Abstract
Primary cilia are non-motile organelles implicated in signaling and sensory functions. Understanding how primary cilia assemble could shed light on the many human diseases caused by mutations in ciliary proteins. The centrosomal protein CP110 is known to suppress ciliogenesis through an unknown mechanism. Here, we report that CP110 interacts with CEP290-a protein whose deficiency is implicated in human ciliary disease-in a discrete complex separable from other CP110 complexes involved in regulating the centrosome cycle. Ablation of CEP290 prevents ciliogenesis without affecting centrosome function or cell cycle progression. Interaction with CEP290 is absolutely required for the ability of CP110 to suppress primary cilia formation. Furthermore, CEP290 and CP110 interact with Rab8a, a small GTPase required for cilia assembly. Depletion of CEP290 interferes with localization of Rab8a to centrosomes and cilia. Our results suggest that CEP290 cooperates with Rab8a to promote ciliogenesis and this function is antagonized by CP110.
Keywords: CP110, CEP290, Rab8a, ciliogenesis, primary cilia
Introduction
The centrosome is the major microtubule-organizing center in animal cells (Bettencourt-Dias and Glover, 2007). It consists of two centrioles surrounded by proteinaceous pericentriolar material from which microtubules emanate and elongate. Centrosome duplication takes place during S phase, a period during which two new centrioles (procentrioles) are formed adjacent to the pre-existing mother and the daughter centriole. Procentriole maturation takes place thereafter, such that by the end of G2, a cell possesses two centrosomes containing a total of four centrioles. In early mitosis, the two centrosomes migrate to opposite poles to set up the spindle poles, and each of the two incipient daughter cells receives a single centrosome upon completion of cytokinesis. Defects in any aspect of centrosome function can be deleterious and lead to cell cycle arrest, genome instability, aneuploidy, cancer, and tumor formation (Badano et al., 2005; Nigg, 2002; Sluder and Nordberg, 2004).
In addition to anchoring and organizing microtubules, recent advances have revealed a critical connection between centrosome function and primary cilia formation. Primary cilia are non-motile projections found at the surface of most quiescent vertebrate cells and are involved in differentiation, sensory functions, and signal transduction (Satir and Christensen, 2007; Singla and Reiter, 2006). Upon cell cycle exit, the mother centriole/basal body migrates toward the cell cortex and nucleates the formation of a finger-like structure consisting of a nine-fold array of doublet microtubules, termed the axoneme, enveloped by the plasma membrane. Receptors and signaling molecules are delivered to the primary cilium by means of polarized trafficking via intraflagellar transport proteins (IFT) and motor proteins such as kinesin-2 and dynein (Rosenbaum, 2002; Scholey, 2003). Deficiencies in genes implicated in centrosome and ciliary function give rise to a spectrum of human phenotypes, including cystic kidney disease, diabetes, neurological disorders, retinal degeneration, and polydactyly. Mechanistic details concerning the role of these proteins are not fully understood (Scholey and Anderson, 2006; Singla and Reiter, 2006). Furthermore, our understanding of the regulatory controls governing the conversion of centrosomes to basal bodies, or vice versa, is rudimentary at best.
We have recently identified a centrosomal protein, CP110, required for centrosome and primary cilia function (Chen et al., 2002; Spektor et al., 2007; Tsang et al., 2006). Depletion of CP110 by RNA interference (RNAi) results in premature centrosome separation, abrogates centrosome reproduction in S phase arrested cells, and inhibits Polo-like kinase 4-induced centrosome amplification (Chen et al., 2002; Kleylein-Sohn et al., 2007). In addition, CP110 functionally interacts with two calcium-binding proteins, centrin and calmodulin (CaM), to regulate cytokinesis (Tsang et al., 2006). Loss of CP110 or expression of mutants that are either unable to bind CaM or that are refractory to CDK phosphorylation results in cytokinesis failure (Chen et al., 2002; Tsang et al., 2006). Moreover, loss of CP110 or CEP97, another CP110-interacting protein, leads to aberrant formation of primary cilia in growing cells, and conversely, enforced expression of CP110 suppresses cilium assembly in quiescent cells (Spektor et al., 2007). Together, these data suggest that CEP97 and CP110 collaborate to inhibit a ciliogenesis program and indicate that cilia formation may be a default pathway engaged in the absence of CP110 function. However, it is currently unknown how CP110 mediates suppression of primary cilia assembly in growing cells.
In an effort to understand how CP110 regulates cilia formation, we initiated studies to identify additional CP110-interacting proteins. We identified CEP290 as a CP110-interacting partner in vivo. CEP290 is a centrosomal protein (Chang et al., 2006; Sayer et al., 2006; Valente et al., 2006), and mutations in CEP290 have been implicated in autosomal recessive disorders, including nephronophthisis (Sayer et al., 2006), Senior-Loken syndrome (Helou et al., 2007), Joubert syndrome (Sayer et al., 2006; Valente et al., 2006), Leber Congenital Amaurosis (den Hollander et al., 2006), Meckel-Gruber syndrome (Baala et al., 2007), and Bardet-Biedl syndrome (BBS) (Leitch et al., 2008). In a mouse model, a CEP290 mutation gives rise to early-onset retinal degeneration and olfactory dysfunction (Chang et al., 2006; McEwen et al., 2007). We have examined the function of CEP290 and the biological significance of the CEP290-CP110 interaction. We show that CEP290 participates strictly in primary cilia formation, and the ability of CP110 to interact with CEP290 is absolutely essential for suppressing cilia formation. Furthermore, CEP290 and CP110 interact with the small GTPase Rab8a, a protein required for ciliary biogenesis. Depletion of CEP290 diminishes the localization of Rab8a to centrosomes and prevents its entry into the cilium. Taken together, our data demonstrate that CP110 suppresses the activity of CEP290, which in turn cooperates with Rab8a to mediate a cilia assembly program.
Materials and Methods
Cell culture and Plasmids
Human 293T, T98G, U2OS, RPE-1 hTERT, and mouse 3T3 cells were grown in DMEM supplemented with 10% FBS at 37°C in a humidified 5% CO2 atmosphere. Flag-tagged CP110 fusion proteins were described previously (Spektor et al., 2007). To generate Flag-tagged CEP290 fusion proteins in vivo, CEP290 fragments encoding residues 1-2479, 1–366, 221-366, 362-822, 816-1207, 580-1695, 1689-2050, and 2037-2479 were amplified by PCR using Pfu Turbo polymerase (Stratagene) and subcloned into the EcoRV and HindIII sites of the mammalian vector pCBF vector (generous gift from M. Cole). All constructs were verified by DNA sequencing.
