Abstract
The stimulation of mitochondrial biogenesis (MB) via cell surface G-protein coupled receptors is a promising strategy for cell repair and regeneration. Here we report the specificity and chemical rationale of a panel of β2-adrenoceptor agonists with regards to MB. Using primary cultures of renal cells, a diverse panel of β2-adrenoceptor agonists elicited three distinct phenotypes: full MB, partial MB, and non-MB. Full MB compounds had efficacy in the low nanomolar range and represent two chemical scaffolds containing three distinct chemical clusters. Interestingly, the MB phenotype did not correlate with reported receptor affinity or chemical similarity. Chemical clusters were then subjected to pharmacophore modeling creating two models with unique and distinct features, consisting of five conserved amongst full MB compounds were identified. The two discrete pharmacophore models were coalesced into a consensus pharmacophore with four unique features elucidating the spatial and chemical characteristics required to stimulate MB.
Keywords: adrenoceptor, biogenesis, mitochondria, pharmacophore, chemical similarity, clustering, renal
The regulation of cellular energy demand is complex and essential for the homeostasis of cellular processes and responses to cellular stress.1, 2 While mitochondria have a number of functions, the synthesis of adenosine triphosphate (ATP) is critical to cellular activities. When mitochondria do not properly function, ATP depletion occurs and redox imbalances result in oxidative stress that can lead to cell death. Ischemic injuries are a primary cause of mitochondrial dysfunction and include acute organ injuries such as kidney, liver and heart, and stroke.3 Mitochondrial dysfunction is also associated with multiple chronic disease states including Alzheimer disease and diabetes.4, 5 Consequently, the discovery of compounds that stimulate mitochondrial biogenesis (MB) may have vast therapeutic potential.
MB is the continuous process to form new mitochondria within the cell. MB is necessary to maintain cellular homeostasis, and can be induced during periods of cellular stress or injury. The recent identification of a few compounds that induce MB, have highlighted the process as an important therapeutic target. To study MB, a phenotypic assay is of particular utility. Cellular O2 consumption rates (OCR) reflect the functional activity of the mitochondria, and are reflective of cellular health. We specifically determined MB by measuring maximal OCR after the addition of the proton ionophore carbonylcyanide p-trifluoromethoxyphenylhydrazone (FCCP).6 FCCP uncouples oxygen consumption from the production of ATP resulting in maximal activity of the mitochondrial electron transport chain. If a chemical induces MB, then the FCCP-uncoupled OCR (FCCP-OCR) increases when compared to diluent controls. We have validated this assay with compounds known to cause MB.7, 8 It should also be noted that the respiratory experiments were conducted in primary cultures of rabbit renal proximal tubule cells (RPTC) grown under improved culture conditions, which are highly dependent on aerobic respiration and similar to that found in vivo.7, 8 Although FCCP-OCR is a one dimensional parameter it is reflective of a complex process and ideal for identifying MB. Furthermore, using primary cultures of RPTC have significant scientific and clinical relevance compared to similar assays in cell lines, due to the reliance of RPTC on aerobic respiration.
The β2-AR represents a major and well-studied receptor responsible for multiple phenotypes including smooth muscle relaxation, increased cardiac chronotropy and ionotropy, increased insulin and renin secretion, and glycogenolysis.9–12 At the molecular level the β2-AR is a classical G-protein coupled receptor that couples to both Gsα and Giα, increasing the scope of potential effects.13, 14 The induction of MB though the β2-AR has been demonstrated but the effect of ligand chemotype is still a major question.15–19 Using RPTC respirometry we showed that the selective β2 agonist formoterol was a potent stimulator of MB while the non-selective β2-AR agonist, isoproterenol, was not. Using formoterol as a template we used Tanimoto coefficient (Tc) similarity clustering to determine chemical similarity and probe the LOPAC library. This method revealed that nisoxetine, a structural conjoiner of epinephrine or formoterol, caused MB and we created an initial pharmacophore model with common chemical elements between formoterol and nisoxetine.15 In this study we expand our work and investigated a panel of chemically diverse but selective β2-AR agonists as well as tomoxetine and nisoxetine in regards to their effects on MB.
