Abstract
The composition of the lipid matrix is critical for function of membrane proteins. Perhaps one of the best studied examples is the function of the G-protein-coupled membrane receptor (GPCR) rhodopsin which is located in membranes with high content of phospholipids with polyunsaturated docosahexaenoic acid chains (DHA, 22:6n-3). Technological advances enabled a more detailed study of structure and dynamics of DHA chains and their interaction with rhodopsin. It was established that polyunsaturated DHA differs from saturated and monounsaturated hydrocarbon chains by far more rapid structural conversions. Furthermore, DHA chains tend to have higher density near the lipid/water interface while density of saturated chains is higher in the bilayer center. The interface of rhodopsin has a small number of sites for tighter interaction with DHA. Polyunsaturated phosphatidylethanolamines accumulate preferentially near the protein. Surprisingly, the high conformational freedom of most DHA chains is not measurably reduced upon interaction with rhodopsin. While some observations point at an involvement of continuum elastic properties of membranes in modulation of rhodopsin function, there is growing evidence for a role of weakly specific DHA-rhodopsin interactions.
Keywords: Docosahexaenoic Acid, G-Protein-coupled membrane receptor, Rhodopsin, Nuclear Magnetic Resonance, Neutron Scattering
1. Conformation and dynamics of DHA
Ever since it was established that the disks of rod outer segments of the retina contain the highly unsaturated docosahexaenoic acid (DHA, 22:6n-3) at concentrations up to 50 mol% of all fatty acids, speculation arose that peculiarities in DHA structure and dynamics may impart unique biophysical properties on membranes that could modulate rhodopsin function. This was confirmed by reconstitution of rhodopsin into bilayers with varying degree of unsaturation followed by a measurement of the metarhodopsin I/metarhodopsin II (MI/MII) ratio after photoactivation [1–3].
How do membranes that are rich in DHA differ from less unsaturated ones? For a long time it is known that lipids in polyunsaturated membranes have low order [4–7]. Initially it was proposed that low order may result from perturbations caused by rigidity and bulkiness of polyunsaturated hydrocarbon chains. Indeed, DHA has a unique chemical structure with six cis-locked double bounds. The number of degrees of freedom of DHA chains is substantially lower which could be indicative for rigidity. Polyunsaturated chains in crystals form highly ordered, elongated structures with angle-iron or helical arrangement of double bonds [8]. Such structures were also observed by molecular mechanics calculations conducted at low temperature [9, 10].
But in the past decade our view of DHA has changed. Early molecular simulations by Rabinovitch and Ripatty [11] suggested that the unique electronic structure of polyunsaturated chains may permit rapid conformational changes. Recent experimental studies at our laboratory as well as quantum chemical calculations and molecular dynamics simulations by our collaborators revealed the full magnitude of conformational flexibility of DHA [12, 13]. High conformational flexibility of DHA chains was reported by Brown and collaborators [14, 15], the Davis [16], as well as the Klein laboratory [17, 18]. Our 13C MAS NMR relaxation experiments have shown that DHA isomerizes on the timescale of 1–100 ps and explore their entire conformational space within 10 ns [19]. Experimental results clearly indicate that low order in bilayers with high DHA content is a direct consequence of high conformational flexibility and of rapid structural conversions of DHA chains themselves. Quantum chemical and molecular mechanical calculations revealed that this flexibility is caused by extremely low potential barriers for changes of dihedral bond angles in vinyl bonds [12]. Low potential barriers permit the polyunsaturated chains to rapidly change conformation without significant energetic penalty.
2. Nonspecific DHA-protein interactions
In continuum models of lipid-protein interaction, the influence of the lipid matrix on protein function can be expressed as lateral pressure that acts on membrane imbedded proteins. It is assumed that the photoisomers of rhodopsin have different shape. When an integral membrane protein changes shape, it performs work against the lateral pressure. As a result, the photointermediates of rhodopsin have free energies that not only reflect the internal energy of the protein but also the work against lateral pressure from the lipid matrix [20].
Lateral pressure varies along the bilayer normal. Pressures are higher at the lipid-water interface where lipids tend to have higher order. Differences in the pressure profile between DHA-containing bilayers and their less unsaturated counterparts could explain the unique influence of DHA on rhodopsin function. Unfortunately, pressure profiles can not be measured directly. But changes of order parameters of lipid hydrocarbon chains may serve as qualitative indicators for changes in the pressure profile induced by DHA [21]. Also the distribution of hydrocarbon chain densities along the bilayer normal has been linked to peculiarities in the pressure profile [13, 22].
