Two independent studies, one of them using a computational approach, identified CHRONO, a gene shown to modulate the activity of circadian transcription factors and alter circadian behavior in mice.
Abstract
Circadian rhythms are controlled by a system of negative and positive genetic feedback loops composed of clock genes. Although many genes have been implicated in these feedback loops, it is unclear whether our current list of clock genes is exhaustive. We have recently identified Chrono as a robustly cycling transcript through genome-wide profiling of BMAL1 binding on the E-box. Here, we explore the role of Chrono in cellular timekeeping. Remarkably, endogenous CHRONO occupancy around E-boxes shows a circadian oscillation antiphasic to BMAL1. Overexpression of Chrono leads to suppression of BMAL1–CLOCK activity in a histone deacetylase (HDAC) –dependent manner. In vivo loss-of-function studies of Chrono including Avp neuron-specific knockout (KO) mice display a longer circadian period of locomotor activity. Chrono KO also alters the expression of core clock genes and impairs the response of the circadian clock to stress. CHRONO forms a complex with the glucocorticoid receptor and mediates glucocorticoid response. Our comprehensive study spotlights a previously unrecognized clock component of an unsuspected negative circadian feedback loop that is independent of another negative regulator, Cry2, and that integrates behavioral stress and epigenetic control for efficient metabolic integration of the clock.
Author Summary
The circadian clock has a fundamental role in regulating biological temporal rhythms in organisms, and it is tightly controlled by a molecular circuit consisting of positive and negative regulatory feedback loops. Although many of the clock genes comprising this circuit have been identified, there are still some critical components missing. Here, we characterize a circadian gene renamed Chrono (Gm129) and show that it functions as a transcriptional repressor of the negative feedback loop in the mammalian clock. Chrono binds to the regulatory region of clock genes and its occupancy oscillates in a circadian manner. Chrono knockout and Avp-neuron-specific knockout mice display longer circadian periods and altered expression of core clock genes. We show that Chrono-mediated repression involves the suppression of BMAL1–CLOCK activity via an epigenetic mechanism and that it regulates metabolic pathways triggered by behavioral stress. Our study suggests that Chrono functions as a clock repressor and reveals the molecular mechanisms underlying its function.
Introduction
Circadian rhythms with a period of approximately 24 h endow organisms with the ability to adapt to changes of solar light following earth's rotation. The mammalian circadian clock system consists of inputs from light and feeding, a core pacemaker located in a paired nuclei, called suprachiasmatic nucleus (SCN), and outputs including, but not limited to, cycles of locomotor activity, sleep–awake, and hormonal secretion. Disturbance of the biological clock causes not only sleep rhythm disruptions but also various pathological conditions such as cancer, metabolic, and psychiatric disorders [1]–[5].
The clock gene, period, was first identified in fly [6]–[8] and later in various organisms [9]. The molecular mechanism of circadian transcription was then found to be based on interconnected transcription–translation feedback loops (TTFL), conserved from prokaryotes to humans [10]–[12]. In mammals, the complex of positive elements BMAL–CLOCK (NPAS2) activates PER and CRY that repress their own transcription to form a negative feedback loop. An accessory feedback loop involves ROR and REV–ERBα, which regulate BMAL1 transcription positively and negatively, respectively, whereas BMAL1 activates REV–ERBα expression. More members of the circadian clock components have been identified [13]–[16]. Because of the complexity of circadian timekeeping, mathematical modeling has emerged as an important tool to understand data and make novel predictions [17]–[19]. In particular, a recently published mathematical model reproduces much of the known data on circadian timekeeping (e.g., mutant phenotypes) and correctly predicts the pharmacological manipulation of circadian rhythms [20],[21].