To expand our understanding of β2-agonist-stimulated MB we tested β2-AR agonists and similar ligands. RPTC were exposed to increasing concentrations (10–3,000 nM) of these compounds and 24 h later FCCP-OCR was determined.20, 21 Chemical evaluation revealed that fourteen compounds contain a phenethylamine core common to many biogenic amine ligands that are β-AR specific agonists (Fig 1). The OCR measurements were performed using a Seahorse Bioscience XF-96 instrument according to the protocol outlined in Beeson et al.21 Each experimental plate was treated with vehicle controls (DMSO<0.5%), a positive control (SRT1720, 10 μM), blank controls, and the appropriate concentration of the compound of interest. Based on preliminary studies the biogenic threshold value was < 1.15 for the mean ratio of (chemical treatment FCCP-OCR/ vehicle control FCCP-OCR). This threshold is ≥ 1 S.D. above the historic mean for the vehicle control. Chemicals were classified as MB based on a mean ratio of: (chemical-treated FCCP-OCR)/(vehicle control FCCP-OCR) > 1.15. This value is ≥1 standard deviation (SD) from the mean ratio for the vehicle control. From our analysis we identified nine compounds that increased FCCP-uncoupled OCR compared to the vehicle control at 24 hr (Fig. 2).
Figure 1.
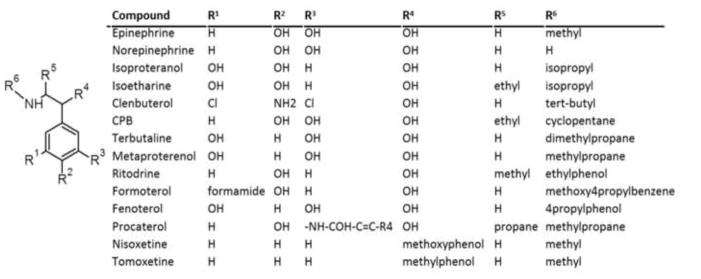
Generalized chemotype of MB stimulating β2-AR agonists and similar compounds.
Figure 2.
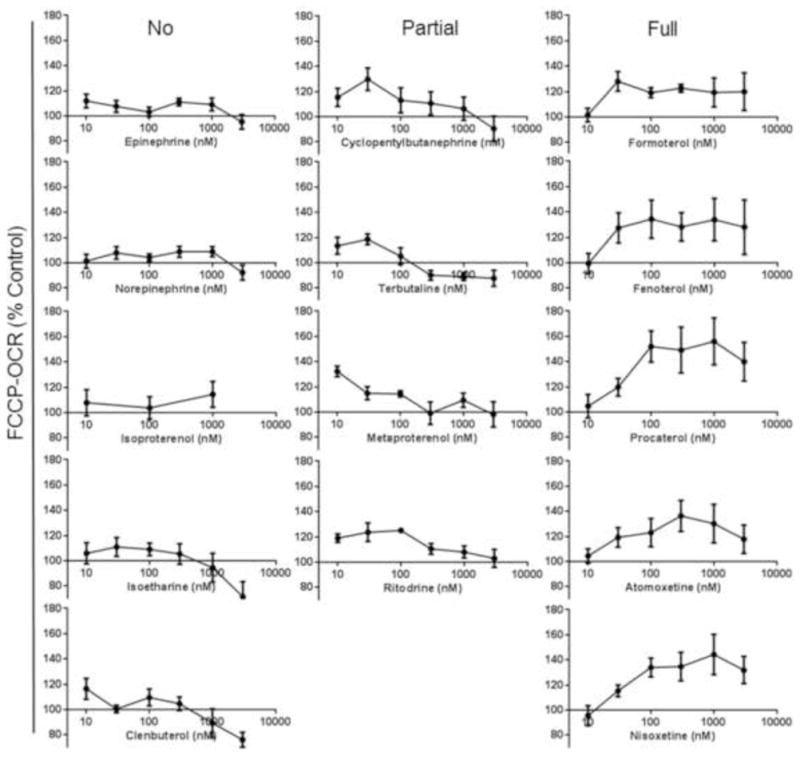
Representative β2-AR agonists and similar compounds induce concentration-responsive increases in FCCP-uncoupled OCR after 24 h. Values indicate a percent of fold change relative to DMSO controls. Data is represented a mean ± s.e.m., N=4.