The distribution of saturated stearic acid (18:0) and polyunsaturated DHA in the mixed chain phosphatidylcholine 18:0-22:6n-3-PC was measured by neutron scattering [23]. Lipids were synthesized with either perdeuterated sn-1 stearic acid chains (18:0d35), or perdeuterated DHA chains (22:6n-3d11) or both protonated stearic acid and DHA chains. Data were acquired on the advanced neutron diffractometer/reflectometer at the NIST Center for Neutron Research, Gaithersburg, MD. Due to the big differences in scattering length densities for neutrons between protonated and deuterated hydrocarbon chains, it is straightforward to calculate scattering length densities of stearic acid and DHA chains over the hydrophobic core of bilayers (see Fig. 3) [23]. It was observed that DHA segments are located with higher probability near the lipid/water interface, while the segments of saturated chains locate with higher probability in the bilayer center.
Fig. 3.
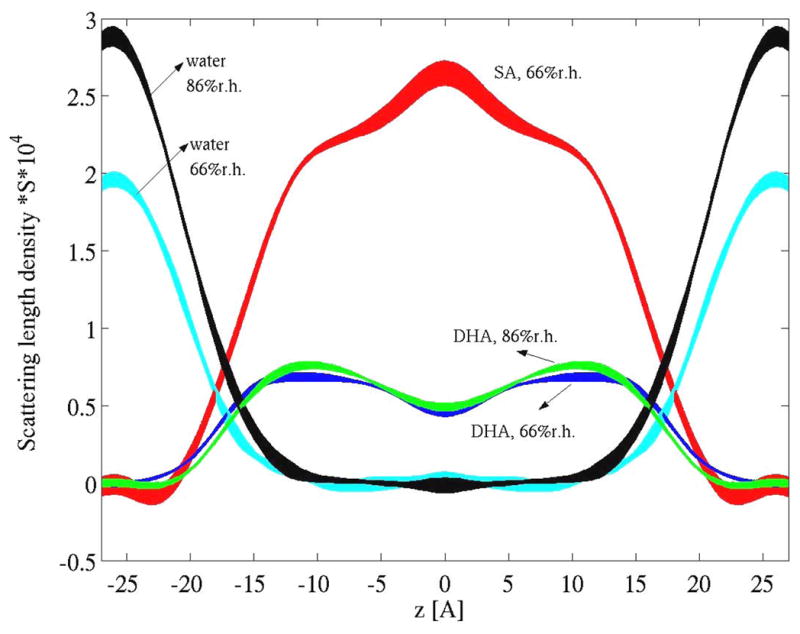
Scattering length density profiles of stearic acid, DHA and water in fluid 18:0-22:6n-3-PC bilayers determined by neutron scattering measured at two levels of relative humidity (86% r.h. and 66% r.h.). It is visible that DHA chain density tends to be higher near the lipid/water interface while stearic acid density is higher in the bilayer center. Water forms continuous layers outside the hydrophobic core of lipid bilayers. DHA scattering length density is generally lower than stearic acid density because of the lower content of deuterons in DHA (see [23] for details).
The results of the neutron scattering experiments corroborated earlier conclusions about an uneven distribution of polyunsaturated chain density from 2H NMR experiments [13]. The DHA distribution causes lower order parameters near the methyl terminal end of the saturated hydrocarbon chains with which they are paired in 18:0-22:6n-3-PC [6, 13, 15, 24]. Lower order stems from the wider space that saturated chains occupy in the bilayer center.
The uneven distribution of hydrocarbon chains, the low order of DHA chains, and their rapid structural conversions result in altered continuum properties of the lipid matrix. Bilayer thickness of mixed chain, DHA-containing bilayers is similar to the thickness of saturated and monounsaturated lipid species with sixteen to eighteen carbon atoms per chain [13, 25]. Consistent with the smaller bilayer thickness, lateral area per molecule of lipids with DHA chains is larger. The increased area could be a consequence of the weakly polar nature of the double bonds of DHA. This may explain why polyunsaturated chains prefer to locate somewhat closer to the lipid/water interface. It may also explain the higher permeability of polyunsaturated bilayers for water that we reported earlier [26].
