endothelial cells, has an apparent function in neurological diseases, and has in vitro immunomodulator functions, decreasing lymphocyte proliferation after mitogenic stimulus30. This alipoprotein has been reported to act against ischemia, by acting as an antioxidant, protecting cells from different tissues, such as the endothelium against the toxic effect of hydrogen peroxide31-34. Considering the activities of Apoe in different tissues, it is possible to construct a hypothesis on the fact there is a protecting response against the harmful effects of ROS on the heart after oxidative stress on our experimental model.
Rag2 gene (recombination-activating gene 2) encodes one of the peroxidases that play an important role in rearranging and recombining immunoglobulin and T cell molecular receptor genes27. It is a complex multiprotein, which mediates the DNA cleavage phase during recombination. RAG1 and RAG2 proteins are crucial for the maturation of bursal (B) and timic (T) lymphocytes. Both cell types are fundamental for immune system adaptation35. Peroxidases are known to influence oxidative stress modulation35,36. Rag2 hyper-expression can be associated with the need of adjustments in cardiac cell replication due to the presence of excessive ROS, especially hydrogen peroxide and organic hydroperoxides, showing a protecting action against the harmful effects of ROS.
The protein encoded by Scd1 gene (Stearoyl-Coenzyme A desaturase 1) catalyzes unsaturated fatty acid synthesis and its major byproduct is oleic acid, formed by stearic acid desaturation, and it is part of the enzyme family related to reactive oxygen species37. Scd1 deficiency increases fatty acid oxidation rate in the liver and skeletal muscle. The higher this rate is, the higher the chances that Scd1 deficiency changes cardiac metabolism, influencing in the proportion of energetic substrate among fatty acids and glucose available to the heart38. Incompatibility between the absorption and the use of long chain fatty acids by the myocardium results in abnormally increased concentration of intracellular fatty acid, inducing myocardium dysfunction37. In this research paper, hyper-expression of the gene in the cardiac tissue may be related to the need of a greater use of fats in the lieu of the glucose dependent oxidizable substrate, in an attempt to repair the cardiac metabolic unbalance.
The protein encoded by Slc38a1 gene (Solute carrier family 38) is an amino acid carrier predominantly present in the heart, brain and placenta. Solute carriers are proteins from the eukaryotic membrane that control the absorption and the outflow of different solutes, including amino acids, sugars and drugs39. Amino acids are necessary for different important cell biological processes, such as in the production of thiol glutathione (GSH) catalyzed by the presence of cysteine, which is also necessary to other reactions in the myocardial tissue and is provided by the Slc38a1 carrier protein40. The duration of the oxidative stress stimulates its uptaking, causing increased Slc38a1 expression in the myocardium of mammals, consequently with a higher expression of GSH39,40. In this study, hyper expression of Slc38a1 corroborates with the indication of similar action in other tissues and shows that its presence can implicate in a protecting action in the energy transportation mechanism of the myocardium cell.
The rationale of this research paper was developing the gene response profile of the cardiac tissue when subjected to stimuli from distant organs affected by oxidative stress. In the light of the unprecedented nature of the results and the complexity of the gene expression, the results can only be confirmed with a subsequent proteomic analysis, where protein identification and quantification in the tissue may provide resources to determine whether the gene expression acted as a real stimulus for protein encoding or whether the gene expression is related to signaling or modulation of other oxidative stress response metabolic pathways. As a starting point to future research, the mapping activity performed allowed eight genes to be identified, which showed to be more responsive to oxidative stress and can, therefore, be initially targeted as new study subject. This research line is very promising, because gene expression understanding may allow evolution monitoring procedures and treatment of multiple organ dysfunction syndrome, especially concerning the participation of the cardiac tissue in this phenomenon.
Conclusions
The gene response profile associated to oxidative stress has identified eight genes in the cardiac tissue that respond in a hyper-expressive way when the intestine is subjected to ischemia and reperfusion. This study presents the perspective of creating a monitoring protocol in ischemia and reperfusion scenarios from real-time gene expression.
Acknowledgments
The authors would like to thank Professor Dr. Ismael Dale Cotrim Guerreiro da Silva, UNIFESP Molecular Gynecology Department, for his logistic and lab support. Professor Dr. Marcos Azevedo Junior for his support and gene expression procedures. Professor Manuel de Jesus Simões for his support in histological assessment procedures.
