Summary
RasGRP proteins are activators of Ras and other related small GTPases by the virtue of functioning as guanine nucleotide exchange factors (GEFs). In vertebrates, four RasGRP family members have been described. RasGRP-1 through −4 share many structural domains but there are also subtle differences between each of the different family members. Whereas SOS RasGEFs are ubiquitously expressed, RasGRP proteins are expressed in distinct patterns, such as in different cells of the hematopoietic system and in the brain. Most studies have concentrated on the role of RasGRP proteins in the development and function of immune cell types because of the predominant RasGRP expression profiles in these cells and the immune phenotypes of mice deficient for Rasgrp genes. However, more recent studies demonstrate that RasGRPs also play an important role in tumorigenesis. Examples are skin- and hematological-cancers but also solid malignancies such as melanoma or prostate cancer. These novel studies bring up many new and unanswered questions related to the molecular mechanism of RasGRP-driven oncogenesis, such as new receptor systems that RasGRP appears to respond to as well as regulatory mechanism for RasGRP expression that appear to be perturbed in these cancers. Here we will review some of the known aspects of RasGRP biology in lymphocytes and will discuss the exciting new notion that RasGRP Ras exchange factors play a role in oncogenesis downstream of various growth factor receptors.
Keywords: Ras, signaling, lymphocytes, cancer, RasGRP, receptor
Preface
Guanine nucleotide exchange factors (GEFs) such as RasGRPs (Ras guanine nucleotide releasing proteins) specifically control the exchange of GDP for GTP on the small GTPase Ras. Activated Ras is a signaling branch point and is involved in many cellular responses to receptor signal input, such as proliferation, differentiation and apoptosis (Chang and Karin, 2001; Starr et al., 2003). In addition to the RasGRP family with four members, there are four more RasGEFs in two additional families, namely SOS-1 and SOS2 (Son of sevenless) and RasGRF-1 and RasGRF-2 (Ras Guanine Nucleotide Releasing Factor). For specifics on SOS and RasGRF we point you other reviews (Mor and Philips, 2006; Yasuda and Kurosaki, 2008; Vigil et al., 2010; Stone, 2011; Jun et al., 2013; Kortum et al., 2013) here we will focus solely on RasGRPs.
Cloning of RasGRP1 by Jim Stone and colleagues in 1998 (Ebinu et al., 1998) provided a long-sought explanation for how exposure of cells to agents such as phorbol esters (TPA or PMA, diacylglycerol analogs) could lead to cellular Ras activation. We now know that RasGRP1 contains a stereotypical REM-Cdc25 catalytic unit and a diacylglycerol (DAG) binding domain. Subsequent studies on RasGRP1 and other RasGRPs, particularly the characterization of Rasgrp1 deficient mouse model with its severely impaired T cell development (Dower et al., 2000), created the false impression that RasGRPs are RasGEFs selective to the hematopoietic system. However, RasGRP1 was cloned in a classical fibroblast transformation assay (Ebinu et al., 1998), suggesting very early on that RasGRPs may also play a role in cancer. It is satisfying to see that recent studies on RasGRPs and cancer have revisited this concept and have started to characterize the mechanisms of oncogenic RasGRP signaling.
RasGRPs: structural domains and biochemical functions
Ras is a membrane-bound small GTPase that needs to be in a GTP-associated active state to exert its pleiotropic cell biological effects (Chang and Karin, 2001). GTP hydrolysis aided by RasGAPs (Ras GTPase activating proteins) ensures deactivation to Ras•GDP, the GDP-bound inactive state of Ras. Nucleotides are very tightly bound to Ras (Vetter and Wittinghofer, 2001; Rajalingam et al., 2007; Ahearn et al., 2012) and RasGRPs need to loosen the grip of Ras on bound GDP, resulting in release of nucleotide-free Ras that can associates with GTP (Bos et al., 2007).
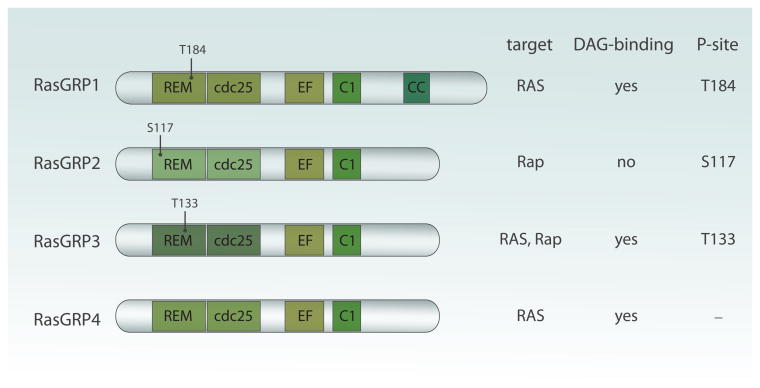
RasGRP1, RasGRP2, RasGRP3, and RasGRP4 are multi domain proteins that all contain the REM (Ras exchange motif) -Cdc25 unit (Fig. 1). The Cdc25 domain contains a helical hairpin that removes GDP form Ras when Ras binds the catalytic pocket of RasGEFs. RasGRP1, RasGRP3, and RasGRP4 can all activate Ras and although RasGRP2 also contains the REM-Cdc25 core and early studies indicated RasGEF activity (Clyde-Smith et al., 2000), it is now generally agreed on that RasGRP2 functions as a GEF for the small GTPase Rap (Kawasaki et al., 1998).
Figure 1.
Domain structure of RasGRP proteins. REM (Ras exchange motif) and Cdc25 domain form the catalytic core and catalyze GDP to GTP exchange on Ras and Rap GTPases. Two EF hands bind calcium ions and may be important for proper localization and/or GEF regulation. C1 domain of all family members but RasGRP2 binds diacylglycerol and that is crucial for anchoring at the plasma membrane. Finally, RasGRP1 uniquely possess C-terminal tail whose function is unknown but is likely to mediate protein–protein interactions. Conserved phosphorylation sites thought to be important for RasGRPs activation are indicated. All illustrations in this review were made by Anna Hupalowska.
Most mechanistic work has been performed on RasGRP1, which we will use as a framework for the other RasGRPs to discuss the protein domain structures. In addition to the catalytic REM-Cdc25 core, we have a fairly extensive understanding of RasGRP1's C1 domain and the role of DAG in RasGRP1 and RasGRP3 regulation. RasGRP1's C1 domain has been shown to bind DAG and its synthetic analogs such as PMA or PdbU with high affinity, similar to classical C1 domain found in PKCα (Ebinu et al., 1998; Lorenzo et al., 2000). Binding of C1 domain to DAG anchors RasGRP1 at plasma membrane where its substrate, RasGDP is present (Ebinu et al., 1998; Ebinu et al., 2000). A similar mode of action has been described for RasGRP3 and RasGRP4 (Lorenzo et al., 2001; Reuther et al., 2002; Yang et al., 2002; Teixeira et al., 2003). Intriguingly, RasGRP2's C1 domain does not bind to DAG so RasGRP2 is probably targeted to the plasma membrane by a different mechanism (Lorenzo et al., 2001; Reuther et al., 2002; Yang et al., 2002; Johnson et al., 2007). Of note, it is still a debate as to where, under physiological conditions, RasGRP1-mediated activation of Ras takes place. The most probable site is at the plasma membrane where signals from the T cell receptor (TCR) and B cell receptor (BCR) are initially transmitted, however some studies suggested that it could also occur at endomembranes such as Golgi apparatus (Bivona et al., 2003; Daniels et al., 2006). For a discussion on this debate as well as some of the technical details and challenges with the FRET-based Ras reporter assays, we refer you to a different review (Jun et al., 2013).
RasGRP1 and RasGRP3 activity is also regulated by phosphorylation, triggered in an indirect way by DAG. Threonine 184 (T184) becomes phosphorylated after TCR engagement or after stimulation with the DAG analog PMA, whereas RasGRP3 is phosphorylated on the analogous site, T133, in BCR-stimulated B cells (Aiba et al., 2004; Roose et al., 2005; Zheng et al., 2005) (Fig. 1). These studies also demonstrated that such phosphorylation very likely occurs through PKC kinases, which themselves can get recruited to the membrane by DAG (Aiba et al., 2004; Roose et al., 2005; Zheng et al., 2005). It is unknown how, at the molecular level, phosphorylation of RasGRP1 and RasGRP3 regulates their exchange activity. Phosphorylation appears to have an enhancing effect but is not absolutely required and alanine mutants of T184 or T133 in RasGRP1 or RasGRP3 demonstrate impaired, but not absent, stimulus-dependent Ras activation (Aiba et al., 2004; Zheng et al., 2005). Mass spec analysis has revealed several phosphorylation sites in RasGRP2 in stimulated CD8-positive cytotoxic T cells (Navarro et al., 2011). It is of interest to note that one of these, the serine 117, is at a similar position in RasGRP2 as T184 in RasGRP1 and T133 in RasGRP3 (Fig. 1). It is conceivable that the negative charge of the phosphate group aids the catalytic activity of RasGRP1, RasGRP2, and RasGRP3 in an analogous manner.
Sandwiched in between the REM-Cdc25 unit and the C1 domains exist a pair of EF hands in all RasGRPs. EF hands are motifs that typically come in pairs or concatamers and often bind calcium ions (Grabarek, 2006; Gifford et al., 2007). Binding of calcium induces conformational change by altering the directional vectors of the two α-helices in each EF hand, which enables regulation of the protein function, such as for calmodulin (Grabarek, 2006; Gifford et al., 2007). However, not all EF hands bind calcium and whereas sequence homologies predict that RasGRP1 has two EF hands, it was recognized early on that this RasGEF binds only one calcium ion in vitro (Ebinu et al., 1998). Sequence alignment of the EF hands of the four RasGRPs also reveals significant divergence (not shown), arguing that there may be differences in the role of calcium for each of these four family members. For instance, EF hands often contain aspartic acids at strategic positions that help bind and orient the calcium ion. RasGRP4 shows rather weak homology in this area and contains proline residues at some of the positions where aspartic acids are expected (Reuther et al., 2002). How binding of calcium ion(s) impact RasGRP function is an active debate that we will discuss next.
Early calcium chelation studies described that calcium was either critical for RasGRP1-driven Ras activation (Bivona et al., 2003), only modestly important (Izquierdo et al., 1992), or was entirely dispensable (Ebinu et al., 2000; Tazmini et al., 2009). A difficult parameter to assess is the completeness of calcium chelation. Robert Kay and colleagues were the first to more definitively identify a link between calcium and RasGRP1. They showed that RasGRP1's first EF hand (EF1) is important for membrane localization after receptor stimulation since truncation or point mutation of this motif resulted in decreased protein targeting to the plasma membrane after BCR triggering in chicken DT40 cell lines (Tazmini et al., 2009). The action of EF1 was subsequently linked to the C-terminal tail of RasGRP1 in that it enabled a plasma membrane recruitment function of this tail (Beaulieu et al., 2007; Zahedi et al., 2011). It should be noted that these studies relied on overexpression of GFP-tagged molecules in cell line models that also express endogenous RasGRP1 and RasGRP3. In addition, the described membrane recruitment enhancing function of this tail appeared only robust in B cells, but was modest in T cells and negligible in fibroblasts (Beaulieu et al., 2007). Future research will be required to identify the exact mechanism of calcium linking to RasGRP's EF hands but the Kay group paved the way.
What domains are in RasGRP1's C-terminal tail and is there conservation among the other RasGRPs? The in vivo importance of this domain was recently demonstrated in knock-in mice which express RasGRP1 lacking this part of the protein and which have perturbed T cell development (Fuller et al., 2012). The roughly 200-amino acid long C-terminal tail has no obvious domains except for one; a predicted leucine zipper motif (Ebinu et al., 1998; Beaulieu et al., 2007; Zahedi et al., 2011). Interestingly, this structural feature is unique to RasGRP1 and absent in other family members (Fig. 1). Its function is unknown but it is likely to participate in protein–protein interactions. It has also been suggested that, through electrostatic interactions, it could mediate plasma membrane targeting of RasGRP1 after B cell receptor (BCR) stimulation (Beaulieu et al., 2007; Zahedi et al., 2011).
RasGRP proteins are expressed in specific patterns
RasGRPs are not ubiquitously expressed but display restricted and overlapping expression patterns, for instance in the brain and in cells of the immune system. For example, RasGRP1 is expressed in cerebellum, cerebral cortex and amygdala whereas RasGRP2 is expressed in striatum (Kawasaki et al., 1998). It should be noted that the expression studies thus far are by no means complete and recent cancer studies have revealed unexpected RasGRP's expression in specific tumors. However, some of the expression patterns help explain understanding the phenotypes of the various Rasgrp deficient mouse models. Therefore, we provide a short summary here.
In immune cells, different family members show distinct albeit sometimes overlapping distribution. RasGRP1 is highly expressed in mature and developing T cells and to a lesser extent in B-, NK- and mast- cells (nu et al., 1998; Dower et al., 2000; EbiLee et al., 2009). Within the hematopoietic system, RasGRP2's expression is restricted to platelets and their precursors, megakaryocytes as well as neutrophils (Crittenden et al., 2004; Cifuni et al., 2008; Carbo et al., 2010). RasGRP3 is highly expressed in B cells (Yamashita et al., 2000; Teixeira et al., 2003) but is also expressed in T cells, macrophages and endothelial cells (Yamashita et al., 2000; Teixeira et al., 2003; Roberts et al., 2004; Botelho et al., 2009). Finally, RasGRP4 is the only family member, which is does not appear to be expressed in the brain (Reuther et al., 2002). It is relatively mast cell specific, although RasGRP4 is also expressed in thymocytes as well neutrohils (Yang et al., 2002; Suire et al., 2012; Zhu et al., 2012).
RasGRP1 and T cell receptor stimulation
Both RasGRP1 and RasGRP3 have been shown to couple to antigen receptors, such as T cell and B-cell receptors (TCR/BCR) in a manner that is non-redundant with SOS (Dower et al., 2000; Aiba et al., 2004; Brodie et al., 2004; Coughlin et al., 2005; Roose et al., 2005; Roose et al., 2007; Limnander et al., 2011). In the next two paragraphs we will discuss the mechanisms involved in the activation of RasGRP1/3 downstream of those receptors as well as functional outcomes of engaging TCR/BCR-RasGRP1/3 signals toward the canonical RasGTP-RAF-MEK-ERK kinase pathway for T and B cell biology.
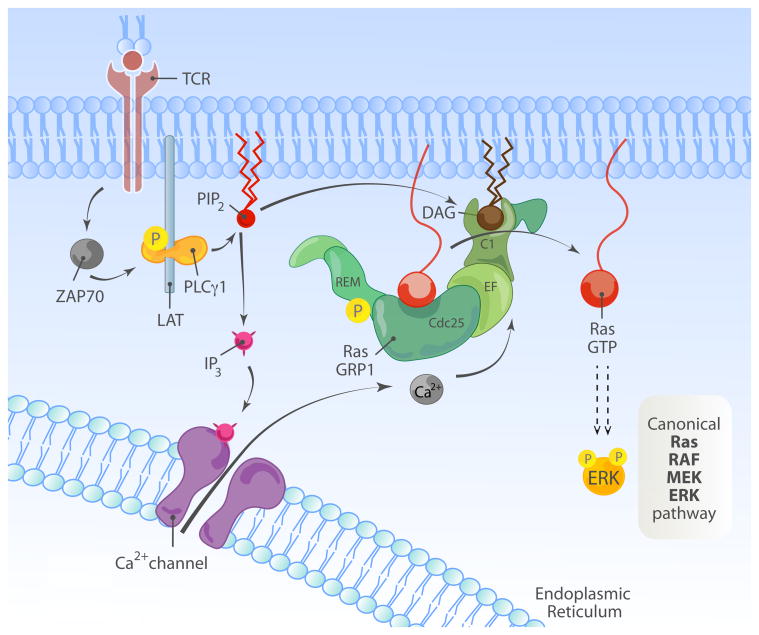
The successful activation of T cells, through their TCR, is a crucial event during immunological response. The TCR recognizes specific peptide antigen presented on MHC molecules (pMHC) by antigen-presenting cells (APCs) such as dendritic cells and others. However, unlike tyrosine kinase receptors, the TCR does not contain an intrinsic kinase domain. The CD3 molecules, which associate with the TCR, contain immunoreceptor tyrosine-based activation motifs (ITAMs) within their C-terminal cytoplasmic domains and are signaling units by virtue of providing docking sites for SH2-containing kinases. Src family kinases, such as Lck, phosphorylate ITAMs, which then serve as docking site for the tyrosine kinase, ZAP70. This step leads to the activation of ZAP70 and subsequent phosphorylation of target proteins, such as the adaptor proteins LAT (linker for activation of T cells) and SLP76 (SH2 domain containing leukocyte protein of 76kDa). LAT contains nine tyrosines, which are phosphorylated upon TCR stimulation and which recruit phospholipase Cγ1 (PLCγ1) along with other molecules (Fig. 2A). Activated PLCγ1 hydrolyzes phosphatidylinositol-(4,5)-bisphosphate (in short PI(4,5)P2 or PIP2) present in the plasma membrane to IP3 (inositol-1,4,5-trisphosphate) and DAG (diacylglycerol), two second messengers crucial in initiating signal transduction (reviewed in (Feske, 2007; Smith-Garvin et al., 2009). The accumulation of PLCγ1-produced DAG leads to the membrane recruitment of RasGRP1 where it can reach its substrate, Ras (Ebinu et al., 2000). As discussed in the previous chapter, the inducible plasma membrane localization of RasGRP1 leads also to its phosphorylation by PKCθ (Roose et al., 2005). Once activated, RasGTP initiates a canonical signaling cascade involving RAF-1 and other serine-threonine kinases. This ultimately results in the activation of ERK1/2 and transcription of genes involved in cell proliferation and other cellular processes (Fig. 2A). T cells also express the SOS RasGEF family members. It has been demonstrated, through biochemical and genetic studies that, downstream of the TCR, RasGRP is dominant over SOS (Dower et al., 2000; Ebinu et al., 2000; Roose et al., 2007). For readers interested in more details on the interplay between the RasGRP and SOS RasGEFs we refer to two recent reviews on that subject (Jun et al., 2013; Kortum et al., 2013).
