Abstract
Currently, there is no standardized panel for immunophenotyping myeloid cells in mouse spleen using flow cytometry. Markers such as CD11b, CD11c, F4/80, Gr-1, Ly6C, and Ly6G have long been used to identify various splenic cell myeloid populations. Flow cytometry and fluorescence-activated cell sorting (FACS) analysis demonstrated that Ly6G/Ly6C markers are superior to Gr-1 for identifying splenic neutrophils, eosinophils, and subsets of monocytes/macrophages. Moreover, these experiments showed that F4/80 is not required for identifying these myeloid subsets and that many of the commercially available preparations of anti-F4/80 antibodies stain poorly for this antigen in spleen. Taken together, we have now developed an informative flow cytometry panel that can be combined with other cell markers to further delineate subpopulations of mouse splenic myeloid cells. This panel will be highly useful to investigators in the flow cytometry field, as there is a critical need to standardize the analysis of myeloid cell subsets.
Keywords: Macrophage, monocyte, F4/80, immunophenotyping
Introduction
The mouse spleen contains several distinct populations of myeloid cells with varying immune functions, including neutrophils, eosinophils, monocytes, macrophages, and dendritic cells. Although previous studies have suggested that these cells are readily identified by flow cytometry based on their cell surface staining characteristics, many of these cells share common expression patterns for myeloid specific antigens. Therefore, a single antibody is not sufficient for demarcation of various myeloid subsets and a need for standardized markers to phenotype mouse myeloid cells in the spleen is required, which is particularly true for monocytes/macrophages.
Under steady-state conditions, most macrophage populations within murine lymphoid tissues are believed to originate from blood monocytes. Based on the expression of cell surface markers, mouse monocytes can be divided into at least two main subsets: classical (Ly6C++CD43CCR2+CD62L+CX3CR1Low) and non-classical (Ly6C-CD43+CCR2-CD62L-CX3CR1Hi) (1). Mouse macrophages have also typically been divided into two subsets based on the expression of Gr-1 or Ly6C antigens. The Ly6C (or Gr-1)Hi subset has been termed “classical” or “inflammatory” while Ly6C (or Gr-1)Low-neg cells are termed “nonclassical” or “resident” (2,3). Both of these subpopulations express the 125 kDa transmembrane adhesion glycoprotein F4/80, (4) which is not essential for macrophage function (5). Antibodies to the F4/80 antigen were originally derived by fusing splenocytes from a rat hyperimmunized with cultured thioglycollate-induced mouse peritoneal macrophages with a mouse myeloma cell line (4). It has generally been assumed to be a macrophage-specific marker, yet other cell types, such as skin Langerhans cells (6) and eosinophils (7), also express F4/80.
To achieve a panel for immunophenotyping splenic myeloid cells, various cell surface markers were tested by flow cytometry and fluorescence-activated cell sorting (FACS) analysis. Compared to Gr-1, Ly6C/Ly6G markers were better for identifying neutrophils, eosinophils, and both subsets of monocytes/macrophages in mouse spleen. Detailed investigations using the antigen F4/80 revealed that myeloid cell subsets could be readily identified without the use of this marker. Furthermore, many of the commercially available anti-F4/80 antibodies stained weakly for this antigen. Herein, a splenic myeloid cell immunophenotyping panel that can be used independently or in combination with other markers is provided. Adoption of this improved panel will be imperative for standardizing the investigation of myeloid cell subsets in mouse spleen.
Materials and Methods
Animals
C57BL/6 (B6) mice and B6.129P-Cx3cr1tm1Litt/J (further referred to as CX3CR1-GFP/GFP) mice were initially obtained from the Jackson Laboratory (Bar Harbor, ME, USA). Only CX3CR1-GFP/+ animals were used in our studies. Animals were bred and maintained in a pathogen free barrier facility within the Center for Comparative Medicine at Northwestern University. All experiments involving mice were approved by the IACUC at Northwestern University.
Cell Preparation
Spleens were harvested and pooled from 6-8 week old B6 or CX3CR1-GFP/+ mice in RPMI 1640 (Mediatech, Manassus, VA, USA). Cell suspensions were prepared by dicing spleens with a razor blade, digesting with a solution containing 0.1 mg/mL DNase I (Roche, Indianapolis, IN, USA) and 1 mg/mL Collagenase D (Roche, Indianapolis, IN, USA) in HBSS (Cellgro, Manassus, VA, USA) for 30 minutes at 37°C, followed by passage through a 40 μM Nylon filter (BD Falcon, Bedford, MA, USA). Red blood cell lysis was performed using 2 mL/spleen of 1x BD Pharm Lyse solution (BD Biosciences, Sparks, MD, USA). Cells were then washed in either MACS buffer (Miltenyi Biotech, Auburn, CA, USA) or staining buffer (Ca2+ and Mg2+ free PBS (BioWhittaker, Wakersville, MD, USA) containing 5% heat-inactivated fetal bovine serum (Atlas, Fort Collins, CO, USA), 0.09% sodium azide (Sigma-Aldrich, St. Louis, MO, USA), and 5 mM EDTA (Acros Organics, Geel, Belgium) and counted using a Countess automated cell counter (Invitrogen, Carlsbad, CA, USA). For flow cytometry immunophenotyping experiments, 3 × 106 cells per tube were stained as described below. For FACS analyses, 2 × 108 cells per cocktail were prepared in MACS buffer (Miltenyi Biotech), rather than staining buffer.
