Abstract
The reprogramming of somatic cells into induced pluripotent stem (iPS) cells upon overexpression of the transcription factors Oct4, Sox2, Klf4 and cMyc is extremely inefficient. It has been assumed that the somatic differentiation state provides a barrier for efficient reprogramming; however, direct evidence for this notion is lacking. Here, we have tested the susceptibilities of hematopoietic cells at different stages of differentiation to be reprogrammed into iPS cells. Surprisingly, hematopoietic stem and progenitor cells give rise to iPS cells up to 300 times more efficiently compared with terminally differentiated B and T cells, yielding reprogramming efficiencies of up to 28%. Our data provide evidence that the differentiation stage of the starting cell has a critical influence on the efficiency of reprogramming into iPS cells. Moreover, we identify adult hematopoietic progenitors as an attractive cell type for applications of iPS technology in research and therapy.
Introduction
Transcription factor-induced reprogramming of somatic cells into iPS cells has been achieved in mouse 1–4, rat 5,6, monkey 7 and human 8–11. It involves introducing the four transcription factors Oct4, Sox2, cMyc and Klf4, or the alternative set Oct4, Sox2, Lin28 and Nanog, into cells by retrovirus-mediated gene delivery, giving rise to pluripotent cells that are highly similar to embryonic stem (ES) cells. A major bottleneck of induced pluripotency in research and therapy, however, is its extremely low efficiency; upon infection of diverse primary adult cells such as fibroblasts, keratinocytes, liver cells and pancreatic β cells with retro- or lentiviruses expressing Oct4, Sox2, cMyc and Klf4, only between 0.01% and 0.2% of transduced cells give rise to iPS cells 12. Reprogramming efficiencies are even lower when using transient gene delivery approaches 13–15. The development of so-called “secondary systems” has enabled the generation of iPS cells at higher efficiencies and without the need for direct infection. In this approach, “primary” iPS cells that have been generated with doxycycline-inducible versions of the reprogramming factors are first differentiated into somatic cells; exposure of these cells to doxycycline then results in the homogeneous re-expression of the factors and the generation of “secondary” iPS cells 16–18.
During reprogramming, the epigenetic state of a somatic cell has to be reset to a state compatible with pluripotentiality 19. It has been assumed, therefore, that the differentiation state of the starting cell might influence the efficiency of reprogramming by providing a barrier for efficient epigenetic remodeling of the genome. Consistent with this notion, the treatment of differentiated cells undergoing reprogramming with small compounds that inhibit DNA or histone methylation as well as histone acetylation, give rise to iPS cells more efficiently 6,20–23. In a potential therapeutic setting, however, small compounds may have unpredictable long-term consequences. Identifying cell types that are most amenable to reprogramming would provide an alternative to the use of chemicals and can teach us about the molecular barriers underlying nuclear reprogramming.
Somatic stem and progenitor cells in the adult share some features with pluripotent stem cells, such as the capacity to differentiate into different cell types and certain transcriptional regulators 24,25. It is conceiveable, therefore, that the genome of adult progenitor cells is more amenable to reprogramming than that of differentiated cells. Previous results from nuclear transfer experiments indeed indicated that neural and keratinocyte progenitors give rise to cloned embryos or mice more efficiently than differentiated cell types 26,27. Surprisingly, in the hematopoietic system, an inverse correlation between the differentiation stage of donor cells and cloning efficiency has been reported 28,29; differentiated granulocytes were found to be more susceptible to reprogramming into cloned blastocysts than hematopoietic progenitor and stem cells. These experiments have recently been challenged, however, as scoring the number of cloned preimplantation embryos can be misleading and may not reflect true reprogramming potential 30.
The role of the differentiation stage of the starting cell during transcription factor-mediated reprogramming has not yet been rigorously addressed. A recent study suggested that neural progenitors give rise to iPS cells at high efficiency using direct viral infection (3.6%) 31. In contrast, two independent reports found no major differences or even a lower efficiency, respectively, in the iPS formation efficiency of neural progenitors compared with fibroblasts 32,33. Notably, the expression of the four reprogramming factors in terminally differentiated B lymphocytes was insufficient to generate iPS cells and required the viral introduction of an additional transcription factor, CEBPα 34, while mature pancreatic β cells converted into iPS cells by the ectopic expression of the four reprogramming factors alone 35. Thus, the question remains if the differentiation stage of the starting cell has an influence on the efficiency of reprogramming into iPS cells.
Here, we addressed this question by generating iPS cells from prospectively isolated hematopoietic stem cells (HSCs), myeloid and lymphoid progenitors and several mature blood cell types using a genetically homogeneous “secondary system” to express the four reprogramming factors. Our results show that all hematopoietic cell types tested, including terminally differentiated lymphocytes, can be reprogrammed into iPS cells by the four transcription factors alone. Moreover, we demonstrate that the differentiation stage of cells has a strong impact on the efficiency and kinetics of reprogramming.
Results
Development of a “secondary system” to generate iPS cells from the hematopoietic lineage
In order to determine the reprogramming potentials of different blood cells, we generated a “secondary system”, which allows expression of reprogramming factors in a controllable fashion in a genetically homogenous population of cells 16–18. Specifically, we derived iPS cells from neonatal tail-tip fibroblasts carrying an Oct4-GFP reporter 36 as well as the ROSA-rtTA transactivator 37 (Figure 1a). Infection of such fibroblasts with lentiviruses expressing Oct4, Sox2, Klf4 and cMyc under the control of doxycycline-inducible promoters 38 gave rise to primary iPS (1° iPS) cells that activated Oct4-GFP and could be propagated in the absence of doxycycline, indicating reprogramming into a transgene-independent pluripotent state (data not shown).
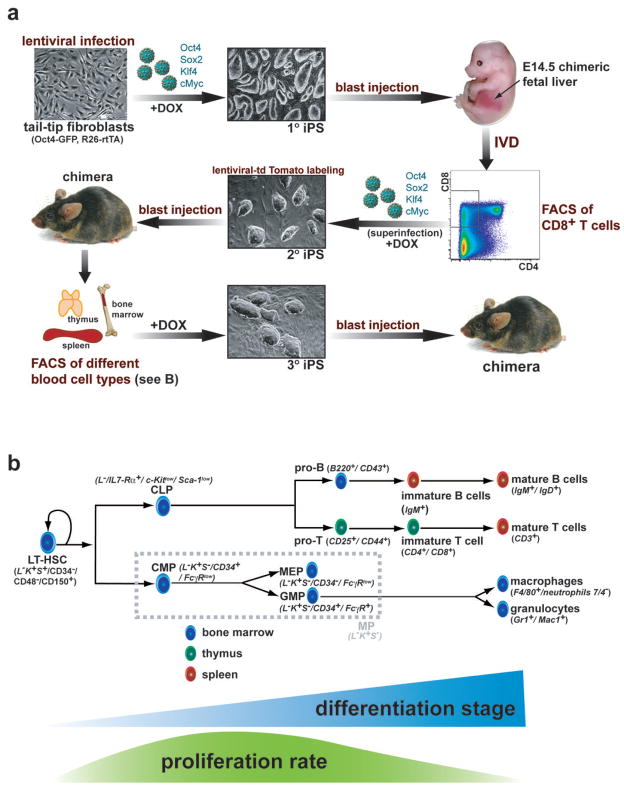
Figure 1. Development of a transgenic system for inducible expression of Oct4, Sox2, Klf4 and cMyc in the murine hematopoietic system.
(a) Strategy used to reprogram different cell types of the hematopoetic system. Tail tip fibroblasts heterozygous for the ROSA26-M2rtTA and Oct4-GFP knock-in alleles were infected with four different doxcycline (dox) inducible lentiviruses encoding Oct4, Sox2, Klf4 and cMyc. Resulting primary (1°) iPS cells were injected into murine blastocytes and fetal liver was isolated from E14.5 embryos, in vitro differentiated (IVD) by stromal co-culture and sorted for emerging CD8 positive (+) T cells by fluorescent-activated cell sorting (FACS). CD8+ T cells were optionally superinfected with the four dox-inducible lentiviruses to ensure strongest possible transgene expression and subsequently cultured on dox for 12 days. Resulting secondary (2°) iPS cells were labeled with a lentivirus constitutively expressing tdTomato and injected into blastocytes to generate adult chimeras. tdTomato+ hematopoietic cells at different stages of differentiation (see Figure 1B) were isolated by FACS from spleen, thymus and bone marrow (BM), induced with dox and resulting tertiary (3°) iPS cell lines were used for molecular and functional analyses including rearrangement and pluripotency assays. (b) Scheme of hematopoiesis with emphasis on the cell populations used in this study. Blue triangle indicates progressive differentiation from LT-HSCs over progenitors to terminally differentiated cells. Green curve illustrates proliferation rate, which is highest in progenitors and lowest in quiescent LT-HSCs and most terminally differentiated cells. LT-HSCs, long-term hematopoietic stem cells; MPs, myeloid progenitors (consisting of CMP, common myeloid progenitors, MEP, megakaryocyte/erythrocyte progenitors and GMPs, granulocyte/macrophage progenitors); CLP, common lymphoid progenitors. Surface markers used for FACS purification are displayed in italic script in brackets, L−K+S+, lin c-Kit+Sca-1+.