Figure 2.
Activation of RasGRP1/3 downstream of TCR/BCR receptor.
(A) Overview of TCR-induced RasGRP1-Ras-MAPK cascade. Recognition of the cognate peptide by TCR results in the activation of tyrosine kinase ZAP70, which phosphorylates multiple downstream targets. One of them, the adaptor protein LAT participates in the assembly of a signaling complex containing PLCγ1. PLCγ1 hydrolyses PIP2 present in the plasma membrane into IP and DAG. IP is essential for the release of calcium from internal stores, whereas DAG activates RasGRP1, a GEF for Ras GTPase and initiates MAPK cascade. Note that the composition of the TCR chains and the proximal TCR signaling event are simplified here for clarity.
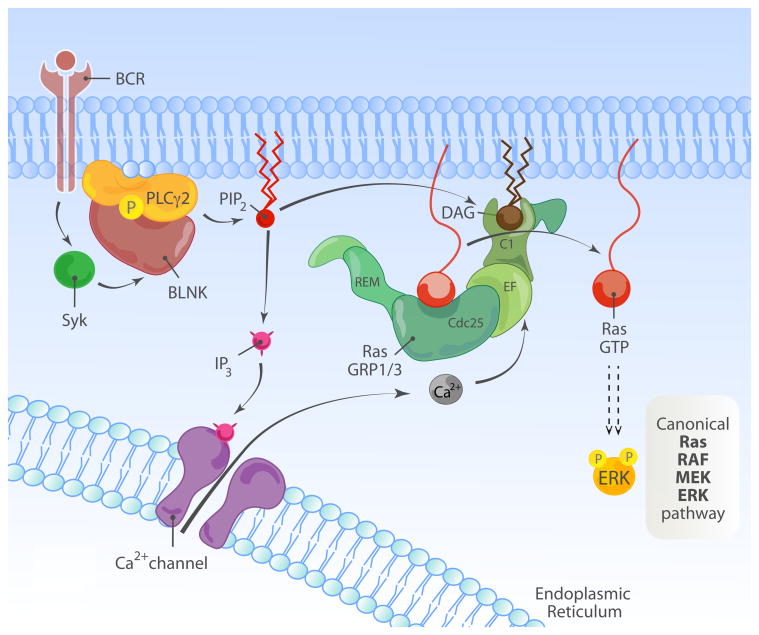
(B) Overview of BCR-induced RasGRP3-Ras-MAPK cascade.Antigen engagement on the BCR leads to activation of the tyrosine kinase Syk. Activated Syk phosphorylates multiple downstream targets, including the adaptor BLNK/SLP-65, which nucleates a signaling complex containing PLCγ2. This leads to hydrolysis of PIP2 into IP3 and DAG. IP3 binds the IP3 receptor on the ER, leading to release of intracellular calcium stores and store-operated calcium entry, while DAG recruits RasGRP1 and RasGRP3 to the plasma membrane and initiates MAPK signaling.
RasGRP3 and B cell receptor signaling
As mentioned above, RasGRP1 and 3 can couple to the B cell antigen receptor (BCR) as well as the TCR (Fig. 2B). Analogies between TCR and BCR signaling are inevitable. Both receptors lead to production of DAG and IP3 downstream of PLCγ proteins (PLCγ2 in B cells) and activation of the RasGRP/Ras/ERK pathway. However, most of the signaling intermediates downstream of these receptors are distinct to each cell type and there are important distinctions that need to be recognized. First, while the TCR recognizes peptide/MHC complexes, the BCR recognizes native antigen. This has enormous implications in the development and function of these cell types, which we discuss in the sections below. Like the TCR, the BCR has no intrinsic kinase activity and instead associates with other molecules that recruit kinases. In the case of the BCR, the Iga and Igb molecules (CD79a and CD79b) provide this function, as they contain ITAMs that become phosphorylated by Src family kinases and initiate signaling. The tyrosine kinase Syk then docks onto these phosphorylated ITAMs and Syk itself is phosphorylated and activated. Syk is essential to couple antigen recognition via the BCR to downstream signaling pathways, and much of this is accomplished by the phosphorylation of the BLNK/SLP-65 adaptor protein. Like LAT and SLP-76 in T cells, BLNK nucleates a large signaling complex by recruiting and facilitating the activation of several proteins including PLCγ2 (reviewed in (Kurosaki, 1999; DeFranco, 2000; Dal Porto et al., 2004)) (Fig. 2B)
Because of the parallels with TCR signaling, the discovery of RasGRP1 as an essential activator of Ras in T cells prompted the investigation of the role of RasGRP proteins in signaling downstream of the BCR. Initial studies found that RasGRP3 was more abundant in B cells than RasGRP1 and studies using chicken DT40 B cells lacking RasGRP3, RasGRP1 and 3, or SOS1 and 2 initially suggested that RasGRPs, but not SOS proteins, are essential for activation of Ras/ERK signaling downstream of the BCR (Oh-hora et al., 2003; Teixeira et al., 2003). It was subsequently shown that RasGRPs are more dominant in this pathway but that SOS proteins increase the amplitude of the responses (Roose et al., 2007). Conversely, SOS proteins appear more potent than RasGRP proteins in the activation of Ras/ERK downstream of the EGF receptor, implying that different receptor systems utilize unique mechanisms for activation of these RasGEFs in B cells. Additional studies showed that, analogous to the phosphorylation of T184 in RasGRP1 downstream of the TCR, T133 in RasGRP3 is phosphorylated downstream of BCR stimulation, and phosphorylation of this residue is important for robust Ras/ERK signaling in B cell lines (Aiba et al., 2004; Zheng et al., 2005). Physiologically, although RasGRP1- and RasGRP3-deficient mice revealed more subtle phenotypes in B cells than was observed for T cell development, it is becoming increasingly clear that these proteins play important roles in B cell development. The role of RasGRP proteins in coupling BCR engagement with functional responses to antigen will be discussed in a later section.
RasGRP1 is uniquely critical for proper T lymphocyte development
The physiological relevance of RasGRP1 downstream of the T cell receptor was first demonstrated with the generation of Rasgrp1 deficient mice, which showed a severe block in T cell development. Before we describe details on how RasGRP1 affects different aspects of T cell development we will quickly review major steps, which lead to the generation of mature and functional T cell repertoire able to mount appropriate immune response.
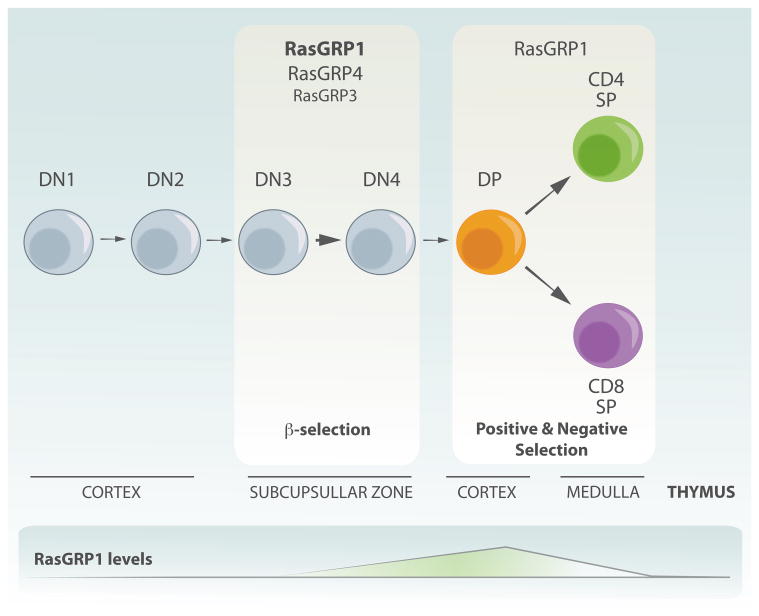
T cells complete their maturation program in the thymus (Fig. 3). During development from thymocytes to mature T lymphocytes, cells acquire specific T cells markers including TCR, CD3, CD4 or CD8. Specific stages in T cell development are characterized by changes in the expression of these cell-surface markers. Upon arrival in the thymus the progenitors do not express either CD4 or CD8 and are termed double negative (DN). Subsequently, cells that successfully rearranged their TCRβ gene receive survival, proliferative and differentiation signals. This process is called β-selection and the signals mainly come from the immature form of the TCR (preTCR) but also from other receptor input, such as IL7 (interleukin 7), Notch1, and CXCR4. Once cells pass this checkpoint, they start to express both CD4 and CD8 coreceptors to become double positive (DP) cells.
Figure 3.
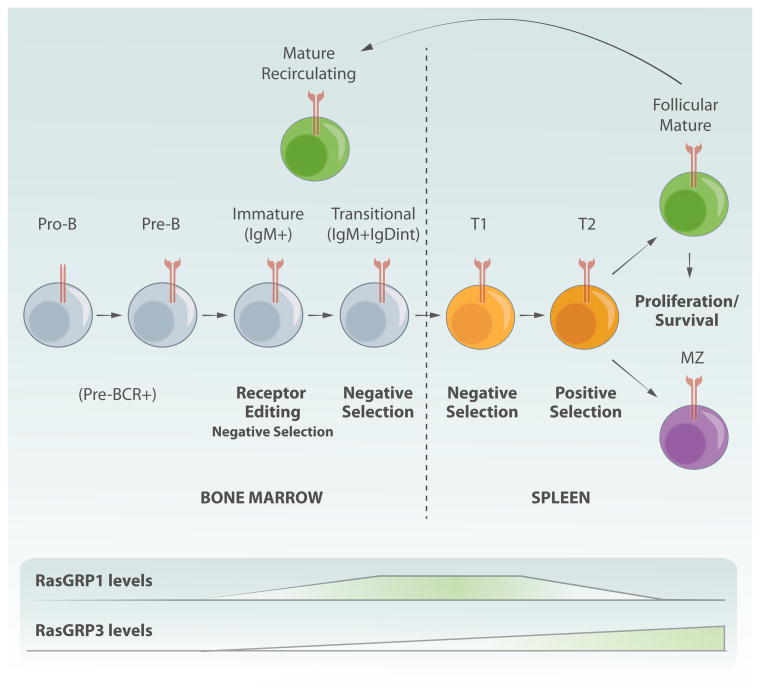
Overview of T cell development and the role of RasGRPs. The expression of CD4 and CD8 marks different stages of T cell development. Early progenitors do not express CD4 or CD8 and are termed double negative (DN). Depending on the expression of other markers those cells are further subdivided into 4 different subsets (DN1–4). DN3 thymocytes express an immature form of the TCR, pre-TCR. Signals from this receptor results in survival, burst of proliferation and differentiation into double positive cells, which express both CD4 and CD8. This process is called β-selection process. Subsequently, signaling from the TCR leads to positive and negative selection, a process depending on avidity of binding to self-peptide-MHC complexes presented by stromal cells in the thymus. Cells differentiate into single positive CD4 or CD8 thymocytes. These cells leave the thymus and migrate to secondary lymphoid organs. RasGRP1 is important for both signaling downstream of pre-TCR as well as for the negative and positive selection. RasGRP4 and 3 also contribute to signaling downstream of preTCR. Note that distinct stages of T cell development are taking places in different anatomical parts of thymus. Also, dynamic patterns of RasGRP1 expression are highlighted-RasGRP1's expression is low in early subsets (DN cells), increases significantly in DP cells and peaks in SP thymocytes to drop again in peripheral T cells.
At the double positive stage thymocytes rearrange TCRα gene, which subsequently will pair with the TCRβ chain to form a mature form of the T cell receptor. At this point, specificity and binding strength of the newly assembled TCRαβ complex with peptide-MHC is functionally checked in a processed called thymocyte selection. Thymocytes with intermediate avidity for self-peptide MHC receive survival and differentiation signals and are positively selected (Starr et al., 2003). Given the random nature of TCR rearrangements, most cells will fail to express a TCR on the surface and the lack of an interaction with pMHC will result in death by neglect (reviewed in (Klein et al., 2009). Surviving thymocytes differentiate to either CD4+ CD8− or CD4−CD8 + single positive cells (SP). Subsequently, in a process called negative selection, cells with high affinity TCR are eliminated, preventing generation of autoreactive T cells. Finally, fully mature T cells leave the thymus to seed secondary lymphoid organs such as lymph nodes or spleen (reviewed in (Klein et al., 2009)).
As discussed in the previous paragraph, RasGRP1 couples to the T cell receptor to activate Ras. Given that and the involvement of Ras pathway in T cell development (reviewed in (Alberola-Ila et al., 1996), it came as no surprise that loss of RasGRP1 has profound effects on T cell development (Fig. 3). The initial study showed that loss of RasGRP1 leads to a decreased thymus size and a severe block in T cell development between DP to SP stage so that there are only very few mature T cells in the periphery (Dower et al., 2000). The severe block in thymocytes maturation in Rasgrp1 deficient mice is accompanied by defects in Ras activation as well as impaired ERK phosphorylation (phospho-ERK; pERK) responses after PMA or TCR triggering. As compared to the wild-type controls, Rasgrp1 knockout thymocytes fail to proliferate in response to TCR stimulation (Dower et al., 2000). Subsequently, it has been shown that RasGRP1 has a non-redundant role in positive selection and neither other family members like RasGRP4/3 or other RasGEFs like SOS can efficiently compensate for the loss of RasGRP1 (Priatel et al., 2002; Starr et al., 2003; Kortum et al., 2012; Zhu et al., 2012; Golec et al., 2013). Whether the uniqueness of RasGRP1 for positive selection of thymocytes is a reflection of a distinct biochemical feature or a reflection of the relatively high RasGRP1 expression levels at this T cell developmental stage (discussed later) are still open-ended questions.
At first, RasGRP1 appeared dispensable at earlier stages of T cell development. However, a series of recent studies demonstrated that Rasgrp1 deficient thymocytes have a mild defect in β-selection. This defect is even more pronounced in mice that also lack RasGRP4 or SOS1 (Kortum et al., 2012; Zhu et al., 2012). These data indicate that RasGRP1 may also transduce signals downstream of pre-TCR receptor. Alternatively, RasGRP1 could operate downstream of the earlier-mentioned IL7R-, Notch1-, and CXCR4-systems and contribute to cellular outcomes at this first selection checkpoint during T cells development downstream of any of these receptors. Which exact cellular outcomes at β-selection are RasGRP1-dependent remain to be determined. It appears that deficiency of Rasgrp1 does not affect proliferation at this stage since a-CD3 induced proliferation was intact (Kortum et al., 2011). It would be of interest to know if other cellular outcomes such as survival are RasGRP1/4 dependent. In addition, given that thymocytes also receive signals from CXCR4 at this checkpoint and that SDF-1 induced proliferation depends on Ras-PI3K (p110γ) interaction (Janas and Turner, 2011), it would be informative to investigate if chemokine-induced proliferation at early thymocytes stages depends on RasGRP1/4 and/or Sos1. Finally, although initial reports indicated that RasGRP1 is not important for negative selection, recent research demonstrated that RasGRP1 together with Sos1 are both important for this process (Priatel et al., 2002; Kortum et al., 2012) and both of those GEFs are essential to transduce signals from TCR to induce apoptosis in order to eliminate self-autoreactive T cells.
It has been appreciated early on that RasGRP1 levels are dynamically and tightly regulated during T cell development (Fig. 3). Its expression is low in early subsets (DN cells), increases significantly in DP cells and peaks in SP thymocytes to drop again in peripheral T cells (Priatel et al., 2002; Kortum et al., 2012). Deregulation of normal expression pattern, either too little or too much of the protein, may tip the balance and result in disrupted T cell development. As has been described above, loss of RasGRP1 expression affects different aspects of T cell development. On the other hand, increased levels of RasGRP1 during thymocytes maturation can lead to development of T cell leukemia (Klinger et al., 2005; Berquam-Vrieze et al., 2011; Oki et al., 2011; Hartzell et al., 2013). We will discuss oncogenic properties of elevated RasGRP1 in the context of T cell leukemogenesis in later sections of the review.
