Abstract
Background
Immune activation is one of the main features of HIV/Hepatitis C virus (HCV) infections and has been linked to the disturbance of the gut-associated lymphoid tissue (GALT). In chronic HIV infection, loss of GALT integrity results in translocation of microbial products and chronic immune activation. We explored the relationship between bacterial translocation and specific colonic proteins, including liver expressed antimicrobial peptide (LEAP 2) which may play a role in modulating the bacterial translocation process.
Methods
A total of 40 subjects (10 HIV/HCV, 10 HIV, 10 HCV-infected patients and 10 controls) were enrolled and underwent serum and colonic tissue sampling. The levels of immune activation were evaluated by measuring plasma sCD27, and the levels of selected proinflammatory, Th2 and regulatory cytokines in both the plasma and supernatant of CD3-stimulated intraepithelial lymphocytes. We also evaluated LEAP-2 expression in the colon biopsies using Affymetrix Human Gene 1.0 ST (HuGene) and fluorescent immunohistochemistry.
Results
Increased levels of sCD27 were observed in HIV/HCV coinfected (p=0.03) and HIV monoinfected (p=0.04) patients compared with controls consistent with the presence of immune activation. The chip array identified LEAP-2 expression as a key marker associated with immune activation. LEAP-2 expression in HIV, HCV and HIV/HCV-infected patients was significantly lower compared with controls, and was significantly negatively correlated (p=0.03, r=−0.44) with sCD27.
Conclusions
Our data suggests that HCV and HIV infections are associated with decreased expression of LEAP-2 in colonic tissue. This may represent a key mechanism for enhanced microbial translocation and immune activation in HIV/HCV-infected patients.
INTRODUCTION
The gut is colonised with ~1014 normal bacterial flora which is important for the normal development of the immune system and efficient metabolic and digestive functions.1 Given the critical functions of the bacterial flora, several mechanisms exist to prevent the invasion of these bacteria or translocation of their products from the intestinal lumen to the systemic circulation. Endothelial cells of the gut, in addition to their important absorptive function, act as a physical and biological barrier to protect the host from microbial invasion. As part of their defensive mechanisms, endothelial cells synthesise and secrete multiple small (<10 KDa) antimicrobial peptides (AMPs) such as defensins,2 cathelicidins3, 4 and liver-expressed antimicrobial peptide-2 (LEAP-2)5 that kill the invading bacteria and promote the immune defence mechanisms against them.
LEAP-2 is a cysteine-rich, cationic AMP secreted mainly by the epithelial cells of the gut, kidney, lung, heart and the hepatocytes.6 LEAP-2 mRNA codes for a precursor protein of 77 amino acid residues and at least four different splice variants. The large native LEAP-2 form of 40 amino acid residues has antimicrobial properties and is generated from the precursor at a putative cleavage site for a furin-like endoprotease.6
AMPs are part of the innate immune system and highly conserved among species. However, during HIV infection, there is a loss of the integrity of the epithelial barrier of the gut.7 Consequently, there is an increase in the gastrointestinal (GI) permeability which is directly associated with microbial translocation8 and chronic immune activation. Additionally, many of the protective mechanisms in the gut during HIV infection are impaired including dysregulation of T cells,9 and defects in the innate immune responses with potential leakage of bacteria and their products to the systemic circulation and induction of chronic immune activation.8, 10, 11
The role of LEAP-2 in microbial translocation in HIV and Hepatitis C virus (HCV) infection is not known. In this study, we examined the levels of expression of LEAP-2 in the colon tissues of controls, HIV, Hepatitis C virus (HCV) and HIV/Hepatitis C virus (HCV)-coinfected patients, and the relationship between LEAP-2 expression to immune activation, and cytokine production.
METHODS
Patients and methods
All participants were recruited from University of Cincinnati clinics. Consent forms were obtained from participating subjects according to a protocol approved by the University of Cincinnati College of Medicine institutional review board. To study the effect of HIV-1 and HCV infection on microbial translocation, 10 non-infected controls, 10 HIV-1 monoinfected, 10 HCV monoinfected, and 10 HIV/HCV-coinfected subjects were enrolled in the study. Inclusion criteria encompassed patients with active HIV and/or HCV of all ages, and without selection by race or gender. Exclusion criteria included any patient with a history of inflammatory bowel diseases (IBD) or suspected IBD, autoimmune diseases including rheumatoid arthritis, and any patients on systemic immunomodulators. Pregnancy was also an exclusionary factor. Controls were non-HCV, non-HIV-infected subjects who were screened for colon cancer, and were all negative for colon cancer.
All HIV-infected patients were receiving different regimens of antiretroviral treatment, including combinations of the following: zidovudine, abacavir, atazanavir, lamivudine, emtricitabine, darunavir, raltegravir, efavirenz and/or ritonavir-boosted protease inhibitor. Additionally, 10 HCV-infected patients were receiving anti-HCV treatment which consisted of pegylated interferon-α and ribavirin. Thirty mL of blood was drawn from each subject. Additionally, rectal biopsies from the distal colon were obtained from patients and controls using flexible sigmoidoscopy (3 biopsies from the descending colon 1–3 mm in size using biopsy forceps (30–45 cm from the anal verge).
Demographic data and characterisation of the enrolled subjects are summarised in table 1. HIV viral loads were determined in patients’ plasma using COBAS AmpliPrep/COBAS taqMan HIV-1 Test V.2.0 (Roche Diagnostics, Indianapolis, Indiana, USA) with a threshold of 40 copies/mL. Peripheral blood mononuclear cells (PBMCs) were isolated from the blood by density gradient using Ficoll-Paque Plus (Histopaque, Sigma, St Louis, Missouri, USA) according to the manufacturer’s instruction. PBMCs were washed twice in RPMI-1640 (Gibco, Carlsbad, California, USA), counted and used immediately, or stored in 10% Dimethyl sulfoxide (DMSO) at −80°C for further analysis.
Table 1.