Although it is widely believed that the major components of the mammalian circadian clock have been identified, the search for additional clock components continues. In our previous study [22] and in others [23],[24], the systematic screening by using ChIP (Chromatin immunoprecipitation)–Chip and ChIP-seq revealed several uncharacterized genome-wide BMAL1 targets. Among strong BMAL1 binding sites, only one novel gene, Gm129, was found besides known clock and clock-controlled genes such as Per1, Per2, Cry1, Cry2, Dbp, and Tef. Gm129 expression shows a robust circadian rhythm antiphasic to Bmal1. In light of its circadian expression, Gm129, now renamed Chrono (ChIP-derived Repressor of Network Oscillator), appeared to be a core clock gene.
Here, we study the functional role of Chrono in the circadian clock. CHRONO binds to the promoters of clock genes and functions as a negative regulatory component of the circadian clock. In vivo loss-of-function of Chrono including an Avp neuron-specific knockout (KO) mouse model displays a longer circadian period of locomotor activity. We demonstrate that Chrono is a core-clock component similar to Cry2, although its repression mechanism operates through an independent pathway. In silico study using a modified Kim–Forger model predicts that the recently identified residual rhythmicity in the Cry1, Cry2 double KO [25], is dependent on Chrono. Remarkably, Chrono is involved in glucocorticoid receptor (GR)–mediated metabolic physiology. We conclude that Chrono is part of the negative feedback loop of the mammalian circadian clock and a potential link between the clock and stress metabolism.
Results
ChIP-Seq Identifies a Novel Clock Gene Regulated by BMAL1
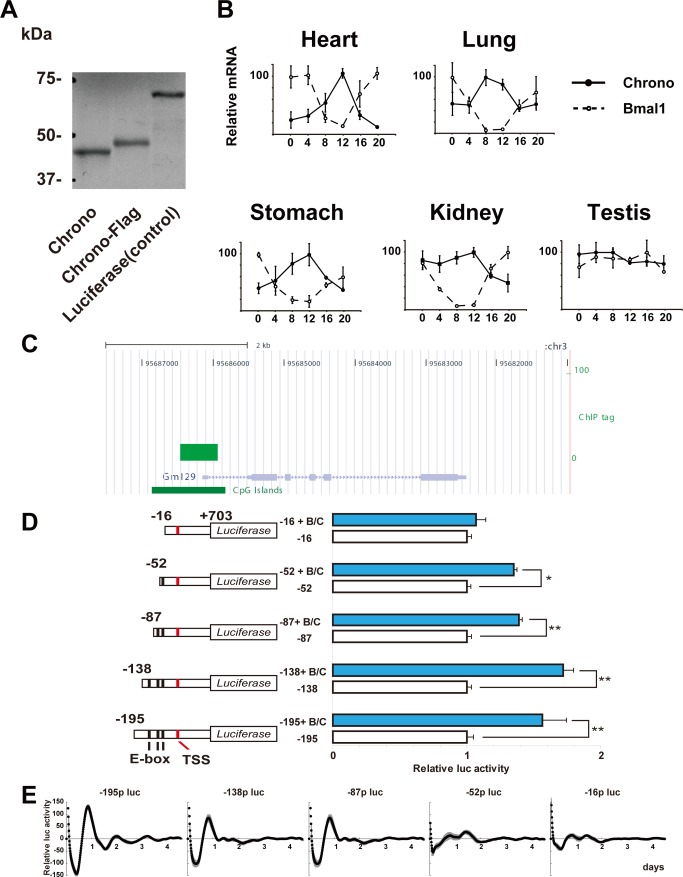
Our previous ChIP-based genome-wide analyses using a core clock transcription factor BMAL1 identified hundreds of target molecules [22]. Among these targets, Gm129, now called Chrono, was one of the groups with the strongest binding including core clock proteins PER and CRY. Another ChIP-seq experiment using in vivo brain samples also identified Chrono as a BMAL1 target. Chrono exists only in mammals, is well conserved among mammals (Figure S1), and consists of 375 amino acids with no functional domains. To examine whether Chrono encodes a polypeptide, we performed an in vitro translation experiment. Bands of approximately 45 kDa (CHRONO) and 46 kDa (CHRONO–FLAG) were observed as an in vitro translation product (Figure 1A).
Figure 1. CHRONO.