Antibodies
Antibodies used in this study included polyclonal rabbit anti-CEP290 (Chang et al., 2006), anti-CP110 (Chen et al., 2002), anti-CEP97 (Spektor et al., 2007), anti-centrin mouse monoclonal 20H5 (generous gift from J. Salisbury), anti-polycystin-2 (generous gift from Y. Cai and S. Somlo), anti-CaM (Upstate), anti-GFP (Roche), anti-Ki-67 (Zymed), anti-pericentin (Covance), anti-α-tubulin, anti-acetylated tubulin, anti-Flag and anti-γ-tubulin (all from Sigma-Aldrich), anti-glutamylated tubulin GT335 (generous gift from C. Janke), anti-polaris (generous gift from B. Yoder), and anti-Rab8a (BD Biosciences and Peranen et al., 1996).
Yeast Two-Hybrid Screen
The Matchmaker Two-Hybrid System 3 (BD Biosciences Clontech) was used to identify clones that interact with CP110. The full-length cDNA of CP110 was cloned into the NcoI and SalI of pGBKT7 bait vector. The resultant plasmid was transformed into the yeast reporter strain AH109. A BD Matchmaker™ pre-transformed human brain cDNA library (BD Biosciences Clontech) provided in yeast strain Y187 was screened by yeast mating following the manufacturer's recommendations. ß-galactosidase activity of colonies growing on stringent selection medium (SD/-Ade/-His/-Leu/-Trp) was assessed by blue/white coloring (X-gal assay) as described by the manufacturer. A total of 480 positive colonies were picked and tested. Eighty-two clones were then selected based on the X-gal assay. Plasmids from these positive clones were extracted and sequenced. The corresponding genes were identified by BLASTn alignment to the NCBI database.
Cell Cycle Synchronization and FACS Analysis
T98G cells were synchronized by serum deprivation and re-stimulation as described (Tsang et al., 2007). FACS analysis was performed as reported previously (Tsang et al., 2007).
Immunoprecipitation, Immunoblotting, and Immunofluorescence Microscopy
Cells were lysed with buffer containing 50 mM Hepes pH 7, 250 mM NaCl, 5 mM EDTA/pH 8, 0.1% NP-40, 1 mM DTT, 0.5 mM PMSF, 2 μg/ml leupeptin, 2 μg aprotinin, 10 mM NaF, 50 mM ß-glycerophosphate, and 10% glycerol at 4°C for 30 minutes. After centrifugation, 2 mg of the resulting supernatant was incubated with an appropriate antibody at 4°C for 1 hour and collected using protein A or G-Sepharose. The resin was washed with lysis buffer, and the bound polypeptides were analyzed by SDS-PAGE and immunoblotting. Typically, 50 μg-100μg of lysate were loaded into the input (IN) lane. For mapping studies, Flag-tagged constructs were transfected into 293T cells. Cells were harvested 48-72 hours after transfection. Flag-beads were incubated with cell extract at 4°C for 2 hours. After washing with lysis buffer, bound proteins were analyzed by SDS-PAGE and immunoblotting. Indirect immunofluorescence was performed as described (Chen et al., 2002). Briefly, cells were grown on glass coverslips, fixed with cold methanol for 2 minutes. The cells were permeabilized with 1% Triton X-100/PBS for 5 minutes. Slides were blocked with 3% BSA in 0.1% Triton X-100/PBS prior to incubation with primary antibodies. Secondary antibodies used were Cy3- or FITC-conjugated donkey anti-mouse or anti-rabbit IgG (Jackson Immunolabs). Cells were then stained with DAPI, and slides were mounted, observed and photographed using a Nikon Eclipse E800 microscope (Nikon; 63X or 100X, NA 1.4) equipped with a Photometrics Coolsnap HQ CCD camera. Images were acquired with MetaMorph7 (Molecular Devices). For analysis of the distance between centrosomes and nuclei, z-sections spaced by 0.2-0.5 μm were taken.
RNAi
Synthetic siRNA oligonucleotides were obtained from Dharmacon. Transfection of siRNAs using Oligofectamine or Lipofectamine 2000 (Invitrogen) was performed according to the manufacturer's instructions. The 21-nucleotide siRNA sequence for the non-specific control was 5’-AATTCTCCGAACGTGTCACGT-3’. The 21-nucleotide siRNA sequences for CEP290 used were 5’-AAATTAAGATGCTCACCGATT-3’ and 5’-GATGAAAGCTCAAGAAGTGTT-3’. The siRNAs for CP110 silencing were described previously (Spektor et al., 2007).
Induction of Primary Cilia
RPE-1 or 3T3 cells transfected with siRNA or plasmid DNA were brought to quiescence by serum starvation for 48-72 hours. Cells were examined for well-established primary cilium markers such as acetylated tubulin or glutamylated tubulin.
Superose 6 Gel Filtration Analysis
2 mg of extract were chromatographed over a Superose 6 gel filtration column (Pharmacia) in lysis buffer (50 mM Hepes pH 7, 250 mM NaCl, 5 mM EDTA/pH 8, 0.1% NP-40, 0.5 mM PMSF, and 10% glycerol). Equal volumes of each fraction were precipitated with TCA and analyzed by SDS-PAGE and immunoblotting.
Statistical Analysis
The statistical significance of the difference between two means was determined using a two-tailed Student's t-test. Differences were considered significant when p<0.01.
Results
Biochemical interaction between CP110 and CEP290
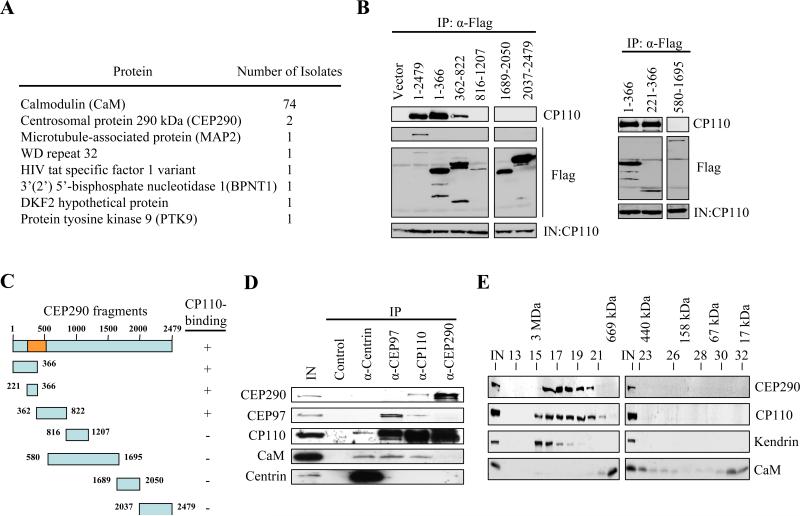
In an effort to understand how CP110 functions during the centrosome cycle and cilium formation, we performed a yeast two-hybrid screen using full-length CP110 as bait. 82 interactors were isolated, and DNA sequencing revealed that 74 clones encoded CaM, a calcium-binding, CP110-interacting protein we had identified previously (Tsang et al., 2006), confirming the success of the screen (Figure 1A). In addition, two clones encoded CEP290 (Figure 1A), spanning regions corresponding to amino acid residues 237-526. To confirm that CEP290 interacts with CP110, we constructed a series of epitope-tagged CEP290 truncation mutants, transfected human cells, and performed immunoprecipitations with anti-epitope antibodies. CP110 was readily detected on western blots after immunoprecipitation of full-length CEP290 (1-2479; Figures 1B and 1C). We found that amino-terminal portions of CEP290 (containing residues 1-366, 221-366, 362-822) were necessary and sufficient to bind CP110, while other fragments did not interact (Figures 1B and 1C). Thus, mapping and yeast two-hybrid data point to CP110 binding near the amino-terminus of CEP290.