Flow Cytometric Cell Staining
Cell viability was assessed by incubation in the amine-reactive dye Aqua (Invitrogen) (1:500) dilution in Ca2+ and Mg2+ free PBS) for 30 minutes in the dark at room temperature (RT), followed by a single wash in 1x PBS. For all experiments, cells were incubated in 0.5 μg Fc Block (BD Biosciences) for 10 minutes at RT. Surface staining was performed in the dark for 30 minutes at 4 °C in staining buffer. Cells were then washed twice with staining buffer followed by fixation in 1% paraformaldehyde (VWR, West Chester, PA, USA). A comprehensive list of surface markers for these experiments includes: CD45R (B220) clone RA3-6B2 PE-Texas Red (1:250, BD Biosciences), CD4 clone RM4-5 PerCP-Cy5.5 (1:160, BD Biosciences), CD8 clone 53-6.7 PerCP-Cy5.5 (1:160, BD Biosciences), CD8 clone 53-6.7 eFluor 450 (1:333, eBioscience), CD11b clone M1/70 eFluor 450 (1:160, eBioscience, San Diego, CA, USA), CD11b clone M1/70 PE-Texas Red (1:500, Invitrogen (Caltag)), CD11c clone HL3 PE-Cy7 (1:125, BD Biosciences), CD16/CD32 clone 2.4G2 PE (1:100, BD Biosciences), CD19 clone 1D3 PerCP-Cy5.5 (1:160, BD Biosciences), CD40 clone 1C10 APC (1:100, eBioscience), CD69 clone H1.2F3 PerCP-Cy5.5 (1:167, BD Biosciences), CD80 clone 16-10A1 PE (1:500, eBioscience), CD86 clone GL1 APC (1:333, eBioscience), CD86 clone GL1 Alexa Fluor 700 (1:100, BD Biosciences), CD115 APC clone AFS98 (1:100, eBioscience), F4/80 clone CI:A3-1 Alexa Fluor 647 (various dilutions, optimized at 1:200, AbD Serotec, Raleigh, NC, USA), F4/80 clone BM8 Alexa Fluor 700 (various dilutions, AbD Serotec), F4/80 clone BM8 APC (various dilutions, optimized at 1:100, eBioscience), F4/80 clone BM8 FITC (various dilutions, eBioscience), F4/80 clone BM8 PE (various dilutions, eBioscience), F4/80 clone BM8 PE-Cy7 (various dilutions, eBioscience), F4/80 clone BM8 PE-Texas Red (various dilutions, eBioscience), F4/80 clone BM8 PerCP-Cy5.5 (various dilutions, eBioscience), Gr-1 clone RB6-8C5 APC-Cy7 (1:160, BD Biosciences), Ly6C clone AL-21 APC-Cy7 (1:500, BD Biosciences), Ly6G clone 1A8 PE (1:416, BD Biosciences), Ly6G clone 1A8 PerCP-Cy5.5 (1:500, BD Biosciences), MHC Class II clone M5/114.15.2 FITC (1:167, eBioscience), MHC Class II clone M5/114.15.2 eFluor 450 (1:500, eBioscience), MHC Class II clone M5/114.15.2 APC-eFluor 780 (1:167, eBioscience), NK1.1 clone PK136 Alexa Fluor 700 (1:160, BD Biosciences), NK1.1 clone PK136 APC (1:286, BD Biosciences), NK1.1 clone PK136 FITC (1:160, BD Biosciences), NK1.1 clone PK136 PerCP-Cy5.5 (1:100, BD Biosciences), mPDCA-1 clone JF05-1C2.4.1 APC (1:10, Miltenyi Biotech), Siglec F clone E50-2440 PE (1:100, BD Biosciences). For flow cytometry immunophenotyping experiments, cells were acquired on an LSR II cytometer (BD Immunocytometry Systems, San Jose, CA, USA) equipped with 405 nm, 488 nm, 561 nm, and 640 nm excitation lasers. The spleen FACS experiments were performed using a FACSAria II instrument (BD Immunocytometry Systems) equipped with 405 nm, 488 nm, or 633 nm lasers located at the University of Chicago Flow Cytometry Core Facility, Chicago, IL, USA. All data collection and sorting were performed using BD FACS Diva software (BD Biosciences) and data analyses were performed using FlowJo software (Tree Star, Ashland, OR, USA). Fluorescence minus one (FMO) controls were used for gating analyses to distinguish positively from negatively staining cell populations. Compensation was performed using single color controls prepared from BD Comp Beads (BD Biosciences) for cell surface staining or Arc Beads (Invitrogen) for Aqua live/dead discrimination. Compensation matrices were calculated and applied using FlowJo software (Tree Star). Biexponential transformation was adjusted manually when necessary.