We began interrogating β2-AR agonist regulation of MB by testing the endogenous catecholamines, epinephrine and norepinephrine (Fig. 1&2). These compounds did not promote MB. A set of three related ligands with greater selectivity for the β2-AR: isoproterenol, clenbuterol, and isoetharine also had no effect on MB. A second set of four ligands (terbutaline, ritodrine, cyclopentylbutanepharine, and metaproteranol) were very potent at low concentrations but were not MB as concentrations increased (Fig. 2). Five of the twelve compounds tested were very potent, had Michaelis-Menten type concentration-response curves and included fenoterol, formoterol, procaterol, nisoxetine and tomoxetine (Fig. 2). Although the mechanism is still not full elucidated, the MB effects of tomoxetine were blocked by the β2-AR antagonist ICI-118,551.15 These data indicate that subtle manipulations of ligand structure can alter potency and efficacy of MB mediated by the β2-AR. The lack of MB with epinephrine and norepinephrine also support that norepinephrine reuptake inhibitor activity is not responsible MB.
Next we analyzed the reported affinities of the β2-AR agonists for a correlation with MB. The panel we tested ranged in affinity (pKd) for the β2-AR from 9.3 to 5.4 at concentrations from 1 to 10,000 nM (Fig. 3). Unexpectedly, there was no strong correlation between affinity and MB efficacy. Compounds with very weak affinity trended to having little efficacy while compounds full MB responses all had a pKd >7. However, compounds with very high affinity like ritodrine, clenbuterol, and CPB only gave a partial MB response, regardless of concentration.
Figure 3.
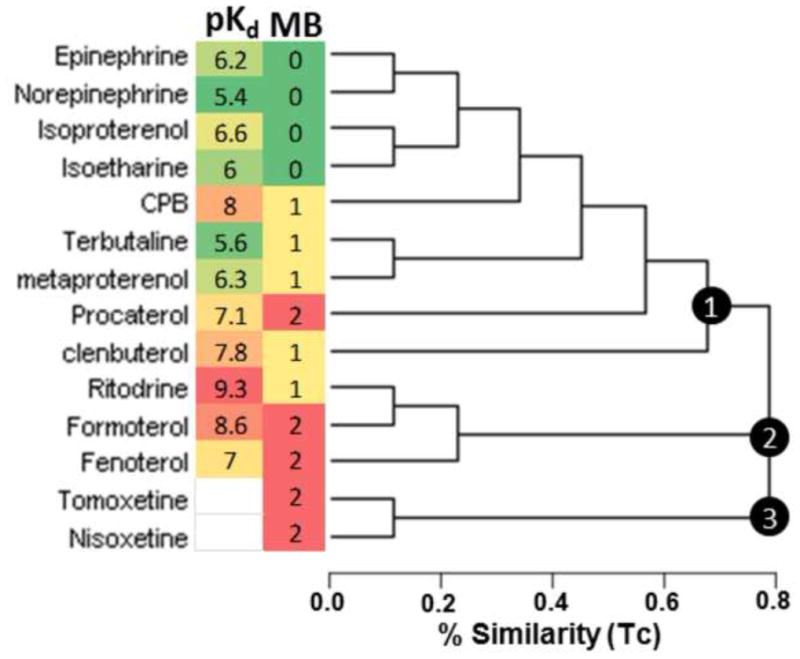
Chemical clustering, pKd and MB activity of β2-AR agonists and similar compounds. MAACS keyed chemical fingerprints were used to cluster compounds based on molecular similarity as measured by Tanimoto Coefficient. Three major clusters were identified and numbered within the chemogram. The MB heat map indicates full (red), partial (yellow), and inactive (green) biogenic compounds as determined by RPTC OCR. The pKd heat map indicates the reported affinity for each ligand to the β2-AR with high affinity (red), intermediate (yellow), and low affinity (green). The chemogram was rendered using Dendroscope.41
Due to the discrepancy in the ability of high affinity and selective β2-AR ligands to stimulate MB, we interrogated the ligand structure using chemical similarity clustering (Fig. 3). Modeling and visualizations were performed using MOE Version 2011.10 (CCG). Compound similarity was measured and visualized using the Tanimoto coefficient metric based on MACCS structural keys using ChemMine single linkage hierarchical clustering.22 Three major clusters were identified. The largest cluster contained most of the classical β2-AR agonists and the endogenous catecholamines. Another cluster contained formoterol, fenoterol and ritodrine. A third cluster segregated nisoxetine and tomoxetine. Again, there was no strong correlation between chemical similarity and MB efficacy as all three clusters contained at least one compound that has a robust MB efficacy profile (Fig. 3). These data reveal that there are very subtle but discrete chemical changes required for promoting MB. To probe these subtle chemical changes, we explored ligand structure in 3D pharmacophoric space.