Figure 1.
CP110 interacts with CEP290 in vivo. (A) The number of times that each annotated protein was identified in the yeast two-hybrid screen is shown. (B) The indicated fragments of Flag-tagged CEP290 were expressed in 293T cells and immunoprecipitated from lysates. Flag-CEP290 fusion proteins and CP110 were detected after western blotting the resulting immunoprecipitates. Input CP110 was detected in lysates from each transfection (IN). (C) Summary of CEP290 truncation mutants and the results of in vivo binding experiments. The orange box denotes the CP110-binding domain based on the yeast two-hybrid screen. (D) Western blotting of endogenous CEP290, CEP97, CP110, CaM, and centrin after immunoprecipitation with anti-Flag (control), anti-centrin, anti-CEP97, anti-CP110, or anti-CEP290 antibodies from 293T cell extracts. IN represents input. (E) Cell extract was chromatographed on a Superose 6 gel filtration column, and the resulting fractions were blotted with antibodies against CP110, CEP290, kendrin, or CaM. Estimated molecular weights are indicated at the top of the panel.
To determine whether CEP290 and CP110 interact in vivo, we performed immunoprecipitations with affinity-purified CEP290 and CP110 antibodies. CEP290 and CP110 were co-immunoprecipitated by CP110 antibodies, and reciprocal immunoprecipitations with CEP290 antibodies confirmed a robust physiological interaction (Figure 1D). CP110 has been shown to exist in two mutually exclusive complexes. One complex, comprised of centrin and CaM, has been implicated in cytokinesis (Tsang et al., 2006). In addition, CP110 interacts with CEP97, and this complex is required to suppress primary cilia formation (Spektor et al., 2007). We therefore tested the ability of CEP290 to interact with a similar cohort of proteins. Although we detected CP110 in both centrin and CEP97 immunoprecipitates, we failed to observe CEP290 (Figure 1D). Moreover, antibodies recognizing CEP290 did not co-precipitate centrin, CEP97, or CaM, although CP110 was detected (Figure 1D), suggesting that CP110 associates with discrete centrin, CEP97, and CEP290 complexes. In addition, we demonstrated by size exclusion chromatography that native CEP290 reproducibly migrated as a high molecular weight complex (with a mass as large as 2-3 MDa) that co-eluted with CP110 (Figure 1E). In contrast, the pericentriolar matrix protein, kendrin, peaks at a somewhat higher molecular weight (Figure 1E and (Tsang et al., 2006)). These observations support a physical association of CEP290 with CP110.
Localization of CEP290
Next, we showed that CEP290 protein levels remain relatively constant throughout the cell cycle (Figure S1A). Although CEP290 is known to localize to centrosomes (Andersen et al., 2003; Chang et al., 2006; Sayer et al., 2006; Valente et al., 2006), its localization during interphase has not been fully investigated. Antibodies against CEP290 stained two or four prominent spots in G1 and G2 phase (Figures S1B, S1C and S1D). CEP290 staining overlapped substantially with that of centrin and CP110 (Figures S1C and S1D), which are located at the distal ends of centrioles (Kleylein-Sohn et al., 2007; Paoletti et al., 1996), and to a lesser extent with γ-tubulin, another centrosomal marker (Figure S1B). In G0, CP110 is present at the daughter centriole but not at the mother centriole/basal body (Figures S1D and S3), consistent with our prior observation that it is extinguished at the basal body in ciliated cells (Spektor et al., 2007). CEP290, on the other hand, is found on both the mother centriole/basal body and the daughter centriole (Figures S1D and 2F). The mother centriole/basal body nucleates the formation of a primary cilium at its distal end and is attached to the daughter centriole at the other, proximal end. Interestingly, CEP290 staining at the mother centriole/basal body is localized to the distal end near the base of the primary cilium (Figure 2F), suggesting that it may be present at the transitional zone and that it could function in cilium assembly. In addition, our data suggests that a portion of CEP290 protein at the mother centriole/basal body remains unassociated with CP110.
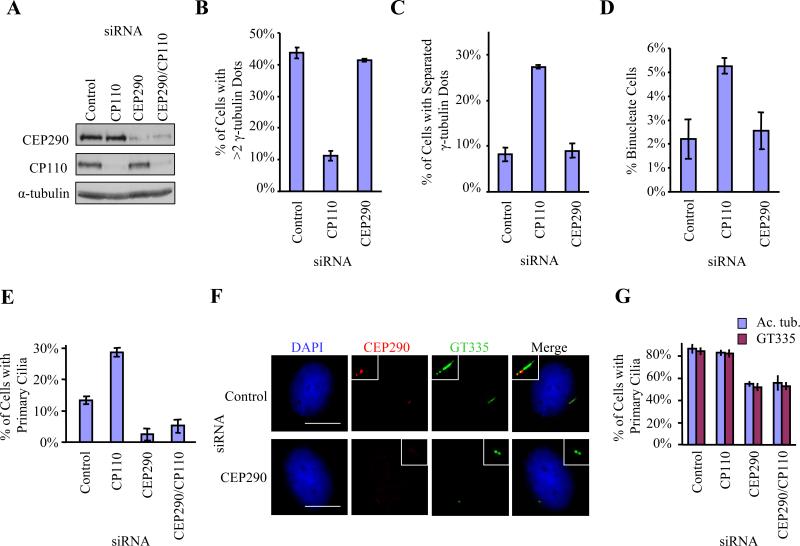
Figure 2.
RNAi-mediated suppression of CEP290 inhibits primary cilia formation. (A) Western blotting of CP110 and CEP290 in RPE-1 cells treated with control, CP110, CEP290, or both siRNAs. α-tubulin was used as a loading control. (B) U2OS cells were arrested at S phase with hydroxyurea for 48 hr after siRNA transfection. The percentages of cells with more than two γ-tubulin dots, indicative of centrosome over-duplication, were determined. (C) The percentages of U2OS cells with well-separated γ-tubulin dots, indicative of premature centrosome separation, were determined. (D) The percentages of binucleate WI38 cells, indicative of cytokinesis failure, were determined. (E) The percentages of growing RPE-1 cells with primary cilia were determined using acetylated tubulin as a marker. (F) RPE-1 cells transiently transfected with control or CEP290 siRNAs, induced to quiescence, stained with antibodies to glutamylated tubulin (GT335, green), CEP290 (red), and with DAPI (blue). Bar: 10 μM; insets: 2 μM. (G) The percentages of quiescent RPE-1 cells with primary cilia were determined using either acetylated tubulin (Ac. tub.) or gltuamylated tubulin (GT335) as a marker. In (B), (C), (D), (E) or (G), average data obtained from three independent experiments were shown. Error bars represent +/- S.D. About 200 cells for each siRNA condition were scored each time.