To assess if defined mature hematopoietic cells derived from these iPS cells can be reprogrammed into secondary iPS cells, we attempted differentiation into T cells, whose maturation stage can be prospectively identified by surface markers and retrospectively by T cell receptor rearrangement analyses. We first generated E14.5 chimeric fetuses by blastocyst injection of 1° iPS cells and isolated fetal liver cells known to contain hematopoietic progenitors (Figure 1a). Co-culture of fetal liver cells with OP9-DL1 stromal feeder cells was performed to induce T cell differentiation 39. After 13 days, mature CD8+ T cells emerged, which were FACS-sorted and explanted on regular irradiated MEFs in the presence of doxycycline to reactivate the viral transgenes. Since previous attempts to reprogram mature lymphocytes with four factors were unsuccesful, we subjected cultured cells to another round of viral infection with the four factors to ensure strongest possible transgene expression. Secondary iPS (2° iPS) colonies became detectable after 12 days that reactivated Oct4-GFP and could be passaged in the absence of doxycycline (Figure 1a and data not shown). Of note, viral superinfection was not essential for generating iPS cells from mature CD8+ T cells as we could also derive them without an additional round of infection (Supplementary Figure 1). These results show that fetal liver-derived CD8+ T cells remain amenable to reprogramming into iPS cells by four factors alone. We will refer to these T cell-derived iPS cells as CD8-iPS.
iPS cells generated from adult mature B and T lymphocytes by only four factors
We next wanted to determine if adult-derived lymphocytes remain equally amenable to reprogramming into iPS cells by the four reprogramming factors. To this end, we labeled the CD8-iPS cell clone with a lentivirus constitutively expressing tdTomato 14 and injected cells into blastocysts to produce chimeric mice (Figure 1a). CD8-iPS cells gave rise to high-degree chimeras as shown by widespread red fluorescence of newborn pups and broad coat color chimerism of surviving adult mice (data not shown).
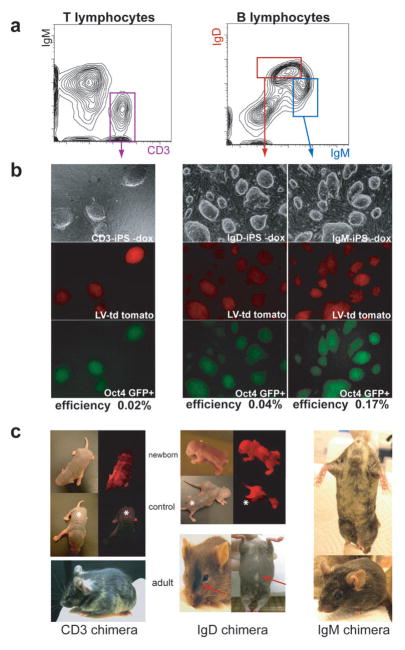
We first attempted to reprogram mature T cells. Mature peripheral splenic T lymphocytes from CD8-iPS chimeric mice were isolated using the surface marker CD3 (Figures 1b and 2a). Upon plating of CD3+ T lymphocytes on feeder cells in ES medium in the presence of doxycycline and the T cell cytokines interleukin-2 (IL-2) and concanavalin A (ConA) (see materials and methods for details), Oct4-GFP expressing tertiary iPS colonies (3° iPS) emerged after 12 days in culture and could be passaged upon discontinuation of doxycycline (Figure 2b, left column); these cells will be referred to as CD3-iPS cells. The efficiency of CD3-iPS formation was roughly 0.02%, which is lower than that observed for heterogeneous tail fibroblasts isolated from the same animal (0.74%).
Figure 2. Generation and characterization of iPS cells derived from mature B and T lymphocytes by four factors.
(a) Sorting strategy of tdTomato+ (gate not shown) mature CD3+ splenic T lymphocytes, immature IgM+ and mature IgMlow IgDhigh B lymphocytes from 10 week-old CD8-iPS chimeras by FACS. (b) ES-like morphology of tdTomato+ Oct4-GFP+ iPS colonies derived from mature B and T cells. (c) (top) Viable newborn chimeras derived from CD3-iPS and IgD-iPS cell lines show red fluorescence originating from the labeled iPS cell lines. Shown are tdTomato+ chimeric pups and non-chimeric littermates (marked by asterisks) under regular light (left) and UV light (right). (bottom) Adult chimeras derived from CD3-T iPS, IgM-B and IgD-B cell iPS show obvious coat color chimerism.
Since previous attempts to generate iPS cells from mature B lymphocytes with only four factors were unsuccessful 34, we next assessed if immature, splenic B cells can give rise to iPS cells. Indeed, plating of IgM+ immature B cells (Figures 1b and 2a) on feeder cells in the presence of doxycycline and the B cell stimulants CpG and lipopolysaccharide (LPS) gave rise to Oct4-GFP positive iPS (“IgM-iPS”) colonies at an average efficiency of 0.17% (Figure 2b, right column). Remarkably, even terminally differentiated B lymphocytes from spleen or lymph nodes, identified by IgM and IgD positivity (IgMlow IgDhigh) (Figures 1b and 2a) and cultured under the same conditions as IgM+ cells, gave rise to Oct4-GFP+ iPS colonies (“IgD-iPS”) at an average frequency of 0.04% (Figure 2b, center column). To exclude the possibility that secondary cells, which have already gone through one round of reprogramming, are more susceptible to reprogramming than primary cells, we also generated iPS cells from terminally differentiated IgMlow IgDhigh B cells isolated from mice carrying the ROSA-rtTA transactivator and directly infected with a polycistronic doxycycline-inducible lentivirus expressing Oct4, Sox2, Klf4 and cMyc 40 (data not shown). Together, these results indicate that adult, terminally differentiated B and T lymphocytes remain amenable to reprogramming into pluripotent stem cells by only four transcription factors. All iPS cell lines gave rise to well-differentiated teratomas consisting of cell types from all three germ layers (Supplementary Figure 2a) and produced chimeric neonatal and adult mice (Figure 2c), thus demonstrating their pluripotency. Moreover, a CD8-iPS derived high-degree chimera gave rise to germline offspring at 100% efficiency when mated with wild-type females (Supplementary Figure 3a).
A previous attempt to reprogram mature B cells required the overexpression of the transcription factor C/EBPα in addition to the four factors to produce iPS cells 34. To test if C/EBPα expression could further enhance the low reprogramming efficiency of B cells in our system, we infected tdTomato positive immature IgM+ and mature IgMlow IgDhigh B cells from a CD8-iPS chimera with a retrovirus expressing C/EBPα as well as a human CD4 epitope to allow isolation of C/EBPα expressing cells by FACS (Supplementary Figure 4a). We observed an up to 20-fold increase in the number of Oct4-GFP positive colonies in IgM+ or IgMlow IgDhigh B cell cultures overexpressing C/EBPα compared with untransduced cells or cells infected with an empty control virus, indicating that the expression of C/EBPα indeed enhances the overall reprogramming efficiency of B lymphocytes (Supplementary Figure 4b and c). In addition, we noticed that C/EBPα-expressing cultures formed faster, grew into larger colonies, downregulated the B cell marker B220 and upregulated Oct4-GFP sooner compared with untransduced cells (Supplementary Figure 4d, e).
