Abstract
Linear chromosomes terminate in specialized nucleoprotein structures called telomeres, which are required for genomic stability and cellular proliferation. Telomeres end in an unusual 3′ single-strand overhang that requires a special capping mechanism to prevent inappropriate recognition by the DNA damage machinery. In Schizosaccharomyces pombe, this protective function is mediated by the Pot1 protein, which binds specifically and with high affinity to telomeric ssDNA. We have characterized the thermodynamics and accommodation of both cognate and noncognate telomeric single-stranded DNA (ssDNA) sequences by Pot1pN, an autonomous ssDNA-binding domain (residues 1–187) found in full-length S. pombe Pot1. Direct calorimetric measurements of cognate telomeric ssDNA binding to Pot1pN show favorable enthalpy, unfavorable entropy, and a negative heat-capacity change. Thermodynamic analysis of the binding of noncognate telomeric ssDNA to Pot1pN resulted in unexpected changes in free energy, enthalpy, and entropy. Chemical-shift perturbation and structural analysis of these bound noncognate sequences show that these thermodynamic changes result from the structural rearrangement of both Pot1pN and the bound oligonucleotide. These data suggest that the ssDNA-binding interface is highly dynamic and, in addition to the conformation observed in the crystal structure of the Pot1pN/d(GGTTAC) complex, capable of adopting alternative thermodynamically equivalent conformations.
Protein recognition of nucleic acids is a key underlying molecular recognition event that guides replication, transcription, recombination, and other essential biological processes. To date, a number of structural, biochemical, and thermodynamic studies have provided a fundamental understanding of how protein/double-stranded DNA (dsDNA)1 recognition occurs. These studies reveal that dsDNA recognition is mediated by direct and indirect protein contacts that form intricate networks of hydrogen bonds with the bases (1–3). Thermodynamically, dsDNA recognition is generally characterized by negative enthalpy, positive entropy (ascribed to water/ion release), and large negative heat-capacity changes (ascribed to surface exclusion of the solvent) (4). Furthermore, the specific recognition of dsDNA typically leads to larger negative heat-capacity changes when compared to its nonspecific counterpart (1, 2). Although these characteristics are common, they can be modulated by many physical processes, including (1) conformational changes of the molecules upon binding (5), (2) redistribution of ion atmospheres, (3) dynamic behavior, and (4) hydration patterns of the molecules (reviewed in ref 6). For example, the binding of a protein to the major groove of dsDNA tends to be an enthalpically driven process, whereas minor groove binding is typically entropically driven because of the different hydration properties of each groove (7).
The characteristics that govern dsDNA binding cannot be directly extrapolated to ssDNA binding because of the differences in the mode of recognition and the extended nature of free ssDNA. Unlike dsDNA, the recognition of ssDNA is dominated by extensive stacking interactions between DNA bases and aromatic amino acids, which are augmented by hydrogen-bonding interactions in the case of specific recognition of ssDNA (8–18). Also, in comparison to dsDNA, ssDNA is inherently more flexible, which provides for a greater number of possible binding configurations. Thermodynamically, the recognition of ssDNA is typically an enthalpically driven process with a negative heat-capacity signature (10, 13, 16, 18, 19). However, unlike dsDNA recognition, our understanding of ssDNA recognition and accommodation of noncognate sequences is less well-developed in terms of correlating structural features with thermodynamics.
A more comprehensive understanding of ssDNA recognition can be achieved from parallel structural and thermodynamic studies within a well-defined system. The telomere end-protection (TEP) family of proteins provides an ideal system to probe the structural and thermodynamic underpinnings of specific ssDNA recognition. Telomeres terminate in a unique 3′ single-stranded overhang that must be distinguished from unnatural ends. TEP proteins provide this essential protective function by binding specifically to this ssDNA overhang, thus preventing unwarranted DNA damage responses at telomere ends (20, 21). To better understand this mode of recognition, structures have been solved of the ssDNA-binding domains (DBDs) of TEBPα/β (22–25) in ciliates, Cdc13 in the Saccharomyces cerevisiae (26), and the Pot1 proteins from Schizosaccharomyces pombe (27) and Homo sapiens (28). These studies reveal that TEP family members share a common protein fold that facilitates ssDNA binding and recognition, the OB (oligonucleotide—oligosaccharide binding) protein superfold (21, 29). Although structurally and functionally homologous, each TEP family member has a unique recognition interface for specifically recognizing and binding ssDNA (reviewed in ref 21), indicating that a common shared mechanism for specific ssDNA recognition has not emerged.
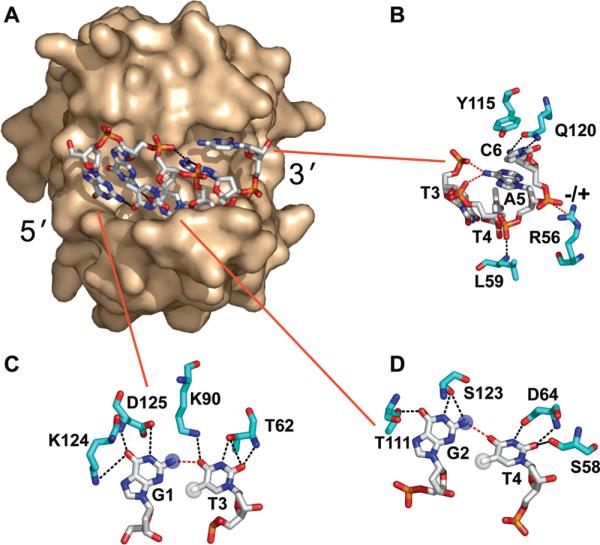
One of the most well-characterized TEP DBDs is the N-terminal 187-residue Pot1pN domain from the Pot1 protein found in S. pombe, which binds tightly (Kd of 83 nM) to a minimal telomeric ssDNA sequence, d(GGTTAC) (30). The high-resolution crystal structure of the Pot1pN/d(GGTTAC) complex reveals that the ssDNA adopts a remarkable compact conformation that is characterized by the self-recognition of nonsequential nucleotides through single hydrogen bonds (27). This compact DNA conformation is in pronounced contrast to the extended DNA conformations found in other TEP family members (21). In the complex, the ssDNA forms two non-Watson–Crick G-T base pairs and extensive intra- and intermolecular base-stacking between bases with hydrophobic extensions into the protein: G1 stacks with G2, which connects with L122, T3 stacks with T4, which stacks onto F88, and A5 stacks with C6, which stacks with Y115 (Figure 1). In addition to these extensive hydrophobic stacking interactions, there is an extensive array of hydrogen bonds formed between the oligonucleotide and the protein.
Figure 1.
(A) Surface representation of the high-resolution crystal structure of the Pot1pN/d(GGTTAC) complex (27) highlighting the compact conformation of bound oligonucleotide, d(GGTTAC). Pot1pN is shown in a surface representation and colored beige, and d(GGTTAC) is shown in a stick representation and colored standard atom designation colors. (B–D) DNA–protein (black dashed lines), DNA–DNA (red dashed lines), and ionic (−/+, red dashes) interactions are highlighted. Missing chemical moieties present in the substituted oligonucleotides are represented as white (CH3) or blue (NH2) opaque spheres (blue, G → I; white, T → U). All figures were made using PyMol version 1.0 (55).
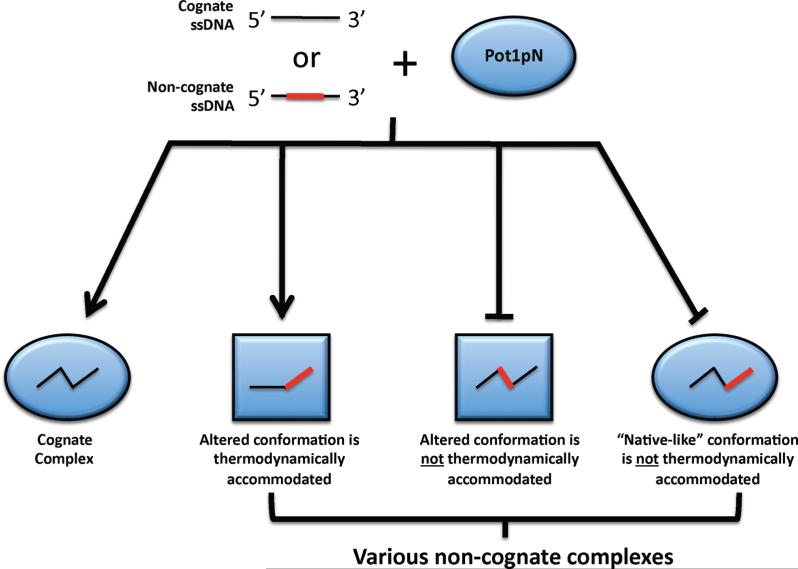
To further our understanding how telomeric ssDNA is recognized by TEP proteins, we have performed a parallel thermodynamic and structural study on the binding of both cognate and noncognate telomeric ssDNA oligonucleotides to Pot1pN. We have determined the thermodynamic signatures for the binding of the cognate telomere sequence, d(GGTTAC), to Pot1pN using isothermal titration calorimetry (ITC). In addition to these studies with cognate ligand, we have also investigated the mechanism of specific recognition exhibited by Pot1pN by measuring the thermodynamic impact of targeted modifications of this cognate sequence. Because of unexpected changes in binding free energy, enthalpy, and entropy for these noncognate oligonucleotides, we probed for structural changes in both the protein and bound oligonucleotide using the nuclear magnetic resonance (NMR) techniques of minimal chemical-shift perturbation (MCSP) mapping and analyzing DNA–DNA contacts via nuclear Overhauser effect (NOE) measurements. These structural studies indicate that the protein and/or bound oligonucleotide undergo structural rearrangements to accommodate each specific mutation and, depending upon the context of the modification, allow for the re-establishment of key thermodynamic contacts. The observed thermodynamic and structural changes indicate that the conformation of protein and oligonucleotide observed in the high-resolution crystal structure of the Pot1pN/d(GGTTAC) complex are not fixed. These studies expand and generalize the nucleotide-shuffling model for noncognate oligonucleotide recognition proposed by Theobald et al. (31) by suggesting that structural changes are not limited to the oligonucleotide but rather significant changes in protein structure also allow for recovery of lost thermodynamic contacts. Finally, these studies suggest that the specific recognition of ssDNA is facilitated by structural accommodation and is not limited to the specific hydrogen-bonding patterns observed in the high-resolution crystal structure of the Pot1pN/d(GGTTAC) complex.