CEP290 depletion prevents primary cilia formation
We examined the consequences of knocking down CEP290, CP110, or both by transfecting cells with small interfering RNAs (siRNAs) targeting the corresponding transcripts. We transfected two distinct CEP290 siRNAs and showed that the levels of CEP290 protein were reduced by ~75% (Figure 2A). Depletion of either CP110 or CEP290 does not affect the levels or localization of the other protein (Figures 2A, S3 and data not shown). Since it is known that CP110 is important for centrosome duplication (Figure 2B and (Chen et al., 2002; Kleylein-Sohn et al., 2007)), for preventing premature centrosome separation (Figure 2C and (Chen et al., 2002)), and for cytokinesis (Figure 2D and (Tsang et al., 2006)), we tested whether CEP290 depletion had any impact on these processes using appropriate assays. Interestingly, although it interacts with CP110 in vivo, CEP290 does not appear to be critical for any of these processes (Figures 2B, 2C and 2D). Furthermore, flow cytometric analysis indicated that suppression of CEP290 did not alter cell cycle progression (Figure S2A).
Given the association with several ciliary diseases, we tested a role for CEP290 in assembly of primary cilia by staining for ciliary markers (glutamylated tubulin or acetylated tubulin). Interestingly, when we ablated CEP290, we noticed a dramatic alteration in the ability of cells to assemble primary cilia in cycling human retinal pigment epithelial cells (RPE-1) (Figure 2E). In contrast, loss of CP110 results in the opposite effect, namely, aberrant formation of primary cilia in growing cells (Figure 2E), consistent with our previous observations (Spektor et al., 2007). Ablation of both CEP290 and CP110 suppressed the phenotype associated with loss of CP110 and mimicked CEP290 ablation (Figure 2E), suggesting that CEP290 and CP110 could functionally interact in antagonistic ways. We also determined the impact of CEP290 ablation in quiescent cells, a period during which the majority of cells (~85%) form primary cilia (Figures 2F and 2G). When we depleted CEP290 protein, we observed a consistent and striking loss of primary cilia (Figures 2F and 2G). These results are consistent with a high throughput screen designed to uncover ciliary proteins (Graser et al., 2007). Ablation of CEP290 abolished glutamylated tubulin staining of the axoneme, although staining at the ciliary base was unaffected (Figure 2F). Combined suppression of CEP290 and CP110 again resulted in a phenotype similar to loss of CEP290 alone (Figure 2G). We could show that the failure to assemble cilia in CEP290-depleted cells was not due to an inability to enter a quiescent state (Figures S2B and S2C). These results provide conclusive evidence that CEP290 is required to promote ciliogenesis in human cells, counteracting the activity of CP110. Furthermore, when the distance between centrosomes and nuclei was examined by immunofluorescence microscopy, we found that 45%±6% and 21%±7% of the centrosomes in control and CEP290-depleted cells, respectively, were well-separated (i.e. located at least 5 μm away) from the nucleus and were localized close to the cell surface (Figure S2D and data not shown). Thus, depletion of CEP290 could disrupt the migration of centrioles to the cell cortex.
Ectopic expression of the CEP290-binding mutant of CP110 does not suppress primary cilia formation
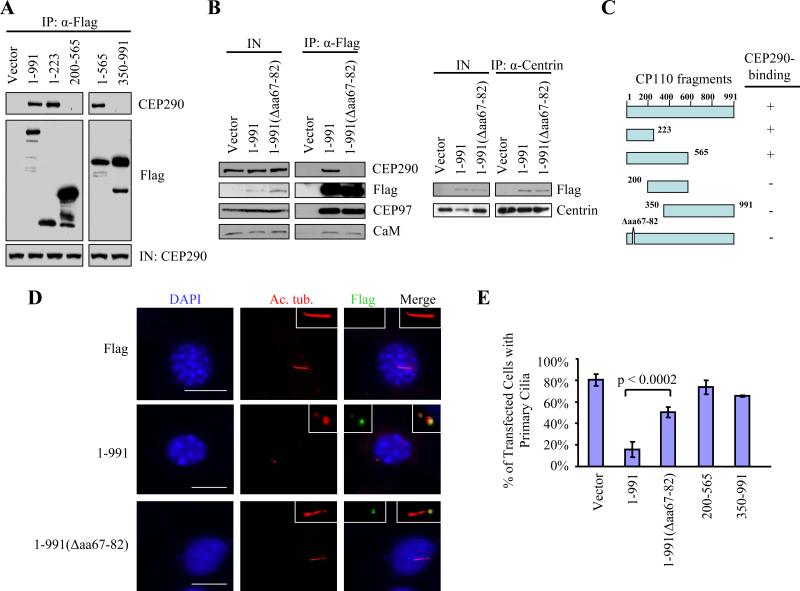
To gain insight into the biological significance of the CP110-CEP290 interaction, we first performed experiments to determine the regions of CP110 involved in binding CEP290. We transfected a set of Flag-tagged CP110 mutants and performed anti-Flag immunoprecipitation and western blotting to detect endogenous CEP290. We found that a region of CP110 encompassing residues 1-223 was required for stable binding to CEP290 (Figures 3A and 3C). We defined the binding domain further by testing a full-length CP110 mutant lacking only residues 67-82 (1-991(Δaa67-82)). Excision of these residues severely compromised its ability to interact with CEP290 (Figures 3B and 3C). Interestingly, 1-991(Δaa67-82) can nevertheless bind to all other known CP110-interacting proteins, including CEP97, centrin, and CaM in vivo (Figure 3B) and in vitro (Tsang et al., 2006).
Figure 3.
A CP110 mutant unable to bind CEP290 fails to inhibit primary cilia formation. (A) The indicated fragments of Flag-tagged CP110 were expressed in 293T cells and immunoprecipitated from lysates. Flag-CP110 fusion proteins and CEP290 were detected after western blotting the resulting immunoprecipitates. Input CEP290 was detected in lysates from each transfection (IN). (B) Left panel: Flag (vector), Flag-tagged full-length CP110 (1-991), and Flag-tagged CEP290-binding mutant of CP110 (1-991(Δaa67-82)) were expressed in 293T cells and immunoprecipitated from lysates. Flag-CP110 fusion proteins, CEP290, CEP97, and CaM were detected after blotting the resulting immunoprecipitates. IN represents input. Right panel: Western blot of Flag-CEP290 and centrin after immunoprecipitation with anti-centrin antibodies using extracts from 293T cells transfected with the indicated constructs. IN represents input. (C) Summary of CP110 truncation mutants and the results of in vivo binding experiments. (D) 3T3 cells transiently transfected with plasmids expressing the indicated constructs, induced to quiescence, and stained with antibodies to Flag (green), Ac. tub. (red), and with DAPI (blue). Bar: 10 μM; insets: 2 μM. (E) The percentages of transfected, quiescent 3T3 cells expressing primary cilia were determined using Ac. tub. as a marker. About 100 transfected cells were scored for each construct, and average data obtained from three independent experiments was shown. Error bars represent +/- S.D.
