Abstract
Objective
To determine whether responsivity of reward circuitry to food predicts future increases in body mass and whether polymorphisms in DRD2 and DRD4 moderate these relations.
Design
The functional magnetic resonance imaging (fMRI) paradigm investigated blood oxygen level dependent activation in response to imagined intake of palatable foods, unpalatable foods, and glasses of water shown in pictures. DNA was extracted from saliva samples using standard salting-out and solvent precipitation methods.
Participants
Forty-four adolescent female high school students ranging from lean to obese.
Main Outcome
Future increases in body mass index (BMI).
Results
Weaker activation of the frontal operculum, lateral orbitofrontal cortex, and striatum in response to imagined intake of palatable foods, versus imagined intake of unpalatable foods or water, predicted future increases in body mass for those with the DRD2 TaqIA A1 allele or the DRD4-7R allele. Data also suggest that for those lacking these alleles, greater responsivity of these food reward regions predicted future increases in body mass.
Discussion
This novel prospective fMRI study indicates that responsivity of reward circuitry to food increases risk for future weight gain, but that genes that impact dopamine signaling capacity moderate the predictive effects, suggesting two qualitatively distinct pathways to unhealthy weight gain based on genetic risk.
Theorists posit that hyper-responsivity of the mesolimbic and mesocortical circuitry that encodes food reward increases risk for obesity (Davis et al., 2004). Functional magnetic resonance imaging (fMRI) studies indicate that obese versus lean individuals show greater activation in the insula, frontal operculum, lateral orbitofrontal cortex (OFC), amygdala, and striatum in response to pictures of palatable foods (Rothmund et al., 2007; Stoeckel et al., 2008) and anticipated receipt of palatable food (Stice et al., 2008b). Data suggest that the insula and frontal operculum are involved in food craving and anticipated reward from food, and further that the OFC, amygdala, and striatum encode the reward value of food (Gottfried et al., 2003; Small et al., 2008). However, because the fMRI studies that have investigated the relation of anticipatory food reward to obesity have been cross-sectional, it is unclear whether hyper-responsivity of this circuitry increases risk for future weight gain or is a result of conditioning from overeating palatable foods. Thus, we tested whether hyper-responsivity of food reward regions increases risk for future increases in body mass.
Other theorists argue that some individuals show hypo-responsivity of reward circuitry, which prompts them to overeat to compensate for this deficiency (Comings and Blum, 2000). Positron emission tomography (PET) studies find that obese versus lean individuals show less D2 receptor binding in the striatum (Volkow et al., 2008; Wang et al., 2001), which suggests that they have fewer D2 receptors. Dopamine is involved in the reinforcing effects of food; feeding results in dopamine release in the dorsal striatum and the degree of pleasure from eating correlates with amount of dopamine release (Small et al., 2003). Dopamine antagonists increase food intake and produce weight gain, whereas dopamine agonists reduce energy intake and produce weight loss (Epstein et al., 2007). Yet, consumption of a high-fat, high-sugar diet may lead to down-regulation of D2 receptors. Repeated intake of sweet and fatty foods results in down-regulation of post-synaptic D2 receptors, increased D1 receptor binding, and decreased D2 sensitivity and μ-opioid receptor binding in animals (Bello et al. 2002; Colantuoni et al., 2001), paralleling neural response to chronic use of drugs that increase dopamine signaling. Thus, it is important to test whether hypo-responsivity of reward circuitry increases risk for future weight gain. One prospective study found that when mice are exposed to a high-fat diet, those with lower D2 receptor density in the putamen showed more weight gain than mice with higher D2 receptor density in this region (Huang et al., 2006). In a prospective fMRI study we found that individuals who showed weaker striatal activation in response to receipt of palatable food were at elevated risk for increases in body mass over a 1-year follow-up, but only if they had an Al allele of the TaqIA SNP (rs1800497) in the DRD2 gene (Stice et al., 2008a). The Taq1A polymorphism has three allelic variants: A1/A1, A1/A2, and A2/A2. Individuals with an A1/A1 or A1/A2 TaqIA genotype have 30–40% fewer striatal D2 receptors than those with the A2/A2 genotype (Ritchie and Noble, 2003; Tupala et al., 2003). The TaqIA A1 genotype is also associated with hypofunctioning of the prefrontal cortex, hypothalamus, striatum, insula, and amygdala (Bowirrat and Oscar-Berman, 2005; Noble, 2003). The DRD2 TaqIA A1 genotype has correlated with body mass in some studies (Spitz et al., 2000; Thomas et al., 2001), but other studies have not found a significant main effect (Jenkinson et al., 2000; Southon et al., 2003).
Given the evidence that weaker striatal activation in response to food intake predicts future increases in body mass if coupled with the DRD2 TaqIA A1 genotype, we used blood oxygen level dependent (BOLD) fMRI to test whether activation of brain reward regions in response to imagined intake of palatable foods predicts future increases in body mass and whether these relations were moderated by genotypes associated with a potential reduction in dopamine signaling. We examined brain activation in response to imagined intake of a range of palatable foods because our previous fMRI paradigm (Stice et al., 2008a) examined only activation in response to chocolate milkshake. In addition to investigating the DRD2 TaqIA gene, we examined the 48-base pair (bp) exon 3 Variable Number Tandem Repeat (VNTR) polymorphism in the DRD4 gene. DRD4 is a postsynaptic receptor that is principally inhibitory of the second messenger adenylate cyclase. D4 receptors are localized in areas innervated by mesocortical projections from the ventral tegmental area, including the prefrontal cortex, cingulate gyrus, and insula (Noaín et al., 2006). The 7-repeat or longer VNTR allele (DRD4-7R) has been associated with compromised dopamine functioning in an in vitro study 25, poorer response to dopamine stimulating drugs (Hamarman et al., 2004), and less dopamine release in the ventral caudate and nucleus accumbens after cigarette use (Brody et al., 2006). Humans with versus without one or more DRD4 long (7R–10R) alleles have shown higher maximum lifetime body mass in samples at risk for obesity (Guo et al., 2007; Kaplan et al., 2008; Levitan et al., 2004), as well as greater food cravings in response to food cues (Sobik et al., 2005), smoking cravings and activation of the superior frontal gyrus and insula in response to smoking cues (McClernon et al., 2007), alcohol cravings and activation in OFC, anterior cingulate gyrus, and striatum in response to alcohol (Hutchison et al., 2002; Filbey et al., 2008), and heroin craving in response to heroin cues (Shao et al., 2006).