The cluster 1 and 2 compounds procaterol, formoterol and fenoterol were aligned by hand based on steric considerations.23 The alignment was then refined using flexible alignment. The final pharmacophore model generated had 100% consensus features with a maximum feature radius of 1.2Å (Fig. 4B). Features F1–F5 represents the phenethylamine core found in many sympathomimetic compounds as well as the endogenous sympathetic agonists. Given features F5 and F6 are present in all of the fully biogenic cluster 1/2 compounds, these features occupy necessary space for MB. However, because non-biogenic compounds (e.g., isoproterenol) contain these features, they are not sufficient for mitochondrial biogenesis. On the other hand, F7 is present in the non-biogenic endogenous catecholamines but not in the cluster 3 compounds, making it neither necessary nor sufficient for mitochondrial biogenesis. Comparing ritodrine to formoterol and fenoterol feature F6, represented by the R6 constituent, needs to contain a propyl group to support MB (Fig. 1 & 4). Comparing procaterol to CPB, terbutaline and metaproterenol there is a unique ring at pharmacophore feature F7, represented by the R3 constituent, and propane at R5 are required to support MB (Fig. 1 & 4).
Figure 4.
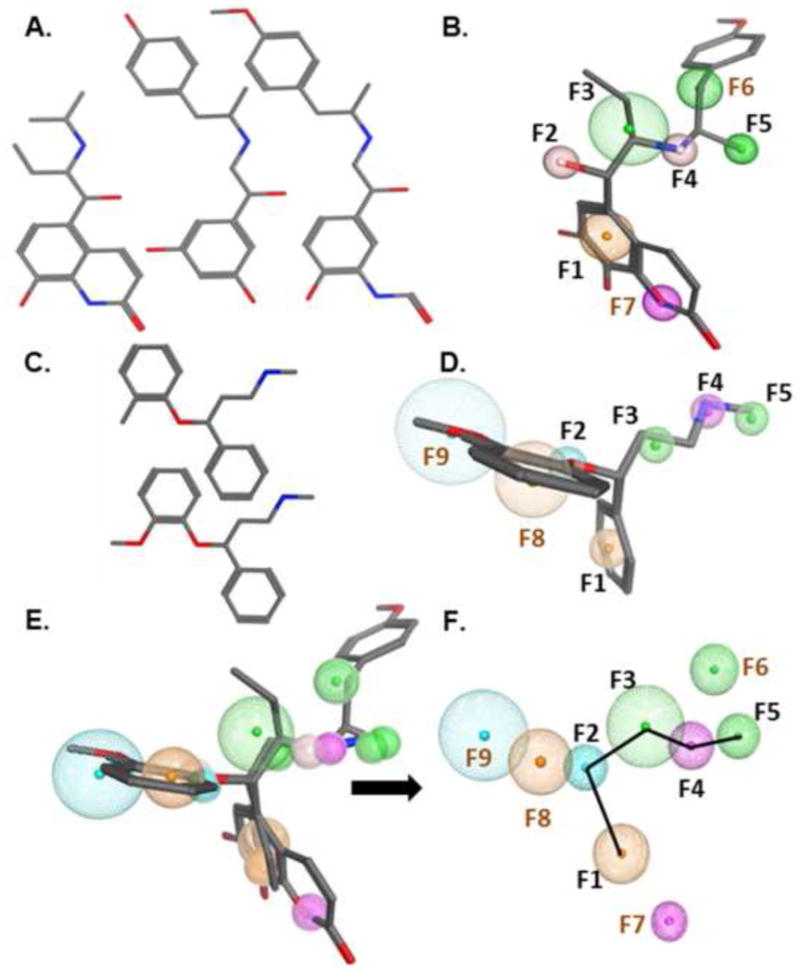
Pharmacophore modeling of phenethylamines. A. Cluster 1/2: procaterol, formoterol and fenoterol. B. Pharmacophore overlay of cluster 1 pharmacophore with cluster 2 pharmacophore. Procaterol, formoterol and fenoterol were flexibly aligned to superimposed chemical features. C. Cluster 3: nisoxetine and tomoxetine. D. Pharmacophore model based on alignment of cluster 3. Nisoxetine and tomoxetine were flexibly aligned to superimposed chemical features. E and F. Pharmacophore overlay of cluster 1/2 pharmacophore with cluster 3 pharmacophore. E. Overlay of 5 compounds with features from cluster 1/2 and cluster 3 pharmacophore models. F. Consensus features derived from both cluster models with consensus features labeled in black and MB specific features in brown. The connectivity path of the phenethylamine core is depicted as a black line. F1 and F8 are aromatic, F2 and F9 are proton donors, F3, F5, and F6 are hydrophobic, F4 and F7 are proton acceptors.
