Abstract
Direct reprogramming of human fibroblasts to a pluripotent state has been achieved through ectopic expression of the transcription factors OCT4, SOX2, and either cMYC and KLF4 or NANOG and LIN28. Little is known, however, about the mechanisms by which reprogramming occurs, which is in part limited by the low efficiency of conversion. To this end, we sought to create a doxycycline-inducible lentiviral system to convert primary human fibroblasts and keratinocytes into human induced pluripotent stem (hiPS) cells. hiPS cells generated with this system were molecularly and functionally similar to human embryonic stem (hES) cells, demonstrated by gene expression profiles, DNA methylation status, and differentiation potential. While expression of the viral transgenes was required for several weeks in fibroblasts, we found that 10 days was sufficient for the reprogramming of keratinocytes. Using our inducible system, we developed a strategy to induce hiPS cell formation at high frequency. Upon addition of doxycycline to hiPS-derived differentiated cells, we obtained “secondary” hiPS cells at a frequency at least 100-fold greater than the initial conversion. The ability to reprogram cells at high efficiency provides a unique platform to dissect the underlying molecular and biochemical processes that accompany nuclear reprogramming.
Introduction
While human fibroblasts and a multitude of mouse somatic cell types can be reprogrammed to pluripotency by ectopic expression of transcription factors (Takahashi and Yamanaka, 2006; Maherali et al, 2007; Okita et al, 2007; Wernig et al. 2007; Takahashi et al, 2007; Yu et al, 2007; Lowry et al, 2007; Aoi et al, 2008; Hanna et al, 2008; Stadtfeld et al, 2008a), the conversion is highly inefficient (~0.01%), making it difficult to examine the underlying molecular events. Further, all hiPS cells to date have been generated with retroviruses and non-inducible lentiviruses, both of which are inefficiently silenced and maintain transgene expression. Another limitation in the generation of hiPS cells is the efficiency of viral gene transduction. While the fraction of cells infected by each individual factor and combinations of factors has not been addressed, only a small proportion of cells likely receive all factors. To overcome these limitations, we have established an inducible lentiviral system to generate “secondary” pluripotent cells, in which hiPS cell clones are differentiated in vitro to yield fibroblast-like cells that harbor the inducible viral transgenes required for reprogramming. Because these cells maintain the same viral integrations that mediated the initial conversion to hiPS cells, this system bypasses the need for direct viral infection and produces a population of cells that can inducibly and homogeneously express the reprogramming factors. Such a system provides a powerful tool for mechanistic analysis, chemical and genetic screening for factors that enhance or block reprogramming, and the optimization of hiPS cell derivation methods.
Results
cDNAs encoding human OCT4, SOX2, cMYC, KLF4, and NANOG were cloned into doxycycline-inducible lentiviral vectors as previously described (Stadtfeld et al, 2008b). In addition, a reverse tetracycline transactivator (rtTA) driven by the ubiquitin promoter was cloned into a lentiviral vector. To generate hiPS cells, we infected neonatal foreskin fibroblasts (BJ) and keratinocytes with lentiviruses containing the rtTA and either four (OCT4, SOX2, cMYC, and KLF4; for fibroblasts only) or five reprogramming factors (4 + NANOG; both fibroblasts and keratinocytes) according to the scheme in Figure 1A. Following infection, cells were plated onto mouse embryonic fibroblast feeder cells (MEFs) under hES cell conditions and induced with doxycycline. From the fibroblast cultures, non-hES-like colonies emerged approximately two weeks after the addition of doxycycline, as previously observed (Takahashi et al, 2007). These colonies contained only Oct4 and Myc integrations and could not be expanded in the absence of doxycycline (data not shown). After 30 days of culture, colonies that resembled hES cells were observed, noted by a high nucleus-to-cytoplasmic ratio, prominent nucleoli, and well-defined phase-bright borders. All hES-like colonies expressed the hES-specific surface antigen Tra-1–81 (data not shown) and could be expanded in the absence of doxycycline. Colonies that did not resemble hES cells did not express Tra-1–81 and could not be passaged independent of doxycycline. Of ~2.5×105 infected fibroblasts seeded, 4 iPS colonies were obtained from each condition (four- or five-factor), representing a frequency of ~0.002%. From the keratinocyte cultures, large non-ES-like colonies appeared within one week; within three weeks, hES-like colonies appeared and could be passaged in the absence of doxycycline. Of ~3×105 cells seeded, 7 colonies emerged, similar to the frequency observed for hiPS derivation from fibroblasts (~0.002%).