Cytospin Preparation and Staining
For cytologic analysis of cell preparations, cells were mounted on slides using a Cytospin centrifuge (Shandon, Ramsey, MN, USA) for 10 minutes at 1000 RPM. Cells were then fixed in methanol for 30 seconds and air-dried. Slides were then submerged in Diff-quick solution II for 30 seconds and drained, followed by immersion in Diff-quick solution I for 30 seconds. The slides were rinsed in tap water for 60 seconds and then rapidly dehydrated in absolute alcohol. Counterstaining with Giemsa was performed for 30 seconds, followed by rinsing with tap water and rapid dehydration in absolute ethanol. Cytospin preparations were imaged using an Olympus (Tokyo, Japan) microscope equipped with an Olympus DP20 imaging system. Independent analysis of the slides was performed by a hematopathologist blinded to the study.
Results
F4/80 staining is highly variable in mouse spleen
F4/80 has long been used to identify monocytes and macrophages in both lymphoid and non-lymphoid tissues. To optimize F4/80 staining by flow cytometry in mouse spleen, we tested the commonly used clone BM8 conjugated to various fluorochromes (Supplemental Figure 1). F4/80 staining was highly variable, with several different fluorochromes conjugated to BM8 giving high background signal that prevented clear delineation of positively and negatively stained cell populations. This was particularly true in the FMO panels. The clone CI:A3-1 conjugated to Alexa Fluor 647 gave the best overall signal-to-noise ratio, allowing for clear distinction of F4/80 positive and negative cells irrespective of CD11b expression (Supplemental Figure 1).
Upon building a splenic phenotyping panel, it became apparent that combinations of the BM8 clone with Gr-1 resulted in poor discrimination of F4/80 positive and negative populations for CD11bHi cells (Supplemental Figure 2, Gates C and E). This effect was also evident, although less robust, when using the CI:A3-1 clone (Supplemental Figure 3, Gates C and E). Combining either F4/80 clone with various other targets including Ly6C, Ly6G, NK1.1, and CD11c did not reproduce this effect (Supplemental Figures 2 and 3 and data not shown). The inability to discriminate F4/80 positive and negative populations may not be a compensation issue, as excitation of various other antigenic targets using the 640 nM laser did not produce a similar result (Supplemental Figures 2 and 3 and data not shown). Pre-incubation with Gr-1 prior to staining with either F4/80 clone corrected this issue, while the converse experiment completely masked the CD11b+F4/80- cell population (Supplemental Figure 4, Gate E and data not shown). Notably, pre-incubation with either F4/80 clone prior to Ly6G staining also obscured the CD11b+F4/80- population. Taken together, these findings suggest that commercially available preparations of anti-F4/80 antibodies vary considerably in their ability to identify F4/80 positive cells in mouse spleen. Another possibility is that there may be a common epitope between F4/80 and the Ly6G component of Gr-1 or steric hindrance between the antibodies, as evidenced by the competition experiments.
Gr-1 and Ly6C/Ly6G markers identify similar populations of monocytes and macrophages in mouse spleen
Given the above findings, we set out to establish a gating strategy for phenotyping mouse myeloid cells in spleen without examining the F4/80 antigen (Figure 1). After exclusion of debris, doublets, nonviable cells, and lineage markers (CD4, CD8, CD19, NK1.1, CD11cHi), CD11b+ cells were sub-divided into 4 distinct populations using either Gr-1 vs SSC or a combination of Ly6C and Ly6G vs SSC markers (Figure 1): A) neutrophils, Ly6GHiSSCInt or Gr-1HiSSCInt, B) eosinophils, Ly6CIntLy6G-SSCHi or Gr-1IntSSCHi, and monocytes and macrophages, C) Ly6CLo-negLy6G-SSCLo or Gr-1Lo-negSSCLo or D) Ly6C+Ly6G-SSCLo or Gr-1+SSCLo. The majority of Ly6CLo-negLy6G-SSCLo or Gr-1Lo-negSSCLo cells were MHC Class II+CD115-, consistent with a macrophage phenotype ((8) and Figure 1). In contrast, Ly6C+Ly6G-SSCLo or Gr-1+SSCLo cells were predominantly MHC Class II-CD115+, suggestive of a classical monocyte phenotype ((9) and Figure 1). Neutrophils and eosinophils were negative for MHC Class II and CD115 staining (Supplemental Figure 5). Identification of the Ly6CIntLy6G-SSCHi eosinophil population was further confirmed by Siglec F staining (Figure 2).
Figure 1. Gr-1 and Ly6C/Ly6G markers identify similar populations of monocytes and macrophages in the mouse spleen.
