Abstract
Precise control of organ size is crucial during animal development and regeneration. In Drosophila and mammals, studies over the past decade have uncovered a critical role for the Hippo tumour-suppressor pathway in the regulation of organ size. Dysregulation of this pathway leads to massive overgrowth of tissue. The Hippo signalling pathway is highly conserved and limits organ size by phosphorylating and inhibiting the transcription co-activators YAP and TAZ in mammals and Yki in Drosophila, key regulators of proliferation and apoptosis. The Hippo pathway also has a critical role in the self-renewal and expansion of stem cells and tissue-specific progenitor cells, and has important functions in tissue regeneration. Emerging evidence shows that the Hippo pathway is regulated by cell polarity, cell adhesion and cell junction proteins. In this review we summarize current understanding of the composition and regulation of the Hippo pathway, and discuss how cell polarity and cell adhesion proteins inform the role of this pathway in organ size control and regeneration.
Organ size regulation is a highly coordinated process involving complex mechanisms in response to physiological cues. On the organismal level, circulating factors such as hormones and insulin-like growth factors (IGF) play important roles in promoting organ size1. In contrast, physiological perturbations, such as prolonged starvation, cause profound reduction of organ size1. Additionally, an intrinsic mechanism limits organ size, which was first demonstrated in salamander limbs by classical transplantation experiments1. The underlying mechanism of organ-autonomous size determination remained largely unknown until the past decade. Extensive research led to the identification of the Hippo tumour-suppressor pathway as a key regulator of organ size in Drosophila and mammals2. It is also known that mutations of genes that are involved in patterning, cell polarity and cell adhesion cause marked alternations of organ size3. Thus, the recent finding that the Hippo pathway is regulated by cell polarity and cell adhesion proteins is a promising basis for the potential crosstalk of the Hippo pathway and cell polarity proteins in the regulation of organ size4. Several studies have also demonstrated important roles for the Hippo pathway in stem cell/progenitor cell expansion and tissue regeneration5–13. These findings will be discussed here.
The Hippo pathway in Drosophila
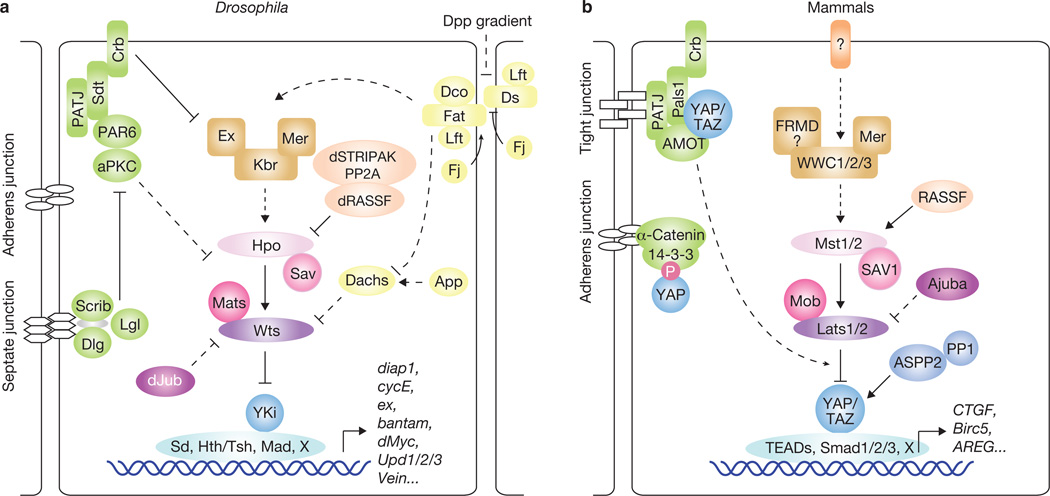
In Drosophila, the first core components of the Hippo pathway to be identified, using genetic mosaic screens, were the tumour-suppressor genes warts (wts)14,15, hippo (hpo)16–20 and salvador (sav)21,22. These genes belong to the hyperplastic group of Drosophila tumour-suppressors. Mutation of these genes results in robust tissue overgrowth without alteration of cell fate determination or cell polarity. Biochemical studies revealed that Hpo directly interacts with Sav to phosphorylate and activate the complex formed by Wts and another core Hippo pathway protein, Mats16,23 (Fig. 1a). The kinase activity of Hpo is antagonized by a PP2A phosphatase complex, dSTRIPAK24. The Hippo pathway is known to limit organ size partly by transcriptional regulation of cyclin E and diap1 (refs 16,17,20,21,23), suggesting the existence of a transcriptional regulator as a downstream effector of the pathway. By performing a yeast two-hybrid screen using Wts as bait, the transcription co-activator Yorkie (Yki) was identified as a potent effector of the Hippo pathway25. Subsequent biochemical studies showed that Wts directly phosphorylates and inhibits Yki26.
Figure 1.
The Hippo pathway in Drosophila and mammals. Corresponding proteins in Drosophila (a) and mammals (b) are indicated by matching colours. Arrowed or blunted ends indicate activation or inhibition, respectively. Dashed lines indicate unknown mechanisms.