RasGRP1 and RasGRP3 in B lymphocyte development
Developing B lymphocytes undergo a series of maturation steps analogous to the ones T cells go through in the thymus –namely the rearrangement of immunoglobulin genes that generate much of the diversity of the antibody repertoire and subsequent selection steps to eliminate potentially pathogenic self-reactive cells. However, unlike T cells, which complete their development in a specialized organ (the thymus), initial B cell development occurs in the bone marrow and is completed in the spleen. These latter stages of development thus occur in the same microenvironment where immune responses are initiated, something that has been likened to B cells “growing up on the streets.” (Townsend et al., 1999)
Committed lymphoid progenitors in the bone marrow transition through a variety of developmental steps accompanied by changes in cell surface molecules (reviewed in (Benschop and Cambier, 1999). During the transition of early pro-B cells to the pre-B cell stage these progenitors rearrange their heavy chain locus, and the resulting heavy chains pair up with the surrogate light chains and the ITAM-containing Iga and Igb chains to form the pre-B cell receptor. Productive signaling through this receptor and the IL-7 receptor are requisite for survival and transitioning to the late pre-B cell stage. Late pre-B cells rearrange their light chain locus, and the resulting light chain can pair with the heavy chain to form the B cell receptor. The expression of this receptor on the cell surface as membrane-bound IgM defines immature B cells in the bone marrow, and tonic signaling through this receptor is essential for survival of B cells throughout development and in mature stages (Torres et al., 1996; Lam et al., 1997). However, it also presents the B cell with the first opportunity to sample antigens in its environment, and it is at this stage that B cells begin a series of antigen-dependent selection checkpoints that shape a healthy B cell repertoire (Fig. 4).
Figure 4.
Overview of B cell development and RasGRP expression. Early B cell progenitors in the bone marrow undergo genetic rearrangement of their immunoglobulin genes, leading to expression of a unique B cell receptor on the surface each B cell. The B cell receptor mediates a series of self antigen-driven checkpoints that progressively eliminate autoreactive clones from the B cell repertoire. As immature B cells progress to a transitional stage, they exit the bone marrow, enter the circulation and migrate to the spleen where their selection and differentiation continues. Splenic B cells differentiate into follicular or marginal zone cells, and mature follicular B cells can recirculate throughout the body and populate the bone marrow and lymph nodes.
Immature B cells in the bone marrow that recognize antigen with strong affinity can re-express RAG enzymes and rearrange the second light chain allele, thus editing the B cell receptor in an attempt to rescue the cells from antigen induced apoptosis (Hertz and Nemazee, 1997). However, as the cells progress through development into a transitional stage where they begin to express IgD as well as high levels of IgM, they lose their ability to receptor edit and instead become highly sensitive to antigen-induced apoptosis (Melamed et al., 1998). Therefore, cells that receive a strong antigenic signal at this stage are deleted from the repertoire (Fig. 4). Transitional B cells exit the bone marrow, enter the circulation and migrate to the spleen, where they enter the periarteriolar lymphoid sheath (PALS) as transitional 1 (T1) cells. T1 cells remain highly susceptible to antigen-induced apoptosis and undergo an additional selection checkpoint as they migrate into the lymphoid follicles and mature into T2 cells, thus deleting additional autoreactive clones (reviewed in (Chung et al., 2003; Su et al., 2004). During this maturation stage the signaling properties of the cells change significantly and the cells become substantially more tolerant to antigenic stimulation (Su et al., 2004). In fact, there is substantial evidence arguing that T2 cells require some level of antigen recognition for positive selection and differentiation into either mature follicular B cells, which mediate T cell-dependent B cell responses, or marginal zone B cells, which participate in T-independent responses (reviewed in (Pillai, 1999; Pillai et al., 2004; Pillai and Cariappa, 2009). Importantly, there is a significant portion of mature B cells that exhibit autoreactivity despite early developmental checkpoints, and the level of autoreactivity influences their development toward a B1, follicular or marginal zone B cell. High levels of antigen exposure of autoreactive follicular B cells leads to a state of unresponsiveness known as anergy, an additional mechanism that prevents autoimmune pathology (Goodnow et al., 1989; Cambier et al., 2007; Zikherman et al., 2012) (Fig. 4).
Several studies demonstrate that Ras/ERK signaling is important at various stages in B cell development, both downstream of the pre-B cell receptor and the BCR (Yasuda et al., 2008; Yasuda et al., 2011). Given that RasGRP proteins are required to activate this pathway in several B cell lines, it was somewhat surprising that the B cell phenotype of RasGRP-deficient mice was considerably less dramatic than the T cell phenotype, where there is a complete block in development at the DP-SP transition in the thymus (Dower et al., 2000; Coughlin et al., 2005). RasGRP1-deficient B cells develop into the periphery in relatively normal numbers, but the mice develop splenomegaly with age and this is partly due to aberrant B cell expansion. However, it remains unclear if this phenotype is B cell intrinsic or if it is secondary to the T cell lymphopenia. Nevertheless, it is interesting that the splenomegaly resolves in mice deficient in both RasGRP1 and RasGRP3, implying that RasGRP3 is required for the expansion of RasGRP1-deficient B cells (Coughlin et al., 2005). Indeed, RasGRP3 is required for splenic B cell proliferation in response to stimulation with IgM, IgM plus IL-4, anti-CD40 or IgM plus anti-CD40, suggesting that RasGRP3 is important in antigen responses in mature B cells (Coughlin et al., 2005). Notably, although these initial studies did not report any obvious defects in peripheral B cell development, they did not address questions related to B cell repertoire selection or B cell development in the bone marrow, and recent studies suggest that RasGRP proteins likely play critical roles in establishing a healthy B cell repertoire.
Activation of Ras/ERK signaling correlates with the induction of apoptosis in immature B cell lines (Lee and Koretzky, 1998), and early studies on the role of RasGRP1 in B cells showed that RasGRP1 overexpression was sufficient to sensitize an immature B cell line to antigen-induced apoptosis (Guilbault and Kay, 2004). More recently, it has become apparent that Ras/ERK signaling mediated by RasGRP proteins sensitizes developing immature and transitional B cells to apoptosis and mediates negative selection checkpoints (Stang et al., 2009; Limnander et al., 2011). In addition, these studies revealed that expression of RasGRP1 and RasGRP3 are differently regulated during B cell development. RasGRP1 and RasGRP3 are not expressed at noticeable levels in early stages of B cell development where VDJ recombination occurs, suggesting that other RasGEFs, such as SOS proteins, may be important in mediating Ras activation downstream of the pre-B cell receptor. However, both RasGRP1 and RasGRP3 expression are upregulated in immature and transitional B cells in the bone marrow concomitant with the expression of surface IgM (Fig. 4). RasGRP1 expression peaks at these transitional stages in development and is then decreases in mature B cells, while RasGRP3 is expressed at lower levels in immature B cells and is vastly upregulated in mature B cells. This pattern of RasGRP protein expression suggests that RasGRP1 may be primarily involved in antigen-dependent selection checkpoints in immature and transitional B cells, whereas RasGRP3 may be more important for mature B cell responses (Limnander et al., 2011; Limnander and Weiss, 2011) (Fig. 4). However, the unique or potentially redundant role of these proteins in B cell development, function and autoimmunity remains incompletely understood and warrants further investigation.
RasGRP2 and RasGRP4 in non-oncogenic settings
As discussed earlier, RasGRP2 can directly activate Ras related GTPases of Rap subfamily. These molecules function downstream of many receptors and one of the cellular consequences of increased RapGTP is the activation of integrins, which are mediators of adhesion in many immune cells (Yamashita et al., 2000).
The generation of RasGRP2 knockout mice allowed for the dissection of its role in the immune cells (Crittenden et al., 2004). This first report showed that RasGRP2 deficient animals had bleeding problems as assessed in a tail-bleed assay. That defect was not due to decreased total numbers of platelets as those were either normal or even slightly elevated. Careful analysis of platelets function showed that bleeding diathesis displayed by those animals was due to defects in platelets aggregation and degranulation, a process, which normally depends on the activation of integrins downstream of many receptors in a process called inside-out signaling. Measurements of activated β3 integrin confirmed that this process is severely impaired in RasGRP2 knockout mice (Crittenden et al., 2004). In addition, it appears that RasGRP2 and PKC cooperate to activate β3 integrin to induce platelets aggregation (Cifuni et al., 2008). Another study showed that RasGRP2 is also important for β1 intergrin activation (Bergmeier et al., 2007). More importantly, in vitro and in vivo assays of thrombi formation showed that RasGRP2 deficient animals have severe defects (Crittenden et al., 2004; Bergmeier et al., 2007). The authors propose a model where various receptors, mainly G-protein coupled receptors (GPCRs) signal through RasGRP2, which can then activate Rap and lead to activation of integrins to mediate adhesion, a crucial step for platelets function (Fig. 5) (Crittenden et al., 2004).
Figure 5.
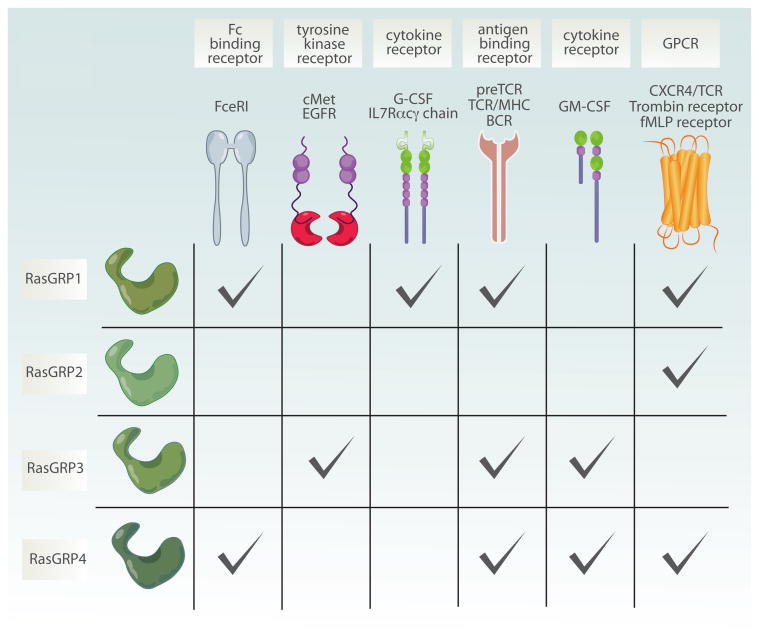
Multiple receptor systems couple to different RasGRP proteins. Scheme is showing different receptors which couple to distinct members of RasGRP family. RasGRP1 signals downstream of antigen receptors such as preTCR, TCR, BCR but also from cytokine receptors including G-CSFR and IL7R. More recently it has also been shown that RasGRP1 also couples to Fc binding receptor as well as to GPCR such as CXCR4. RasGRP2 is mainly activated downstream of GPCR such as thrombin receptor. RasGRP3, similarly to RasGRP1 is engaged after antigen receptor triggering, but can also function downstream of receptor tyrosine kinases such as cMet (HGFR) or EGFR and cytokine receptor (GM-SCF). Finally, fMLP receptor (GPCR), FcR, preTCR and GM-SCF can all activated RasGRP4. preTCR-pre T cell receptor; TCR-T cell receptor; BCR-B cell receptor; G-CSFR- granulocyte colony stimulating factor receptor; GM-CSFR- granulocyte-macrophage colony stimulating factor receptor; EGFR- Epidermal growth factor receptor; HGFR- hepatocyte growth factor receptor; GPCR- G protein coupled receptor.
Those results prompted another group to study the role of RasGRP2 in FcγRIIA-mediated platelets activation and aggregation, which can lead to thrombocytopenia and thrombosis syndromes, a pathological condition for which few safe therapeutical options are available. The results showed that mice deficient for RasGRP2 are protected in a mouse model of immune-mediated thrombocytopenia and thrombosis syndromes (ITT) and suggested that RasGRP2 can be an attractive therapeutic target for treatments of this pathology (Stolla et al., 2011).
Studies of neutrophils lacking RasGRP2 showed that RasGRP2 is also essential for efficient adhesion and chemotaxis of this cell type (Bergmeier et al., 2007; Carbo et al., 2010). Using a variety of approaches the authors showed that RasGRP2 deficient neutrophils have impaired β2/β3 integrins-mediated adhesion and Rap1 activation. In vivo that resulted in impaired adhesion and extravasation. Given that the RasGRP2 mice also show defects in platelets aggregation this mouse model closely resembles leukocyte adhesion deficiency (LAD) syndrome (Bergmeier et al., 2007). Finally, it also has been shown that RasGRP2 is expressed in peripheral human T cells and that chemokine-induced adhesion to ICAM-1 coated surfaces depends on RasGRP2 presence. Interestingly, that was not the case for VLA-4 mediated adhesion (Ghandour et al., 2007). In summary, RasGRP2-dependent Rap1 activation is important for adhesion of different immune cells, including platelets, neutrophils and T cells.
RasGRP4 was the last RasGRP family member to be cloned (Reuther et al., 2002; Yang et al., 2002). It is highly expressed in mast cells but recent data showed that it is also present in developing T cells and neutrophils (Suire et al., 2012; Zhu et al., 2012). We described the role of RasGRP4 in T cell development in previous chapter and here we will highlight novel data pointing out at its role in mast cells and neutrophils function.
The description of a mouse strain expressing defective RasGRP4 variant (without C1 domain) and which are hyporesponsive to methacholine stimulation via the airway prompted speculations that RasGRP4 is important for mast cell function (Li et al., 2003). Subsequently, two independent groups generated RasGRP4 deficient mice and examined its role in mast cells development and function. Both groups demonstrated that RasGRP4 is dispensable for mast cell development and maturation (Adachi et al., 2012; Zhu et al., 2012). The Zhang laboratory showed that RasGRP4 deficiency results in impaired FcεRI-dependent mast cell function such as degranulation and cytokines production. Interestingly, the phenotype was even more severe when RasGRP1 was lost suggesting that these two RasGRPs signal downstream of FcεRI and orchestrate FcεRI-dependent cellular processes (Fig. 5). At a molecular level, deficiency of RasGRP4 lead to decreased phosphorylation of ERK, p38 as well as JNK after FcεRI stimulation. Intriguing, double deficiency of RasGRP1/4 resulted in impaired phosphorylation of PLCγ1 and calcium flux. In contrast to other cellular and receptor systems, it seems that in mast cells and specifically downstream of FcεRI, RasGRPs are upstream of PLCγ1 (Zhu et al., 2012). The Stevens group investigated the consequences of RasGRP4 deficiency in two inflammation models, which involve mast cells. Interestingly, they show that RasGRP4 knockout mice are protected from DSS (dextran sodium sulfate)-induced acute colitis as well as from one type of experimental arthritis (Adachi et al., 2012). However, since both of these models also involve other cell types of innate and adaptive arms of the immune system, it is difficult to point out which cells are affected (and how?) by RasGRP4 deficiency. Therefore, more detailed analysis will be needed to address this question.
Finally, it has been shown that neutrophils, among other RasGEFs, express RasGRP4 (Suire et al., 2012). Neutrophils from RasGRP4 knockout mice were fully differentiated and functional in many aspects. However, when stimulated by fMLP, a chemotactic peptide derived from bacteria, which binds to FPR1, a GPCR, RasGRP4 deficient neutrophils showed a severe decrease in RasGTP levels, which resulted in impaired activation of Erk. Interestingly, fMLP-induced PIP3 production and phospho-Akt were also impaired by lack of RasGRP4, which suggested that GPCR-induced PI3K activation was RasGRP4-dependent In addition, ROS production in response to various stimuli activating GPCRs was also impaired in RasGRP4 knockout neutrophils. Overall, RasGRP4 deficiency closely photocopied the phenotype seen in mice expressing Ras-insensitive PI3Kγ mutant. The authors also showed that deficiency of PLCβ1/β2 has similar effects to RasGRP4 deficiency on fMLP-induced Ras, ERK, Akt activation as well as PIP3 production. Moreover and, as expected, neutrophils from these mice also had impaired DAG production. Overall, the experiments support a model where GPCR-induced activation of PLCβ1/β2 leads to DAG production and RasGRP4 activation (Fig. 5), which results in loading of Ras with GTP. Once activated, Ras initiaties signaling cascades, which, on one hand, involves Raf-MEK-ERK pathway and on another PI3Kγ and production of PIP3 and activation of pleckstrin homology (PH) domain-containing proteins such as Akt. As a consequence, ROS production and migration of neutrophils are increased and those are important for efficient clearing of the pathogens (Suire et al., 2012).
From cancer to lymphocyte biology and back to cancer?
Historically, fibroblast screening assays that measure anchorage-independent growth were standard assays to measure a molecule's transformative potential. As mentioned earlier, the first RasGRP1 clone was originally identified from a cDNA library in such a fibroblast transformation screen (Ebinu et al., 1998). Subsequent comparisons demonstrated that RasGRP1 was fairly efficient in transforming fibroblasts, similar to the transformative capacity of mutated H-Ras (H-RasG12V), whereas RasGRP3 revealed only very modest activity in this assays and RasGRP2 none at all (Yamashita et al., 2000). As sometimes occurs in science, RasGRP research took a sharp left turn toward lymphocyte development and for a while disappeared from the radar screen of cancer biologists. Recent reports on RasGRPs in various cancers are likely to renew the interest of the larger cancer community in RasGRP RasGEFs.