MATERIALS AND METHODS
Chemicals, Reagents, and Proteins
All chemicals and reagents were obtained from Fisher Scientific (Pittsburgh, PA) unless otherwise indicated. ATP (γ-32P) was obtained from Perkin-Elmer Life and Analytical Sciences (Wellesley, MA). Oligonucleotides, d(GGTTAC), d(IGTTAC), d(GITTAC), d(GGUTAC), and d(GGTUAC), were commercially synthesized (Operon, Huntsville, AL) and reversed-phase purified over a semipreparative C18 column (Grace-Vydac, Hesperia, CA) in 1% triethanolamine acetate/H2O and eluted with a 5–40% gradient of 1% triethanolamine acetate/19% H2O/80% acetonitrile. Purified oligonucleotides were dialyzed extensively against 1 M NaCl to remove excess triethanolamine, followed by dialysis against deionized water. Desalted oligonucleotides were stored at 4 °C. All protein chromatography media were purchased from GE Healthcare (Piscataway, NJ). All buffers were filtered through a 0.2 μm membrane (Fisher Scientific) and degassed prior of use. The plasmid encoding Pot1pN was graciously provided by Professor Thomas Cech (30).
Expression and Purification of Pot1pN
The 22 kDa, N-terminal (His)6-Pot1pN was expressed, purified, and stored according to the protocols established by Lei et al. (30) and Croy et al. (32) with a final yield of 60 mg/L. Pot1pN used for NMR backbone assignment studies was expressed in Escherichia coli and purified in the same manner as the nonlabeled Pot1pN protein, except as follows. Expression of uniformly 13C,15N-labeled Pot1pN was accomplished by growing in minimal M9 media [25 mM Na2HPO4, 22 mM KH2PO4, 25 mM NaCl, 1× MEM vitamin solution (Invitrogen, Carlsbad CA), 1 μM FeCl3, 46 nM H3BO4, 102 nM CaCl2, 0.19 nM CoCl2, 0.78 nM CuSO4, 1.02 μM MgCl2, 1 nM MnCl3, 3 pM Na2MoO4, and 1.7 nM ZnCl2], supplemented with 11 mM (15NH4)2SO4 and 10.7 mM 13C-glucose as the sole nitrogen and carbon sources, respectively (33, 34).
Concentration Determination of Oligonucleotides and Protein
Protein and oligonucleotide concentrations were calculated from A280 and A260 values, respectively. The molar extinction coefficient (ε280) for Pot1pN was calculated to be 27 900 M–1 cm–1 by the ±GndHCl method by Pace et al. (35). The molar extinction coefficients (ε260) for d(GGTTAC), G1I, G2I, T3dU, and T4dU, were the estimates of 54 995, 53 204, 53 204, 56 951, and 56 951 M–1 cm–1, respectively, of the manufacturer.
Differential Scanning Calorimetry (DSC) of the Pot1pN/d(GGTTAC) Complex
DSC experiments were performed on a VP-DSC (Microcal LLC, Northampton, MA) with an active cell volume of 0.5 mL at 1.5 atm overpressure using scanning rates of 0.5, 1, and 1.5 °C/min. A total of 70–100 μM of free d(GGTTAC) and Pot1pN and complex (protein/DNA ratio of ~1:1.2) were monitored from 5 to 95 °C in 50 mM Na2HPO4 at pH 7.5, 150 mM NaCl, and 1 mM BME. Apparent thermal transition midpoint temperatures were obtained, because the system was not reversible under the experimental conditions used. Specifically, the post-transition baseline did not reach a steady baseline level but rather drops sharply following denaturation for the first run (Supplemental Figure S1 in the Supporting Information), and there is no transition/excess enthalpy on repetitive runs on the same sample. Visual inspection of each sample following heat denaturation yielded an opaque/milky solution indicative of nonreversibly aggregated/precipitated protein. Measured transition midpoint temperatures show no significant scan rate dependence suggesting that actual values were obtained. Buffer subtraction, concentration normalization, baseline correction, and data analysis were performed using the DSC module in the Origin version 7.0 (Microcal LLC, Northampton, MA).
ITC of the Pot1pN/Oligonucleotide Complexes
ITC studies were performed on a VP-ITC (Microcal LLC, Northampton, MA) with an active cell volume of 1.3 mL and a 250 μL syringe. The instrument was calibrated by titration of 18-crown-6 with BaCl2 (36, 37). Pot1pN and oligonucleotides were dialyzed separately against buffer (50 mM Na2HPO4 at pH 7.5, 150 mM NaCl, and 1 mM BME) using a 3500 Da molecular-weight cutoff (MWCO) Slide-a-Lyzer dialysis cassette (Pierce, Rockford, IL) and a 500 Da MWCO Float-a-Lyzer (Spectrum Laboratories, Rancho Dominguez, CA), respectively, for at least 10 h with 2–3 buffer changes. The last buffer change from the protein dialysis solution was used to dilute protein and oligonucleotide samples and wash the ITC before each run. Samples were thoroughly degassed on a Thermovac (Microcal LLC) for 5 min at ~5 °C lower than the experimental temperature and then titrated at temperatures ranging from 5 to 35 °C (in 5 °C increments). Reference power was set to 12 μcal/s, and the stirring rate was 300 rpm. Typically, each experiment consisted of a single 2 μL injection followed by 19 injections of 4 μL of 115–130 μM d(GGTTAC) into 4–5 μM Pot1pN with injection intervals of 3 min. Reverse titration was performed at 20 °C with 130 μM Pot1pN titrated into 4 μM d(GGTTAC) with the same parameters as above. Titrations of G1I and T3dU were performed at 20 °C and consisted of a single 2 μL injection followed by 24 injections of 4 μL of 100–150 μM DNA into 6 μM Pot1pN with injection intervals of 4 min. Titrations of G2I and T4dU were performed at 20 °C and consisted of a single 2 μl injection followed by 69 injections of 4–5 μL of 700–900 μM DNA into 50 μM Pot1pN with injection intervals of 4 min. Visual inspection after each titration experiment revealed a clear solution at every temperature. Experiments were performed in 3–5 replicates with different preparations of protein and oligonucleotides.
ITC gives reliable thermodynamic data within an affinity regime defined by the c value (c = Ka[cell component] for a 1:1 binding reaction, where Ka is the equilibrium association constant) (38). A high c value gives a steeper binding curve with less data points in the transition region (c > 1000), whereas a low c value (c < 1) gives too shallow of a curve for good estimates of thermodynamic parameters. The c values in this study ([Pot1pN] = 4 μM) are between 1000 at 5 °C and 67 at 35 °C, suggesting that the conditions are within the ideal range for ITC analysis.
Thermodynamic Analysis of ITC Data
Data analysis was performed using the ITC module in the software Origin version 7.0 (Microcal LLC, Northampton, MA). Thermograms were analyzed with a standard 1:1 binding model included in the software to obtain the binding enthalpy (ΔHcal), Kd, and binding stoichiometry (n). The binding free energy (ΔG) was calculated as RT ln(Kd), and the binding entropy (ΔS) was calculated as (ΔH – ΔG)/T after rearrangement of the Gibbs free-energy equation. Errors for n, Kd, ΔG, ΔHcal, and –TΔS represent the standard deviation obtained from the individual fits of replicate data. The molar heat-capacity change (ΔCp) was obtained from plots of the binding enthalpy versus the temperature (ΔH versus T) fitted to the equation ΔH = ΔCp(T – TH), where TH is the reference temperature at which ΔH = 0, using Kaleidagraph version 3.5 (Synergy Software, Reading PA). Errors associated with ΔCp were obtained from the fitting of ΔH versus temperature (K).
NMR Backbone Assignment of the Pot1pN/d(GGTTAC) Complex
Purified, monomeric Pot1pN was exchanged into NMR sample buffer [50 mM KH2PO4 at pH 6.15, 50 mM NaCl, 1 mM deuterated dithiothreitol (DTT)-d, and 10% D2O] through several rounds of concentration and dilution with NMR buffer. Complexes of 1 mM 13C,15N-labeled Pot1pN and high-performance liquid chromatography (HPLC)-purified d(GGTTAC) were formed by adding a 0.5 M excess of ssDNA to ensure that all Pot1pN was completely bound to d(GGTTAC). NMR analysis of the Pot1pN/d(GGTTAC) complex showed that it was in slow chemical exchange with respect to the experimental time scales used. Standard Varian BioPack pulse-derived two- and three-dimensional sensitivity-enhanced, gradient-selected non-TROSY assignment experiments [15N and 13C HSQC, HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, C(CO)NH–TOCSY, HN(CA)CO, and HNCO (39)] were acquired at 30 °C on a Varian Inova 500 MHz spectrometer equipped with a HCN warm probe or a Varian 600 Inova MHz spectrometer equipped with a HCN cold probe. NMR data were processed using the nmrPipe software suite (40), and backbone resonance assignments were made using the ANSIG version 3.3 software package (41) and CCPNMR analysis version 1.0 software package (42).
1H/15N MCSP Analysis of the Pot1pN/Oligonucleotide Complexes
Complexes of 15N-labeled Pot1pN and each HPLC-purified oligonucleotide [d(GGTTAC), G1I, G2I, T3dU, and T4dU] were formed at ~300 μM with a 1:1.5 molar ratio of protein/ssDNA to ensure that all protein present was bound to oligonucleotide. Gradient-selected, sensitivity-enhanced 15N HSQC spectra for each Pot1pN/ssDNA complex were acquired at 30 °C on a Varian Inova 600 spectrometer equipped with a HCN cold probe using Varian BioPack pulse sequences with minor modifications. Acquired spectra were analyzed using CCPNMR analysis version 1.0 software package (42). Values for the MCSPs were calculated according to the following equation:
| (1) |
Assignment and DNA–DNA NOE Analysis of the Pot1pN/Oligonucleotide Complexes
13C,15N-labeled Pot1pN was prepared in 100% D2O containing NMR buffer (50 mM KH2PO4 at pH 6.17, 50 mM NaCl, 1 mM DTT-d, and 100% D2O) as described above. Complexes of 13C,15N-labeled Pot1pN and each HPLC-purified oligonucleotide [d(GGTTAC), G1I, G2I, T3dU, and T4dU] were formed at ~1.5 mM by mixing a 1:0.8 molar ratio of protein/ssDNA to ensure that all ssDNA in solution was bound to Pot1pN. 13C (ω1,ω2) NOESY and 13C (ω1,ω2) TOCSY spectra for each Pot1pN/ssDNA complex were acquired at 25 °C (because of instability of free Pot1pN at 30 °C) on an Varian Inova 900 spectrometer equipped with a HCN warm probe using pulse sequences with minor modifications (43, 44). Assignment and analysis of NOE cross-peaks for each Pot1pN/oligonucleotide complex was performed using CCPNMR analysis version 1.0 software package (42).
