Abstract
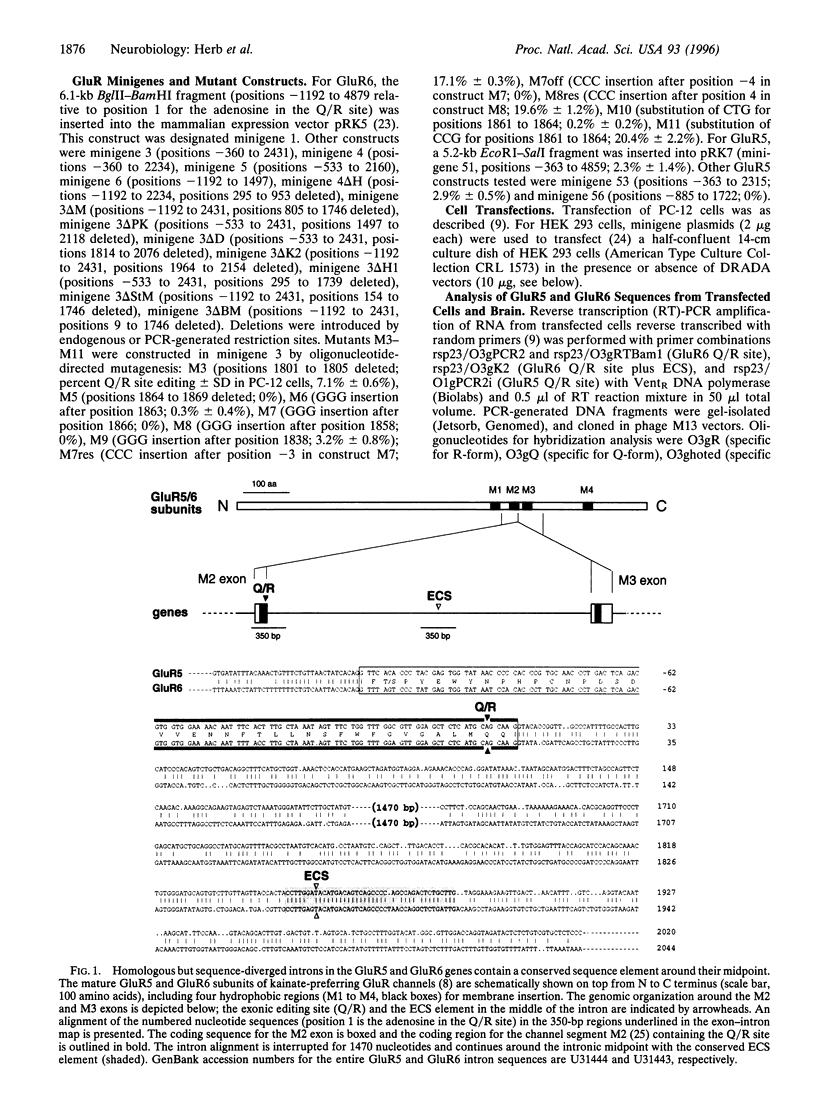
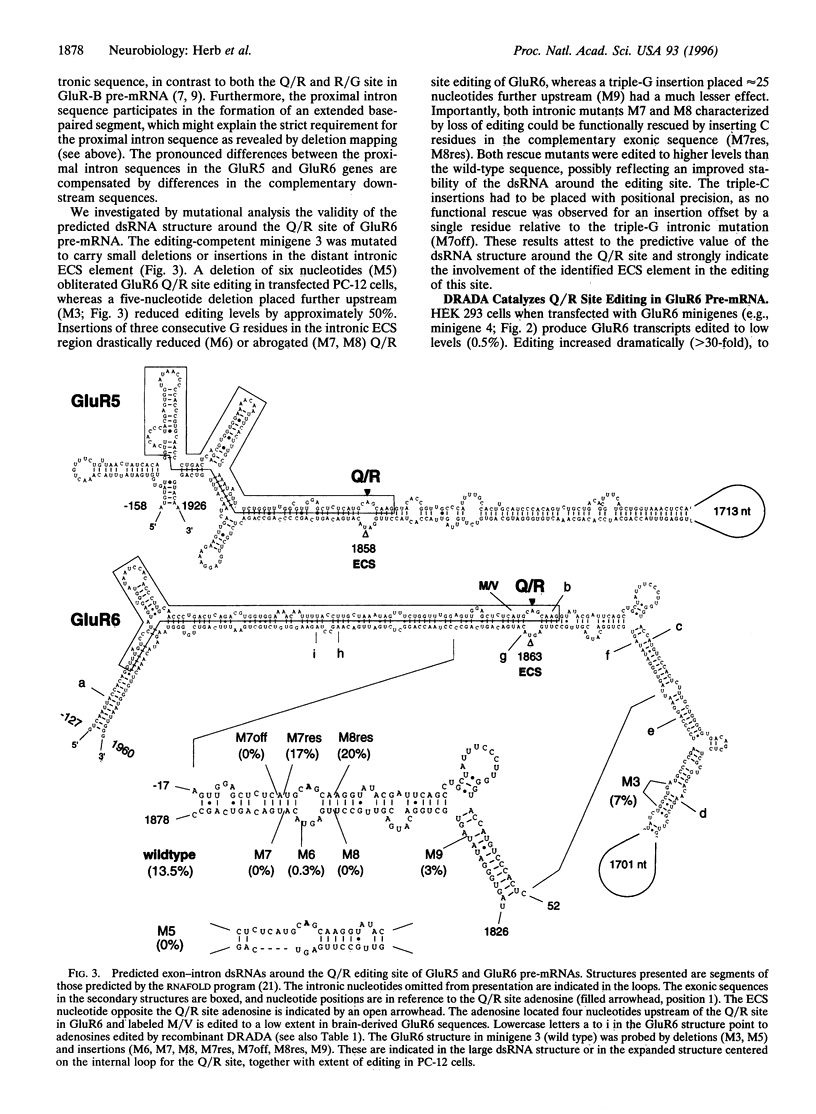
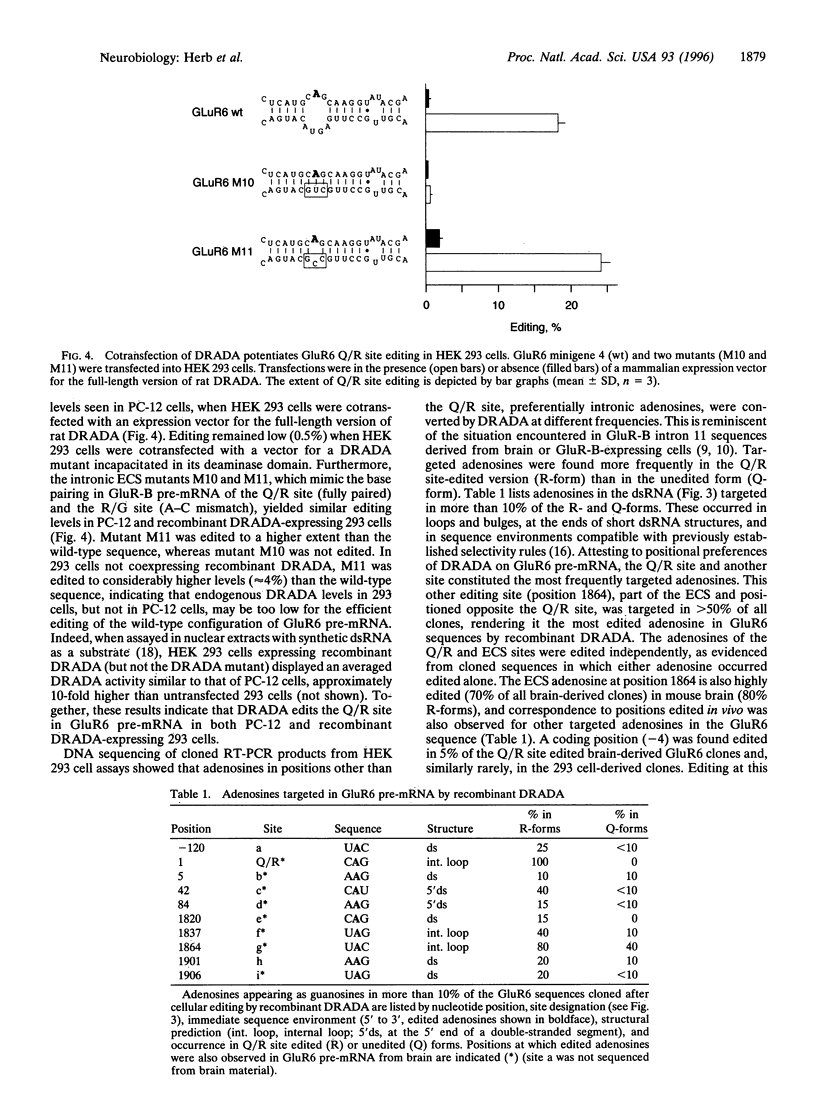
RNA editing by adenosine deamination in brain-expressed pre-mRNAs for glutamate receptor (GluR) subunits alters gene-specified codons for functionally critical positions, such as the channel's Q/R site. We show by transcript analysis of minigenes transiently expressed in PC-12 cells that, in contrast to GluR-B pre-mRNA, where the two editing sites (Q/R and R/G) require base pairing with nearby intronic editing site complementary sequences (ECSs), editing in GluR5 and GluR6 pre-mRNAs recruits an ECS located as far as 1900 nucleotides distal to the Q/R site. The exon-intron duplex structure of the GluR5 and GluR6 pre-mRNAs appears to be a substrate of double-stranded RNA-specific adenosine deaminase. This enzyme when coexpressed in HEK 293 cells preferentially targets the adenosine of the Q/R site and of an unpaired position in the ECS which is highly edited in brain.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bass B. L. RNA editing. An I for editing. Curr Biol. 1995 Jun 1;5(6):598–600. doi: 10.1016/s0960-9822(95)00119-9. [DOI] [PubMed] [Google Scholar]
- Bass B. L., Weintraub H. A developmentally regulated activity that unwinds RNA duplexes. Cell. 1987 Feb 27;48(4):607–613. doi: 10.1016/0092-8674(87)90239-x. [DOI] [PubMed] [Google Scholar]
- Bernard A., Khrestchatisky M. Assessing the extent of RNA editing in the TMII regions of GluR5 and GluR6 kainate receptors during rat brain development. J Neurochem. 1994 May;62(5):2057–2060. doi: 10.1046/j.1471-4159.1994.62052057.x. [DOI] [PubMed] [Google Scholar]
- Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987 Aug;7(8):2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. H., Habib G., Yang C. Y., Gu Z. W., Lee B. R., Weng S. A., Silberman S. R., Cai S. J., Deslypere J. P., Rosseneu M. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987 Oct 16;238(4825):363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- Ellington A. D. RNA ligands: out of shape but fir for recognition. Curr Biol. 1993 Jun 1;3(6):375–377. doi: 10.1016/0960-9822(93)90206-4. [DOI] [PubMed] [Google Scholar]