The role of RasGRP1 in skin cancer– a biochemical Ras-JNK2 pathway?
Patricia Lorenzo has been on the forefront to describe a role for RasGRPs in cancer. Her team first described that RasGRP1 is expressed in primary mouse keratinocytes. The protein mainly localizes in the cytoplasm and shows perinuclear staining in unstimulated cells. Upon addition of TPA, which is a DAG analog, RasGRP1 redistributes to plasma membrane. Overexpression of RasGRP1 had a profound effect on keratinocyte morphology and cell biology as it increased apoptosis and the percentage of hypodiploid cells (Rambaratsingh et al., 2003). In addition, RasGRP1 overexpression could also block calcium-induced keratinocyte differentiation, arguing that RasGRP1 may play a role in lineage differentiation of not only lymphocytes but also of keratinocytes. At a molecular level, elevated amounts of RasGRP1 resulted in increased levels of RasGTP that further increase upon TPA stimulation (Rambaratsingh et al., 2003). This study also demonstrated for the first time that TPA induces degradation of RasGRP1, suggesting that RasGRP1 levels might be regulated via a stimulus-dependent, negative feedback loop. In subsequent study, TPA-induced RasGTP production in keratinocytes was confirmed to be RasGRP1-dependent when the Lorenzo group used Jim Stone's mouse model to show that Rasgrp1 deficient keratinocytes failed to activate both H- and N-Ras (Sharma et al., 2010). Interestingly, the magnitude of phospho-ERK was not affected by Rasgrp1 deficiency, postulating that RasGRP1 in this situation does not signal to the canonical RasGTP-RAF-MEK-ERK pathway. The authors did not investigate the levels of PI3K/Akt pathway activation, another known effector cascade downstream of RasGTP. It was noted though that phospho-JNK2- but not phospho-JNK1- levels were diminished in cells lacking RasGRP1. Conversely, overexpression of RasGRP1 increased pJNK2 levels after TPA stimulation (Sharma et al., 2010). The reason that RasGRP1's differential effect on JNK-1 and -2 is interesting is that phospho-JNK2 has been reported to have pro-tumorigenic effects in mouse keratinocytes, whereas pJNK1 has the opposite effect rising possibility that deregulated levels of RasGRP1 could have oncogenic properties through specific activation of JNK2 (Sharma et al., 2010). Since TPA is a synthetic DAG analog, it remains to be established which of the physiologic stimulus activates RasGRP1 in keratinocytes. Does RasGRP1 in keratinocytes require receptor systems that can couple to PLC to generate DAG, or does RasGRP1 operate in a DAG-independent manner in this cell lineage? Likewise, it will be of interest to determine which canonical and non-canonical signaling pathways are activated in a RasGRP1-dependent manner.
The role of RasGRP1 in skin cancer-mouse models of multi-step carcinogenesis
The Lorenzo group also employed mouse models. Characterization of RasGRP1 transgenic mice, which overexpress RasGRP1 specifically in epidermal keratinocytes (the expression of RasGRP1 is driven by K5 promoter; termed K5-RasGRP1 here), showed that elevated RasGRP1 does not cause any overall disruption of skin architecture in young mice. However, almost 50% of the mouse colony spontaneously developed skin tumors on the dorsal skin as well as on the tail by 7 months of age. The incidence of tumors was higher in animals caged together suggesting that wounding may contribute to the development of those neoplasms. The majority of those tumors were classified as benign papillomas and an only small fraction as malignant squamous cell sarcomas (SCC) (Oki-Idouchi and Lorenzo, 2007). It should be noted that papillomas are not seen in human UVB-induced cutaneous carcinomas. As expected, K5-RasGRP1 tumors expressed RasGRP1 but did not contain oncogenic H-Ras mutations. H-Ras mutations are very frequently found in carcinogen-induced skin cancer (reviewed in (Perez-Losada and Balmain, 2003). It is conceivable that overexpressed RasGRP1 and H-Ras mutations are mutually exclusive molecular events in skin cancer in a manner that is analogous for what we have published on T cell leukemia (Hartzell et al., 2013) and will describe toward the end of this review.
At molecular level, K5-RasGRP1 keratinocytes demonstrated elevated RasGTP (Oki-Idouchi and Lorenzo, 2007) suggesting that increased levels of Ras signals could contribute to the abnormal growth. Since RasGTP can activate numerous effectors pathways, it would be of interest to investigate which of the effectors arms increased RasGRP1 levels affects in vivo. Is JNK2 activation involved? Again, it is unclear if receptor stimulation (and if so, which receptor) plays a role in K5-RasGRP1 keratinocytes. Interestingly, RasGRP1 overexpressing keratinocytes produced more G-CSF in an in vitro wound assay (Oki-Idouchi and Lorenzo, 2007). G-CSF is a growth factor that is known to promote keratinocyte growth. Although no functional data exists to date, it is tempting to speculate that RasGRP1 and G-CSF may operate in an autocrine positive feedback loop enabled by the unknown wound-induced parameter.
The relevance of skin wounding toward the development of skin tumors in the K5-RasGRP1 mice was further substantiated by the same group (Diez et al., 2009). Whereas introduction of skin wounds did not cause tumors in wild-type mice, 50% of K5-RasGRP1 mice developed papillomas or squamous cell carcinomas (SCC) following such insult. The size of tumors was proportionate to the level of RasGRP1 expression -the higher RasGRP1 levels, the bigger the tumors- suggesting that gene dosage may be important in the promotion of the abnormal growth. Serum G-CSF levels were elevated in the K5-RasGRP1 mice 24h post wounding, again pointing to a possible connection between RasGRP1 and G-CSF in promoting tumor development in vivo. However, no functional data, such as blocking G-CSF was performed to explore this further mechanistically.
A multi-stage chemical carcinogenesis model is a widely used protocol to study epithelial cancer initiation and progression. The initiation step is accomplished by treatment of shaved mouse skin with a carcinogen such as 7,12-dimethylbenz[a]anthracene (DMBA) at a very low dose. DMBA promotes stereotypic oncogenic mutations in H-Ras (reviewed in (Perez-Losada and Balmain, 2003)). In the second (promotion) stage, repetitive application of promoting agent such as TPA, takes place. The advantage of this model is that it mimics, to certain extent, repetitive exposure to the mutagene and that it allows for the separation of initiation and progression stages of tumor development (Abel et al., 2009), although it should be noted that cumulative UV exposure rather than chemicals causes most human skin neoplasms (reviewed in (Ratushny et al., 2012)). Using this model, Luke et al. explored the role of deregulated RasGRP1 in skin tumor initiation and progression (Luke et al., 2007). Comparing wild-type to K5-RasGRP1 mice, the authors established that RasGRP1 does not contribute to tumor initiation stage since the incidence and the number of tumors per mouse was similar for both models. The type of tumors (benign papilomas vs. malignant sarcomas) was also similar between two experimental groups. However, increased RasGRP1 levels contributed to tumor progression since higher incidence of animals with bigger and less differentiated tumors were noted in the K5-RasGRP1 group. How, e.g. through which effectors and signaling pathways, RasGRP1 contributes to SCC progression is not unclear and remains largely unexplored so far. Moreover, whereas the discussed studies point out at a possible role of RasGRP1 in the pathogenesis of cutaneous neoplasms, how those observations in murine models correlate to human situation is unclear. Therefore, further research is needed to explore the role of RasGRP1 in human skin cancer.
RasGRP3 in prostate cancer and melanoma
Following the paved path by RasGRP1 and skin neoplasms in the mouse models, RasGRP3 has recently emerged as a player contributing to the prostate cancer as well as melanoma. Significantly, Peter Blumberg and colleagues included analyses of patient samples so that clinical relevance is more apparent here.
We will first discuss the prostate cancer study, which starts with the observation that RasGRP3 mRNA levels are elevated in a subset (around 26%) of patients with prostate cancer as compared to healthy individuals (Yang et al., 2010). Evaluation of RasGRP3 expression in human prostate cell lines showed that androgen-independent cell lines expressed almost 10 times more RasGRP3 as compared to androgen-dependent cell lines. Androgen-independent tumors are typically more aggressive (reviewed in (Feldman and Feldman, 2001)). Subsequent in vitro experiments using siRNA strategies revealed that downregulating RasGRP3 expression decreased basal RasGTP levels, interfered with cell growth, and increased apoptosis. Similar results were obtained in vivo; using a xenograft model the authors revealed that decreasing levels of RasGRP3 expression inhibited growth of subcutaneously injected cell lines (Yang et al., 2010).
To understand better the molecular mechanism by which RasGRP3 promotes tumor growth, the authors investigated activation of signaling molecules downstream of HGF (hepatocyte growth factor), a growth factor know to be important in normal as well as cancerous prostate (Knudsen and Edlund, 2004). Unfortunately, these approaches were only taken in a few cell lines but the results are nevertheless interesting. In one cell line, reduction of RasGRP3 levels caused diminished levels of HGF-induced phospho-Akt and to lesser extent phospho-ERK in one cell line. Thus, RasGRP3 may be downstream of HGF receptor (Fig. 5). Interestingly, the effect on phospho-Akt was also seen in second cell line that is PTEN-deficient and therefore does not rely on growth factor stimulation for the activation of the PI3kinase-Akt pathway-P-Akt is constitutively high in these cells without HGF stimulation but reduced when RasGRP3 is targeted by shRNA (Yang et al., 2010).These results, if generalizable, suggest that RasGRP3 may promote excessive growth and survival through the activation of Akt pathway in the context of prostate cancer, either in response to HGF stimulation or in a constitutive manner when mutations result in active PI3kinase signaling. It would also imply that patients with elevated RasGRP3 levels may potentially benefit from therapies based on PI3K inhibitors. At this point it is not clear if and how RasGRP3 becomes activated after HGF stimulation and how it intersects with the PI3K pathway in the absence of growth factor stimulation. We will come back to this issue in later sections. The authors also perform add-on experiment where they overexpress RasGRP3 in an androgen-dependent cell line, which has low endogenous RasGRP3 protein levels. Elevated levels of RasGRP3 caused phenotypic changes and turned androgen-dependent line into androgen-independent, which is interesting and relevant as it would suggest that deregulated levels of RasGRP3 may facilitate progression from androgen-dependent to more aggressive, androgen-independent phenotype (Yang et al., 2010).
The Blumberg group also explored RasGRP3's contribution to melanoma development and demonstrated that approximately 12% of the samples from melanoma patients expressed RasGRP3 (Yang et al., 2011). For now it is unclear if those patients are developing more aggressive forms of the disease or if they are at risk for relapse. The authors also show that many of the melanoma cell lines but not normal melanocytes express various amounts of RasGRP3. To assess the role of RasGRP3 in those cell lines, the investigators decreased RasGRP3 expression levels in some of them. With few exceptions, in most cell lines interfering with RasGRP3 levels decreased cell proliferation in vitro and similar results were obtained in vivo. The effects of RasGRP3 ablation on signaling pathways were also examined. Knockdown of RasGRP3 expression caused a reduction of both basal and HGF-induced phospho-Akt levels, suggesting that RasGRP3 can contribute to the activation of the HGF pathway pathway in melanoma. The expression of the HGFR (cMet) was diminished, which is an interesting result in the light of more recent data showing that stromal cells can secrete HGF and contribute to the resistance to various kinase inhibitors (Straussman et al., 2012; Wilson et al., 2012). Therefore, co-targeting RasGRP3 could potentially overcome this resistance mechanism.
RasGRPs role in blood cancers
Several lines of evidence suggest that different RasGRP family members contribute to blood neoplasms. The high frequency is perhaps a reflection of the already existing RasGRP expression in normal cells of the hematopoietic system. Distinct RasGRP family members have been implicated in the cell biology of B cell lymphomas, acute myeloid leukemia and T cell acute lymphoblastic leukemia/lymphoma and we will discuss further those findings in the coming paragraphs.
The first indication that RasGRP proteins are contributing to the development of hematological malignancies came from retroviral mutagenesis screens, which looked for new genes involved in B cell lymphoma. Two independent groups demonstrated that both RasGRP1 and RasGRP2 genes, along with other genes implicated in Ras signaling, were found to be common insertional sites (CIS) (Mikkers et al., 2002; Suzuki et al., 2002). Unfortunately, these CIS mapping studies were not validated by mechanistic approaches and it remains to be determined if and how RasGRP1/2 are involved in tumor initiation and/or progression in B cell lymphoma.
There is growing evidence that RasGRPs are also important in acute myeloid leukemia (AML). AML is a blood neoplasm, which is characterized by failure of bone marrow to produce functional myeloid cells; i.e., granulocytes, red blood cells or platelets. It can arise de novo or can result from a progression of myelodysplastic/myeloproliferative disorder (MDS/MPD). An important distinction between the two entities, apart from the percentage of malignant blasts in the bone marrow, is the fact that in AML there is a block in differentiation, which results in the accumulation of immature cells with blast morphology in the bone marrow and decrease of mature, functional cells in the blood. By contrast, in MDS/MPD cells fail to differentiate that is cells mature to certain stage but then instead of achieving next step, they die. A large proportion of patients with MDS/MPD progress to AML (reviewed in (Corey et al., 2007). First indication came from cloning of RasGRP4, which was isolated from patients with AML (Reuther et al., 2002; Watanabe-Okochi et al., 2009). RasGRP4, similarly to RasGRP1 and 3 had some transforming potential when assessed in fibroblast or epithelial cells (Reuther et al., 2002). More importantly, RasGRP4 when overexpressed in 32D myeloid cell lines promotes cytokine-independent growth. Significantly, this effect was dependent on presence of PMA, indicating that RasGRP4 still needs to be activated in a DAG-dependent manner to exert its tumorigenic function. Importantly, PMA alone (without RasGRP4 overexpression) did not cause cytokine-independent growth in those cell lines (Reuther et al., 2002). Transplantation of RasGRP4-transduced bone marrow progenitors resulted in the development of AML-like disease in roughly one fourth of a limited mouse number, providing some in vivo evidence that deregulated RasGRP4 can initiate the disease (Watanabe-Okochi et al., 2009). It is worth noting that the E468K-RasGRP4 variant used in this experiment was found in a patient with myeloproliferative disorder (MPD). It is not clear if wild-type protein can have same tumorigenic potential. It also remains to be established how this E468K mutation – changing negatively charged glutamic acid into arginine with a positive charge - affects RasGRP4 protein function. Nor is it known what the incidence is for carrying this (or other RasGRP4's) mutations/variants in patients with MPD, who are predisposed to develop AML. Those would be interesting questions to be pursued in the future.
In vitro hypersensitivity to GM-SCF is one of the cellular hallmarks found in patients with juvenile myelomonocytic leukemia, implicating aberrant GM-SCF signaling in the pathogenesis of this disease and our recent study investigated the role of RasGRP3 and 4 downstream of this growth factor (Emanuel et al., 1991) reviewed in (Koike and Matsuda, 2008). Collaborating with the teams of Ben Braun and Kevin Shannon and others, we showed that both RasGRP3 and 4, but not RasGRP1, are expressed in bone marrow macrophage progenitors cells (BMMPC) (Diaz-Flores et al., 2013). Significantly, RasGRP3 expression was elevated in BMMPC derived from KRasG12D mice with mutated KRas, as compared to wild-type cells. Biochemical analyses revealed that GM-SCF-induced phospho-ERK is RasGRP4-dependent in wild type cells but depends on both RasGRP3 and RasGRP4 in KRasG12D cells (Fig. 5). The mechanism of and reason for RasGRP3's elevated expression in KRasG12D cells is unclear at this point and requires further investigation. As for many of the other cancer studies previously discussed, in this AML study, it is also unclear how RasGRP3 and RasGRP4 couple to GM-SCF and we will further discuss that issue in the next paragraph.
Hyperactive Ras signaling has been implicated in many cancers, including acute myeloid leukemia (reviewed in (Ward et al., 2012)). Mice deficient for NF1, which functions as RasGAP, develop myeproliferative disorder, which closely models human juvenile myelomonocytic leukemia. Retroviral mutagenesis, achieved through infection of NF1-deficient pups with MOL4070LTR retrovirus, induces progression from MPD to aggressive AML in those mice. This approach can be used to explore genes and pathways that cooperate with NF1 to induce AML, as well as to investigate resistance to clinically relevant MEK inhibitors (Lauchle et al., 2009). Kevin Shannon's group used this model to discover that one of the mechanisms conferring the resistance to MEK inhibitors is through increased expression of RasGRP1. As a result of elevated RasGRP1, resistant leukemias show increased basal RasGTP and spontaneous cytokine-independent colony growth. Importantly, downregulating RasGRP1 levels via shRNA approach could restore sensitivity to MEK inhibitor, proving that resistance to the drug was in fact mediated by increased RasGRP1 levels.
