Abstract
Response surface methodology (RSM) was used to optimize the cultivation conditions for the production of phytase by recombinant Escherichia coli DH5α. The optimum predicted cultivation conditions for phytase production were at 3 hours seed age, a 2.5% inoculum level, an L-arabinose concentration of 0.20%, a cell concentration of 0.3 (as measured at 600 nm) and 17 hours post-induction time with a predicted phytase activity of 4194.45 U/mL. The model was validated and the results showed no significant difference between the experimental and the predicted phytase activity (P = 0.305). Under optimum cultivation conditions, the phytase activity of the recombinant E. coli DH5α was 364 times higher compared to the phytase activity of the wild-type producer, Enterobacter sakazakii ASUIA279. Hence, optimization of the cultivation conditions using RSM positively increased phytase production from recombinant E. coli DH5α.
Keywords: phytase, recombinant, optimization, Escherichia coli, response surface methodology, cultivation conditions
Introduction
Phytate, the salt form of phytic acid (myo-inositol 1, 2, 3, 4, 5, 6-hexakis dihydrogen phosphate), is the primary and major storage form of organically bound phosphorus in many plant-based foods such as cereals and legumes.1 It accounts for 30% of total phosphorus in roots (eg, cassava), and 60%–80% of total phosphorus in plants seeds and cereals (eg, oilseeds, corn, wheat, barley and etc.).2 Phytic acid acts as an anti-nutritional factor in human and animal diets as it can bind to many minerals including phosphorus, calcium, zinc, magnesium and copper, forming insoluble salts.3 It also can form a complex with proteins and cause them to become more resistant to proteolytic enzymes such as pepsin and trypsin. Thus, phytic acid reduces the availability of all minerals, including phytate phosphorus, to simple stomach animals (monogastric animals) since they lack or have an absence of phytase enzymes in their gastrointestinal tracts.4–6
Phosphorus is important in the diet of pigs and poultry because it is needed for maturity and maintenance of the skeletal system. Phosphorus is also involved in carbohydrate and fat metabolism. It is a part of the phospholipid bilayer within cell membranes, and is a component of adenine triphosphate (ATP) and creatinine phosphate for energy metabolism.7 Since phytase (myo-inositol hexakisphosphate phosphohydrolase) is able to hydrolyze phytate complexes (IP6) into lower inositol phosphate (IP5-IP1), inorganic phosphorus and other minerals,8,9 the addition of phytase as an alternative to inorganic phosphate supplementation in monogastric animal diets might reduce fecal phosphate excretion by up to 50%.10
Apparently, the use of phytase in foods has also become of great interest to many scientific communities.11 The supplementation of phytase might increase protein digestibility and mineral availability during digestion in the stomach or during food processing such as soaking, grinding, malting, fermentation, heat treatment and germination.1,12–14
Phytase can be isolated from various sources such as microorganisms, plants and certain animal tissues.6,13 Based on the great potential and value of phytase, many studies have been performed in order to increase phytase production. Response surface methodology (RSM) has been widely and effectively used to study the various factors that affect phytase production, such as the composition of media and induction conditions.15–17 Response surface methodology has an advantage in that it includes interactions between variables compared to the one-variable-at-a-time technique; thus, it signifies the full effect of all parameters on the experimental process.18,19
Recombinant phytase is produced by inserting a phytase gene into a new host strains prior to gene expression using certain promoter system. Type of strain use, substrate, growth conditions and nutrients may affect the production of phytase.20 Basically, E. coli has been widely used as a host for the recombinant protein production including phytase because of its rapid and easy to growth as well as possessed recognized characteristics.21,22 In addition, the promoter system as well influences the selection of an expression host.23 For example, E. coli DH5α can be integrated with the expression vector that been attached with araBAD promoter, such as pBAD-TOPO vectors and E. coli BL21(DE3) with the expression vector contained T7 promoter as an example of pET vectors.
In this study, ES-TOPO plasmid vector was previously constructed by co-researchers at International Islamic University Malaysia (IIUM), Malaysia with stop codon prior to transform into competent E. coli DH5α host cells. The genotype of E. coli DH5α (F− Φ80lacZΔM15 Δ(lacZYA−argF) U169 recA1 endA1 hsdR17 (rK−mK+) phoA supE44 λ− thi-1 gyrA96 relA1) contained many mutations, thus provide many excellent properties to be an expression host. The most significant mutations are lacZDelta M15 mutation that allows for the blue/white screening for recombinant cells, recA1 mutation may reduce homologous recombination for a more stable insert, endA1 mutation may reduce endonuclease digestion of plasmid for higher plasmid yield and hsdR17(rK−mK+) may reduce the activity of EcoK restriction enzyme.24
Thus, E. coli DH5α is a stable host strain for an expression of foreign gene. In addition, the stability of plasmid was observed for more than 16 h after induction with L-arabinose (Nuge, unpublished data). ES-TOPO plasmid vector derived from the pBAD-TOPO vector integrated with phytase gene of E. sakazakii ASUIA279. The ES-TOPO plasmid was incorporated with the araBAD promoter (PBAD) for the regulation of phytase gene expression. In the PBAD system, L-arabinose was used as an inducer while glucose was a repressor for protein expression and the optimum concentration of L-arabinose need to optimize in each strain. Several advantages were obtained when using PBAD promoter system such as the system permit a high level of recombinant protein production and L-arabinose is a low-cost inducer.25 Thus, by using this expression system, it may reduce the cost for commercial application.
Finally, the goal of the current study was to predict the optimum settings of each independent variable: seed age (hours), inoculum level (%, v/v), time of induction (by measuring the cell concentration at 600 nm), L-arabinose concentration (%, w/v) and post-induction time (hours). This occurred under cultivation conditions in order to increase the phytase production of recombinant E. coli DH5α carrying an ES-TOPO plasmid inserted by an E. sakazakii ASUIA279 phytase gene, using the statistical optimization method of RSM on a laboratory scale.
Materials and Methods
Chemicals
Phytic acid as a dodecasodium salt was purchased from Sigma Chemical Co. (St. Louis, MO, USA). All other chemicals and media were products of Merck (Darmstadt, Germany). All reagents were analytical grade.