Lymphocyte-derived iPS cells give rise to a monoclonal immune system
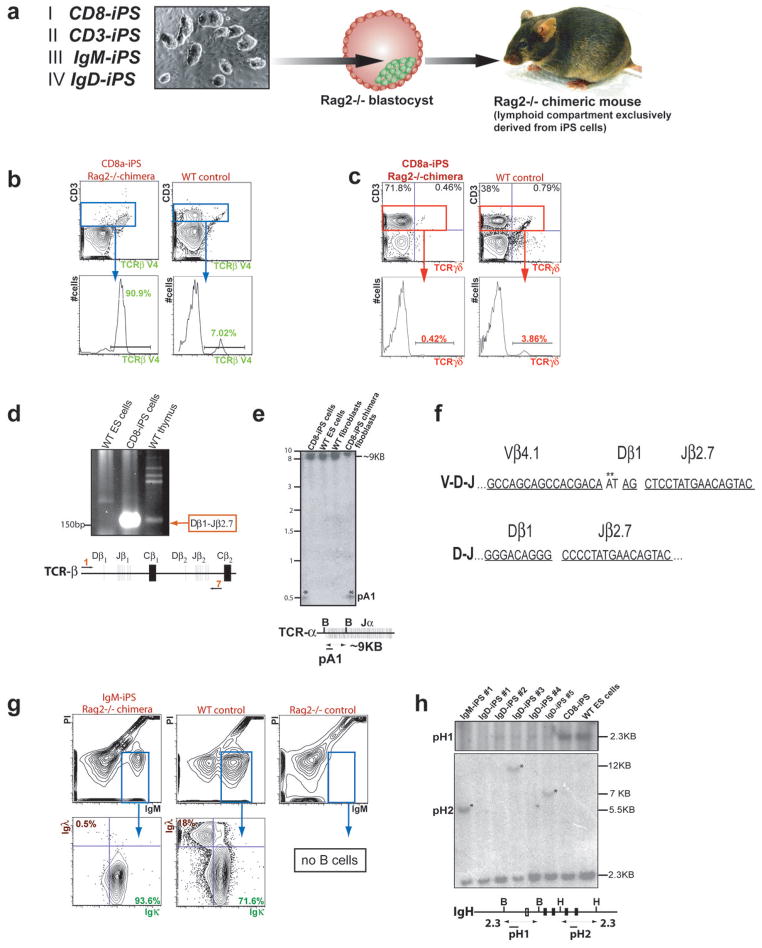
Pluripotent cells derived from terminally differentiated lymphocytes by nuclear transfer have previously been shown to give rise to a monoclonal immune system 41. In order to determine if lymphocyte-derived iPS cells also produce a monoclonal lymphoid compartment, we injected IgM-iPS, IgD-iPS, CD3-iPS and CD8-iPS cells into Rag2-deficient blastocysts (Figure 3a). Rag2 deficiency in mice results in a complete absence of all mature B and T lymphocytes and thus, any B and T cells detected in iPS cell chimeras are exclusively derived from the injected iPS cells 42. Rag2−/− chimeras produced with the CD8-iPS clone had normal spleen, thymus and lymph nodes, indicating that iPS cells can entirely reconstitute different hematopoietic organs. Importantly, T cells expressed only one out of 25 possible viable TCRβ receptors (Vβ4.1) (Chen et al., 2001), demonstrating that the donor T cell had undergone a functional rearrangement that is homogeneously expressed on T cells of Rag2−/− chimeric mice, giving rise to an apparently monoclonal T cell compartment (Figure 3b). Germline offspring from CD8-iPS chimeras also expressed the Vβ4.1 rearrangement on the vast majority of their T cells (Supplementary Figure 3b). Consistent with this, we saw an absence of TCRγ/δ positive T cells in these mice (Figure 3c). In addition to the functionally rearranged TCRβ allele, CD8-iPS cells contained a partially rearranged TCRβ allele comprising a Dβ1 to Jβ2.7 rearrangement (Figure 3d) and Southern blot analysis confirmed that the TCRα locus had also been rearranged (Figure 3e). Sequence analyses of both rearranged TCR alleles from genomic DNA of CD8-iPS cells corroborated the finding that these cells had been derived from a functional mature T cell (Figure 3f).
Figure 3. Monoclonal immune system in Rag2−/− chimeras produced with lymphocyte-derived iPS cells.
(a) Scheme of Rag2 complementation assay to produce chimeras whose lymphoid compartment is entirely derived from iPS cells. (b) Expression of the Vβ4.1 T cell receptor (TCR) on the majority of CD3+ cells derived from CD8-iPS chimeras (90.9% compared to 7.02% in wild-type (WT) controls). C57BL/6xDBA/2 F1 (BDF1) mice served as WT controls. (c) CD3+ splenocytes from CD8-iPS chimeras almost lack TCRγδ-expressing T cells compared to WT controls (0.42 % vs 3.86 %, respectively). (d) Detection of TCRβ D-J rearrangement by PCR on DNA isolated from the CD8-iPS cell line. D-J locus and primer pair (1,7) are shown below the PCR gel. (e) (top) V-D-J-rearrangement band (marked by asterisk) at the TCRα locus as determined by Southern blot analysis of BamHI (B) digested genomic DNA isolated from CD8a-iPS cells and purified tail tip cells from CD8-iPS chimeras. DNA from V6.5 wild type ES cells and BDF1 WT tail tip cells served as controls. Probe (pA1) and fragment length are indicated on the side. (bottom) Schematic map of the TCRα locus including all relevant restriction sites, fragment length (in kilo bases, KB) and the Southern blot probe (maps not drawn to scale). (f) Sequenced joints of V-D-J and D-J rearrangements of both TCRβ alleles of the CD8-iPS cell line as determined by genomic PCR with specific primers. Single elements are underlined and labeled. Asterisks mark P/N nucleotides not encoded in the germline. Results were identical in 10 different bacterial clones. (g) Detection of Igλ and Igκ light chains on IgM+ splenocytes of IgM-iPS Rag2−/− chimeras. Most iPS cell-derived IgM+ B cells express Ig kappa light chain compared with WT control (93.6 % vs 71.6 % WT, respectively), but lack Igλ chain expression (0.5 % vs 18 % WT). Note the complete absence of IgM+ B cells in Rag2−/− control spleen. Representative graphs of 2 experiments are shown. (h) (top) Southern blot analysis on genomic DNA from different IgD-iPS cell lines to detect rearrangements at the immunoglobulin heavy chain (IgH) locus. The following enzyme/probe combinations were used: BamHI (B)/pH1 and HindIII (H)/pH2. (bottom) Schematic map of the IgH locus including all relevant restriction sites, fragment lengths, and Southern blot probes. Black bars denote JH1 to JH4 elements.
Rag2−/− chimeric animals produced from IgM-iPS cells also contained a monoclonal T cell compartment as they were originally derived from CD8-iPS cells (data not shown). In addition, however, these mice contained an apparently monoclonal B cell compartment as suggested by the absence of Igλ and a stronger bias towards Igκ light chains on the great majority of peripheral B cells (Figure 3g). Accordingly, we detected the presence of rearrangement bands at the imunoglobulin heavy chain locus by Southern blot analysis of IgM-iPS cell DNA (Figure 3h). Collectively, these results provide unambiguous genetic and phenotypic evidence for the derivation of iPS cells from terminally differentiated B and T cells by four factors.
High efficiency of iPS cell formation from various lymphoid progenitors
Our observation that IgM+ immature B cells are more efficiently reprogrammed into iPS cells than mature IgMlow IgDhigh B cells (Figure 2b) prompted us to test if more primitive hematopoietic cell types are even more amenable to reprogramming. To address this question we sorted several progenitors from the bone marrow and thymus of chimeras produced by injection of the CD8-iPS clone described above into wild-type blastocysts.
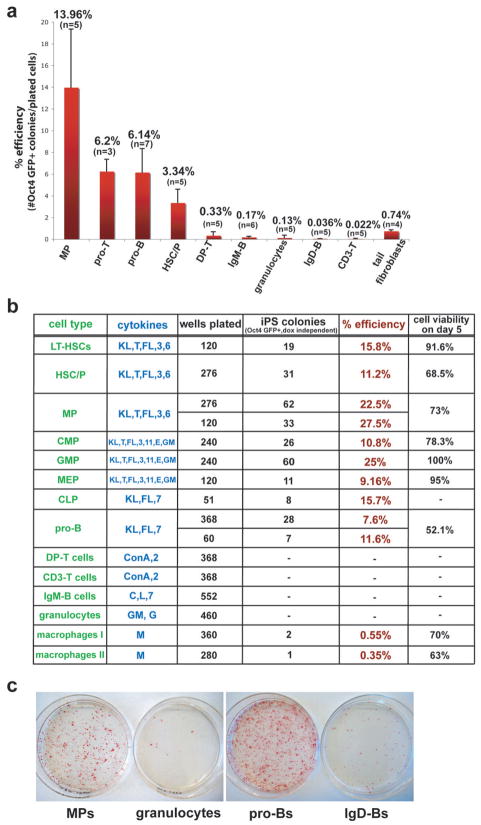
We first determined the efficiencies of deriving iPS cells from bone marrow-derived progenitor (pro-) B and thymic progenitor (pro-) T cells, which are the immediate precursors of B and T cells and can be isolated based on the surface marker combinations B220+ CD43+ for pro-B cells and CD4− CD8− CD44+ CD25+ for pro-T cells (also termed DN2 stage of T cell development) (Figure 1b and Supplementary Figure 2b). Indeed, pro-T cells gave rise to iPS cells at a frequency of ~6.2%, which is almost 20-fold higher than that of immature CD4/CD8-double positive T (DP-T) cells (0.3%) and over 300-fold higher compared with that of mature peripheral T cells (0.02%) (Figure 4a). Likewise, the derivation of iPS cells from pro-B cells was ~6.1%, a 35-fold increase over immature IgM+ B cells (0.17%) and a 150-fold increase over mature IgMlow IgDhigh B cells (0.04%) (Figure 4a). Our results show that at least in these two major lymphoid lineages, cells progressively lose the potential to give rise to iPS cells with increasing differentiation stage.