Demographic and clinical data characteristics of the enrolled subjects
| Demographic characters | Control n=10 |
HIV n=10 |
HCV n=10 |
HIV/HCV n=10 |
|---|---|---|---|---|
| Sex | ||||
| Male | 4 | 10 | 6 | 7 |
| Age | ||||
| Range | 42–64 | 24–54* | 47–67 | 46–54 |
| (Average±SD) | (56±6.8) | (43±10.7) | (56.8±6.6) | (49.1±6.3) |
| Race | ||||
| White | 7 | 7 | 4 | 5 |
| A–A | 3 | 2 | 6 | 5 |
| Others | 0 | 1 | 0 | 0 |
| Antiretroviral treatment | NA | 10 | NA | 10 |
| HCV treatment | NA | NA | 3 | 7 |
| CD4 counts (cells/μL) | ||||
| Range | 420–890 | 310–550† | 520–1060 | 320–960 |
| (Average±SD) | (699±138) | (472±183) | (773±180) | (603±233) |
| HCV viral load (106 IU/mL) | ||||
| Range | NA | NA | 0.09–6.3 | 0–7.5 |
| (Average±SD) | (2.9±2.3) | (2.5±4.3) | ||
| HIV viral load (copies/mL) | ||||
| Range | NA | 0–6670 | NA | 0–110 |
| (Average±SD) | (805±2087) | (43±34) | ||
| Alanine Aminotransferase (ALT) (U/L) | ||||
| Range | 12–35 | 12–46 | 20–97 | 17–82 |
| (Average±SD) | 22.5±7.6 | 23.3±10.1 | 44.8±27.2 | 37.8±18.8 |
| Aspartate Aminotransferase (AST) (U/L) | ||||
| Range | 14–47 | 16–52 | 22–124 | 18–78 |
| (Average±SD) | 24.3±9.7 | 23.5±10.8 | 57.4±33.7 | 40.2±18 |
Alanine Aminotransferase (ALT) and Aspartate Aminotransferase (AST) are significantly different among the groups (p<0.001), and significantly higher (p<0.05) in HCV mono-infected and HCV/HIV coinfected patients compared with controls.
Age: There is a significant difference (p<0.01) between HIV and other groups.
CD4 counts: There are significant differences between CD4 counts in HIV and control or HCV (p<0.05 and p<0.01, respectively).
RNA isolation and microarray analysis
RNA was isolated from one part of colon tissue biopsies using RNeasy Mini Kit (Qiagen Valencia, California, USA). Gene expression patterns were investigated by Chip Selection Human Gene 1.0 ST microarray according to the manufacturer’s instructions. Briefly, the quality of the total RNA was checked by the Agilent 2100 Bioanalyzer using the RNA 6000 Nano Assay. For each sample, the Ambion Whole Transcript (WT) Expression Kit (Life Technologies) synthesises cDNA target from 50 ng of total RNA. Then the GeneChip Whole Transcript (WT) Terminal Labeling Kit (Affymetrix), was used to both chemically fragment and biotin-label the cDNA target. Each sample was hybridised with hybridisation cocktail to a standard Probe Array Cartridge (GeneChip Human Gene 1.0 ST Array–Affymetrix) in the GeneChip Hybridisation Oven 640 (Affymetrix). Probe arrays were washed and stained using the Fluidics Station 450 (Affymetrix). The arrays were scanned with the Affymetrix GeneChip Scanner 3000 7G. Raw data files were created by Command Console, the Affymetrix operating software program. The Affymetrix Expression Console Program was used to examine the Affymetrix Gene Array quality control factors. Data were normalised using the Robust Multi-Array Analysis (RMA) algorithm.
Fluorescent immunohistochemical staining and confocal imaging
LEAP-2 protein expression was evaluated by fluorescent immunohistochemistry as previously described.5 Briefiy, colon biopsy samples were fixed in 10% buffered formalin, dehydrated in ethanol, and embedded in paraffin. Tissue sections (4 μm thickness) were treated with boiling sodium citrate buffer for antigen retrieval, and washed. The non-specific sites were blocked with phosphate buffer saline (PBS) containing 10% normal donkey serum and 3% bovine serum albumin (BSA) for 2 h at room temperature. The slides were then treated with rabbit antihuman LEAP-2 (Cat No H-075-40, Phoenix Pharmaceuticals, Burlingame, California, USA) at 1 : 500 dilution and incubated overnight at 4°C. The slides were washed and then treated with the secondary antibody Alexa Fluor 594 donkey antirabbit IgG (Jackson Immunoresearch, West Grove, Pennsylvania, USA) diluted 1 : 100 for 1 h at room temperature. 4′,6-diamidino-2-phe-nylindole (DAPI) diluted 1:1000 was used for nuclear staining. The slides were air dried for 5 min, cover-slipped with Prolong Gold Antifade (Life Technologies, Grand Island, New York, USA), and examined by Zeiss LASER scanning confocal microscopy (LSM510) with high-power fields (1HPF=0.237 mm2) LEAP-2 expression in colon tissue from HIV, HCV, or HIV/HCV-infected patients and matched normal healthy controls, as well as from archived positive control samples from ulcerative colitis and Crohn’s diseases12 were examined.
Evaluation of markers of immune activation
The level of plasma sCD27 was measured using ELISA (Pelikine, RDI) as previously described,13 and specified by the manufacturer. sCD27 Levels were measured in duplicate using 25–50 μL of sample for each individual. The lower limit of detection of the assay was 0.2 U/mL. The concentrations of sCD27 were determined by extrapolation from the appropriate standard curves.
Measuring cytokines levels
Intraepithelial lymphocytes (IEL) was separated using a dithiothreitol (DDT)/EDTA/collagenase method, as described,14 with modification.