Bacterial culture and expression system
A glycerol stock of E. coli DH5α that was previously transformed with an ES-TOPO plasmid carrying the phytase gene from E. sakazakii ASUIA279 was provided by the Department of Biotechnology Engineering, International Islamic University Malaysia (IIUM) and stored at −80 °C. This culture was used as the expression host for phytase production (U/mL). The phytase was expressed as an intracellular enzyme from the ES-TOPO plasmid by arabinose induction under the regulation of PBAD.
Fermentation conditions for phytase production
E. coli DH5α cells were incubated for 24 hours at 37 °C on a Luria-Bertani (LB) agar plate supplemented with 100 μg/mL ampicillin, using a Memmert incubator (Schwabach, Germany). The plate was incubated in the inverted position to prevent condensation, which can affect bacterial growth. After the incubation period, a single colony (Fig. 1) was inoculated into 10 mL LB broth supplemented with 100 μg/mL ampicillin in an Erlenmeyer flask using a sterile inoculation loop and was aerobically grown at 37°C with agitation at 200 rpm for the saturated culture preparation, using an incubator shaker, model SI-600 (JEIO TECH, Seoul, Korea). Following this, 10% (v/v) of this culture was sub-cultured into a new LB broth supplemented with 100 μg/mL ampicillin in an Erlenmeyer flask and grown under the same conditions as above for the 3 to 11 h seed age preparation. Then, 2.5% to 7.5% (v/v) of this culture was used as an inoculum for the fermentation conditions. The bacterial culture was grown in a 500 mL Erlenmeyer flask containing 100 mL fermentation medium supplemented with 100 μg/mL ampicillin. Fermentation media used for the phytase production was prepared according to Nuge, (unpublished data). The culture was induced with different levels of L-arabinose concentrations: 0.002, 1% and 2%, respectively when the cell concentration of the growing bacterial culture reached ODs of 0.3, 0.5 and 0.7 (equivalent to 3.8 × 108 − 1.0 × 109 CFU/mL cells), as assessed by measuring at 600 nm. Finally, the cultures were harvested at different time intervals between 2.5 and 17.5 h after induction.
Figure 1.
Colonies development of E. coli DH5α after 24 h incubation at 37 °C.
Enzyme extraction using ultrasonication
The bacterial cells were harvested by centrifugation of the fermentation media at 11,500 rpm for 20 minutes (min) at 4°C using a Sigma 3–18 K centrifuge (Sartorius Stedim, Göttingen, Germany). Then the bacterial pellet was collected and dissolved in 100 mM of sodium acetate buffer, pH 5, and the cells were disrupted using a 150 V/T ultrasonic homogenizer (Biologics Inc., Manassas, Virginia, USA) equipped with a stepped titanium microtip, 3.9 mm in diameter and 255.8 mm in length. The cells were disrupted for 30 seconds (sec) with 30 sec cooling periods for 1 min with 30 Watt acoustic power and a 50% duty cycle at 20 kHz. The samples were kept in an ice bath during the ultrasonication process to prevent overheating, thus preventing the proteins from denaturing.26,27 Following this, the sonicated cells were centrifuged at 11,000 rpm, 4 °C, for 30 min to remove the cell debris. This was done with the aid of a Sigma 3–18 K centrifuge (Sartorius Stedim, Göttingen, Germany). The supernatant was collected and assayed for phytase activity (U/mL). The suitable processing volume for this probe ranged from 300 μL to 15 mL.
Measurement of phytase activity
The assay mixture consisted of 299 μL of 100 mM sodium acetate buffer (pH 5) and 100 μL of 3.6 mM sodium phytate.28 The assay mixture was pre-incubated at 50°C for 5 min using a shaking water bath (PROTECH®, Selangor, Malaysia). Furthermore, the enzymatic reaction was started by adding 1 μL of the enzyme solution that was previously diluted in 100 mM of sodium acetate buffer, pH 5. The assay mixture was further incubated at 50°C for 30 min. Then, the inorganic liberated phosphate was measured according to an ammonium molybdate method29 with some modifications.30 A 1.5 mL of a freshly prepared stop solution consisting of acetone: 5N H2SO4:10 mM ammonium molybdate (2:1:1 v/v) was added to the assay mixture before adding 100 μL of 1 M citric acid. Any cloudiness was removed by centrifugation prior to measuring the absorbance at 355 nm. In order to calculate the enzyme activity, a calibration curve was produced over the range of 500– 600 mmoL phosphate (ɛ = 8.7 cm2/nmoL). Activity (units) was expressed as 1 μmoL inorganic phosphate liberated in 1 min under assay conditions.31,32 Stop solution was added to the assay mixture prior to adding the enzyme for the blank run. The reaction was performed in triplicate.
Response surface methodology (RSM)
The face-centered central composite design (FCCCD) under the response surface methodology (RSM) was used to optimize the cultivation conditions for intracellular phytase production (U/mL) from recombinant E. coli DH5α involving five independent variables, namely: seed age (hours), inoculum level (%, v/v), L-arabinose concentration (%, w/v), induction time (cell concentration by measuring at 600 nm) and post-induction time (hours). Minitab™ version 15.0 software (Minitab Inc., PA, USA) was used for the experimental design and statistical analysis. The average value of phytase activity (U/mL) represented by γ̂ was measured as a response throughout the experiment. The independent variables and the range were chosen based on previous studies performed by Sunitha et al16 and Pan et al,33 with some modifications.34 The independent variables and the ranges were first screened using a fractional factorial design. Based on the screening study, factors including seed age (x1), inoculums level (x2), induction time (by measuring the cell concentration at 600 nm; x3), L-arabinose concentration (x4) and post-induction time (harvesting time; x5) were shown to be highly significant and largely contributed to phytase production (U/mL) produced from recombinant E. coli DH5α (unpublished data). Thus, the levels of each independent variable were further optimized using FCCCD under RSM. Table 1 shows the range of both the coded and original values of the five independent variables measured in the experiment. The face- centered central composite design (FCCCD) was used at three levels; low, central and high (−1, 0, and +1), leading to a total of 32 experimental runs together with six replicates at the central point (Table 2) to predict the optimum cultivation conditions for phytase production (U/mL). Replication at the central point is important in order to determine the curvature and the pure error sum of squares. The experimental runs were performed at random across three replications. The experiment aimed to predict the optimum settings for each independent variable that yielded the desirable response and to determine the interaction effects of each variable.35
Table 1.
