Summary
Induced pluripotent stem cells (iPSCs) have been generated by enforced expression of defined sets of transcription factors in somatic cells. It remains controversial whether iPSCs are molecularly and functionally equivalent to blastocyst-derived embryonic stem cells (ESCs). By comparing genetically identical mouse ESCs and iPSCs, we show here that the overall mRNA and miRNA expression patterns of these cell types are indistinguishable with the exception of a few transcripts and miRNAs encoded on chromosome 12qF1. Specifically, maternally expressed imprinted genes in the Dlk1-Dio3 cluster including Gtl2, Rian and Mirg as well as a larger number of miRNAs encoded within this region were aberrantly silenced in the majority of iPSC clones, irrespective of their cell type of origin. Consistent with a developmental role of the Dlk1-Dio3 gene cluster, iPSC clones with repressed Gtl2 contributed poorly to chimeras and failed to support the development of entirely iPSC-derived animals (“all-iPSC mice”). In contrast, iPSC clones with normal expression levels of these genes contributed to high-grade chimeras and generated viable all-iPSC mice. Importantly, treatment of an iPSC clone that had silenced Dlk1-Dio3 and failed to give rise to all-iPSC animals with a histone deacetylase inhibitor reactivated the locus and rescued its ability to support full-term development of exclusively iPSC-derived mice. Thus, the expression state of a single imprinted gene cluster distinguishes most murine iPSCs from ESCs and allows for the prospective identification of iPSC clones that have the full development potential of ESCs.
Induced pluripotent stem cells (iPSCs), generated by overexpression of transcription factors such as Oct4, Sox2, Klf4 and c-Myc in somatic cells1, have enormous therapeutic potential as they enable the derivation of patient-specific pluripotent cell lines to study and possibly treat degenerative diseases. However, it remains debated if iPSCs are molecularly and functionally equivalent to blastocyst-derived ESCs, the gold standard for pluripotent cells. For example, recent studies have reported major mRNA and miRNA expression differences between ESCs and iPSCs in both mouse and human2,3,4. At a functional level, many iPSC clones give rise to low-grade chimeras after injection into blastocysts, indicating a poorer developmental potential of iPSCs compared with ESCs. Nevertheless, three recent reports claimed the generation of all-iPSC mice, demonstrating that at least some iPSCs are functionally indistinguishable from ESCs5,6,7.
We took advantage of genetically matched mouse ESCs and derivative iPSCs to screen for possible molecular and functional differences between these two pluripotent cell types. Briefly, a polycistronic cassette expressing Oct4, Klf4, Sox2, and c-Myc under the control of a doxycycline-inducible promoter was inserted into the Col1a1 locus of ESCs cells expressing the reverse tetracycline-dependent transactivator (rtTA) from the ROSA26 promoter8. These ESCs (designated Collagen-OKSM ESCs) were then used to generate mice from which different somatic cell types were isolated and induced with doxycycline to derive genetically matched iPSCs for molecular and functional comparisons (Figure 1a,b).
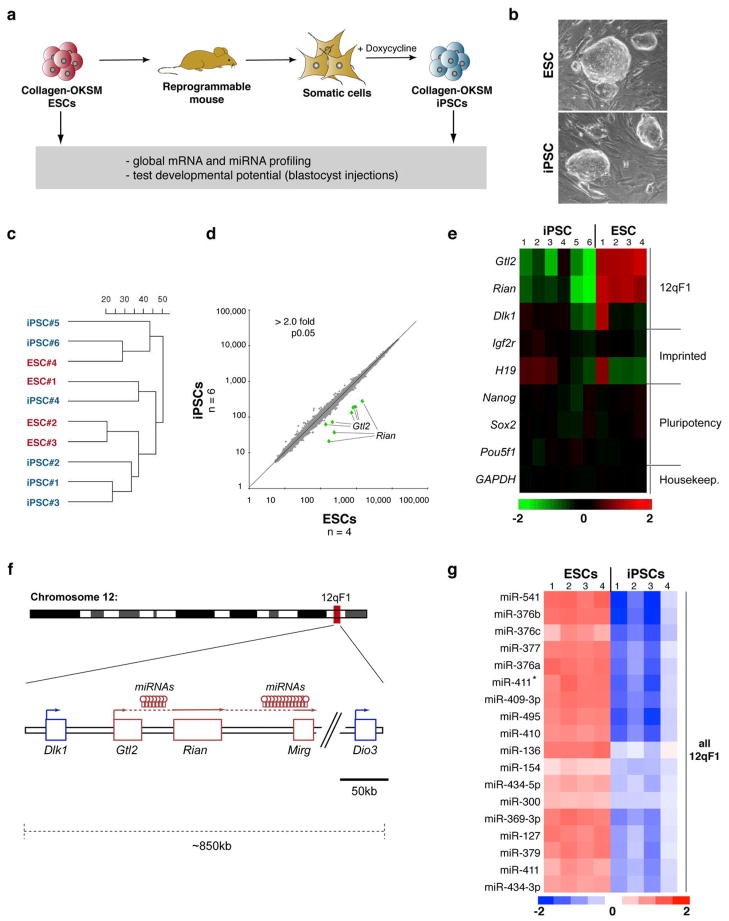
Figure 1. Aberrant silencing of the Dlk1-Dio3 gene cluster in mouse iPSCs.
(a) Strategy for comparing genetically matched ESCs and iPSCs using “reprogrammable mice” harboring a doxycycline-inducible polycistronic reprogramming cassette (OKSM) in the Col1a1 (Collagen) locus. (b) Morphology of Collagen-OKSM ESCs and iPSCs. (c) Unsupervised clustering of four ESC and six iPSC lines based on microarray expression data. (d) Scatterplot of microarray data comparing iPSCs and ESCs with differentially expressed genes highlighted in green (2-fold, p0.05, t-test with Benjamini-Hochberg correction). (e) Heatmap showing relative expression levels of selected mRNAs in ESCs and iPSCs, covering in addition to Gtl2 and Rian other imprinted genes (Dlk1, Igf2r and H19) and pluripotency-associated transcripts (Nanog, Sox2 and Pou5f1). (f) Schematic representation of mouse chromosome 12 with position of the Dlk1-Dio3 gene cluster highlighted. Maternally-expressed and paternally-expressed transcripts are shown in red and blue, respectively. (g) Heatmap showing miRNAs that are differentially expressed between ESCs and iPSCs (2-fold, p0.01, t-test).