Figure 4. Reprogramming potentials of different hematopoietic cell types into iPS cells.
(a) Average reprogramming efficiencies into iPS cells of different hematopoietic cell types explanted on 10cm dishes on feeder cells in the presence of ES medium supplemented with dox and cytokines. Efficiencies were determined by dividing the number of Oct4-GFP+ colonies that grew in the absence of doxycycline by the number of seeded cells (for details see material and method section). Numbers above each bar represent the mean value for that sample. Bars represent mean ± standard deviation (SD). “n” denotes the number of independent experiments for that particular cell type. Note that the progenitors from different hematopoietic lineages show higher reprogramming potentials compared with any differentiated cell type shown. See figure 1B and legend for details of cell types. (b) Reprogramming efficiencies of different cell types after single cell sorting into 96-well plates. Reprogramming efficiencies were determined by counting Oct4 GFP+ colonies at day 18, 3 days after doxycycline withdrawal. Cell viabilities of individual populations were determined by scoring uninduced cells sorted in 96-well or terasaki plates. “Macrophage I” denotes macrophages directly sorted from BM into 96-well plates while “Macrophage II” denotes macrophages arising from total BM cultures for 5 days in ES medium with M-CSF. KL, Kit-ligand; T, TPO; FL, Flt3-ligand; E, EPO; GM, GM-CSF; G, G-CSF; M, M-CSF; C, CpG; L, LPS; 3, IL-3; 6, IL-6; 7, IL-7; 11, IL-11. (c) AP staining of iPS-like colonies obtained from selected progenitors and mature cell types. Bone marrow MPs (1.5×10^3 cells per 10cm dish), proBs (2×10^3 cells per 10cm dish), granulocytes (1.5×10^3 cells per 10cm dish) and spleen IgD+ mature B cells (1×10^6 cells per 10cm dish) were plated on feeder cells in ES cell media supplemented with dox and cytokines. After 12 days, dox was removed and plates were stained for AP activity at day 14.
To exclude the possibility that differences in the expression levels of the viral transgenes between different cell populations may account for the observed differences in reprogramming efficiency, we compared the expression levels of the four doxycycline-induced viral transgenes between pro-B cells and IgMlow IgDhigh -B cells, which showed a 150-fold difference in reprogramming efficiency (Figure 4a). No significant differences in the pattern of transgene reactivation between these cell types were observed (Supplementary Figure 5a).
Since most hematopoietic cells initially grew in suspension culture, some reprogrammed colonies cells might have originated from identical founder cells, which spread and formed satellite colonies on the plate. To rule out this possibility, we sorted single mature T and B cells (spleen), pro-T cells, pro-B cells as well as their common precursor cells, common lymphoid progenitors (CLPs) (bone marrow), into single wells of multiple 96 well plates and counted the number of emerging Oct4-GFP positive colonies after 18 days, 3 days following dox withdrawal. While the efficiency of reprogramming T and B cells was too low to be detected in this experiment (200–500 wells analyzed per cell type), pro-B cells gave rise to iPS cells at a similar frequency (7.6–11.6%) as when plated on 10cm plates (3.1–8.8%). Interestingly, the more primitive CLPs yielded colonies at an efficiency of 15.7%, which is higher than that observed for pro-B and pro-T cells (Figure 4b).
HSCs and myeloid progenitors produce iPS cells more efficiently than mature granulocytes and macrophages
We next evaluated if our observations in the lymphoid lineage could be extended to other cell types of the hematopoietic system and thus determined if myleoid progenitors give rise to iPS cells more efficiently than mature granulocytes and macrophages. Specifically, we isolated myeloid progenitor cells (MPs) from CD8-iPS chimeras using the surface marker combination, lin−, c-Kit+, Sca-1− (L−K+S−) (Figure 1b and Supplementary Figure 2b) as well as mature granulocytes using the Gr-1+ Mac-1+ surface antigens and macrophages, isolated by the F4/80+ Neutrophil 7/4− marker combination, and plated defined numbers of cells on feeders in the presence of doxycycline. MPs gave rise to iPS cells at an average frequency of ~14% (Figure 4a) and single cell sorting into 96-well plates yielded an even higher efficiency of up to 27.5% (Figure 4b). MPs are a heterogeneous population of cells comprising common myeloid progenitors (CMPs) and derivative megakaryocyte/erythrocyte progenitors (MEPs) and granulocyte/macrophage progenitors (GMPs) (Figure 1b). Single cell sorting of CMPs, GMPs and MEPs into 96-well plates on feeder cells in ES medium supplemented with doxycycline and cytokines (see Figure 4b) gave rise to iPS cells at efficiencies of 10.8%, 25% and 9.2%, respectively, demonstrating that each progenitor cell type within the MP population has a high reprogramming potential. In contrast, mature granulocytes and macrophages gave rise to iPS cells at frequencies of only 0.13% (>200-fold difference to MPs) and 0.45%, (>60-fold difference to MPs), respectively. We obtained similar results when MPs and granulocytes from ROSA26-rtTA/Oct4-GFP animals were directly infected with concentrated lentivirus expressing Oct4, Sox2, Klf4 and cMyc from a polycistronic construct 40, thus excluding the possibility that the observed differences were specific to the secondary system used (Supplementary Figure 5b). The slightly lower efficiencies of the whole-plate counting approach over the single cell sorting approach are likely due to the conservative scoring approach; we counted only large iPS colonies that appeared on the 10cm plate while small colonies were ignored as they could have been satellite colonies (see materials and methods).
No significant differences were observed when studying the reprogramming potential of a mixed, more primitive population of cells containing HSCs and progenitor cells (HSC/Ps), identified by lin− c-Kit+ Sca1+ (L−K+S+) surface markers (Supplementary Figure 2b). iPS cells from HSC/Ps were generated at efficiencies of 3.3% and 11.2% when explanted on whole plates or 96-well dishes, respectively, in ES medium supplemented with doxycycline and the cytokines Kit-ligand (KL), TPO, Flt3-ligand (FL), IL-3, IL-6 (Figure 4a–c). Similarly, pure HSCs, isolated with the most stringent marker combination available (L−K+S+, CD48−, CD150+, CD34−) 43,44 and grown under the same culture conditions gave rise to iPS colonies at a comparable efficiency (15.8%) (Figure 4b). Both MP- and HSC/P-derived iPS cells produced differentiated teratomas, indicating their pluripotency (Supplementary Figure 2a). Together, these data indicate that immature cell populations (HSCs, HSC/Ps and MPs) in general give rise to iPS cells at significantly higher efficiencies than terminally differentiated cell types.
To test if human hematopoietic progenitors are amenable to reprogramming into iPS cells, we infected CD34+ cord blood progenitors with a human polycistronic vector expressing Oct4, Sox2, Klf4 and cMyc under doxycycline control as well as with a lentiviral construct expressing rtTA (Supplementary Figure 6). iPS colonies emerged after roughly 12 days, which could be expanded in the absence of doxycycline, stained positive for the pluripotency markers Tra1-81 and Sox2, differentiated into AFP+ endodermal and Tuj1+ ectodermal derivatives within embryoid bodies and gave rise to teratomas consisting of ectodermal, mesodermal and endodermal derivatives. This experiments provides a proof-of-principle for the derivation of iPS cells from human cord blood progenitors.