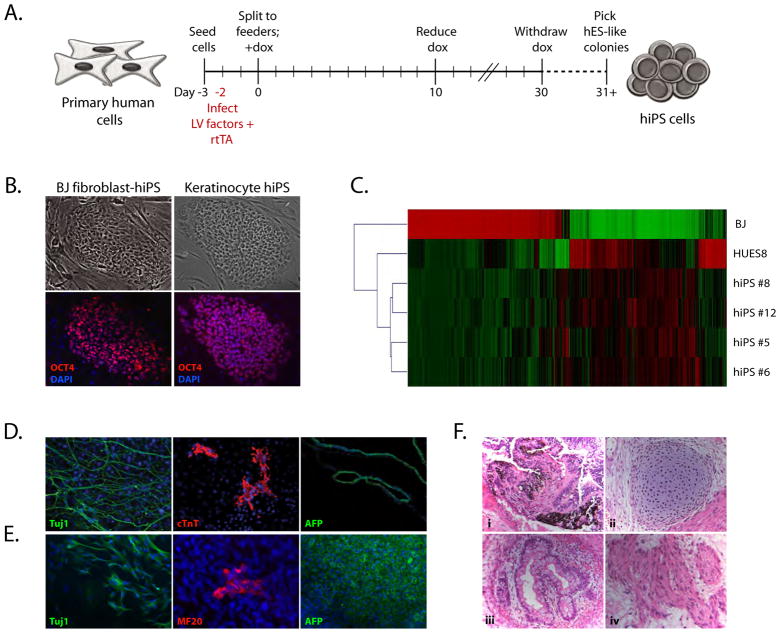
Figure 1. Generation of hiPS cells using inducible lentiviruses.
A. Experimental scheme for the generation of hiPS cells. Primary human fibroblasts and keratinocytes were infected with separate lentiviruses (LV) containing a constitutively active rtTA and doxycycline-inducible reprogramming factors. After infection, cells were seeded to feeders and doxycycline (dox) was applied for 30 days. hiPS clones were picked based on hESC-like morphology and doxycycline-independent growth.
B. Morphology and marker expression in hiPS colonies. Doxycycline-independent fibroblast- and keratinocyte-derived hiPS cells express OCT4 protein.
C. Microarray analysis of gene expression in hiPS cells. Genes with greater than twofold expression level between HUES8 hES cells and BJ fibroblasts were analyzed. Shown are BJ fibroblasts, HUES8 hES cells, and BJ fibroblast-derived hiPS clones.
D. In vitro differentiation of fibroblast-derived hiPS cells into lineages from all three germ layers. Immunostaining for i) Tuj1 (neuronal), ii) cardiac troponin T (cTnT; cardiac muscle), and iii) alpha-fetoprotein (AFP; epithelial, early endodermal).
E. In vitro differentiation of keratinocyte-derived hiPS cells into lineages from all three germ layers. Immunostaining for i) Tuj1, ii) skeletal muscle (MF20), and iii) alpha-fetoprotein.