The lateral pressure profile of 18:0-22:6n-3-PC bilayers was calculated by molecular simulations [27, 28]. It was observed that the shift of DHA chain density to the lipid/water interface is correlated with the migration of lateral stresses within the bilayer which in turn may explain the shifts in the MI/MII equilibrium [27]. Another indicator for migration of lateral stresses is formation of non-lamellar lipid phases. We reported earlier that DHA chains in phosphatidylethanolamines facilitate formation of inverted hexagonal phases [29]. Lipid monolayers of phosphatidylethanolamines have a smaller lateral area in the headgroup region and lager area in the region of hydrocarbon chains. The monolayers are under negative curvature elastic stress when forced into a bilayer arrangement. Levels of stress in 18:0-22:6n3-PE membranes are similar to 18:1n-9-18:1n-9-PE, the golden standard for lipids with negative curvature stress [29]. Brown and colleagues reported that 18:1n-9-18:1n-9-PE in bilayers favors formation of the G-protein activation competent MII photointermediate of rhodopsin [30]. Since the 18:0-22:6n-3-PE is a very common lipid, in particular in neuronal membranes, it is reasonable to assume that those membranes are under considerable negative curvature elastic stress as well.
3. Specific DHA-protein interactions
An alternative hypothesis to explain the shifts in rhodopsin function is to assume that the free energy of rhodopsin is under influence from weakly specific interactions with DHA, similar to ligand-protein interactions. In recent years, several examples of specific interactions between proteins and polyunsaturated chains were reported, e.g. the interaction of arachidonic acid with prostaglandin synthase [31], the interaction of DHA with the binding pocket of human fatty acid binding protein [32], the interaction of the arachidonyl inhibitor with the binding pocket of fatty acid amide hydrolase [33], the interaction of fatty acids with the retinoic acid-related orphan receptor β [34], and last but not least, the interaction of retinal with rhodopsin [35].
In all instances, binding of polyunsaturated chains to proteins was stabilized by major contributions from polar, non-covalent interactions like salt bridges, direct hydrogen bonding between hydroxyl or carbonyl groups to amino acids, or indirect hydrogen bonding via water molecules. But those interactions do not explain selectivity to the number and location of double bounds in the chains. The attractive π-π interactions that may occur between double bonds of polyunsaturated fatty acids and aromatic side chains of amino acids may be the missing link. Indeed, it is remarkable that the fatty acid binding pockets in the proteins that are listed above are lined by a very high density of the aromatic amino acids phenylalanine, tyrosine, and tryptophan.
Recent NMR results obtained at our lab provide some evidence for involvement of specific interactions with DHA in rhodopsin function. In experiments on rhodopsin that was reconstituted into membranes with variable content of phosphatidylcholine, phosphatidylethanolamine, and phosphatidylserine, all with DHA hydrocarbon chains, it was observed that DHA chains compete for contact with a small number of weakly specific sites on rhodopsin [36]. While specificity was caused by DHA, the lipid headgroups modulated interaction strength. Interactions were strongest for PE, followed by PS, and PC [36]. Monounsaturated lipid species have preferential contact with a different site on rhodopsin. The lifetimes of associations between lipids and rhodopsin are short, indicating that mutual attraction has modest strength. According to our NMR measurements, all lipids that surround the protein in a first layer are replaced within microseconds or even shorter times.
The conclusion is that rhodopsin has sites for preferential interaction with particular lipid species. Molecular simulations suggest as well that the surface of rhodopsin is inhomogeneous in terms of lipid-rhodopsin interaction (see Fig. 4 [37]). Does DHA-rhodopsin interaction depend on the state of photoactivation of rhodopsin? To answer this question, rhodopsin was bleached during the NMR studies. Experiments were conducted at high and low pH to yield the photointermediates MI or MII, respectively. Neither of those two photointermediates showed differences in the interaction with DHA compared to dark adapted rhodopsin. But the photointermediate MIII had stronger contacts with DHA [36]. Interaction of opsin with DHA was similar to dark adapted rhodopsin.
Fig. 4.