Experimental range and levels of the independent variables used for the optimization of phytase production from recombinant E. coli DH5α.
| Variable | Symbol coded | Level | ||
|---|---|---|---|---|
|
| ||||
| −1 | 0 | 1 | ||
| Seed age (hours) | x1 | 3 | 7 | 11 |
| Inoculums level (%, v/v) | x2 | 2.5 | 5 | 7.5 |
| L-arabinose concentration (%, w/v) | x3 | 0.002 | 1 | 2 |
| Induction time (cell concentration at 600 nm) | x4 | 0.3 | 0.5 | 0.7 |
| Harvesting time (hours) | x5 | 2.5 | 10 | 17.5 |
Table 2.
Face centered central composite design (FCCCD) for the optimization of phytase production from recombinant E. coli DH5α showing the corresponding experimental and predicted phytase activity (U/mL).
| Run order | Seed age (h) | Inoculums level (%, v/v) | L-arabinose conc. (%, w/v) | Cell conc. at 600 nm | Harvesting time (h) | Phytase activity (U/mL) | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Experimental | Predicted | ||||||
| 1 | 3 (−1) | 7.5 (+1) | 2 (+1) | 0.7 (+1) | 2.5 (−1) | 533.65 | 662.38 |
| 2 | 7 (0) | 5 (0) | 0.002 (−1) | 0.5 (0) | 10 (0) | 2277.57 | 2433.86 |
| 3 | 7 (0) | 5 (0) | 2 (+1) | 0.5 (0) | 10 (0) | 1031.55 | 696.46 |
| 4 | 3 (−1) | 7.5 (+1) | 0.002 (−1) | 0.3 (−1) | 2.5 (−1) | 571.95 | 833.81 |
| 5 | 3 (−1) | 2.5 (−1) | 0.002 (−1) | 0.7 (+1) | 2.5 (−1) | 1363.48 | 1568.10 |
| 6* | 7 (0) | 5 (0) | 1.001 (0) | 0.5 (0) | 10 (0) | 1657.11 | 1565.16 |
| 7 | 7 (0) | 5 (0) | 1.001 (0) | 0.5 (0) | 17.5 (+1) | 1437.53 | 2049.57 |
| 8* | 7 (0) | 5 (0) | 1.001 (0) | 0.5 (0) | 10 (0) | 1271.56 | 1565.16 |
| 9 | 3 (−1) | 5 (0) | 1.001 (0) | 0.5 (0) | 10 (0) | 1289.43 | 1423.60 |
| 10* | 7 (0) | 5 (0) | 1.001 (0) | 0.5 (0) | 10 (0) | 1953.30 | 1565.16 |
| 11 | 7 (0) | 5 (0) | 1.001 (0) | 0.7 (+1) | 10 (0) | 1271.56 | 1662.90 |
| 12 | 7 (0) | 5 (0) | 1.001 (0) | 0.3 (−1) | 10 (0) | 1835.85 | 1467.43 |
| 13 | 11 (+1) | 7.5 (+1) | 2 (+1) | 0.3 (−1) | 2.5 (−1) | 824.73 | 827.28 |
| 14 | 3 (−1) | 2.5 (−1) | 0.002 (−1) | 0.3 (−1) | 17.5 (+1) | 5017.30 | 4620.39 |
| 15* | 7 (0) | 5 (0) | 1.001 (0) | 0.5 (0) | 10 (0) | 1445.19 | 1565.16 |
| 16 | 7 (0) | 5 (0) | 1.001 (0) | 0.5 (0) | 2.5 (−1) | 194.82 | 249.56 |
| 17 | 11 (+1) | 2.5 (−1) | 0.002 (−1) | 0.7 (+1) | 17.5 (+1) | 4123.63 | 4213.63 |
| 18 | 11 (+1) | 7.5 (+1) | 0.002 (−1) | 0.7 (+1) | 2.5 (−1) | 1833.29 | 1235.18 |
| 19 | 7 (0) | 7.5 (+1) | 1.001 (0) | 0.5 (0) | 10 (0) | 1618.81 | 2205.27 |
| 20* | 7 (0) | 5 (0) | 1.001 (0) | 0.5 (0) | 10 (0) | 1598.39 | 1565.16 |
| 21 | 11 (+1) | 5 (0) | 1.001 (0) | 0.5 (0) | 10 (0) | 2106.50 | 1706.73 |
| 22 | 11 (+1) | 7.5 (+1) | 2 (+1) | 0.7 (+1) | 17.5 (+1) | 1440.08 | 1375.03 |
| 23 | 11 (+1) | 7.5 (+1) | 0.002 (−1) | 0.3 (−1) | 17.5 (+1) | 3916.81 | 4287.46 |
| 24* | 7 (0) | 5 (0) | 1.001 (0) | 0.5 (0) | 10 (0) | 1710.73 | 1565.16 |
| 25 | 3 (−1) | 2.5 (−1) | 2 (+1) | 0.7 (+1) | 17.5 (+1) | 773.66 | 745.35 |
| 26 | 11 (+1) | 2.5 (−1) | 2 (+1) | 0.3 (−1) | 17.5 (+1) | 1613.71 | 1718.38 |
| 27 | 3 (−1) | 7.5 (+1) | 2 (+1) | 0.3 (−1) | 17.5 (+1) | 1013.67 | 819.18 |
| 28 | 3 (−1) | 2.5 (−1) | 2 (+1) | 0.3 (−1) | 2.5 (−1) | 280.87 | 197.60 |
| 29 | 11 (+1) | 2.5 (−1) | 0.002 (−1) | 0.3 (−1) | 2.5 (−1) | 788.98 | 770.40 |
| 30 | 7 (0) | 2.5 (−1) | 1.001 (0) | 0.5 (0) | 10 (0) | 2259.70 | 2340.03 |
| 31 | 11 (+1) | 2.5 (−1) | 2 (+1) | 0.7 (+1) | 2.5 (−1) | 1514.13 | 1561.58 |
| 32 | 3 (−1) | 7.5 (+1) | 0.002 (−1) | 0.7 (+1) | 17.5 (+1) | 4769.63 | 4277.03 |
Note:
Central point.