Briefly, immediately after resection, the tissue is transported to the laboratory in ice-cold calcium and magnesium-free Hank’s balanced salt solution (HBSS-CMF), and washed twice with Calcium and Magnesium-free Hank’s balanced salt solution (HBSS-CMF), and incubated at 4°C for 30 min with 1 mmol/L DDT (Sigma Chemical) in HBSS supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, gentamycin (50 μg/mL) and 25 μg/mL amphotericin B to remove the mucus layer. Then, the tissue was incubated with 0.75 mmol/L EDTA in Calcium and Magnesium-free Hank’s balanced salt solution (HBSS-CMF), at 37°C for 1 h to remove epithelial cells. The tissue was then washed twice with Calcium and Magnesium-free Hank’s balanced salt solution (HBSS-CMF) and incubated at 37°C in a 24-well plate with 1 mL of media containing 10% autologous plasma, RPMI-1640 (Biofluids) containing sodium pyruvate (Hazleton); HEPES buffer (Sigma), L-glutamine (Sigma), penicillin/streptomycin, gentamycin (Sigma), and amphotericin B (2.5 μg/mL). Anti-CD3 (Mabtech Cat No 3605-1-50) was added at a concentration of 100 ng/mL. After 48 h, 0.5 mL of the supernatant was aspirated from the wells and was divided into four aliquots. The aliquots were stored at −80°C until used for measuring the cytokines level. Evaluation of the levels of regulatory cytokines secreted by the intraepithelial lymphocytes was measured using LINCOplex Multiplex according to the manufacturer’s instructions. Briefly, supernatant and plasma cytokine levels for proinflammatory cytokines (IL-2, IL-12p40, and tumour necrosis factor (TNF)-α), Th2 cytokines (IL-4) and regulatory cytokines (IL-10 and IL-17) were measured simultaneously using the LINCOplex Multiplex Immunoassay (Millipore Corporation, Billerica, Massachusetts, USA). IL-22 levels were measured using ELISA assay (R&D system, Minnesota, USA) according to the manufacturer’s instructions. Cytokine levels were measured in duplicate using 25–50 μL of sample for each individual. The concentrations of cytokines were determined by extrapolation from the appropriate standard curves. Normal plasma cytokines levels (table 3) were extracted from manufacturer’s instruction of cytokines ELISA and Multiplex kits.
Table 3.
Cytokine levels measured in the plasma of enrolled subjects
| Cytokine levels (pg/mL) Normal range | Control N=10 |
HIV N=10 |
HCV N=10 |
HIV/HCV N=10 |
p Value* |
|---|---|---|---|---|---|
| IL-2 5.13±2.31 |
2.7±5.8 | 1.1±1.7 | 11.6±21.9 | 14.5±30.1 | 0.5 |
| IL-4 8±4.7 |
8.4±15.7 | 3.9±4.2 | 3.7±5.5 | 8.6±13.1 | 0.7 |
| IL-10 3.6±2.53 |
7.3±8.7 | 2.8±1.4 | 2.9±2.8 | 4.4±4.7 | 0.6 |
| TNF-a 3.12±4.04 |
9.3±5.6 | 10.6±5.5 | 8.5±3.4 | 19.3±14.8 | 0.03* |
| IL12p40 8±5 |
14.8±26.2 | 1.8±5.7 | 9.9±21.9 | 18.9±28.3 | 0.2 |
| IL-17 6.53±7.42 |
3.5±5.3 | 3.9±9.9 | 0.6±0.9 | 3.1±6.3 | 0.6 |
| IL-22 63.01±27.22 |
25.8±50.9 | 492±1313 | 28.1±66.3 | 105.7±262.6 | 0.4 |
p<0.05.
TNF, tumour necrosis factor.
Statistical analysis
Data were given as range (minimum, maximum); mean ±SD. Student t test was used to examine the difference between two groups with a significance value at p≤0.05. One-way analysis of variance test was used to examine the difference between groups of the enrolled subjects. Correlations between parameters measured were calculated using Spearman’s correlation coefficient for patients and controls.
RESULTS
Characterization of the enrolled subjects
Forty subjects were enrolled in the study, 10 in each group. Demographic and clinical characteristics of the enrolled subjects are shown in table 1. HIV-infected subjects were significantly younger (p<0.01) than the other groups, and all of them were male. Additionally, CD4 counts were significantly lower in HIV-infected patients compared with controls or HCV groups (p<0.05 and p<0.01, respectively). Liver enzymes (ALT and AST) were significantly higher (p<0.05) in HCV monoinfected and HIV/HCV coinfected patients, compared with the controls.
Gene array of colon tissues
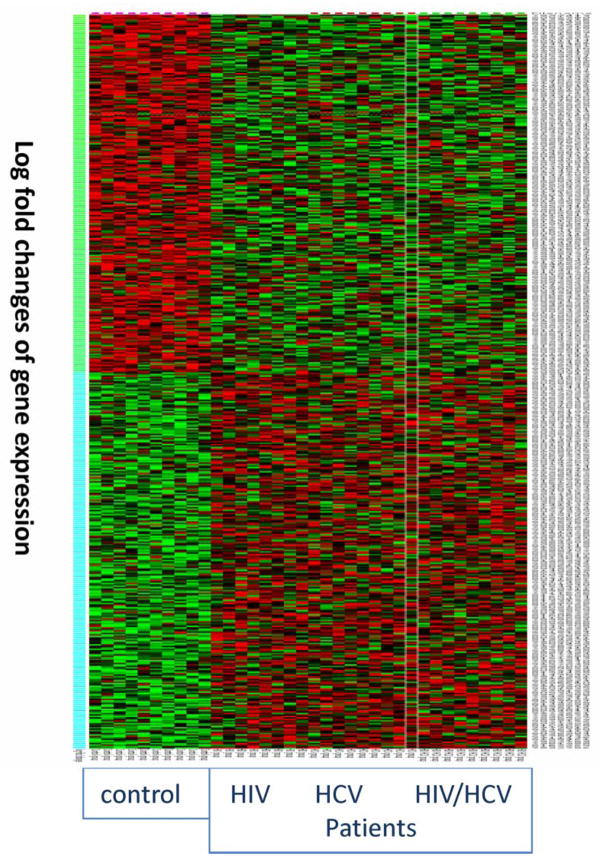
Gene array of the colon tissues of HIV, HCV and HIV/HCV-coinfected patients showed significant differences between gene expressions of the colon tissues from control subjects compared with patients as shown in the heat map of the gene array (figure 1). More than 390 genes were significantly different in expression levels between patients and control groups. Table 2 summarises some of the gene groups that were different in the colon tissues from control and patients groups. Further analysis revealed that 77 and 307 genes were down-regulated in HIV and HIV/HCV-coinfected patients, respectively, compared with controls. Additionally, 173 and 167 genes were upregulated in HIV and HIV/HCV coinfected patients, respectively, compared with controls (data not shown). Significant differences were observed in the expression of genes related to receptor binding (30 genes, p=2.34E-8), extracellular matrix (9 genes, p=4.1E-8), histocompatibility complex genes (3 genes, p<0.001), and chemokine ligands (2 genes, p<0.001). Differential expression was carefully evaluated to determine targets for further evaluation, in relation to our interest in bacterial translocation and immune activation. This evaluation led to further study of LEAP-2 gene expression.