Research in the past years has uncovered many proteins that act upstream in the Drosophila Hippo pathway. Two apical cytoskeleton-binding proteins, Merlin (Mer) and Expanded (Ex)27, and their interacting protein Kibra28–30, were found to activate the Hippo pathway. The Fat protocadherin, a cell-surface molecule, was also identified as an upstream regulator of the Hippo pathway31–35. Fat activity is regulated by binding to another protocadherin, Dachsous (Ds)36, and is modulated by several proteins, such as the casein kinase Discs overgrown (Dco)37,38, the Golgiresident kinase Four-jointed (Fj)39–41 and the Fat/Ds-interacting protein Lowfat (Lft)42. Fat/Hippo pathway activity may also be influenced by Decapentaplegic (Dpp) and Wingless (Wg) morphogen gradients40,41,43, which affect the expression of Fj and Ds. It has been proposed that Fat activates the Hippo pathway by regulating the protein level and localization of the protein Ex31–33,35. Another study suggests that Fat may control the abundance of Wts through Dachs34,44. Recently, dJub, a LIM-domain-containing protein that physically interacts with Wts and Sav, was shown to negatively regulate Hippo signalling, although the detailed mechanism has not been delineated45. A number of proteins that determine cell polarity were also found to regulate the Hippo pathway. These include the Scribble (Scrib)–Discs large (Dlg)–Lethal giant larvae (Lgl) complex, atypical protein kinase C (aPKC) and Crumbs (Crb)46–49, indicating a role of cell polarity in the regulation of Hippo signalling.
The Hippo pathway in mammals
The core components and downstream effectors of the Drosophila Hippo pathway are highly conserved in mammals: Mst1/2 (homologues of Hpo), Sav1 (Sav homologue), Lats1/2 (Wts homologues), MOBKL1A and MOBKL1B (collectively referred to as Mob1; homologues of Mats), and YAP and its paralogue TAZ (also called WWTR1; homologues of Yki) (Fig. 1b). Expression of human YAP, Lats1, Mst2 and Mob1 can rescue the phenotypes of their corresponding Drosophila mutants in vivo16,23,25,50. The core components Mst1/2 are pro-apoptotic kinases that are activated by caspase cleavage under apoptotic stress51. Sav1 interacts with Mst1/2 through the SARAH domains present in both Sav1 and Mst1/2 (ref. 52). Although Sav1 has been shown to activate Mst1/2, the underlying mechanism is unclear, but might involve regulation of Mst1 nuclear translocation53. Mst1/2 is also activated by binding to Ras association domain family (RASSF) proteins54, possibly owing to alteration of Mst1/2 subcellular localization55. In Drosophila, however, dRASSF inhibits Hpo possibly through competition with Sav for Hpo binding56 and through recruitment of the dSTRIPAK–PP2A complex24. Activation of Mst1/2 leads to phosphorylation and activation of their direct substrates, Lats1/2 (ref. 57). Mob1, which forms a complex with Lats1/2, is also phosphorylated by Mst1/2, resulting in an enhanced Lats1/2–Mob1 interaction58. Activated Lats1/2 in turn phosphorylate and inhibit YAP/TAZ transcription co-activators26,59–62.
Functions of the Hippo pathway in organ size determination and tumour suppression have been confirmed in genetically engineered mouse models. For instance, liver-specific overexpression of YAP results in enlarged livers that return to their normal size after cessation of YAP expression12,26. However, sustained YAP overexpression leads to tumour formation26. Genomic amplification of YAP is also observed in human cancers and a mouse model of breast cancer63,64. Furthermore, elevated YAP protein levels and nuclear localization have been observed in multiple human cancers59,63,65, and the alterations of YAP may have prognostic value for certain human cancers66. Overexpression of TAZ, the paralogue of YAP, has been noted in human breast cancer samples and non-small-cell lung-cancer cell lines67,68. Ablation of the Hippo pathway components Mer and Sav and double knockout of Mst1/2 in mice also result in liver enlargement and tumour formation69–74. Remarkably, loss of one or both copies of YAP can suppress liver expansion and tumorigenesis induced by Mer deficiency69. Aberrant Mst1/2 and Lats1/2 expression and Lats2, Sav1 and Mob1 mutation were also observed in human cancers or cancer cell lines2. Together, these studies highlight a significant role of the Hippo pathway in organ size regulation and tumorigenesis.
Mechanisms of YAP/TAZ/Yki inhibition
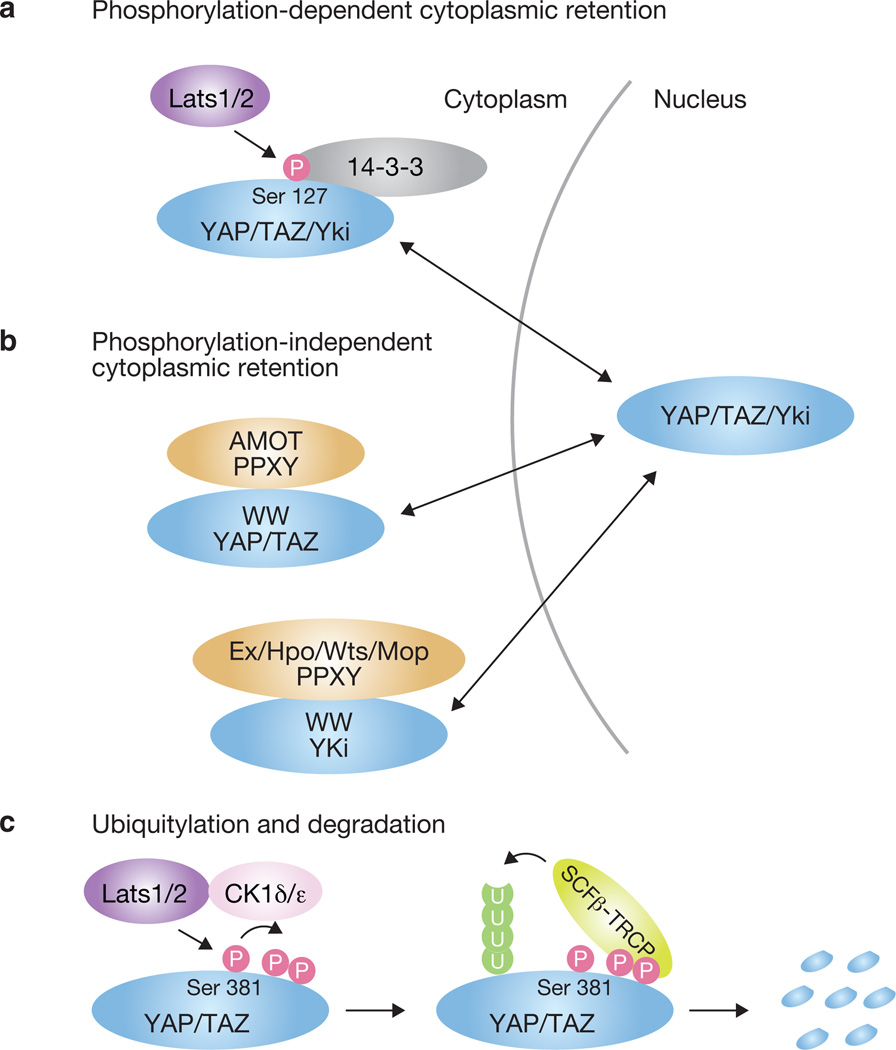
Activation of the Hippo pathway leads to phosphorylation and inhibition of YAP, TAZ and Yki transcription co-activators. In mammals, YAP and TAZ are phosphorylated by Lats1/2 in vitro and in vivo59,60,75. The mechanism of inhibition by Hippo signalling involves phosphorylation of Ser 127 in YAP or the corresponding sites in TAZ and Yki, which promotes 14-3-3 binding and subsequent cytoplasmic sequestration and inactivation26,59,60,62,76 (Fig. 2a). Indeed, mutation of Ser 127 and disruption of 14-3-3 binding activate YAP59, confirming the inhibitory nature of this phosphorylation. In Drosophila, Yki phosphorylation on two other sites by Wts similarly results in Yki inhibition, although the mechanism is yet to be determined77.