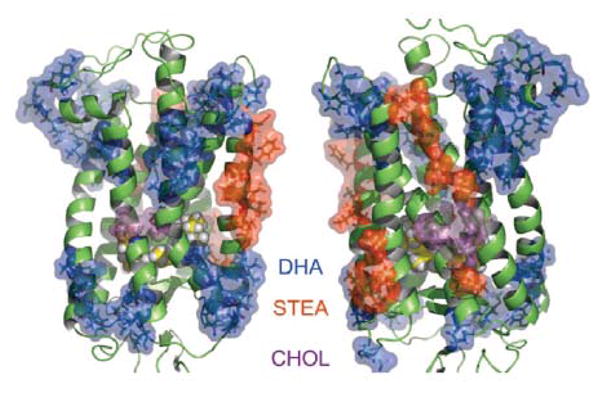
Sites of rhodopsin in a 18:0-22:n-3-PC/PE/cholesterol bilayer that have been proposed to interact preferentially with DHA (blue) stearic acid (STEA, red) or cholesterol (CHOL, purple). Sites of preferential interaction were identified in a molecular dynamics simulation by Grossfield at al. [37] (Copyright (2006) National Academy of Sciences, U.S.A.).
Specificity in lipid-rhodopsin interaction may shift lipid composition in the protein annulus compared to the bulk of the lipid matrix. Lipids that have higher affinity to rhodopsin will be enriched near the protein, despite the rapid exchange of lipids. Indeed, by NMR saturation transfer from protein to lipids, we obtained evidence that polyunsaturated phosphatidylethanolamines are partially enriched near rhodopsin [36].
How may specific interactions between lipids and protein influence protein function? The double bonds of polyunsaturated chains could be attracted to aromatic amino acids of GPCR that stabilize a particular conformation of the protein. Polyunsaturated hydrocarbon chains may interfere with interactions inside the GPCR if the chains could reach into the protein. There is evidence for such events from molecular simulations [38]. Interaction with phospholipids may also interfere with hydrogen bonding and salt bridges that form between transmembrane helices, as well as with the ubiquitous van der Waals interactions.
4. Summary
Recent NMR studies have improved our understanding of the role of polyunsaturated fatty acids like DHA in lipid bilayers. The DHA chains are special because they interconvert between conformations more rapidly, they prefer to locate in the hydrophobic core of bilayers near the lipid/water interface, and they are capable of engaging in partially specific interactions with GPCR like rhodopsin. Bilayers rich in DHA may alter protein function both by a change of general membrane properties as well as by specific interactions with particular regions of the protein. Experiments that are currently underway at our laboratory suggest that GPCR adjust their structure far more nimbly to the lipid environment than generally assumed. It is not just the lipid matrix that deforms in response to the needs of the protein, but the protein may adjust structurally to the lipid matrix as well [39]. The latter aspect deserves more attention in future investigations.
Fig. 1.
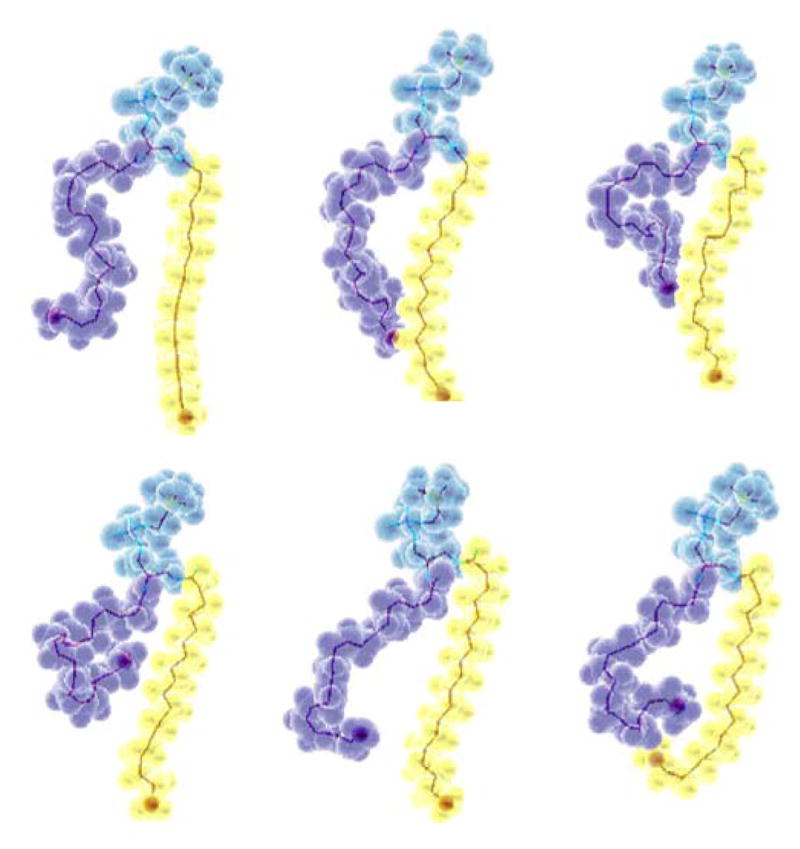
Snapshots of an 18:0-22:6n-3-PC molecule in a lipid bilayer taken from a 500 ps-window of a 10 ns-long molecular dynamics simulation by Feller et al. [12]. The polyunsaturated chain is shown in purple, the saturated chain in yellow and the phosphocholine headgroup and glycerol region of the lipid in blue. The images demonstrate the rapid structural conversions of DHA chains on the subnanosecond timescale. The tendency of DHA to have higher density near the lipid water interface is visible as well.