- Higuchi M., Single F. N., Köhler M., Sommer B., Sprengel R., Seeburg P. H. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993 Dec 31;75(7):1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- Hollmann M., Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- Kim U., Nishikura K. Double-stranded RNA adenosine deaminase as a potential mammalian RNA editing factor. Semin Cell Biol. 1993 Aug;4(4):285–293. doi: 10.1006/scel.1993.1034. [DOI] [PubMed] [Google Scholar]
- Kim U., Wang Y., Sanford T., Zeng Y., Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler M., Burnashev N., Sakmann B., Seeburg P. H. Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron. 1993 Mar;10(3):491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- Lomeli H., Mosbacher J., Melcher T., Höger T., Geiger J. R., Kuner T., Monyer H., Higuchi M., Bach A., Seeburg P. H. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994 Dec 9;266(5191):1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- Melcher T., Maas S., Higuchi M., Keller W., Seeburg P. H. Editing of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR-B pre-mRNA in vitro reveals site-selective adenosine to inosine conversion. J Biol Chem. 1995 Apr 14;270(15):8566–8570. doi: 10.1074/jbc.270.15.8566. [DOI] [PubMed] [Google Scholar]
- O'Connell M. A., Krause S., Higuchi M., Hsuan J. J., Totty N. F., Jenny A., Keller W. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol. 1995 Mar;15(3):1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen W., Hedreen J. C., Ross C. A. RNA editing of the glutamate receptor subunits GluR2 and GluR6 in human brain tissue. J Neurochem. 1994 Nov;63(5):1596–1602. doi: 10.1046/j.1471-4159.1994.63051596.x. [DOI] [PubMed] [Google Scholar]