We first compared the abilities of parental Collagen-OKSM ESCs and iPSCs derived from mouse embryonic fibroblasts (MEFs) that had been isolated from ESC-chimeric fetuses, to support the development of all-iPSC mice using tetraploid (4n) embryo complementation9,10. In this assay, iPSCs or ESCs are injected into 4n host blastocysts, which can only give rise to extra-embryonic tissues, whereas the injected pluripotent cells generate the entire mouse conceptus. Two tested ESC lines gave rise to neonatal and adult mice at expected frequencies (13–20%)10, demonstrating that the OKSM transgene per se does not affect the developmental potential of these cells (Supplemental Table 1). In contrast, all four tested iPSC lines repeatedly failed to support the development of all-iPSC mice, indicating qualitative differences between ESCs and these iPSC clones (Supplemental Table 1).
We reasoned that a transcriptional comparison of the iPSC lines, which failed in the 4n complementation assay, with their parental ESC lines that supported the development of all-ESC mice, might reveal molecular changes that could explain the developmental deficits of iPSCs. Global mRNA profiling showed striking similarities in the overall transcriptional patterns of four Collagen-OKSM ESCs and six iPSCs and did not separate these cells using unsupervised clustering or principal component analysis (Figure 1c and data not shown). In fact, only two transcripts were identified as differentially expressed (>2-fold difference, t-test, p<0.05) between ESCs and iPSCs. These were the non-coding cDNA Gtl2 (also know as Meg3) and the small nucleolar RNA (snoRNA) Rian (Figure 1d, e).
Gtl2 and Rian localize to the imprinted Dlk1-Dio3 gene cluster on mouse chromosome 12qF1 and are maternally expressed in mammals (Figure 1f)11. Of note, both genes were strongly repressed in iPSC clones compared to ESC clones while expression of pluripotency and housekeeping genes remained unaffected (Figure 1e). Quantitative PCR (qPCR) analysis of Gtl2, Rian and Mirg, another maternally expressed imprinted gene in the Dlk1-Dio3 cluster, confirmed transcriptional silencing in iPSCs (Supplemental Figure 1a).
Interestingly, expression of the paternally expressed Dlk1 gene, that also localizes to chromosome 12qF1, and of other imprinted genes including H19 and Igf2r, showed clone-to-clone variations, as was seen previously for ESCs12, but no consistent expression differences between ESCs and iPSCs. This shows that imprinted gene silencing is not a genome-wide phenomenon in iPSCs (Figure 1e and Supplemental Table 2). Moreover, none of the almost 300 genes that had previously been reported to be differentially expressed between iPSCs and ESCs2 was changed in Collagen-OKSM iPSCs (Supplemental Figure 2a). These data indicate that a relatively small set of transcripts distinguishes genetically matched iPSCs and ESCs and suggest that the majority of previously seen differences are likely due to variations in genetic background or viral transgene insertions.
Imprinting of the Dlk1-Dio3 locus is accompanied by differential expression of about 50 miRNAs that are also encoded within the gene cluster (Figure 1f)13,14. To evaluate if miRNAs are differentially expressed between ESCs and iPSCs, we performed genome-wide miRNA profiling on the same samples as analyzed for mRNA expression. Of 336 miRNAs detected, 21 (6.3%) were differentially expressed between all ESC and iPSC clones analyzed (Figure 1g and Supplemental Table 3). Remarkably, all of these miRNAs localized to chromosome 12qF1 and were silenced in iPSC, thus corroborating the notion that most iPSCs show aberrant silencing of this major imprinting domain.
To determine the generality of Gtl2 silencing in iPSCs, we analyzed its expression in 61 additional iPSC lines derived from hematopoietic stem cells (HSC, 11 lines), granulocyte-macrophage progenitors (GMP, 11 lines), granulocytes (Gran, 9 lines), peritoneal fibroblasts (PF, 6 lines), tail tip fibroblasts (TTF, 6 lines) and keratinocytes (18 lines) (Figure 2a and Supplemental Figure 1b,c). Only four of these lines (5.8%), all originating from either peritoneal or tail tip fibroblasts, showed Gtl2 expression levels similar to ESCs (termed “Gtl2on clones”). It remains to be determined if the observed low expression levels of Gtl2 in hematopoietic cells (Supplemental Figure 1d) affects silencing of the locus in resultant iPSCs, and whether iPSCs derived from distinct cell types exhibit discernible global gene expression patterns. However, the finding that the vast majority of iPSC clones derived from different somatic cell types showed partial or complete suppression of Gtl2 expression (termed “Gtl2off clones”) demonstrates that silencing of this locus occurs in iPSCs regardless of their cell of origin. In agreement with our data, analyses of published microarray datasets comparing ESCs and iPSCs derived from mouse fibroblasts, neural and bone marrow cells also showed repression of maternally expressed 12qF1 transcripts (Supplemental Figure 2b–e), supporting the notion that silencing of this cluster is common upon factor-mediated reprogramming.