Cluster 3 contained nisoxetine and tomoxetine, which are characterized as norepinephrine reuptake inhibitors (NRIs) and represent non-classical but chemically similar compounds to β2-AR agonists that stimulate MB. Nisoxetine and tomoxetine were aligned in first two and then three dimensions. As in cluster 1/2, the final pharmacophore model generated had 100% consensus features with a maximum feature radius of 1.4Å (Fig. 4D). Features F1–F5 in this pharmacophore also represents the phenethylamine core. However, unlike the cluster 1/2 pharmacophore, there were no further hydrophobic substituents on the carbon alpha to the nitrogen hydrogen bond donor group. This indicates that features F8–F9 (aromatic and hydrophobic acceptor, respectively), which are absent from the cluster 1/2 pharmacophore, are sufficient to stimulate MB when coupled with the phenethylamine core. Comparing nisoxetine and tomoxetine to the rest of the agonists indicated pharmacophore features F8 and F9, representing the R4 constituent, minimally requires a methylphenol group to support MB (Fig. 1 & 4).
We then determined potential overlap of cluster 1/2 and cluster 3 (Fig. 4E/F). The primary positioning parameter was the overlap of corresponding features with hydrogen-bonding interactions. The resulting similar feature volumes were merged into a consensus feature to create a single larger volume that encompassed both the discrete features for each model. Five features had both chemical and spatial overlap. Two of the conserved features represent the phenethylamine core and we found three more features conserved between clusters 1/2 and cluster 3 compounds. This left four discrete features (F6–F9) derived from the two primary models.
These data indicate that all of the MB compounds share a portion of a similar binding mode, but each cluster utilizes at least two discrete interactions that are required outside of the phenethylamine core to support full MB. In the consensus pharmacophore and consistent with the discrete models, F1–F5 represent the phenethylamine core. The hydrophobic feature F5 is present in all clusters, but absent or minimal in the endogenous catecholamines, is therefore a necessary addition to the phenethylamine core in stimulating MB. Of note, F7 is a donor feature in the meta position to the ethanamine portion of the core and is unique to the cluster 1/2 pharmacophore. Because it is not found in the other clusters and is also present in non-biogenic compounds, it is neither necessary nor sufficient for MB. However, it may play a role in stabilizing receptor interactions of individual compounds to generate a conformational change that induces MB. The presence of the hydrophobic F6 may provide the necessary additional feature to stabilize a specific MB stimulating confirmation. The combined features of the phenyl F8 and donor F9 together are also sufficient to support MB. In both cases the addition of two features is required to the canonical five features of the phenethylamine core.