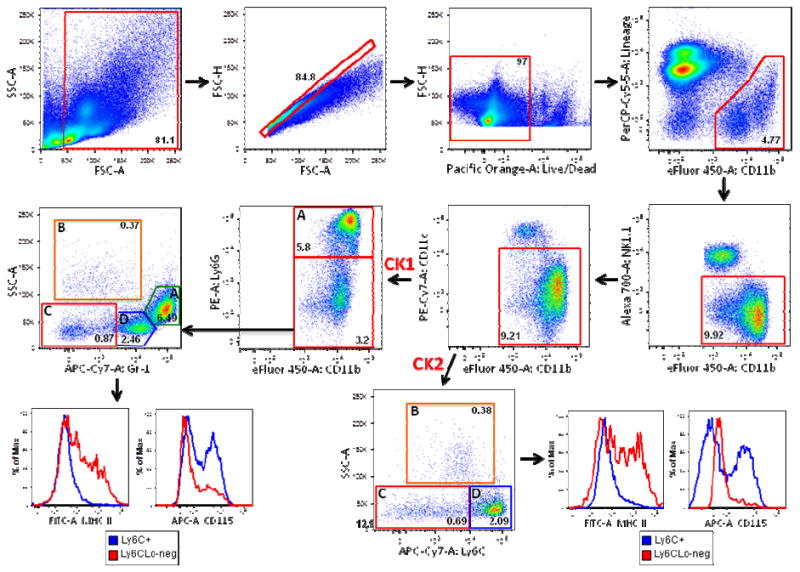
Total mouse splenocytes from C57BL/6 mice were prepared as described in Materials and Methods. Debris (SSC-A vs. FSC-A) and doublets (FSC-H vs. FSC-A) were excluded and live/dead discrimination was determined using the amine reactive dye Aqua (FSC-H vs. Pacific Orange-A Live/Dead). CD11b+Lineage- cells (gated out using a CD4, CD8, CD19 PerCP-Cy5.5 dump channel) were then sub-gated on CD11b+NK1.1- cells, followed by excluding CD11cHi cells. For the first cocktail (CK1), 4 subpopulations were identified: A) Gr-1HiSSCInt cells (neutrophils) and B) Gr-1Lo-negSSCHi cells (eosinophils) were negative for MHC Class II and CD115 staining (Supplemental Figure 5) C) Gr-1Lo-negSSCLo cells (monocytes/macrophages) were MHC Class II+/-CD115- (red histogram) D) Gr-1+SSCLo cells (monocytes/macrophages) were MHC Class II-CD115+/- (blue histogram). For the second cocktail (CK2), 4 subpopulations were identified: A) Ly6GHiCD11b+ cells (neutrophils) and B) Ly6CLo-negLy6G-SSCHi cells (eosinophils) were negative for MHC Class II and CD115 (Supplemental Figure 5) C) Ly6CLo-negLy6G-SSCLo cells (monocytes/macrophages) were MHCII+/-CD115- (red histogram) D) Ly6C+Ly6G-SSCLo cells (monocytes/macrophages) were MHC Class II-CD115+/- (blue histogram). Frequencies of cells in each sub-gate (after debris and doublet exclusion) are expressed as a percentage of live cells.
Figure 2. Gr-1Lo-negSSCHi mouse splenocytes are eosinophils.
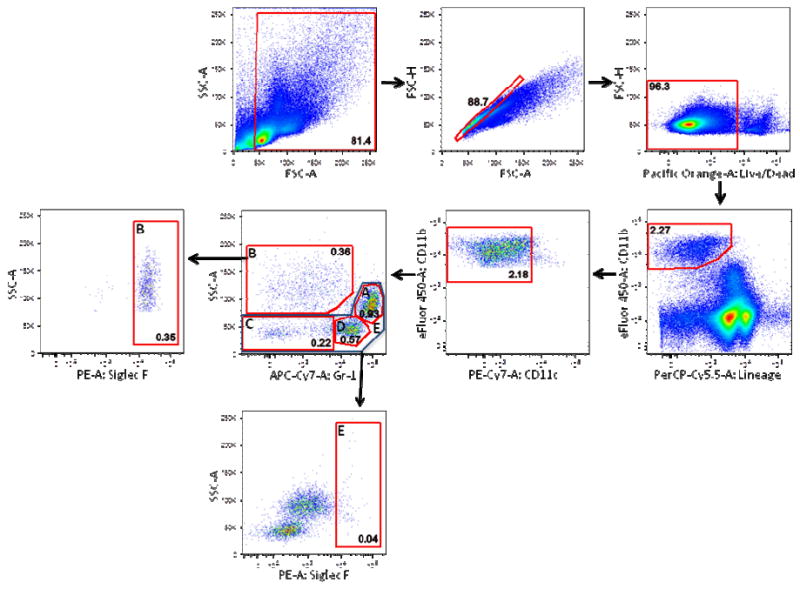
As in Figure 1, debris, doublets, and nonviable cells were excluded from total C57BL/6 mouse splenocytes. Live cells were sub-gated on CD11b+Lineage- cells (gated out using a CD4, CD8, CD19, NK1.1 PerCP-Cy5.5 dump channel). After gating out CD11cHi cells, 4 subpopulations of cells were identified: A) Gr-1HiSSCInt cells (neutrophils) B) Gr-1Lo-negSSCHi cells (eosinophils) were positive for Siglec F staining on sub-gate analysis C) Gr-1Lo-negSSCLo and D) Gr-1+SSCLo monocytes/macrophages were also identified. Gate E, which encompasses gates A, C, and D, was negative for Siglec F staining. Frequencies of cells in each sub-gate (after debris and doublet exclusion) are expressed as a percentage of live cells.