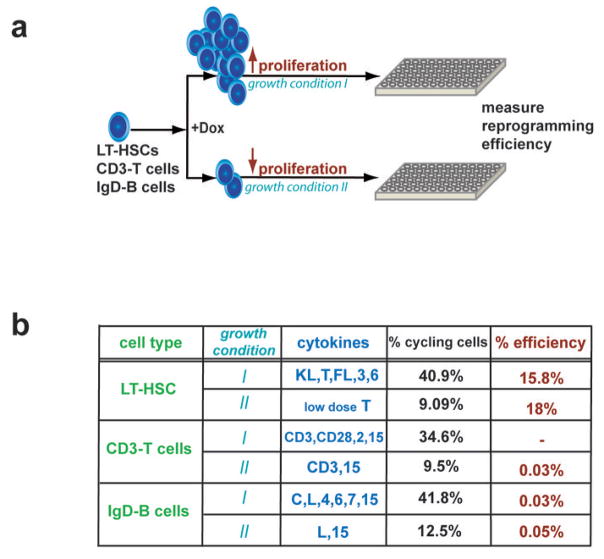
Reprogramming potential correlates with differentiation stage, not with proliferation rate
Proliferation has been assumed to be critical for successful reprogramming 45. HSCs and differentiated blood cells are in a low proliferative or postmitotic state, respectively, whereas progenitor cells are highly proliferative 44 (Figure 1b, Supplementary Figure 7 and data not shown). To directly test if differentiation stage or the actual proliferation rate of a given cell type affect reprogramming potential, we evaluated the reprogramming susceptibilities of HSCs and mature B and T cells grown under conditions that either slow or induce proliferation (Figure 5a). Growth of HSCs in low dose of TPO (0.5ng/ml) kept the majority of cells in a quiescent undifferentiated state 46 while exposure to the full cytokine cocktail (KL, TPO, IL-3, IL-6, Flt3L) resulted in the induction of vigorous proliferation (Figure 5b). No differences in the potentials to produce iPS cells were observed under these two growth conditions, suggesting that the differentiation stage rather than the actual proliferation rate of HSCs influences reprogramming. Likewise, the reprogramming potentials of B and T cells cultured under conditions that promote proliferation (B cells: CpG, IL-4, IL-6, IL-7, IL-15, LPS; T cells: α-CD3, α-CD28, IL-15, IL-2) or slow proliferation (B cells: LPS, IL-15; T cells: α-CD3, IL-15) were comparable. Together, these data indicate that the differentiation stage of immature and mature hematopoietic cells rather than their actual proliferation rate are critical parameters influencing reprogramming potential.
Figure 5. Effect of proliferation rate of hematopoietic cells on reprogramming potential.
(a) Experimental design: HSCs, B and T cells were grown under either proliferation-inducing or – survival-promoting conditions at the time of transgene expression. The formation of iPS colonies was scored 18 days later, 3 days after doxycycline discontinuation. (b) Table summarizing the different growth factor conditions for HSCs, B and T cells, the resultant fraction of cycling cells (measured by Hoechst or PI staining) and efficiency of iPS formation. Note that reprogramming potential correlates with differentiation stage but not with proliferation rate. The TPO concentration differed in the high (20ng/ml) and low (0.5ng/ml) proliferation condition. Efficiencies of HSCs were determined by single-cell sorting into 96-wells into multiple 96-well plates. Efficiencies of B and T cells were assessed by plating 1×10^5 cells into 12-well dishes in duplicates or triplicates.
KL, Kit-ligand; T, TPO; FL, Flt3-ligand; C, CpG; L, LPS; CD3, αCD3 antibody; CD28, αCD28 antibody; 3, IL-3; 6, IL-6; 7, IL-7; 11, IL-11; 15, IL-15.
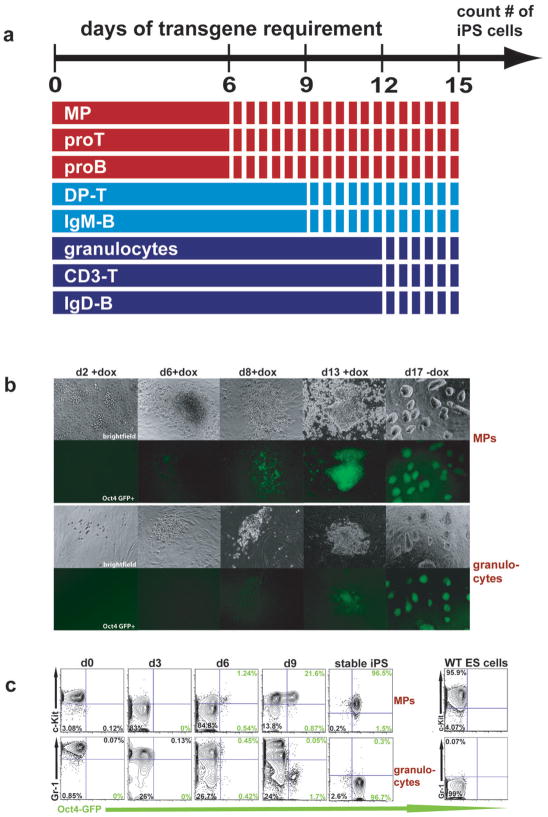
Progenitors reprogram faster than differentiated cells
We next wanted to test if differences in reprogramming efficiencies manifest as differences in reprogramming kinetics. We first evaluated if progenitor-derived iPS cells become independent of the viral transgenes sooner than differentiated cells by withdrawing doxycycline from the individual cultures at 6, 9 and 12 days after induction and scoring for Oct4-GFP positive colonies at day 15. MPs, pro-B cells and pro-T cells gave rise to stable iPS colonies after only 6 days of transgene expression whereas mature granulocytes, B cells and T cells required 12 days to become stably reprogrammed into a pluripotent state. Immature B (IgM+) and DP-T (CD4+/CD8+) cells required 9 days of transgene expression (Figure 6a), suggesting a direct link between differentiation stage and the length of transgene requirement.
Figure 6. Transgene requirement and reprogramming kinetics in progenitors and mature cells.
(a) Temporal requirement for viral transgene expression in individual cell populations undergoing reprogramming. Doxycycline was added to each cultured cell type (solid lines) and withdrawn at days 6, 9 and 12, followed by growth in regular ES cell medium (dotted lines) until day 15 when Oct4-GFP positive colonies were scored. Note that all progenitors (MPs, Pro-Ts, Pro-Bs) are transgene independent and develop into stabe iPS lines by day 6 while more committed DP-thymocytes and IgM-splenocytes become independent of transgene expression by day 9 and mature granulocytes, IgD-B cells and CD3-T cells by day 12. (b) Time course of morphological changes and Oct4-GFP expression in MPs and granulocytes undergoing reprogramming. Note appearance of weak Oct4-GFP positive cells by day 6 in MP cultures compared with day 8 in granulocytes. (c) Time course of surface marker expression and Oct4-GFP activity in MPs and granulocytes undergoing reprogramming by FACS analysis. Note that MPs reactivate Oct4-GFP sooner (day 6) than granulocytes (day 9). Oct4-GFP+ cells emerge from within the c-Kit+ population. Granulocytes gradually downregulate Gr-1 before activating Oct4-GFP within the Gr-1− population. Representative graphs of 3 experiments are shown. Numbers shown in quadrants are percentages of gated populations.
Next, we followed the temporal changes in surface marker expression on MPs and granulocytes undergoing reprogramming (Figure 6b, c). Consistent with results from the requirements of transgene expression, explanted MPs gave rise to ES-like colonies containing Oct4-GFP+ cells as early as 6 days after transgene induction whereas granulocytes required at least 9 days to assume ES-like morphology and Oct4-GFP expression (Figure 6b and c). FACS analysis for the MP marker c-Kit, which is also expressed on ES cells, showed that a subpopulation of c-Kit+ cells reactivated Oct4-GFP as early as 6 days after transgene activation and rapidly expanded over the next few days (Figure 6c). In contrast, granulocytes gradually downregulated the granulocyte marker Gr-1 and then reactivated Oct4-GFP beginnning at day 9, indicating that reprogramming is delayed compared with MPs (Figure 6c). Consistent with this, the number of Oct4-GFP+ cells was 10-fold higher in MP-derived cultures compared with granulocyte-derived cultures at day 9 (Figure 6c). Tail-tip fibroblasts obtained from chimeric animals showed a similar Oct4-GFP reactivation kinetics as differentiated hematopoietic cells (Supplementary Figure 8).
To test if differences in the reprogramming potentials of progenitors and mature cell populations persist at later stages of reprogramming, we sorted individual Oct4-GFP+ cells derived from either MPs or granulocytes at day 9 into 96 well plates in the continuous presence of doxycycline and counted the number of wells with iPS colonies at day 20. We detected no significant differences in the potentials of Oct4-GFP+ MPs and granulocytes to produce iPS colonies (12% vs. 9%), suggesting that upon reactivation of the endogenous Oct4 locus, both progenitors and mature cells have acquired the same abilities to become stable iPS cells (data not shown).