Figure 1.
Heat map of gene array data of colon biopsies from control, HIV, HCV and HIV/HCV-coinfected patients. Log-fold changes of gene expressions (Y axis) for the colon tissues from control, HIV, HCV and HIV/HCV-coinfected patients are plotted in a heat map graph. Red represents up-regulated gene expression, while green represents down-regulated gene expression. Degree of colour reflects the levels of changes.
Table 2.
Comparison between the gene expression in colon tissues from control and HIV/HCV-infected patients
| Functions | HIV/HCV compared with controls* Gene family |
p Value | Number of genes involved |
|---|---|---|---|
| Molecular function | Receptor binding | 2.34E-08 | 30 |
| Extracellular matrix structural constituent | 4.16E-08 | 9 | |
| Collagen binding | 5.39E-06 | 6 | |
| Polysaccharide binding | 3.95E-05 | 9 | |
| Pattern binding | 3.95E-05 | 9 | |
| Biological process | Extracellular matrix organisation | 1.18E-11 | 14 |
| Cell adhesion | 1.39E-10 | 30 | |
| Biological adhesion | 1.39E-10 | 30 | |
| Extracellular structure organisation | 2.42E-10 | 16 | |
| Response to wounding | 1.10E-09 | 31 | |
| Cellular component | Extracellular region part | 3.47E-20 | 48 |
| Extracellular space | 4.93E-17 | 39 | |
| Proteinaceous extracellular matrix | 7.33E-15 | 24 | |
| Extracellular matrix | 3.17E-14 | 25 | |
| Extracellular matrix part | 2.19E-11 | 14 | |
| Pathways | ECM-receptor interaction | 2.14E-10 | 12 |
| Focal adhesion | 8.54E-08 | 14 | |
| Genes involved in integrin cell surface interactions | 4.64E-06 | 8 | |
| Syndecan-2-mediated signalling events | 3.89E-05 | 5 | |
| Integrin signalling pathway | 1.05E-04 | 9 | |
| Gene family | Histocompatibility complex genes | 6.31E-04 | 3 |
| Chemokine ligands | 5.84E-03 | 2 | |
| CD molecules | 2.28E-02 | 4 |
Date were presented as group of genes according to their functions, and the numbers of genes in each group.
Expression data of LEAP-2
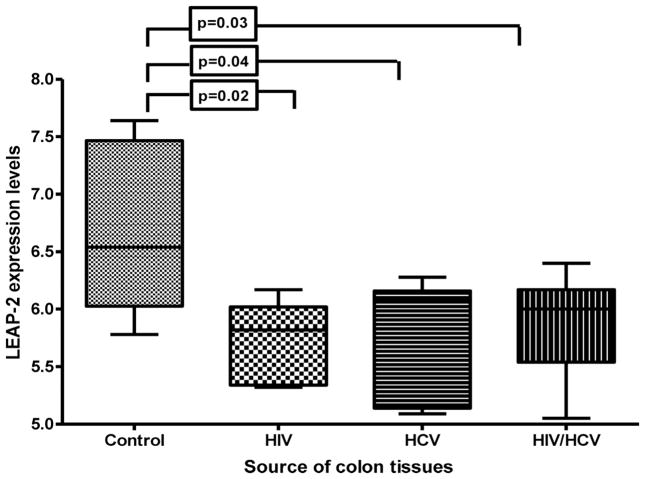
LEAP-2 is expressed by both hepatocytes and enterocytes of the colon and has antimicrobial effects.5, 6, 15, 16 LEAP-2 expression was significantly higher (p<0.05) in controls (mean±SD: 6.9 ±0.6) compared with HIV-monoinfected (5.80±0.4), HCV-monoinfected (5.84±0.5), and HIV/HCV-coinfected (5.84 ±0.5) patients (figure 2). Immunohistochemical staining of the colon biopsies by anti-LEAP-2 antibody revealed comparable results to gene array where colon tissues with higher level of LEAP-2 gene expression (controls) were associated with high levels of LEAP-2 protein expression in the colon tissues (figure 3A,B).
Figure 2.
Liver expressed antimicrobial peptide-2 (LEAP-2) gene expression in colon tissue. Relative expression of LEAP-2 mRNA was measured using gene array as described in the methods. The levels of LEAP-2 expressions were compared between the enrolled groups using the Student t test. Data was presented using vertical box and whiskers graph. The horizontal lines represent the median values. The vertical lines represent the SDs. The box represents the 75% percentile of the data. LEAP-2 expression is significantly higher (p<0.05) in control groups compared with HIV-monoinfected, HCV-monoinfected and HIV/HCV-coinfected patients (mean±SD: 6.9±0.6, 5.80±0.4, 5.84±0.5, and 5.84±0.5, respectively). The differences between the controls and HIV-monoinfected, HCV-monoinfected and HIV/HCV-coinfected patients were significant (p=0.02, p=0.04, and p=0.03, respectively). No significant difference was observed among patient groups.
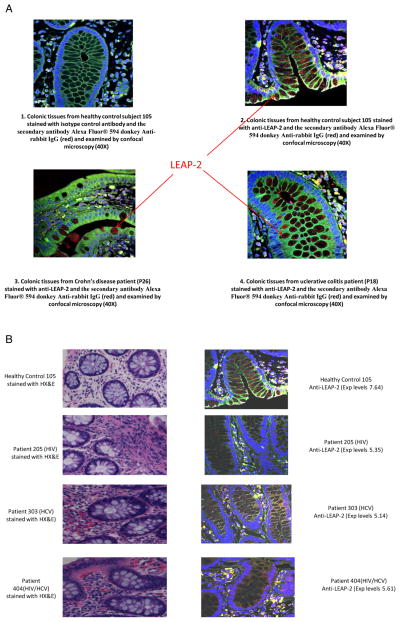
Figure 3.