Figure 2.
Mechanisms of YAP/TAZ/Yki inhibition by the Hippo pathway. (a) Phosphorylation-dependent cytoplasmic retention. Phosphorylation of YAP on Ser 127 by Lats1/2 induces 14-3-3 binding and cytoplasmic retention of YAP. The mechanism is conserved in TAZ and Yki. (b) Phosphorylation-independent cytoplasmic retention. Through WW domain–PPXY motif interactions, Yki binds to Mop, Ex, Hpo and Wts, and YAP/TAZ binds to AMOT family proteins. These interactions physically sequester Yki and YAP/TAZ in the cytoplasm. (c) Phosphorylation-induced ubiquitylation and degradation. Phosphorylation of YAP on Ser 381 by Lats1/2 primes further phosphorylation of YAP by CK1δ/ε, which induces interaction with SCFβ–TRCP and finally leads to YAP ubiquitylation and degradation. The mechanism is conserved in TAZ.
Phosphorylation of YAP can also induce its degradation. Lats1/2 phosphorylates YAP at Ser 381, which primes YAP for subsequent phosphorylation by another kinase, possibly casein kinase 1 (CK1δ/ε), activating a phosphorylation-dependent degradation motif termed a phosphodegron. Subsequently, the E3 ubiquitin ligase SCFβ–TRCP is recruited to YAP, leading to its polyubiquitylation and degradation75 (Fig. 2c). Consistently, decreased YAP phosphorylation in sparsely cultured NIH-3T3 cells, as well as in Mst1/2-deficient mouse liver, correlates with increased YAP protein levels73,75. This mechanism is conserved in TAZ but not in Yki78, which lacks a residue equivalent to Ser 381.
YAP, TAZ and Yki can also be inhibited through protein–protein interactions that result in their cytoplasmic sequestration (Fig. 2b). Yki contains two WW domains that can interact with PPXY motifs present in Mop79 and the Hippo pathway components Ex, Wts and Hpo80,81. Recently, YAP/TAZ and angiomotin (AMOT) family proteins were shown to interact82–85, resulting in YAP/TAZ localization to tight junctions and inhibition through phosphorylation-dependent and -independent mechanisms82. In addition, YAP and TAZ interact with another tight junction protein ZO-2, which was reported to increase nuclear localization of YAP and tight-junction localization of TAZ86,87. It will be important to investigate the relationship between phosphorylation and these physical interactions in YAP regulation, and whether disruption of these interactions alters organ growth.
Transcriptional regulation of Hippo pathway target genes by YAP, TAZ and Yki
The TEAD family transcription factors were found to be critical partners of YAP and TAZ in the regulation of gene expression (the Drosophila TEAD homologue Scalloped (Sd) is partner of Yki)88–92. Knockdown of TEADs or disruption of the YAP–TEAD interaction abolishes YAP-dependent gene transcription and largely diminishes YAP-induced cell proliferation, oncogenic transformation and the epithelial-to-mesenchymal transition (EMT)88. In Drosophila, Sd was shown to genetically interact with Yki and to be required for Yki-induced target gene expression in vivo88,89,91,92. Intriguingly, a mutation of TEAD1 Tyr 406, which forms a hydrogen bond with YAP, results in loss of interaction with YAP and leads to the human genetic disease Sveinsson’s chorioretinal atrophy93–96. Precise regulation of YAP–TEAD interaction is therefore important in maintaining normal physiology.
Several direct target genes of YAP–/TAZ–TEAD and Yki–Sd have been identified, including CTGF and Cyr61 in mammalian cells88,97, and diap1 and dMyc in Drosophila89,91,98,99. CTGF was shown to have an important role in YAP-induced proliferation and anchorage-independent growth88. In Drosophila, diap1 is essential for Yki-induced overgrowth, but is not sufficient to explain all Yki phenotypes. Recently, Yki–Sd was shown to induce transcription of dMyc, a potent promoter of ribosome biogenesis and cell growth98,99. dMyc expression also mediates a cell phenomenon induced by imbalance of Hippo pathway activity, referred to as cell competition — wherein the contact between fast- and slow-growing cells in genetic mosaics favours the positive selection and clonal expansion of fast-dividing cells at the expense of slow-dividing cells98,99. YAP also induces Myc in transgenic mouse liver26, although the mechanism remains to be investigated.