Previous studies have utilized anti-Gr-1 antibodies or a combination of anti-Ly6C/anti-Ly6G antibody staining to characterize myeloid cell populations in mouse spleen. Direct comparison of sorted populations using Gr-1 or Ly6C/Ly6G surface markers revealed that Gr-1HiSSCInt cells included neutrophils, monocytes, and macrophages (Supplemental Figure 6A) while Ly6GHiSSCInt cells were entirely neutrophils and Ly6CLo-negLy6G-SSCLo and Ly6C+Ly6G-SSCLo cells were monocytes and macrophages (Supplemental Figure 6B). Because anti-Gr-1 antibody recognizes both Ly6C and Ly6G antigens (10,11) and differences in intensity of the staining between neutrophils and classic monocytes/macrophages may not be sufficient for their accurate separation, our data suggest the use of anti-Ly6C/anti-Ly6G antibodies for delineating splenic myeloid cells populations.
Staining for the F4/80 antigen is not required to identify myeloid cell subsets in mouse spleen
The requirement of anti-F4/80 antibody for flow cytometric immunophenotyping of monocytes and macrophages in spleen is not known. To directly test this hypothesis, splenocytes were sorted followed by cytospin preparation and staining with Diff-quick and Giemsa (Figure 3). Samples were stained concurrently with two different cocktails. The first cocktail contained CD11b, Ly6C, Ly6G, CD11c, and F4/80 (Figure 3a) while the second cocktail contained CD11b, Ly6C, Ly6G, CD11c, and NK1.1 (Figure 3b). After gating out debris, doublets, and nonviable cells, 4 sub-populations of CD11b+ cells were sorted. Both sorts yielded identical results independent of F4/80 being included in the cocktail. Ly6GHiSSCInt cells were exclusively neutrophils (Figures 3a and 3b, Gate A). The Ly6CIntLy6G-SSCHi population consisted almost entirely of eosinophils with very rare monocytes/macrophages (Figures 3a and 3b, Gate B). Ly6CLo-negLy6G-SSCLo and Ly6C+Ly6G-SSCLo cells were all monocytes and macrophages. Taken together, these data indicate that F4/80 is not required for identifying mouse splenic myeloid cell sub-populations.
Figure 3. Monocytes/macrophages in the mouse spleen are readily identified without using the F4/80 antigen.
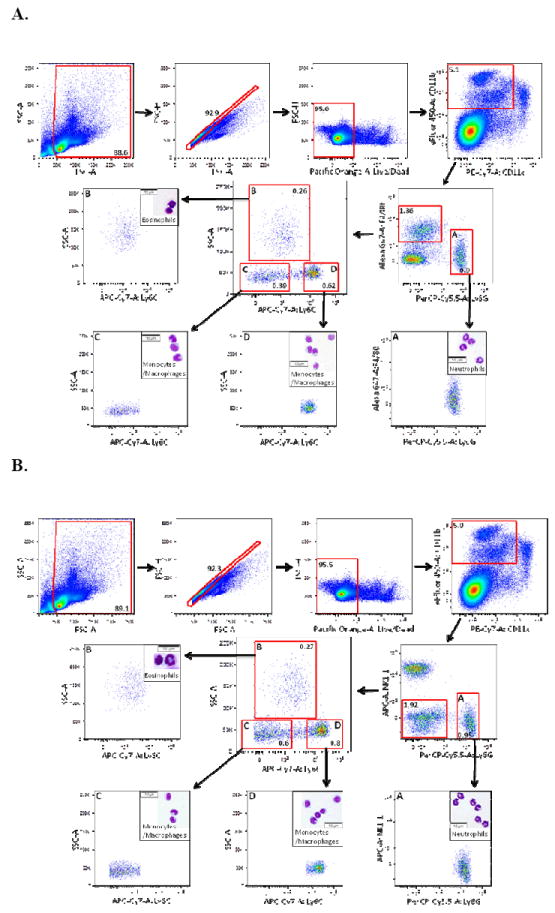
A) Debris, doublets, and dead cells were excluded from total C57BL/6 mouse splenocytes as in Figures 1-2. CD11cHi cells were excluded from the CD11b+ population, followed by gating into F4/80+Ly6G- and F4/80-Ly6GHi (A) subpopulations. F4/80+Ly6G- cells were divided further into subpopulations B, C, and D based on Ly6C versus SSC. Populations A-D were then isolated by FACS. Cytospin preparations from the sorted cells were stained with Diff-Quick and Giemsa. Subpopulation A (F4/80-Ly6GHi) contained solely neutrophils. Subpopulation B (Ly6CLo-negSSCHi) contained eosinophils while subpopulations C (Ly6CLo-negSSCLo) and D (Ly6C+SSCLo) contained monocytes/macrophages. B) Cells were prepared and gated as in Figure 3A except that NK1.1 was substituted for F4/80 staining to distinguish NK1.1-Ly6G- and NK1.1-Ly6GHi (A) subpopulations. NK1.1-Ly6G- cells were divided into subpopulations B-D based on Ly6C versus SSC. Subpopulations A-D were then sorted and cytospin preparations were prepared as above. Subpopulation A (Ly6GHiNK1.1-) contained exclusively neutrophils. Subpopulation B (Ly6CLo-negSSCHi) contained eosinophils while subpopulations C (Ly6CLo-negSSCLo) and D (Ly6C+SSCLo) contained monocytes/macrophages. For all analyses, frequencies of cells in each sub-gate (after debris and doublet exclusion) are expressed as a percentage of live cells.