Methods
Participants
Participants were 44 adolescent girls (M age = 15.6; SD = .96; 2.3% African Americans, 84.1% European Americans, 4.5% Native Americans, and 9.1% mixed racial heritage) recruited from a larger study of female high schools students who reported body image concerns to participate in a study on the neural response to presentation of food (see also Stice et al., 2008b). Those who reported binge eating or compensatory behavior in the past 3 months, any use of psychotropic medications or illicit drugs, head injury with a loss of consciousness, or current Axis I psychiatric disorder (including anorexia or bulimia nervosa) were excluded. Data from 5 participants were not analyzed because they showed excessive head movement during the scan; 2 showed such pronounced head movement that the scans were terminated and head movement for 3 other subjects exceeded 2 mm (M = 2.8 mm, range 2.5–5 mm). This resulted in a final sample of 39 participants (M age = 15.5, SD = .94; M BMI = 24.6; SD = 5.35, BMI range = 17.3–38.9). We recruited participants who ranged in BMI from lean to obese to ensure that we would not have a restriction in range for BMI for the analyses. The local Institutional Review Board approved this project. Participants and parents provided written consent.
Measures
Body Mass
The body mass index (BMI = kg/m2) was used to reflect adiposity. After removal of shoes and coats, height was measured to the nearest millimeter using a stadiometer and weight was assessed to the nearest 0.1 kg using a digital scale. Two measures of height and weight were obtained and averaged at baseline and at 6- and 12-month follow-up assessments. BMI correlates with direct measures of total body fat such as dual energy x-ray absorptiometry (r = .80 to .90) and with health measures including blood pressure, adverse lipoprotein profiles, atherosclerotic lesions, serum insulin levels, and diabetes mellitus in adolescent samples (Dietz and Robinson, 1998).
Genotyping
Participants were asked to provide saliva, from which epithelial cells were collected, using a commercial product, Oragene® (DNAgenotek, Ottawa, ON, Canada), though only 32 of the 39 participants consented and provided a saliva sample, which was requested at 6-month follow-up rather than at baseline. DNA was extracted from the samples using standard salting-out and solvent precipitation methods, yielding an average of 45 μg of DNA. The DRD2 TaqIA assay was done using a fluorogenic 5′ nuclease (Taqman®, ABI, Foster City, CA) method (Haberstick and Smolen, 2004) on an ABI Prism® 7000 Sequence Detection System using the allelic discrimination mode (Livak, 1999). Reactions containing 20 ng of DNA were performed in 10 μl reactions with TaqMan® Universal PCR Master Mix using the standard cycling conditions. Sequences of the primers and probes are: Forward Primer: 5′-GTGCAGCTCACTCCATCCT-3′; Reverse Primer: 5′-GCAACACAGCCATCCTCAAAG-3′; A1 Probe: 5′-VIC-CCTGCCTTGACCAGC-NFQMGB-3′; A2 Probe: 5′-FAM-CTGCCTCGACCAGC-NFQMGB-3′. Each 96 well plate included non-template and DNA standards of known genotype. Two investigators independently scored each genotype. DRD2 was coded A1/A1 or A1/A2 versus A2/A2; 13 subjects had the A1 allele of the DRD2 gene and 19 subjects did not. The DRD2 TaqIA site is now known to reside in exon 8 of the ANKK1 gene on the opposite strand. This SNP results in a glutamate-to-lysine (E713K) substitution within the eleventh ankyrin repeat of ANKK1, prompting the suggestion that changes in ANKK1 activity may be responsible for some of the associations attributed to DRD2 (Neville et al, 2004). With this caveat, we refer to the polymorphism as DRD2 TaqIA throughout the text. The assay for the 48-base pair (bp) exon 3 VNTR polymorphism in the DRD4 gene was a modification (Anchordoquy et al., 2003) of the method of (Lerman et al., 1998). The primer sequences were forward: 5′-VIC-GCTCATGCTGCTGCTCTACTGGGC-3′; and reverse: 5′-CTGCGGGTCTGCGGTGGAGTCTGG-3′, which yield PCR products from 279 (2R) to 519 (7R) bp. Following PCR, the amplicons were analyzed on an ABI PRISM® 3130xl Genetic Analyzer (Foster City, CA). Based on studies suggesting that the 7 repeat allele confers a functional difference in D4 receptors (Asghari et al., 1995), participants were classified as having at least one 7R variant or none; 11 subjects had the 7R variant of the DRD4 gene and 21 subjects did not. None of the subjects had DRD4 alleles longer than 7R. The correlation between TaqIA and DRD4 allele status (r = −.20) did not differ significantly from zero.
fMRI paradigm
Participants were asked to consume a typical breakfast or lunch, but to refrain from eating or drinking (except water) for 4–6 hours immediately preceding their imaging session for standardization purposes. We selected this deprivation period to capture the hunger state that most individuals experience as they approach their next meal, which is a time when individual differences in food reward would logically impact caloric intake. Most participants completed the paradigm between 16:00 and 18:00 (approximately 5 hours after eating a typical lunch), but a subset completed scans between 11:00 and 13:00 (approximately 5 hours after eating a typical breakfast). Before the imaging session, participants were familiarized with the fMRI paradigm. The imagine paradigm was designed to examine brain responses to imagined consumption of palatable foods, unpalatable foods, and glasses of water shown in pictures. Because there is evidence that obese individuals show a positive implicit attitude to both palatable and unpalatable foods (Craeynest et al., 2005), we included a contrast involving imagined intake of a non-caloric substance (water), as it seemed possible that overweight individuals would show hyper-responsivity of reward circuitry to both palatable and unpalatable foods. Before the scanning session, they rated how appetizing they found various foods shown in 103 pictures. Pictures included processed foods (e.g., cheeseburger, cupcakes), fruits (e.g., grapes, peaches), and vegetables (e.g., cauliflower, eggplant). During the fMRI paradigm, each participant was exposed to the 20 pictures of food they rated as the most appetizing and the 20 pictures of food they rated as the least appetizing, as well as 20 pictures of glasses of water. Stimuli were presented in 2 separate scanning runs. There were 3 events of interest in the paradigm: (1) imagined intake of appetizing foods, 2) imagined intake of unappetizing foods, and 3) imagined intake of water shown in pictures. The images were presented for 5 seconds using Presentation (Version 9.81, www.neuro-bs.com) run from Windows. Participants were asked to imagine tasting and eating the pictured food, in attempt to activate brain regions associated with anticipatory reward from the pictured foods. Order of presentation was randomized. Each run consisted of 60 events (20 images each from the same category; appetizing, unappetizing, water). A blank screen with a cross-hair at the center for fixation (to eliminate random eye-movement) was presented for 2 seconds between each stimulus picture.