F. Hematoxylin and eosin stain of teratomas generated from fibroblast-derived hiPS cells. Differentiated structures from all three germ layers were present. i) Pigmented epithelium (ectoderm), ii) cartilage (mesoderm), iii) gut-like epithelium (endoderm), and iv) muscle (mesoderm).
hiPS cell colonies stained positive for OCT4 protein and the hES cell-specific surface antigen Tra 1–81 (Figure 1B and data not shown). Further, these cells showed expression of pluripotency genes from the endogenous loci and lacked expression of the viral transgenes (Supplementary Figure 1A). To assess whether hiPS cells were molecularly similar to hES cells, we examined promoter methylation and performed global transcriptional analysis. The NANOG and OCT4 promoters in fibroblast-derived hiPS cells were demethylated to a similar extent as in hES cells, in contrast to the highly methylated promoters in BJ fibroblasts (Supplementary Figure 1B), thus demonstrating epigenetic reprogramming in hiPS cells. Global analysis of gene expression in fibroblast-derived hiPS cells, hES cells, and fibroblasts was conducted by microarray through comparison of differentially-expressed genes between fibroblasts and hES cells (Figure 1C), indicating that hiPS cells had repressed the fibroblast program of gene expression and reactivated an embryonic program of transcription.
Pluripotency of both fibroblast- and keratinocyte-derived hiPS cells was examined in vitro through embryoid body (EB) formation. After 7 days in suspension culture, EBs were explanted and gave rise to well-defined neuronal outgrowths and beating cardiomyocyte structures (data not shown). Immunofluorescence analysis confirmed the presence of neurons, cardiomyocytes, skeletal muscle cells, and epithelial structures (Figure 1D, E), thus demonstrating multi-lineage differentiation. As a more stringent test of pluripotency, fibroblast-derived hiPS cells were injected either subcutaneously or under the kidney capsule of immunodeficient SCID mice to assay for teratoma formation. Tumors were recovered after 10 weeks and contained well-defined structures arising from all three embryonic germ layers, including pigmented cells, cartilage, skeletal muscle, and gut-like epithelium (Figure 1F). These results indicate that hiPS cells generated with an inducible system strongly resemble hES cells and fulfill all criteria for pluripotency.
Noting that keratinocyte-derived hiPS colonies appeared faster than fibroblast-derived hiPS cells, we sought to determine the minimum amount of time required to convert keratinocytes to hiPS cells. To test this, keratinocytes were infected with rtTA and five factors (OCT4, SOX2, cMYC, KLF4, NANOG), and doxycycline was withdrawn at different time-points throughout the reprogramming process. The number of hES-like colonies was counted at day 30 and plotted against the day of doxycycline withdrawal (Supplemental Figure 1C). hES-like colonies first appeared after 18 days when doxycycline had been withdrawn after 10 days. The frequency of reprogramming appeared to decline with the length of doxycycline exposure, which may reflect unfavorable culture conditions at later time points or adverse effects of continued transgene expression.
To establish the system of “secondary” hiPS cells, we differentiated several fibroblast-derived hiPS clones to fibroblast-like cells in vitro according to the scheme in Figure 2A. hiPS cell colonies were placed in suspension culture for one week and the resulting EBs were then plated to adherent conditions. Outgrowths of fibroblast-like cells were picked and passaged a minimum of three times prior to experimental manipulation to ensure that no residual pluripotent cells were present. Quantitative RT-PCR analysis confirmed a lack of pluripotency gene expression in these populations (Supplementary Figure 2A).