Calculation of the Solvent-Accessible Area for Pot1pN
GetArea (45) was used to calculate the polar and apolar accessible surface area (ASA) present in the complexes and the free states. Because only the complex structures are known, free-state structures were first approximated assuming no conformational changes upon binding. The ASAs for free states were subtracted from the complex values and heat-capacity changes calculated using the approach by Spolar and Record (46) with the equation ΔCp = 0.32S – 0.14P, where S is the apolar ΔASA and P is the polar ΔASA. To obtain a more realistic estimate of the surface exclusion upon binding, we recalculated the accessible surfaces areas in a randomized model of free d(GGTTAC) generated with software X-PLOR with standard parameter settings containing no DNA–DNA constraints (47).
RESULTS
Thermodynamic Characterization of the Pot1pN/ssDNA Interaction
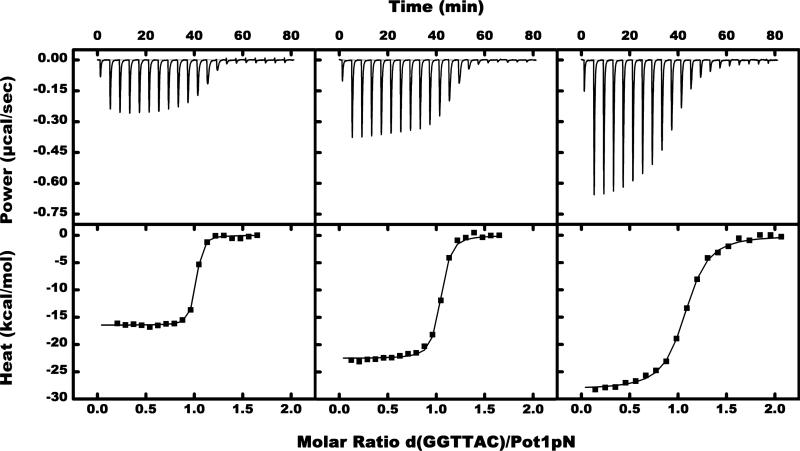
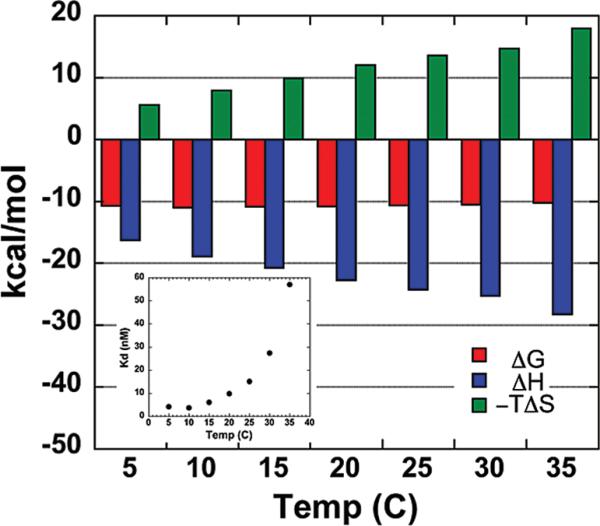
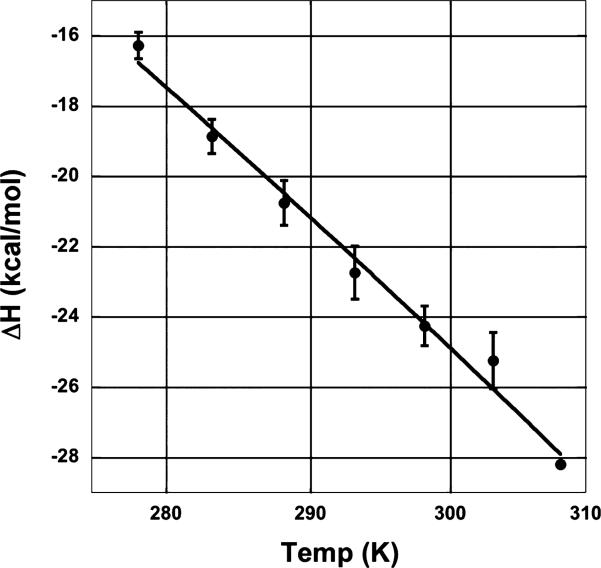
Enthalpic and entropic contributions to the binding free energy of Pot1pN binding to d(GGTTAC) were determined from ITC experiments measuring the total heat released upon titration of d(GGTTAC) into Pot1pN. Thermograms collected over the temperature range of 5–35 °C in 5 °C increments (Figure 2 and Table 1) were fit at all temperatures and yielded stoichiometric values close to unity, consistent with previously published X-ray crystallographic (27) and NMR titration studies (data not shown), indicating a 1:1 complex. DSC data show a midpoint transition temperature of 52.9 °C for free Pot1pN and 57.2 °C for the complex (Supplemental Figure S1 in the Supporting Information), demonstrating that there is no unfolded protein influencing the binding parameters at the experimental ITC temperatures (DSC data are further discussed in the Supporting Information). The ITC results were not dependent upon the order of reagent addition, because ITC values obtained with a reverse titration [Pot1pN titrated into d(GGTTAC)] at 20 °C gave similar results as d(GGTTAC) titrated into Pot1pN (data not shown). Formation of the Pot1pN/d(GGTTAC) complex was found to be exothermic, and the heat released increased with a higher temperature (from –16.3 to –28.2 kcal mol–1) (Figure 3 and Table 1). The negative molar heat-capacity change (ΔCp = –372 ± 19 cal K–1 mol–1, with TH = 233) obtained from fitting a plot of the binding enthalpy versus temperature to the equation ΔH = ΔCp(T – TH), where TH is the reference temperature at which ΔH = 0 (Figure 4), is consistent with previously reported negative heat-capacity changes for protein–ds/ssDNA interactions (1, 2, 13, 18, 46). Our ITC measurements of the binding affinity of Pot1pN for d(GGTTAC) were in reasonable agreement with previously measured values using filter-binding techniques (30).
Figure 2.
Representative thermograms (upper part of panel) and binding isotherms (lower part of panel) obtained by ITC measurements at 5 (left), 20 (middle), and 35 (right) °C. A total of 20 injections of 130 μM d(GGTTAC) was titrated into 5 μM Pot1pN in 50 mM Na2HPO4 at pH 7.5, 150 mM NaCl, and 1 mM BME. Resulting data from the exothermic binding reactions were nonlinear-fitted to a 1:1 model, yielding stoichiometry values close to unity (see Table 1).
Table 1.
Thermodynamic Parameters for Pot1pN Binding to d(GGTTAC) by ITC at Indicated Temperatures in 50 mM Na2HPO4 at pH 7.5, 150 mM NaCl, and 1 mM BME
| temperature (°C) | n | Kd (nM) | ΔG (kcal/mol) | ΔHcal (kcal/mol) | –TΔS (kcal/mol) |
|---|---|---|---|---|---|
| 5 | 1.02 ± 0.06 | 4.1 ± 1.6 | –10.7 ± 0.2 | –16.3 ± 0.4 | 5.6 ± 0.6 |
| 10 | 0.94 ± 0.02 | 3.6 ± 1.0 | –11.0 ± 0.2 | –18.9 ± 0.5 | 7.9 ± 0.6 |
| 15 | 0.97 ± 0.09 | 6.0 ± 1.4 | –10.9 ± 0.1 | –20.8 ± 0.6 | 9.9 ± 0.8 |
| 20 | 1.02 ± 0.03 | 9.7 ± 1.1 | –10.8 ± 0.1 | –22.8 ± 0.8 | 12.0 ± 0.8 |
| 25 | 1.03 ± 0.07 | 15.0 ± 3.1 | –10.7 ± 0.1 | –24.3 ± 0.6 | 13.6 ± 0.7 |
| 30 | 1.02 ± 0.05 | 27.3 ± 4.5 | –10.5 ± 0.1 | –25.3 ± 0.8 | 14.8 ± 0.9 |
| 35 | 1.06 ± 0.02 | 57.0 ± 4.1 | –10.2 ± 0.04 | –28.2 ± 0.1 | 18.0 ± 0.04 |
| ΔCpcal (cal mol–1 K–1) | –372 ± 19 | ||||
| TH (K) | 233 ± 3 | ||||
| TS (K) | 338 |
Figure 3.
Thermodynamic signatures obtained by ITC for Pot1pN binding to d(GGTTAC) at various temperatures. Data reveal a narrow binding free-energy range (ΔG, red bars) composed of a favorable binding enthalpy (ΔH, blue bars) and an unfavorable binding entropy (–TΔS, green bars). The inset shows a 14-fold decrease in the equilibrium dissociation constant (Kd) over the considered temperature interval.
Figure 4.
Binding enthalpy from ITC for Pot1pN binding to d(GGTTAC) as a function of the temperature. A linear fit of the data yielded a binding ΔCp of –372 cal mol–1 K–1. Errors were obtained as described in the Materials and Methods.
The equilibrium dissociation constant for Pot1pN binding to d(GGTTAC) is temperature-dependent and ranged from 4 to 60 nM (from –10.7 to –10.2 kcal mol–1) (Table 1 and inset in Figure 3), with the tightest binding observed between 10 and 15 °C (Figure 3 and Table 1). The entropic term (–TΔS) for binding was found to be unfavorable (positive) and varied in magnitude with an increasing temperature (Figure 3 and Table 1). The entropic and enthalpic terms make compensatory contributions to the binding free energy, resulting in a narrow Gibbs free-energy range. This phenomenon is commonly observed in biological (and other) systems composed of many weak interactions and can be ascribed to statistical thermodynamic laws (48, 49).