These data argue that CP110 binds to CEP290 independently of other known CP110 interacting proteins, consistent with our biochemical observations (Figure 1D), thus allowing us to functionally separate and dissect these independent activities. Next, we tested the ability of these CP110 mutants to inhibit cilia assembly by expressing this short internal deletion mutant and other CP110 fragments in quiescent 3T3 cells (Figures 3D and 3E). Like RPE-1, 3T3 fibroblasts are a well-established model for cilia formation, as they readily develop primary cilia upon entry into quiescence. We confirmed expression and proper localization of each of these proteins by immunofluorescence and found that only the full-length protein and the internal deletion mutant localize to the centrosome (Figure 3D and data not shown). Cells transfected with control plasmids developed primary cilia upon induction of quiescence, as expected (Figures 3D and 3E). On the other hand, enforced expression of full-length CP110 suppressed primary cilia formation (Figures 3D and 3E), in line with previous observations (Spektor et al., 2007). In striking contrast, loss of CEP290 binding led to a 3-fold reduction in inhibitory activity (Figures 3D and 3E), strongly supporting the notion that CP110-binding to CEP290 is necessary to prevent a CEP290-mediated cilia assembly program. Indeed, expression of any CP110 truncation mutant unable to bind CEP290 (200-565 and 350-991) fails to inhibit cilia assembly (Figures 3C and 3E). Interestingly, CP110 fragments (1-223 and 1-565) that are nevertheless able to interact with CEP290 also fail to suppress ciliogenesis, most likely because they do not localize to centrosomes (Figure 3C and data not shown). Similar findings were also observed in RPE-1 cells (data not shown). These results establish a clear and important mechanistic link between CEP290-binding to CP110 and primary cilia formation.
CEP290 depletion affects localization of Rab8a to centrosomes and to primary cilia
These findings prompted us to obtain further insights into the molecular mechanisms underlying the role of CEP290 in ciliogenesis, and we examined several different well-established centrosome/ciliary markers in CEP290-depleted quiescent cells by immunofluorescence. CP110 weakly but specifically localizes to the daughter centriole in serum-deprived cells (Figure S3). Centrin and pericentrin are essential for centrosome and cilia function and exhibit differential localizations at the primary cilium (Jurczyk et al., 2004; Laoukili et al., 2000; Taulman et al., 2001). Centrin is detected at the daughter centriole and at the mother centriole/basal body (Figure S3), while pericentrin, a pericentriolar matrix protein, is found at the periphery of the two centrioles and exists as one dot (Figure S3). We observed no difference in the staining patterns of CP110, centrin, or pericentrin in control and CEP290-siRNA treated cells (Figure S3), suggesting that these proteins are targeted to centrosomes in the absence of CEP290. This is consistent with our results indicating that centrosomal function is not compromised in growing cells depleted of CEP290 (Figures 2B, 2C and 2D). Next, we examined the localization of an IFT protein, Polaris/IFT88, found at the distal end of the mother centriole/basal body and along the axoneme (Figure S3 and (Taulman et al., 2001)). While we did not observe axonemal staining of polaris owing to an inhibition of cilia formation upon suppression of CEP290, polaris was nevertheless targeted to one of the two centrioles (Figure S3). Similarly, the localization of another IFT protein, IFT20 (Jurczyk et al., 2004; Yoshimura et al., 2007), and a calcium cation channel, polycystin-2 (Jurczyk et al., 2004) were not affected upon CEP290 depletion (Figure S3 and data not shown). Since IFT20 and polycystin-2 localize primarily to the basal body, these data suggest that localization of at least a subset of proteins to the basal body is not perturbed in the absence of CEP290.
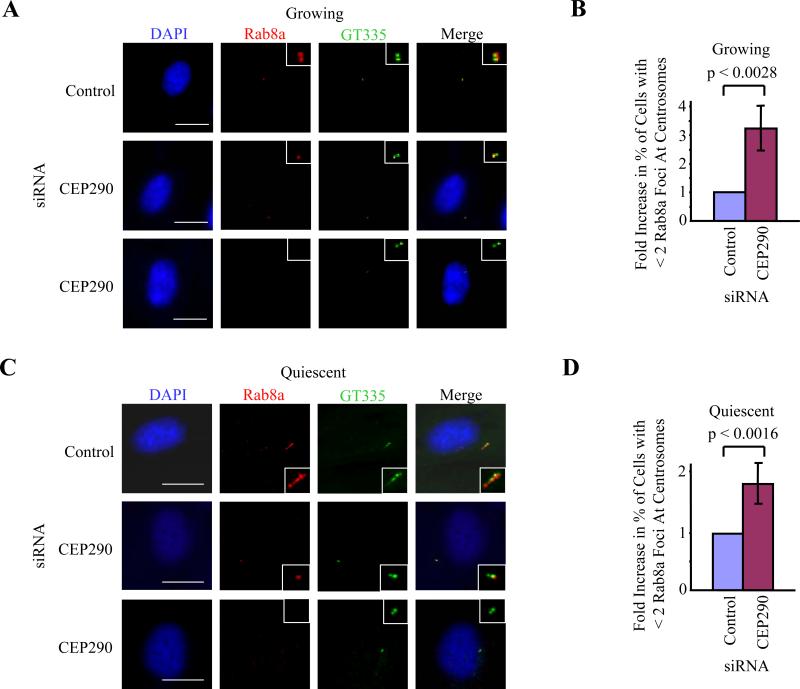
We next asked whether other components critical for cilia assembly might be affected by loss of CEP290. The Rab family of proteins, comprising at least thirty different members, plays a critical role in vesicular trafficking (Zerial and McBride, 2001). Rab8a is the sole Rab family member enriched at primary cilia and is absolutely essential for their formation (Yoshimura et al., 2007). It is believed that Rab8a regulates cilia assembly by targeting and promoting fusion of vesicles near the ciliary membrane, which envelops the cilium and is continuous with the plasma membrane. A molecular function for Rab8a in ciliogenesis has recently been demonstrated, wherein it cooperates with the GTP exchange factor, Rabin8, and a core complex of BBS proteins to promote ciliary membrane biogenesis (Nachury et al., 2007). Rab8a localizes to the centrosome in growing cells and to the ciliary membrane in quiescent cells (Figures 4A and 4C). Remarkably, depletion of CEP290 led to a significant reduction of Rab8a at centrosomes in growing cells (Figure 4B). These CEP290-depleted cells often exhibit one or no detectable Rab8a dots (Figure 4A, second and third rows), in contrast with two Rab8a foci that co-localize with polyglutamylated tubulin in control cells (Figure 4A, first row). Similar reductions in the number of Rab8a foci were observed in serum-deprived cells that were depleted of CEP290 and lacked primary cilia (Figures 4C and 4D). In contrast, using similar criteria, we found that centrosomal localization of Rab8a, (as judged by the number of Rab8a foci) was not affected by over-expression or RNAi-mediated depletion of CP110 (data not shown). Our data suggest that Rab8a localization to the centrosome is dependent on CEP290 and that ciliogenesis is regulated by a CEP290-Rab8a-dependent mechanism.