Despite a major role for TEADs in YAP/TAZ function, other transcription factors containing PPXY motifs are known to interact with the WW domains of YAP/TAZ. These include Smad1, RUNX, ErbB4 and p73 for YAP100–104, and RUNX, PPARγ, Pax3, TBX5 and TTF-1 for TAZ105–109. The interaction of YAP with Smad1 is believed to be important for BMP-mediated maintenance of pluripotency of mouse embryonic stem cells104. YAP and TAZ also bind Smad2/3 through the coiled-coil region, and this interaction is believed to dictate the subcellular localization of Smad2/3 (refs 85,110). YAP also interacts with p73, a p53 family pro-apoptotic transcription factor, to induce expression of genes such as Bax, puma and PML111. However, there are contradictory reports on the role of the Hippo pathway in activating112 or inhibiting61 this activity. Recently, YAP was also shown to interact with β-catenin and induce expression of canonical Wnt target genes such as Sox2 and Snai2 in mouse heart tissue113.
bantam microRNA is a target gene of the Hippo pathway and promotes cell survival and proliferation114,115. Homothorax (Hth) and Teashirt (Tsh) are two transcription factors mediating bantam expression anterior to the morphogenetic furrow116. In addition, the expression of bantam is also directly induced by a transcriptional complex formed by Yki and Mad, an effector of the Drosophila Dpp signalling pathway117. The existence of a bantam counterpart and the functions of Hth and Tsh homologues in the Hippo pathway in mammals remain to be investigated.
YAP, TAZ and Yki also induce many other genes directly or indirectly. In Drosophila, Yki induces: cycE (ref. 21) and E2F1 (ref. 92), which may be involved in cell-autonomous regulation of cell proliferation; the EGFR (epidermal growth factor receptor) ligands Vein, Keren and Spitz11,118 and the Jak–Stat pathway ligands Unpaired1/2/3 (Upd1/2/3)8–11, which might mediate non-cell-autonomous functions of the Hippo pathway; and the Hippo pathway genes Ex, Kibra, Crb, and Fj27,29,34,119, which may constitute a signal feedback loop. In mammals, YAP and TAZ also induce the expression of AREG118 and FGF1 (ref. 60), which may also mediate non-cell-autonomous functions of the Hippo pathway. However, the mechanisms underlying the induction of these genes, including the responsible transcription factors, are mostly unclear.
Regulation of the Hippo pathway by cell polarity and cell adhesion complexes
In Drosophila, mutations of several genes that are involved in cell polarity and cell junction lead to massive overgrowth. The Dlg–Lgl–Scrib protein complex localizes to the basal–lateral membrane of epithelial cells, where it is required for the lateral exclusion of apical proteins, including the Par3–Par6–aPKC complex and the Crb–Stardust (Sdt)–PATJ complex. Interestingly, Lgl mutations lead to nuclear translocation of Yki and upregulation of Hippo pathway target genes in Drosophila epithelium47. Expression of dominant-negative aPKC rescued the tissue overgrowth in Lgl-mutant tissues47. In zebrafish, Scrib was shown to interact genetically with and suppress the activity of the YAP homologue during embryonic kidney development120. The tumour-suppressor function of the Dlg–Lgl–Scrib complex is possibly conserved in mammals, as depletion of Scrib in mammary epithelium results in disruption of apoptosis inhibition by cell polarity, and induction of dysplasia in vivo that progresses to tumours after long latency121. It would be interesting to determine whether the mammalian Hippo pathway mediates the tumour-suppressor function of the Dlg–Lgl–Scrib complex.
Crb is another cell polarity protein that regulates the Drosophila Hippo pathway46,48,49. The intracellular domain of Crb contains a juxtamembrane FERM-binding motif (FBM) and a carboxy-terminal PDZ-binding motif (PBM). The PBM is important for polarity formation122, whereas the FBM regulates Hippo-pathway-dependent proliferation and apoptosis by promoting apical localization of the upstream component Ex46,48,49. Thus, Crb regulates cell polarity and tissue growth through distinct mechanisms. In addition, it seems that the functions of the Dlg–Lgl–Scrib complex in cell polarity and tissue growth are also separable47,123. It is therefore important to determine whether, and how, the two functions of these proteins are coupled to regulate tissue homeostasis.
In mammalian cells, Hippo pathway activation is triggered in part by cell–cell contact. In tissue culture, high cell density induces YAP phosphorylation and cytoplasmic translocation59. And in mouse blastocysts, YAP is nuclear in outer layer cells, and cytoplasmic in the inner blastocyst layer cells124. Consistently, it has been observed that disruption of cell–cell junctions in epithelium results in the nuclear localization of YAP and TAZ85. Collectively, these studies suggest that maintenance of cell–cell junctions is important for mammalian Hippo pathway function.
Recent studies shed some light into the mechanisms of YAP/TAZ regulation by cell–cell contact. First, a tight-junction protein complex, composed of the AMOT family proteins, PALS1, PATJ/MPDZ and Lin7, was found to interact with YAP and TAZ82–85. This interaction inhibits YAP and TAZ by promoting their localization to tight junctions and their phosphorylation by the Hippo pathway. In addition, α-catenin was shown to interact with YAP125,126, possibly through a 14-3-3 protein, in a phosphorylation-dependent manner126. This interaction may prevent YAP dephosphorylation by PP2A and results in YAP inhibition126. Thus, it is possible that the tight junction and adherens junction are critically important for relaying cell contact signals to the Hippo pathway. Such a hypothesis needs to be further investigated.