We tested 14 chemically similar compounds for their ability to promote increases in RPTC OCR at low concentrations 24 hr after exposure. We observed there are three distinct OCR phenotypes including full MB, partial MB, and non-MB activities amongst the agonists and similar compounds. These compounds were parsed into three major chemical clusters, two cluster-specific pharmacophore models, and a consensus pharmacophore model. MB efficacy did not correlate with any particular chemical cluster using this approach nor reported pKd for the β2-AR (Fig. 3). We have rationally expanded the chemical space available for MB to include two norepinephrine/serotonin reuptake inhibitors (NRIs). NRIs represent a vast chemical space, but nisoxetine and tomoxetine represent a distinct and limited class of NRIs.5 Other NRIs lack the features we describe here and found necessary for supporting β2-AR mediated MB. Common features found in many other NRIs including tricyclic systems (desipramine, mazindol, tandamine, ciclazindol) or substituted indenes (amedalin, daledalin, talopram, and talsupram) would not be supportive of MB, and are structurally more dissimilar form the β2 type compounds we and others describe.
Our initial supposition was that the MB potential of the compounds would correlate with their reported affinity. Even though most the compounds studied here are β2-AR selective and seven of the partial and full compounds are efficacious at 10–30 nM, there appears to be no relationship between pKd and MB potential (Fig. 3). Alternatively, other properties including duration of signaling and receptor occupancy could explain the observed effect. It is therefore interesting to note how dissimilar the absolute chemical structures are between compounds like formoterol, procaterol and tomoxetine are but their 3D chemical features are nonetheless in tight correlation in chemical space (Fig. 1 & 3 versus Fig. 4). The consensus pharmacophore model derived from the discrete pharmacophore models allows for the precise spatial orientation of the chemical moieties and revealed there are four discrete interactions that govern MB.
The partial and non-biogenic compounds have classical architecture incorporating both ethanolamine and either catechol or resorcinol moieties while none of the full biogenics have a catechol. From a chemistry perspective isoetharine and procaterol are bioisosteres of one another, with procaterol having a quinoline system rather than a catechol. However, procaterol is strongly biogenic, while isoetharine is not, indicating that the space occupied by the quinoline ring is also important for the MB phenotype. Furthermore, CPB exhibits a partial biogenic response while differing from isoetharine by only two carbons. This indicates that, in addition to the stronger hydrogen bond interactions with the receptor, weaker hydrophobic interactions play an important role in differentiating phenotypic responses in the β2-AR. There were more hydrophobic interactions in the full biogenics in our simulations compared to the other two classes of compounds.
The mechanisms and utility of β2-AR regulated MB are of great interest. The regulation of adenylyl cyclases and cAMP by the β2-AR are well studied and there is evidence in the literature to suggest cAMP directly and indirectly regulates mitochondrial function by activating diverse molecules like PKA and Epac that lead to PGC1α activation.16, 24–29 However, other β2-AR regulated pathways (i.e. Ca2+ and CamKKβ or AMPK) may also be involved.30–33 Future studies are needed to elucidate the specific β2-AR -mediated signaling pathway(s) of MB.
The ability of structurally related ligands to induce a spectrum of receptor conformations allows for multiple signaling states and the ability to manipulate pharmacotherapy.27, 34–36 In the case of the MB β2-AR agonists, the non-overlapping features can be inferred to stabilize different conformations of the receptor.37–40 By utilizing informed chemical modeling and phenotypic assays in the process of compound development, it should be possible to attenuate negative effects in addition to enhancing desired pathways. Our therapeutic paradigm for usage of MB agents would be short term and low dose in order to boost mitochondrial function to stimulate cellular/organ repair and regeneration. The pharmacophore models presented can be utilized to develop novel compounds with biogenic potential that minimize the deleterious effects sometimes associated with activation of the β2-AR.
Acknowledgments
RBC is funded by T32 GM08716. LPW is funded by the National Institutes of Health grant T32 CA119945-04 and F32 ES020103-01. RGS is funded by the National Institutes of Health grants ES012878, DK071997 and GM084147, and the Biomedical Laboratory Research and Development Program of the Department of Veterans Affairs [1BX000851]. The authors also thank Kathryn Appleton and Louis Luttrell for thoughtful discussions and Gyda Beeson for technical assistance.