Figure 4.
CEP290 recruits Rab8a to centrosomes. RPE-1 cells transfected with control or CEP290 siRNAs were (A, B) grown asynchronously in the presence of serum or (C, D) induced to quiescence, and stained with antibodies to gltuamylated tubulin (GT335, green), Rab8a (red), and with DAPI (blue). Bar: 10 μM; insets: 2 μM. (B, D) Histograms quantifying the fold increase in the percentage of cells with fewer than two Rab8a foci at centrosomes. About 100 cells were scored for each siRNA condition, and the histograms showed the averages of three independent experiments. Error bars represent +/- S.D. The average percentage of cells with fewer than two Rab8a foci after transfection with control siRNA was 16.5% in quiescent cells and 9.8% in growing cells.
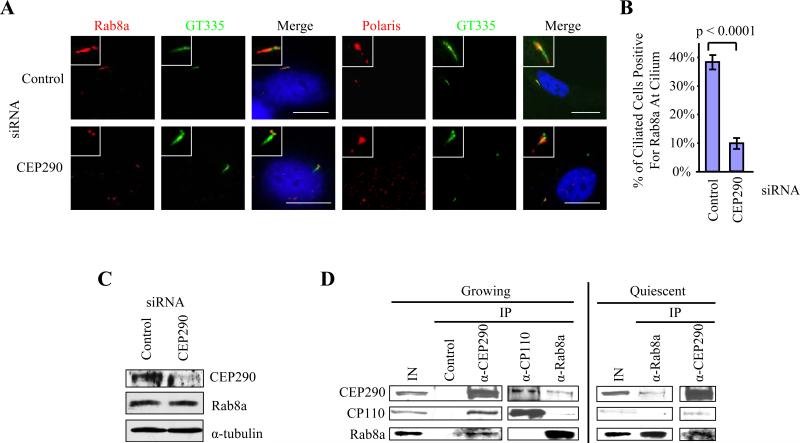
We next addressed whether CEP290 has a role in the entry of Rab8a into the primary cilium. In a quiescent population treated with control siRNA, 38±3% of cells have cilia that are Rab8a positive (Figures 5A and 5B). Strikingly, of the population of CEP290 siRNA-treated cells that have retained their cilia, only 10±2% were positive for Rab8a at the cilium (Figures 5A and 5B). Interestingly, loss of CEP290 does not affect the entry of polaris into the cilium. The percentages of ciliated cells that stained positive for polaris at the cilium in control versus CEP290 knockdown were 76% and 75%, respectively. Next, we asked whether Rab8a protein levels were altered upon knocking down CEP290. We showed that there was no difference in the abundance of total Rab8a between control and CEP290-depleted cells (Figure 5C). Taken together, our data suggest that CEP290 plays a role in recruiting Rab8a to the centrosome and in regulating the entry of Rab8a into the cilium during assembly of this organelle.
Figure 5.
CEP290 regulates entry of Rab8a into the primary cilium and interacts with Rab8a in vivo. (A) RPE-1 cells transfected with control or CEP290 siRNAs, induced to quiescence, and stained with antibodies to glutamylated tubulin (GT335, green), Rab8a or polaris (red), and with DAPI (blue). A ciliated cell is shown in each case. Bar: 10 μM; insets: 2 μM. (B) A histogram quantifying the percentage of ciliated cells that were positive for Rab8a at the cilium. About 100 cells from the ciliated population of cells treated with control or CEP290 siRNA were scored, and the averages of four independent experiments were shown. Error bars represent +/- S.D. (C) Western blotting of CEP290 and Rab8a in RPE-1 cells treated with control or CEP290 siRNAs. α-tubulin was used as a loading control. (D) Western blotting of endogenous CEP290, CP110 and Rab8a after immunoprecipitation with anti-Flag (control), anti-CEP290, anti-CP110, or anti-Rab8a antibodies from growing or quiescent 3T3 cell extracts. IN, input.
To determine whether CEP290 and CP110 interact with Rab8a in vivo, we performed immunoprecipitations with CEP290, CP110, and Rab8a antibodies. CEP290, CP110, and Rab8a were immunoprecipitated with Rab8a antibodies (Figure 5D), and reciprocal immunoprecipitations with CEP290 antibodies confirmed this interaction with Rab8a in growing cells (Figure 5D). Similarly, the interaction between CEP290 and Rab8a was also observed in quiescent cells. However, both proteins associated weakly with CP110, whose levels fall precipitously in this setting (Figure 5D). Considered together, our data indicate that CEP290 and Rab8a interact with one another in growing and quiescent cells, while their interactions with CP110 are mostly restricted to growing cells. These interactions could provide proper cues for cilia assembly.
Ectopic expression of certain CEP290 fragments prevents primary cilia formation
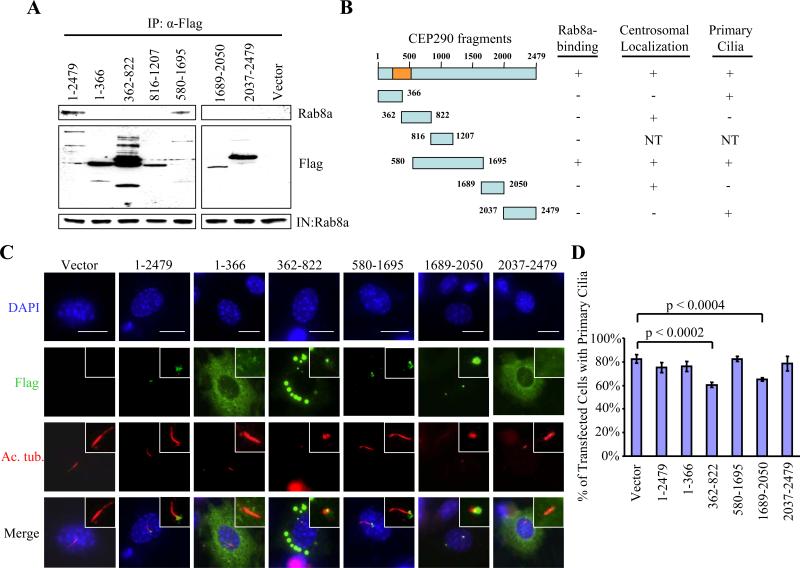
We next sought to determine which regions of CEP290 are involved in binding Rab8a and the effect of ectopically expressing our CEP290 truncation mutants (Figure 1C) in quiescent 3T3 cells. We found that a fragment spanning residues 580-1695 interacted with Rab8a, and use of additional deletion mutants suggested that residues 1208-1695 were required for stable binding to Rab8a (Figures 6A and 6B). Thus, CP110 and Rab8a bind to mutually distinct portions of CEP290. The truncation mutant containing residues 580-1695 is properly targeted to the centrosome, and it supports the ability of cells to form cilia (Figures 6B, 6C and 6D). In contrast, expression of fragments containing residues 362-822 or 1689-2050 leads to a significant decrease in the number of ciliated cells (Figures 6B, 6C and 6D). Interestingly, both of these mutants localize to the centrosome, but they fail to interact with Rab8a (Figures 6A, 6B and 6C), suggesting that they could exert their dominant negative effect by displacing the endogenous CEP290 and Rab8a from the centrosome. Thus, our data clearly demonstrate a strong correlation between Rab8a binding to CEP290 and cilia assembly.