Thermodynamic Consequences of DNA Substitutions
The unusual self-recognition DNA motif observed in the Pot1pN crystal structure has been investigated biochemically by making targeted nucleotide substitutions that probe the importance of specific hydrogen-bonding and hydrophobic interactions to the overall binding affinity (27). We have extended these studies to further understand the thermodynamic origins of these targeted mutations on the overall binding free energy (Figure 5). Specifically, the individual substitution of G1 and G2 with inosine eliminates the self-recognition hydrogen bonds formed between G1/T3 and G2/T4, respectively, by eliminating the C-2 NH2 group and hence removing the hydrogen-bond donor that contacts the thymine C-4 carbonyl oxygen (blue spheres in parts C and D of Figure 1). Furthermore, individual substitution of T3 and T4 with deoxyuracil (dU) eliminates the hydrophobic packing interactions of the C-5 CH3 group in the protein/DNA interface (white spheres in parts C and D of Figure 1).
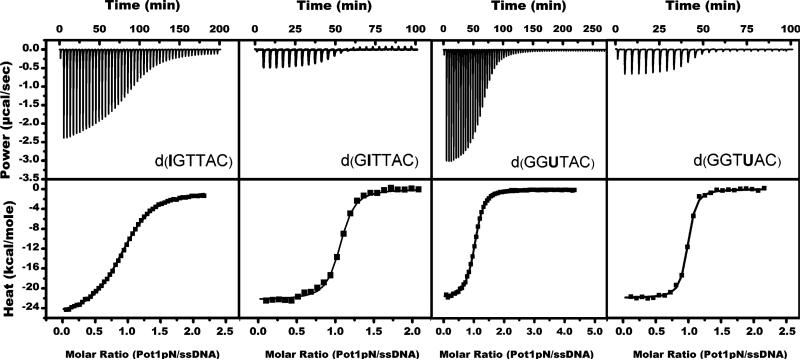
Figure 5.
Representative ITC thermograms (upper) and isotherms (lower) for Pot1pN titrated with noncognate oligonucleotides in 50 mM Na2HPO4 at pH 7.5, 150 mM NaCl, and 1 mM BME at 20 °C. d(IGTTAC), 25 injections of 99 μM G1I titrated into 6 μM Pot1pN; d(GITTAC), 60 injections of 500 μM G2I titrated into 43 μM Pot1pN; d(GGUTAC), 25 injections of 144 μM T3dU titrated into 6 μM Pot1pN; and d(GGTUAC), 70 injections of 880 μM T4dU titrated into 43 μM Pot1pN.
Substitution of G1 with inosine results in very little change in the overall binding affinity for Pot1pN (decreased less than 2-fold) (Table 2). Thermodynamically, elimination of this hydrogen bond impacts both the enthalpy and entropy of binding in a compensatory fashion. In comparison to d(GGTTAC), binding of G1I exhibits a +2.5 kcal/mol (unfavorable) change in enthalpy and a compensating –2.2 kcal/mol (favorable) change in entropy, netting a negligible change in binding free energy [0.3 kcal/mol, relative to d(GGTTAC)]. Conversely, the substitution of G2 with inosine results in a drastic 115-fold loss of binding affinity to 2.7 μM (ΔΔG = 2.8 kcal/mol) at 20 °C. Although the same molecular interaction present in G1I was disrupted in G2I, the thermodynamic signatures for these two mutants differ substantially. Instead of the compensating enthalpic and entropic changes observed in G1I, we find that removal of the same hydrogen-bond interaction in G2I results in a favorable enthalpic change (ΔHG2I–d(GGTTAC) = –1.7 kcal/mol) and a larger unfavorable entropic change [Δ(–TΔS)G2I–d(GGTTAC) = 4.4 kcal/mol]. Thus, the thermodynamic origin of the reduced binding affinity is not due to unfavorable enthalpic changes associated with the loss of the hydrogen bond but rather results from unexpected unfavorable entropic changes.
Table 2.
Thermodynamic Parameters of ssDNA Recognition by Pot1pN by ITC at 20 °C in 50 mM Na2HPO4 at pH 7.5, 150 mM NaCl, and 1 mM BME
| DNA sequence | n | Kd (nM) | ΔG (kcal/mol) | ΔHcal (kcal/mol) | –TΔS (kcal/mol) |
|---|---|---|---|---|---|
| d(GGTTAC) | 1.0 ± 0.01 | 23.5 ± 2.3 | –10.3 ± 0.04 | –23.7 ± 0.2 | 13.5 ± 0.1 |
| d(IGTTAC) | 1.04 ± 0.06 | 39.5 ± 9.3 | –10.0 ± 0.10 | –21.3 ± 1.3 | 11.3 ± 1.3 |
| d(GITTAC) | 0.97 ± 0.08 | 2730 ± 232 | –7.5 ± 0.05 | –25.4 ± 0.1 | 17.9 ± 0.1 |
| d(GGUTAC) | 0.96 ± 0.07 | 31.7 ± 7.7 | –10.1 ± 0.15 | –22.8 ± 0.1 | 12.7 ± 1.0 |
| d(GGTUAC) | 1.06 ± 0.03 | 1070 ± 96 | –8.0 ± 0.05 | –21.9 ± 0.03 | 13.9 ± 0.03 |
Substitution of T3 with deoxyuracil leads to minimal changes in the overall binding affinity (a decrease of less than 0.5-fold) (Table 2). The loss of the T3 C-5 CH3 group, which is more solvent-exposed in the structure than the T4 C-5 CH3 group, results in a modest unfavorable enthalpic change of 1.0 kcal/mol. This change in enthalpy is completely compensated by a favorable entropic change of –1.0 kcal/mol leading to a negligible net change in binding free energy [decreased 0.2 kcal/mol relative to d(GGTTAC)]. However, unlike T3, the C-5 CH3 group of T4 is buried in a hydrophobic pocket formed by the DNA and the protein and upon removal results in a 45-fold binding affinity drop to 1.1 μM (ΔΔG = 2.3 kcal/mol) at 20 °C. This loss in binding affinity is due in large part to a sizable unfavorable enthalpic change (ΔΔHT4dU–d(GGTTAC) of 1.8 kcal/mol), as well as a modest unfavorable entropic change [0.4 kcal/mol relative to d(GGTTAC) binding]. All of the measured binding affinities for these targeted mutants are in agreement with previous measurements obtained by filter-binding methods (30).
Backbone Atom Assignment of Pot1pN Bound to d(GGTTAC)
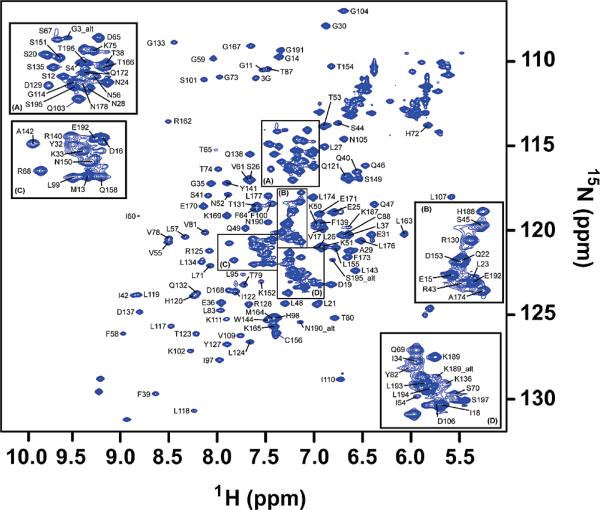
The variable range of thermodynamic changes observed in our noncognate oligonucleotide binding studies led us to investigate whether the structure of Pot1pN and/or the bound oligonucleotide was altered from that of the high-resolution crystal structure, particularly when bound to the noncognate oligonucleotides. To assess these changes using NMR, the backbone atoms (1HN, 15N, 13CO, 13Cβ, and 13Cα) of Pot1pN bound to d(GGTTAC) were assigned using standard triple-resonance experiments (Table S2 in the Supporting Information) and are plotted onto the 15N HSQC spectrum of the Pot1pN/d(GGTTAC) complex (Figure 6). The construct of Pot1pN used in these experiments contains 189 observable residues, of which 155 (82%) have been completely assigned (assigned in 1HN, 15N, 13CO, and 13Cα) and 8 (4%) have been partially assigned (assigned only in 13CO, 13Cα, and where applicable, 13Cβ). Chemical-shift indexing (CSI) (50) of composite resonances (13Cα, 13CO, 13Cβ, and 15NH) are consistent with the secondary-structure elements present in the high-resolution crystal structure of the Pot1pN/d(GGTTAC) complex (27). A total of 12 peaks could not be assigned in the 15N HSQC spectrum of the Pot1pN/d(GGTTAC) complex because of their absence in the triple-resonance data and/or because of intervening proline residues. All but one of these unassigned residues clusters either to the N or C termini or to a region of the protein directly adjacent to the ssDNA-binding interface.
Figure 6.
Assigned 15N HSQC spectrum of the Pot1pN/d(GGTTAC) complex collected on a Varian 600 MHz spectrometer equipped with a HCN cold probe at 30 °C. Peaks representing the individual amino acids present in Pot1pN have been labeled with their corresponding residue assignments. For clarity, selected regions of the 15N HSQC spectrum have been enlarged (A–D) to display selected assignments.
Assignment of Sugar and Base Protons in Free and Pot1pN-Bound d(GGTTAC)
Differences in the conformation of both cognate and noncognate oligonucleotides bound to Pot1pN were also probed using NMR techniques. Assignments for nonexchangeable sugar (1H1′) and base (1H5/6/8/7*) protons in both free and 13C,15N-labeled Pot1pN-bound d(GGTTAC) were made using standard 1H–1H NOESY/TOCSY experiments and 13C (ω1,ω2) double half-filtered NOESY experiments, respectively (Table S3 in the Supporting Information). The NOE data obtained for free d(GGTTAC) reveals six distinct spin systems that contain very few sequential NOE cross-peaks and no nonsequential NOEs, indicating a single extended conformation. Unlike the extended state of free d(GGTTAC), upon binding to Pot1pN, d(GGTTAC) adopts a ordered conformation as indicated by the large number of sequential and nonsequential NOEs that are formed upon binding (Table 3). Because of the proton-poor nature of Pot1pN-bound d(GGTTAC), the chemical-shift degeneracy present in the H2′/H2″, H3′, H4′, and H5′/H5″ protons, and the large amount of conformational space allotted to d(GGTTAC), we were unable to calculate a high-resolution de novo structure of d(GGTTAC) bound to Pot1pN. However, analysis of the DNA-DNA NOEs reveals that the observed internucleotide interactions are consistent with the high-resolution crystal structure of the Pot1pN/d(GGTTAC) complex (27).