Statistical analysis
The experimental data were subjected to a regression analysis using the statistical software mentioned above. The general form of the second order polynomial regression model used to explain phytase production was as follows:
| (1) |
where γ̂ is the predicted response (phytase production), β0 is a constant, and βi, βii and βij are the linear, quadratic and interaction term estimated coefficients, respectively. Theoretically, based on the estimated regression coefficient table, the final reduced model should only consist of the significant terms (P < 0.05). However, in some cases, non-significant (P > 0.05) variables are also included in the final reduced model. For example, the main terms are included in the final reduced model if the higher-order terms of this variable are found to be statistically significant.35 The analysis of variance (ANOVA) including the F-value, the lack of fit test (LOF) and the coefficient of determination, R2, were used to determine the appropriate model. The ‘response optimizer’ in Minitab™ version 15.0 software (Minitab Inc., PA, USA) was used to predict the optimum setting for each independent variable that contributed to the optimum predicted response.36 In addition, response surface plots were also used to represent the interaction effects of the significant variables represented by the model’s equation.
Validation of the statistical model
A validation study for the regression analysis of the experimental data was performed by running the experiment in triplicate under the optimized conditions predicted by the face-centered central composite design (FCCCD) by using the method described above.33,37,38 The enzyme was extracted and assayed for phytase activity (U/mL).
Results and Discussion
Response surface methodology (RSM): optimization of the cultivation conditions for phytase production
The FCCCD was used to determine the estimated optimum settings of each variable for phytase production (U/mL) from recombinant E. coli and to determine the interaction effects among those variables. The results for the central composite design are shown in Table 2. A total of 32 experimental runs and six replicates at the central point, together with experimental (U/mL) and predicted phytase activity (U/mL), were included. The experimental data were analyzed using a multiple regression analysis. The regression coefficient shown in Table 3 was used to present the polynomial regression equation. The final reduced model of the polynomial regression equation was used to estimate phytase production, as shown below:
Table 3.
Estimated regression coefficient by face centered central composite design (FCCCD) using coded level of independent variables used for phytase production from recombinant E. coli DH5α.
| Term | Coefficient | Std. error coefficient | T-value | P-value |
|---|---|---|---|---|
| Constant | 1565.16 | 99.81 | 15.682 | 0.000 |
| x1 | 141.57 | 84.63 | 1.673 | 0.109 |
| x2 | −67.38 | 84.63 | −0.796 | 0.435 |
| x3 | −868.70 | 84.63 | −10.265 | 0.000* |
| x4 | 97.74 | 84.63 | 1.155 | 0.261 |
| x5 | 900.01 | 84.63 | 10.635 | 0.000* |
| x2x2 | 707.48 | 192.13 | 3.682 | 0.001* |
| x5x5 | −415.60 | 192.13 | −2.163 | 0.042* |
| x1x3 | 240.65 | 89.76 | 2.681 | 0.014* |
| x3x5 | −723.87 | 89.76 | −8.064 | 0.000* |
| x4x5 | −202.03 | 89.76 | −2.251 | 0.035* |
Note:
P-value < 0.05, indicates the significant of the coefficient to the model.
| (2) |
where γ̂ represents the predicted response (phytase production) while x1, x2, x3, x4, and x5 are the coded values of the five independent variables.
Based on the results displayed in Table 3, the significant terms from the main effects for phytase productions are x3 (L-arabinose concentration) and x5 (post-induction time). In addition, the significant quadratic terms for phytase production are x22 (inoculum level, inoculum level) and x52 (post-induction time, post-induction time). Meanwhile, the interaction terms x1x3 (seed age, L-arabinose concentration), x3x5 (L-arabinose concentration, post-induction time) and x4x5 (induction time, by measuring the cell concentration at 600 nm, post-induction time) were also significant. These significant factors are very important, as phytase production will be affected by even the smallest variation in their values. In this study, the significant terms included in the final reduced model of the second order polynomial regression equation were selected using a backward elimination method. Starting with full quadratic terms, it involved a stepwise elimination of the non-significant terms where the most non-significant term will be eliminated first. The next subsequent steps followed the same procedure until there remained only significant terms (P < 0.05). The final reduced model of the second order polynomial regression equation (2) should only consist of the significant terms (P < 0.05). However, the non-significant main terms (P > 0.05): x1, x2 and x4 were also included as their higher order terms are found to be statistically significant in this study.
The analysis of variance (ANOVA) was used to fit the appropriate model (Table 4). The F-value of 31.49 with a very small significant P-value (P < 0.001) shows that the respective model fits the data. Also, the non-significant lack-of-fit (LOF) value, which was 0.130 (P > 0.05), indicates that the experimental data fit the model. Moreover, the high value of the coefficient of determination (adjusted R2 = 0.908%) also shows the significance of the model. This value was used to refer to the sample variance in phytase production due to seed age (hours), inoculum level (%, v/v), L-arabinose concentration (%, w/v), induction time (by measuring the cell concentration at 600 nm) and post-induction time (hours). Theoretically, an R2 value close to 1 indicates a better correlation between the experimental and predicted values.39,40 Thus, in this study, the high value of the adjusted R2 implied a good agreement between the experimental and predicted values of phytase activity (U/mL), as shown in Table 2.
Table 4.
ANOVA table for the phytase production from recombinant E. coli DH5α using face centered central composite design (FCCCD).
| Source | DF | Sum of square | Mean square | F-value | P-value |
|---|---|---|---|---|---|
| Regression | 10 | 40588723 | 4058872 | 31.49 | 0.000* |
| Linear | 5 | 28778185 | 5755637 | 44.65 | 0.000* |
| Square | 2 | 1847046 | 923523 | 7.16 | 0.004* |
| Interaction | 3 | 9963492 | 3321164 | 25.76 | 0.000* |
| Residual error | 21 | 2707151 | 128912 | ||
| Lack-of-fit | 16 | 2435183 | 152199 | 2.80 | 0.130 |
| Pure error | 5 | 271968 | 54394 | ||
| Total | 31 | 271968 |
Notes: Coefficient of determination, R2 = 0.938, R2 (adjusted) = 0.908.
P-value < 0.05.
Abbreviation: DF, degree of freedom.