Figure 6.
Ecotpic expression of certain CEP290 fragments prevents primary cilia formation. (A) The indicated fragments of Flag-tagged CEP290 were expressed in 293T cells and immunoprecipitated from lysates. Flag-CEP290 fusion proteins and Rab8a were detected after western blotting the resulting immunoprecipitates. Input Rab8a was detected in lysates from each transfection (IN). (B) Summary of CEP290 truncation mutants and the results of in vivo binding experiments. The orange box denotes the CP110-binding domain based on the yeast two-hybrid screen. NT denotes “not tested.” (C) 3T3 cells transiently transfected with plasmids expressing Flag-CEP290 truncation mutants, induced to quiescence, and stained with antibodies to Flag (green), Ac. tub. (red), and with DAPI (blue). Bar: 10 μM; insets: 2 μM. (D) The percentages of transfected, quiescent 3T3 cells expressing primary cilia were determined using Ac. tub. as a marker. About 100 transfected cells for each construct were scored, and average data obtained from three independent experiments were shown. Error bars represent +/- S.D.
Discussion
We and others have previously described CP110 as a centrosomal protein that localizes to the distal end of centrioles (Chen et al., 2002; Kleylein-Sohn et al., 2007). Here we show that CP110 interacts with CEP290, which localizes primarily to centrioles and basal bodies. CP110 is extinguished from the mother centriole/basal body upon entry into quiescence, while CEP290 is found on the daughter centriole and the mother centriole/basal body of a primary cilium. These observations point to mechanistic differences in their regulation and function and they suggest that there is a pool of CEP290 that might function independently of CP110. Indeed, CP110 suppresses cilia growth and formation in growing cells, while CEP290 promotes primary cilia assembly. Depletion of CEP290 has no noticeable effect on the cell cycle or the centrosome cycle in growing cells. Furthermore, ablation of CP110 does not enhance the ciliary phenotype observed after CEP290 knockdown. These data suggest that CEP290 functions exclusively in ciliary biogenesis and that CEP290 and CP110 functionally interact. IFT20, a component of the IFT particle involved in anterograde transport, is localized to the centrosome and is required only for cilia assembly (Follit et al., 2006; Yoshimura et al., 2007). CEP290 and IFT20 may therefore represent a new class of proteins that are strictly required for primary cilia but not centrosome cycle functions yet are assembled and targeted to the centrosome prior to the execution of the cilia assembly program in quiescent cells.
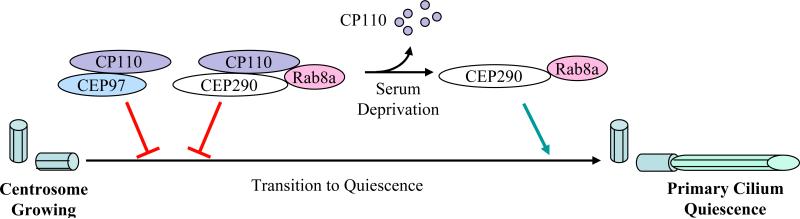
Detailed molecular and cellular descriptions of the events leading to the conversion of centrosomes to primary cilia have not been obtained. Here, we identified a CP110 complex comprised of CP110 and CEP290 that is distinguishable from CP110/centrin/CaM and CP110/CEP97 complexes. In addition, CP110 and Rab8a bind to mutually distinct portions of CEP290, and studies involving over-expression of CP110 suggest that CP110 does not compete with Rab8a for binding to CEP290 (data not shown). Thus, CP110, CEP290 and Rab8a likely exist as one distinct complex. It seems plausible that the main role of CP110 in this complex is to keep CEP290 inactive in growing cells until cells are ready to undergo ciliogenesis as they transit into the quiescent state (Figure 7). Intriguingly, our preliminary studies have shown that depletion of CEP290 disrupts the migration of mother centrioles to the cell cortex, although additional, higher resolution studies will be required. In this context, CP110 could act primarily on CEP290 to prevent migration of centrioles to the cell surface. Upon transition into the quiescent state, CP110 is extinguished from the mother centriole and its levels dramatically decrease (Figure 7). This could free up CEP290 and allow for proper migration and insertion of mother centrioles into the cell cortex and subsequent implementation of a cilia assembly program involving Rab8a as an effector. CEP290 is present at the basal body and perhaps the transitional zone, a structure believed to regulate transport of protein cargos into and out of the cilium. Depletion of CEP290 results in a significant decrease of Rab8a at the centrosome and at the cilium, raising the possibility that CEP290 first recruits Rab8a through direct protein-protein interactions to the centrosome in cycling cells and later promotes ciliogenesis by allowing the entry of Rab8a into the cilium. Entry of Rab8a into the cilium promotes the docking and fusion of membranous vesicles thought to be critical for cilia formation. Consistent with a role of CEP290 in mediating protein trafficking, a hypomorphic mutation in the retinal degeneration 16 mouse (rd16) mouse model is associated with redistribution of the ciliary protein Retinitis Pigmentosa GTPase Regulator in photoreceptors (Chang et al., 2006) and of G proteins in olfactory sensory neurons (McEwen et al., 2007). Experiments are currently underway to further delineate precisely how CEP290 promotes ciliogenesis and to identify additional molecular components involved in this process.
Figure 7.
A model depicting the role of CEP290 in mediating ciliogenesis. In growing cells, CP110 antagonizes the action of CEP290, which in turn prevents CEP290-dependent Rab8a ciliogenesis. Another complex containing CP110 and CEP97 also serves to suppress primary cilia assembly. When cells exit the cell cycle, the levels of CP110 diminish dramatically. This frees up CEP290, which in turn cooperates with Rab8a to promote primary cilia formation.