Imaging and statistical analysis
Scanning was performed by a Siemens Allegra 3 Tesla head-only MRI scanner. A standard birdcage coil was used to acquire data from the entire brain. A thermo foam vacuum pillow and additional padding was used to restrict head motion. In total, 720 brain volumes were collected during both functional runs. Functional scans used a T2* weighted gradient single-shot echo planar imaging (EPI) sequence (TE=30 ms, TR = 2000 ms, flip angle=80°) with an in plane resolution of 3.0 × 3.0 mm2 (64 × 64 matrix; 192 × 192 mm2 field of view). To cover the whole brain, 32 4mm slices (interleaved acquisition, no skip) were acquired along the AC-PC transverse, oblique plane as determined by the midsagittal section. Structural scans were collected using an inversion recovery T1 weighted sequence (MP-RAGE) in the same orientation as the functional sequences to provide detailed anatomic images aligned to the functional scans. High-resolution structural MRI sequences (FOV = 256 × 256 mm2, 256 × 256 matrix, thickness = 1.0 mm, slice number ≈ 160) were acquired.
Data were pre-processed and analyzed using SPM5 software (Wellcome Department of Imaging Neuroscience, London, UK) in MATLAB Mathworks, Inc., Sherborn, MA) (Worsley and Friston, 1995). The images were time-acquisition corrected to the slice obtained at 50% of the TR. All functional images were then realigned to the mean. The images (anatomical and functional) were normalized to the standard MNI template brain implemented in SPM5 (ICBM152, based on an average of 152 normal MRI scans). Normalization resulted in a voxel size of 3 mm3 for functional images and a voxel size of 1 mm3 for structural images. Functional images were smoothed with a 6 mm FWHM isotropic Gaussian kernel. Condition specific effects at each voxel were analyzed using the conjoint use of the general linear model and Gaussian random field theory. The response to events (i.e., indicated by stimulus onsets) were modeled by a canonical hemodynamic response function (HRF), consisting of a mixture of 2 gamma functions that emulate the early peak at 5 seconds and the subsequent undershoot. We included temporal derivatives of the hemodynamic function to obtain a better model of the data (Henson et al., 2002). A 128 second high-pass filter (per SPM5 convention) was used to remove low-frequency noise and slow drifts in the signal. To identify brain regions activated in response to the appetizing food condition, we contrasted BOLD responses during viewing pictures of appetizing food versus viewing pictures of unappetizing food (appetizing food – unappetizing food) and BOLD responses during viewing pictures of appetizing food versus viewing pictures of glasses of water (appetizing food – water) at an individual level.
We performed small volume correction analyses within the somatosensory cortex, gustatory cortex, and striatum with activation peaks from previous independent fMRI studies (Rothmund et al., 2007; Stoeckel et al., 2008) as centroids to define 10-mm diameter spheres. Reference coordinates from these prior studies are shown in Table 1. Peaks within these regions were considered significant at P < .05, false-discovery rate (FDR) corrected across the small volume. Additional exploratory inferences were made at the P<.001, uncorrected for multiple comparisons. To test whether the genotypes moderated the cross-sectional relations between BMI and brain responses, we conducted multiple regression models using SPSS. Independent variables included BMI scores, genotype (DRD2, DRD4), and the interaction between genotype and BMI scores. For the longitudinal analyses, we conducted mixed models analyses using SAS PROC MIXED that included BMI at 6- and 12-month follow-ups as the outcomes; independent variables included baseline BMI, genotype, brain response, and time as well as all two- and three-way interactions between genotype, brain responses, and time. We examined model assumptions by inspecting residual by predicted values and normal probability plots. Visual examination indicated a random distribution around zero in the residuals and a straight line for the residual probability plot. We evaluated outliers with the criteria recommended by (Neter et al., 1996): a Cook’s distance F probability value greater than .2. In only one case was this criterion met and because it did not alter the pattern of results, we retained that case in the data set. These results suggest that no univariate or multivariate outliers contributed unduly to the observed main and interactive effects reported herein.1
Table 1.
Regions Responding during the Appetizing Food Condition of the Imagine Paradigm as a Function of Concurrent Body Mass Index (N = 39)
| Reference coordinates1 | X2 | y | z | Cluster | Z | P value <.05 FDR corrected | P <.001 (uncorrected) | Effect size r | |
|---|---|---|---|---|---|---|---|---|---|
| Appetizing food – unappetizing food | |||||||||
| Putamen | −16, −2, 4 | −15 | 6 | 3 | 12 | 3.59 | .011 | .000 | .56 |
| Lateral OFC | 38, 20, −20 | 36 | 27 | −15 | 9 | 3.27 | .018 | .001 | .51 |
| Superior frontal gyrus | 6 −3 |
33 36 |
54 51 |
39 | 4.22 4.02 |
.15 .15 |
.0003 .0003 |
.62 .60 |
|
| Frontal operculum | −39 33 |
27 27 |
6 0 |
7 9 |
4.21 3.51 |
.15 .24 |
.0003 .0003 |
.62 .55 |
|
| Rolandic operculum | 39 | −18 | 33 | 8 | 3.66 | .21 | .0003 | .56 | |
| Appetizing food – water | |||||||||
| Lateral OFC | −32, 22, −18 −32, 22, −18 |
33 −36 |
27 21 |
−12 −12 |
22 14 |
4.01 3.26 |
.002 .035 |
.000 .001 |
.60 .50 |
| Frontal operculum | −38, −14, −8 44, 28, −4 |
−39 33 |
27 27 |
3 0 |
6 18 |
3.72 3.51 |
.017 .013 |
.000 .000 |
.56 .55 |
| Superior frontal gyrus | 6 −6 |
33 36 |
51 51 |
26 | 4.05 4.02 |
.25 .25 |
.0003 .0003 |
.60 .60 |
|
| Ventrolateral prefrontal cortex | 45 | 45 | 9 | 6 | 4.03 | .25 | .0003 | .60 | |
| Parietal operculum | 54 | 24 | 18 | 4 | 3.46 | .58 | .0003 | .53 | |
| Frontal operculum | −42 | 24 | 9 | 3 | 3.36 | .60 | .0003 | .52 |
Reference coordinates from Stoeckel et al., 2008 or Rothemund et al., 2007.
X, Y, and Z coordinates shown are peak coordinates of the clusters.
Activation clusters with t-maps thresholded at P uncorrected <.001 (cluster criterion of 3 voxels).