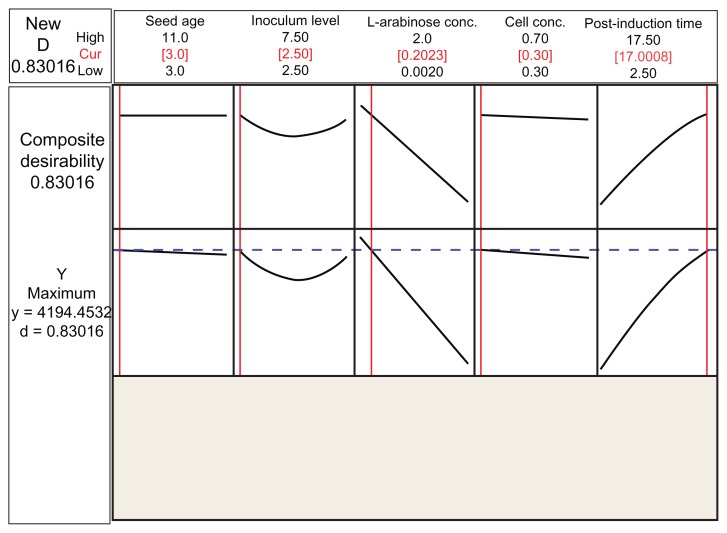
The value of P < 0.05 supports the significance of the model coefficient in the response surface model. By applying the regression analysis on Equation 2 using the ‘response optimizer’ in Minitab™ version 15.0 software (Minitab Inc., PA, USA), the optimum operating conditions for phytase production were predicted as a 3 h seed age, 2.5% inoculum level, 0.2% L-arabinose concentration, and cell concentration of 0.3, as measured at 600 nm, and 17 hours post-induction time, with a predicted phytase activity of 4194.45 U/mL (Fig. 2).
Figure 2.
Optimization plot derived from ‘response optimizer’ showed the optimum settings of each factor with the maximum predicted phytase activity of 4194.45 U/mL.
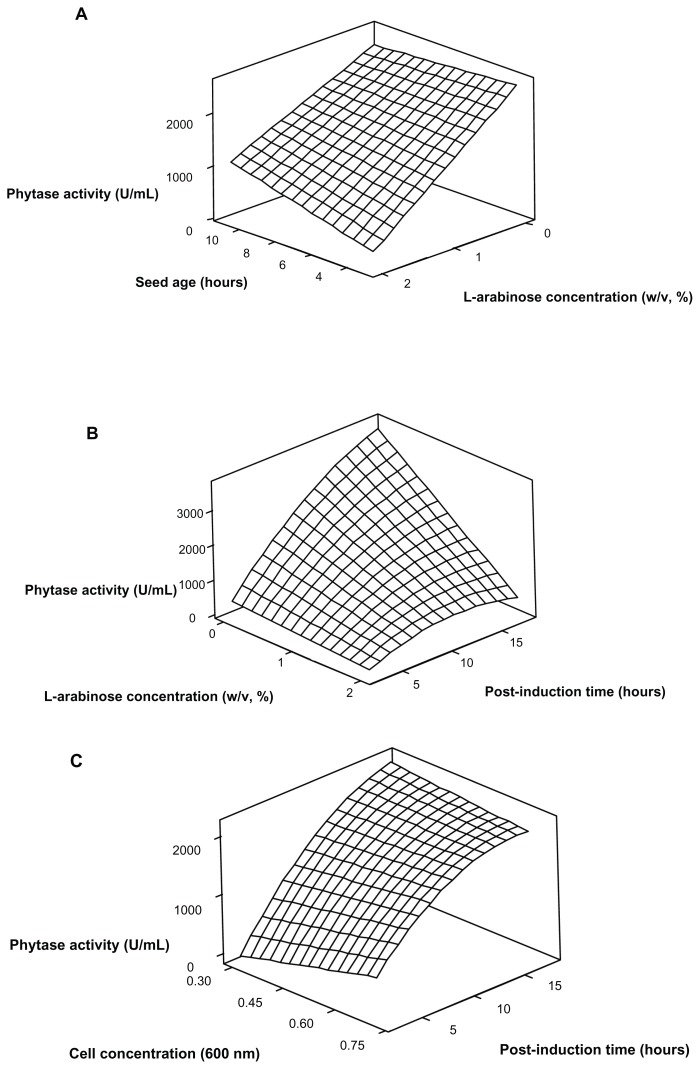
Furthermore, the interaction effects of the significant variables were determined by plotting and analyzing response surface graphs.40 Figure 3A shows the interaction effect of seed age and L-arabinose concentration on the phytase activity with other factors were kept at their central values. The highest phytase activity occurred at the lower level of seed age and L-arabinose concentration. Basically, it was suggested that the effect of seed age and inoculum level are related to the length of the lag phase in the cultivation culture. Additionally, the seed age and inoculum level was considered as interconnected factors, and they are supposed to be optimized as one for the optimum level determination of each factor.41,42 In this study, it was suggested that the youngest culture of E. coli cells reached an exponential phase of the growth cycle and resulted in a short lag phase during its growth in a newly inoculated fermentation medium. Meanwhile, the higher seed age culture may have caused the slow adaptation of E. coli when inoculated in a new fermentation medium, causing a long lag phase during the cultivation, and possibly causing suppression of phytase activity.
Figure 3.
Response surface graphs for the phytase production by optimizing the cultivation condition: (A) Interaction between seed age and L-arabinose concentration while other factors were kept at their central level. (B) Interaction between L-arabinose concentration and post-induction time (h) while other factors were kept at their central level. (C) Interaction between induction time (by measuring cell concentration at 600 nm) and post-induction time while other factors were kept at their central level.
Figure 3A shows the interaction effect of L- arabinose concentration and seed age. Figure 3B shows the interaction effect of L-arabinose concentration and post-induction time on phytase activity at L-arabinose concentrations of 0.002%, 1.001% and 2%. The optimization of L-arabinose concentration is necessary because the PBAD that was incorporated in the expression vector is a highly regulated promoter, which means that its level of expression is effectively modulated by L-arabinose concentration.25,43,44 As shown in Figure 3A, the phytase activity was increased with a decrease in the L-arabinose concentration at any levels of seed age. Meanwhile, the phytase activity was increased with a decrease in the L-arabinose concentration and with increasing of post-induction time (Fig. 3B). Moreover, a significant increase in phytase activity was observed at the lower L- arabinose concentration and higher post-induction time. This shows that a lower level of L-arabinose concentration is probably sufficient to induce the phytase gene expression, and results in a high phytase production at a higher post-induction time. Meanwhile, the higher levels of L-arabinose concentration may cause retardation of cell growth and decrease the phytase production.45
The effect of induction time (by measuring the cell concentration at 600 nm) and post-induction time on phytase activity at cell concentration levels of 0.3, 0.5 and 0.7 is presented in Figure 3C. It shows that at the later of post-induction time (17.5 h), phytase activity increased as the cell concentration decreased (as measured at 600 nm). This could mean that induction of the E. coli DH5α culture with L-arabinose at the early exponential phase increased the phytase activity. In bacterial growth, the cell concentration in the exponential phase is normally between 0.3–1.5 (as measured at 600 nm).46 However, phytase activity was decreased with decreasing cell concentration (as measured at 600 nm) at the earlier post-induction time (2.5 h). This may due to having had inadequate time for the protein expression. Overall, this shows that the highest phytase activity occurred at a lower level of cell concentration and at a higher level of post-induction time.