Footnotes
Potential Conflict of Interest
No potential conflict of interest relevant to this article was reported.
Sources of Funding
This study was partially funded by Universidade Federal da Grande Dourados- UFGD - Mato Grosso do Sul.
Study Association
This article is part of the thesis of master submitted by Frederico Somaio Neto from Universidade Federal de São Paulo.
Author contributions: Conception and design of the research: Somaio Neto F, Fagundes DJ; Acquisition of data, Analysis and interpretation of the data and Writing of the manuscript: Somaio Neto F, Ikejiri AT, Bertoletto PR, Chaves JCB, Teruya R, Fagundes DJ, Taha MO; Statistical analysis: Somaio Neto F, Teruya R, Fagundes DJ; Obtaining funding: Somaio Neto F, Ikejiri AT, Bertoletto PR, Chaves JCB, Taha MO; Critical revision of the manuscript for intellectual content: Fagundes DJ.
References
- 1.Pashkow FJ. Oxidative stress and inflammation in heart disease: do antioxidants have a role in treatment and/or prevention? Int J Inflamm. 2011;2011:514623. doi: 10.4061/2011/514623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohen R, Nyska A. Oxidation of biological systems: oxidative stress phenomena, antioxidants, redox reactions, and methods for their quantification. Toxicol Pathol. 2002;30(6):620–650. doi: 10.1080/01926230290166724. [DOI] [PubMed] [Google Scholar]
- 3.Ondiveeran HK, Fox-Robichaud A. New developments in the treatment of ischemia/reperfusion injury. Curr Opin Investig Drugs. 2001;2(6):783–791. [PubMed] [Google Scholar]
- 4.Chamoun F, Burne M, O'Donnell M, Rabb H. Pathophysiologic role of selectins and their ligands in ischemia reperfusion injury. Front Biosci. 2000;5:E103–E109. doi: 10.2741/chamoun. [DOI] [PubMed] [Google Scholar]
- 5.Zweier JL, Talukder MA. The role of oxidants and free radicals in reperfusion injury. Cardiovasc Res. 2006;70(2):181–190. doi: 10.1016/j.cardiores.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 6.Cantor EJ, Mancini EV, Seth R, Yao XH, Netticadan T. Oxidative stress and heart disease: cardiac dysfunction, nutrition, and gene therapy. Curr Hypertens Rep. 2003;5(3):215–220. doi: 10.1007/s11906-003-0023-z. [DOI] [PubMed] [Google Scholar]
- 7.Asyali MH, Colak D, Demirkaya O, Inan MS. Gene expression profile classification: a review . Current Bioinformatics. 2006;1(1):55–73. [Google Scholar]
- 8.Santos-Silva MA, Nagato AC, Trajano ET, Alves JN, Bandeira AC, Porto LC, et al. A resposta oxidativa em corações de camundongos é modulada por background genético. Arq Bras Cardiol. 2013;100(2):157–163. doi: 10.5935/abc.20130029. [DOI] [PubMed] [Google Scholar]
- 9.Konstantinov IE, Arab S, Li J, Coles JG, Boscarino C, Mori A, et al. The remote ischemic preconditioning stimulus modifies gene expression in mouse myocardium. J Thorac Cardiovasc Surg. 2005;130(5):1326–1332. doi: 10.1016/j.jtcvs.2005.03.050. [DOI] [PubMed] [Google Scholar]
- 10.Bertoletto PR, Ikejiri AT, Somaio F, Neto, Chaves JC, Teruya R, Bertoletto ER, et al. Oxidative stress gene expression profile in inbred mouse after ischemia/reperfusion small bowel injury. Acta Cir Bras. 2012;27(11):773–782. doi: 10.1590/s0102-86502012001100006. [DOI] [PubMed] [Google Scholar]
- 11.Huda R, Chung DH, Mathru M. Ischemic preconditioning at a distance: altered gene expression in mouse heart and other organs following brief occlusion of the mesenteric artery. Heart Lung Circ. 2005;14(1):36–43. doi: 10.1016/j.hlc.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Colombo J, Rahal P. A tecnologia de microarray no estudo do câncer de cabeça e pescoço. R Bras Bioci. 2010;8(1):64–72. [Google Scholar]