Fig. 2.
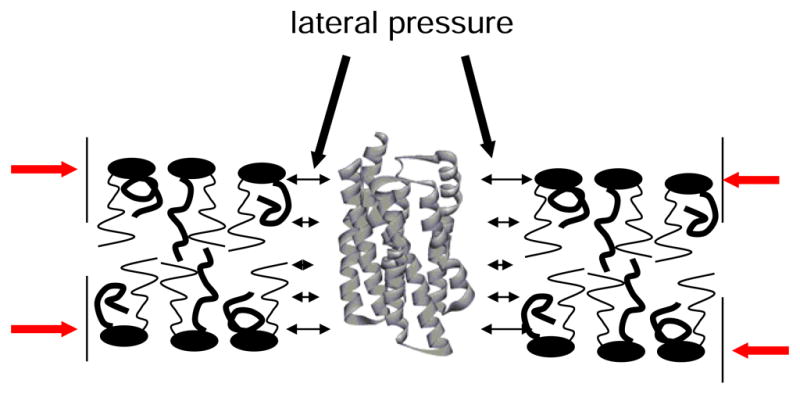
Membrane proteins sense the pressure profile of a lipid bilayer. Lipids are compressed laterally by the hydrophobic effect (red arrows). When the protein undergoes a conformational change it performs work against membrane lateral pressure which is a contribution to free energy of protein conformers.
Acknowledgments
This work was supported by the Intramural Research Program of NIAAA, NIH. The neutron diffraction studies were conducted on the AND/R instrument, constructed by the Cold Neutrons for Biology and Technology (CNBT) partnership, supported by the National Institute of Standards and Technology, the Regents of the University of California, and by a grant RR14812 from the National Institute for Research Resources awarded to the University of California at Irvine.
References
- 1.Niu SL, Mitchell DC, Lim SY, Wen ZM, Kim HY, Salem N, Jr, Litman BJ. Reduced G protein-coupled signaling efficiency in retinal rod outer segments in response to n-3 fatty acid deficiency. J Biol Chem. 2004;279:31098–31104. doi: 10.1074/jbc.M404376200. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell DC, Niu SL, Litman BJ. DHA-rich phospholipids optimize G-Protein-coupled signaling. J Pediatr. 2003;143:S80–86. doi: 10.1067/s0022-3476(03)00405-0. [DOI] [PubMed] [Google Scholar]
- 3.Gibson NJ, Brown MF. Lipid headgroup and acyl chain composition modulate the MI-MII equilibrium of rhodopsin in recombinant membranes. Biochemistry. 1993;32:2438–2454. doi: 10.1021/bi00060a040. [DOI] [PubMed] [Google Scholar]
- 4.Stubbs CD, Smith AD. The modification of mammalian membrane polyunsaturated fatty acid composition in relation to membrane fluidity and function. Biochim Biophys Acta. 1984;779:89–137. doi: 10.1016/0304-4157(84)90005-4. [DOI] [PubMed] [Google Scholar]
- 5.Salmon A, Dodd SW, Williams GD, Beach JM, Brown MF. Configurational statistics of acyl chains in polyunsaturated lipid bilayers from 2 H NMR. J Am Chem Soc. 1987;109:2600–2609. [Google Scholar]
- 6.Holte LL, Peter SA, Sinnwell TM, Gawrisch K. 2 H nuclear magnetic resonance order parameter profiles suggest a change of molecular shape for phosphatidylcholines containing a polyunsaturated acyl chain. Biophys J. 1995;68:2396–2403. doi: 10.1016/S0006-3495(95)80422-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell DC, Litman BJ. Molecular order and dynamics in bilayers consisting of highly polyunsaturated phospholipids. Biophys J. 1998;74:879–891. doi: 10.1016/S0006-3495(98)74011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ernst J, Sheldrick WS, Fuhrhop JH. Structures of the essential unsaturated fatty acids -crystal structure of linoleic acid and evidence for the crystal structures of alpha-linolenic acid and arachidonic acid. Z Naturforsch. 1979;34B:706–711. [Google Scholar]
- 9.Applegate KR, Glomset JA. Computer-based modeling of the conformation and packing properties of docosahexaenoic acid. J Lipid Res. 1986;27:658–680. [PubMed] [Google Scholar]
- 10.Applegate KR, Glomset JA. Effect of acyl chain unsaturation on the conformation of model diacylglycerols: a computer modeling study. J Lipid Res. 1991;32:1635–1644. [PubMed] [Google Scholar]