Figure 3B presents the effect of post-induction time and L-arabinose concentration on phytase activity at the central level of seed age (7 h), inoculums level (5%) and induction time (by measuring the cell concentration at 600 nm; 0.5). Meanwhile, Figure 3C shows the effect of post-induction time and induction time (by measuring the cell concentration at 600 nm) on phytase activity at central level of seed age (7 h), inoculum level (5%) and L-arabinose concentration (1.001%). Along the L-arabinose concentration, phytase activity was increased with increasing of post-induction time. Indeed, it is clearly shown that the highest phytase activity occurred at the lower L-arabinose concentration and higher post-induction time (Fig. 3B). Under good fermentation conditions, the longer post-induction time might provide an extended time for the protein synthesis to occur, thus producing high phytase activity. However, the lower phytase activity achieved at the lower post-induction time perhaps due to insufficient time for protein formation. In addition, Figure 3C shows that the phytase activity increased with increasing of post-induction time (hours) along with the induction time (by measuring the cell concentration at 600 nm). Basically, the optimum post-induction time/harvesting time is highly influenced by the other factors such as inducer concentration and induction time during the fermentation.
Validation of the statistical model
A student’s t-test was performed to determine the significance difference between the experimental and predicted levels of phytase activity (U/mL). The experimental data were obtained from triplicate experiments performed under optimized cultivation conditions, while the predicted phytase activity (U/mL) was the result of regression model. The value of P = 0.305 indicates that there was no significant difference between the experimental (3890.43 ± 386 U/mL) and predicted levels of phytase activity (4194.4532 U/mL).
The results showed that by optimizing these variables, the production of phytase (4194.4532 U/mL) from recombinant E. coli DH5α was higher compared to phytase production under optimized conditions of 39.7°C incubation temperature, pH 7.1, 13.6% (w/v) rice bran, agitation speed and aeration at 320 rpm and 0 vvm in E. sakazakii ASUIA279 (11.511 U/mL). This condition also may reduce the fermentation time, as the highest phytase activity was found at 17 h post-induction time compared to 5 days in E. sakazakii ASUIA279.47 The fermentation medium used in this study allowed the bacterial culture to maintain its growth phase for a long duration, thus enabling its growth and induction overnight before the cell was harvested in the early hours of the following day.48 Using this method, the culture can be freshly used without the need to freeze and thaw it first, as was necessary with the Luria-Bertani (LB) medium, from which the cells were harvested 3 to 6 hours after induction.49 Also, based on the preliminary study, this medium showed high phytase activity compared to the commercial LB medium. The results obtained were comparable with studies performed by other researchers33,16 who reported that after optimizing seed age, inoculum level, substrate concentration, cell concentration and post-induction time, phytase production from the recombinant E. coli BL21 (DE3) was successfully increased. Thus, these variables play an important role in the production of phytase from recombinant Escherichia coli. Other studies also showed that RSM is a valuable tool for optimizing phytase production.15–17
Previously, E. sakazakii ASUIA279 has been demonstrated to be the most promising strain for producing phytase as compared to other isolates, including Pantoea stewartii ASUIA271 and Bacillus cereus ASUIA260.28 These were newly isolated strains from the endophyte area of maize plantation in Malaysia. To our knowledge, not much information is reported on the expression of phytase genes derived from E. sakazakii in E.coli. Extracellular phytase produced by E. sakazakii ASUIA279 has been characterized and shows potential properties as feed additives or soil nutrient enhancement.50 Since the optimization of phytase production in wild-type E. sakazakii ASUIA279 produces low activity,47 phytase production in E. coli DH5α was aimed to overcome this problem. In addition, Enterobacter sp. was commonly known as a strain of pathogens that can cause serious illness such as neonatal meningitis.51 Therefore, the integration of the E. sakazakii ASUIA279 phytase gene into E. coli DH5α may be able to prevent the risk of pathogenic effects Enterobacter sp. Results presented here showed that E. coli DH5α was successfully used for the improvement of phytase production under a lab scale. Thus, study of the E. sakazakii ASUIA279 phytase gene may have great potential for the Malaysian enzyme industry.
Conclusions
This was the first attempt to optimize the cultivation conditions for phytase production from recombinant E. coli DH5α carrying the phytase gene from E. sakazakii ASUIA279, isolated from Malaysian maize roots. The RSM was successfully used to predict the optimum settings for each independent variable, which all contributed to the high level of predicted phytase activity of 4194.45 U/mL. This indicated that optimization of the cultivation conditions for E. coli DH5α produced a high level of phytase production compared to the wild-type producer E. sakazakii ASUIA279. By using the expression system in E. coli DH5α, high phytase activity was obtained only after induction for 17.0 hours compared with 5 days as reported for E. sakazakii ASUIA279. Therefore, production of phytase in a recombinant E. coli DH5α system under optimum cultivation conditions helps to reduce the fermentation time. The fermentation media also influenced bacterial growth and product formation. By using optimum cultivation conditions obtained in this study, phytase production by E. coli DH5α may be taken to a greater scale using large-scale bioreactor system for industrial purposes, by taking other parameters into consideration. Overall, this study successfully increased the production of phytase by recombinant E. coli DH5α.
Acknowledgements
This project was supported by the BIOTEK Malaysia, Ministry of Science, Technology and Innovation, Malaysia.