Distinguishing monocytes from macrophages in mouse spleen
Swirski and colleagues (9) have previously shown that the mouse spleen serves as a reservoir for monocytes that can be mobilized in response to inflammatory signals. These investigators defined monocytes as Lin-CD11bHiCD11cLoMHC Class II-F4/80Lo cells, which were further split into Ly6CLo and Ly6CHi subpopulations. To address whether splenic monocytes and macrophages can reliably be distinguished based on expression of cell surface markers, we performed flow cytometric immunophenotyping on CX3CR1-GFP/+ mice (12). After exclusion of debris, doublets, dead cells, NK cells, dendritic cells, neutrophils, and eosinophils, CD11b+ cells were divided into CD11b+F4/80- or CD11b+F4/80+ populations. Each of these subsets was then further segregated into Ly6CLo-negSSCLo and Ly6CHiSSCLo populations (Supplemental Figure 7). In both the F4/80- and F4/80+ populations, the majority of CD11b+Ly6CLo-neg cells were MHC Class II+CD115- while CD11b+Ly6CHi cells were MHC Class II-CD115+. CX3CR1 expression, as measured by GFP fluorescence, was higher among CD11b+F4/80+Ly6CLo-neg cells compared to CD11b+F4/80-Ly6CLo-neg cells. In contrast, levels of CX3CR1 were similar in CD11b+F480+Ly6CHi and CD11b+F480-Ly6CHi cells.
Since the markers used thus far could not reliably identify splenic monocytes, we further phenotyped Ly6CLo-neg and Ly6CHi monocytes/macrophages for various cell surface antigens (Figure 4). The activation markers CD40, CD80, and CD86 were all more highly expressed on Ly6CLo-neg cells compared to Ly6CHi cells. Ly6CLo-neg cells also expressed greater levels of CX3CR1 and MHC Class II, heterogeneous levels of FcγR, and lower levels of CD115 relative to their Ly6CHi counterparts. Together, these data suggest that expression of F4/80 cannot be used to discriminate between mouse splenic monocytes and macrophages. In contrast, Ly6CLo-neg and Ly6CHi monocytes/macrophages differentially express various phenotypic markers, which could be indicative of distinct maturation stages between these cell populations.
Figure 4. Immunophenotyping monocyte/macrophage subsets in mouse spleen.
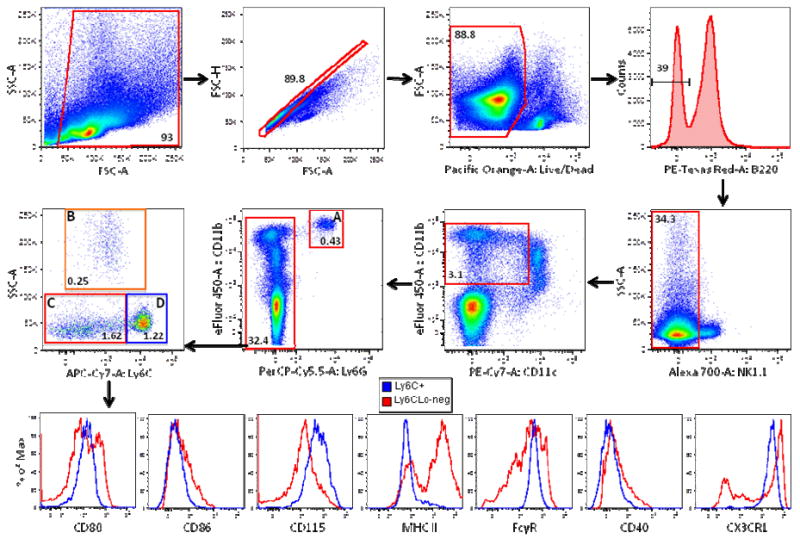
Debris, doublets, and dead cells were excluded from mouse splenocytes of CX3CR1-GFP/+ mice as described in Figure 1. B220+ cells were gated out followed by exclusion of NK1.1+ cells. CD11cHi cells were then gated out from the CD11b+ population and the remaining CD11b+ cells were separated into Ly6G- and (A) Ly6GHi (neutrophils) subpopulations. Ly6G- cells were divided further into subpopulations B (eosinophils), C, and D based on their Ly6C versus SSC properties. Monocyte/macrophage subpopulations C (red histograms) and D (blue histograms) were then further characterized for expression of various cell surface markers including CD80, CD86, CD115, MHC Class II (MHC II), FcγR, CD40, and CX3CR1 as indicated.