Expression of liver expressed antimicrobial peptide-2 (LEAP-2) protein in the colon tissues. Expression of LEAP-2 protein in the colonic tissue was evaluated using fluorescent immunohistochemistry. (A) Represents the expression of LEAP-2 protein in colonic tissue from normal healthy control subject using anti-LEAP-2 antibody (A2), positive control colon biopsy samples from a patient with Crohn’s disease (A3) or patient with ulcerative colitis (A4) or isotype control (A1) and the secondary antibody Alexa Fluor 594 donkey antirabbit IgG. LEAP-2 appears as red particles in intraepithelial and interstitial tissue of the colon. (B) Shows representatives data of the comparative evaluation of LEAP-2 expression in colonic tissues from a healthy control subject (#105), HIV-monoinfected (#205), HCV-monoinfected (#303) and HIV/HCV-coinfected (#404) patients. The relative expression levels of LEAP-2 for control (#105), HIV patient (#205), HCV-monoinfected patient (#303), and HIV/HCV-coinfected patient (#404) were 7.64, 5.35, 5.14 and 5.61, respectively. The corresponding haematoxylin and eosin (H&E) staining for colonic tissues of the subjects demonstrated an increase in infiltrating inflammatory cells (markers of immune activation) in colonic tissues of patients compared with control.
Correlation between LEAP-2 gene expression and immune activation
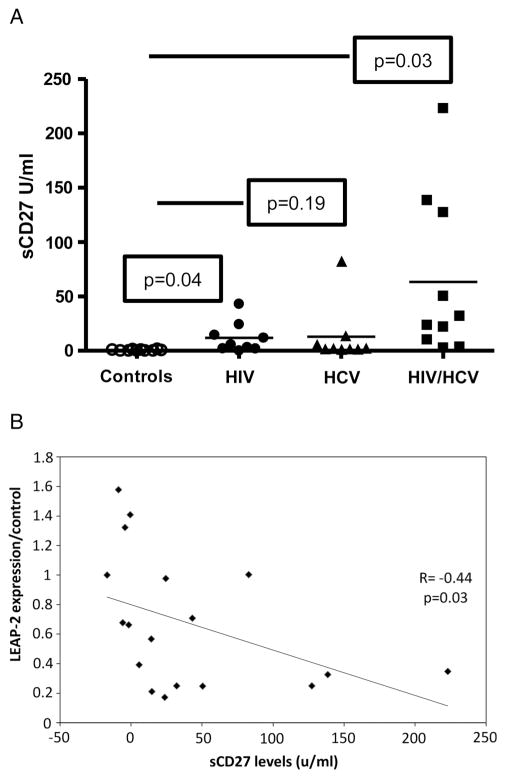
To further analyse the role of LEAP-2 expression on microbial translocation and immune activation which are commonly associated with HIV infection, we measured makers of immune activation in the plasma and correlated it with the levels of LEAP-2 expression. sCD27 Is one of the reliable markers of T cell activation which correlates strongly with immune activation. The plasma levels of sCD27 in the controls were significantly lower than the levels in HIV-monoinfected (p=0.04) and HIV/HCV-coinfected (p=0.03) patients (mean ± SD: 0.5±0.2, 11.9 ±4.7 and 63.4±23.5 U/mL, respectively), but not significantly different from the levels of sCD27 in HCV-monoinfected subjects (p=0.19, mean ± SD: 12.9±8.8 U/mL) figure 4A. Additionally, there was a significant negative correlation (p=0.03, r=−0.44) between the levels of plasma sCD27 in all the enrolled subjects and LEAP-2 gene expression (figure 4B).
Figure 4.
Levels of sCD27 in the plasma of enrolled subjects. (A) Plasma levels of sCD27 were measured by ELISA as described in methods. Data are presented using vertical scatterplot graph. The horizontal lines represent the mean values with each symbol represents one subject; controls (open circle), HIV-monoinfected (filled circle), HCV-monoinfected (filled triangle), and HIV/HCV-coinfected (filled rectangle) patients. The levels of sCD27 in the control were significantly lower than the levels in HIV-monoinfected (p=0.04), and HIV/HCV-coinfected patients (p=0.03) (mean±SD: 0.5±0.2, 11.9±4.7 and 63.4±23.5 U/mL, respectively), but not significantly different from the level of sCD27 in the HCV-monoinfected patients (p=0.19, mean±SD: 12.9±8.8 U/mL). (B) A significant negative correlation (p=0.03 r= −0.44) between plasma sCD27 levels of the patients with their relative expression of liver expressed antimicrobial peptide-2 in colon tissue compared with controls. The data was evaluated using Spearman’s correlation coefficient.
Correlation between LEAP-2 gene expression and cytokines
To identify the status of immune activation in the enrolled subjects, inflammatory cytokines (IL-2, TNF-α and IL12p40), Th2 cytokine (IL-4, IL-10), and regulatory cytokines (IL-17 and IL-22) were measured in the plasma and the supernatant of the IEL in vitro stimulated with anti-CD3. As shown in tables 3 and 4, significant differences in the levels of TNF-α were observed both in the plasma and the supernatant of stimulated IEL (p=0.03 and p=0.02, respectively). IL-4 levels were significantly different in the supernatant (p=0.02) but not in the plasma (p=0.7). However, there was no significant correlation between the TNF-α or IL-4 levels and LEAP-2 expressions (data not shown). No significant differences were observed with other cytokines (tables 3 and 4).
Table 4.
Cytokine levels in the supernatant of CD3-stimulated intraepithelial lymphocytes of enrolled subjects
| Cytokines levels (pg/mL) | Control n=10 |
HIV n=10 |
HCV n=10 |
HIV/HCV n=10 |
p Value |
|---|---|---|---|---|---|
| IL-2 | 0.9±0.6 | 2.6±1.6 | 1.1±1.3 | 0.5±0.1 | 0.1 |
| IL-4 | 11.5±22.1 | 6.5±3.6 | 7.9±6.6 | 2.4±0.9 | 0.02* |
| IL-10 | 8.5±6.5 | 0 | 11.9±0.1 | 8.2±8.6 | 0.08 |
| TNF-α | 3.6±2.8 | 4.9±3.9 | 1.3±4 | 4.6±5.1 | 0.02* |
| IL12p40 | 15.3±5.8 | 12.5±2.1 | 10.8±5.8 | 14.3±14.2 | 0.3 |
| IL-17 | 0 | 4.7±8.1 | 0 | 0.1±0.3 | 0.2 |
| IL-22 | 49.2±75.5 | 422.9±732.5 | 741.5±1714 | 1474±3581 | 0.8 |
Measured using one-way analysis of variance test comparing the differences in the levels of cytokines between the four groups.