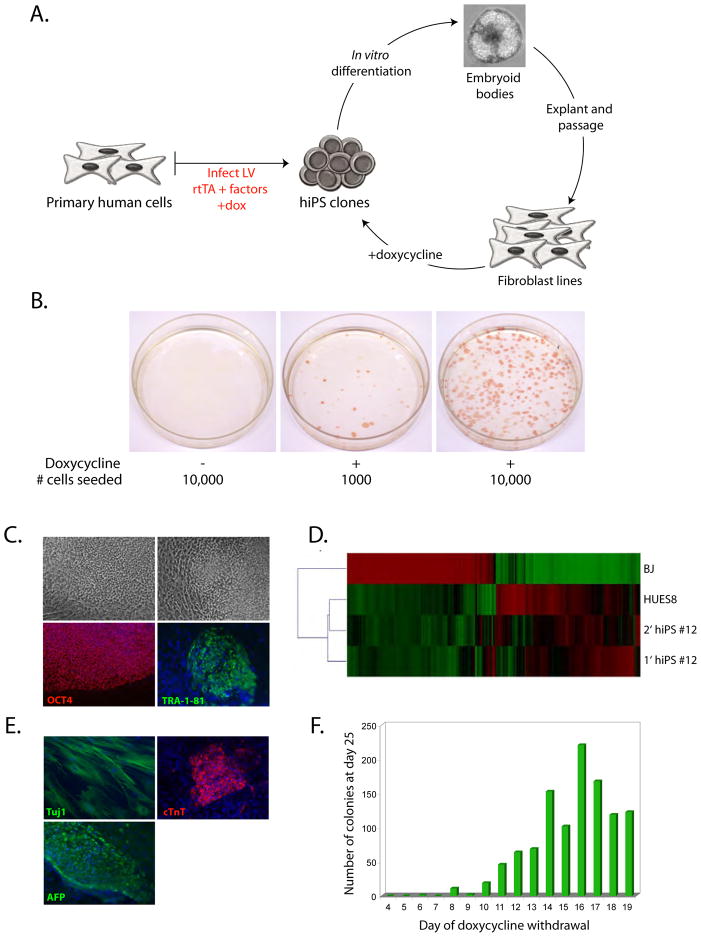
Figure 2. Generation of secondary hiPS cells.
A. Experimental scheme depicting the generation of secondary hiPS cells. hiPS cells were differentiated in vitro as embryoid bodies for 7 days, then plated to adherent conditions. Fibroblast-like colonies were picked and expanded for at least three passages prior to undergoing re-induction by doxycycline.
B. Alkaline phosphatase staining of reprogrammable cells grown in the absence or presence of doxycycline. Doxycycline was withdrawn at day 21, and colonies were stained and counted at day 30.
C. Morphology and expression of OCT4 and Tra-1–81 in doxycycline-independent secondary hiPS cells.
D. Microarray analysis of gene expression between BJ fibroblasts, HUES8 hES cells, primary fibroblast-derived hiPS cells, and a resulting secondary hiPS clone. Shown are genes with >2-fold expression value between BJ fibroblasts and HUES8 hES cells.
E. In vitro differentiation of secondary hiPS cells into lineages from all three germ layers. Immunostaining for i) Tuj1, ii) cardiac troponin T, and iii) alpha-fetoprotein.
F. Temporal requirement of factor expression in hiPS-derived fibroblast-like cells. 104 cells were plated per time-point and doxycycline was withdrawn daily from days 4 through 19. The number of hES-like colonies that expressed Tra-1–81 were counted at day 25.
We assessed the ability of hiPS-derived cells to generate “secondary” hiPS cells through doxycycline addition and transfer to hiPS derivation conditions. Fibroblast-like cells derived from two hiPS clones demonstrated reprogramming in the presence, but not absence, of doxycycline (Figure 2B). The frequency of conversion ranged from 1–3%. Re-induction of viral transgenes in hiPS-derived fibroblasts clones was also assessed by quantitative RT-PCR (Supplementary Figure 2B), demonstrating a correlation between factor reactivation and the ability of the clone to produce secondary hiPS. Clones that gave rise to secondary hiPS cells showed reactivation of all factors (BJ hiPS #11 and #12), while those that did not lacked re-expression of a factor (BJ hiPS #5) or re-expressed the factors at levels that are likely not permissive for the induction of pluripotency (BJ hiPS #8).