Immunophenotyping dendritic cell populations in mouse spleen
To expand the analysis of splenic myeloid cells, we incorporated the markers CD8α and mPDCA-1 into our panel to identify subsets of dendritic cells in CX3CR1-GFP/+ mice (Supplemental Figure 8). After exclusion of debris, singlets, and nonviable cells, splenocytes were gated into B220+ and B220- populations. B220+CD11cIntmPDCA-1+ plasmacytoid dendritic cells (pDC) and B220-CD11cHiCD8- and B220-CD11cHiCD8+ conventional dendritic cells (cDC) were easily identified. Further analysis revealed that both subsets of cDC expressed higher amounts of CD80 and MHC Class II and lower levels of CD69 compared to pDC. CX3CR1 expression was found to be greatest in CD8- cDC. High levels of CX3CR1 expression was also detected in a small subpopulation of CD8+ cDC, which are believed to be more closely related to pDC (13). Therefore, a minimal panel of core markers including B220, CD11c, mPDCA-1, and CD8α can be used to phenotype DC populations in mouse spleen.
Discussion
F4/80 has generally been considered to be a marker specific for both circulating monocytes and tissue-residing monocytes and macrophages, yet its expression is not limited to these cell populations (6,7). Here, we have demonstrated that F4/80 is not necessary to phenotype myeloid cells in mouse spleen. Moreover, we found that several commercially available fluorochrome-conjugated anti-F4/80 antibodies are not suitable for flow cytometry use in spleen due to potentially high levels of non-specific staining or increased background. This increased background fluorescence may be due to the high intrinsic autofluorescence of myeloid cell populations (14). The significance of these findings for other lymphatic and non-lymphatic tissues under various conditions should be the subject of future investigations.
Upon constructing an improved immunophenotyping cocktail, we uncovered further evidence that F4/80 should not be included in mouse spleen panels. An interaction between anti-F4/80 and anti-Gr-1 (or anti-Ly6G) was revealed when sequentially adding antibodies to the panel. Combinations of either anti-F4/80 clone (BM8 or CI:A3-1) with anti-Gr-1 antibody lead to poor discrimination of F4/80 positive and negative cells within the CD11bHi cell population. Co-incubation of anti-F4/80 antibody with other markers conjugated to the same fluorochrome did not reproduce this effect. Pre-incubation with anti-Gr-1, anti-Ly6C, or anti-Ly6G antibodies prior to addition of anti-F4/80 antibody corrected this problem. However, the converse experiment completely abrogated identification of the F4/80-CD11bHi subset when staining first with either anti-Ly6G or anti-Gr-1 antibodies, but not anti-Ly6C antibody. These results indicate that Ly6G and Gr-1 may share a common epitope with F4/80, albeit with dissimilar binding affinities. Another explanation for these findings is that there may be steric hindrance with concurrent use of anti-Gr-1 (or anti-Ly6G) and anti-F4/80 antibodies in the same staining panel.
Anti-Gr-1 antibody, or alternatively, the combination of anti-Ly6C and anti-Ly6G antibodies, has been used extensively to phenotype murine blood and tissue myeloid cells including the spleen. While either of these marker selections identify similar percentages of neutrophils, eosinophils, and monocytes/macrophages by flow cytometric analysis, we have demonstrated by FACS that Ly6C/Ly6G staining allows more accurate identification of splenic neutrophils than Gr-1. Eosinophils and monocyte/macrophage populations were equivalently discerned by flow cytometry or FACS analysis whether utilizing anti-Gr-1 antibody or anti-Ly6C and anti-Ly6G antibodies. Thus, Ly6C/Ly6G markers should be favored over Gr-1 whenever phenotyping mouse splenic neutrophils is desired.