Table 3.
Comparison of Normalized NOE Cross-peak Intensities Comparison of Pot1pN-Bound Cognate and Non-cognate Telomeric ssDNA Sequences
| NOEa connectivity | d(GGTTAC) NOE peak intensityb | G1I NOE peak intensityb | G2I NOE peak intensityb | T3dU NOE peak intensityb | T4dU NOE peak intensityb |
|---|---|---|---|---|---|
| 1H1′–2H1′ | 0.10 | (---)c | 0.06 | (---)c | 0.09 |
| 1H1′–2H8 | 0.22 | (---)c | 0.19 | 0.33 | 0.23 |
| 1H1′–3H7* | 0.74 | (---)c | 0.45 | N/A | 0.66 |
| 1H8–2H8 | 0.31 | 0.40 | 0.21 | (---)c | 0.31 |
| 1H8–2H1′ | 0.05 | 0.82 | 0.04 | (---)c | 0.08 |
| 1H8–3H7* | 0.19 | 0.10 | 0.17 | N/A | 0.23 |
| 2H1′–3H6 | 0.12 | 0.13 | 0.07 | (---)c | 0.11 |
| 2H1′–3H7* | 0.54 | 0.65 | 0.42 | N/A | 0.69 |
| 2H1′–4H6 | 0.09 | 0.29 | 0.25 | 0.14 | (---)c |
| 2H1′–4H7* | 0.52 | 0.41 | 0.52 | (---)c | N/A |
| 2H8–3H7* | 0.13 | 0.14 | 0.12 | N/A | 0.23 |
| 2H8–4H7* | 0.13 | 0.09 | 0.08 | 0.16 | N/A |
| 3H1′–4H1′ | 0.18 | 0.14 | 0.18 | 0.23 | 0.17 |
| 3H1′–4H7* | 0.30 | 0.20 | 0.26 | 0.96 | N/A |
| 3H6–4H1′ | 0.13 | 0.14 | 0.12 | 0.08 | 0.12 |
| 3H6–4H6 | 0.06 | 0.28 | 0.10 | (---)c | (---)c |
| 3H5–1H1′ | N/Ad | N/A | N/A | 0.10 | N/A |
| 3H5–4H7* | N/A | N/A | N/A | 0.65 | N/A |
| 3H6–4H7* | 0.49 | 0.45 | 0.49 | 0.33 | N/A |
| 3H7*–4H1′ | 0.11 | 0.11 | 0.10 | N/A | (---)c |
| 3H7*–4H7* | 1.0 | 1.2 | 1.0 | N/A | N/A |
| 3H7*–4H6 | 0.10 | 3.20 | 0.35 | N/A | 0.12 |
| 4H1′–6H1′ | 0.14 | (---)c | (---)c | (---)c | (---)c |
| 4H5–3H1′ | N/A | N/A | N/A | N/A | 0.07 |
| 4H7*–6H1′ | 0.11 | (---)c | (---)c | (---)c | N/A |
NOE connectivity is given as nucleotide position and atom to nucleotide position and atom, with H1′ representing nonexchangeable sugar proton present at position 1′ and H5/H6 representing nonexchangeable base protons present at positions 5 and 6, respectively.
All NOE intensities have been normalized to the individual distance-fixed H5–H6 NOE present in each respective oligonucleotide.
No NOE was detected for these proton pairs.
N/A represents NOE cross-peaks that are specific to the targeted mutation and are not observed in the other sequences.
Chemical-Shift Perturbation Mapping of Pot1pN/Oligonucleotide Complexes
To assess structural changes occurring in each Pot1pN/oligonucleotide complex, we used NMR to monitor changes in the local chemical environment surrounding the individual backbone amino acids in Pot1pN. Because backbone assignments were available for the Pot1pN/d(GGTTAC) complex and not for the Pot1pN/noncognate oligonucleotide complexes, the use of MSCP (51) represents the most conservative calculation of change relative to that of the Pot1pN/d(GGTTAC) complex. MCSP values (see eq 1 in the Materials and Methods) were obtained by comparing the minimal change in the individual amide peak positions present in the 15N HSQC spectra for each Pot1pN/noncognate oligonucleotide complex with the assigned reference position in the 15N HSQC spectrum of the Pot1pN/d(GGTTAC) complex. A comparative analysis of the MCSP data for each Pot1pN/noncognate oligonucleotide complex relative to the Pot1pN/d(GGTTAC) complex follows below.
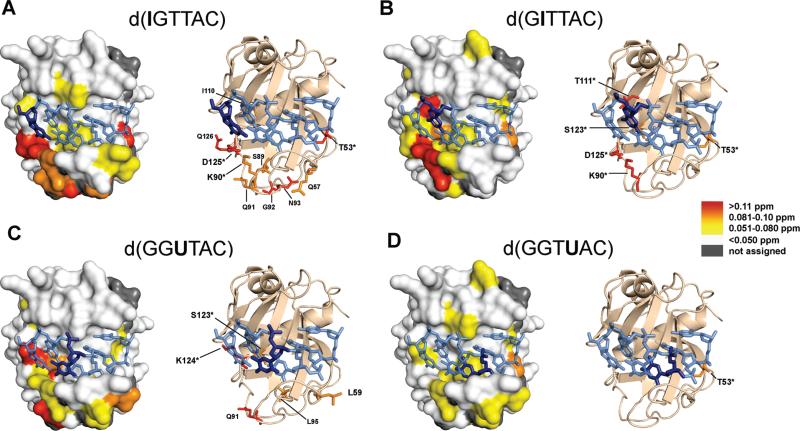
Substitution of G1 with inosine leads to perturbation of residues throughout the ssDNA-binding interface, with a majority of the strong and moderate changes occurring in the binding pocket of G1/G2 and residues that make direct contact with T3 (Figure 7A). The widespread scope of these changes do not reflect the perturbations expected from the simple removal of the C-2 NH2 group from G1, which, on the basis of the conformation in the Pot1pN/d(GGTTAC) complex, would only lead to perturbations of the directly contacting residues K90 and D125 (27). Instead, a total of 26 residues (21% of the total assigned residues) undergo perturbation, of which five (T53, G92, N93, D125, and Q126) are strongly perturbed (Δppm ≥ 0.11), five (Q57, S89, K90, I110, and Q91) are moderately perturbed (Δppm = 0.081–0.1), and 16 are weakly perturbed (Δppm = 0.051–0.080). The strongly and moderately perturbed residues were found to cluster to the irregularly structured region of β-strand 5 (β5) and loop 3–4 (L3–4), which contact the nucleotides G1 and T3, as well as to loop 1–2 (L1–2), which makes contact with the nucleotides A5 and C6 (21, 27) (Figure 7A). Of these residues that are perturbed in Pot1pN, three are known to make direct contact with the bound nucleotides (21, 27) (T53 → C6, D125 → G1, and K90 → T3). The remaining residues that are perturbed surround these ssDNA-contacting residues.
Figure 7.
(Left) Surface representation of the high-resolution crystal structure of the Pot1pN/d(GGTTAC) complex (27) highlighting the MCSPs resulting from the binding of each substituted oligonucleotide, G1I, G2I, T3dU, and T4dU (site of variation highlighted in dark blue on a stick model of DNA). (Right) Cartoon/ribbon model of the Pot1pN/d(GGTTAC) complex highlighting the side chains of specific residues undergoing moderate and strong perturbations in each Pot1pN/substituted nucleotide complex. Residues that make direct contact with the oligonucleotide in the Pot1pN/d(GGTTAC) complex are designated with an asterisk. (A) Moderate and strongly perturbed residues in Pot1pN/d(IGTTAC) complex occur on loop1–2 (T53* and Q57), loop3–4 (S89, K90*, G92, and N93), β4 (I110), and β5 (D125* and Q126). (B) Moderate and strongly perturbed residues in the Pot1pN/d(GITTAC) complex occur in loop1–2 (T53*), loop3–4 (K90*), β4 (I111*), and β5 (S123* and D125*). (C) Moderate and strongly perturbed residues in the Pot1p/d(GGUTAC) complex occur in loop1–2 (L59), loop3–4 (Q91 and L95), and β5 (S123* and K124*). (D) The only moderately perturbed residue in the Pot1pN/d(GGTUAC) complex is found on loop1–2 (T53*).
Substitution of G2 with inosine also leads to the perturbation of residues throughout the ssDNA-binding interface (Figure 7B). These widespread changes are more than expected for the simple removal of the C-2 NH2 group from G2, which, on the basis of the crystal structure of the Pot1pN/d(GGTTAC) complex is expected to impact the directly contacting residues F88, L122, and S123 in the absence of conformational changes (27). We instead find a total of 24 (15% of the total assigned residues) residues are perturbed: 3 (K90, T111, and D125) are strongly perturbed (Δppm ≥ 0.11), 2 (T53 and S123) are moderately perturbed (Δppm = 0.081–0.1), and 19 are weakly perturbed (Δppm = 0.051–0.080). Similar to G1I, the majority of strongly and moderately perturbed residues were found to cluster to the unstructured regions of β5 and L3–4. In addition to this region, there is a single residue that is perturbed in β4 (T111). Furthermore, every strongly and moderately perturbed residue is known to make direct contact with the ssDNA oligonucleotide in the Pot1pN/d(GGTTAC) complex crystal structure (T53 → C6, K90 → T3, T111 → G2, S123 → G2, and D125 → G1) (21, 27).