Discussion
Using nine different primitive hematopoietic cell populations (LT-HSCs, HSC/Ps, MPs, GMPs, MEPs, CMPs, CLPs, pro-Bs, pro-Ts) and their differentiated progeny (B cells, T cells, macrophages, granulocytes), we have shown that immature cells of the hematopoietic lineage are generally more susceptible to reprogramming than differentiated cell types, providing the first direct link between differentiation stage and reprogramming efficiency into iPS cells. The efficiencies at which progenitors converted into iPS cells was up to 2 orders of magnitude higher (7–28%) than that of differentiated blood cell types (0.02–0.6%) or fibroblasts (0.74%) and constitutes the highest reprogramming efficiencies reported so far.
Our data are consistent with the notion that HSCs and hematopoietic progenitors are more efficiently reprogrammed because their epigenetic state is more amenable to transcription factor-induced reprogramming. Accordingly, HSC/Ps, MPs, pro-B and pro-T cells become independent of transgene expression sooner than any differentiated cell types tested. Moreover, stem and progenitor cells lack expression of lineage specific genes, which can be inhibitory for reprogramming 22, and share expression of at least one marker, c-Kit, with ES cells, which may contribute to the enhanced reprogramming of immature blood cells.
Importantly, reprogramming into iPS cells was independent of the proliferation rate of cells but rather correlated with their differentiation stage. That is, quiescent and cytokine-activated HSCs as well as proliferative progenitors reprogrammed most efficiently, whereas differentiated B and T lymphocytes reprogrammed least efficiently, irrespective of their proliferation rate (see Figure 1b). We failed to detect an increased efficiency of HSCs compared with progenitor cells, suggesting that within immature cell populations, there may not be a strict correlation between differentiation stage and reprogramming efficiency. Alternatively, our failure to detect a higher reprogramming efficiency of stem cells over progenitor cells and of the more primitive CMPs over GMPs/MEPs may be due to technical issues such as slight variations in transgene expression or plating efficiencies of individual cell populations.
The observation that HSCs and hematopoietic progenitors are more susceptible to reprogramming into iPS cells than any tested differentiated cell type is consistent with nuclear transfer experiments of neural and keratinocyte stem cells 26,27. However, our data are in contrast to results obtained by Sung et al., who showed that HSC and progenitor populations give rise to cloned blastocysts less efficiently than granulocytes 29. Differences in efficiency may be due to the different reprogramming methods used, i.e. nuclear transfer vs. transcription factor-mediated reprogramming. Another reason may be differences in measuring reprogramming efficiency. Sung et al. measured reprogramming efficiency by counting the number of cloned blastocysts derived from oocytes injected with individual nuclei, whereas we measured the number of transgene-independent, Oct4-GFP expressing iPS colonies derived from a clonal population of viable cells.
Our results may provide an explanation for the previously reported failure to produce iPS cells from mature B cells by the expression of Oct4, Sox2, Klf4 and cMyc alone 34. In that study, Hanna et al. ectopically expressed the myeloid transcription factor C/EBPα in addition to the four reprogramming factors to generate B cell derived iPS cells. As discussed by the authors, their failure to generate iPS cells may be due to the low overall efficiency of reprogramming B cells, which is overcome by C/EBPα expression. Our observation that C/EBPα expression further enhances the reprogramming efficiency of B cells is consistent with this idea.
These data also corroborate the previous observations that genetically defined, terminally differentiated cell populations remain amenable to reprogramming into iPS cells 34,35,47. Because mature cell types appear slightly less efficient than heterogeneous cell populations such as fibroblasts and drastically less efficienct than isolated progenitors, as shown here, our findings raise the possibility that many iPS cells produced from explanted tissues may in fact be derived from resident stem or progenitor cells rather than from differentiated cells as assumed. It will certainly be interesting to test if somatic stem and progenitor cells from other tissues are more amenable to reprogramming than their mature progeny.
Our findings that adult progenitor cells are efficiently converted into iPS cells without additional chemical treatments or genetic manipulation have implications for research and medicine. The use of progenitor cells will make it feasible to perform biochemical and genetic analyses on cells undergoing reprogramming. Moreover, understanding the molecular differences between progenitors and differentiated cells can further teach us about the epigenetic and transcriptional barriers that seem inherent to nuclear reprogramming. On the clinical side, the use of somatic progenitors from adult tissues should make the derivation of patient-specific iPS cell lines more efficient and thus affordable. In addition, neonatal and adult progenitor cells, such as cord blood cells, have likely accrued fewer, if any, genetic aberrations and may thus be a safer source for iPS cells.
Materials and Methods
Viral production and infection
Viral infections were performed with replication-defective doxycycline-inducible lentiviral vectors and a lentiviral vector constitutively expressing the reverse tetracycline-controlled transactivator (rtTA) as decribed previously 38. To produce infectious viral particles, 293T cells cultured on 10 cm dishes were transfected with the LV-tetO vectors (11 μg) together with the packaging plasmids VSV- G (5.5 μg) and Δ8.9 (8.25 μg) using Fugene. Viral supernatants were harvested on 3 consecutive days starting 24 hours after transfection, yielding a total of ~30 ml of supernatant per viral vector. Viral supernatant was concentrated approximately 100-fold by ultracentrifugation at 20,000 rpm for 1.5 hours at 4°C and resuspension in 300 μl PBS. Infections were carried out in 1 ml ES medium using 5 μl of each viral concentrate per 35mm plate.
To assess reprogramming efficiencies of MPs and granulocytes after direct viral infection, 5×10^4 MPs or granulocytes from a ROSA26-rtTA+/− Oct4-GFP+/− animal were infected for 24h in a round bottom 96-well plate with a concentrated polycistronic lentivirus expressing Oct4, Sox2, Klf4 and cMyc.
Differentiation of fetal liver cells into T cells
The tail tip fibroblast-derived Oct4-GFP iPS clone #5 was injected into BDF1 blastocysts and the fetal liver was isolated from an E14.5 embryo, minced and co-cultured on OP9-DL1 (delta-like1) fibroblasts in αMEM medium (Invitrogen) containing 20% FBS (selected FBS for OP-9 cells), penicillin/streptomycin, 10 ng/ml murine Flt3-ligand and 10 ng/ml murine interleukin-7 (IL-7) (R&D Systems, USA) to induce differentiation into mature T cells as described elsewhere 39. After 13 days, cells were sorted based on CD8 and CD4 expression. Sorted cells were optionally superinfected with all four doxycycline-inducible lentiviral vectors in 96-well format over night and then plated on irradiated MEFs in cytokine-conditioned medium (described below) in the presence of 2 μg/ml puromycin and 1 μg/ml doxycyclin (dox). After 15 days on dox, emerging ES-like colonies were picked and cultured on MEFs in regular ES medium without dox. iPS cell clone CD8 was expanded and used for blast injection to generate CD8-iPS chimeras.
Hematopoietic cell culture and induction of iPS cell formation
All cell types were cultured in 5% CO2 at 37°C in ES cell medium. LT-HSCs (L−K+S+ CD34− CD48− CD150+), HSC/Ps (L−K+S+) and MPs (L−K+S−) were cultured in the presence of 50 ng/ml murine Kit-ligand (KL), 20 ng/ml human Thrombopoietin (TPO), 10 ng/ml murine Interleukin-3 (IL-3), 10 ng/ml murine Interleukin-6 (IL-6), and 10 ng/ml murine Flt3-ligand (FL) (R&D Systems, USA). To sustain LT-HSCs in a low proliferative state, they were cultured in low TPO (0.5 ng/ml) to promote survival 46. Granulocytes were cultured in the presence of 10 ng/ml G-CSF and 5 ng/ml GM-CSF. Primary and in vitro differentiated macrophages were cultured in the presence of 5 ng/ml M-CSF. CMPs, MEPs and GMPs were cultured in the presence of 25 ng/ml murine KL, 25 ng/ml human TPO, 10 ng/ml murine IL-3, 25 ng/ml murine Interleukin-11 (IL-11), 25 ng/ml murine FL, 2.5 U/ml EPO and 10 ng/ml GM-CSF (R&D Systems, USA). CLPs, pro-B cells as well as pro-T/Double Negative-2 (DN2) T cell progenitors were cultured in the presence of 50 ng/ml murine KL, 10 ng/ml murine FL and 10 ng/ml murine IL-7. Mature CD3+ T cells as well as CD4+CD8+ Double Positive (DP) T cell precursors were cultured in the presence of 10 ng/ml murine Interleukin-2 (R&D Systems, USA) and 2.5 μg/ml Concanavalin A (ConA) (Sigma). IgM+ and IgM+IgD+ mature B cells were cultured in the presence of 2 μM CpG (Invivogen) and 10 ng/ml LPS (Sigma Aldrich) and 10 ng/ml murine IL-7 (R&D Systems, USA). Immediately following FACS, all cell types were counted and dead cells excluded by trypan blue staining, plated on irradiated MEFs and cultured in ES cell medium supplemented with the individual cytokines in the presence or absence of dox (1 μg/ml). For the high and low proliferative T cell conditions, CD3/Cd28 Dynabeads T cell expander (Invitrogen) was used in a cell to bead ratio of 1:1.