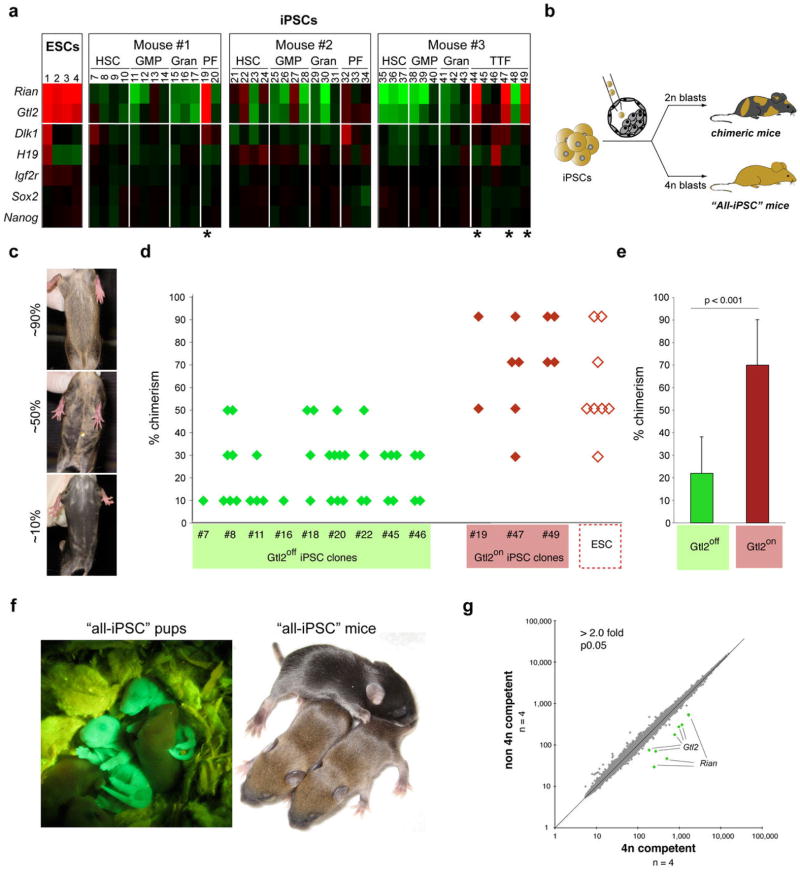
Figure 2. Full developmental potential of Gtl2on iPSCs.
(a) Heatmap showing relative expression levels of Gtl2, Rian, other selected imprinted genes (Dlk1, H19 and Igf2r) and pluripotency-associated transcripts (Sox2 and Nanog) in ESCs and iPSCs derived from hematopoietic stem cells (HSC), granulocyte-macrophage progenitors (GMP), granulocytes (Gran), peritoneal fibroblasts (PF) and tail-tip fibroblasts (TTFs), isolated from three individual reprogrammable mice. Four iPSC clones expressing ESC-like levels of Gtl2 and Rian were identified (highlighted by asterisks) (for technical reasons, iPSC clone #18 could not be analyzed by microarray but instead was evaluated by qPCR. See Supplemental Figure 1b). (b) Strategy for assessing the developmental potential of iPSC clones by injection into diploid (2n) and tetraploid (4n) blastocysts to produce chimeric or all-iPSC mice, respectively. (c) Images of representative coat color chimeras with agouti indicating iPSC origin. (d) Coat color chimerism in mice derived from indicated Gtl2off (green diamonds), Gtl2on iPSC clones (red diamonds) and ESCs (open diamonds). (e) Statistical analysis of coat color chimerism in mice derived form Gtl2off and Gtl2on iPSC clones. (f) Image of two GFP+ all-iPSC pups (left panel) and two agouti all-iPSC mice (right). (g) Scatterplot showing intensity levels of all probesets covered by microarray analysis with those highlighted in green that were significantly different between 4n complementation-competent iPSCs (clones #19, #44, #47 and #49) and non-4n complementation-competent iPSCs (clones #18, #20, #45 and #48) (2-fold, p0.05, t-test with Benjamini-Hochberg correction).
It remains unclear whether similar expression abnormalities are seen in human iPSCs. While a preliminary evaluation of published expression data15 did not indicate aberrant expression of the human Gtl2 homolog MEG-3 in human iPSCs compared with ESCs, our observations in mouse cells suggest that a meaningful answer to this question requires the establishment of genetically matched human ESCs and iPSCs.
Dysregulation of genes within the Dlk1-Dio3 cluster can be detrimental during pre-and postnatal mouse development16,17,18,19. To assess whether the expression status of Gtl2 and associated transcripts correlates with the developmental potential of iPSC, we injected a total of nine Gtl2off clones (3 HSC-iPSC, 1 GMP-iPSC, 2 PF-iPSC, 2 TTF-iPSC) into diploid blastocysts, which gave rise to 38 adult chimeras that exhibited low to medium degree (10–50%) coat color chimerism (Figure 2b–e and Supplemental Table 4). In contrast, three Gtl2on iPSC clones (1 PF-iPSC, 2 TTF-iPSC) injected into diploid blastocysts yielded 11 adult mice with a coat color chimerism ranging from 70–100%, similar to the chimerism seen with ESCs (Figure 2d and Supplemental Table 4). Importantly, all four Gtl2on iPSC clones supported the development of neonatal all-iPSC mice upon injection into 4n blastocysts at efficiencies similar to those seen with ESCs (7–19% for iPSCs compared with 13–20% for ESCs) (Supplemental Table 1). We confirmed that these mice were entirely iPSC-derived by PCR for strain-specific polymorphisms (Supplemental Figure 3), by detection of homogenous GFP fluorescence of all-iPSC neonates, originating from a ROSA26-EGFP allele that had been introduced into the parental ESCs, and by uniform agouti coat color of adolescent all-iPSC mice (Figure 2f). To our knowledge, this is the first demonstration of animals produced entirely from adult-derived iPSCs. In contrast to Gtl2on iPSC clones, injection of ten different Gtl2off iPSC clones (4 MEF-iPSC, 1 HSC-iPSC, 1 GMP-iPSC, 1 PF-iPSC, 3 TTF-iPSC) into 4n blastocysts consistently failed to produce all-iPSC pups but instead resulted in resorptions (Supplemental Table 1). Thus, the expression status of Gtl2 in iPSCs predicts their developmental potential into chimeric and all-iPSC mice. It remains to be tested whether 4n-competent iPSC clones can be derived from somatic cells other than fibroblasts.
To test whether Gtl2on and Gtl2off iPSCs could be distinguished by the expression of other genes, we performed global mRNA and miRNA expression profiling of four fibroblast-derived non-4n complementation-competent and four 4n complementation-competent iPSC lines. This analysis identified only Gtl2, Rian and a total of 26 miRNAs, which all localize to the Dlk1-Dio3 cluster, as differentially expressed (Figure 2g and Supplemental Table 5). The conclusion that the activation status of maternally expressed genes on chromosome 12qF1 is a strong indicator of the developmental potential of iPSCs was further supported by analysis of two published array datasets showing that Gtl2 was expressed in ESCs and 4n complementation-competent iPSC lines but was downregulated in non-4n complementation-competent iPSC lines5,7 (Supplemental Figure 4).