Results
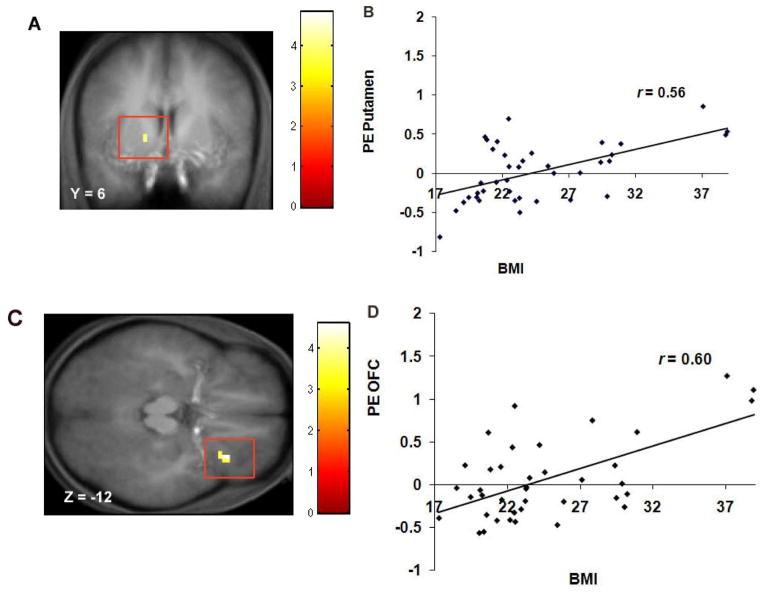
Baseline BMI correlated positively with activation in the left putamen (r = .56; p<.001 Figure 1A–B) and right lateral OFC (r = .51, p <.01) during exposure to appetizing versus unappetizing foods and with activation in the OFC (r = .50, p <.01; r = 60; p <.001 Figure 1C–D) and frontal operculum (r = .55 and .56) during exposure to appetizing food versus water (Table 1). Presence of the A1 allele (Figure 2) significantly moderated the relation between baseline BMI and increased response in the right lateral OFC (r = −.33, p < .05) during exposure to appetizing versus unappetizing food and the relation between baseline BMI and increased response in the right frontal operculum (r = −.52, p < .01) during exposure to appetizing food versus water. Presence of the DRD4-7R allele significantly moderated the relation between baseline BMI and increased response in the right frontal operculum (r = −.37, p = .02) during exposure to appetizing food versus water.
Figure 1.
A. Coronal section of increased activation in a region of the putamen (−15, 6, 3, Z = 3.59, P < 0.05 FDR corrected) during appetizing food - unappetizing food as a function of BMI with B. the graph of parameter estimates (PE) from that region. C. Axial section of increased activation in the lateral OFC (33, 27, −12, Z = 4.01, P < 0.05 FDR corrected) during appetizing food – water as a function of BMI with D. the graph of PE from that region.
Figure 2.
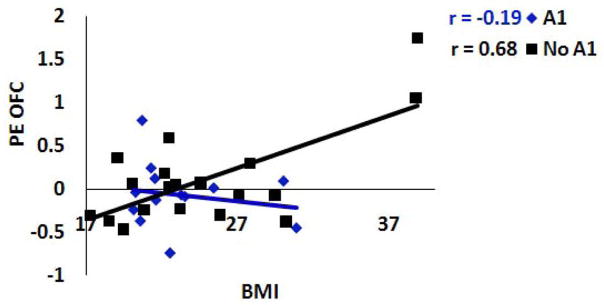
A1 allele of the DRD2 attenuates the relation between concurrent Body Mass Index (BMI) and response in a region of the OFC (36, 27, −15, Z = 3.27, P < 0.05 FDR corrected) during appetizing food – unappetizing food2.
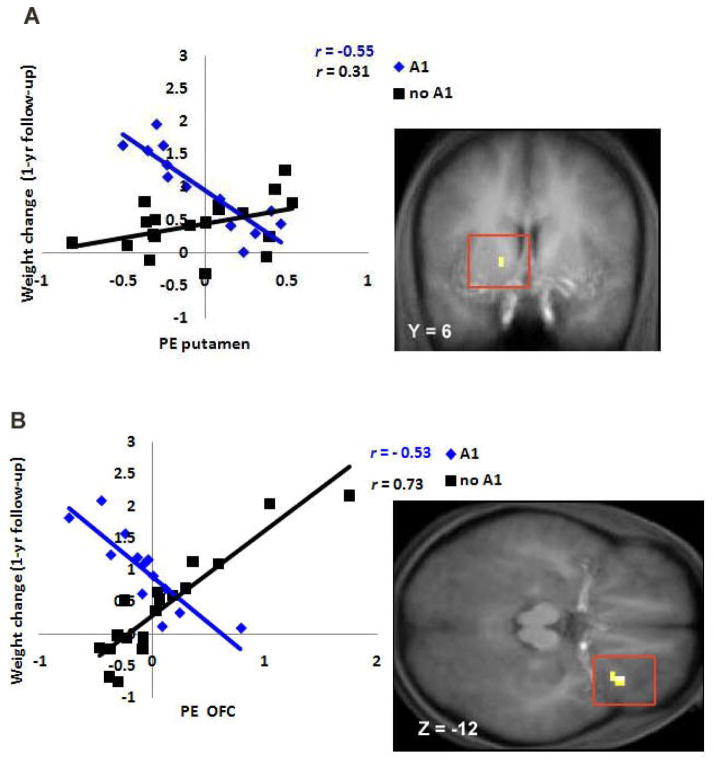
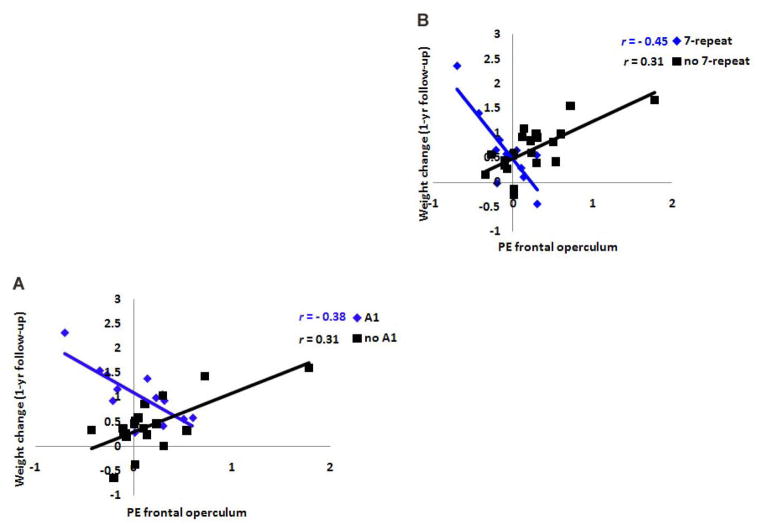
Mean BMI was 24.6 at baseline, 24.7 at 6-month follow-up, and 24.8 at 1-year follow-up. The average increase in BMI over the 1-year follow-up period was 0.23 (SD = .56, range −0.97 to 1.42), suggesting sufficient variation in weight change over the follow-up period. Although BOLD activation did not show significant main effects on risk for future increases in BMI over the 1-year follow-up, the TaqIA1 allele of the DRD2 gene significantly moderated the relations of responses in the left putamen (r = .33, p = .042) and right lateral OFC (r = .60, p < .001) (Figure 3) during exposure to appetizing versus unappetizing food to risk for increases in BMI over the 1-year follow-up. Further, both the TaqIA1 allele (r = .36, p < .031) and the DRD4-7R allele (r = .42, p < .012) (Figure 4) significantly moderated the relation between response in the right frontal operculum during exposure to appetizing food versus water and risk for future increases in BMI.