Around 35% of AML patients have a mutation in nucleophosmin protein, a nuclear chaperone protein, which is important in several cellular processes such as ribosome biogenesis and centrosome duplication (reviewed in (Grisendi et al., 2006). Around 30% of mice, which harbor mutation in the nucleophosmin, develop AML with delayed latency suggesting that cooperating mutations are needed as well. A Bis-functional PiggyBac/Sleeping beauty transposon approach was used to identify additional mutations that can cooperate with mutant nucleophosmin in inducing AML. The Sleeping beauty model has the particular advantage of being capable to both activate and disrupt genes (Vassiliou et al., 2011). One of the most common insertional sites (CIS) was found in the gene encoding for GM-SCF, the growth factor previously linked, as previously mentioned to the pathogenesis of JMML, but also of AML (Young and Griffin, 1986; Dührsen et al., 1990; Rogers et al., 1994). Other CIS were found in genes regulating/activating Ras proteins, including RasGRP1, Flt3 (FMS- related tyrosine kinase 3, also known as Flk2 or CD135) and KRas itself. Interestingly, all latter three CIS were mutually exclusive with CIS in GM-SCF suggesting that they are within the same pathway or provide similar oncogenic signals. Flt3 is a receptor tyrosine kinase known to regulate hematopoiesis and is frequently mutated in human AML cases (Yamamoto et al., 2001). The fact that RasGRP1 insertions cooperate with mutant nucleophosmin indicate that it can provide alternative proliferation and/or survival signals that complement mutant nucleophosmin. It remains to be determined what kind of cellular signals are under RasGRP1's control in these settings. It is also worth noting that RasGRP1 insertions were never found in wild-type mice subjected to same mutagenesis protocol, suggesting that CIS in RasGRP1 alone are not sufficient to drive AML. In contrast, insertions in Flt3 or NF1 were found in both genotypes. In addition RasGRP1 CISs were also mutually exclusive with insertions in KRas, indicating that these CIS types may have similar cell biologic impacts.
Multiple receptor systems couple to RasGRP proteins
On the basis of the RasGRP biology described in the previous sections, it appears that different RasGRP proteins appear to couple to multiple receptor systems in addition to the well-accepted role of RasGRPs downstream of the TCR and BCR (Fig. 5). For many of these receptors, it is still an open question if these engage phospholipase C γ and lead to DAG production, or if there are alternative manners of RasGRP membrane recruitment. The cancer studies described thus far added the receptor tyrosine kinases such as HGFR (cMet) and EGFR to the list of BCR, preTCR, TCR, and CXCR4. For the latter it was shown that RasGRP1 also couples to CXCR4 (receptor for chemokine SDF-1), which activates phospholipase C β. CXCR4-induced activation of Ras and phospho-ERK was RasGRP1-dependent and resulted in increased migration of Jurkat cells in chemotaxis assays. Curiously, CXCR-4-induced activation of phosho-ERK was dependent on the presence of RasGRP1's C1 domain but CXCR-4-induced relocalization of RasGRP1 was not (Kremer et al., 2011). This result suggests that RasGRP1's localization after CXCR4 stimulation is mediated by a different mechanism than binding of membrane DAG, as it is the case for e.g. antigen receptors. At the same time RasGRP1's function downstream of CXCR4 still depends on the intact C1 domain and presumably DAG binding. As intriguing those results are, these studies need to be confirmed in primary cells and with use of carefully designed C1 domain mutant instead of truncation mutation.
The conventional view on receptor-induced RasGRP's activation is that DAG needs to be generated in order to coordinate proper localization and/or activation of the protein. However, there is growing evidence that RasGRPs can also couple to receptors which are not known to trigger activation of phospholipase C and therefore likely do not engage active DAG production. As mentioned above, RasGRP3 and 4 can potentially couple to GM-CSF in macrophage progenitors and orchestrate activation of phospho-ERK (Fig. 5) (Diaz-Flores et al., 2013). A second report suggested that RasGRP1 couples to a related cytokine, G-CSF, to induce phospho-ERK that is important for neutrophil development (de la Luz Sierra et al., 2010). More recently, our group demonstrated that IL-7-induced RasGTP is RasGRP1-dependent in T cell leukemic cells (Hartzell et al., 2013). In the next paragraph, we will discuss the specific example of this novel IL7R-RasGRP1 pathway in more detail, but it is likely that future studies will mechanistically link the RasGRPs to receptor systems in addition to the TCR and BCR (Fig. 5).
RasGRP1 and oncogenic KRas in T cell acute lymphoblastic leukemia/lymphoma (T-ALL)
T cell acute lymphoblastic leukemia/lymphoma (T-ALL) is an aggressive blood cancer, which can affect children as well as adults. T-LL (T cell lymphoblastic leukemia/lymphoma) has very similar features as T-ALL, except for the presence of mediastinal masses, which cause respiratory distress in patients. Here we refer to both T-ALL and T-LL as T-ALL. T-ALL accounts for around 15%–20% of all acute leukemia cases and is associated with poor prognosis. In T-ALL, abnormally proliferating cells arise from developing thymocytes, which then spread out through out the body. Another feature that complicates the treatment is dissemination to the central nervous system. The current line of treatment involves multi-agent chemotherapy regiments, which are known to have many side effects (reviewed in (Aifantis et al., 2008).
Genetic make up of T-ALL have been elusive for many years but the past decade has been quite fruitful in elucidating this disease. Molecular analyses showed that the oncogenic mutations are mostly present in genes regulating T cell differentiation, preTCR/TCR signaling and also cytokine signaling (reviewed in (Grabher et al., 2006; Aifantis et al., 2008; Gutierrez et al., 2009; Zhang et al., 2012). Those include Notch mutations, aberrant activation of PI3K pathway through various mechanisms, as well as the Ras pathway. In this chapter we wish to discuss in more detail the role and contribution of Ras signaling in T-ALL.
Early studies investigating the prevalence of oncogenic N- or K- Ras mutations in childhood T-ALL showed that these are relatively rare and present in only 4%–10% of patients, which is much lower than in other tumor types such as pancreatic or colorectal cancer (Ahuja et al., 1990; Yokota et al., 1998; Kawamura et al., 1999; Friday and Adjei, 2005). Significantly, it has also been shown that roughly 50% of patients have increased Ras activation levels in bone marrow cells from punctures or in peripheral blood cells, as compared to healthy controls (von Lintig et al., 2000). This result underscored the importance of aberrantly activated Ras signaling in T-ALL but at the same time it pointed out that there must be alternative mechanisms, other than mutations in Ras genes themselves, which can lead to increased Ras signals. Several possibilities that would result in increased RasGTP levels can be envisioned, such as through increased receptor input, via increased activity of positive regulators, such as GEFs, or through the loss of negative regulators such as GAPs. For unknown reasons, mutations in the RasGAP NF-1 are very rare in T-ALL and are present in only around 3% of patients (Balgobind et al., 2008). By contrast, both Kitamura group as well as our group uncovered that deregulated RasGRP1 is a very common feature in T-ALL and able to drive aberrant Ras signaling (Oki et al., 2011; Hartzell et al., 2013). We will further discuss those findings.
The first study implicating RasGRP1 in T-ALL pathogenesis came once again from retroviral mutagenesis screens where insertions in the RasGRP1 gene were one of the most commonly found (Kim et al., 2003). Transgenically engineered mice corroborated these CIS studies and Robert Kay and colleagues reported that overexpression of RasGRP1 in thymocyte leads to thymic lymphomas even in the absence of input from T cell receptor (Klinger et al., 2005). The penetrance of the phenotype was dependent on the transgenic line and the highest rate was 50% with a latency of 130 days. The immunophenotype of expanding blasts was also variable and included a mix of DN, DP and/or SP thymocytes (Fig. 3). Transformed cells disseminated to other lymphoid organs, notably to the spleen, lymph nodes and/or bone marrow. Since expansion of different thymocyte subsets occurred before the onset of the disease, the authors speculated that one of the mechanism by which inappropriate high levels of RasGRP1 could be oncogenic is through increased pool of proliferating immature thymocytes. An independent group more recently made very similar observations by using retroviral overexpression of RasGRP1 in a bone marrow transfer model to induce leukemia (Oki et al., 2011). One important contribution of this report was to show that aberrant expression of RasGRP1 cooperates with Notch1 in initiation and/or progression of T-ALL.
As much as those aforementioned studies were helpful at pointing out at the possible involvement of RasGRP1 in T-ALL, none of them addressed the relevance to human disease or the molecular mechanism of oncogenic RasGRP1. We have addressed some of those points in our recently published study. First, we have demonstrated that deregulated expression of RasGRP1 is a frequent event in both pediatric T-ALL patients and in mouse models where T-ALL is driven by mouse leukemia virus insertions (Hartzell et al., 2013). Secondly, we were able to show that elevated levels of RasGRP1 as well as classical oncogenic mutations in K-Ras are two distinct mechanisms to trigger oncogenic Ras signaling in T-ALL (Fig. 6). Those events are mutually exclusive and significantly differ in terms of biochemistry. K-RasG12D T-ALLs critically depend on uncoupling strong, constitutive Ras signal from a P53-P21 cell cycle arrest program to aggressively grow. By contrast, dysregulated RasGRP1 T-ALL with modest overexpression of RasGRP1 display low constitutive Ras signals that do not trigger cell cycle arrest. Instead, we find that these RasGRP1 T-ALLs efficiently cooperate with receptor signal input from cytokines such as IL-7 that are typically produced by stromal cells in the bone marrow. Signaling through IL-7 receptor has already been shown to be important for maintaining survival and proliferation of leukemic blasts (Barata et al., 2004a, 2004b, 2005; Silva et al., 2011; Zenatti et al., 2011). At this point it is not clear how RasGRP1 couples to IL-7 receptor and future studies are needed to dissect this novel signaling pathway. It is also not clear if that pathway operates in normal thymocytes as well or only in leukemic cells.
Figure 6.
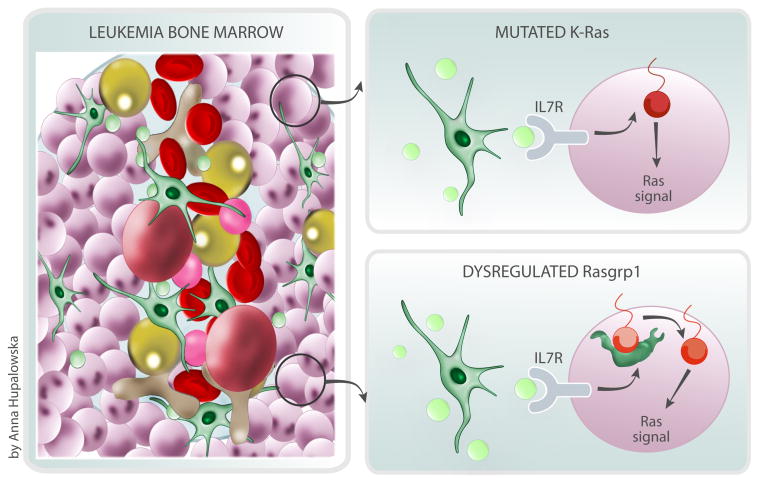
Dysregulated RasGRP1 cooperates with cytokine receptor input in T cell leukemogenesis. T cell leukemia cells expand in the bone marrow in response to growth factors like interleukin 7 (IL7 in green) and take over the space in the cavity (uniform purple cells in the illustration). This expansion leads to a loss of the variety in bone marrow cells, such as blood stem cells (in pink), red blood cells (in red) and fat cells (in yellow) that are normally seen in the bone marrow. The Hartzell et al. study describes how two related, but distinct, genetic alterations in T cell leukemia cells, mutated K-Ras or dysregulated Rasgrp1, both lead to T cell leukemia by responding to IL7 and other signals in different manners.
A perplexing and clinically relevant finding when considering molecularly targeted therapy was the observation that clonal T-ALL lines (with KRas mutation of RasGRP1 overexpression) exhibited a great deal of biochemical heterogeneity, that is in terms of activation of effector pathways downstream of Ras. In addition, when RasGRP1 levels were decreased (in vitro or in vivo), cells could “switch” from using one pathway to another suggesting that there is a great deal of plasticity in the usage of signaling pathways which give similar cellular outcomes (Chakraborty and Roose, 2013). We will elaborate on that point at the end of our review.
A special role for RasGRP1; expression or biochemical features?
Both, human data and mouse models pointed out at that RasGRP1 is unique among other RasGEFs in its capability in induce T cell leukemogenesis (Hartzell et al., 2013). We hypothesize that two of its features, RasGRP1's expression pattern and its biochemical properties, could contribute to this phenomenon.
Genetic studies showed that RasGRP1 is the most important RasGEF during T cell development. RasGRP1 KO mice have profound T cell developmental block and, as discussed in the previous chapters, RasGRP1 is essential for β-selection, positive and negative selection. Deficiency in other thymocytes-specific RasGEFs such as RasGRP3, RasGRP4, RasGRF2 or Sos has either no phenotype or much milder phenotype as compared to RasGRP1 KO mice (Ruiz et al., 2007; Kortum et al., 2011; Zhu et al., 2012; Golec et al., 2013). The role of RasGRP1 in T cell development can be explained to some extent by its expression pattern (Fig. 3). RasGRP1 mRNA and protein levels are dynamically regulated: they are low in DN thymocytes, then they rise sharply at the DP stage to be downregulated again once cells reach SP stage (Priatel et al., 2002; Norment et al., 2003; Kortum et al., 2011). RasGRP1's mRNA levels are also regulated in response to TCR triggering (Diehn et al., 2002) and the DAG analog TPA induced RasGRP1 degradation in mouse keratinocytes (Rambaratsingh et al., 2003). These features of RasGRP1 suggest that the regulation of mRNA and protein abundance is one of the mechanisms to control its activity and function.
Currently, we do not fully understand the mechanisms behind regulation of RasGRP1 levels during T cell development. A candidate regulator is a transcription factor Gfi1. It has been shown that Gfi1 knockout mice have deficiency in RasGRP1 mRNA and protein levels (de la Luz Sierra et al., 2010). Interestingly, a recent report by the Moroy group demonstrated that Gfi1 mRNA levels are increased in a subset of human T-ALL patients, which had a Notch1-like gene expression signature (Khandanpour et al., 2013). Similarly to what has been shown for RasGRP1, Gfi1 levels are low in ETP T-ALLs (Zhang et al., 2012). It is not known if RasGRP1 abundance correlates with increased levels of Gfi1 in T-ALLs. It is also a conundrum how Gfi1 regulates RasGRP1 transcripts levels since putative RasGRP1 promoter does not contain conserved Gfi1 binding sequences. Therefore, it is possible that it does so in an indirect way by regulating some, yet to be determined, negative regulators. Future studies should shed light on that question.
Careful analysis of biochemical properties of RasGRP1 and Sos in normal lymphocytes revealed that downstream of the antigen receptors, RasGRP1 is dominant over Sos (Roose et al., 2007). The reason for this is because RasGRP1-activated RasGTP not only activates its downstream effectors such as Raf-MEK-Erk pathway but also binds to the allosteric pocket of Sos to increase its GEF activity and therefore engage positive feedback loop (Boykevisch et al., 2006; Roose et al., 2007; Das et al., 2009). Therefore, RasGRP1 promotes activation of Sos. Our study showed that this dominance is maintained downstream of cytokine receptors. In fact, decreased Sos levels did not change cytokine-induced RasGTP levels in T-ALL cell lines and conversely lowering RasGRP1 levels resulted in significant decrease RasGTP loading (Hartzell et al., 2013). As mentioned in earlier paragraph, RasGRP1, but not other RasGRP proteins, contains the C-terminal tail and although the function of this domain is still not very well understood, it is believed that it mediates protein–protein interactions (Ebinu et al., 1998; Beaulieu et al., 2007; Zahedi et al., 2011). Therefore, RasGRP1, but not other family members, would have very unique property of binding distinct partners and could potentially modulate different signaling pathways.
In summary, although the mechanism underlying RasGRP1 specificity in T-ALL are still elusive and need further investigation, the mRNA and protein expression pattern as well as biochemical properties are likely to contribute.
Developmental heterogeneity in T-ALL
T-ALL is a heterogenous disease in terms of immunophenotype, cytogenic abnormalities and response to treatment. Unlike for B-ALL, age, leukocyte count at the diagnosis, or immunophenotype do not predict response to treatment (Pieters and Carroll, 2010). Similarly, cytogenic abnormalities or gene expression profiles, although sometimes correlate with the outcome, are not used as a basis of stratification protocols (Graux et al., 2006).
A classical view is that the immunophenotype of malignant blasts reflect the developmental stages of normal thymocytes (Graux et al., 2006). Microarray data suggested that certain gene expression signatures found in leukemic cells can help categorize T-ALL to different stages of normal thymocyte development (Ferrando et al., 2002). That view has been strengthened by description of subtype of pediatric T-ALL with the properties similar to murine early T cell progenitors (ETPs) (Coustan-Smith et al., 2009). ETPs are early immigrants from the bone marrow to the thymus with some stem cell signature and dual lymphoid and myeloid potential (Bell and Bhandoola, 2008; Wada et al., 2008). Human ETP T-ALLs show similar gene signature and immunophenotype to normal murine ETPs suggesting that the normal ETP are the target cell for transformation (Coustan-Smith et al., 2009). This group of patients responds poorly to lymphoid-cell-directed therapies and it has been suggested that those patients would benefit from myeloid-cell targeted therapies such as those used for patients with AML. More recently, whole genome sequencing data comparing ETP and non-ETP patients became available (Zhang et al., 2012). This study showed that the majority (almost 70%) of patients with ETP T-ALL had mutations in genes affecting cytokine receptor and Ras signaling although the type and genes affected were extremely heterogeneous between individuals. Overall, the spectrum of mutations found in this group of T-ALLs resembled mutations found in patients with myeloid malignancies. Moreover, the analysis of gene expression profiles showed that ETP T-ALL resemble more normal human hematopoietic stem cells and granulocyte macrophage progenitors rather than normal human ETPs suggesting that ETP T-ALL is a poorly differentiated stem cell-like leukemia.