TNF, tumour necrosis factor.
DISCUSSION
The colon contains a large population of diverse bacterial species which consist of ~1012 micro-organisms per gram of colonic content. The bacterial flora is composed of approximately 1000 species of bacteria.17, 18 The relationships between the bacterial flora and the host is complex and involves interactions among the bacterial flora, the local and systemic innate and adaptive immune response and enterocytes, and their secreted products, such as mucus, AMPs and enzymes.
During both HIV19, 20 and HCV21, 22 infections, there is an increase in translocation of microbial products into peripheral circulation. Different mechanisms have been proposed regarding microbial translocation in HIV and HCV infections, including alteration in the integrity of the GI barrier,18, 20 increased GI inflammation with abundant proinflammatory cytokines production,23, 24 dysregulation of T cell subsets,9, 10 and decreased microbial clearance.18
In this study, we examined the colonic tissues and the peripheral blood of 10 healthy controls, and compared them with 30 HIV, HCV and HIV/HCV-coinfected patients. Genetic analysis of the colon tissues of the enrolled groups revealed a remarkable difference in gene expression in colon tissues from the controls compared with patients (table 2 and figure 1). Though subjects with HIV were somewhat younger than other groups, and all of them were male, this is unlikely to be a factor in the observed gene expression outcomes. To further characterise immune-related genes, we analysed the up-regulated and down-regulated genes in HIV and HIV/HCV compared with controls. Compared with controls, approximately 77 and 307 genes were significantly (>1.5 folds) down-regulated in HIV and HIV/HCV, respectively, while 173 and 163 immune-related genes were significantly (>1.5 folds) up-regulated in HIV and HIV/HCV-coinfected patients respectively. Although HCV has been recognised mainly as a hepatotropic virus, our data indicated that HIV/HCV coinfection is associated with dramatic changes in gene expression in the colonic tissues compared with HIV alone. These changes in gene expression may be due to HCV replication in the colon tissues which has been described previously,25–28 or due to modulation of cytokines/chemokine production by remote infection and injury in the liver and elsewhere. Since all the HIV patients were under treatment, we were unable to investigate the effect of antiretroviral treatment or HIV viral RNA levels on LEAP-2 expression. For HCV treatment, no significant effect of anti-HCV treatment was observed on LEAP-2 expression. Further detection of negative strand HCV RNA in colon tissue by in situ hybridisation and immunohistochemistry may be able to differentiate between these two possibilities, and is currently under investigation in our lab.
One of the candidate genes which is expressed by both hepatocytes and enterocytes and showed significant differences (p<0.05) between the levels of expressions in the control and patients is LEAP-2 (figure 2). Immunohistochemical analysis of LEAP-2 showed decreases of LEAP-2 protein expression in the colonic tissues of HIV, HCV and HIV/HCV-coinfected patients compared with the controls which correlate with gene expression levels (figure 2). The differences in genes’ expression were specific for LEAP-2 and not observed in other AMPs such as defensins (see online supplement 2).
To investigate the relationship between LEAP-2 expression and immune activation, the plasma levels of sCD27, which is a marker of T cell activation,13, 29 was measured. CD27 is type II transmembrane glycoprotein, which belongs to the TNF receptor family, and is expressed on the surface of antigen-experienced B cells,30 most T cells,31 and natural killer cells.32 Surface CD27 can be cleaved through the action of matrix metalloproteases33 forming soluble CD27.34 This cleavage may occur in activated T cells after triggering of the T cell receptor complex.35 During HIV-1 infection, plasma levels of sCD27 have been used as an independent marker to monitor the effect of antiretroviral treatment on HIV-1-induced immune activation.13, 29 In the present study, plasma sCD27 was investigated and correlated its level with LEAP-2 gene expression. Our data indicated that there are significant differences in the levels of sCD27 in the plasma of HIV and HIV/HCV-coinfected patients compared with the controls (p=0.04 and p=0.03, respectively), but not with the levels in HCV-infected patients (p=0.19) (figure 3A). The absence of significant differences in sCD27 between HCV-infected patients and controls may be due to the limited effects of HCV alone on immune activation. However, there is significant negative correlation between the levels of sCD27 and LEAP-2 expression (p=0.03, r=−0.44) (figure 3B), which supports the possible role of LEAP-2 expression in prevention of immune activation in HIV and HCV infections.
Immune activation and microbial translocation pathogenesis could be influenced by the levels of proinflammatory cytokines (IL-2, IL-12p40 and TNF-α), Th2 cytokines (IL-4), or regulatory cytokines (IL-10 and IL-17) either in the plasma or the microenvironment of the gut. Therefore, we analysed the cytokines profiles in the plasma as markers of systemic immune activation, and in the supernatant of anti-CD3 stimulated IEL in vitro as surrogate markers for the inflammatory conditions in the microenvironment of the colonic tissues. Our data indicated that TNF-α is the only cytokine which differs significantly, especially in HIV/HCV-infected and HIV-infected patients, in both the plasma levels and the supernatant of stimulated IEL (p=0.03 and p=0.02, respectively) (tables 3 and 4). TNF-α is secreted by activated macrophages and lymphocytes and induces diverse responses, including inflammation and apoptosis.36 Accumulating data indicate that TNF-α is a key player in HIV pathogenesis.36 HIV proteins have been shown to target TNF-α receptor activation pathways, leading both to apoptosis of uninfected T cells and to sustained viral replication in infected T cells. IL-4 which is Th2 and an anti-inflammatory cytokine, is significantly lower in the supernatant of the HIV/HCV groups compared with the other groups (p=0.02) which indicated the dominant status of immune activation in those patients.37–39 The absence of differences in the plasma levels of IL-4 may reflect the dilution of IL-4 with blood plasma and the low levels of systemic secretion of IL-4.