Imprinting of the Dlk1-Dio3 cluster is regulated by differentially methylated regions (DMRs) that become epigenetically modified in the germline. These include an intergenic DMR (IG-DMR), located between the Dlk1 and Glt2 genes19, and a DMR spanning the Gtl2 promoter (Gtl2 DMR)15. To determine whether aberrant DNA methylation might be responsible for the transcriptional silencing seen in Gtl2off iPSC lines, we compared the methylation status of the IG-DMR and Gtl2 DMR as well as that of three other CpG-rich regions on chromosome 12qF1 in ESCs, Gtl2on iPSCs, Gtl2off iPSCs and their parental tail-tip fibroblasts (Figure 3a). As expected for germline-imprinted regions, approximately 50% of CpGs within the IG-DMR and Gtl2 DMR were methylated in fibroblasts, ESCs and Gtl2on iPSCs, whereas close to 100% of CpGs within these DMRs were methylated in Gtl2off iPSC lines (Figure 3b and Supplemental Figure 5). The other CpG-rich regions analyzed remained unaffected (Supplemental Figure 5). Imprinting of the Dlk1-Dio3 cluster is also regulated by histone acetylation20 and chromatin immunoprecipitation experiments indeed revealed a significant decrease in activation marks such as methylated H3K4 and acetylated H3 and H4 in Gtl2off iPSC lines compared with Gtl2on iPSC lines and ESCs (Figure 3c). Together, these observations demonstrate that the normally expressed maternal Gtl2 allele has acquired an aberrant paternal-like silenced state in Gtl2off iPSC clones.
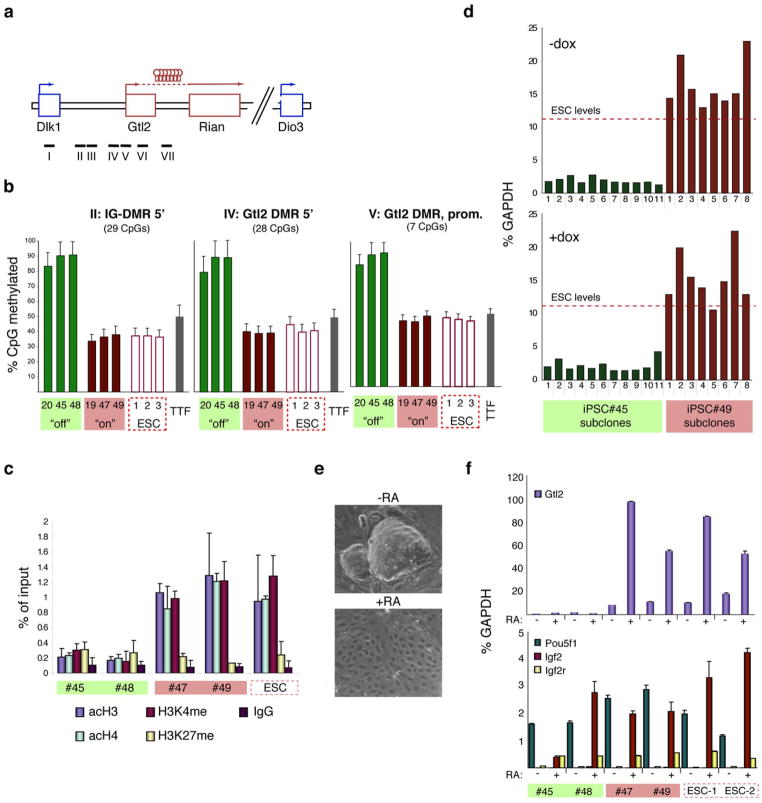
Figure 3. Epigenetic silencing of the Gtl2 locus in iPSCs.
(a) Structure of the Dlk1-Dio3 locus with the position of the genomic regions analyzed by pyrosequencing indicated by black bars. (b) Degree of DNA methylation at IG-DMR and Gtl2 DMR in three Gtl2off iPSC clones (green bars), three Gtl2on iPSC clones (red bars), three ESCs clones (red open bars), as well as parental tail-tip fibroblasts (TTFs, grey bars). The methylation status of the other regions is shown in Supplemental Figure 5. (c) Prevalence of activation-associated (acH3, acH4 and H3K4me) and repression-associated (H3K27me) chromatin marks at the Gtl2 promoter in two Gtl2off iPSC clones, two Gtl2on iPSCs clones and ESCs. (d) Gtl2 expression levels as measured by qPCR in subclones derived from Gtl2off clone #45 and Gtl2on clone #49 in the absence (upper panel) or presence (lower panel) of doxycycline (dox). (e) Representative brightfield images of iPSCs culture in the absence or presence of all-trans retinoic acid (RA). (f) Expression levels of Gtl2, other imprinted genes (Igf2, Igf2r) and the pluripotency marker Pou5f1 in cells cultured with (+) or without (-) retinoic acid (RA). Note that the two Gtl2off clones fail to activate Gtl2, but show normal expression levels of the other imprinted genes.
Imprinted gene expression is unstable in murine ESCs12,21. To evaluate if silencing of the Dlk1-Dio3 locus in iPSCs is maintained, we derived subclones from Gtl2off and Gtl2on iPSCs and assessed Gtl2 expression by qPCR. The Gtl2 locus remained silent in all Gtl2off iPSC clones and continued to be expressed in all Gtl2on iPSC clones, demonstrating stability of the Gtl2 expression state in undifferentiated iPSCs (Figure 3d, top). This pattern was not altered if doxycycline was adminstered during the subcloning procedure (Figure 3d, bottom), thus indicating that overexpression of the reprogramming factors in established iPSCs is insufficient to induce silencing.