More recently, an elegant study using Sleeping Beauty transposon suggested that some T-ALL blasts might not follow the normal developmental path and be simply abnormally proliferating arrested thymocytes but can instead dedifferentiate (Berquam-Vrieze et al., 2011). This conclusion was based on the observation that some leukemias in which mutagenesis through the activity of Sleeping Beauty transposon was initiated at the DP stage (using CD4Cre system) showed ETP-like gene signature. This result is provocative and the underlying mechanism is still an unresolved puzzle. In support of the validity of the mouse model data, roughly half of human ETPs show TCR genes rearrangements suggesting that they arise from at least DN2/DN3 stage (Coustan-Smith et al., 2009). Interestingly, insertions in the RasGRP1 locus mostly occurred in early hematopoietic subsets, similarly to those in Notch1 locus suggesting that deregulation of RasGRP1 may be an early event during tumorigenesis and that early hematopoietic cells may be more sensitive and susceptible to inappropriate levels of this Ras GEF.
Certainly, there is still a lot to understand about initiation and progression of T-ALL and recent data point out that this disease is more complex that we have previously anticipated complicating the design of effective therapies. New tools such as recently described single-mass cytometry (CyTOF) would allow in the near future for more detailed analysis of normal as well perturbed hematopoiesis (Chakraborty and Roose, 2013)
Biochemical hetereogenity and plasticity in T-ALL- implications for therapeutic intervention
The genomic landscape of T-ALL, similarly to other cancer types, is extremely complex due to many, so-called “driver” and “passenger” mutations. In addition, heterogeneity within tumor as well as between the patients adds another layer of complexity and unpredictability (Zhang et al., 2012; Vogelstein et al., 2013).
Modern cancer therapies target signal transduction pathways, which drive aberrant proliferation and survival of cancer. There are fairly few true successes with this approach, such as BCR-ABL inhibitors used for CML or B-Raf inhibitors for melanoma. In general, it appears to be extremely difficult to predict how genetic alterations affect signaling networks. Together with Kevin Shannon's group, we demonstrated that mouse T-ALL lines with the same “driving” mutation/alteration such as KRasG12D or high RasGRP1 and with similar growth characteristics can use different (and to variable degree) signaling pathways (Dail et al., 2010; Hartzell et al., 2013). Similar observations were obtained by the examination of human T-ALL cell lines, which showed very variable usage of PI3K signaling pathway (Maser et al., 2007; Palomero et al., 2007). Although the biochemical heterogeneity have been mostly observed in the cell line systems, it is highly likely that human primary samples also share that feature.
One of the inherit properties of kinase signaling networks are the presence of feedback loops and ability of cells to rewire these networks (Pawson and Linding, 2008). Cancer cells, including T-ALLs, seem to have similar mechanism as their normal counterparts and seem to explore modulation of signaling networks to avoid oncogene-induced senescence or achieve resistance to small inhibitors targeting particular signaling pathway. For example, we observed that T-ALL lines, which grow subcutaneously, can “switch” from using NF-κB pathway to Akt pathway in response to shRNA-mediated RasGRP1 knockdown (Hartzell et al., 2013). Signaling “plasticity” of particular cancer type has potentially tremendous consequences for targeted therapies and could be a major deal breaker.
A very elegant recent study by Lee and colleagues demonstrated that careful analysis of normal- and oncogenic- signaling network rewiring in response to drugs can lead to a design of the protocol treatment that specifically and with higher efficiency targets tumor cells (Lee et al., 2012). The authors combined mathematical modeling, high-throughput measurements of signaling events, gene expression profiling and cell responses (growth vs. apoptosis) to demonstrate that in triple negative breast cancer cells time-staggered inhibition of EGFR rather than simultaneous administration, sensitizes cancer cells to genotoxic drugs. It remains to be determined if that is true for other cancer cell types or maybe the rules are different. Finally, it would be exciting to prove the efficacy of such approach in the clinic.
As for T-ALL, targeted therapies are not available at the moment and patients are treated with chemotherapy, known for its severe side effects. Therefore, there is unmet need to improve treatment for T-ALL patients with more efficient therapies and with fewer side effects. Few attempts had so far limited successes. Prompted by studies which showed that Notch1 is mutated in 50% of patients with T-ALL and encouraged by in vitro results in human T-ALL cell lines showing that Notch1 inhibition affects cells growth and proliferation, a clinical trial was launched with a γ secratase inhibitor, which affects processing and activation of Notch1 (Weng et al., 2004; Palomero et al., 2006a, 2006b, 2007; Weng et al., 2006; Chan et al., 2007;). However, it had to be stopped due to severe gastrointenstinal toxicity and limited anti-leukemic efficacity (DeAngelo, 2006). The development of safer inhibitors could still prove that this strategy is a therapeutic option. Another attractive candidate to target is Ras signaling since, as it has been discussed in previous chapters, it is hyperactivated in around 50% of patients and overexpression of RasGRP1 is a frequent event, at least in pediatric T-ALLs. Targeting Ras itself proved to be unfeasible so far due to the nature of oncogenic mutation and it remains to be determined if small molecules against RasGRP1 could be developed and effective. For now then, targeting downstream effectors such as MEK-Erk pathway or PI3K/Akt/mTOR could prove to be a valid therapeutic option. Indeed, some groups showed that inhibiting PI3K and mTOR or PI3K only seem to be effective in inducing apoptosis and growth arrest of human T-ALL cell lines (Chiarini et al., 2009; Subramaniam et al., 2012). It remains to be determined if that strategy proves to be efficacious in patients.
One has to keep in mind that the aforementioned developmental and biochemical heterogeneity as well as plasticity are likely to complicate the design of effective molecular therapy for T-ALL. Systems-levels analysis, which combine “signaling/phospho- signatures,” together with genomic and gene expression data as well as mathematical modeling will be instrumental in the design of better targeted therapies, not only for T-ALL, which seem to be particularly challenging, but also for other tumor types.
Concluding remarks
The course of specific scientific findings has taken RasGRP research from cancer to lymphocytes and back to cancer. Whereas many of the molecular details still need to be uncovered over the coming years, it is clear that RasGRPs play roles in receptors systems other than the TCR and BCR. Likewise, future research will surely establish more definitive functions for the RasGRP RasGEFs outside the hematopoietic system.
Acknowledgments
We thank Anna Hupalowska for generating all the illustrations for this review and all the members of the Roose laboratory for useful insights. Olga Ksionda, Andre Limnander, Jeroen Roose, and the research in the Roose laboratory is supported by a Sandler Program in Basic Science (start-up JPR), NIH-NCI Physical Science Oncology Center grant U54CA143874 (JPR), NIH grant 1P01AI091580-01 (JPR), a Gabrielle's Angel Foundation grants (JPR), a UCSF PBBR/Sanofi Leap to Innovation for Therapeutics and Technology (LIFTT) Program ant (JPR), and a NIH grant 1R03AR062783-01A1 (Al).
Footnotes
Olga Ksionda, Andre Limnander, and Jeroen Roose declare that they have no conflict of interest.
Compliance with ethics guidelines: This manuscript is a review article and does not involve a research protocol requiring approval by the relevant institutional review board or ethics committee.
References
- Abel EL, Angel JM, Kiguchi K, DiGiovanni J. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat Protoc. 2009;4(9):1350–1362. doi: 10.1038/nprot.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi R, Krilis SA, Nigrovic PA, Hamilton MJ, Chung K, Thakurdas SM, Boyce JA, Anderson P, Stevens RL. Ras guanine nucleotide-releasing protein-4 (RasGRP4) involvement in experimental arthritis and colitis. J Biol Chem. 2012;287(24):20047–20055. doi: 10.1074/jbc.M112.360388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol. 2012;13(1):39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja H, Foti A, Bar-Eli M, Cline M. The pattern of mutational involvement of RAS genes in human hematologic malignancies determined by DNA amplification and direct sequencing. Blood. 1990;75:1684–1690. [PubMed] [Google Scholar]
- Aiba Y, Oh-hora M, Kiyonaka S, Kimura Y, Hijikata A, Mori Y, Kurosaki T. Activation of RasGRP3 by phosphorylation of Thr-133 is required for B cell receptor-mediated Ras activation. Proc Natl Acad Sci USA. 2004;101(47):16612–16617. doi: 10.1073/pnas.0407468101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8(5):380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- Alberola-Ila J, Hogquist KA, Swan KA, Bevan MJ, Perlmutter RM. Positive and negative selection invoke distinct signaling pathways. J Exp Med. 1996;184(1):9–18. doi: 10.1084/jem.184.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balgobind BV, Van Vlierberghe P, van den Ouweland AMW, Beverloo HB, Terlouw-Kromosoeto JNR, van Wering ER, Reinhardt D, Horstmann M, Kaspers GJL, Pieters R, Zwaan CM, Van den Heuvel-Eibrink MM, Meijerink JP. Leukemia-associated NF1 inactivation in patients with pediatric T-ALL and AML lacking evidence for neurofibromatosis. Blood. 2008;111(8):4322–4328. doi: 10.1182/blood-2007-06-095075. [DOI] [PubMed] [Google Scholar]
- Barata JT, Cardoso AA, Boussiotis VA. Interleukin-7 in T-cell acute lymphoblastic leukemia: an extrinsic factor supporting leukemogenesis? Leuk Lymphoma. 2005;46(4):483–495. doi: 10.1080/10428190400027852. [DOI] [PubMed] [Google Scholar]
- Barata JT, Keenan TD, Silva A, Nadler LM, Boussiotis VA, Cardoso AA. Common gamma chain-signaling cytokines promote proliferation of T-cell acute lymphoblastic leukemia. Haematologica. 2004a;89(12):1459–1467. [PubMed] [Google Scholar]
- Barata JT, Silva A, Brandao JG, Nadler LM, Cardoso AA, Boussiotis VA. Activation of PI3K is indispensable for interleukin 7-mediated viability, proliferation, glucose use, and growth of T cell acute lymphoblastic leukemia cells. J Exp Med. 2004b;200(5):659–669. doi: 10.1084/jem.20040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu N, Zahedi B, Goulding RE, Tazmini G, Anthony KV, Omeis SL, de Jong DR, Kay RJ. Regulation of RasGRP1 by B cell antigen receptor requires cooperativity between three domains controlling translocation to the plasma membrane. Mol Biol Cell. 2007;18(8):3156–3168. doi: 10.1091/mbc.E06-10-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452(7188):764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- Benschop RJ, Cambier JC. B cell development: signal transduction by antigen receptors and their surrogates. Curr Opin Immunol. 1999;11(2):143–151. doi: 10.1016/S0952-7915(99)80025-9. [DOI] [PubMed] [Google Scholar]
- Bergmeier W, Goerge T, Wang HW, Crittenden JR, Baldwin ACW, Cifuni SM, Housman DE, Graybiel AM, Wagner DD. Mice lacking the signaling molecule CalDAG-GEFI represent a model for leukocyte adhesion deficiency type III. J Clin Invest. 2007;117(6):1699–1707. doi: 10.1172/JCI30575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berquam-Vrieze KE, Nannapaneni K, Brett BT, Holmfeldt L, Ma J, Zagorodna O, Jenkins NA, Copeland NG, Meyerholz DK, Knudson CM, Mullighan CG, Scheetz TE, Dupuy AJ. Cell of origin strongly influences genetic selection in a mouse model of T-ALL. Blood. 2011;118(17):4646–4656. doi: 10.1182/blood-2011-03-343947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivona TG, Pérez De Castro I, Ahearn IM, Grana TM, Chiu VK, Lockyer PJ, Cullen PJ, Pellicer A, Cox AD, Philips MR. Phospholipase Cgamma activates Ras on the Golgi apparatus by means of RasGRP1. Nature. 2003;424(6949):694–698. doi: 10.1038/nature01806. [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129(5):865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Botelho RJ, Harrison RE, Stone JC, Hancock JF, Philips MR, Jongstra-Bilen J, Mason D, Plumb J, Gold MR, Grinstein S. Localized diacylglycerol-dependent stimulation of Ras and Rap1 during phagocytosis. J Biol Chem. 2009;284(42):28522–28532. doi: 10.1074/jbc.M109.009514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boykevisch S, Zhao C, Sondermann H, Philippidou P, Halegoua S, Kuriyan J, Bar-Sagi D. Regulation of ras signaling dynamics by Sos-mediated positive feedback. Curr Biol. 2006;16(21):2173–2179. doi: 10.1016/j.cub.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Brodie C, Steinhart R, Kazimirsky G, Rubinfeld H, Hyman T, Ayres JN, Hur GM, Toth A, Yang D, Garfield SH, Stone JC, Blumberg PM. PKCdelta associates with and is involved in the phosphorylation of RasGRP3 in response to phorbol esters. Mol Pharmacol. 2004;66(1):76–84. doi: 10.1124/mol.66.1.76. [DOI] [PubMed] [Google Scholar]
- Cambier JC, Gauld SB, Merrell KT, Vilen BJ. B-cell anergy: from transgenic models to naturally occurring anergic B cells? Nat Rev Immunol. 2007;7(8):633–643. doi: 10.1038/nri2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbo C, Duerschmied D, Goerge T, Hattori H, Sakai J, Cifuni SM, White GC, 2nd, Chrzanowska-Wodnicka M, Luo HR, Wagner DD. Integrin-independent role of CalDAG-GEFI in neutrophil chemotaxis. J Leukoc Biol. 2010;88(2):313–319. doi: 10.1189/jlb.0110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty AK, Roose JP. Biochemical heterogeneity and developmental varieties in T-cell leukemia. Cell Cycle. 2013;12(10):1480–1481. doi: 10.4161/cc.24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ. Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood. 2007;110(1):278–286. doi: 10.1182/blood-2006-08-039883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410(6824):37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- Chiarini F, Falà F, Tazzari PL, Ricci F, Astolfi A, Pession A, Pagliaro P, McCubrey JA, Martelli AM. Dual inhibition of class IA phosphatidylinositol 3-kinase and mammalian target of rapamycin as a new therapeutic option for T-cell acute lymphoblastic leukemia. Cancer Res. 2009;69(8):3520–3528. doi: 10.1158/0008-5472.CAN-08-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JB, Silverman M, Monroe JG. Transitional B cells: step by step towards immune competence. Trends Immunol. 2003;24(6):343–349. doi: 10.1016/S1471-4906(03)00119-4. [DOI] [PubMed] [Google Scholar]
- Cifuni SM, Wagner DD, Bergmeier W. CalDAG-GEFI and protein kinase C represent alternative pathways leading to activation of integrin alphaIIbbeta3 in platelets. Blood. 2008;112(5):1696–1703. doi: 10.1182/blood-2008-02-139733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde-Smith J, Silins G, Gartside M, Grimmond S, Etheridge M, Apolloni A, Hayward N, Hancock JF. Characterization of RasGRP2, a plasma membrane-targeted, dual specificity Ras/Rap exchange factor. J Biol Chem. 2000;275(41):32260–32267. doi: 10.1074/jbc.M006087200. [DOI] [PubMed] [Google Scholar]
- Corey SJ, Minden MD, Barber DL, Kantarjian H, Wang JCY, Schimmer AD. Myelodysplastic syndromes: the complexity of stem-cell diseases. Nat Rev Cancer. 2007;7(2):118–129. doi: 10.1038/nrc2047. [DOI] [PubMed] [Google Scholar]
- Coughlin JJ, Stang SL, Dower NA, Stone JC. RasGRP1 and RasGRP3 regulate B cell proliferation by facilitating B cell receptor-Ras signaling. J Immunol (Baltimore, Md: 1950) 2005;175(11):7179–7184. doi: 10.4049/jimmunol.175.11.7179. [DOI] [PubMed] [Google Scholar]
- Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, Cheng C, Su X, Rubnitz JE, Basso G, Biondi A, Pui CH, Downing JR, Campana D. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10(2):147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden JR, Bergmeier W, Zhang Y, Piffath CL, Liang Y, Wagner DD, Housman DE, Graybiel AM. CalDAG-GEFI integrates signaling for platelet aggregation and thrombus formation. Nat Med. 2004;10(9):982–986. doi: 10.1038/nm1098. [DOI] [PubMed] [Google Scholar]
- Dail M, Li Q, McDaniel A, Wong J, Akagi K, Huang B, Kang HC, Kogan SC, Shokat K, Wolff L, Braun BS, Shannon K. Mutant Ikzf1, KrasG12D, and Notch1 cooperate in T lineage leukemogenesis and modulate responses to targeted agents. Proc Natl Acad Sci USA. 2010;107(11):5106–5111. doi: 10.1073/pnas.1001064107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Porto JM, Gauld SB, Merrell KT, Mills D, Pugh-Bernard AE, Cambier J. B cell antigen receptor signaling 101. Mol Immunol. 2004;41(6-7):599–613. doi: 10.1016/j.molimm.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Holländer GA, Gascoigne NRJ, Palmer E. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444(7120):724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- Das J, Ho M, Zikherman J, Govern C, Yang M, Weiss A, Chakraborty AK, Roose JP. Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell. 2009;136(2):337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Luz Sierra M, Sakakibara S, Gasperini P, Salvucci O, Jiang K, McCormick PJ, Segarra M, Stone J, Maric D, Zhu J, Qian X, Lowy DR, Tosato G. The transcription factor Gfi1 regulates G-CSF signaling and neutrophil development through the Ras activator RasGRP1. Blood. 2010;115(19):3970–3979. doi: 10.1182/blood-2009-10-246967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelo DJ. A phase I clinical trial of the notch inhibitor MK-0752 in patients with T-cell acute lymphoblastic leukemia (T-ALL) and other leukemias. J Clin Oncol. 2006;24(18 Suppl):6585. [Google Scholar]
- DeFranco AL. B-cell activation 2000. Immunol Rev. 2000;176:5–9. [PubMed] [Google Scholar]
- Diaz-Flores E, Hana Goldschmidt, Philippe Depeille, Victor Ng, Kimberly Krisman, Michael Crone, Burgess Michael R, Williams Olusegun, Houseman Benjamin, Kevan Shokat, et al. PLCγ and PI3 kinase link cytokine stimulation to ERK activation in primary hematopoietic cells expressing normal and oncogenic Kras. Science Signaling. 2013 doi: 10.1126/scisignal.2004125. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Alizadeh AA, Rando OJ, Liu CL, Stankunas K, Botstein D, Crabtree GR, Brown PO. Genomic expression programs and the integration of the CD28 costimulatory signal in T cell activation. Proc Natl Acad Sci USA. 2002;99(18):11796–11801. doi: 10.1073/pnas.092284399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez FR, Garrido AA, Sharma A, Luke CT, Stone JC, Dower NA, Cline JM, Lorenzo PS. RasGRP1 transgenic mice develop cutaneous squamous cell carcinomas in response to skin wounding: potential role of granulocyte colony-stimulating factor. Am J Pathol. 2009;175(1):392–399. doi: 10.2353/ajpath.2009.090036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower NA, Stang SL, Bottorff DA, Ebinu JO, Dickie P, Ostergaard HL, Stone JC. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat Immunol. 2000;1(4):317–321. doi: 10.1038/79766. [DOI] [PubMed] [Google Scholar]
- Dührsen U, Stahl J, Gough NM. In vivo transformation of factor-dependent hemopoietic cells: role of intracisternal A-particle transposition for growth factor gene activation. EMBO J. 1990;9(4):1087–1096. doi: 10.1002/j.1460-2075.1990.tb08214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebinu JO, Bottorff DA, Chan EY, Stang SL, Dunn RJ, Stone JC. RasGRP, a Ras guanyl nucleotide- releasing protein with calcium- and diacylglycerol-binding motifs. Science. 1998;280(5366):1082–1086. doi: 10.1126/science.280.5366.1082. [DOI] [PubMed] [Google Scholar]
- Ebinu JO, Stang SL, Teixeira C, Bottorff DA, Hooton J, Blumberg PM, Barry M, Bleakley RC, Ostergaard HL, Stone JC. RasGRP links T-cell receptor signaling to Ras. Blood. 2000;95(10):3199–3203. [PubMed] [Google Scholar]
- Emanuel PD, Bates LJ, Castleberry RP, Gualtieri RJ, Zuckerman KS. Selective hypersensitivity to granulocyte-macrophage colony-stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood. 1991;77(5):925–929. [PubMed] [Google Scholar]
- Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1(1):34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, Behm FG, Pui CH, Downing JR, Gilliland DG, Lander ES, Golub TR, Look AT. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1(1):75–87. doi: 10.1016/S1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- Feske S. Calcium signalling in lymphocyte activation and disease. Nat Rev Immunol. 2007;7(9):690–702. doi: 10.1038/nri2152. [DOI] [PubMed] [Google Scholar]
- Friday BB, Adjei AA. K-ras as a target for cancer therapy. Biochimica et Biophysica Acta (BBA) -Rev Can. 2005;1756:127–144. doi: 10.1016/j.bbcan.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Fuller DM, Zhu M, Song X, Ou-Yang CW, Sullivan SA, Stone JC, Zhang W. Regulation of RasGRP1 function in T cell development and activation by its unique tail domain. PLoS ONE. 2012;7(6):e38796. doi: 10.1371/journal.pone.0038796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandour H, Cullere X, Alvarez A, Luscinskas FW, Mayadas TN. Essential role for Rap1 GTPase and its guanine exchange factor CalDAG-GEFI in LFA-1 but not VLA-4 integrin mediated human T-cell adhesion. Blood. 2007;110(10):3682–3690. doi: 10.1182/blood-2007-03-077628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem J. 2007;405(2):199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- Golec DP, Dower NA, Stone JC, Baldwin TA. RasGRP1, but not RasGRP3, is required for efficient thymic β-selection and ERK activation downstream of CXCR4. PLoS ONE. 2013;8(1):e53300. doi: 10.1371/journal.pone.0053300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnow CC, Crosbie J, Jorgensen H, Brink RA, Basten A. Induction of self-tolerance in mature peripheral B lymphocytes. Nature. 1989;342(6248):385–391. doi: 10.1038/342385a0. [DOI] [PubMed] [Google Scholar]
- Grabarek Z. Structural basis for diversity of the EF-hand calcium-binding proteins. J Mol Biol. 2006;359(3):509–525. doi: 10.1016/j.jmb.2006.03.066. [DOI] [PubMed] [Google Scholar]
- Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6(5):347–359. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- Graux C, Cools J, Michaux L, Vandenberghe P, Hagemeijer A. Cytogenetics and molecular genetics of T-cell acute lymphoblastic leukemia: from thymocyte to lymphoblast. Leukemia. 2006;20(9):1496–1510. doi: 10.1038/sj.leu.2404302. [DOI] [PubMed] [Google Scholar]
- Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6(7):493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- Guilbault B, Kay RJ. RasGRP1 sensitizes an immature B cell line to antigen receptor-induced apoptosis. J Biol Chem. 2004;279(19):19523–19530. doi: 10.1074/jbc.M314273200. [DOI] [PubMed] [Google Scholar]
- Gutierrez A, Sanda T, Grebliunaite R, Carracedo A, Salmena L, Ahn Y, Dahlberg S, Neuberg D, Moreau LA, Winter SS, Larson R, Zhang J, Protopopov A, Chin L, Pandolfi PP, Silverman LB, Hunger SP, Sallan SE, Look AT. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114(3):647–650. doi: 10.1182/blood-2009-02-206722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell C, Ksionda O, Lemmens E, Coakley K, Yang M, Dail M, Harvey RC, Govern C, Bakker J, Lenstra TL, Ammon K, Boeter A, Winter SS, Loh M, Shannon K, Chakraborty AK, Wabl M, Roose JP. Dysregulated RasGRP1 responds to cytokine receptor input in T cell leukemogenesis. Sci Signal. 2013;6(268):ra21. doi: 10.1126/scisignal.2003848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz M, Nemazee D. BCR ligation induces receptor editing in IgM+IgD− bone marrow B cells in vitro. Immunity. 1997;6(4):429–436. doi: 10.1016/S1074-7613(00)80286-1. [DOI] [PubMed] [Google Scholar]
- Izquierdo M, Downward J, Graves JD, Cantrell DA. Role of protein kinase C in T-cell antigen receptor regulation of p21ras: evidence that two p21ras regulatory pathways coexist in T cells. Mol Cell Biol. 1992;12(7):3305–3312. doi: 10.1128/mcb.12.7.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas ML, Turner M. Interaction of Ras with P110 is required for thymic -selection in the mouse. J Immunol (Baltimore, Md: 1950) 2011;187:4667–4675. doi: 10.4049/jimmunol.1101949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Goulding RE, Ding Z, Partovi A, Anthony KV, Beaulieu N, Tazmini G, Cornell RB, Kay RJ. Differential membrane binding and diacylglycerol recognition by C1 domains of RasGRPs. Biochem J. 2007;406(2):223–236. doi: 10.1042/BJ20070294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JE, Rubio Ignacio, Roose JP. Regulation of Ras exchange factors and cellular localization of Ras activation by lipid messengers in T cells. Fronit Immunol. 2013 doi: 10.3389/fimmu.2013.00239. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura M, Ohnishi H, Guo SX, Sheng XM, Minegishi M, Hanada R, Horibe K, Hongo T, Kaneko Y, Bessho F, Yanagisawa M, Sekiya T, Hayashi Y. Alterations of the p53, p21, p16, p15 and RAS genes in childhood T-cell acute lymphoblastic leukemia. Leuk Res. 1999;23(2):115–126. doi: 10.1016/S0145-2126(98)00146-5. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Springett GM, Toki S, Canales JJ, Harlan P, Blumenstiel JP, Chen EJ, Bany IA, Mochizuki N, Ashbacher A, Matsuda M, Housman DE, Graybiel AM. A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc Natl Acad Sci USA. 1998;95(22):13278–13283. doi: 10.1073/pnas.95.22.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandanpour C, Phelan JD, Vassen L, Schütte J, Chen R, Horman SR, Gaudreau MC, Krongold J, Zhu J, Paul WE, Dührsen U, Göttgens B, Grimes HL, Möröy T. Growth factor independence 1 antagonizes a p53-induced DNA damage response pathway in lymphoblastic leukemia. Cancer Cell. 2013;23(2):200–214. doi: 10.1016/j.ccr.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R, Trubetskoy A, Suzuki T, Jenkins NA, Copeland NG, Lenz J. Genome-based identification of cancer genes by proviral tagging in mouse retrovirus-induced T-cell lymphomas. J Virol. 2003;77(3):2056–2062. doi: 10.1128/JVI.77.3.2056-2062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. 2009;9(12):833–844. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- Klinger MB, Guilbault B, Goulding RE, Kay RJ. Deregulated expression of RasGRP1 initiates thymic lymphomagenesis independently of T-cell receptors. Oncogene. 2005;24(16):2695–2704. doi: 10.1038/sj.onc.1208334. [DOI] [PubMed] [Google Scholar]
- Knudsen BS, Edlund M. Prostate cancer and the met hepatocyte growth factor receptor. Adv Cancer Res. 2004;91:31–67. doi: 10.1016/S0065-230X(04)91002-0. [DOI] [PubMed] [Google Scholar]
- Koike K, Matsuda K. Recent advances in the pathogenesis and management of juvenile myelomonocytic leukaemia. Br J Haematol. 2008;141(5):567–575. doi: 10.1111/j.1365-2141.2008.07104.x. [DOI] [PubMed] [Google Scholar]
- Kortum RL, Rouquette-Jazdanian AK, Samelson LE. Ras and extracellular signal-regulated kinase signaling in thymocytes and T cells. Trends Immunol. 2013;34(6):1–10. doi: 10.1016/j.it.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortum RL, Sommers CL, Alexander CP, Pinski JM, Li W, Grinberg A, Lee J, Love PE, Samelson LE. Targeted Sos1 deletion reveals its critical role in early T-cell development. Proc Natl Acad Sci USA. 2011;108(30):12407–12412. doi: 10.1073/pnas.1104295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortum RL, Sommers CL, Pinski JM, Alexander CP, Merrill RK, Li W, Love PE, Samelson LE. Deconstructing Ras signaling in the thymus. Mol Cell Biol. 2012;32(14):2748–2759. doi: 10.1128/MCB.00317-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer KN, Kumar A, Hedin KE. G i2 and ZAP-70 mediate RasGRP1 membrane localization and activation of SDF-1-induced T cell functions. J Immunol(Baltimore, Md: 1950) 2011;187:3177–3185. doi: 10.4049/jimmunol.1100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T. Genetic analysis of B cell antigen receptor signaling. Annu Rev Immunol. 1999;17(1):555–592. doi: 10.1146/annurev.immunol.17.1.555. [DOI] [PubMed] [Google Scholar]
- Lam KP, Kühn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90(6):1073–1083. doi: 10.1016/S0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- Lauchle JO, Kim D, Le DT, Akagi K, Crone M, Krisman K, Warner K, Bonifas JM, Li Q, Coakley KM, Diaz-Flores E, Gorman M, Przybranowski S, Tran M, Kogan SC, Roose JP, Copeland NG, Jenkins NA, Parada L, Wolff L, Sebolt-Leopold J, Shannon K. Response and resistance to MEK inhibition in leukaemias initiated by hyperactive Ras. Nature. 2009;461(7262):411–414. doi: 10.1038/nature08279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Koretzky GA. Extracellular signal-regulated kinase-2, but not c-Jun NH2-terminal kinase, activation correlates with surface IgM-mediated apoptosis in the WEHI 231 B cell line. J Immunol. 1998;161(4):1637–1644. [PubMed] [Google Scholar]
- Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, Yaffe MB. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149(4):780–794. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Yun S, Lee J, Kim MJ, Piao ZH, Jeong M, Chung JW, Kim TD, Yoon SR, Greenberg PD, Choi I. RasGRP1 is required for human NK cell function. J Immunol (Baltimore, Md: 1950) 2009;183:7931–7938. doi: 10.4049/jimmunol.0902012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yang Y, Wong GW, Stevens RL. Mast cells in airway hyporesponsive C3H/HeJ mice express a unique isoform of the signaling protein Ras guanine nucleotide releasing protein 4 that is unresponsive to diacylglycerol and phorbol esters. J Immunol (Baltimore, Md: 1950) 2003;171:390–397. doi: 10.4049/jimmunol.171.1.390. [DOI] [PubMed] [Google Scholar]
- Limnander A, Depeille P, Freedman TS, Liou J, Leitges M, Kurosaki T, Roose JP, Weiss A. STIM1, PKC-δ and RasGRP set a threshold for proapoptotic Erk signaling during B cell development. Nat Immunol. 2011;12(5):425–433. doi: 10.1038/ni.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limnander A, Weiss A. Ca-dependent Ras/Erk signaling mediates negative selection of autoreactive B cells. Small GTPases. 2011;2(5):282–288. doi: 10.4161/sgtp.2.5.17794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo PS, Beheshti M, Pettit GR, Stone JC, Blumberg PM. The guanine nucleotide exchange factor RasGRP is a high -affinity target for diacylglycerol and phorbol esters. Mol Pharmacol. 2000;57(5):840–846. [PubMed] [Google Scholar]
- Lorenzo PS, Kung JW, Bottorff DA, Garfield SH, Stone JC, Blumberg PM. Phorbol esters modulate the Ras exchange factor RasGRP3. Cancer Res. 2001;61(3):943–949. [PubMed] [Google Scholar]
- Luke CT, Oki-Idouchi CE, Cline JM, Lorenzo PS. RasGRP1 overexpression in the epidermis of transgenic mice contributes to tumor progression during multistage skin carcinogenesis. Cancer Res. 2007;67(21):10190–10197. doi: 10.1158/0008-5472.CAN-07-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, O'Neil J, Gutierrez A, Ivanova E, Perna I, Lin E, Mani V, Jiang S, McNamara K, Zaghlul S, Edkins S, Stevens C, Brennan C, Martin ES, Wiedemeyer R, Kabbarah O, Nogueira C, Histen G, Aster J, Mansour M, Duke V, Foroni L, Fielding AK, Goldstone AH, Rowe JM, Wang YA, Look AT, Stratton MR, Chin L, Futreal PA, DePinho RA. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447(7147):966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed D, Benschop RJ, Cambier JC, Nemazee D. Developmental regulation of B lymphocyte immune tolerance compartmentalizes clonal selection from receptor selection. Cell. 1998;92(2):173–182. doi: 10.1016/S0092-8674(00)80912-5. [DOI] [PubMed] [Google Scholar]
- Mikkers H, Allen J, Knipscheer P, Romeijn L, Hart A, Vink E, Berns A, Romeyn L. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet. 2002;32(1):153–159. doi: 10.1038/ng950. [DOI] [PubMed] [Google Scholar]
- Mor A, Philips MR. Compartmentalized Ras/MAPK signaling. Annu Rev Immunol. 2006;24(1):771–800. doi: 10.1146/annurev.immunol.24.021605.090723. [DOI] [PubMed] [Google Scholar]