- 11.Rabinovich AL, Ripatti PO. On the conformational, physical properties and functions of polyunsaturated acyl chains. Biochim Biophys Acta. 1991;1085:53–62. doi: 10.1016/0005-2760(91)90231-6. [DOI] [PubMed] [Google Scholar]
- 12.Feller SE, Gawrisch K, MacKerell AD. Polyunsaturated fatty acids in lipid bilayers: Intrinsic and environmental contributions to their unique physical properties. J Am Chem Soc. 2002;124:318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 13.Eldho NV, Feller SE, Tristram-Nagle S, Polozov IV, Gawrisch K. Polyunsaturated docosahexaenoic vs docosapentaenoic acid - Differences in lipid matrix properties from the loss of one double bond. J Am Chem Soc. 2003;125:6409–6421. doi: 10.1021/ja029029o. [DOI] [PubMed] [Google Scholar]
- 14.Huber T, Rajamoorthi K, Kurze VF, Beyer K, Brown MF. Structure of docosahexaenoic acid-containing phospholipid bilayers as studied by H-2 NMR and molecular dynamics simulations. Journal of the American Chemical Society. 2002;124:298–309. doi: 10.1021/ja011383j. [DOI] [PubMed] [Google Scholar]
- 15.Petrache HI, Salmon A, Brown MF. Structural properties of docosahexaenoyl phospholipid bilayers investigated by solid-state H-2 NMR spectroscopy. Journal of the American Chemical Society. 2001;123:12611–12622. doi: 10.1021/ja011745n. [DOI] [PubMed] [Google Scholar]
- 16.Everts S, Davis JH. H-1 and C-13 NMR of multilamellar dispersions of polyunsaturated (22 : 6) phospholipids. Biophys J. 2000;79:885–897. doi: 10.1016/S0006-3495(00)76344-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saiz L, Klein ML. Influence of highly polyunsaturated lipid acyl chains of biomembranes on the NMR order parameters. J Am Chem Soc. 2001;123:7381–7387. doi: 10.1021/ja003987d. [DOI] [PubMed] [Google Scholar]
- 18.Saiz L, Klein ML. Structural properties of a highly polyunsaturated lipid bilayer from molecular dynamics simulations. Biophys J. 2001;81:204–216. doi: 10.1016/S0006-3495(01)75692-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soubias O, Gawrisch K. Docosahexaenoyl chains isomerize on the sub-nanosecond time scale. J Am Chem Soc. 2007;129:6678–6679. doi: 10.1021/ja068856c. [DOI] [PubMed] [Google Scholar]
- 20.Cantor RS. Lateral pressures in cell membranes: a mechanism for modulation of protein function. J Phys Chem B. 1997;101:1723–1725. [Google Scholar]
- 21.Gawrisch K, Holte LL. NMR investigations of non-lamellar phase promoters in the lamellar phase state. Chem Phys Lipids. 1996;81:105–116. [Google Scholar]
- 22.Binder H, Gawrisch K. Effect of unsaturated lipid chains on dimensions, molecular order and hydration of membranes. J Phys Chem B. 2001;105:12378–12390. [Google Scholar]
- 23.Mihailescu M, Gawrisch K. The structure of polyunsaturated lipid bilayers important for rhodopsin function - a neutron diffraction study. Biophys J. 2006;90:L4–6. doi: 10.1529/biophysj.105.071712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Binder H, Gawrisch K. Dehydration induces lateral expansion of polyunsaturated 18:0-22:6 phosphatidylcholine in a new lamellar phase. Biophys J. 2001;81:969–982. doi: 10.1016/S0006-3495(01)75755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koenig BW, Strey HH, Gawrisch K. Membrane lateral compressibility