Footnotes
YKP, RBC, RET and LPW conducted the experiments. YKP, RBC, and RGS co-wrote the manuscript. All authors aided in the analysis and interpretation of the data. YKP, CCB and RGS designed and managed the overall project.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hirsch T, Susin SA, Marzo I, Marchetti P, Zamzami N, Kroemer G. Cell Biol and Tox. 1998;14:141–145. doi: 10.1023/a:1007486022411. [DOI] [PubMed] [Google Scholar]
- 2.Birch-Machin MA, Turnbull DM. In: In Methods in Cell Biology. Pon Liza A, EAS, editors. Academic Press; 2001. pp. 97–117. [DOI] [PubMed] [Google Scholar]
- 3.Wallace KB, Starkov AA. Ann Rev Pharm and Tox. 2000;40:353–388. doi: 10.1146/annurev.pharmtox.40.1.353. [DOI] [PubMed] [Google Scholar]
- 4.Baloyannis SJ, Costa V, Michmizos D. Am J Alzheimers Dis Other Demen. 2004;19:89–93. doi: 10.1177/153331750401900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Civitarese AE, Ravussin E. Endocrinology. 2008;149:950–954. doi: 10.1210/en.2007-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrick DA, Neilson A, Beeson C. Drug Discovery Today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Nowak G, Schnellmann RG. Am J Physiol Cell Physiol. 1995;268:C1053–1061. doi: 10.1152/ajpcell.1995.268.4.C1053. [DOI] [PubMed] [Google Scholar]
- 8.Nowak G, Schnellmann RG. Am J Physiol Cell Physiol. 1996;271:C2072–2080. doi: 10.1152/ajpcell.1996.271.6.C2072. [DOI] [PubMed] [Google Scholar]
- 9.Penn RB, Benovic JL. Proc Am Thorac Soc. 2008;5:47–57. doi: 10.1513/pats.200705-054VS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goligorsky MS, Osborne D, Howard T, Hruska KA, Karl IE. Am J Physiol Renal Physiol. 1987;253:F802–809. doi: 10.1152/ajprenal.1987.253.5.F802. [DOI] [PubMed] [Google Scholar]
- 11.Hall RA, Premont RT, Chow CW, Blitzer JT, Pitcher JA, Claing A, Stoffel RH, Barak LS, Shenolikar S, Weinman EJ, Grinstein S, Lefkowitz RJ. Nature. 1998;392:626–630. doi: 10.1038/33458. [DOI] [PubMed] [Google Scholar]
- 12.Granier S, Kobilka B. Nat Chem Biol. 2012;8:670–673. doi: 10.1038/nchembio.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harding SE, Gong H. Congest Heart Fail. 2004;10:181–185. doi: 10.1111/j.1527-5299.2004.02052.x. quiz 186–187. [DOI] [PubMed] [Google Scholar]
- 14.Xiao RP. Sci STKE. 2001;2001:re15. doi: 10.1126/stke.2001.104.re15. [DOI] [PubMed] [Google Scholar]
- 15.Wills LP, Trager RE, Beeson GC, Lindsey CC, Peterson YK, Beeson CC, Schnellmann RG. J Pharmacol Exp Ther. 2012;342:106–118. doi: 10.1124/jpet.112.191528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia MV, Hernandez-Berciano R, Lopez-Mediavilla C, Orfao A, Medina JM. Exp Cell Res. 1997;237:403–409. doi: 10.1006/excr.1997.3804. [DOI] [PubMed] [Google Scholar]
- 17.Chiang MC, Lin H, Cheng YC, Yen CH, Huang RN, Lin KH. J Neurosci Methods. 2012;207:130–136. doi: 10.1016/j.jneumeth.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 18.De Rasmo D, Gattoni G, Papa F, Santeramo A, Pacelli C, Cocco T, Micelli L, Sardaro N, Larizza M, Scivetti M, Milano S, Signorile A. Eur J Pharmacol. 2011;652:15–22. doi: 10.1016/j.ejphar.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Yan B, Huo Z, Liu Y, Xu J, Sun Y, Liang D, Peng L, Zhang Y, Zhou ZN, Shi J, Cui J, Chen YH. J Physiol. 2010;588:2987–2998. doi: 10.1113/jphysiol.2010.190900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.All experiments were performed in accordance with the national and institutional guidelines for animal welfare and adhering to protocols approved by the institutional subcommittee on research animal care.