In vitro differentiation of bone marrow cells into macrophages
Total bone marrow cells were cultured for 6 days in the presence of 5ng/ml M-CSF in ES cell medium and then sorted as single cells into the same medium with dox onto 96 wells. These cells correspond to the “Macrophage II” cells described in Figure 4b.
Calculation of iPS cell formation efficiency
Efficiencies were determined by dividing the number of Oct4-GFP positive colonies following dox withdrawal by the initial number of plated cells. HSC/Ps, MPs, pro-Bs, pro-Ts, DP-T cells and granulocytes were plated at a densities between 1×10^3 and 2.5×10^3 cells per 10cm dish. IgM, IgD and CD3+ cells were seeded at densities between 1×10^5 and 4×10^5 cells per 6-well dish. Each cell type was plated in duplicates; the total number of experiments for each cell type is indicated in the figure. To exclude the scoring of satellite colonies or non-iPS colonies, early ES-like colonies were marked on the dish and counted as an iPS colony only when positive for Oct4-GFP after dox withdrawal. Efficiencies using the 96-well approach were determined by counting wells with iPS colonies on day 18, 3 days after dox withdrawal. Efficiencies of iPS formation upon direct infection were determined as described previously in Eminli et al., 2008.
Transgene requirement assay
Each sorted hematopoietic cell type was plated right after sorting in triplicates into individual wells of 24-wells on MEFs in ES cell medium supplemented with cell-specific cytokines and dox (described in material methods section - hematopoietic cell culture). In detail, 1×103 MPs, 1×103 pro-Bs, 1×103 pro-T cells, 2×104 DP-T cells, 4×104 CD3-T cells, 2×105 IgM+ B cells, 4×105 IgD+ B cells and 1×105 granulocytes were plated in triplicates on MEFs. To determine the length of transgene requirement for each cell type, dox was withdrawn on days 6, 9 and 12 and medium was changed to normal ES cell medium without cytokines and dox. Each well was scored for the presence of dox-independent iPS cell colonies on day 15.
Flow cytometric analysis and cell sorting
Immunostaining was performed as previously described 48. Antibody conjugates and matched isotype controls were obtained from BD Biosciences (San Jose, CA) or eBioscience Inc. (San Diego, CA), unless otherwise indicated: anti-human CD4 (OKT4, APC, biotin), CD4 (GK1.5, biotin), CD8a (53-6, APC-Alexa Fluor 750), CD3ε (145-2C11, biotin, APC), TCRβ (H54-597, APC), TCRγδ (eBioGL3, biotin), Mouse Vβ TCR screening panel (FITC), CD44 (IM7, FITC), CD25 (PC61, APC), B220 (RA3-6B2, APC), IgM (II/41, biotin), IgD (11–26, Alexa Fluor-647), Ig kappa light chain (187.1, FITC; BD Pharmingen), Ig lambda light chain (RML-42, APC; BioLegend, San Diego, CA), CD43 (S7, Alexa fluor-488), Gr1 (RB6-8C5, APC), Mac-1 (M1/70, FITC), F4/80 (MCA497, FITC, AbD Serotec), neutrophils 7/4 (MCAA771, Alexa Fluor 647, AbD Serotec), SSEA-1 (eBioMC-480), CD48 (HM48.1, APC), CD150 (TC15-12F12.2, PE, Alexa488; Biolegend), CD34 (RAM34, FITC, pacific blue), CD117 (c-Kit, 2B8, APC, APC-Alexa Fluor 750), Sca-1 (D7, APC, Alexa fluor-488; BioLegend). Biotinylated antibodies were further subjected to either Streptavidin-APC or Streptavidin-Pacific Blue (Molecular Probes). For cell sorting experiments of LT-HSCs HSC/Ps and MPs, lineage-positive cells were depleted prior to flow-cytometry using Streptavidin-coupled Dynabeads (M-280; Invitrogen, Carlsbad, CA) after labeling with a cocktail consisting of biotinylated anti-Gr-1, Mac-1, B220, CD4, CD8, and Ter-119 monoclonal antibodies. Subsequently, cells were stained with appropriate fluorochrome-conjugated L−S+K+ markers as well as streptavidin-PE-Cy5 to detect residual biotinylated primary antibodies. Propodium iodide (1 μg/ml; Molecular Probes) was used to exclude dead cells at all times. A modified FACS ARIA flow cytometer with five lasers (UV 300 nm, violet 405 nm, blue 488 nm, green 532 nm, red 633 nm) running FACS Diva software was used for sorting. Analyses were performed using FACS Diva as well as FlowJo (Tree Star Inc., San Carlos, CA) software.
For single-cell sorting experiments into 96-well plates, MEFs were seeded into the inner 60 wells of a 96well plate the day before FACS. The medium of the wells was exchanged to ES cell medium supplemented with cell specific cytokines and doxycycline prior to FACS.
VDJ rearrangement analyses
VDJ rearrangements at the TCRα locus and rearrangements at the IgH locus were determined by Southern Blot analysis as previously described 41. DJ and VDJ rearrangements at the TCRβ locus of the CD8-iPS clone were amplified by PCR analysis 49. PCR products were then cloned into the TOPO vector system (INVITROGEN) and at least 10 bacterial clones, corresponding to DJ and VDJ rearrangements, were sequenced. Sequences were analyzed using the NCBI/BLAST/blastn suite and aligned to sequences of the murine TCRβ locus taken from GenBank accession numbers AE000663, AE000664, and AE000665.
Alkaline phosphatase staining
Alkaline phosphatase staining was performed with the Vector Red substrate kit (Vector Labs) according to manufacturer’s instructions.
Generation of teratomas and chimeras
For teratoma induction, 2×10^6 cells of each iPS cell line were injected subcutaneously into the dorsal flank of isoflurane-anesthetized SCID mice. Teratomas were recovered 3–4 weeks postinjection, fixed overnight in 10% formalin, paraffin embedded and sectioned. Sections were stained with hematoxylin and eosin and imaged using LEICA DMI4000B camera. For chimera production, female BDF1 or Rag2−/− mice were superovulated with PMS (pregnant mare serum) and hCG (human chorion gonadotropin) and mated to BDF1 stud males or Rag2−/− stud males, respectively. Zygotes were isolated from plugged females 24 h after hCG injection. After 3 days of in vitro culture in KSOM media, blastocysts were injected with iPS cells, and transferred into d2.5 pseudopregnant recipient females. C-sections were performed 17 days later and pups were fostered with lactating Swiss females.
C/EBPα retroviral infection
The mouse C/EBPα retrovirus has been previously described50. Retroviral stocks were prepared by transient transfection of PlatE-Eco cells using Fugene (Roche), and supernatants were harvested 48h later and concentrated by Centricon spin columns (Millipore). For infections, purified B cell subsets were resuspended in ES cell medium supplemented with CpG (2μM, Invivogen), lipopolysaccharide (LPS) (10ng/ml, Sigma Aldrich) and polybrene (8μg/ml), Sigma). Then, 1ml aliquots were plated on 24-well plate pre-coated with retronectin (Takara) and 50μl of concentrated retrovirus was added to each 24-well. The plates were incubated at 37°C and 24h later the medium was replaced by supplemented B cell ES medium described above. 48–72h later cells were sorted by FACS into C/EBPα expressing and non-expressing cells. Both populations of cells were plated on MEFs, in ES medium supplemented with cytokines and dox (1μg/ml).