The Hippo pathway in tissue regeneration, and stem cell self-renewal and expansion
The Hippo pathway was initially thought to limit organ size by inhibiting proliferation and promoting apoptosis16–20. However, emerging evidence suggests that the Hippo pathway may also regulate stem cell and progenitor cell self-renewal and expansion. For instance, YAP and TAZ regulate embryonic stem cell self-renewal in response to TGFβ/BMP (transforming growth factor beta/bone morphogenetic protein) signalling104,110. In addition, YAP is inactivated during mouse embryonic stem cell differentiation and activated in induced pluripotent stem (iPS) cells5. YAP knockdown in mouse embryonic stem cells leads to loss of pluripotency, whereas ectopic expression of YAP prevents embryonic stem cell differentiation5.
Additionally, the Hippo pathway also regulates tissue-specific progenitor cells. YAP expression is generally restricted to the progenitor cells in normal mouse intestines, and transgenic expression of YAP in mouse intestines causes a marked expansion of the progenitor cell compartment12. Activation of YAP–TEAD also results in the expansion of neural progenitor cells in a chicken neural tube model13. Similarly, YAP expression expands basal epidermal progenitors in mouse skin and inhibits their terminal differentiation127. In contrast, conditional knockout of YAP or knock-in of a TEAD-binding-deficient YAP mutant in mouse skin leads to decreased proliferation of basal cells, thinner epidermis and failure of skin expansion126. Consistently, adult liver stem cells known as oval cells accumulate in Mst1/2-, Sav1- and Mer-knockout mice liver70–73. It should be noted that these genetic manipulations are applied at the whole organ level and not specifically to the progenitor cell compartment. However, the contribution of progenitor cell expansion in YAP-induced organ overgrowth is likely to be tissue-dependent. For instance, overgrown hearts induced by Sav1 knockout showed excessive proliferation in cardiomyocytes but normal proliferation level of cardiac progenitors113. In addition, in certain cancers, such as a subtype of medulloblastomas, YAP expression is highly elevated in the perivascular cancer stem cell compartment128.
The Hippo pathway was recently shown to be involved in tissue regeneration. In the Drosophila midgut, Yki expression is largely restricted to intestinal stem cells (ISC)10. Under resting conditions, Yki is mostly localized to the cytoplasm and seems to be inactive10. In contrast, Yki displays increased nuclear localization and reporter activity, and has an important and cell-autonomous role in ISC proliferation in response to injury9,10. Interestingly, the Hippo pathway also has a non-cell-autonomous function during regeneration8,9,11. In response to damage, the Hippo pathway is inactivated in enterocytes, a differentiated cell type in the Drosophila midgut, resulting in Yki activation and subsequent expression of Upd1/2/3 (refs 8,9,11), as well as EGFR ligands11. This results in increased ISC proliferation in a non-cell-autonomous manner. Yki activation in enterocytes and in wing discs (where Yki also plays a role in regeneration6) seems to involve JNK signalling8,129.
In mammals, there is also evidence for a role of YAP in tissue regeneration. Intestinal damage markedly induces YAP expression, and loss of YAP severely impairs dextran sodium sulfate-induced intestinal regeneration7. In the mouse liver, Yap knockout causes a defect in bile duct development69. Interestingly, most adult mouse biliary ductal epithelial cells express Sox9 and these cells make a significant contribution to liver regeneration after injury as shown by lineage tracing130. It remains to be determined whether ablation of YAP also results in compromised liver regeneration, and more importantly, whether the Hippo pathway activity is regulated during regeneration in mammals.
Conclusions and perspectives
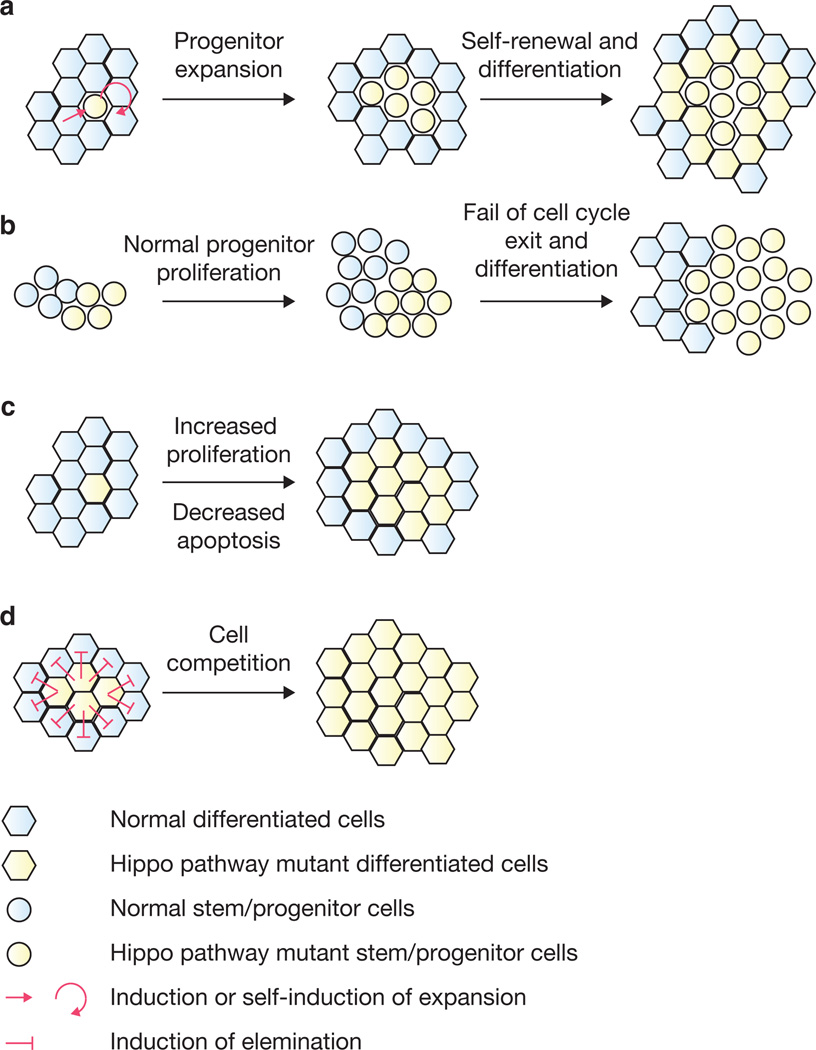
Extensive studies in the past decade have elucidated the importance of the Hippo pathway in organ size control and regeneration in both Drosophila and mammals. Several mechanisms have been proposed, and it is clear that cell adhesion and polarity complexes play a key role in Hippo pathway regulation. YAP and Yki may promote organ size and regeneration by inducing stem cell and progenitor cell proliferation through both cell-autonomous and non-cell-autonomous mechanisms (Fig. 3a). In addition, inactivation of the Hippo pathway may block cell-cycle exit, leading to hyperplasia and differentiation defects53 (Fig. 3b). The Hippo pathway can also inhibit proliferation and promote apoptosis in non-stem cells/non-progenitor cell types (Fig. 3c). Lastly, an imbalance of Hippo pathway activity in neighbouring cells may induce cell competition through differential expression of dMyc in Drosophila98,99 (Fig. 3d). How these mechanisms fit into organ size regulation and regeneration in vivo is yet to be determined.