In conclusion, we have shown for the first time a relationship between the AMP, LEAP-2, from enterocytes and the level of immune activation in HIV-infected and HCV-infected patients. Decreased gene expression and production of LEAP-2 represent a potential mechanism for microbial translocation and subsequent immune activation in HIV-infected and HCV-infected patients.
Supplementary Material
Take-home messages.
Immune activation is one of the main features of HIV/HCV infections and it is linked to the disturbance of the gut-associated lymphoid tissue in HIV/HCV.
Plasma sCD27, and tumour necrosis factor-α are correlated with the presence of immune activation in HIV-infected and HIV/HCV-infected patients.
Liver expressed antimicrobial peptide-2 gene and protein expressions by enterocytes were significantly lower in HIV, HCV and HIV/HCV-infected patients compared with controls, and may play a role in microbial translocation and immune activation experienced during HIV and HCV infections
Acknowledgments
We would like to thank the entire participants in this study, particularly the patients. We thank Mrs Diane Daria and Mrs Suzan Sibert for their assistance in enrolling the subjects. We also thank the Cincinnati Children’s Hospital medical Center (CCHMC) Luminex cytokines core facility, the Pathology Core facility at the Digestive Health Center, the Gene Expression Microarray Core, and Bioinformatic services at CCHMC. Also, we thank Dr Jason Blackard and Mrs Susan Rouster for their help in editing the manuscript.
Funding This investigation was supported by Merck Investigator Initiated Studies [IISP #: 38879 (Shata)]; National Institute of Health Grant [K24DK070528 (Sherman)]; and was supported in part by Public Health Service Grant [P30 DK078392 (The Gene Expression Microarray Core Cincinnati Children’s Hospital Medical Center)]; National Institute of Health Grant [NIH P30 DK078392 (Bioinformatics Core of the Digestive Disease Research Core Center in Cincinnati)].
Footnotes
This work was partially presented at the 19th International Symposium of Hepatitis C Virus and Related Viruses, in Venice, Italy, 5–9 October 2012. The manuscript explores a new mechanism involved the role of LEAP-2 secreted by enterocytes in microbial translocation and immune activation in HIV-infected and HCV-infected patients.
Contributors MTMS designed the study, analysed the data and wrote the manuscript. EAA-H: prepared the RNA from samples, did the gene array experiments, and analysed the data for the gene array. She also evaluated the plasma cytokines of the samples and helped in analysing the data and prepared the manuscript. HFH stained the tissue biopsies slides with the appropriate antibodies, examined the slides with confocal laser-scanning microscopy, analysed the data and evaluated the levels of expression of LEAP-2 in tissues. KES: helped in the design of the study, and collecting the biopsies from enrolled subjects. He also helped in analysed the data and writing and editing the manuscript.
Patient consent Obtained.
Ethics approval University of Cincinnati College of Medicine Institutional Review Board.
Provenance and peer review Not commissioned; internally peer reviewed.
Data sharing statement We are happy to share our data with researchers and discuss with them any potential collaborations.
Additional material is published online only. To view please visit the journal online (http://dx.doi.org/10.1136/jclinpath-2013-201581).
References
- 1.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alp S, Skrygan M, Schlottmann R, et al. Expression of beta-defensin 1 and 2 in nasal epithelial cells and alveolar macrophages from HIV-infected patients. Eur J Med Res. 2005;10:1–6. [PubMed] [Google Scholar]
- 3.Mukherjee S, Vaishnava S, Hooper LV. Multi-layered regulation of intestinal antimicrobial defense. Cell Mol Life Sci. 2008;65:3019–27. doi: 10.1007/s00018-008-8182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menard S, Forster V, Lotz M, et al. Developmental switch of intestinal antimicrobial peptide expression. J Exp Med. 2008;205:183–93. doi: 10.1084/jem.20071022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howard A, Townes C, Milona P, et al. Expression and functional analyses of liver expressed antimicrobial peptide-2 (LEAP-2) variant forms in human tissues. Cell Immunol. 2010;261:128–33. doi: 10.1016/j.cellimm.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Krause A, Sillard R, Kleemeier B, et al. Isolation and biochemical characterization of LEAP-2, a novel blood peptide expressed in the liver. Protein Sci. 2003;12:143–52. doi: 10.1110/ps.0213603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nazli A, Chan O, Dobson-Belaire WN, et al. Exposure to HIV-1 directly impairs mucosal epithelial barrier integrity allowing microbial translocation. PLoS Pathog. 2010;6:e1000852. doi: 10.1371/journal.ppat.1000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estes JD, Harris LD, Klatt NR, et al. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. 2010;6:e1001052. doi: 10.1371/journal.ppat.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klatt NR, Estes JD, Sun X, et al. Loss of mucosal CD103+ DCs and IL-17+ and IL-22+ lymphocytes is associated with mucosal damage in SIV infection. Mucosal Immunol. 2012;5:646–57. doi: 10.1038/mi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon SN, Cervasi B, Odorizzi P, et al. Disruption of intestinal CD4+ T cell homeostasis is a key marker of systemic CD4+ T cell activation in HIV-infected individuals. J Immunol. 2010;185:5169–79. doi: 10.4049/jimmunol.1001801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharp TM, Estes MK. An inside job: subversion of the host secretory pathway by intestinal pathogens. Curr Opin Infect Dis. 2010;23:464–9. doi: 10.1097/QCO.0b013e32833dcebd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arijs I, De Hertogh G, Lemaire K, et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PloS One. 2009;4:e7984. doi: 10.1371/journal.pone.0007984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Milito A, Aleman S, Marenzi R, et al. Plasma levels of soluble CD27: a simple marker to monitor immune activation during potent antiretroviral therapy in HIV-1-infected subjects. Clin Exp Immunol. 2002;127:486–94. doi: 10.1046/j.1365-2249.2002.01786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuss IJ, Becker C, Yang Z, et al. Both IL-12p70 and IL-23 are synthesized during active Crohn’s disease and are down-regulated by treatment with anti-IL-12 p40 monoclonal antibody. Inflamm Bowel Dis. 2006;12:9–15. doi: 10.1097/01.mib.0000194183.92671.b6. [DOI] [PubMed] [Google Scholar]