To assess if silencing of Gtl2 might be resolved during differentiation, we exposed Gtl2off and Gtl2on iPSCs as well as ESCs to the differentiation-stimulating agent retinoic acid (RA) for 5 days. Dramatic changes in cellular morphology and downregulation of Pou5f1 in all RA-treated clones indicated successful differentiation (Figure 3e,f). Whereas Gtl2on iPSCs and ESCs readily upregulated Gtl2 (Figure 3f, top) and Rian (Supplemental Figure 6) during differentiation, Gtl2off iPSCs showed stable silencing of these genes, demonstrating that in vitro differentiation fails to reactivate maternally imprinted genes in the Dlk1-Dio3 cluster. The expression of imprinted genes outside of chromosome 12qF1 was not affected (Figure 3f, bottom, and Supplemental Figure 6).
Because Gtl2off iPSC clones failed to produce viable all-iPSC mice, we next sought to determine if they could autonomously support development into early embryos. Injection of Gtl2off and Gtl2on iPSC clones into 4n blastocysts gave rise to normal-appearing embryos at midgestation (E11.5) (Figure 4a). However, the number of living E11.5 embryos obtained from Gtl2off iPSC clones was reduced compared with embryos obtained from Gtl2on iPSC clones (Figure 4b), suggesting that Gtl2off mice die around this developmental stage. This phenotype resembles that of mice with paternal uniparental disomy of distal chromosome 1222, which die before E16.5, but is distinct from that of maternal Gtl2 knock-out mice (Gtl2mKO), which die perinatally16. The less severe phenotype of Gtl2mKO embryos compared with Gtl2off embryos might be due to the comparably modest reduction in maternally expressed 12qF1 genes seen in Gtl2mKO mice16. For example, Rian and Mirg transcripts were low but detectable in Gtl2mKO MEFs (Figure 4c). In contrast, these genes were almost completely silenced in MEFs and different tissues derived from Gtl2off all-iPSC embryos (Figure 4d,e). Notably, expression of the Dlk1 gene, which is reciprocally imprinted to Gtl223, was upregulated in Gtl2off MEFs but not in Gtl2mKO MEFs (Figure 4c), further supporting the observation that the maternal Dlk1-Dio3 cluster has acquired a paternal-like expression state. Accordingly, the IG-DMR and Gtl2-DMR were hypermethylated in Gtl2off MEFs but remained unaffected in Gtl2mKO MEFs (Figure 4f). Together, these observations are in agreement with the notion that stable transcriptional repression of the Dlk1-Dio3 locus is the cause for the developmental failure of Gtl2off all-iPSC embryos.
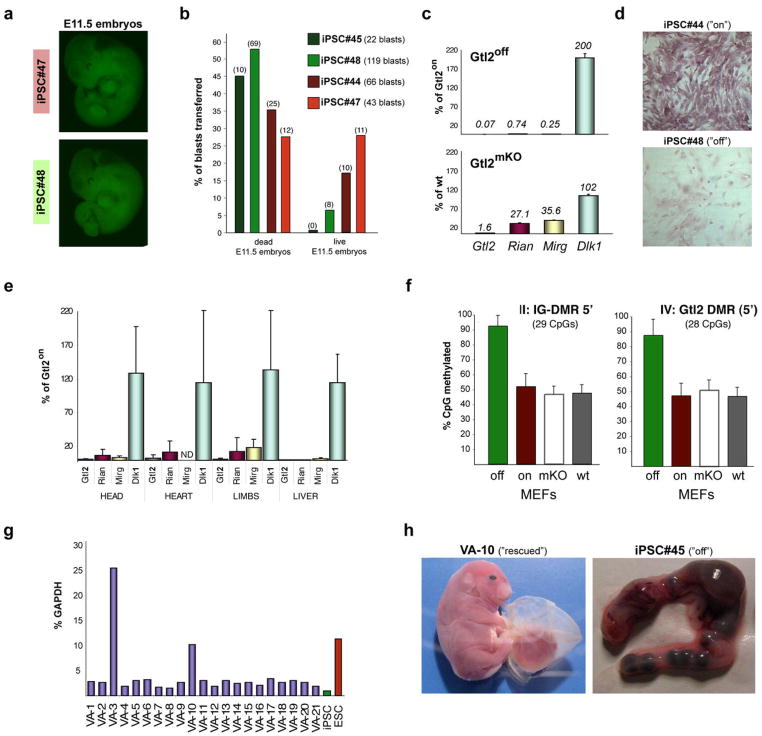
Figure 4. Developmental defects in embryos derived from Gtl2off iPSCs.
(a) Green fluorescence images of “all-iPSC” E11.5 embryos obtained with Gtl2on clone #47 (upper panel) and Gtl2off clone #48 (lower panel), both of which express EGFP from the ubiquitous ROSA26 locus. (b) Frequency of dead and living all-iPSC embryos obtained with two Gtl2on (red bars) and two Gtl2off (green bars) iPSC clones upon 4n blastocyst injection. Numbers of blastocysts transferred per clone and numbers of embryos recovered are indicated in brackets. (c) Expression of Glt2, Rian, Mirg and the paternally expressed gene Dlk1 in Gtl2off MEFs relative to Gtl2on MEFs (upper panel) as well as in Gtl2mKO MEFs relative to MEFs isolated from wildtype embryos (lower panel). (d) In situ hybridization against Gtl2 mRNA in MEFs derived from all-iPSC embryos generated with either Gtl2on clone #44 or Gtl2off clone #48. (e) Expression levels of Gtl2, Rian, Mirg and Dlk1 in the indicated tissues isolated from all-iPSC embryos made with Gtl2off iPSCs relative to the levels seen in tissues derived from Gtl2on iPSCs. (f) Degree of DNA methylation at the indicated regions in Gtl2off, Gtl2on, Gtl2mKO and wildtype MEFs. (g) Gtl2 expression levels in iPSC lines derived by subcloning Gtl2off clone #45 in the presence of valproic acid (VA). (h) Images of a fully developed stillborn pup (left) and a uterus filled with resorptions (right) derived after 4n blastocyst injections with either VA-10 or the parental iPSC clone #45, respectively.