Figure 3.
Mechanisms of the Hippo pathway in regulation of organ size and regeneration. Hexagons denote differentiated cells and circles denote stem/progenitor cells. Blue colour indicates wild-type and yellow colour indicates Hippo-pathway mutant cells. (a) Hippo pathway inactivation leads to stem/progenitor cell expansion in both cell-autonomous and non-autonomous manners. (b) Hippo pathway inactivation leads to cell cycle exit defects in some cellular contexts. (c) Hippo pathway mutations promote proliferation and decrease apoptosis in non-stem/progenitor cells. (d) Imbalance of Hippo pathway activity in neighbouring cells induces cell competition.
Despite these insights into the critical role of this pathway in stem cell expansion and tissue regeneration, many important questions await answers. These include the role and mechanism of cell polarity and cell adhesion proteins in sensing organ size to regulate the Hippo pathway and the position of the Hippo pathway in the known signalling networks regulating cell proliferation, apoptosis and stem cell function. In addition, the mechanism by which upstream regulators of the Hippo pathway are integrated to initiate or terminate signalling is not yet fully understood. Importantly, Hippo pathway dysregulation in cancer remains to be fully elucidated. The Hippo–YAP pathway holds great promise as a target in cancer therapy and regenerative medicine. Insights into the upstream regulators and downstream targets of this pathway, and their mechanism of regulation, are crucial in translating our basic knowledge of this pathway into therapeutic designs.
ACKNOWLEDGEMENTS
We apologize for those primary works that we could not cite due to space constraints. Research in the lab of K.L.G. is supported by grants from NIH and CIRM.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
Contributor Information
Bin Zhao, Life Sciences Institute, Zhejiang University, Hangzhou, Zhejiang 310058, China; Department of Pharmacology and Moores Cancer Center, University of California at San Diego, La Jolla, California 92093-0815, USA.
Karen Tumaneng, Department of Pharmacology and Moores Cancer Center, University of California at San Diego, La Jolla, California 92093-0815, USA.
Kun-Liang Guan, Email: kuguan@ucsd.edu, Department of Pharmacology and Moores Cancer Center, University of California at San Diego, La Jolla, California 92093-0815, USA.
References
- 1.Stanger BZ. Organ size determination and the limits of regulation. Cell Cycle. 2008;7:318–324. doi: 10.4161/cc.7.3.5348. [DOI] [PubMed] [Google Scholar]
- 2.Pan D. The Hippo signaling pathway in development and cancer. Dev. Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leevers SJ, McNeill H. Controlling the size of organs and organisms. Curr. Opin. Cell Biol. 2005;17:604–609. doi: 10.1016/j.ceb.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lian I, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grusche FA, Degoutin JL, Richardson HE, Harvey KF. The Salvador/Warts/Hippo pathway controls regenerative tissue growth in Drosophila melanogaster. Dev. Biol. 2010;350:255–266. doi: 10.1016/j.ydbio.2010.11.020. [DOI] [PubMed] [Google Scholar]
- 7.Cai J, et al. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010;24:2383–2388. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staley BK, Irvine KD. Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr. Biol. 2010;20:1580–1587. doi: 10.1016/j.cub.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaw RL, et al. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development. 2010;137:4147–4158. doi: 10.1242/dev.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karpowicz P, Perez J, Perrimon N. The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development. 2010;137:4135–4145. doi: 10.1242/dev.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren F, et al. Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc. Natl Acad. Sci. USA. 2010;107:21064–21069. doi: 10.1073/pnas.1012759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camargo FD, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 13.Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 15.Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 16.Wu S, Huang J, Dong J, Pan D. Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 17.Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 18.Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- 19.Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, Hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 20.Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tapon N, et al. salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 22.Kango-Singh M, et al. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- 23.Lai ZC, et al. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 24.Ribeiro PS, et al. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell. 2010;39:521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamaratoglu F, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 28.Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev. Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 29.Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J, et al. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and expanded. Dev. Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyler DM, Baker NE. Expanded and Fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev. Biol. 2007;305:187–201. doi: 10.1016/j.ydbio.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willecke M, et al. The Fat cadherin acts through the Hippo tumor-suppressor pathway to regulate tissue size. Curr. Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene Fat controls tissue growth upstream of expanded in the Hippo signaling pathway. Curr. Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Cho E, et al. Delineation of a Fat tumor suppressor pathway. Nat. Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 35.Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr. Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 36.Matakatsu H, Blair SS. Separating the adhesive and signaling functions of the Fat and Dachsous protocadherins. Development. 2006;133:2315–2324. doi: 10.1242/dev.02401. [DOI] [PubMed] [Google Scholar]