- 13.Moore-Olufemi SD, Olufemi SE, Lott S, Sato N, Kozar RA, Moore FA, et al. Intestinal Intestinal ischemic preconditioning after ischemia/reperfusion injury in rat intestine: profiling global gene expression patterns. Dig Dis Sci. 2010;55(7):1866–1877. doi: 10.1007/s10620-009-0980-4. [DOI] [PubMed] [Google Scholar]
- 14.O'Donnell CJ, Nabel EG. Genomics of cardiovascular disease. N Engl J Med. 2011;365(22):2098–2109. doi: 10.1056/NEJMra1105239. [DOI] [PubMed] [Google Scholar]
- 15.Deepak SA, Kottapalli KR, Rakwal R, Oros G, Rangappa KS, Iwahashi H, et al. Real-time PCR: revolutionizing detection and expression analysis of genes. Curr Genomics. Curr Genomics. 2007;8(4):234–251. doi: 10.2174/138920207781386960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sacksteder KA, Biery BJ, Morrell JC, Goodman BK, Geisbrecht BV, Cox RP, et al. Identification of the alpha-aminoadipic semialdehyde synthase gene, which is defective in familial hyperlysinemia. Am J Hum Genet. 2000;66(6):1736–1743. doi: 10.1086/302919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Struys EA, Jakobs C. Metabolism of lysine in alpha-aminoadipic semialdehyde dehydrogenase-deficient fibroblasts: evidence for an alternative pathway of pipecolic acid formation. FEBS Lett. 2010;584(1):181–186. doi: 10.1016/j.febslet.2009.11.055. [DOI] [PubMed] [Google Scholar]
- 18.Seminotti B, Leipnitz G, Amaral AU, Fernandes CG, Silva Lde B, Tonin AM, et al. Lysine induces lipid and protein damage and decreases reduced glutathione concentrations in brain of young rats. Int J Dev Neurosci. 2008;26(7):693–698. doi: 10.1016/j.ijdevneu.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Cleveland BM. The effect of alpha-aminoadipate delta-semialdehyde synthase knockdown on the lysine requirement and urate oxidase knockdown on oxidative stress in a murine hepatic cell line. [thesis] Morgantown, (WV): West Virginia University; 2007. [Google Scholar]
- 20.Buetler TM, Leclerc E, Baumeyer A, Latado H, Newell J, Adolfsson O, et al. N (epsilon)-carboxymethyllysine-modified proteins are unable to bind to RAGE and activate an inflammatory response. Mol Nutr Food Res. 2008;52(3):370–378. doi: 10.1002/mnfr.200700101. [DOI] [PubMed] [Google Scholar]
- 21.Mason RP, Kalyanaraman B, Tainer BE, Eling TE. A carbon-centered free radical intermediate in the prostaglandin synthetase oxidation of arachidonic acid. Spin trapping and oxygen uptake studies. J Biol Chem. 1980;255(11):5019–5022. [PubMed] [Google Scholar]
- 22.Gilroy DW, Colville-Nash PR, Willis D, Chivers J, Paul-Clark MJ, Willoughby DA. Inducible cyclooxygenase may have anti-inflammatory properties. Nat Med. 1999;5(6):698–701. doi: 10.1038/9550. [DOI] [PubMed] [Google Scholar]
- 23.Wong SC, Fukuchi M, Melnyk P, Rodger I, Giaid A. Induction of cyclooxygenase-2 and activation of nuclear factor-kappaB in myocardium of patients with congestive heart failure. Circulation. 1998;98(2):100–103. doi: 10.1161/01.cir.98.2.100. [DOI] [PubMed] [Google Scholar]
- 24.Yueh MF. Identification and characterization of flavin-containing monooxygenase isoform 2 (FMO2) in Rhesus monkey and examination of a human FMO2 polymorphism [thesis] Corvallis (OR): Oregon State University; 1999. [Google Scholar]
- 25.Krueger SK, Siddens LK, Martin SR, Yu Z, Pereira CB, Cabacungan ET, et al. Differences in FMO2*1 allelic frequency between Hispanics of Puerto Rican and Mexican descent. Drug Metab Dispos. 2004;32(12):1337–1340. doi: 10.1124/dmd.104.001099. [DOI] [PubMed] [Google Scholar]