In comparison to the G1I and G2I mutants, substitutions of T3 with deoxyuracil lead to fewer overall perturbations in the protein chemical environment (Figure 7C). Although fewer total perturbations were observed, they were unexpected, because none of the residues make direct contact with the C-5 CH3 group of T3 in the crystal structure of the Pot1pN/d(GGTTAC) complex (27). Of the total 16 residues (10% of the total assigned residues) that undergo perturbation, 2 (Q91 and K124) are strongly perturbed (Δppm ≥ 0.11), 3 (L59, S123, and L95) are moderately perturbed (Δppm = 0.081–0.1), and 11 are weakly perturbed (Δppm = 0.051–0.080). Similar to the G1I and G2I mutations, the majority of strongly and moderately perturbed residues localize to the irregularly structured region of β5 and L3–4, even though the region lies in a distinct part of the binding interface (Figure 7C). Of these moderately and strongly perturbed residues, three are known to make contact with the ssDNA in the Pot1pN/d(GGTTAC) crystal structure (K124 → G1, S123 → G2, and L59 → A5) (21, 27), with the remaining noncontact residues neighboring these contact residues.
Substitution of T4 with deoxyuracil lead to the smallest perturbations in protein chemical environment when compared to the Pot1pN/d(GGTTAC) complex (Figure 7D). Although modest, these perturbations are more than would be expected upon removal of the C-5 CH3 group of T4 based on the Pot1pN/d(GGTTAC) structure, because only two residues make direct contact with the oligonucleotide: F88 and L122 (27). Of the 17 (11% of the total assigned residues) residues that were perturbed, only 1 (T65) was moderately perturbed (Δppm = 0.081–0.1), while the remaining 16 perturbations were weakly perturbed, including F88 (Δppm = 0.051–0.080). The lone moderately perturbed residue, T65, resides in L1–2 and makes contact with the C6 nucleotide in the Pot1pN/d(GGTTAC) complex crystal structure (21, 27). The T4dU mutant in contrast to G1I, G2I, and T3dU showed very minor perturbations of the unstructured region of β5 and L3–4 (Figure 7D).
Probing Conformational Changes in Pot1pN-Bound Cognate and Noncognate Oligonucleotides
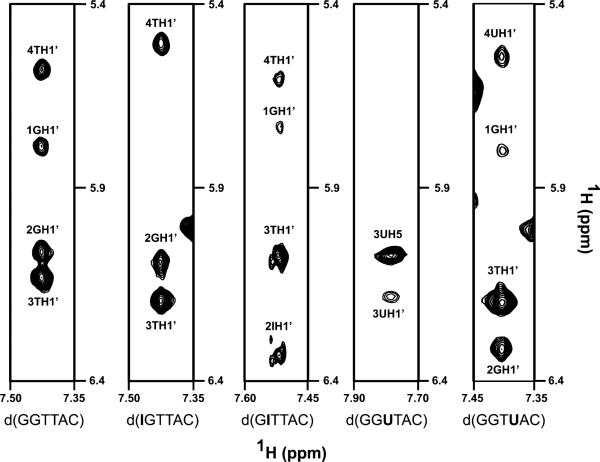
The MCSP data cannot distinguish between conformational changes occurring in Pot1pN or a restructuring of the bound ssDNA oligonucleotide. Therefore, we employed 13C (ω1,ω2) double half-filtered NOESY experiments of the 13C,15N-labeled Pot1pN/d(GGTTAC) complex (43) to directly probe the changes occurring in the Pot1pN-bound noncognate oligonucleotides (Figure 8). Assignments for the nonexchanged sugar proton (H1′) and base proton (H5/6/7*) in the Pot1pN-bound noncognate oligonucleotides were made in the same fashion as that of the Pot1pN-bound d(GGTTAC) using 13C (ω1,ω2) double half-filtered NOESY and TOCSY experiments (Table S3 in the Supporting Information). The comparative DNA-DNA NOE analysis of each individually Pot1pN-bound noncognate oligonucleotide with that of Pot1pN-bound d(GGTTAC) follows below.
Figure 8.
Individual strip plots taken from the 13C (ω1,ω2) double half-filtered NOESY experiments for each Pot1pN/oligonucleotide complex. NOEs between the H8/H6 base proton with the H1′ sugar proton of various nucleotides are highlighted. Individual NOE cross-peaks have been labeled with their corresponding H1′ sugar proton assignment, and each strip plot has been contoured to the same level to facilitate direct comparisons of NOE intensity for each different Pot1pN/oligonucleotide complex. Data are quantitated in Table 3.
Substitutions of G1 with inosine lead to significant changes in the NOE patterns observed near the site of mutation (at the 5′ end). Relative to the Pot1pN-bound d(GGTTAC), NOE cross-peaks connecting the sugar proton of I1 to the sugar proton of G2, the sugar proton of I1 to the base proton of G2, the sugar proton of I1 to the base proton of T3, and the sugar and base proton of T4 to sugar proton of C6 are completely eliminated in the GI1-substituted oligonucleotide (Table 3). Additionally, we observe substantial increases in the intensity of the NOE connecting the base proton of I1 to the sugar proton of G2 (~17-fold) and moderate increases in NOE intensities connecting the base proton of T3 and the base proton of T4 (~5-fold), as well as the base proton of T4 to the sugar proton of G2 (~3-fold) (Table 3). Taken together, the MCSP and DNA–DNA NOE changes indicates significant differences in the conformation of both the bound oligonucleotide and Pot1pN, specifically in the binding pocket of G1/G2 and key residues that make direct contact with T3, relative to that of the Pot1pN/d(GGTTAC) complex. These structural changes in the absence of changes in the binding affinity (Table 2) imply that both the protein and DNA undergo a significant reconfiguration, resulting in an alternate yet thermodynamically equivalent conformation as found in the Pot1pN/d(GGTTAC) complex (Figure 1).
Substitution of G2 with inosine lead to no major changes in the NOE cross-peak pattern relative to Pot1pN-bound d(GGTTAC), with the exception of the NOE connecting the sugar and base proton of T4 with the sugar proton of C6 (Table 3). Additionally, no substantial changes in NOE intensity were observed in this noncognate oligonucleotide, but rather very modest increases in the NOEs connecting the sugar proton of I2 to the base proton of T4 (~4-fold) and the methyl base proton of T3 to the base proton of T4, (~3-fold) were observed (Table 3). The lack of changes in the NOE patterns coupled with the very modest changes in NOE intensity suggest that the bound conformation of the G2I mutant is very similar to that of Pot1pN-bound d(GGTTAC). Although conformation of Pot1pN-bound d(GITTAC) remains similar to that of Pot1pN-bound d(GGTTAC), MCSP data suggest that Pot1pN undergoes significant rearrangement within the G1/G2-binding pocket (Figure 7B). These changes, unlike those that occur in G1I, do not allow for the formation of an equivalent thermodynamic binding configuration, suggesting that the complex cannot completely readjust to compensate for the loss in key thermodynamic contacts (Table 2).
Substitution of T3 with deoxyuracil lead to a pronounced loss of NOE cross-peaks when compared to Pot1pN-bound d(GGTTAC). There is a noticeable absence of NOE cross-peaks, extending from the 5′ end, that connect the sugar proton of G1 to the sugar proton of G2, the base proton of G1 to the base proton of G2, the base proton of G1 to the sugar proton of G2, the sugar proton of G2 to the base proton of dU3, the sugar proton of G2 to the methyl proton of T4, the base proton of dU3 to the base proton of T4, and the sugar and base protons of T4 to the sugar proton of C6 (Table 3). In addition to these absent NOEs, we also observed a modest increase in the intensity of the NOE cross-peak connecting the sugar proton of dU3 to the methyl base proton of T4 (~3-fold). Taken together, the MCSP and NOE analysis suggests that coupled local environment changes in both dU3 and G1 lead to the reconfiguration of the G1/G2-binding pocket (Figure 7C). Similar to the G1I mutant, these conformational changes in oligonucleotide and protein allow for the reformation of key thermodynamic contacts that result in little change observed in the overall binding affinity of Pot1pN for T3dU (Table 2).
Substitution of T4 with deoxyuracil also leads to pronounced changes in the NOE patterns and intensities relative to that of Pot1pN-bound d(GGTTAC). We observed a loss of NOE cross-peaks that clustered around the site of mutation, involving the connections of dU4 with neighboring nucleotides: the sugar proton of G1 to the base proton of dU4, the base proton T3 to base proton of dU4, the base proton of T3 with the sugar proton of dU4, and sugar and base protons of dU4 to the sugar proton of C6 (Table 3). In addition to these absent NOEs, we also detected a subtle increase in the intensity of the NOE connecting the base proton of G2 to methyl base proton T3 (~2-fold) (Table 3). Taken together, the changes detected by NOE and MCSP indicate a local conformational change at the site of mutation in dU4, which does not significantly impact the chemical environment of residues in Pot1pN, relative to that observed in the Pot1pN/d(GGTTAC) complex (27). Similar to the G2I mutant, these conformational changes do not allow for the full re-establishment of key thermodynamic contacts, resulting in the reduced binding affinity observed (27) (Table 2).
DISCUSSION
Binding of Pot1pN to Cognate Telomeric ssDNA Is an Enthalphically Driven Process with a Moderately Negative Heat-Capacity Change
Pot1pN binding to a cognate telomeric ssDNA, d(GGTTAC), is exothermic and characterized by favorable enthalpic and nonfavorable entropic contributions. The exothermic nature of Pot1pN binding to d(GGTTAC) indicates that a large net number of hydrogen bonds and stacking/van der Waals interactions are formed upon binding, with electrostatic interactions playing a negligible role (4). These observations are consistent with the Pot1pN/d(GGTTAC) crystal structure, which shows an intricate network of inter- and intramolecular hydrogen bonds as well as protein/DNA and DNA/DNA base stacking in addition to van der Waals interactions formed upon binding (27). The negative enthalpy (around –22 kcal/mol at room temperature) observed for Pot1pN binding to d(GGTTAC) likely arises from a combination of the 5 stacking interactions and the 20 hydrogen bonds formed between both the protein and ssDNA as well within the ssDNA itself. The overall favorable enthalpic nature of Pot1pN binding to d(GGTTAC) is similar to that observed for other specific ssDNA-binding systems, because the majority of the systems characterized show favorable enthalpic contributions to the binding free energy between –3 and –25 kcal/mol at 20–25 °C (Table S4 and references therein in the Supporting Information). Finally, the nonfavorable entropic contributions suggest that any contributions to the binding free energy from water, ion release, or the hydrophobic effect are surpassed by unfavorable entropy contributions from ssDNA ordering and loss of rotational and translational degrees of freedom upon complex formation.