- 37.Sopko R, et al. Phosphorylation of the tumor suppressor Fat is regulated by its ligand Dachsous and the kinase discs overgrown. Curr. Biol. 2009;19:1112–1117. doi: 10.1016/j.cub.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng Y, Irvine KD. Processing and phosphorylation of the Fat receptor. Proc. Natl Acad. Sci. USA. 2009;106:11989–11994. doi: 10.1073/pnas.0811540106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon MA, Xu A, Ishikawa HO, Irvine KD. Modulation of Fat:Dachsous binding by the cadherin domain kinase Four-jointed. Curr Biol. 2010;20:811–817. doi: 10.1016/j.cub.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Dev. Cell. 2008;15:309–321. doi: 10.1016/j.devcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willecke M, Hamaratoglu F, Sansores-Garcia L, Tao C, Halder G. Boundaries of Dachsous cadherin activity modulate the Hippo signaling pathway to induce cell proliferation. Proc. Natl Acad. Sci. USA. 2008;105:14897–14902. doi: 10.1073/pnas.0805201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao Y, Kucuk B, Irvine KD. Drosophila lowfat, a novel modulator of Fat signaling. Development. 2009;136:3223–3233. doi: 10.1242/dev.036152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zecca M, Struhl G. A feed-forward circuit linking wingless, Fat–Dachsous signaling, and the Warts-Hippo pathway to Drosophila wing growth. PLoS Biol. 2010;8:e1000386. doi: 10.1371/journal.pbio.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng Y, Irvine KD. Fat and expanded act in parallel to regulate growth through warts. Proc. Natl Acad. Sci. USA. 2007;104:20362–20367. doi: 10.1073/pnas.0706722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das Thakur M, et al. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr. Biol. 2010;20:657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen CL, et al. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc. Natl Acad. Sci. USA. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr. Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 48.Ling C, et al. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc. Natl Acad. Sci. USA. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein expanded. Curr. Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tao W, et al. Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat. Genet. 1999;21:177–181. doi: 10.1038/5960. [DOI] [PubMed] [Google Scholar]
- 51.Graves JD, et al. Caspase-mediated activation and induction of apoptosis by the mammalian Ste20-like kinase Mst1. EMBO J. 1998;17:2224–2234. doi: 10.1093/emboj/17.8.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Callus BA, Verhagen AM, Vaux DL. Association of mammalian sterile twenty kinases, Mst1 and Mst2, with hSalvador via C-terminal coiled-coil domains, leads to its stabilization and phosphorylation. FEBS J. 2006;273:4264–4276. doi: 10.1111/j.1742-4658.2006.05427.x. [DOI] [PubMed] [Google Scholar]
- 53.Lee JH, et al. A crucial role of WW45 in developing epithelial tissues in the mouse. EMBO J. 2008;27:1231–1242. doi: 10.1038/emboj.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oh HJ, et al. Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res. 2006;66:2562–2569. doi: 10.1158/0008-5472.CAN-05-2951. [DOI] [PubMed] [Google Scholar]
- 55.Khokhlatchev A, et al. Identification of a novel Ras-regulated proapoptotic pathway. Curr. Biol. 2002;12:253–265. doi: 10.1016/s0960-9822(02)00683-8. [DOI] [PubMed] [Google Scholar]
- 56.Polesello C, Huelsmann S, Brown NH, Tapon N. The Drosophila RASSF homolog antagonizes the Hippo pathway. Curr. Biol. 2006;16:2459–2465. doi: 10.1016/j.cub.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan EH, et al. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- 58.Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 2008;18:311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 61.Oka T, Mazack V, Sudol M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP) J. Biol. Chem. 2008;283:27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- 62.Lei QY, et al. TAZ promotes cell proliferation and epithelial–mesenchymal transition and is inhibited by the Hippo pathway. Mol. Cell Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zender L, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Overholtzer M, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl Acad. Sci. USA. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steinhardt AA, et al. Expression of Yes-associated protein in common solid tumors. Hum. Pathol. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu MZ, et al. Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer. 2009;115:4576–4585. doi: 10.1002/cncr.24495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chan SW, et al. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–2598. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- 68.Zhou Z, et al. TAZ is a novel oncogene in non-small cell lung cancer. Oncogene. 2011;30:2181–2186. doi: 10.1038/onc.2010.606. [DOI] [PubMed] [Google Scholar]
- 69.Zhang N, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benhamouche S, et al. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 2010;24:1718–1730. doi: 10.1101/gad.1938710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee KP, et al. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size and liver tumorigenesis. Proc. Natl Acad. Sci. USA. 2010;107:8248–8253. doi: 10.1073/pnas.0912203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu L, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl Acad. Sci. USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song H, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc. Natl Acad. Sci. USA. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou D, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCFβ-TRCP. Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ren F, Zhang L, Jiang J. Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev. Biol. 2009;337:303–312. doi: 10.1016/j.ydbio.2009.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu CY, et al. The Hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCFβ-TRCP E3 ligase. J. Biol. Chem. 2010;285:37159–37169. doi: 10.1074/jbc.M110.152942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gilbert MM, Tipping M, Veraksa A, Moberg KH. A screen for conditional growth suppressor genes identifies the Drosophila homolog of HD-PTP as a regulator of the oncoprotein Yorkie. Dev. Cell. 2011;20:700–712. doi: 10.1016/j.devcel.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Badouel C, et al. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev. Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 81.Oh H, Reddy BV, Irvine KD. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev. Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao B, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2001;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang W, Huang J, Chen J. Angiomotin-like proteins associate with and negatively regulate YAP1. J. Biol. Chem. 2010;286:4364–4370. doi: 10.1074/jbc.C110.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chan SW, et al. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J. Biol. Chem. 2011;286:7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Varelas X, et al. The crumbs complex couples cell density sensing to Hippo-dependent control of the TGF–β–SMAD pathway. Dev. Cell. 2010;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 86.Oka T, et al. Functional complexes between YAP2 and ZO-2 are PDZ domain-dependent, and regulate YAP2 nuclear localization and signalling. Biochem. J. 2010;432:461–472. doi: 10.1042/BJ20100870. [DOI] [PubMed] [Google Scholar]