(A) Cell-free synthesis of CHRONO (GM129). Autoradiogram of in vitro translated CHRONO, CHRONO–FLAG, and luciferase (control). Protein was synthesized in the presence of [35S]methionine and resolved on a 10% SDS-PAGE gel. (B) Temporal mRNA expression of Chrono (solid lines with black circles) and Bmal1 (dotted lines with white circles) in mouse peripheral tissues. The abscissa represents time in CT and the ordinate the mRNA amounts. The relative levels of mRNA were normalized to the corresponding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels. The maximum mRNA amount was set to 100. Plots and error bars represent mean ± S.E.M of triplicate samples. (C) BMAL1 ChIP-seq tag enrichments in the whole brain sample were located on the Chrono promoter region in the UCSC genome browser view. (D) The effects of overexpression of BMAL1 and CLOCK (B/C) on the Chrono promoter modification were evaluated by using a luciferase assay. The left scheme shows each construct on a black line indicating the position of the E-box element and a red line indicating the TSS. The basal transcriptional activity of each Chrono promoter was set to 1. Horizontal bars represent means ± S.E.M. of four samples (*p<0.005, **p<0.0005, Student's t test). (E) E-boxes of the Chrono promoter required for transcriptional oscillation. The cell-culture-based bioluminescent reporter assay was performed with the each construct in (D). The abscissa indicates the day in culture, and the ordinate the relative bioluminescence intensity (kcpm, 1,000 photon counts per minute). Shaded area indicates ± S.E.M of four samples.
Chrono Transcripts Display Robust Circadian Expression
Our previous study showed robust circadian oscillation of Chrono mRNA in the mouse SCN and liver [22]. We further examined its expression in five different mouse peripheral tissues (heart, lung, stomach, kidney, and testis) by quantitative RT-PCR. After entrainment of mice housed for 2 wk under a 12–12 h light–dark (LD) cycle, samples were collected every 4 h starting at circadian time (CT) 0 (n = 3 at each time point) in the third dark–dark (DD) cycle. The temporal expression of Chrono transcripts in all tested tissues except testis displayed robust circadian rhythms peaking at approximately CT 12 (Figure 1B), which were antiphasic to Bmal1. This result supports that Chrono encodes a component of the circadian clock loops [26]. The ChIP-seq experiment using brain samples revealed that BMAL1 strongly binds to CpG islands on the Chrono promoter in vivo (Figure 1C). Studying differently sized Chrono promoter constructs (−195, −138, −87, −52, and −16 bp from the transcriptional start site (TSS) of Chrono/PGL3B) showed that the closest E-box to the TSS is necessary to generate circadian oscillation of Chrono in NIH3T3 cells and that all of the three E-boxes contribute to robust circadian rhythms (Figure 1D and E). These results suggest that BMAL1 strongly binds to the E-boxes on the Chrono promoter and regulates circadian expression of Chrono, making Chrono a novel clock gene.
We next asked whether CHRONO is also expressed rhythmically at the protein level. We prepared liver samples at CT 2, 8, 14, and 20. We raised a specific antibody against the CHRONO protein. CHRONO showed circadian rhythm antiphasic to BMAL1 as in the mRNA expression (Figure S2A and C). This oscillation was observed in both the mouse CHRONO antibody we generated and the human C1orf51 antibody (ab106120, Abcam) (Figure S2B).