The binding of Pot1pN to d(GGTTAC) results in a moderately negative heat-capacity change (ΔCp,exp) of –372 cal K–1 mol–1 in the temperature interval studied (5–35 °C). Negative heat-capacity changes are common in protein–nucleic acid interactions (Table S4 in the Supporting Information) and are believed to arise from the burial of a nonpolar surface area (46). One strategy for predicting theoretical heat-capacity changes is based on solvation changes of the polar and nonpolar groups found in the ssDNA-binding interface (46, 52). Using these methods, we calculate a theoretical heat-capacity change (ΔCp,theo = of –193 cal K–1 mol–1 (Table S4 in the Supporting Information), which significantly underestimates the ΔCp,exp of –372 cal K–1 mol–1. Differences between ΔCp,exp and ΔCp,theo may result from the choice of binding parameters used in these calculations; as parameters used in protein–protein interactions may not be suitable for accurate estimations of heat-capacity changes in nucleic acid/protein interactions. Additionally, observed differences may result from other binding events, such as conformational change, protonation/deprotonation, ion exchanges, DNA base–base unstacking, and vibrational mode changes, all of which can impact the heat capacity. Indeed, we find that the differences between ΔCp,exp and ΔCp,theo do not result from ion release or protonation events, because the binding of Pot1pN to d(GGTTAC) shows only a modest sensitivity to NaCl concentrations [(30) and data not shown] and minimal uptake/release of protons (data not shown). Furthermore, large conformational changes in Pot1pN upon binding d(GGTTAC) is also unlikely to be a major contributor, because 15N HSQC patterns of free and d(GGTTAC)-bound Pot1pN do not suggest a global conformational change (data not shown). Rather, we propose that these differences are most likely the result of the conformational changes occurring in d(GGTTAC) from a dynamic extended state to a single fixed, compacted state upon binding, as indicated by NOE data. These differences between ΔCp,exp and ΔCp,theo based on changes in solvation are not limited to our studies, and analysis of structure and thermodynamics data from other protein/ssDNA interactions also result in similar discrepancies (Table S4 in the Supporting Information). Taken together, these results suggest that heat-capacity changes based solely on the dehydration of nucleic acids and solvent-accessible surface area cannot accurately calculate the expected heat-capacity changes for protein/nucleic acid interactions (53).
A larger negative heat-capacity change and a lower salt dependence commonly separate specific and nonspecific dsDNA recognition (1, 54), and these trends might apply to ssDNA recognition as well. Indeed TEP proteins, which bind specifically to ssDNA, show a modest salt dependence upon binding ssDNA (18, 30, 32), while nonspecific ssDNA-binding proteins, such as E. coli SSB protein, show a strong salt dependence (11). This presumably arises from more DNA backbone interactions/ionic interactions, protein–anion interactions, and a lack of hydrogen-bond formation in the later system. It is more difficult to discern any general difference in the magnitude of the heat-capacity changes upon binding within this limited experimental data set. Rather, it appears that both specific and nonspecific ssDNA recognition are characterized by significant negative heat capacities potentially originating from major negative contributions from larger structural ordering of ssDNA compared to dsDNA.
Targeted Nucleotide Substitution of d(GGTTAC) Induces Structural Changes in Both Pot1pN and Bound Oligonucleotide
To gain a better understanding of how the specific recognition of ssDNA is facilitated by Pot1pN, a panel of noncognate oligonucleotides were studied that target the unusual hydrogen bond formed between G1/T3 and G2/T4, as well the hydrophobic contacts made by the methyl group found on T3 and T4. Consistent with previous studies (27), our ITC results show that mutations G1I and T3dU do not affect binding affinity (<2-fold change), while mutations G2I and T4dU show dramatic increases in the binding affinity (~100- and ~50-fold, respectively) (Table 2). The thermodynamic signatures of these noncognate oligonucleotides were more varied than would have been expected on the basis of changes introduced by the targeted mutations (Table 2). These results immediately suggested an altered structural configuration present in these Pot1pN/noncognate oligonucleotide complexes relative to the Pot1pN/d(GGTTAC) complex crystal structure. Consistent with this hypothesis, MCSP mapping and NOE analysis indicate structural rearrangements in each Pot1pN/noncognate oligonucleotide complex studied. Interestingly, parallel mutations (G1I and G2I) that target the similar molecular interactions (loss of an DNA-DNA hydrogen bond) appear to have differential effects with regard to conformational change and accommodation by Pot1pN. Mutation of G1I leads to a rearrangement of both the protein and bound oligonucleotide, which together result in the formation of a thermodynamically equivalent binding configuration, relative to the Pot1pN/d(GGTTAC) complex (27). Although similar in nature, the G2I mutation results in a markedly different response, in which changes in the bound oligonucleotide conformation are not observed, but rather conformational changes are limited to Pot1pN (Figure 7B). Unlike G1I, these changes do not allow for the recapturing of key thermodynamic contacts and losses in binding affinity are observed (Table 2). Similar differences in conformational change are observed in T3dU and T4dU, which eliminate the hydrophobic contacts made by the CH3 group in the thymine nucleotides. T3dU undergoes a local conformational change at the site of mutation (dU3) that induces a conformational change in Pot1pN away from the site of mutation (Figure 7C). Similar to G1I, these changes are sufficient for the formation of a thermodynamically equivalent conformation relative to that in the Pot1pN/d(GGTTAC) complex. Conversely, T4dU appears only to undergo changes in the bound oligonucleotide conformation and not Pot1pN, resulting in an overall conformation that is incapable of reforming necessary thermodynamic contacts. Contrary to our expectations, it should be noted that conformational changes were observed in all oligonucleotide mutants studied and were not limited to those mutations that resulted in decreases in binding affinity.
The number of thermodynamically equivalent conformations that are accommodated by Pot1pN provides insight into how ssDNA is specifically recognized. The mechanisms classically thought to drive the specific recognition of ssDNA in the TEP family is the hydrogen-bonding interactions of basic, acidic, and (occasionally) aromatic amino acids with individual nucleotide bases (21). In addition to specificity being determined by these intermolecular interactions, our studies suggest that the principles that dictate specificity should be expanded to include the ability of the interface to adjust and accommodate altered states. This fluidity at the interface allows for a structurally distinct but thermodynamically equivalent conformation to be achieved. These observations highlight the fact that the ssDNA-binding interface present in Pot1pN is a dynamic surface that can actively accommodate a number of different oligonucleotide structures that are altered from that observed in the Pot1pN/d(GGTTAC) complex (30). Conformational rearrangement in response to nucleotide mutation has been previously observed in the TEBPαβ/ssDNA complex (31) and has been termed “nucleotide shuffling”. In these studies, the oligonucleotide, in response to mutation undergoes changes in conformation, while the protein remains fairly static, to reestablish the thermodynamic contacts lost (31), a situation observed in the Pot1pN/d(GGTUAC) complex. In an elaboration of these studies, we find that the bound oligonucleotide, as well as Pot1pN, can undergo significant conformational adjustments to restore the thermodynamic contacts lost to mutation. Furthermore, we find that gross nucleotide mutation (i.e., nucleotide addition or removal) is not necessary to invoke radical changes in the protein and/or bound oligonucleotide, but rather subtle changes to the chemical nature of the individual nucleotides (i.e., the loss of a hydrogen-bond donor) can invoke such changes. Our studies also indicate that nucleotide shuffling is not limited to TEBPR but rather may be a general feature that is shared among TEP family members.
Finally, the changes that we observe in the context of these targeted mutations may extend to the differences that we have observed between Pot1pN and Pot11–389. In Pot11–389, we have previously observed changes in binding specificity when an additional putative OB-fold is added to Pot1pN to form Pot11–389 (32). One possibility is that there is a conformational switch between a self-recognition and more extended mode of binding, a state that can be achieved in Pot1pN with small changes in the substrate. The flexibility of these ssDNA-binding sites might indicate an adaptation for the recognition of an inherently conformationally flexible and heterogeneous ligand, suggesting a fluid mechanism of accommodation within the ssDNA-binding interface.
Supplementary Material
Figure 9.
Cartoon model highlighting the malleable nature of the Pot1pN ssDNA-binding interface and the thermodynamic accommodation of multiple distinct complex configurations.
ACKNOWLEDGMENT
We thank Professor LaToya Jones (UCDHSC, Denver, CO) for access to DSC and Professor Robert Batey (UC Boulder) for helpful comments on the manuscript. We also thank the Rocky Mountain 900 MHz NMR Facility and Dr. Geoff Armstrong for aiding in the collection of the NMR data used in this paper.
Footnotes
We acknowledge the Wenner–Gren Foundation Postdoctoral Fellowship (to J.L.F.), National Research Service Award (NRSA) Postdoctoral Fellowship GM-071257 (to J.E.C.), and National Institutes of Health (NIH) GM-059414 (to D.S.W.), and National Science Foundation (NSF) (to D.S.W.) (MCB-0617956) for funding this research.
Abbreviations: BME, 2-betamercapthoethanol; DBD, DNA-binding domain; DSC, differential scanning calorimetry; dsDNA, double-stranded DNA; G1I, d(IGTTAC); G2I, d(GITTAC); HSQC, hetero-nuclear single-quantum coherence; ITC, isothermal titration calorimetry; MCSP, minimal chemical-shift perturbation; MWCO, molecular-weight cutoff; NMR, nuclear magnetic resonance; NOE, nuclear Overhauser effect; OB-fold, oligonucleotide/oligosaccharide fold; Pot1, protection of telomeres 1; Pot1pN, residues 1–187 of full-length S. pombe Pot1 with N-terminal His6 tag; ssDNA, single-stranded DNA; TEP, telomere end protection; T3dU, d(GGUTAC); T4dU, d(GGTUAC).