ESCs derived from cloned embryos are transcriptionally identical with ESCs produced from fertilized embryos and also support the development of all-ESC mice, regardless of donor cell identity24, indicating that nuclear transfer (NT) generates faithfully reprogrammed pluripotent cells (Supplementary Figure 7a). In agreement with this observation, Gtl2 is expressed in 4n complementation-competent control ESC and NT ESC lines derived from fibroblasts and hematopoietic cells (Supplemental Figure 7b). We therefore tested whether NT could reverse the aberrant silencing of genes within the Dlk1-Dio3 cluster in Gtl2off iPSCs and rescue their ability to support the development of all-iPSC mice (Supplemental Figure 7c). To this end, we derived nine NT ESC lines from Gtl2off iPSCs that had been generated from TTFs and fetal liver cultures using adenoviral vectors25 or from hematopoietic stem cells and granulocytes using the Collagen-OKSM system. Some of these iPSCs were germline competent25, indicating that they were genetically normal, but failed to give rise to all-iPSC mice (Supplemental Table 6). Global transcriptome analysis showed no consistent differences in mRNA and miRNA expression profiles between NT ESCs and the donor iPSC clones. Most importantly, Gtl2 and Rian remained repressed in all NT ESCs (Supplemental Figure 7d). Accordingly, these cells failed to generate all-iPSC mice (Supplemental Table 6), suggesting that NT cannot reset the aberrant gene expression patterns and rescue the limited developmental potential acquired during iPSC generation. This notion is consistent with the previous finding that aberrant genomic imprints present in somatic donor cells cannot be restored in cloned animals following nuclear transfer12.
Given that Gtl2off iPSC clones showed reduced histone acetylation at the Gtl2 locus (Figure 3c), we wondered whether treatment of Gtl2off iPSC clones with the histone deacetylase inhibitor valproic acid (VA) could reactivate the silenced gene cluster. Indeed, two out of 21 subclones treated with VA exhibited increased Gtl2 expression with one iPSC clone showing expression levels comparable to ESCs (Figure 4g). Consistent with transcriptional reactivation of the cluster, we found re-appearance of H3K4 methylation and H3 acetylation at the Gtl2 locus in this rescued clone (Supplemental Figure 8). Injection of this clone into 4n blastocysts gave rise to apparently normal midgestation (E11.5) embryos at frequencies similar to those seen with Gtl2on iPSC clones (Figure 4b and Supplemental Figure 9a). These embryos expressed Gtl2, Rian and Mirg at significantly higher levels compared with embryos produced with Gtl2off iPSC clones (Supplemental 10a) and also showed normal expression levels of tissue-specific marker genes such as Mash-1 and Hes-5 that were repressed in Gtl2off embryos and thus might represent direct or indirect targets of one of the miRNAs encoded in Dlk1-Dio3 (Supplemental Figure 10b). Importantly, the rescued clone supported the development of several full-term pups, which was not seen with the parental iPSCs or any other Gtl2off clone (Figure 4h and Supplemental 9b). These pups were severely overgrown, however, and hence non-viable. We surmise that the observed overexpression of Dlk1 in the rescued iPSC clone (Supplemental Figure 10a), which causes neonatal lethality due to fetal overgrowth19, is responsible for this phenotype. Alternatively, VA treatment may have caused the dysregulation of other genes even though we found no aberrant expression of several candidate imprinted genes implicated in growth control (Supplemental Figure 10c).
Our data show that the expression of a surprisingly small number of transcripts and miRNAs, which localize to a single cluster in the genome, distinguishes most mouse iPSCs from ESCs and is predictive for their developmental potential. It remains to be tested whether human iPSCs show a similar dysregulation of genes, which may affect their utility in drug screening and therapy. Understanding the causes for the specific silencing of the Dlk1-Dio3 cluster during factor-mediated reprogramming will shed light on the molecular mechanisms of reprogramming as well as on the epigenetic regulation of this particular locus. Such studies may also lead to improved reprogramming strategies that faithfully establish a fully pluripotent state in somatic cells.
Methods
Generation of OKSM ESCs
A polycistronic cassette encoding Oct4, Klf4, Sox2 and c-Myc was cloned into the shuttle plasmid pBS31 using NotI/ClaI digestion. The resulting plasmid was electroporated into KH2 ESCs26 together with a plasmid driving expression of Flp recombinase. Correctly targeted clones were isolated by hygromycin selection and confirmed by Southern blot analysis as previously described26. Individual OKSM ESC subclones were gene targeted with ROSA26-EGFP as has been described before27 to facilitate tracking of ESC-derived cells after blastocyst injection. OKSM ESCs and derivative mice are described in detail elsewhere8.
Cell culture
ESCs and iPSCs were cultured in ESC medium (DMEM with 15% FBS, L-Glutamin, penicillin-streptomycin, non-essential amino acids, β-mercaptoethanol and 1000 U/ml LIF) on irradiated feeder cells. Mouse embryonic fibroblasts (MEFs) were isolated by trypsin-digestion of midgestation (E14.5) ESC-chimeric embryos followed by culture in fibroblast medium (DMEM with 10% FBS, L-Glutamin, penicillin-streptomycin, non-essential amino acids and β-mercaptoethanol). 2 μg/ml puromycin was added to these cultures for five days to selected for ESC-derived cells. Tail-tip fibroblast (TTF) cultures were established by trypsin digestion of tail-tip biopsies taken from newborn (3–8 days of age) chimeric mice derived after blastocyst injection of ROSA26-EGFP targeted ESCs. ESC-derived cells were isolated based on GFP expression and maintained in fibroblast medium. For the establishment of peritoneal fibroblast (PF) cultures, adult OKSM strain mice were euthanized and roughly 1 square centimeter of peritoneal muscle isolated and chopped into small pieces in 0.25% Trypsin/EDTA in a 35mm cell culture vessel. After five minutes of incubation at 37°C, 6 ml fibroblast medium was added and the tissue resuspended several times through a pipette. PF cultures were maintained and propagated like MEF and TTF cultures. Hematopoietic cells were isolated from peripheral blood and bone marrow as previously described28. Briefly, freshly isolated bone marrow cells were isolated by FACS using the following surface marker combinations: CD150+CD48−ckit+Sca-1+lineage− for HSCs, FcγR+CD34+ckit+Sca-1−lineage− for GMPs and CD11bhighGr-1highckit− for granulocytes. Sorted cells were immediately plated on top of irradiated feeder layers in ESC medium containing doxycycline. For HSCs and GMPs, the medium was supplemented with Flt3-ligand (10 ng μl−1), SCF (10 ng μl−1) and TPO (10 ng μl−1). Doxycycline was withdrawn from all cells after two weeks and colonies picked and expanded using standard ESC culture techniques.