- 26.Chen F, Haigh S, Barman S, Fulton DJ. From form to function: the role of Nox4 in the cardiovascular system. Front Physiol. 2012;3:412–412. doi: 10.3389/fphys.2012.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin FF, Bailey S, Innis CA, Ciubotaru M, Kamtekar S, Steitz TA, et al. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat Struct Mol Biol. 2009;16(5):499–508. doi: 10.1038/nsmb.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basuroy S, Bhattacharya S, Leffler CW, Parfenova H. Nox4 NADPH oxidase mediates oxidative stress and apoptosis caused by TNF-alpha in cerebral vascular endothelial cells. Am J Physiol Cell Physiol. 2009;296(3):C422–C432. doi: 10.1152/ajpcell.00381.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos CX, Anilkumar N, Zhang M, Brewer AC, Shah AM. Redox signaling in cardiac myocytes. Free Radic Biol Med. 2011;50(7):777–793. doi: 10.1016/j.freeradbiomed.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou S, Wu H, Zeng C, Xiong X, Tang S, Tang Z, et al. Apolipoprotein E protects astrocytes from hypoxia and glutamate-induced apoptosis. FEBS Lett. 2013;587(2):254–258. doi: 10.1016/j.febslet.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Laskowitz DT, Sheng H, Bart RD, Joyner KA, Roses AD, Warner DS. Apolipoprotein E-deficient mice have increased susceptibility to focal cerebral ischemia. J Cereb Blood Flow Metab. 1997;17(7):753–758. doi: 10.1097/00004647-199707000-00005. [DOI] [PubMed] [Google Scholar]
- 32.Miyata M, Smith JD. Apolipoprotein E allele-specific antioxidant activity and effects on cytotoxicity by oxidative insults and beta-amyloid peptides. Nat Genet. 1996;14(1):55–61. doi: 10.1038/ng0996-55. [DOI] [PubMed] [Google Scholar]
- 33.Tarnus E, Wassef H, Carmel JF, Rondeau P, Roche M, Davignon J, et al. Apolipoprotein E limits oxidative stress-induced cell dysfunctions in human adipocytes. FEBS Lett. 2009;583(12):2042–2048. doi: 10.1016/j.febslet.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 34.Yue L, Bian JT, Grizelj I, Cavka A, Phillips SA, Makino A, et al. Apolipoprotein E enhances endothelial-NO production by modulating caveolin 1 interaction with endothelial NO synthase. Hypertension. 2012;60(4):1040–1046. doi: 10.1161/HYPERTENSIONAHA.112.196667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arbuckle JL, Rahman NS, Zhao S, Rodgers W, Rodgers KK. Elucidating the domain architecture and functions of non-core RAG1: the capacity of a non-core zinc-binding domain to function in nuclear import and nucleic acid binding. BMC Biochem. 2011;12:23–23. doi: 10.1186/1471-2091-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorsuch WB, Chrysanthou E, Schwaeble WJ, Stahl GL. The complement system in ischemia-reperfusion injuries. Immunobiology. 2012;217(11):1026–1033. doi: 10.1016/j.imbio.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsui H, Yokoyama T, Sekiguchi K, Iijima D, Sunaga H, Maniwa M, et al. Stearoyl-CoA desaturase-1 (SCD1) augments saturated fatty acid-induced lipid accumulation and inhibits apoptosis in cardiac myocytes. PLoS One. 2012;7(3): doi: 10.1371/journal.pone.0033283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobrzyn P, Sampath H, Dobrzyn A, Miyazaki M, Ntambi JM. Loss of stearoyl-CoA desaturase 1 inhibits fatty acid oxidation and increases glucose utilization in the heart. Am J Physiol Endocrinol Metab. 2008;294(2):E357–E364. doi: 10.1152/ajpendo.00471.2007. [DOI] [PubMed] [Google Scholar]
- 39.Schlessinger A, Matsson P, Shima JE, Pieper U, Yee SW, Kelly L, et al. Comparison of human solute carriers. Protein Sci. 2010;19(3):412–428. doi: 10.1002/pro.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King N, Lin H, Suleiman MS. Oxidative stress increases SNAT1 expression and stimulates cysteine uptake in freshly isolated rat cardiomyocytes. Amino Acids. 2011;40(2):517–526. doi: 10.1007/s00726-010-0664-6. [DOI] [PMC free article] [PubMed] [Google Scholar]