Secondary hiPS cells were molecularly and functionally similar to primary hiPS and hES cells. They stained positive for OCT4 and Tra-1–81 (Figure 2C), had a similar gene expression profile to hES cells (Figure 2D), and demonstrated pluripotency in vitro, generating cell types from all three embryonic germ layers (Figure 2E).
To determine the temporal requirement of transgene expression for secondary hiPS cell formation, we seeded 104 fibroblast-like cells per time point under hiPS derivation conditions and withdrew doxycycline daily from days 4 through 19. The final number of Tra-1–81+ hES-like colonies was counted at day 25 (Figure 2F). hiPS colonies began to appear after withdrawal of doxycycline at day 6 (one colony), and the number of colonies increased with the time of doxycycline exposure, reaching a maximum after withdrawal at 16 days (frequency of ~2%). This increase in frequency with length of doxycycline exposure has also been reported in the reprogramming of mouse cells (Wernig et al, 2008).
Transformed granular colonies did not appear during the reprogramming process, in contrast to the early colonies that appeared during the primary induction with direct viral infection. The lack of these background colonies coupled with the high efficiency of the secondary system enabled us to monitor the progression of colonies during reprogramming. We tracked individual colonies on a daily basis with different time points of doxycycline withdrawal (Supplementary Figure 3), and observed that induced cells transited through non-ES-like structures prior to acquiring a hiPS cell phenotype. Not all colonies developed fully to hiPS cells; those that did not undergo successful reprogramming began to regress two days after doxycycline withdrawal and ultimately formed fibroblast-like structures. The colonies that successfully generated hiPS cells also showed some regression after withdrawal of doxycycline, however, hES-like outgrowths gradually appeared, which could be expanded into stable hiPS cell lines.
Discussion
Here we have described the use of an inducible lentiviral system for the reprogramming of human somatic cells. Using this system, we converted neonatal foreskin fibroblasts and keratinocytes to a pluripotent state molecularly and functionally similar to hES cells. This system also enabled us to establish the temporal requirement of factor expression for cells undergoing reprogramming. While fibroblasts relied on transgene expression for several weeks, keratinocytes required only 10 days of factor expression to revert to a state that was poised to become pluripotent. It is unknown why keratinocytes are more amenable to reprogramming. Keratinocytes, like hES cells, represent an epithelial cell type and in contrast to fibroblasts may not need to undergo a mesenchymal-epithelial transition during reprogramming (Yang and Weinberg, 2008). Differences in reprogrammability between fibroblasts and keratinocytes may also be explained by differences in the cell cycle status or viral infectivity. Moreover, keratinocytes express much higher levels of endogenous MYC and KLF4 than fibroblasts (Supplementary Figure 1A), which may accelerate their conversion to hiPS cells. The fast kinetics of reprogramming observed for keratinocytes suggests that these cells would be useful for the development and optimization of methods to reprogram cells by transient delivery of factors.
Controlled expression of the reprogramming factors provided an inherent selection strategy by eliminating cells that were dependent upon viral transgene expression and conferring a growth advantage to fully reprogrammed cells that were doxycycline-independent. This is in contrast to the constitutive retro- and lentiviral systems that have so far been used to reprogram human cells, where the viral transgenes maintain expression in the hiPS cell state. The persistence of viral gene expression could have deleterious effects in clinical applications such as in vitro disease modeling; for example, the overexpression of Oct4 or Sox2 has been shown to promote the differentiation of mouse ES cells (Niwa et al, 2000; Kopp et al, 2008), suggesting that their continued expression during in vitro differentiation of hiPS cells may bias the resulting cell fate.