Mutations in CEP290 have been implicated in a broad spectrum of disease phenotypes, including BBS (Leitch et al., 2008). Like CEP290, the BBS proteins, comprising a core complex with at least seven highly conserved members, are essential for primary cilia formation. The ciliogenic function of the BBS proteins appears to be mediated in part by Rab8a (Nachury et al., 2007). Here, we have shown that CEP290 interacts with Rab8a and that Rab8a binding to CEP290 is required for ciliogenesis. Thus, interference with Rab8a function at the centrosome and/or at the cilium could represent the molecular basis for many ciliary defects found in human patients. While no human mutations in Rab8a have been found so far, disruption of Rab8a function recapitulates the BBS phenotype in zebrafish (Nachury et al., 2007). Given the significant overlap of clinical manifestations in patients with BBS and CEP290 mutations and the connection between BBS proteins and CEP290 with Rab8a, we hypothesize that BBS proteins may associate with CEP290. Although we and others have not been able to detect an interaction between BBS4 and CEP290 (McEwen et al., 2007 and data not shown), further studies will be needed to address whether CP110 and CEP290 are directly linked to Rab8a via other BBS proteins.
Supplementary Material
Acknowledgments
We wish to thank M. Takahaski and Y. Ono for the gift of anti-kendrin antibody, J. Salisbury for anti-centrin antibody (20H5), Y. Cai and S. Somlo for anti-polycystin2 antibody (YCC2), C. Janke for anti-polyglutamylated tubulin antibody (GT335), B. Yoder for anti-polaris antibody, M. Cole for the gift of pCBF plasmid DNA, F. Hildebrandt, J. Gleeson, C. Wilkinson and E. Nigg for CEP290 plasmid DNA, and A. Khodjakov for providing the RPE-1 hTERT cell line. We thank I. Sanchez, A. Spektor and S. Vijayakumar for critical reading of the manuscript. We are especially grateful to P. Asp in our laboratory for invaluable assistance with FPLC purification. We thank all members of the Dynlacht laboratory for constructive advice and encouragement. This work was supported in part by an Irma T. Hirschl Career Scientist Award to B.D.D., and by grants from the National Institutes of Health (EY007961) and Midwest Eye Banks and Transplantation Center to A.S. and H.K. C.B. was supported by a Human Frontier Science Program postdoctoral fellowship. W.Y.T. was supported by an Alberta Heritage Foundation for Medical Research full-time postdoctoral fellowship.
Abbreviations
- RNAi
RNA interference
- siRNA
small interfering RNA
- CaM
calmodulin
- IFT
intraflagellar transport
- BBS
Bardet-Biedl syndrome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Baala L, Audollent S, Martinovic J, Ozilou C, Babron MC, Sivanandamoorthy S, Saunier S, Salomon R, Gonzales M, Rattenberry E, et al. Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am. J. Hum. Genet. 2007;81:170–179. doi: 10.1086/519494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano JL, Teslovich TM, Katsanis N. The centrosome in human genetic disease. Nat. Rev. Genet. 2005;6:194–205. doi: 10.1038/nrg1557. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- Chang B, Khanna H, Hawes N, Jimeno D, He S, Lillo C, Parapuram SK, Cheng H, Scott A, Hurd RE, et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Shou C. Molecular cloning of a tumor-associated antigen recognized by monoclonal antibody 3H11. Biochem. Biophys. Res. Commun. 2001;280:99–103. doi: 10.1006/bbrc.2000.4087. [DOI] [PubMed] [Google Scholar]
- Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- den Hollander AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KE, Zonneveld MN, Strom TM, Meitinger T, Brunner HG, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helou J, Otto EA, Attanasio M, Allen SJ, Parisi MA, Glass I, Utsch B, Hashmi S, Fazzi E, Omran H, et al. Mutation analysis of NPHP6/CEP290 in patients with Joubert syndrome and Senior-Loken syndrome. J. Med. Genet. 2007;44:657–663. doi: 10.1136/jmg.2007.052027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyk A, Gromley A, Redick S, San Agustin J, Witman G, Pazour GJ, Peters DJ, Doxsey S. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J. Cell Biol. 2004;166:637–643. doi: 10.1083/jcb.200405023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Perret E, Middendorp S, Houcine O, Guennou C, Marano F, Bornens M, Tournier F. Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J. Cell Sci. 2000;113(Pt 8):1355–1364. doi: 10.1242/jcs.113.8.1355. [DOI] [PubMed] [Google Scholar]
- Leitch CC, Zaghloul NA, Davis EE, Stoetzel C, Diaz-Font A, Rix S, Al-Fadhel M, Lewis RA, Eyaid W, Banin E, et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat. Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- McEwen DP, Koenekoop RK, Khanna H, Jenkins PM, Lopez I, Swaroop A, Martens JR. Hypomorphic CEP290/NPHP6 mutations result in anosmia caused by the selective loss of G proteins in cilia of olfactory sensory neurons. Proc. Natl. Acad. Sci. U S A. 2007;104:15917–15922. doi: 10.1073/pnas.0704140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, Jackson PK. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- Paoletti A, Moudjou M, Paintrand M, Salisbury JL, Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 1996;109(Pt 13):3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- Peranen J, Auvinen P, Virta H, Wepf R, Simons K. Rab8 promotes polarized membrane transport through reorganization of actin and microtubules in fibroblasts. J. Cell Biol. 1996;135:153–167. doi: 10.1083/jcb.135.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J. Intraflagellar transport. Curr. Biol. 2002;12:R125. doi: 10.1016/s0960-9822(02)00703-0. [DOI] [PubMed] [Google Scholar]
- Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- Sayer JA, Otto EA, O'Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV, et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- Scholey JM. Intraflagellar transport. Annu Rev Cell Dev Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- Scholey JM, Anderson KV. Intraflagellar transport and cilium-based signaling. Cell. 2006;125:439–442. doi: 10.1016/j.cell.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Singla V, Reiter JF. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- Sluder G, Nordberg JJ. The good, the bad and the ugly: the practical consequences of centrosome amplification. Curr. Opin. Cell Biol. 2004;16:49–54. doi: 10.1016/j.ceb.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- Taulman PD, Haycraft CJ, Balkovetz DF, Yoder BK. Polaris, a protein involved in left-right axis patterning, localizes to basal bodies and cilia. Mol. Biol. Cell. 2001;12:589–599. doi: 10.1091/mbc.12.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Spektor A, Luciano DJ, Indjeian VB, Chen Z, Salisbury JL, Sanchez I, Dynlacht BD. CP110 cooperates with two calcium-binding proteins to regulate cytokinesis and genome stability. Mol. Biol. Cell. 2006;17:3423–3434. doi: 10.1091/mbc.E06-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Wang L, Chen Z, Sanchez I, Dynlacht BD. SCAPER, a novel cyclin A-interacting protein that regulates cell cycle progression. J. Cell Biol. 2007;178:621–633. doi: 10.1083/jcb.200701166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Silhavy JL, Brancati F, Barrano G, Krishnaswami SR, Castori M, Lancaster MA, Boltshauser E, Boccone L, Al-Gazali L, et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.