Proliferation and apoptosis assays
All cell types were cultured in 5% CO2 at 37°C in ES cell medium supplemented with their specific growth factors as described in the methods. After 5 days, aliquots of each sample were diluted with trypan blue and viable cells were counted using a hemacytometer. To determine the rate of apoptosis, aliquots were stained with Annexin-V-FITC and 7-Aminoactinomycin-D according to the manufacturer’s recommendations (BD Biosciences) and analyzed on a BD LSRII flow cytometer as described in the methods. The proliferation rate of individual cell populations was measured either by Hoechst 33342 staining as described previously51 or by propidium iodide (PI)-based cell cycle analysis. For PI-based analyses, cells are first permeabilized by fixation in 70 % ice cold ethanol for 30 min at 4 °C. After two washes in wash buffer (PBS supplemented with 0.2 % BSA), pellets are resuspended in cell cycle buffer (3.8 mM sodium citrate, 40 μg/ml propidium iodide, 1 μg/ml RNase A) and stained for 30 min at room temperature. Samples were run on a FACScan and analyzed with FlowJo.
Quantitative PCR (qPCR)
Following cell sorting by flow cytometry, all cell types were cultured in the absence and presence of dox in cell specific medium (as described in the methods). After three days, cells were harvested for mRNA preparation. Real-time quantitative PCR reactions were set up in triplicates with the Brilliant II SYBR Green QPCR Master Mix (Stratagene) and run on a Mx3000P QPCR System (Stratagene) (see Supplementary Table 1).
Derivation of human iPS (hiPS) cells from CD34+ cord blood progenitors
Human umbilical CD34+ cord blood cells were purchased from STEMCELL Technologies and were cultured (after thawing) for 24 hours in Stemspan SFEM medium supplemented with STEMSPAN CC100 cytokine cocktail (both from STEMCELL Technologies). 1×10^5 cells were infected for 24h in a round bottom 96-well plate with a concentrated polycistronic lentivirus expressing human cDNAs for Oct4, Sox2, Klf4 and cMyc, similar to the mouse polycistronic lentivirus vector published recently52 and then plated on Mefs in Stemspan SFEM medium and STEMSPAN CC100 cytokine cocktail supplemented with human bFGF (10 ng/ml) and dox. Human bFGF and dox were added to the cultures daily. After dox withdrawal on day 12 the medium was switched to human ES cell medium (see below).
Cell Culture of hiPS cells on feeder cells and on matrigel and chemical defined medium mTESR
Irradiated fibroblasts were grown in DMEM with 10% FBS, hiPS were grown on feeder cells in KO-DMEM, KOSR, nonessential amino acids, glutamine, β-mercaptoethanol and 10 ng/ml human bFGF as described previously53. hiPS cells were also grown in feeder free conditions on culture dishes treated with Matrigel, in medium mTESR.
In vitro differentiation of hiPS cells derived from human CD34+ cord blood cells
For in vitro differentiation, hiPS colonies were dissociated using dispase and placed in suspension culture with either growth-medium DMEM with 10 % FBS or neuronal induction medium DMEM/F12 +Glutamax +N2 supplement +B27 supplement to initiate embryoid body formation. After 1 week, embryoid bodies were plated to adherent conditions with gelatin and treated with described media. For teratoma formation, 1×10^7 hiPSCs were pelleted and injected into SCID mice, either subcutaneously or underneath the kidney capsule. Tumors were harvested after 7–13 weeks and processed for histological analysis.
Immunostaining
Immunostaining was performed using the following antibodies: SOX2 (Abcam ab15830), α-Tra-1-81 (MAB4381, Millipore), α-beta-III tubulin (T2200, Sigma), and α-AFP (sc-15375, Santa Cruz Biotech).
Supplementary Material
Acknowledgments
We thank Dr. Huafeng Xie for sharing the C/EBPα overexpression vector, Dr. Raul Mostoslavsky for critical reading of the manuscript and Laura Prickett and Kat Folz-Donahue for help with FACS and Patti Follett for help with blastocyst injections. A.F was the recipient of the Lady Tata Memorial Trust Award. A Postdoctoral Fellowship from Schering Foundation supported M.S. This work was supported by a contribution from the Ellison Foundation to MGH start-up funds for H.H., an ASH Scholar Award to H.H., and the Harvard Stem Cell Institute. Support to K.H. came from the NIH Director’s Innovator Award, the Harvard Stem Cell Institute, the Kimmel Foundation and the V Foundation.
Footnotes
Author contributions
S.E. Conception and design, provision of study material, collection and assembly of data, data analysis and interpretation, manuscript writing
A.F. Conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing
M.S. Provision of study material, collection of data
N.M. Provision of study material
T.A. Collection and assembly of data
G.M. Provision of study material
H.H. Conception and design, financial support, data analysis and interpretation, manuscript writing
K.H. Conception and design, financial support, administrative support, data analysis and interpretation, manuscript writing
References
- 1.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 2.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–7. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–24. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 5.Liao J, et al. Generation of Induced Pluripotent Stem Cell Lines from Adult Rat Cells. Cell Stem Cell. 2008 doi: 10.1016/j.stem.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Li W, et al. Generation of Rat and Human Induced Pluripotent Stem Cells by Combining Genetic Reprogramming and Chemical Inhibitors. Cell Stem Cell. 2008 doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Liu H, et al. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–90. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Lowry WE, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–8. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–6. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 12.Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–23. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–53. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 14.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–9. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou H, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–4. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hockemeyer D, et al. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–53. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maherali N, et al. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–5. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–24. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061–7. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- 20.Huangfu D, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–7. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huangfu D, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–75. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 22.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y, et al. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–74. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Ellis P, et al. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26:148–65. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- 25.Galan-Caridad JM, et al. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129:345–57. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blelloch R, et al. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells. 2006;24:2007–13. doi: 10.1634/stemcells.2006-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Greco V, Guasch G, Fuchs E, Mombaerts P. Mice cloned from skin cells. Proc Natl Acad Sci U S A. 2007;104:2738–43. doi: 10.1073/pnas.0611358104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue K, et al. Inefficient reprogramming of the hematopoietic stem cell genome following nuclear transfer. J Cell Sci. 2006;119:1985–91. doi: 10.1242/jcs.02913. [DOI] [PubMed] [Google Scholar]
- 29.Sung LY, et al. Differentiated cells are more efficient than adult stem cells for cloning by somatic cell nuclear transfer. Nat Genet. 2006;38:1323–8. doi: 10.1038/ng1895. [DOI] [PubMed] [Google Scholar]
- 30.Hochedlinger K, Jaenisch R. On the cloning of animals from terminally differentiated cells. Nat Genet. 2007;39:136–7. doi: 10.1038/ng0207-136. author reply 137–8. [DOI] [PubMed] [Google Scholar]
- 31.Kim JB, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–50. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 32.Eminli S, Utikal J, Arnold K, Jaenisch R, Hochedlinger K. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells. 2008;26:2467–74. doi: 10.1634/stemcells.2008-0317. [DOI] [PubMed] [Google Scholar]
- 33.Shi Y, et al. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–8. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 34.Hanna J, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–64. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stadtfeld M, Brennand K, Hochedlinger K. Reprogramming of pancreatic beta cells into induced pluripotent stem cells. Curr Biol. 2008;18:890–4. doi: 10.1016/j.cub.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lengner CJ, et al. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–77. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 38.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–40. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramsdell F, Zuniga-Pflucker JC, Takahama Y. In vitro systems for the study of T cell development: fetal thymus organ culture and OP9-DL1 cell coculture. Curr Protoc Immunol. 2006;Chapter 3(Unit 3):18. doi: 10.1002/0471142735.im0318s71. [DOI] [PubMed] [Google Scholar]
- 40.Sommer CA, et al. iPS Cell Generation Using a Single Lentiviral Stem Cell Cassette. Stem Cells. 2008;3:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hochedlinger K, Jaenisch R. Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature. 2002;415:1035–8. doi: 10.1038/nature718. [DOI] [PubMed] [Google Scholar]
- 42.Chen J, Lansford R, Stewart V, Young F, Alt FW. RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proc Natl Acad Sci U S A. 1993;90:4528–32. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–21. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 44.Foudi A, et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–82. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sitnicka E, et al. The effect of thrombopoietin on the proliferation and differentiation of murine hematopoietic stem cells. Blood. 1996;87:4998–5005. [PubMed] [Google Scholar]
- 47.Aoi T, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 48.Hock H, et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003;18:109–20. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- 49.Whitehurst CE, Chattopadhyay S, Chen J. Control of V(D)J recombinational accessibility of the D beta 1 gene segment at the TCR beta locus by a germline promoter. Immunity. 1999;10:313–22. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]
- 50.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–76. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 51.Foudi A, et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sommer CA, et al. iPS Cell Generation Using a Single Lentiviral Stem Cell Cassette. Stem Cells. 2008 doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cowan CA, et al. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med. 2004;350:1353–6. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.