Figure 3.
A. Activation in a region of the putamen (−15, 6, 3, Z = 3.59, P < 0.05 FDR corrected) during appetizing food - unappetizing food was negatively related to predicted values of future weight gain for participants with the A1 allele, but positively related to predicted values of future weight gain for participants without the A1 allele. B. Activation in the lateral OFC (36, 27, −15, Z = 3.27, P < 0.05 FDR corrected) during appetizing food – unappetizing food was negatively related to predicted values of future weight gain for participants with the A1 allele, but positively related to predicted values of future weight gain for participants without the A1 allele.
Figure 4.
Activation in a region of the frontal operculum (33, 27, −12, Z = 4.01, P < 0.05 FDR corrected) was A negatively related to predicted values of future weight gain for participants with the A1 allele, but positively related to predicted values of future weight gain for participants without the A1allele and B negatively related to predicted values of future weight gain for participants with the 7-repeat allele of the DRD4, but positively related to predicted values of future weight gain for participants without the 7-repeat allele3.
Based on the evidence that reward-related neural function in women is heightened during the mid-follicular phase (Dreher et al., 2007), we created a dichotomous variable that reflected whether participants completed the fMRI scans during the midfollicular phase (days 4–8 after onset of menses; n = 12) or not (n = 27). When we controlled for this variable in all analyses, the activation in the reported regions remained significant.
Discussion
The prospective interactive effects suggest that individuals who show weaker activation of brain reward circuitry in response to imagined intake of palatable foods are at elevated risk for future weight gain if they possess the TaqIA1 allele of the DRD2 gene or the exon 3 7-repeat allele of the DRD4 gene, presumably because they have reduced dopamine signaling. These findings dovetail with evidence that weaker striatal activation in response to receipt of palatable food increases risk for weight gain among individuals with the TaqIA1 allele of the DRD2 gene (Stice et al., 2008a). The present results extend the earlier findings in several ways. First, they suggest that blunted activation of food reward circuitry to food cues, in addition to actual intake of food, increases risk for weight gain if coupled with genetic risk for attenuated dopamine signaling in reward circuitry. Collectively results suggest that a hypo-responsivity to both consummatory and anticipatory food reward increases risk for weight gain. Second, whereas the effect for blunted activation in the putamen echoes the earlier findings regarding blunted striatal activation, the present study observed similar effects for the frontal operculum and OFC, regions that play a role in encoding the taste and reward value of foods. Third, whereas the finding for the TaqIA1 allele echoes the results seen in the earlier report, the present study also suggests that the DRD4-7R allele, which is associated with compromised dopamine functioning (Noaín et al., 2006), increases risk for weight gain if coupled with blunted responsivity of reward circuitry. Thus, data imply that genotypes thought to be associated with reduced signaling capacity of dopamine-based reward circuitry may increase risk for weight gain. These findings align with the thesis that individuals who experience less activation of reward circuitry from food may overeat to compensate for this reward deficit (Comings and Blum, 2000). Yet, Figures 3 and 4 imply that individuals who show elevated responsivity of these food reward regions may be at increased risk for weight gain if they are not at genetic risk for compromised dopamine signaling, which also echoes our previous results (Stice et al., 2008a). These findings hint at two possible pathways to unhealthy weight gain and appear to provide support for both the hypo-responsivity model and the hyper-responsivity model. Indeed, this may explain why prior findings have appeared to provide support for both of these contrasting theories. This pattern of findings suggests that experiencing too little or too much reward from food may both paradoxically increase risk for obesity and that experiencing moderate reward from food might serve as a protective factor. This interpretation echoes a previous suggestion that there may be a quadratic relation between dopamine activity and reward sensitivity (Kirsch et al., 2006), wherein individuals with too little or too much dopamine activity show disturbances in reward sensitivity.
Other aspects of the present findings are also noteworthy. First, the cross-sectional analyses revealed a positive association between baseline BMI and activation in the putamen. This finding is important because it suggests that overweight individuals do not show a general reduced responsivity of the striatum to food stimuli. It was also reassuring that the cross-sectional main effects converge with previous evidence that obese versus lean individuals show greater activation of reward regions to food images (Rothmund et al., 2007; Stoeckel et al., 2008). Further, there was a positive relation between BMI and activation in the opercular regions (i.e., frontal operculum, Rolandic operculum, parietal operculum). As these regions have been associated with subjective reward from food intake as well as anticipated intake (Small et al., 2003; Stice et al., 2008b; Yamamoto, 2006), these findings may also imply that overweight and obese individuals experience greater food reward relative to lean individuals. There was also a positive relation between BMI and superior frontal gyrus activation, an area implicated in working memory (Postle et al, 2000), in response to palatable foods versus unpalatable foods or water. This result converges with those from Rothemund and colleagues (2007). We also found that overweight versus lean individuals showed preferential activity in the ventrolateral prefrontal cortex (VLPFC) during exposure to palatable foods, which converges with our previous finding that BMI is positively correlated with activation in the VLPFC in response cues predicting the receipt of a chocolate milkshake (Stice et al., 2008b). This region has projections to both sensory and reward processing areas (e.g., Pertrides & Pandya, 2002). The VLPFC is thought to use object information to guide goal-directed behavior (Kennerley & Wallis, 2009) and to play an important role in attentional control (e.g., Roth et al., 2006). That is, VLPFC is responsible for detecting behaviorally relevant events and redirecting attention appropriately. Thus, results may imply that overweight individuals show increased attention to palatable food and find it more rewarding. However, the fact that dopamine-related genes moderated the relations implies that it is important to focus on relations that are conditional on DRD2 and DRD4 status. Lastly, findings also illustrate the importance of conducting prospective brain imaging studies to document that potential risk factors temporally precede weight gain and provide a more rigorous test of etiologic processes.