The extremely low efficiency of reprogramming led us to develop a system of “secondary” pluripotent cells in which we could reprogram hiPS-derived differentiated cells at a high frequency. The >100-fold increase we observed was likely attributed to the ability to reactivate all factors within a given cell, thus enabling efficient reprogramming. The kinetics of this process were faster than that of primary fibroblasts but similar to keratinocytes, with the highest efficiency occurring after 16 days of factor expression. In vitro-derived fibroblasts appeared to be more amenable to reprogramming than primary fibroblasts as previously observed (Yu et al, 2007; Park et al, Nature), which may reflect a lack of exposure to potent differentiation cues in vitro. However, four lines of evidence suggest that the reprogramming of hiPS-derived differentiated cells is representative of the process that occurs in primary cells: 1) hiPS-derived fibroblast-like cells lacked detectable expression of key pluripotency genes, 2) no colonies formed in the absence of doxycycline, 3) clones that did not reactivate all factors could not successfully form secondary hiPS cells, and 4) visual tracking of colonies demonstrated that cells transit through a non-hES-like state prior to becoming bona fide hiPS colonies. These data collectively support the use of a secondary system as a platform for mechanistic dissection of the reprogramming process.
Despite the fact that viral transgenes were reactivated in most of the hiPS-derived cells, the frequency of reprogramming remained quite low, ranging from 1–3%. The basis for the low efficiency is poorly understood but may reflect a multitude of factors such as the starting cell type, cell cycle status and the ability to undergo replication, variability in factor reactivation, and other stochastic events. Epigenetic events have been implicated in the reprogramming process; for example, it has recently been shown that valproic acid, which acts primarily as a histone deacetylase inhibitor, can enhance the efficiency of reprogramming (Huangfu et al, 2008). Also, DNA methylation and the activation of differentiation pathways have been shown to impede the reprogramming of mouse fibroblasts (Mikkelsen et al, 2008). However, these events account only for a small proportion of cells and it will be interesting to further define the limiting factors that constrain the efficiency of reprogramming.
The secondary system of hiPS cells presents an attractive model for which to study the molecular events that underlie the reversion of human somatic cells to a pluripotent state. By providing a homogeneous population of cells that harbor all the viral transgenes, the cultures are not subject to the background of cells that do not receive all factors, allowing proper analysis of the reprogramming process. The secondary system will enable chemical and genetic screening efforts to identify key molecular constituents of reprogramming, as well as important obstacles in this process, and will ultimately lend itself as a powerful tool in the development and optimization of methods to produce hiPS cells.
Experimental Procedures
Virus production
Vectors were constructed as previously described (Stadtfeld et al, 2008b). To generate virus, 293T cells were transfected at 30% confluence using Fugene 6 reagent (Roche). For a 10cm plate, 560uL DMEM, 27uL Fugene, and 12ug DNA (4:3:2 vector:Δ 8.9:vsv-g) was used. Virus was harvested over 3 days and concentrated 300-fold. For a standard infection in a 35mm dish (~105 cells), 10uL rtTA + 5uL factors (OCT4, SOX2, KLF4, NANOG) + 2uL cMYC was used in an overnight infection supplemented with 6ug/mL polybrene.
Cell culture and hiPS cell generation
Fibroblasts were grown in DMEM with 10% FBS, non-essential amino acids, glutamine, and β-mercaptoethanol; keratinocytes were cultured on collagen IV in keratinocyte serum-free medium and growth supplement (Invitrogen). Human ES and iPS cells were cultured as previously described (Cowan et al, NEJM, 2004). hiPS cells were generated as follows: Day 0, to 1μg/mL dox in hES media +2% defined FBS (Gibco) for fibroblasts, 1% FBS for keratinocytes; Day 2, hES +1μg/mL dox +1% FBS (fibroblasts) or hES +dox (keratinocytes); Day 4, hES +1μg/mL dox; Day 10, hES +0.5μg/mL dox (continued until colonies appeared).
Differentiation
For in vitro differentiation, hiPS cell colonies were mechanically picked and placed in suspension culture with fibroblast media. After one week, embryoid bodies were plated to adherent conditions with gelatin. For teratoma formation, ~107 hiPS cells were pelleted and injected into SCID mice, either subcutaneously above the dorsal flank or underneath the kidney capsule. Tumors were harvested after 10–12 weeks and processed for histological analysis.