Footnotes
Author Contributions
Conceived and designed the experiments: RMA, ASMH. Analysed the data: RMA. Wrote the first draft of the manuscript: RMA. Contributed to the writing of the manuscript: RMA, AS, ASMH. Agree with manuscript results and conclusions: AF, MYAM, AI, AK, ASMH. Jointly developed the structure and arguments for the paper: RMA, AF, MYAM, AI, AK, AS, ASMH. Made critical revisions and approved final version: AF, MYAM, AI, AK, AS, ASMH. All authors reviewed and approved of the final manuscript.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
References
- 1.Reddy NR, Sathe SK, Salunkhe DK. Phytates in legumes and cereals. Adv Food Res. 1982;28:1–92. doi: 10.1016/s0065-2628(08)60110-x. [DOI] [PubMed] [Google Scholar]
- 2.Haefner S, Knietsch A, Scholte E, Braun J, Lohscheidt M, Zelder O. Biotechnological production and applications of phytases. Appl Microbiol Biotechnol. 2005;68(5):588–97. doi: 10.1007/s00253-005-0005-y. [DOI] [PubMed] [Google Scholar]
- 3.Xiong AS, Yao QH, Peng RH, et al. High level expression of a synthetic gene encoding Peniophora lycii phytase in methylotrophic yeast Pichia pastoris. Appl Microbiol Biotechnol. 2006;72(5):1039–47. doi: 10.1007/s00253-006-0384-8. [DOI] [PubMed] [Google Scholar]
- 4.Ozusaglam MA, Ozcan N. Cloning of phytase gene in probiotic bacterium Bacillus coagulans. Adv Studies Biol. 2009;1(1):15–24. [Google Scholar]
- 5.Singh PK. Significance of phytic acid and supplemental phytase in chicken nutrition: a review. World’s Poultry Sci J. 2008;64(4):553–80. [Google Scholar]
- 6.Vohra A, Satyanarayana T. A cost-effective cane molasses medium to enhance cell-bound phytase production by Pichia anomala. J Appl Microbiol. 2004;97(3):471–6. doi: 10.1111/j.1365-2672.2004.02327.x. [DOI] [PubMed] [Google Scholar]
- 7.Pandey A, Szakacs G, Soccol CR, Leon JA, Soccol VT. Production, purification and properties of microbial phytases. Bioresour Technol. 2001;77(3):203–14. doi: 10.1016/s0960-8524(00)00139-5. [DOI] [PubMed] [Google Scholar]
- 8.Lei XG, Porres JM. Phytase enzymology, applications, and biotechnology. Biotechnol Lett. 2003;25(21):1787–94. doi: 10.1023/a:1026224101580. [DOI] [PubMed] [Google Scholar]
- 9.Liu B-L, Rafiq A, Tzeng Y-M, Rob A. The induction and characterization of phytase and beyond. Enz Microbial Technol. 1998;22(5):415–24. [Google Scholar]
- 10.Walz OP, Pallauf J. Microbial phytase combined with amino acid supplementation reduces P and N excretion of growing and finishing pigs without loss of performance. Int J Food Sci Technol. 2002;37(7):835–48. [Google Scholar]
- 11.Konietzny U, Greiner R. Phytic acid: Nutritional impact. In: Caballero B, Trugo L, Finglas P, editors. Encyclopedia of Food Sciences and Nutrition. London: Elsevier; 2003. pp. 4555–63. [Google Scholar]
- 12.Reddy NR, Pierson MD, Sathe SK, Salunkhe DK. Phytates in cereals and legumes. Boca Raton: CRC Press; 1989. [Google Scholar]
- 13.Tijskens LM, Greiner R, Biekman ES, Konietzny U. Modeling the effect of temperature and pH on activity of enzymes: the case of phytases. Biotechnol Bioeng. 2001;72(3):323–30. doi: 10.1002/1097-0290(20010205)72:3<323::aid-bit9>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 14.Sandberg AS, Andlid T. Phytogenic and microbial phytases in human nutrition. Int J Food Sci Technol. 2002;37(7):823–33. [Google Scholar]
- 15.Vohra A, Satyanarayana T. Statistical optimization of the medium components by response surface methodology to enhance phytase production by Pichia anomala. Proc Biochem. 2002;37(9):999–1004. [Google Scholar]
- 16.Sunitha K, Kim YO, Lee JK, Oh TK. Statistical optimization of seed and induction conditions to enhance phytase production by recombinant Escherichia coli. Biochem Eng. 2000;5(1):51–6. [Google Scholar]
- 17.Sunitha K, Lee JK, Oh TK. Optimization of medium components for phytase production by E. coli using response surface methodology. Bioproc Eng. 1999;21(6):477–81. [Google Scholar]
- 18.Bas D, Boyasi IH. Modeling and optimization 1: Usability of response surface methodology. J Food Eng. 2007;78(3):836–45. [Google Scholar]
- 19.Chang Y, Huang J, Lee C, Lee C, Shih I, Tzeng YM. Use of response surface methodology to optimize culture medium for production of lovastatin by Monascus rubber. Enz and Microbial Technol. 2002;30(7):889–94. [Google Scholar]
- 20.Vats P, Banerjee UC. Production studies and catalytic properties of phytases (myo-inositolhexakisphosphate phosphohydrolases): an overview. Enzyme Microb Tech. 2004;35(1):3–14. [Google Scholar]
- 21.Sharma SS, Blattner FR, Harcum SW. Recombinant protein production in an Escherichia coli reduced genome strain. Metabolic Eng. 2007;9:133–41. doi: 10.1016/j.ymben.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi JH, Lee SY. Secretory and extracellular production of recombinant proteins using Escherichia coli. Appl Microb Biotechnol. 2004;64:625–35. doi: 10.1007/s00253-004-1559-9. [DOI] [PubMed] [Google Scholar]
- 23.Bell PA. E. coli Expression Systems. In: Gerstein AS, editor. Molecular Biology Problem Solver: A Laboratory Guide. New York: Wiley-Liss; 2001. pp. 462–86. [Google Scholar]
- 24.Life Technologies. Genotypes of Invitrogen™ Competent cells. [Accessed Nov 24, 2012]. Available at: http://www.invitrogen.com/site/us/en/home/Products-and-Services/Applications/Cloning/Transformation/Trans-Misc/Learn_More_About_Choosing_Competent_Cells/Genotypes_of_Competent_Cells.html.