- 15.Henriques ST, Tan CC, Craik DJ, et al. Structural and functional analysis of human liver-expressed antimicrobial peptide 2. Chembiochem. 2010;11:2148–57. doi: 10.1002/cbic.201000400. [DOI] [PubMed] [Google Scholar]
- 16.Hocquellet A, Odaert B, Cabanne C, et al. Structure-activity relationship of human liver-expressed antimicrobial peptide 2. Peptides. 2010;31:58–66. doi: 10.1016/j.peptides.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Klatt NR, Funderburg NT, Brenchley JM. Microbial translocation, immune activation, and HIV disease. Trends Microbiol. 2013;21:6–13. doi: 10.1016/j.tim.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brenchley JM, Douek DC. Microbial translocation across the GI tract. Ann Rev Immunol. 2012;30:149–73. doi: 10.1146/annurev-immunol-020711-075001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brenchley JM, Douek DC. The mucosal barrier and immune activation in HIV pathogenesis. Curr Opin HIV AIDS. 2008;3:356–61. doi: 10.1097/COH.0b013e3282f9ae9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brenchley JM, Price DA, Schacker TW, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–71. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 21.Caradonna L, Mastronardi ML, Magrone T, et al. Biological and clinical significance of endotoxemia in the course of hepatitis C virus infection. Curr Pharm Des. 2002;8:995–1005. doi: 10.2174/1381612024606983. [DOI] [PubMed] [Google Scholar]
- 22.Neuman MG, Sha K, Esguerra R, et al. Inflammation and repair in viral hepatitis C. Dig Dis Sci. 2008;53:1468–87. doi: 10.1007/s10620-007-0047-3. [DOI] [PubMed] [Google Scholar]
- 23.Prasad S, Mingrino R, Kaukinen K, et al. Inflammatory processes have differential effects on claudins 2, 3 and 4 in colonic epithelial cells. Lab Invest. 2005;85:1139–62. doi: 10.1038/labinvest.3700316. [DOI] [PubMed] [Google Scholar]
- 24.Walsh SV, Hopkins AM, Nusrat A. Modulation of tight junction structure and function by cytokines. Adv Drug Deliv Rev. 2000;41:303–13. doi: 10.1016/s0169-409x(00)00048-x. [DOI] [PubMed] [Google Scholar]
- 25.Deforges S, Evlashev A, Perret M, et al. Expression of hepatitis C virus proteins in epithelial intestinal cells in vivo. J Gen Virol. 2004;85(Pt 9):2515–23. doi: 10.1099/vir.0.80071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miglioresi L, Riva E, Antonelli G, et al. Localization of hepatitis C virus in gastrointestinal mucosa: a possible reservoir for relapse. Hepatology. 2003;38:775. doi: 10.1053/jhep.2003.50322. [DOI] [PubMed] [Google Scholar]
- 27.De Vita S, De Re V, Sansonno D, et al. Gastric mucosa as an additional extrahepatic localization of hepatitis C virus: viral detection in gastric low-grade lymphoma associated with autoimmune disease and in chronic gastritis. Hepatology. 2000;31:182–9. doi: 10.1002/hep.510310127. [DOI] [PubMed] [Google Scholar]
- 28.Tursi A, Brandimante G, Chiarelli F, et al. Detection of HCV RNA in gastric mucosa-associated lymphoid tissue by in situ hybridization: evidence of a new extrahepatic localization of HCV with increased risk of gastric malt lymphoma. Am J Gastroenterol. 2002;97:1802–6. doi: 10.1111/j.1572-0241.2002.05848.x. [DOI] [PubMed] [Google Scholar]
- 29.Atlas A, Thanh Ha TT, Lindstrom A, et al. Effects of potent antiretroviral therapy on the immune activation marker soluble CD27 in patients infected with HIV-1 subtypes A-D. J Med Virol. 2004;72:345–51. doi: 10.1002/jmv.20006. [DOI] [PubMed] [Google Scholar]
- 30.Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med. 1998;188:1679–89. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Lier RA, Borst J, Vroom TM, et al. Tissue distribution and biochemical and functional properties of Tp55 (CD27), a novel T cell differentiation antigen. J Immunol. 1987;139:1589–96. [PubMed] [Google Scholar]
- 32.Sugita K, Robertson MJ, Torimoto Y, et al. Participation of the CD27 antigen in the regulation of IL-2-activated human natural killer cells. J Immunol. 1992;149:1199–203. [PubMed] [Google Scholar]
- 33.Kato K, Chu P, Takahashi S, et al. Metalloprotease inhibitors block release of soluble CD27 and enhance the immune stimulatory activity of chronic lymphocytic leukemia cells. Exp Hematol. 2007;35:434–42. doi: 10.1016/j.exphem.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 34.Loenen WA, De Vries E, Gravestein LA, et al. The CD27 membrane receptor, a lymphocyte-specific member of the nerve growth factor receptor family, gives rise to a soluble form by protein processing that does not involve receptor endocytosis. Eur J Immunol. 1992;22:447–55. doi: 10.1002/eji.1830220224. [DOI] [PubMed] [Google Scholar]
- 35.Hintzen RQ, de Jong R, Hack CE, et al. A soluble form of the human T cell differentiation antigen CD27 is released after triggering of the TCR/CD3 complex. J Immunol. 1991;147:29–35. [PubMed] [Google Scholar]
- 36.Herbein G, Khan KA. Is HIV infection a TNF receptor signalling-driven disease? Trends Immunol. 2008;29:61–7. doi: 10.1016/j.it.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 37.Li JR, Gong RY, Tian KL, et al. Study on the blood-borne virus co-infection and T lymphocyte subset among intravenous drug users. World J Gastroenterol. 2007;13:2357–62. doi: 10.3748/wjg.v13.i16.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antonucci G, Goletti D, Lanini S, et al. HIV/HCV co-infection: putting the pieces of the puzzle together. Cell Death Differ. 2003;10(Suppl 1):S25–26. doi: 10.1038/sj.cdd.4401164. [DOI] [PubMed] [Google Scholar]
- 39.Page EE, Nelson M, Kelleher P. HIV and hepatitis C coinfection: pathogenesis and microbial translocation. Curr Opin HIV AIDS. 2011;6:472–7. doi: 10.1097/COH.0b013e32834bbc71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.