- Polson A. G., Bass B. L. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994 Dec 1;13(23):5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell L. M., Wallis S. C., Pease R. J., Edwards Y. H., Knott T. J., Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987 Sep 11;50(6):831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Rueter S. M., Burns C. M., Coode S. A., Mookherjee P., Emeson R. B. Glutamate receptor RNA editing in vitro by enzymatic conversion of adenosine to inosine. Science. 1995 Mar 10;267(5203):1491–1494. doi: 10.1126/science.7878468. [DOI] [PubMed] [Google Scholar]
- Schall T. J., Lewis M., Koller K. J., Lee A., Rice G. C., Wong G. H., Gatanaga T., Granger G. A., Lentz R., Raab H. Molecular cloning and expression of a receptor for human tumor necrosis factor. Cell. 1990 Apr 20;61(2):361–370. doi: 10.1016/0092-8674(90)90816-w. [DOI] [PubMed] [Google Scholar]
- Sommer B., Köhler M., Sprengel R., Seeburg P. H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991 Oct 4;67(1):11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- Wagner R. W., Yoo C., Wrabetz L., Kamholz J., Buchhalter J., Hassan N. F., Khalili K., Kim S. U., Perussia B., McMorris F. A. Double-stranded RNA unwinding and modifying activity is detected ubiquitously in primary tissues and cell lines. Mol Cell Biol. 1990 Oct;10(10):5586–5590. doi: 10.1128/mcb.10.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wo Z. G., Oswald R. E. Unraveling the modular design of glutamate-gated ion channels. Trends Neurosci. 1995 Apr;18(4):161–168. doi: 10.1016/0166-2236(95)93895-5. [DOI] [PubMed] [Google Scholar]
- Yang J. H., Sklar P., Axel R., Maniatis T. Editing of glutamate receptor subunit B pre-mRNA in vitro by site-specific deamination of adenosine. Nature. 1995 Mar 2;374(6517):77–81. doi: 10.1038/374077a0. [DOI] [PubMed] [Google Scholar]
- Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]