- 25.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bact. 1995;177:4121–30. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho CW, Chew TK, Ling TC, Kamaruddin S, Tan WS, Tey BT. Efficient mechanical cell disruption of Escherichia coli by an ultrasonicator and recovery of intracellular hepatitis B core antigen. Proc Biochem. 2006;41(8):1829–34. [Google Scholar]
- 27.Feliu JX, Cubarsi R, Villaverd A. Optimized released of recombinant protein by ultrasonication of E. coli cells. Biotechnol and Bioeng. 1998;58(5):536–40. doi: 10.1002/(sici)1097-0290(19980605)58:5<536::aid-bit10>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 28.Meor Hussin AS, Farouk AE, Ali AM, Greiner R. Production of phytate-degrading enzyme from Malaysian soil bacteria using rice bran containing media. J Agrobiotechnol. 2011;1(1):17–28. [Google Scholar]
- 29.Heinonen JK, Lahti RJ. A new and convenient colorimetric determination of inorganic orthophosphate and its application to the assay of inorganic pyrophosphatase. Anal Biochem. 1981;113(2):313–7. doi: 10.1016/0003-2697(81)90082-8. [DOI] [PubMed] [Google Scholar]
- 30.Greiner R, Farouk A-El. Purification and characterization of bacterial phytase whose make it exceptionally useful as a feed supplement. Protein J. 2007;26(7):467–74. doi: 10.1007/s10930-007-9086-z. [DOI] [PubMed] [Google Scholar]
- 31.Lee J, Choi Y, Lee P-C, Kang S, Bok J, Cho J. Recombinant production of Penicillium oxalicum PJ3 phytase in Pichia Pastoris. World J Microbiol Biotechnol. 2007;23(3):443–6. [Google Scholar]
- 32.Quan C-S, Fan S-D, Zhang L-H, Wang Y-J, Ohta Y. Purification and properties of a phytase from Candida krusei WZ-001. J Biosci Bioeng. 2002;94(5):419–25. [PubMed] [Google Scholar]
- 33.Pan H, Xie Z, Bao W, Zhang J. Optimization of culture conditions to enhance cis-epoxysuccinate hydrolase production in Escherichia coli by response surface methodology. Biochem Eng J. 2008;42(2):133–8. [Google Scholar]
- 34.Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, Keasling JD. Homogeneous expression of the PBAD promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiol. 2001;147(12):3241–7. doi: 10.1099/00221287-147-12-3241. [DOI] [PubMed] [Google Scholar]
- 35.Mirhosseini H, Tan CP, Hamid NSA, Yusof S. Modeling the relationship between the main emulsion components and stability, viscosity, fluid behavior, ζ-potential, and electrophoretic mobility of orange beverage emulsion using response surface methodology. J Agri Food Chem. 2007;55(19):7659–66. doi: 10.1021/jf071061k. [DOI] [PubMed] [Google Scholar]
- 36.Pan H, Bao W, Xie Z, Zhang J. Optimization of medium composition for cisepoxysuccinate hydrolase production in Escherichia coli by response surface methodology. African J Biotechnol. 2012;9(9):1366–73. [Google Scholar]
- 37.Singh B, Satyanarayana T. Phytase production by a thermophilic mould Sporotrichum thermophile in solid state fermentation and its potential applications. Biores Technol. 2008;99(8):2824–30. doi: 10.1016/j.biortech.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 38.Chadha SB, Harmeet G, Mandeep M, Saini SH, Singh N. Phytase production by the thermophilic fungus Rhizomucor pusillus. World J Microbiol and Biotechnol. 2004;20(1):105–9. [Google Scholar]
- 39.Salihu A, Alam MZ, Abdulkarim MI, Salleh HM. Optimization of lipase production by Candida cylindracea in palm oil mill effluent based medium using statistical experimental design. J Mol Catal B: Enz. 2011;69:66–73. [Google Scholar]
- 40.Gangadharan D, Sivaramakrishnan S, Nampoothiri MK, Sukumaran KR, Pandey A. Response surface methodology for the optimization of alpha amylase production by Bacillus amyloliquefaciens. Biores Technol. 2008;99(11):4597–602. doi: 10.1016/j.biortech.2007.07.028. [DOI] [PubMed] [Google Scholar]
- 41.Xia Z, Wu S. Cell number as an important variable in optimising inoculum age and size in yeast cultivation. African J Biotechnol. 2012;11(4):919–22. [Google Scholar]
- 42.Panesar PS. Production of β-galactosidase from whey using Kluyveromyces marxianus. Res J Microbiol. 2008;3(1):24–9. [Google Scholar]
- 43.Geertsma ER, Groeneveld M, Slotboom DJ, Poolman B. Quality control of overexpressed membrane proteins. PNAS. 2008;105(15):5722–7. doi: 10.1073/pnas.0802190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgan-Kiss RM, Caryn W, Cronan JE. Long-term and homogeneous regulation of the Escherichia coli araBAD promoter by use of a lactose transporter of relaxed specificity. PNAS. 2002;99(11):7373–7. doi: 10.1073/pnas.122227599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muntari B, Amid A, Mel M, Jami MS, Salleh HM. Recombinant bromelain production in Escherichia coli: process optimization in shake flask culture by response surface methodology. AMB Express. 2012;2(12):1–9. doi: 10.1186/2191-0855-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flick K, Ahuja S, Chene A, Bejarano MT, Chen Q. Optimized expression of Plasmodium falciparum erythrocyte membrane protein 1 domains in Escherichia coli. Malaria J. 2004;3:50. doi: 10.1186/1475-2875-3-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meor Hussin AS, Farouk A, El Salleh H, Ali AM, Ideris A. Optimization of cultivation conditions for the production of bacterial phytase from Enterobacter sakazakii ASUIA279 newly isolated from Malaysian maize root. In: Waldron KW, Moates GK, Faulds CB, editors. Total Food: Sustainability of the Agri-Food Chain. London: Royal Society of Chemistry; 2009. pp. 96–104. [Google Scholar]
- 48.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif. 2005;41(1):207–34. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 49.Mihasan M, Ungureanu E, Artenie V. Optimum parameters for overexpression of recombinant protein from tac promotors on autoinducible medium. Romanian Biotechnol Lett. 2007;12(6):3473–82. [Google Scholar]
- 50.Farouk AE, Greiner R, Hussin AS. Purification and properties of a phytate-degrading enzyme produced by Enterobacter sakazakii ASUIA279. J Biotec Biodivers. 2012;3(1):1–9. [Google Scholar]
- 51.Pagotto FJ, Nazarowec-White M, Bidawid S, Farber JM. Enterobacter sakazakii: infectivity and enterotoxin production in vitro and in vivo. J Food Prot. 2003;66(3):370–5. doi: 10.4315/0362-028x-66.3.370. [DOI] [PubMed] [Google Scholar]