When interpreting these results it is important to note that BOLD response reflects relative amount of oxygenated hemoglobin rather than dopamine signaling; thus we can only infer that our results may arise because of differences in dopamine functioning. It is possible that the differences in activation are a product of some other neurotransmitter. Further, when we re-estimated the relation of the interaction between DRD2 and BMI to activation in the OFC (see Figure 2) with the two participants with the greatest BMI removed from the data set, the interaction became non-significant. Given this and the fact that this is one of the first prospective fMRI studies to predict future increases in body mass, results should be considered provisional until replicated in a larger sample. When we re-estimated the relation of the interaction between DRD2 and BMI to activation in the OFC (see Figure 2) with the two participants with the greatest BMI removed from the data set, the interaction became non-significant. In addition, because we only studied females, results may not generalize to males. Moreover, because hypo-responsivity of reward circuitry has been linked to both obesity and substance abuse (Comings and Blum, 2000), it follows that there must be unmeasured moderators that determine whether at-risk individuals develop obesity, substance abuse, or some other motivated behavior problem. Interestingly, there is evidence that obese individuals show a decreased risk of substance use and abuse (Simon et al., 2006; Warren, Fronst-Pindea, & Gold, 2005), suggesting that routinely engaging in one behavior that produces dopamine release may reduce the probability of turning to another behavior for this end.
In sum, the present results suggest that individuals who show weaker responsivity of food reward circuitry are at increased risk for unhealthy weight gain if they are at genetic risk for reduced dopamine signaling. Yet, data also suggest that greater activation in these brain regions predicted elevated future weight gain for those not at genetic risk for reduced dopamine signalling, suggesting two distinct pathways to unhealthy weight gain that arise from abnormal responsivity of reward circuitry and genetic risk.
Acknowledgments
Funding/Support: This research was supported by a Road Supplement for Interdisciplinary Research in Behavioral and Biological Sciences (R1MH64560A)
Footnotes
It should be noted that the skew for baseline BMI was only 1.28 and the skew for the slope in BMI, reflecting change over the follow-up period, was only −0.30, which confirms that the outcomes met the assumption of univariate normal distributions.
Despite the lack of evidence for influential outliers, we re-estimated the test of the interaction between DRD2 and activation in the OFC in the prediction of concurrent BMI with the two participants with the greatest BMI removed from the data set; the interaction became non-significant (r = .04, p>.05), suggesting that this particular interaction was strongly influenced by the 5% of participants who had the highest initial BMI values.
We re-estimated the test of the interactions between DRD2 allele and activation in the frontal operculum and between DRD4 and activation in the frontal operculum in the prediction of future increases in BMI excluding the single case that showed the greatest increase in BMI over the follow-up period; both interactions remained statistically significant (r = .54, p <.01 and r = .61, p<.001, respectively) suggesting that these interactions were not strongly driven by this potential outlier.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anchordoquy HC, McGeary C, Liu L, Krauter KS, Smolen A. Genotyping of three candidate genes following whole genome preamplification of DNA collected from buccal cells. Behav Genet. 2003;33(1):73–78. doi: 10.1023/a:1021007701808. [DOI] [PubMed] [Google Scholar]
- Asghari V, Sanyal S, Buchwaldt S, Paterson A, Jovanovic V, Van Tol HH. Modulation of intracellular cyclic AMP levels by different human dopamine D4 receptor variants. J Neurochem. 1995;65(3):1157–1165. doi: 10.1046/j.1471-4159.1995.65031157.x. [DOI] [PubMed] [Google Scholar]
- Bello NT, Lucas LR, Hajnal A. Repeated sucrose access influences dopamine D2 receptor density in the striatum. Neuroreport. 2002;13(12):1557–1578. doi: 10.1097/00001756-200208270-00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody AL, Mandelkern MA, Olmstead RE, Scheibal D, Hahn E, Shiraga S, Zamora-Paja E, Farahi J, Saxena S, London ED, McCracken JT. Gene variants of brain dopamine pathways and smoking-induced dopamine release in the ventral caudate/nucleus accumbens. Arch Gen Psychiatry. 2006;63(7):808–816. doi: 10.1001/archpsyc.63.7.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowirrat A, Oscar-Berman M. Relationship between dopaminergic neurotransmission, alcoholism, and Reward Deficiency syndrome. Am J Med Genet B. 2005;132B(1):29–37. doi: 10.1002/ajmg.b.30080. [DOI] [PubMed] [Google Scholar]
- Colantuoni C, Schwenker J, McCarthy J, Rada P, Ladenheim B, Cadet JL, Schwartz GJ, Moran TH, Hoebel BG. Excessive sugar intake alters binding to dopamine and mu-opioid receptors in the brain. Neuroreport. 2001;12(16):3549–3552. doi: 10.1097/00001756-200111160-00035. [DOI] [PubMed] [Google Scholar]
- Comings DE, Blum K. Reward deficiency syndrome: genetic aspects of behavioral disorders. Prog Brain Res. 2000;126:325–341. doi: 10.1016/S0079-6123(00)26022-6. [DOI] [PubMed] [Google Scholar]
- Craeynest M, Crombez G, De Houwer J, Deforche B, Tanghe A, De Bourdeaudhuij I. Explicit and implicit attitudes towards food and physical activity in childhood obesity. Behaviour Research and Therapy. 2005;43:1111–1120. doi: 10.1016/j.brat.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Davis C, Strachan S, Berkson M. Sensitivity to reward: Implications for overeating and obesity. Appetite. 2004;42(2):131–138. doi: 10.1016/j.appet.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Dietz WH, Robinson TN. Use of body mass index (BMI) as a measure of overweight in children and adolescents. J Pediatr. 1998;132(2):191–193. doi: 10.1016/s0022-3476(98)70426-3. [DOI] [PubMed] [Google Scholar]