Reprogramming into iPSCs
Collagen-OKSM MEFs, TTFs and PFs were counted and seeded in fibroblast media at the desired density onto gelatin-coated plates that contained a layer of irradiated feeder cells. The next day, ES medium containing 2 μg/ml doxycycline was added and replenished every 3 days. Upon doxycycline withdrawal, cultures were washed twice with PBS and then continued in standard ESC medium until colonies were picked.
RNA isolation
ESCs and iPSCs grown on 35mm dishes were harvested when they reached about 50% confluency and preplated on non-gelatinized T25 flasks for 45 minutes to remove feeder cells. Cells were spun down and the pellet used for isolation of total RNA using the miRNeasy Mini Kit (QIAGEN) without DNase digestion. RNA was eluted from the columns using 50 μl RNase-free water or TE buffer, pH7.5 (10 mM Tris-HCl and 0.1 mM EDTA) and quantified using a Nanodrop (Nanodrop Technologies).
Quantitative PCR
cDNA was produced with the First Strand cDNA Synthesis Kit (Roche) using 1 μg of total RNA input. Real-time quantitative PCR reactions were set up in triplicate using 5 μl of cDNA (1:100 dilution) with the Brilliant II SYBR Green QPCR Master Mix (Stratagene) and run on a Mx3000P QPCR System (Stratagene). Primer sequences are listed in Supplemental Table 6.
mRNA profiling
Total RNA samples (RIN > 9) were subjected to transcriptomal analyses using Affymetrix U-133plus2.0 mRNA expression microarray as previously described29. Hierarchical clustering was performed using Cluster and Treeview software30 as well as the GeneSifter server (Geospiza, Seattle).
miRNA profiling
Total RNA was subjected to quality control consisting of RNA measurement on the Nanodrop (OD260/230 and OD260/280 had to be greater than 1.8) and a run on the Agilent Bioanalyser2100 (RIN values had to be higher than 7). The samples were then labeled using the miRCURY™ Hy3™/Hy5™ power labeling kit (Exiqon) and hybridized on the miRCURY™ LNA Array (v.11.0) (Exiqon). Labeling was determined to be successful when all capture probes for the control spike-in oligo nucleotides produced signals in the expected range. The quantified signals were normalized using the global Lowess (LOcally WEighted Scatterplot Smoothing) regression algorithm.
Blastocyst injections
2n and 4n blastocyst injections were performed as described before10. Briefly, female BDF1 mice were superovulated by intraperitoneal injection of PMS and hCG and mated to BDF1 stud males. Zygotes were isolated from females with a vaginal plug 24 hour after hCG injection. Zygotes for 2n injections were in vitro cultured for 3 days in vitro in KSOM media, blastocysts were identified, injected with ESCs or iPSCs and transferred into pseudopregnant recipient females. For 4n injections, zygotes were cultured overnight until they reached the 2-cell stage, at which point they were electrofused. One hour later, 1-cell embryos were carefully identified and separated from embryos that had failed to fuse, cultured in KSOM for another 2 days and then injected.
Nuclear transfer
Nuclear transfer was performed as previously described31. Briefly, donor iPSCs were cultured in collagen-coated dishes without a feeder layer for 3 days in standard ESC medium. To synchronize cells at metaphase, the cultures were cultured for 2 h in a medium containing 0.4 μg/ml nocodazole (Sigma-Aldrich), a microtubule polymerization inhibitor. Cells floating in the medium were collected. While being sucked into a transfer pipette, only the cells arrested at metaphase were selected and used as nuclear donors. The recipient oocytes were collected from mature B6CBF1 female mice. Micromanipulations were performed in M2 medium containing 5 μg/ml cytochalasin B (Sigma) and 1 μg/ml nocodazole in a micromanipulation chamber. Explantation of cloned blastocysts and ESC-derivation was done as described previously31.
Chromatin immunoprecipitation
20 million iPSCs, ESCs or MEFs were fixed with 1% formaldeyde for 10 minutes at room temperature (RT) and then lysed in 1ml lysis buffer (50mM Tris-HCl, pH 8.0, 10mM EDTA, 1% SDS, protease inhibitors) for 20 minutes on ice. The lysate was split into three tubes and sonicated using Bioruptor for five times five minutes at high intensity, 30 sec on −-30 sec off. After 10 minutes centrifugation, the supernatant was precleared for 1 hour at 4°C with agarose beads preblocked with BSA (1mg BSA for 10ml beads) in IP Buffer (50mMM Tris-HCl, pH8, 150mM NaCl, 2mM EDTA, 1% NP-40, 0.5% Sodium Deoxycholate, 0.1% SDS, protease inhibitors). 100ml of precleared chromatin per reaction diluted in 1ml IP Buffer in presence of 2ug antibody were used for each immunoprecipitation reaction according to manufacturer’s protocol. The antibodies used for this study were: anti-acH3 (06–599 Millipore), anti-acH4 (06–866, Millipore), anti-dimethyl K4 of H3 (07–030, Millipore), anti-trimethyl K27 of H3 (ab6002, Abcam) and normal rabbit IgG (Millipore). The precipitate was purified using Qiaquick PCR purification kit and was analyzed by qPCR using Brilliant II SYBR Green qPCR Master Mix (600828, Agilent Technologies) using the sequence specific primer sets. Gtl2: 5′-AGCCCCTGACTGATGTTCTG-3′ (FWD) and 5′-TGGAAGGGCGATTGGTAGAC-3′ (REV) and Pou5f1: 5′-GGAGGTGCAATGGCTGTCTTGTCC-3′ (FWD) and 5′-CTGCCTTGGGTCACCTTACACCTCAC-3′ (REV).