- Navarro MN, Goebel J, Feijoo-Carnero C, Morrice N, Cantrell DA. Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes. Nat Immunol. 2011;12(4):352–361. doi: 10.1038/ni.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norment AM, Bogatzki LY, Klinger M, Ojala EW, Bevan MJ, Kay RJ. Transgenic expression of RasGRP1 induces the maturation of double-negative thymocytes and enhances the production of CD8 single-positive thymocytes. J Immunol (Baltimore, Md: 1950) 2003;170:1141–1149. doi: 10.4049/jimmunol.170.3.1141. [DOI] [PubMed] [Google Scholar]
- Oh-hora M, Johmura S, Hashimoto A, Hikida M, Kurosaki T. Requirement for Ras guanine nucleotide releasing protein 3 in coupling phospholipase C-gamma2 to Ras in B cell receptor signaling. J Exp Med. 2003;198(12):1841–1851. doi: 10.1084/jem.20031547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki T, Kitaura J, Watanabe-Okochi N, Nishimura K, Maehara A, Uchida T, Komeno Y, Nakahara F, Harada Y, Sonoki T, Harada H, Kitamura T. Aberrant expression of RasGRP1 cooperates with gain-of-function NOTCH1 mutations in T-cell leukemogenesis. Leukemia. 2011 doi: 10.1038/leu.2011.328. [DOI] [PubMed] [Google Scholar]
- Oki-Idouchi CE, Lorenzo PS. Transgenic overexpression of RasGRP1 in mouse epidermis results in spontaneous tumors of the skin. Cancer Res. 2007;67(1):276–280. doi: 10.1158/0008-5472.CAN-06-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T, Barnes KC, Real PJ, Glade Bender JL, Sulis ML, Murty VV, Colovai AI, Balbin M, Ferrando AA. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006a;20(7):1279–1287. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O'Neil J, Neuberg D, Weng AP, Aster JC, Sigaux F, Soulier J, Look AT, Young RA, Califano A, Ferrando AA. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Nat Acad Sci U S A. 2006b;103(48):18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, Caparros E, Buteau J, Brown K, Perkins SL, Bhagat G, Agarwal AM, Basso G, Castillo M, Nagase S, Cordon-Cardo C, Parsons R, Zúñiga-Pflücker JC, Dominguez M, Ferrando AA. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13(10):1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Linding R. Network medicine. FEBS Lett. 2008;582(8):1266–1270. doi: 10.1016/j.febslet.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Perez-Losada J, Balmain A. Stem-cell hierarchy in skin cancer. Nat Rev Cancer. 2003;3(6):434–443. doi: 10.1038/nrc1095. [DOI] [PubMed] [Google Scholar]
- Pieters R, Carroll WL. Biology and treatment of acute lymphoblastic leukemia. Pediatric Clinics of NA. 2008;24:1–20. doi: 10.1016/j.pcl.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Pillai S. The chosen few? Positive selection and the generation of naive B lymphocytes. Immunity. 1999;10(5):493–502. doi: 10.1016/S1074-7613(00)80049-7. [DOI] [PubMed] [Google Scholar]
- Pillai S, Cariappa A. The follicular versus marginal zone B lymphocyte cell fate decision. Nat Rev Immunol. 2009;9(11):767–777. doi: 10.1038/nri2656. [DOI] [PubMed] [Google Scholar]
- Pillai S, Cariappa A, Moran ST. Positive selection and lineage commitment during peripheral B-lymphocyte development. Immunol Rev. 2004;197(1):206–218. doi: 10.1111/j.0105-2896.2003.097.x. [DOI] [PubMed] [Google Scholar]
- Priatel JJ, Teh SJ, Dower NA, Stone JC, Teh HS. RasGRP1 transduces low-grade TCR signals which are critical for T cell development, homeostasis, and differentiation. Immunity. 2002;17(5):617–627. doi: 10.1016/S1074-7613(02)00451-X. [DOI] [PubMed] [Google Scholar]
- Rajalingam K, Schreck R, Rapp UR, Albert S. Ras oncogenes and their downstream targets. Biochim Biophys Acta. 2007;1773(8):1177–1195. doi: 10.1016/j.bbamcr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Rambaratsingh RA, Stone JC, Blumberg PM, Lorenzo PS. RasGRP1 represents a novel non-protein kinase C phorbol ester signaling pathway in mouse epidermal keratinocytes. J Biol Chem. 2003;278(52):52792–52801. doi: 10.1074/jbc.M308240200. [DOI] [PubMed] [Google Scholar]
- Ratushny V, Gober MD, Hick R, Ridky TW, Seykora JT. From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J Clin Invest. 2012;122(2):464–472. doi: 10.1172/JCI57415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuther GW, Lambert QT, Rebhun JF, Caligiuri MA, Quilliam LA, Der CJ. RasGRP4 is a novel Ras activator isolated from acute myeloid leukemia. J Biol Chem. 2002;277(34):30508–30514. doi: 10.1074/jbc.M111330200. [DOI] [PubMed] [Google Scholar]
- Roberts DM, Anderson AL, Hidaka M, Swetenburg RL, Patterson C, Stanford WL, Bautch VL. A vascular gene trap screen defines RasGRP3 as an angiogenesis-regulated gene required for the endothelial response to phorbol esters. Mol Cell Biol. 2004;24(24):10515–10528. doi: 10.1128/MCB.24.24.10515-10528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SY, Bradbury D, Kozlowski R, Russell NH. Evidence for internal autocrine regulation of growth in acute myeloblastic leukemia cells. Exp Hematol. 1994;22(7):593–598. [PubMed] [Google Scholar]
- Roose JP, Mollenauer M, Gupta VA, Stone J, Weiss A. A diacylglycerol-protein kinase C-RasGRP1 pathway directs Ras activation upon antigen receptor stimulation of T cells. Mol Cell Biol. 2005;25(11):4426–4441. doi: 10.1128/MCB.25.11.4426-4441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose JP, Mollenauer M, Ho M, Kurosaki T, Weiss A. Unusual interplay of two types of Ras activators, RasGRP and SOS, establishes sensitive and robust Ras activation in lymphocytes. Mol Cell Biol. 2007;27(7):2732–2745. doi: 10.1128/MCB.01882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S, Santos E, Bustelo XR. RasGRF2, a guanosine nucleotide exchange factor for Ras GTPases, participates in T-cell signaling responses. Mol Cell Biol. 2007;27(23):8127–8142. doi: 10.1128/MCB.00912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Luke CT, Dower NA, Stone JC, Lorenzo PS. RasGRP1 is essential for ras activation by the tumor promoter 12-O-tetradecanoylphorbol-13-acetate in epidermal keratinocytes. J Biol Chem. 2010;285(21):15724–15730. doi: 10.1074/jbc.M109.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A, Laranjeira ABA, Martins LR, Cardoso BA, Demengeot J, Yunes JA, Seddon B, Barata JT. IL-7 contributes to the progression of human T-cell acute lymphoblastic leukemias. Cancer Res. 2011;71(14):4780–4789. doi: 10.1158/0008-5472.CAN-10-3606. [DOI] [PubMed] [Google Scholar]
- Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27(1):591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stang SL, Lopez-Campistrous A, Song X, Dower NA, Blumberg PM, Wender PA, Stone JC. A proapoptotic signaling pathway involving RasGRP, Erk, and Bim in B cells. Exp Hematol. 2009;37(1):122–134. 134.e2. doi: 10.1016/j.exphem.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21(1):139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- Stolla M, Stefanini L, André P, Ouellette TD, Reilly MP, McKenzie SE, Bergmeier W. CalDAG-GEFI deficiency protects mice in a novel model of Fcγ RIIA-mediated thrombosis and thrombocytopenia. Blood. 2011;118(4):1113–1120. doi: 10.1182/blood-2011-03-342352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JC. Regulation and Function of the RasGRP Family of Ras Activators in Blood Cells. Genes & Cancer. 2011;2:320–334. doi: 10.1177/1947601911408082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, Cooper ZA, Chapman PB, Solit DB, Ribas A, Lo RS, Flaherty KT, Ogino S, Wargo JA, Golub TR. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487(7408):500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT, Guo B, Wei B, Braun J, Rawlings DJ. Signaling in transitional type 2 B cells is critical for peripheral B-cell development. Immunol Rev. 2004;197(1):161–178. doi: 10.1111/j.0105-2896.2004.0102.x. [DOI] [PubMed] [Google Scholar]
- Subramaniam PS, Whye DW, Efimenko E, Chen J, Tosello V, De Keersmaecker K, Kashishian A, Thompson MA, Castillo M, Cordon-Cardo C, Davé UP, Ferrando A, Lannutti BJ, Diacovo TG. Targeting nonclassical oncogenes for therapy in T-ALL. Cancer Cell. 2012;21(4):459–472. doi: 10.1016/j.ccr.2012.02.029. [DOI] [PubMed] [Google Scholar]
- Suire S, Lécureuil C, Anderson KE, Damoulakis G, Niewczas I, Davidson K, Guillou H, Pan D, Clark J, Hawkins PT, Stephens L. GPCR activation of Ras and PI3Kγ in neutrophils depends on PLCβ2/β3 and the RasGEF RasGRP4. EMBO J. 2012;31(14):3118–3129. doi: 10.1038/emboj.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, Naiman DQ, Jenkins NA, Copeland NG. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32(1):166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- Tazmini G, Beaulieu N, Woo A, Zahedi B, Goulding RE, Kay RJ. Membrane localization of RasGRP1 is controlled by an EF-hand, and by the GEF domain. Biochim Biophys Acta. 2009;1793(3):447–461. doi: 10.1016/j.bbamcr.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Teixeira C, Stang SL, Zheng Y, Beswick NS, Stone JC. Integration of DAG signaling systems mediated by PKC-dependent phosphorylation of RasGRP3. Blood. 2003;102(4):1414–1420. doi: 10.1182/blood-2002-11-3621. [DOI] [PubMed] [Google Scholar]
- Torres RM, Flaswinkel H, Reth M, Rajewsky K. Aberrant B cell development and immune response in mice with a compromised BCR complex. Science. 1996;272(5269):1804–1808. doi: 10.1126/science.272.5269.1804. [DOI] [PubMed] [Google Scholar]
- Townsend SE, Weintraub BC, Goodnow CC. Growing up on the streets: why B-cell development differs from T-cell development. Immunol Today. 1999;20(5):217–220. doi: 10.1016/S0167-5699(98)01440-6. [DOI] [PubMed] [Google Scholar]
- Vassiliou GS, Cooper JL, Rad R, Li J, Rice S, Uren A, Rad L, Ellis P, Andrews R, Banerjee R, Grove C, Wang W, Liu P, Wright P, Arends M, Bradley A. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat Genet. 2011;43(5):470–475. doi: 10.1038/ng.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294(5545):1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10(12):842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lintig FC, Huvar I, Law P, Diccianni MB, Yu AL, Boss GR. Ras activation in normal white blood cells and childhood acute lymphoblastic leukemia. Clin Cancer Res. 2000;6(5):1804–1810. [PubMed] [Google Scholar]
- Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452(7188):768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- Ward AF, Braun BS, Shannon KM. Targeting oncogenic Ras signaling in hematologic malignancies. Blood. 2012;120(17):3397–3406. doi: 10.1182/blood-2012-05-378596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe-Okochi N, Oki T, Komeno Y, Kato N, Yuji K, Ono R, Harada Y, Harada H, Hayashi Y, Nakajima H, Nosaka T, Kitaura J, Kitamura T. Possible involvement of RasGRP4 in leukemogenesis. Int J Hematol. 2009;89(4):470–481. doi: 10.1007/s12185-009-0299-0. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JP, 4th, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, Li Y, Wolfe MS, Shachaf C, Felsher D, Blacklow SC, Pear WS, Aster JC. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes & Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J, Ribas A, Li J, Moffat J, Sutherlin DP, Koeppen H, Merchant M, Neve R, Settleman J. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487(7408):505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, Miyawaki S, Asou N, Kuriyama K, Yagasaki F, Shimazaki C, Akiyama H, Saito K, Nishimura M, Motoji T, Shinagawa K, Takeshita A, Saito H, Ueda R, Ohno R, Naoe T. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97(8):2434–2439. doi: 10.1182/blood.V97.8.2434. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Mochizuki N, Ohba Y, Tobiume M, Okada Y, Sawa H, Nagashima K, Matsuda M. CalDAG-GEFIII activation of Ras, R-ras, and Rap1. J Biol Chem. 2000;275(33):25488–25493. doi: 10.1074/jbc.M003414200. [DOI] [PubMed] [Google Scholar]
- Yang D, Kedei N, Li L, Tao J, Velasquez JF, Michalowski AM, Tóth BI, Marincsák R, Varga A, Bíró T, Yuspa SH, Blumberg PM. RasGRP3 contributes to formation and maintenance of the prostate cancer phenotype. Cancer Res. 2010;70(20):7905–7917. doi: 10.1158/0008-5472.CAN-09-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Tao J, Li L, Kedei N, Tóth ZE, Czap A, Velasquez JF, Mihova D, Michalowski AM, Yuspa SH, Blumberg PM. RasGRP3, a Ras activator, contributes to signaling and the tumorigenic phenotype in human melanoma. 2011;30:4590–4600. doi: 10.1038/onc.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li L, Wong GW, Krilis SA, Madhusudhan MS, Sali A, Stevens RL. RasGRP4, a new mast cell-restricted Ras guanine nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Identification of defective variants of this signaling protein in asthma, mastocytosis, and mast cell leukemia patients and demonstration of the importance of RasGRP4 in mast cell development and function. J Biol Chem. 2002;277(28):25756–25774. doi: 10.1074/jbc.M202575200. [DOI] [PubMed] [Google Scholar]
- Yasuda T, Kometani K, Takahashi N, Imai Y, Aiba Y, Kurosaki T. ERKs induce expression of the transcriptional repressor Blimp-1 and subsequent plasma cell differentiation. Sci Signal. 2011;4(169):ra25. doi: 10.1126/scisignal.2001592. [DOI] [PubMed] [Google Scholar]
- Yasuda T, Kurosaki T. Regulation of lymphocyte fate by Ras/ERK signals. Cell Cycle. 2008;7(23):3634–3640. doi: 10.4161/cc.7.23.7103. [DOI] [PubMed] [Google Scholar]
- Yasuda T, Sanjo H, Pagès G, Kawano Y, Karasuyama H, Pouysségur J, Ogata M, Kurosaki T. Erk kinases link pre-B cell receptor signaling to transcriptional events required for early B cell expansion. Immunity. 2008;28(4):499–508. doi: 10.1016/j.immuni.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Yokota S, Nakao M, Horiike S, Seriu T, Iwai T, Kaneko H, Azuma H, Oka T, Takeda T, Watanabe A, Kikuta A, Asami K, Sekine I, Matsushita T, Tsuhciya T, Mimaya J, Koizumi S, Miyake M, Nishikawa K, Takaue Y, Kawano Y, Iwai A, Ishida Y, Matsumoto K, Fujimoto T. Mutational analysis of the N-ras gene in acute lymphoblastic leukemia: a study of 125 Japanese pediatric cases. Int J Hematol. 1998;67(4):379–387. doi: 10.1016/S0925-5710(98)00015-2. [DOI] [PubMed] [Google Scholar]
- Young DC, Griffin JD. Autocrine secretion of GM-CSF in acute myeloblastic leukemia. Blood. 1986;68(5):1178–1181. [PubMed] [Google Scholar]
- Zahedi B, Goo HJ, Beaulieu N, Tazmini G, Kay RJ, Cornell RB. Phosphoinositide 3-kinase regulates plasma membrane targeting of the Ras-specific exchange factor RasGRP1. J Biol Chem. 2011;286(14):12712–12723. doi: 10.1074/jbc.M110.189605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenatti PP, Ribeiro D, Li W, Zuurbier L, Silva MC, Paganin M, Tritapoe J, Hixon JA, Silveira AB, Cardoso BA, Sarmento LM, Correia N, Toribio ML, Kobarg J, Horstmann M, Pieters R, Brandalise SR, Ferrando AA, Meijerink JP, Durum SK, Yunes JA, Barata JT. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat Genet. 2011;43(10):932–939. doi: 10.1038/ng.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, Lu C, Chen SC, Wei L, Collins-Underwood JR, Ma J, Roberts KG, Pounds SB, Ulyanov A, Becksfort J, Gupta P, Huether R, Kriwacki RW, Parker M, McGoldrick DJ, Zhao D, Alford D, Espy S, Bobba KC, Song G, Pei D, Cheng C, Roberts S, Barbato MI, Campana D, Coustan-Smith E, Shurtleff SA, Raimondi SC, Kleppe M, Cools J, Shimano KA, Hermiston ML, Doulatov S, Eppert K, Laurenti E, Notta F, Dick JE, Basso G, Hunger SP, Loh ML, Devidas M, Wood B, Winter S, Dunsmore KP, Fulton RS, Fulton LL, Hong X, Harris CC, Dooling DJ, Ochoa K, Johnson KJ, Obenauer JC, Evans WE, Pui CH, Naeve CW, Ley TJ, Mardis ER, Wilson RK, Downing JR, Mullighan CG. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Liu H, Coughlin J, Zheng J, Li L, Stone JC. Phosphorylation of RasGRP3 on threonine 133 provides a mechanistic link between PKC and Ras signaling systems in B cells. Blood. 2005;105(9):3648–3654. doi: 10.1182/blood-2004-10-3916. [DOI] [PubMed] [Google Scholar]
- Zhu M, Fuller DM, Zhang W. The role of Ras guanine nucleotide releasing protein 4 in Fc epsilonRI-mediated signaling, mast cell function, and T cell development. J Biol Chem. 2012;287(11):8135–8143. doi: 10.1074/jbc.M111.320580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikherman J, Parameswaran R, Weiss A. Endogenous antigen tunes the responsiveness of naive B cells but not T cells. Nature. 2012;489(7414):160–164. doi: 10.1038/nature11311. [DOI] [PMC free article] [PubMed] [Google Scholar]