- Dreher JC, Schmidt PJ, Kohn P, Furman D, Rubinow D, Berman KF. Menstrual cycle phase modulates reward-related neural function in women. Proc Natl Acad Sci U S A. 2007;104(7):2465–2470. doi: 10.1073/pnas.0605569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein LJ, Temple JL, Neaderhiser BJ, Salis RJ, Erbe RW, Leddy JJ. Food reinforcement, the dopamine D2 receptor genotype, and energy intake in obese and nonobese humans. Behav Neuroscience. 2007;121(5):877–886. doi: 10.1037/0735-7044.121.5.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filbey FM, Ray L, Smolen A, Claus ED, Audette A, Hutchison KE. Differential neural response to alcohol priming and alcohol taste cues is associated with DRD4 VNTR and OPRM1 genotypes. Alcohol Clin Exp Res. 2008;32(7):1113–1123. doi: 10.1111/j.1530-0277.2008.00692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottfried JA, O’Doherty J, Dolan RJ. Encoding predictive reward value in human amygdala and orbitofrontal cortex. Science. 2003;301(5636):1104–1107. doi: 10.1126/science.1087919. [DOI] [PubMed] [Google Scholar]
- Guo G, North KE, Gorden-Larsen P, Bulik CM, Choi S. Body mass, DRD4, physical activity, sedentary behavior, and family socioeconomic status: The Add Health Study. Obesity. 2007;15(5):1199–1206. doi: 10.1038/oby.2007.640. [DOI] [PubMed] [Google Scholar]
- Haberstick BC, Smolen A. Genotyping of three single nucleotide polymorphisms following whole genome preamplification of DNA collected from buccal cells. Behav Genet. 2004;34(5):541–547. doi: 10.1023/B:BEGE.0000038492.50446.25. [DOI] [PubMed] [Google Scholar]
- Hamarman S, Fossella J, Ulger C, Brimacombe M, Dermody J. Dopamine receptor 4 (DRD4) 7-repeat allele predicts methylphenidate dose response in children with attention deficit hyperactivity disorders: A pharmacogentic study. J Child Adolesc Psychopharmacol. 2004;14(4):564–574. doi: 10.1089/cap.2004.14.564. [DOI] [PubMed] [Google Scholar]
- Henson RN, Price CJ, Rugg MD, Turner R, Friston KJ. Detecting latency differences in event related BOLD responses: application to words versus nonwords and initial versus repeated face presentations. Neuroimage. 2002;15(1):83–97. doi: 10.1006/nimg.2001.0940. [DOI] [PubMed] [Google Scholar]
- Huang XF, Zavitsanou K, Huang X, Yu Y, Wang H, Chen F, Lawrence AJ, Deng C. Dopamine transporter and D2 receptor binding densities in mice prone or resistant to chronic high fat diet-incuded obesity. Behav Brain Res. 2006;175(2):415–419. doi: 10.1016/j.bbr.2006.08.034. [DOI] [PubMed] [Google Scholar]
- Hutchison KE, LaChance H, Niaura R, Bryan AD, Smolen A. The DRD4 VNTR polymorphism influences reactivity to smoking cues. J Abnormal Psychol. 2002;111(1):134–142. doi: 10.1037//0021-843x.111.1.134. [DOI] [PubMed] [Google Scholar]
- Jenkinson CP, Hanson R, Cray K, Wiedrich C, Knowler WC, Bogardus C, Baier L. Association of dopamine D2 receptor polymorphisms Ser311Cys and TaqIA with obesity or type 2 diabetes mellitus in Pima Indians. Int J Obes Relat Metab Disord. 2000;24(10):1233–1238. doi: 10.1038/sj.ijo.0801381. [DOI] [PubMed] [Google Scholar]
- Kaplan AS, Levitan RD, Yilmaz Z, Davis C, Tharmalingam S, Kennedy JL. A DRD4/BDNF gene-gene interaction associated with maximum BMI in women with bulimia nervosa. Int J Eat Disord. 2008;41:22–28. doi: 10.1002/eat.20474. [DOI] [PubMed] [Google Scholar]
- Kennerley SW, Wallis JD. Reward-dependent modulation of working memory in lateral prefrontal cortex. J Neurosci. 2009;29:3259–3270. doi: 10.1523/JNEUROSCI.5353-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch P, Reuter M, Mier D, Lonsdorf T, Stark R, Gallhofer B, et al. Imaging gene-substance interactions: The effect of the DRD2 TaqIA polymorphism and the dopamine agonists bromocriptine on the brain activation during the anticipation of reward. Neuroscience Letters. 2006;405:196–201. doi: 10.1016/j.neulet.2006.07.030. [DOI] [PubMed] [Google Scholar]
- Lerman C, Caporaso N, Main D, Audrain J, Boyd NR, Bowman ED, Shields PG. Depression and self-medication with nicotine: the modifying influence of the dopamine D4 receptor gene. Health Psychol. 1998;17(1):56–62. doi: 10.1037//0278-6133.17.1.56. [DOI] [PubMed] [Google Scholar]
- Levitan RD, Masellis M, Lam RW, Muglia P, Basile VS, Jain U, Kaplan AS, Tharmalingam S, Kennedy SH, Kennedy JL. Childhood in-attention and dysphoria and adult obesity associated with the dopamine D4 receptor gene in overeating women with seasonal affective disorder. Neuropsychopharmacology. 2004;29(1):179–186. doi: 10.1038/sj.npp.1300314. [DOI] [PubMed] [Google Scholar]
- Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14(5–6):143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- McClernon FJ, Hutchison KE, Rose JE, Kozink RV. DRD4 VNTR polymorphism is associated with transient fMRI-BOLD responses to smoking cues. Psychopharmacology. 2007;194(4):433–441. doi: 10.1007/s00213-007-0860-6. [DOI] [PubMed] [Google Scholar]
- Neter J, Kutner MH, Nachtsheim CJ, Wasserman W. Applied linear statistical models. 4. Boston: McGraw-Hill; 1996. [Google Scholar]
- Neville MJ, Johnstone EC, Walton RT. Identification and characterization of ANKK1: a novel kinase gene closely linked to DRD2 on chromosome band 11q23.1. Hum Mutat. 2004;23:540–545. doi: 10.1002/humu.20039. [DOI] [PubMed] [Google Scholar]
- Noble EP. D2 dopamine receptor gene in psychiatric and neurologic disorders and its phenotypes. Am J Med Genet A. 2003;116B(1):103–125. doi: 10.1002/ajmg.b.10005. [DOI] [PubMed] [Google Scholar]