In situ hybridization
MEFs grown on coverslips were fixed with 4% formaldeyde/5% acetic acid in PBS for 15 minutes at RT. After extensive PBS washes, they were dehydrated in 70% ethanol and left overnight at 4°C. The next day, they were rehydrated in a series of ethanol dilutions and incubated in hybridization buffer (50% formamide-5X SSC-RNase inhibitors) for 1 hour at 65°C. The hybridization was done overnight in a humidified chamber using 400ng of sense or anti-sense Gtl2 specific probe/ml of hybridization buffer. The sense and antisense probes were synthesized by in vitro transcription with DIG RNA labeling mix (Roche) and SP6 and T7 polymerase, respectively, using Gtl2 cDNA amplified with the primers 5′-CTCTCGGGACTCCTGGCTCCAC-3′ (FWD) and 5′-GGGTCCAGCATGTCCCACAGGA-3′ (REV). The cells were serially washed and stained with an anti-DIG AP conjugated FAB fragment (1:2000 in blocking buffer) for 1 hour at RT. The detection was performed with NBT/BCIP reagent.
Pyrosequencing
Genomic DNA was isolated using the DNeasy Blood & Tissue Kit (QIAGEN). ESCs and iPSCs were preplated onto cell culture vessels for 45 minutes after harvesting to remove feeder cells. Genomic DNA was bisulfite- converted using the EpiTect Bisulfite Kit (QIAGEN) with 400 ng of input DNA. DNA was eluted with 10 ml and 1 ml of it was used for PCR. PCR products were sequenced using the Pyrosequencing PSQ96 HS System (Biotage AB) following the manufacturer’s instrunctions. The methylation status of each locus was analyzed using QCpG software (Biotage).
Supplementary Material
Acknowledgments
We are grateful to Hugh Arnold for assistance with GeneSifter analysis, Kat Coser for technical support doing Affymetrix expression profiling, S. Sato and M. Machida for technical assistance and Steffen Schubert for advice on miRNA isolation. We sincerely thank Jose Polo and members of the Hochedlinger lab for valuable discussions and suggestions and A. Umezawa for helpful discussions and generous support. M.S. was supported by a postdoctoral fellowship from the Schering Foundation, E.A. was supported by a Jane Coffin Childs postdoctoral fellowship and K.H. was supported by a NIH Director’s Innovator Award as well as by funds provided by the Harvard Stem Cell Institute and MGH.
References
- 1.Takahashi K, Yamanaka S. Cell. 2006;126(4):663. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Chin MH, Mason MJ, Xie W, et al. Cell stem cell. 2009;5(1):111. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchetto MC, Yeo GW, Kainohana O, et al. PloS one. 2009;4(9):e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson KD, Venkatasubrahmanyam S, Jia F, et al. Stem cells and development. 2009;18(5):749. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao XY, Li W, Lv Z, et al. Nature. 2009 [Google Scholar]
- 6.Boland MJ, Hazen JL, Nazor KL, et al. Nature. 2009 [Google Scholar]
- 7.Kang L, Wang J, Zhang Y, et al. Cell stem cell. 2009;5(2):135. doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Stadtfeld M, Maherali N, Borkent M, et al. Nature methods. 7(1):53. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy A, Gocza E, Diaz EM, et al. Development (Cambridge, England) 1990;110(3):815. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 10.Eggan K, Akutsu H, Loring J, et al. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(11):6209. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Rocha ST, Edwards CA, Ito M, et al. Trends Genet. 2008;24(6):306. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Humpherys D, Eggan K, Akutsu H, et al. Science (New York, NY. 2001;293(5527):95. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 13.Seitz H, Youngson N, Lin SP, et al. Nature genetics. 2003;34(3):261. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- 14.Seitz H, Royo H, Bortolin ML, et al. Genome research. 2004;14(9):1741. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J, Hu K, Smuga-Otto K, et al. Science (New York, NY. 2009;324(5928):797. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi N, Okamoto A, Kobayashi R, et al. Human molecular genetics. 2009;18(10):1879. doi: 10.1093/hmg/ddp108. [DOI] [PubMed] [Google Scholar]
- 17.Lin SP, Coan P, da Rocha ST, et al. Development (Cambridge, England) 2007;134(2):417. doi: 10.1242/dev.02726. [DOI] [PubMed] [Google Scholar]
- 18.Steshina EY, Carr MS, Glick EA, et al. BMC genetics. 2006;7:44. doi: 10.1186/1471-2156-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lin SP, Youngson N, Takada S, et al. Nat Genet. 2003;35(1):97. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 19.da Rocha ST, Charalambous M, Lin SP, et al. PLoS genetics. 2009;5(2):e1000392. doi: 10.1371/journal.pgen.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carr MS, Yevtodiyenko A, Schmidt CL, et al. Genomics. 2007;89(2):280. doi: 10.1016/j.ygeno.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dean W, Bowden L, Aitchison A, et al. Development (Cambridge, England) 1998;125(12):2273. doi: 10.1242/dev.125.12.2273. [DOI] [PubMed] [Google Scholar]
- 22.Tevendale M, Watkins M, Rasberry C, et al. Cytogenet Genome Res. 2006;113(1–4):215. doi: 10.1159/000090835. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt JV, Matteson PG, Jones BK, et al. Genes & development. 2000;14(16):1997. [PMC free article] [PubMed] [Google Scholar]
- 24.Brambrink T, Hochedlinger K, Bell G, et al. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(4):933. doi: 10.1073/pnas.0510485103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stadtfeld M, Nagaya M, Utikal J, et al. Science (New York, NY. 2008;322(5903):945. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beard C, Hochedlinger K, Plath K, et al. Genesis. 2006;44(1):23. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 27.Hochedlinger K, Yamada Y, Beard C, et al. Cell. 2005;121(3):465. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 28.Eminli S, Foudi A, Stadtfeld M, et al. Nature genetics. 2009;41(9):968. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coser KR, Chesnes J, Hur J, et al. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(24):13994. doi: 10.1073/pnas.2235866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eisen MB, Spellman PT, Brown PO, et al. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(25):14863. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono Y, Kono T. Biology of reproduction. 2006;75(2):210. doi: 10.1095/biolreprod.105.049171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.