CHRONO Forms a Complex with Other Clock Components
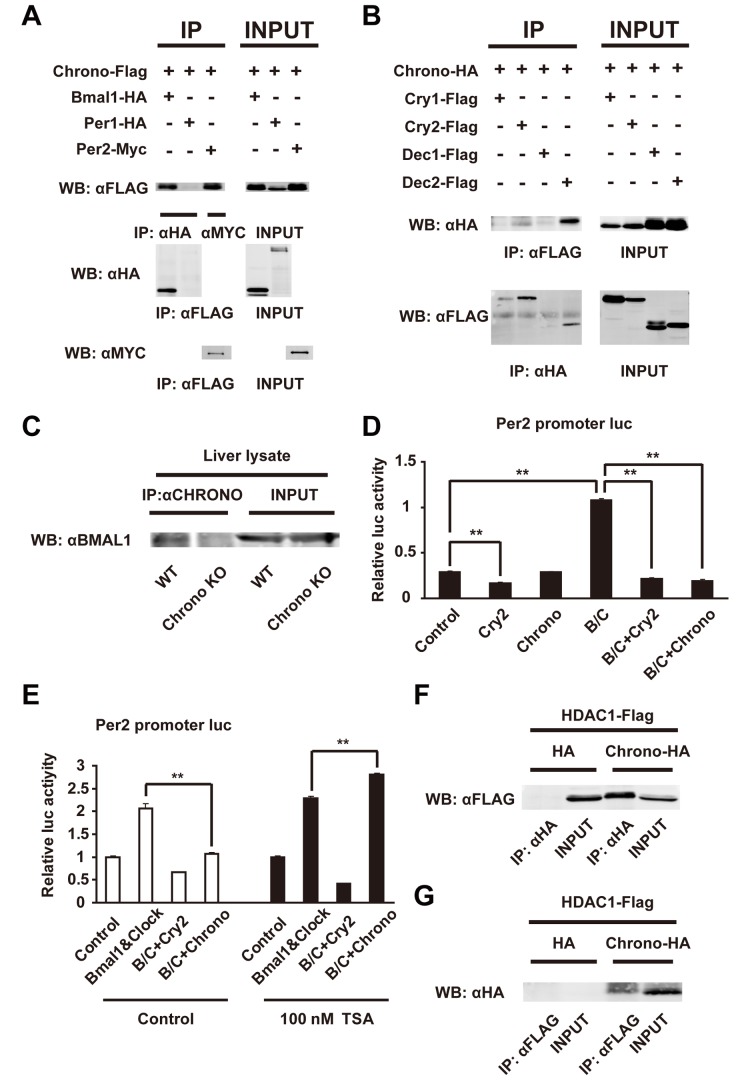
Because CHRONO showed a similar rhythmic expression profile to other core clock proteins, we asked if CHRONO binds directly with clock proteins. Various clock proteins with tags were expressed in COS7 cells and the expression was assessed by immunoprecipitation (IP) and blotting with anti-tag antibodies. CHRONO bound to BMAL1, PER2, CRY2, and DEC2 but not to PER1, CRY1, and DEC1 (Figure 2A and B and Figure S2D). Among these interactions, we asked if CHRONO–BMAL1 binding occurs endogenously in vivo. We observed a band of BMAL1 in the CHRONO antibody IP from mouse liver lysate that was absent in the IP from Chrono-deficient mouse liver (Figure 2C). These results suggest that CHRONO endogenously binds to BMAL1 in vivo.
Figure 2. CHRONO's interaction and the role of CHRONO as a transcriptional repressor.
(A and B) Immunoblots show expression of proteins in COS7 cells transfected with the indicated plasmids after IP with antibodies. INPUT indicates immunoblotting results of total cell lysates. (C) Endogenous BMAL1 and CHRONO interaction in the WT and Chrono KO mice liver. (D) Effects of Chrono expression on the Per2 promoter luciferase activity. Overexpressed CHRONO repressed transactivation by exogenous expression of BMAL1 and CLOCK (B/C) in the Per2 promoter in a similar way to Cry2. Bars represent means ± S.E.M. of four samples (**p<0.001, Student's t test). (E) An HDAC inhibitor (100 nM TSA) restored the Chrono-derived repression of transactivation with BMAL1 and CLOCK (B/C) on the Per2 promoter. The basal transcriptional activity of Per2 promoter in each experiment was set to 1. Data are means ± S.E.M. of four samples (**p<0.001, Student's t test). (F and G) CHRONO and HDAC1 interaction in vitro. Co-IP assays of COS7 cells using HA-tagged CHRONO and FLAG-tagged HDAC1 as indicated. HA protein was used as negative control.