- Noaín D, Avale ME, Wedemeyer C, Calvo D, Peper M, Rubinstein M. Identification of brain neurons expressing the dopamine D4 receptor gene using BAC transgenic mice. Eur J Neurosci. 2006;24(9):2429–2438. doi: 10.1111/j.1460-9568.2006.05148.x. [DOI] [PubMed] [Google Scholar]
- Petrides M, Pandya DN. Comparative cytoarchitectonic analysis of the human and the macaque ventrolateral prefrontal cortex and cortico-cortical connection patters in the monkey. Eur J Neurosci. 2002;16:409–423. doi: 10.1046/j.1460-9568.2001.02090.x. [DOI] [PubMed] [Google Scholar]
- Postle BR, Stern CE, Rosen BR, Corkin S. An fMRI investigation of cortical contributions to spatial and nonspatial visual working memory. Neuroimage. 2000;11:409–23. doi: 10.1006/nimg.2000.0570. [DOI] [PubMed] [Google Scholar]
- Ritchie T, Noble EP. Association of seven polymorphisms of the D2 dopamine receptor gene with brain receptor-binding characteristics. Neurochem Res. 2003;28(1):73–82. doi: 10.1023/a:1021648128758. [DOI] [PubMed] [Google Scholar]
- Roth JK, Serences JT, Courtney SM. Neural system for controlling the contents of object working memory in humans. Cereb Cortex. 2006;16(11):1595–1603. doi: 10.1093/cercor/bhj096. [DOI] [PubMed] [Google Scholar]
- Rothemund Y, Preuschhof C, Bohner G, Bauknecht H, Klingebiel R, Flor H, Klapp BF. Differential activation of the dorsal striatum by high-calorie visual food stimuli in obese individuals. Neuroimage. 2007;37(2):410–421. doi: 10.1016/j.neuroimage.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Shao C, Li Y, Jiang K, Zhang D, Xu Y, Lin L, Wang Q, Zhao M, Jin L. Dopamine D4 receptor polymorphism modulates cue-elicited heroin craving in Chinese. Psychopharmacology (Berl) 2006;186:185–190. doi: 10.1007/s00213-006-0375-6. [DOI] [PubMed] [Google Scholar]
- Simon G, von Korff M, Saunders K, Miglioretti D, Crane P, et al. Association between obesity and psychiatric disorders in the US population. Archives of General Psychiatry. 2006;63:824–830. doi: 10.1001/archpsyc.63.7.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small DM, Jones-Gotman M, Dagher A. Feeding-induced dopamine release in dorsal striatum correlates with meal pleasantness ratings in healthy human volunteers. Neuroimage. 2003;19(4):1709–1715. doi: 10.1016/s1053-8119(03)00253-2. [DOI] [PubMed] [Google Scholar]
- Small DM, Veldhuizen MG, Felsted J, Mak YE, McGlone F. Separable substrates for anticipatory and consummatory chemosensation. Neuron. 2008;57(5):786–797. doi: 10.1016/j.neuron.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobik L, Hutchison K, Craighead L. Cue-elicited craving for food: a fresh approach to the study of binge eating. Appetite. 2005;44(3):253–261. doi: 10.1016/j.appet.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Southon A, Walder K, Sanigorski AM, Zimmet P, Nicholson GC, Kotowicz MA, Collier G. The TaqIA and Ser311 Cys polymorphisms in the dopamine D2 receptor gene and obesity. Diabetes Nutr Metab. 2003;16(1):72–76. [PubMed] [Google Scholar]
- Spitz MR, Detry MA, Pillow P, Hu Y, Amos CI, Hong WK, Wu X. Variant alleles of the D2 dopamine receptor gene and obesity. Nutr Res. 2000;20(3):371–380. [Google Scholar]
- Stice E, Spoor S, Bohon C, Small DM. Relation between obesity and blunted striatal response to food is moderated by TaqlA1 DRD2 gene. Science. 2008a;322(5900):449–452. doi: 10.1126/science.1161550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stice E, Spoor S, Bohon C, Veldhuizen M, Small DM. Relation of reward from food intake and anticipated intake to obesity: A functional magnetic resonance imaging study. J Abnorm Psychology. 2008b;117(4):924–935. doi: 10.1037/a0013600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckel LE, Weller RE, Cook EW, Twieg DB, Knowlton RC, Cox JF. Widespread reward-system activation in obese women in response to pictures of high-calorie foods. Neuroimage. 2008;41(2):636–647. doi: 10.1016/j.neuroimage.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Thomas GN, Critchley JA, Tomlinson B, Cockram CS, Chan JC. Relationships between the TaqI polymorphism of the dopamine D2 receptor and blood pressure in hyperglycaemic and normoglycaemic Chinese subjects. Clin Endocrinol (Oxf) 2001;55(5):605–611. doi: 10.1046/j.1365-2265.2001.01404.x. [DOI] [PubMed] [Google Scholar]
- Tupala E, Hall H, Bergström K, Mantere T, Räsänen P, Särkioja R, Tiihonen J. Dopamine D2 receptors and transporters in type 1 and 2 alcoholics measured with human whole hemisphere autoradiography. Hum Brain Mapp. 2003;20(2):91–102. doi: 10.1002/hbm.10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Telang F, Fowler JS, Thanos PK, Logan J, Alexoff D, Ding YS, Wong C, Ma Y, Pradhan K. Low dopamine striatal D2 receptors are associated with prefrontal metabolism in obese subjects: Possible contributing factors. Neuroimage. 2008;42(4):1537–1543. doi: 10.1016/j.neuroimage.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Ding YC, Flodman JR, Kidd JR, Kidd KK, Grady DL, Ryder OA, Spence MA, Swanson JM, Moyzis RK. The genetic architecture of selection at the human dopamine receptor D4 (DRD4) gene locus. Am J Human Genet. 2004;74(5):931–944. doi: 10.1086/420854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GJ, Volkow ND, Logan J, Pappas NR, Wong CT, Zhu W, Netusil N, Fowler JS. Brain dopamine and obesity. Lancet. 2001;357(9253):354–357. doi: 10.1016/s0140-6736(00)03643-6. [DOI] [PubMed] [Google Scholar]
- Warren M, Frost-Pineda K, Gold M. Body mass index and marijuana use. Journal of Addictive Diseases. 2005;24:95–100. doi: 10.1300/J069v24n03_08. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Friston KJ. Analysis of fMRI time-series revisited – again. Neuroimage. 1995;2(3):173–181. doi: 10.1006/nimg.1995.1023. [DOI] [PubMed] [Google Scholar]
- Yamamoto T. Neural substrates for the processing of cognitive and affective aspects of taste in the brain. Archives of Histology and Cytology. 2006;69(4):243–255. doi: 10.1679/aohc.69.243. [DOI] [PubMed] [Google Scholar]