Bisulfite sequencing
Genomic DNA was converted using the Epitect bisulfite kit (Qiagen). OCT4 and NANOG promoters were PCR amplified using primers listed in Supplementary Table 2. PCR products were cloned into TOPO vectors and sequenced.
Immunostaining
Immunostaining was performed using the following antibodies: α-Oct4 (sc-8628, Santa Cruz Biotech), α-Tra-1–81 (MAB4381, Millipore), α–β-III Tubulin (T2200, Sigma), α-cardiac troponin T (clone 13–11, Neomarkers), α-myosin heavy chain (clone MF20, Developmental Studies Hybridoma Bank), α-AFP (sc-15375, Santa Cruz Biotech).
qPCR
RNA was extracted using a Qiagen RNeasy kit, then converted to cDNA with the Superscript III First-Strand synthesis system (Invitrogen) using oligo-dT primers. qRT-PCRs were carried out using Brilliant II SYBR Green mix (Stratagene) and run on a Stratagene MXPro400. Reactions were carried out in duplicate with –RT controls, and data was analyzed using the delta-delta Ct method. Primer sequences are listed in Supplementary Table 2.
Whole genome expression analysis
Total RNA was isolated using an RNeasy kit (Qiagen). Samples were processed as independent triplicates. RNA probes for microarray hybridization were prepared and hybridized to Affymetrix HGU133 plus 2 oligonucleotide microarrays. Data was extracted and analyzed using Rosetta Resolver system. During importation, the data was subjected to background correction, intrachip normalization, and the Rosetta Resolver Affymetrix GeneChip error model (Weng et al, 2006). For the generation of intensity plots, genes that showed greater than a two-fold difference in expression value (p<0.01) in HUES8 hES cells and BJ fibroblasts were noted (19,663 probes) and their expression analyzed. A hierarchical clustering was performed.
Supplementary Material
Acknowledgments
We thank the Stowers Institute for Medical Research Microarray Group, Christopher Seidel, and Reddy Gali (Harvard FAS Life Sciences Research Computing) for assistance with transcriptional profiling and analysis; Jennifer Shay for technical assistance; and Matthias Stadtfeld and Jose Polo for critical reading of the manuscript. N.M. is supported by a graduate scholarship from the Natural Sciences and Engineering Research Council of Canada and a Sir James Lougheed Award from Alberta Scholarships. J.U. is supported by the Dr. Mildred Scheel Foundation for Cancer Research. C.C. is supported by the Harvard Stem Cell Institute and the Stowers Institute for Medical Research. K.H. is supported by an NIH Director’s Innovator Award, the Harvard Stem Cell Institute, the Kimmel Foundation, and the V Foundation.
References
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of Pluripotent Stem Cells from Adult Mouse Liver and Stomach Cells. Science. 2008 doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–7. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, Ormsbee BD, Desler M, Rizzino A. Small increases in the level of sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells. 2008;26:903–911. doi: 10.1634/stemcells.2007-0951. [DOI] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic reprogramming and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008a;2:230–40. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Brennand K, Hochedlinger K. Reprogramming of Pancreatic beta Cells into Induced Pluripotent Stem Cells. Curr Biol. 2008b;18:890–4. doi: 10.1016/j.cub.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Weng L, Dai H, Zhan Y, He Y, Stepaniants SB, Bassett DE. Rosetta error model for gene expression analysis. Bioinformatics. 2006;22:1111–1121. doi: 10.1093/bioinformatics/btl045. [DOI] [PubMed] [Google Scholar]
- Wernig M, Lengner CJ, Hanna J, Lodato MA, Steine EJ, Foreman R, Staerk J, Markoulaki S, Jaenisch R. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008 doi: 10.1038/nbt1483. AOP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development tumor metastasis. Dev Cell. 2008;14:818–29. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.