SUPPORTING INFORMATION AVAILABLE
Calorimetric stability studies of free and Pot1pN-bound d(GGTTAC) using DSC, the polar and apolar surface areas and dehydration heat-capacity changes for specific ssDNA-binding proteins, and NMR resonance assignment tables for the backbone atoms of Pot1pN and selected nonexchangeable sugar and base protons present in free and Pot1pN-bound d(GGTTAC). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Oda M, Nakamura H. Thermodynamic and kinetic analyses for understanding sequence-specific DNA recognition. Genes Cells. 2000;5:319–326. doi: 10.1046/j.1365-2443.2000.00335.x. [DOI] [PubMed] [Google Scholar]
- 2.Jen-Jacobson L, Engler LE, Jacobson LA. Structural and thermodynamic strategies for site-specific DNA binding proteins. Struct. Fold Des. 2000;8:1015–1023. doi: 10.1016/s0969-2126(00)00501-3. [DOI] [PubMed] [Google Scholar]
- 3.Mandel-Gutfreund Y, Schueler O, Margalit H. Comprehensive analysis of hydrogen bonds in regulatory protein DNA-complexes: In search of common principles. J. Mol. Biol. 1995;253:370–382. doi: 10.1006/jmbi.1995.0559. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien R, Haq I. Applications of Biocalorimetry: Binding, Stability and Enzyme Kinetics. Wiley; New York: 2004. [Google Scholar]
- 5.Milev S, Gorfe AA, Karshikoff A, Clubb RT, Bosshard HR, Jelesarov I. Energetics of sequence-specific protein–DNA association: Binding of integrase Tn916 to its target DNA. Biochemistry. 2003;42:3481–3491. doi: 10.1021/bi0269355. [DOI] [PubMed] [Google Scholar]
- 6.Hard T, Lundback T. Thermodynamics of sequence-specific protein–DNA interactions. Biophys. Chem. 1996;62:121–139. doi: 10.1016/s0301-4622(96)02197-7. [DOI] [PubMed] [Google Scholar]
- 7.Privalov PL, Dragan AI, Crane-Robinson C, Breslauer KJ, Remeta DP, Minetti CASA. What drives proteins into the major or minor grooves of DNA? J. Mol. Biol. 2007;365:1–9. doi: 10.1016/j.jmb.2006.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochkarev A, Bochkareva E. From RPA to BRCA2: Lessons from single-stranded DNA binding by the OB-fold. Curr. Opin. Struct. Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Mol. Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 10.Kozlov AG, Lohman TM. Adenine base unstacking dominates the observed enthalpy and heat capacity changes for the Escherichia coli SSB tetramer binding to single-stranded oligoadenylates. Biochemistry. 1999;38:7388–7397. doi: 10.1021/bi990309z. [DOI] [PubMed] [Google Scholar]
- 11.Kozlov AG, Lohman TM. Calorimetric studies of E. coli SSB protein-single-stranded DNA interactions: Effects of monovalent salts on binding enthalpy. J. Mol. Biol. 1998;278:999–1014. doi: 10.1006/jmbi.1998.1738. [DOI] [PubMed] [Google Scholar]
- 12.Larkin C, Datta S, Harley MJ, Anderson BJ, Ebie A, Hargreaves V, Schildbach JF. Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure. 2005;13:1533–1544. doi: 10.1016/j.str.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 13.Stern JC, Anderson BJ, Owens TJ, Schildbach JF. Energetics of the sequence-specific binding of single-stranded DNA by the F factor relaxase domain. J. Biol. Chem. 2004;279:29155–29159. doi: 10.1074/jbc.M402965200. [DOI] [PubMed] [Google Scholar]
- 14.Stern JC, Schildbach JF. DNA recognition by F factor TraI36: Highly sequence-specific binding of single-stranded DNA. Biochemistry. 2001;40:11586–11595. doi: 10.1021/bi010877q. [DOI] [PubMed] [Google Scholar]
- 15.Tanner JJ, Komissarov AA, Deutscher SL. Crystal structure of an antigen-binding fragment bound to single-stranded DNA. J. Mol. Biol. 2001;314:807–822. doi: 10.1006/jmbi.2001.5178. [DOI] [PubMed] [Google Scholar]
- 16.Komissarov AA, Deutscher SL. Thermodynamics of Fab–ssDNA interactions: Contribution of heavy chain complementarity determining region 3. Biochemistry. 1999;38:14631–14637. doi: 10.1021/bi991347l. [DOI] [PubMed] [Google Scholar]
- 17.Buczek P, Horvath MP. Structural reorganization and the cooperative binding of single-stranded telomere DNA in Sterkiella nova. J. Biol. Chem. 2006;281:40124–40134. doi: 10.1074/jbc.M607749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buczek P, Horvath MP. Thermodynamic characterization of binding Oxytricha nova single-strand telomere DNA with the α protein N-terminal domain. J. Mol. Biol. 2006;359:1217–1234. doi: 10.1016/j.jmb.2006.02.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerr ID, Wadsworth RIM, Cubeddu L, Blankenfeldt W, Naismith JH, White MF. Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of Sulfolobus SSB protein. EMBO J. 2003;22:2561–2570. doi: 10.1093/emboj/cdg272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 21.Croy JE, Wuttke DS. Themes in ssDNA recognition by telomere-end protection proteins. Trends. Biochem. Sci. 2006;31:516–525. doi: 10.1016/j.tibs.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Peersen OB, Ruggles JA, Schultz SC. Dimeric structure of the Oxytricha nova telomere end-binding protein α-subunit bound to ssDNA. Nat. Struct. Mol. Biol. 2002;9:182–187. doi: 10.1038/nsb761. [DOI] [PubMed] [Google Scholar]
- 23.Horvath MP, Schultz SC. DNA G-quartets in a 1.86 Å resolution structure of an Oxytricha nova telomeric protein-DNA complex. J. Mol. Biol. 2001;310:367–377. doi: 10.1006/jmbi.2001.4766. [DOI] [PubMed] [Google Scholar]
- 24.Classen SC, Ruggles JA, Schultz S. Crystal structure of the N-terminal domain of Oxytricha nova telomere end-binding protein α subunit both uncomplexed and complexed with telomeric ssDNA. J. Mol. Biol. 2001;314:1113–1125. doi: 10.1006/jmbi.2000.5191. [DOI] [PubMed] [Google Scholar]
- 25.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 26.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 27.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–204. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 28.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 29.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei M, Baumann P, Cech TR. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry. 2002;41:14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- 31.Theobald DL, Schultz SC. Nucleotide shuffling and ssDNA recognition in Oxytricha nova telomere end-binding protein complexes. EMBO J. 2003;22:4314–4324. doi: 10.1093/emboj/cdg415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Croy JE, Podell ER, Wuttke DS. A new model for Schizosaccharomyces pombe telomere recognition: The telomeric single-stranded DNA-binding activity of Pot11–389. J. Mol. Biol. 2006;361:80–93. doi: 10.1016/j.jmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Anderson EM, Halsey WA, Wuttke DS. Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomuces cerevisiae Cdc13. Nucleic Acids Res. 2002;30:4305–4313. doi: 10.1093/nar/gkf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eldridge AM, Halsey WA, Wuttke DS. Identification of the determinants for the specific recognition of single-strand telomeric DNA by Cdc13. Biochemistry. 2006;45:871–879. doi: 10.1021/bi0512703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Briggner L-E, Wadsö I. Test and calibration processes for microcalorimeters, with special reference to heat conduction instruments used with aqueous systems. J. Biochem. Biophys. Methods. 1991;22:101–118. doi: 10.1016/0165-022x(91)90023-p. [DOI] [PubMed] [Google Scholar]
- 37.Buschmann H-J, Schollmeyer E. A test reaction from macrocyclic chemistry for calorimetric titrations. Thermochim. Acta. 1999;333:49–53. [Google Scholar]
- 38.Turnbull WB, Daranas AH. On the value of c: Can low affinity systems be studied by isothermal titration calorimetry? J. Am. Chem. Soc. 2003;125:14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- 39.Bennett T, Farmer BT, II, Müeller L. Unambiguous resonance assignments in 13C, 15N-labelled nucleic acids by 3D triple resonance NMR. J. Am. Chem. Soc. 1993;115:11040–11041. [Google Scholar]
- 40.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 41.Kraulis PJ. ANSIG: A program for the assignment of protein 1H 2D NMR spectra by interactive computer graphics. J. Magn. Reson. 1989;84:627–633. [Google Scholar]
- 42.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 43.Otting G, Wüthrich K. Extended heteronuclear editing of 2D 1H NMR spectra of isotope labeled proteins, using the X(w1,w2) double half filter. J. NMR Res. 1989;85:586–594. [Google Scholar]
- 44.Ikura M, Bax A. Isotope-filtered 2D NMR of a protein-peptide complex: Study of a skeletal muscle myosin light chain kinases fragment bound to calmodulin. J. Am. Chem. Soc. 1992;114:2433–2440. [Google Scholar]
- 45.Fraczkiewicz R, Werner B. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 1998;19:319–333. [Google Scholar]
- 46.Spolar RS, Record MT., Jr. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 47.Brunger AT. X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. Yale University Press; New Haven, CT: 1993. [Google Scholar]
- 48.Sharp K. Entropy–enthalpy compensation: Fact or artifact? Protein Sci. 2001;10:661–667. doi: 10.1110/ps.37801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooper A, Johnson CM, Lakey JH, Nollmann M. Heat does not come in different colours: Entropy-enthalpy compensation, free energy windows, quantum confinement, pressure perturbation calorimetry, solvation and the multiple causes of heat capacity effects in biomolecular interactions. Biophys. Chem. 2001;93:215–230. doi: 10.1016/s0301-4622(01)00222-8. [DOI] [PubMed] [Google Scholar]
- 50.Wishart DS, Sykes BD. The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 51.Farmer BT, II, Constantine KL, Goldfarb V, Friedrichs MS, Wittekind M, Yanchunas J, Jr., Robertson JG, Mueller L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat. Struct. Mol. Biol. 1996;3:995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
- 52.Spolar RS, Livingstone JR, Record MT., Jr. Use of liquid hydrocarbon and amide transfer data to estimate contributions to thermodynamic functions of protein folding from the removal of nonpolar and polar surface from water. Biochemistry. 1992;31:3947–3955. doi: 10.1021/bi00131a009. [DOI] [PubMed] [Google Scholar]
- 53.Madan B, Sharp KA. Hydration heat capacity of nucleic acid constituents determined from the random network model. Biophys. J. 2001;81:1881–1887. doi: 10.1016/S0006-3495(01)75839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalodimos CG, Boelens R, Kaptein R. Toward an integrated model of protein–DNA recognition as inferred from NMR studies on the Lac-repressor system. Chem. Rev. 2004;104:3567–3586. doi: 10.1021/cr0304065. [DOI] [PubMed] [Google Scholar]
- 55.DeLano WL. The PyMOL Molecular Graphics System, DeLano Scientific. San Carlos, CA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.