- 87.Remue E, et al. TAZ interacts with zonula occludens-1 and -2 proteins in a PDZ-1 dependent manner. FEBS Lett. 2010;584:4175–4180. doi: 10.1016/j.febslet.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 88.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang L, et al. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang H, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial–mesenchymal transition. J. Biol. Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 92.Goulev Y, et al. SCALLOPED interacts with YORKIE, the nuclear effector of the Hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 93.Li Z, et al. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24:235–240. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kitagawa M. A Sveinsson’s chorioretinal atrophy-associated missense mutation in mouse Tead1 affects its interaction with the co-factors YAP and TAZ. Biochem. Biophys. Res. Commun. 2007;361:1022–1026. doi: 10.1016/j.bbrc.2007.07.129. [DOI] [PubMed] [Google Scholar]
- 95.Chen L, et al. Structural basis of YAP recognition by TEAD4 in the Hippo pathway. Genes Dev. 2010;24:290–300. doi: 10.1101/gad.1865310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fossdal R, et al. A novel TEAD1 mutation is the causative allele in Sveinsson’s chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration) Hum. Mol. Genet. 2004;13:975–981. doi: 10.1093/hmg/ddh106. [DOI] [PubMed] [Google Scholar]
- 97.Lai D, Ho KC, Hao Y, Yang X. Taxol resistance in breast cancer cells is mediated by the Hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 2011;71:2728–2738. doi: 10.1158/0008-5472.CAN-10-2711. [DOI] [PubMed] [Google Scholar]
- 98.Neto-Silva RM, de Beco S, Johnston LA. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev. Cell. 2010;19:507–520. doi: 10.1016/j.devcel.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ziosi M, et al. dMyc functions downstream of Yorkie to promote the supercompetitive behavior of Hippo pathway mutant cells. PLoS Genet. 2010;6:e1001140. doi: 10.1371/journal.pgen.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Strano S, et al. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J. Biol. Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- 102.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 103.Omerovic J, et al. Ligand-regulated association of ErbB-4 to the transcriptional co-activator YAP65 controls transcription at the nuclear level. Exp. Cell Res. 2004;294:469–479. doi: 10.1016/j.yexcr.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 104.Alarcon C, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hong JH, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 106.Murakami M, Nakagawa M, Olson EN, Nakagawa O. A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc. Natl Acad. Sci. USA. 2005;102:18034–18039. doi: 10.1073/pnas.0509109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cui CB, Cooper LF, Yang X, Karsenty G, Aukhil I. Transcriptional coactivation of bone-specific transcription factor Cbfa1 by TAZ. Mol. Cell Biol. 2003;23:1004–1013. doi: 10.1128/MCB.23.3.1004-1013.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Park KS, et al. TAZ interacts with TTF-1 and regulates expression of surfactant protein-C. J. Biol. Chem. 2004;279:17384–17390. doi: 10.1074/jbc.M312569200. [DOI] [PubMed] [Google Scholar]
- 109.Murakami M, et al. Transcriptional activity of Pax3 is co-activated by TAZ. Biochem. Biophys. Res. Commun. 2006;339:533–539. doi: 10.1016/j.bbrc.2005.10.214. [DOI] [PubMed] [Google Scholar]
- 110.Varelas X, et al. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- 111.Lapi E, et al. PML, YAP, and p73 are components of a proapoptotic autoregulatory feedback loop. Mol. Cell. 2008;32:803–814. doi: 10.1016/j.molcel.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 112.Matallanas D, et al. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol. Cell. 2007;27:962–975. doi: 10.1016/j.molcel.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Heallen T, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 115.Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the Hippo tumor-suppressor pathway. Curr. Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- 116.Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–2319. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Oh H, Irvine KD. Cooperative regulation of growth by Yorkie and Mad through bantam. Dev. Cell. 2011;20:109–122. doi: 10.1016/j.devcel.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang J, et al. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat. Cell Biol. 2009;11:1444–1450. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Genevet A, et al. The Hippo pathway regulates apical-domain size independently of its growth-control function. J. Cell Sci. 2009;122:2360–2370. doi: 10.1242/jcs.041806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Skouloudaki K, et al. Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc. Natl Acad. Sci. USA. 2009;106:8579–8584. doi: 10.1073/pnas.0811691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhan L, et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hong Y, Stronach B, Perrimon N, Jan LY, Jan YN. Drosophila Stardust interacts with Crumbs to control polarity of epithelia but not neuroblasts. Nature. 2001;414:634–638. doi: 10.1038/414634a. [DOI] [PubMed] [Google Scholar]
- 123.Grzeschik NA, Amin N, Secombe J, Brumby AM, Richardson HE. Abnormalities in cell proliferation and apico-basal cell polarity are separable in Drosophila lgl mutant clones in the developing eye. Dev. Biol. 2007;311:106–123. doi: 10.1016/j.ydbio.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nishioka N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 125.Silvis MR, et al. α-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci. Signal. 2011;4:ra33. doi: 10.1126/scisignal.2001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schlegelmilch K, et al. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang H, Pasolli HA, Fuchs E. Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc. Natl Acad. Sci. USA. 2011;108:2270–2275. doi: 10.1073/pnas.1019603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fernandez LA, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–2741. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sun G, Irvine KD. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev. Biol. 2011;350:139–151. doi: 10.1016/j.ydbio.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Furuyama K, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat. Genet. 2010;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]