Swirski and colleagues (9) recently identified a pool of splenic monocytes that are distinct from tissue-resident macrophages and can readily mobilize to participate in inflammatory responses. In agreement with their findings, we have shown that under steady state conditions the majority of CD11b+Ly6C+SSCLo or CD11b+Gr-1HiSSCLo splenocytes are CD115+MHC Class II- (Figures 1, 4), which is suggestive of a classical monocyte phenotype (1). Consistent with this notion, we have also demonstrated that these cells express lower levels of activation markers and CX3CR1 compared to CD11b+Ly6CLo-negSSCLo monocytes/macrophages (Figure 4). It is possible that Ly6C+ (or Gr-1Hi) and Ly6CLo-neg (or Gr-1Lo-neg) cells represent different stages of maturation, with Ly6C+ (or Gr-1Hi) cells being less differentiated. Alternatively, these two splenic monocyte/macrophage subsets may arise from distinct precursors (2). Despite extensive phenotypic analysis of splenocytes from CX3CR1-GFP/+ mice (Figure 4), we were unable to reliably distinguish between monocytes and macrophages based solely on the expression of surface antigens, including F4/80 (Supplemental Figure 8). Thus, the search for an elusive phenotypic marker to better distinguish between splenic monocytes and macrophages will be an important subject for future studies.
Based upon our data, we propose a simplified panel for identifying myeloid cell populations by multicolor flow cytometry in mouse spleen. Key markers to be included in this panel are B220, CD11b, CD11c, Ly6C, Ly6G, NK1.1, mPDCA-1 and a live/dead cell discriminating dye. Lineage markers, such as CD3, CD4, CD8, and CD19, may be incorporated individually or concurrently in a dump channel, but are nonessential. More restricted markers, such as CD115, Siglec F, and MHC Class II, can be built into the panel as required. We believe that this panel will be highly informative for delineating subpopulations of mouse splenic myeloid cells and provide more consistent results among investigators in the myeloid cell biology field.
Supplementary Material
Acknowledgments
Grant Support:
NIH Loan Repayment Grant (SR). NIH/NIAMS T32 training grant AR07611 (SR). NIH/NIAID R21 AI092490 (HP). NIH/NIAMS R01 AR050250 (HP). NIH/NIAMS R01 (HP). NIH/NIAMS R01 AR054796 (HP).
Literature Cited
- 1.Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, Leenen PJ, Liu YJ, MacPherson G, Randolph GJ, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116:e74–80. doi: 10.1182/blood-2010-02-258558. [DOI] [PubMed] [Google Scholar]
- 2.Yona S, Jung S. Monocytes: subsets, origins, fates and functions. Current Opinion in Hematology. 2010;17:53–9. doi: 10.1097/MOH.0b013e3283324f80. [DOI] [PubMed] [Google Scholar]
- 3.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–69. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Austyn JM, Gordon S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. European Journal of Immunology. 1981;11:805–15. doi: 10.1002/eji.1830111013. [DOI] [PubMed] [Google Scholar]
- 5.Schaller E, Macfarlane AJ, Rupec RA, Gordon S, McKnight AJ, Pfeffer K. Inactivation of the F4/80 glycoprotein in the mouse germ line. Molecular and Cellular Biology. 2002;22:8035–43. doi: 10.1128/MCB.22.22.8035-8043.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hume DA, Robinson AP, MacPherson GG, Gordon S. The mononuclear phagocyte system of the mouse defined by immunohistochemical localization of antigen F4/80. Relationship between macrophages, Langerhans cells, reticular cells, and dendritic cells in lymphoid and hematopoietic organs. J Exp Med. 1983;158:1522–36. doi: 10.1084/jem.158.5.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGarry MP, Stewart CC. Murine eosinophil granulocytes bind the murine macrophage-monocyte specific monoclonal antibody F4/80. J Leukoc Biol. 1991;50:471–8. doi: 10.1002/jlb.50.5.471. [DOI] [PubMed] [Google Scholar]
- 8.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–64. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 9.Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, Figueiredo JL, Kohler RH, Chudnovskiy A, Waterman P, et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. 2009;325:612–6. doi: 10.1126/science.1175202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleming TJ, Fleming ML, Malek TR. Selective expression of Ly-6G on myeloid lineage cells in mouse bone marrow. RB6-8C5 mAb to granulocyte-differentiation antigen (Gr-1) detects members of the Ly-6 family. Journal of Immunology. 1993;151:2399–408. [PubMed] [Google Scholar]
- 11.Hestdal K, Ruscetti FW, Ihle JN, Jacobsen SE, Dubois CM, Kopp WC, Longo DL, Keller JR. Characterization and regulation of RB6-8C5 antigen expression on murine bone marrow cells. Journal of Immunology. 1991;147:22–8. [PubMed] [Google Scholar]
- 12.Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Molecular and Cellular Biology. 2000;20:4106–14. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bar-On L, Birnberg T, Lewis KL, Edelson BT, Bruder D, Hildner K, Buer J, Murphy KM, Reizis B, Jung S. CX3CR1+ CD8alpha+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107:14745–50. doi: 10.1073/pnas.1001562107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell AJ, Pradel LC, Chasson L, Van Rooijen N, Grau GE, Hunt NH, Chimini G. Technical advance: autofluorescence as a tool for myeloid cell analysis. J Leukoc Biol. 2010;88:597–603. doi: 10.1189/jlb.0310184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