- 21.Beeson CC, Beeson GC, Schnellmann RG. Anal Biochem. 2010;404:75–81. doi: 10.1016/j.ab.2010.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Backman TW, Cao Y, Girke T. Nucleic Acids Res. 2011;39:W486–491. doi: 10.1093/nar/gkr320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The pharmacophore models were generated by manual alignment of compounds and rigid refinement. Molecules were added to the pharmacophore with successive adjustment of the alignment and modification of the features. A generalized pharmacophore was created by generating features that encompassed the set of corresponding features from each individual pharmacophore. If there was overlap in a feature of type X in pharmacophores A, B, and C (features XA, Xb, and Xc, respectively), a feature of type X would be created for the generalized pharmacophore and its sphere would be expanded to contain X¬A, XB, and X¬c to give feature Xgen. Molecules were rigidly aligned manually, and then subjected to flexible body refinement (configuration limit 100, alpha 1, gradient test 0.01, RMSD tolerance 0.5, maximum steps 500). Initial alignments were performed using the molecules’ respective energy minima.
- 24.Kaya AI, Onaran HO, Ozcan G, Ambrosio C, Costa T, Balli S, Ugur O. J Biol Chem. 2012;287:6362–6374. doi: 10.1074/jbc.M111.301820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasman J, Kukkonen JP, Ammoun S, Akerman KE. Biochem Pharmacol. 2001;62:913–922. doi: 10.1016/s0006-2952(01)00730-4. [DOI] [PubMed] [Google Scholar]
- 26.Pines M, Santora A, Gierschik P, Menczel J, Spiegel A. Bone Miner. 1986;1:15–26. [PubMed] [Google Scholar]
- 27.Stallaert W, Dorn JF, van der Westhuizen E, Audet M, Bouvier M. PLoS One. 2012;7:e29420. doi: 10.1371/journal.pone.0029420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao RP, Avdonin P, Zhou YY, Cheng H, Akhter SA, Eschenhagen T, Lefkowitz RJ, Koch WJ, Lakatta EG. Circ Res. 1999;84:43–52. doi: 10.1161/01.res.84.1.43. [DOI] [PubMed] [Google Scholar]
- 29.Insel PA, Zhang L, Murray F, Yokouchi H, Zambon AC. Acta Physiol. 2012;204:277–287. doi: 10.1111/j.1748-1716.2011.02273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sosroseno W, Sugiatno E. Acta Biomed. 2008;79:110–116. [PubMed] [Google Scholar]
- 31.Kloster MM, Naderi EH, Haaland I, Gjertsen BT, Blomhoff HK, Naderi S. Int J Oncol. 2013 doi: 10.3892/ijo.2013.1853. [DOI] [PubMed] [Google Scholar]
- 32.Carlucci A, Lignitto L, Feliciello A. Trends Cell Biol. 2008;18:604–613. doi: 10.1016/j.tcb.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Iguchi H, Mitsui T, Ishida M, Kanba S, Arita J. Endocr J. 2011;58:747–759. doi: 10.1507/endocrj.k11e-104. [DOI] [PubMed] [Google Scholar]
- 34.Galandrin S, Bouvier M. Mol Pharmacol. 2006;70:1575–1584. doi: 10.1124/mol.106.026716. [DOI] [PubMed] [Google Scholar]
- 35.Kenakin T, Miller LJ. Pharmacol Rev. 2010;62:265–304. doi: 10.1124/pr.108.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luttrell LM, Kenakin TP. Methods Mol Biol. 2011;756:3–35. doi: 10.1007/978-1-61779-160-4_1. [DOI] [PubMed] [Google Scholar]
- 37.Sauliere A, Bellot M, Paris H, Denis C, Finana F, Hansen JT, Altie MF, Seguelas MH, Pathak A, Hansen JL, Senard JM, Gales C. Nat Chem Biol. 2012;8:622–630. doi: 10.1038/nchembio.961. [DOI] [PubMed] [Google Scholar]
- 38.Audet M, Bouvier M. Nat Chem Biol. 2008;4:397–403. doi: 10.1038/nchembio.97. [DOI] [PubMed] [Google Scholar]
- 39.Reynolds KA, Katritch V, Abagyan R. J Comput Aided Mol Des. 2009;23:273–288. doi: 10.1007/s10822-008-9257-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seifert R, Wenzel-Seifert K, Gether U, Kobilka BK. J Pharmacol Exp Ther. 2001;297:1218–1226. [PubMed] [Google Scholar]
- 41.Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]