determined by NMR and X-ray diffraction: Effect of acyl chain polyunsaturation. Biophys J. 1997;73:1954–1966. doi: 10.1016/S0006-3495(97)78226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huster D, Jin AJ, Arnold K, Gawrisch K. Water permeability of polyunsaturated lipid membranes measured by 17O NMR. Biophys J. 1997;73:855–864. doi: 10.1016/S0006-3495(97)78118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carillo-Tripp M, Feller SE. Evidence for a mechanism by which ω-3 polyunsaturated lipids may affect membrane protein function. Biochemistry. 2005;44:10164–10169. doi: 10.1021/bi050822e. [DOI] [PubMed] [Google Scholar]
- 28.Cantor RS. Lipid composition and the lateral pressure profile in bilayers. Biophys J. 1999;76:2625–2639. doi: 10.1016/S0006-3495(99)77415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teague WE, Fuller NL, Rand RP, Gawrisch K. Polyunsaturated lipids in membrane fusion events. Cell Mol Biol Lett. 2002;7:262–264. [PubMed] [Google Scholar]
- 30.Botelho AV, Gibson NJ, Thurmond RL, Wang Y, Brown MF. Conformational energetics of rhodopsin modulated by nonlamellar-forming lipids. Biochemistry. 2002;41:6354–6368. doi: 10.1021/bi011995g. [DOI] [PubMed] [Google Scholar]
- 31.Malkowski MG, Ginell SL, Smith WL, Garavito RM. The productive conformation of arachidonic acid bound to prostaglandin synthase. Science. 2000;289:1933–1937. doi: 10.1126/science.289.5486.1933. [DOI] [PubMed] [Google Scholar]
- 32.Balendiran GK, Schnutgen F, Scapin G, Borchers T, Xhong N, Lim K, Godbout R, Spener F, Sacchettini JC. Crystal structure and thermodynamic analysis of human brain fatty acid-binding protein. J Biol Chem. 2000;275:27045–27054. doi: 10.1074/jbc.M003001200. [DOI] [PubMed] [Google Scholar]
- 33.Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF. Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science. 2002;298:1793–1796. doi: 10.1126/science.1076535. [DOI] [PubMed] [Google Scholar]
- 34.Stehlin C, Wurtz JM, Steinmetz A, Greiner E, Schule R, Moras D, Renaud JP. X-ray structure of the orphan nuclear receptor ROR-βeta ligand-binding domain in the active conformation. EMBO J. 2001;20:5822–5831. doi: 10.1093/emboj/20.21.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le TI, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 36.Soubias O, Teague WE, Gawrisch K. Evidence for specificity in lipid-rhodopsin interactions. J Biol Chem. 2006;281:33233–33241. doi: 10.1074/jbc.M603059200. [DOI] [PubMed] [Google Scholar]
- 37.Grossfield A, Feller SE, Pitman MC. A role for direct interactions in the modulation of rhodopsin by omega-3 polyunsaturated lipids. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4888–4893. doi: 10.1073/pnas.0508352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feller SE, Gawrisch K, Woolf TB. Rhodopsin exhibits a preference for solvation by polyunsaturated docosohexaenoic acid. J Am Chem Soc. 2003;125:4434–4435. doi: 10.1021/ja0345874. [DOI] [PubMed] [Google Scholar]
- 39.Soubias O, Niu SL, Mitchell DC, Gawrisch K. Lipid-rhodopsin hydrophobic mismatch alters rhodopsin helical content. J Am Chem Soc. 2008;130:12465–12471. doi: 10.1021/ja803599x. [DOI] [PMC free article] [PubMed] [Google Scholar]