Chrono Is Involved in HDAC-Dependent Transcriptional Repression
Next, we asked how Chrono is involved in circadian transcription. The luciferase activity of the Per2 promoter (∼−2,817+110 bp from TSS/PGL3B) in NIH3T3 cells was repressed by co-expression with Cry2 and Dec2 (Figures 2D and S3A). The basic transcription activity of Per2 was increased by Bmal1 and Clock co-expression, and this activation was repressed by Chrono as well as Cry2. This repression was also seen in the Chrono promoter (−1,333 bp from TSS/PGL3B) (Figure S3B). Moreover, overexpression of Chrono reduced the transcriptional amplitude of expression on the Dbp promoter, just as Cry2 (Figure S3C and D). These results suggest that Chrono functions as a negative element of circadian transcription, similar to Cry2.
Histone modification by histone deacetylase (HDAC) is one of the mechanisms of transcriptional regulation [27]. HDAC is often recruited during the transcriptional repression process. We hypothesized that CHRONO is in a repressor complex that includes CRY2. To investigate the potential role of HDAC in Chrono-mediated transcriptional repression, we treated cells with trichostatin A (TSA), an HDAC inhibitor, in the reporter assay. Chrono, as well as Cry2, repressed the enhanced luciferase activity of the Per2 promoter. However, in the presence of TSA, Chrono did not repress activity but rather enhanced activity, whereas Cry2 did not change its repression (Figure 2E). To confirm the involvement of HDAC in Chrono-mediated transcriptional repression, we investigated the interaction of CHRONO with HDAC1. We expressed and showed co-IP of CHRONO–HA and HDAC1–FLAG in COS7 cells, indicating that CHRONO is bound with HDAC1 (Figure 2F and G).
CHRONO Rhythmically Binds the E-Boxes on Circadian Gene Promoters
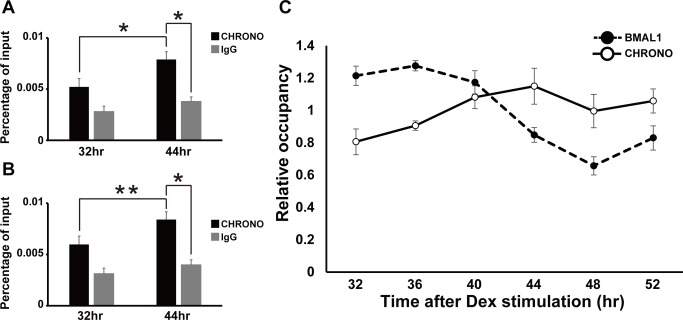
We then asked if endogenous CHRONO participates in clock function as a core clock component. A ChIP experiment with the CHRONO antibody in NIH3T3 cells after induction with dexamethasone showed endogenous binding of CHRONO to the E-boxes on Per2 (Figure 3A) and Dbp (Figure 3B) promoters. The levels of chromatin occupancy around the E-boxes on Per2 and Dbp were significantly different between 32 h and 44 h after dexamethasone stimulation (Figure 3A and B, *p<0.05, **p<0.01). Moreover, endogenous CHRONO occupancy showed circadian oscillation antiphasic to BMAL1 (Figures 3C and S3G). These results strongly suggest that Chrono behaves as an auto-regulated clock component.
Figure 3. Endogenous CHRONO rhythmically binds to the E-box on circadian gene promoter.
(A and B) ChIP analysis for CHRONO and IgG (negative control). The CHRONO occupancies at the endogenous E-boxes of Per2 (A) and Dbp (B) promoters were detected in the two different NIH3T3 cells 32 or 44 h after induction with dexamethasone. The graphs showed relative real-time PCR values compared to input. Data are means ± S.E.M. of three samples (*p<0.05, **p<0.01, Student's t test). (C) ChIP abundance of CHRONO and BMAL1 at the Per2 promoter E-box in NIH3T3 cells after 100 nM dexamethasone stimulation. The graph showed relative real-time PCR values. The data were plotted as percentages relative to the average (1.0). Data are means ± S.E.M. of 3–4 samples.
Chrono KO Mice Have a Lengthened Circadian Period
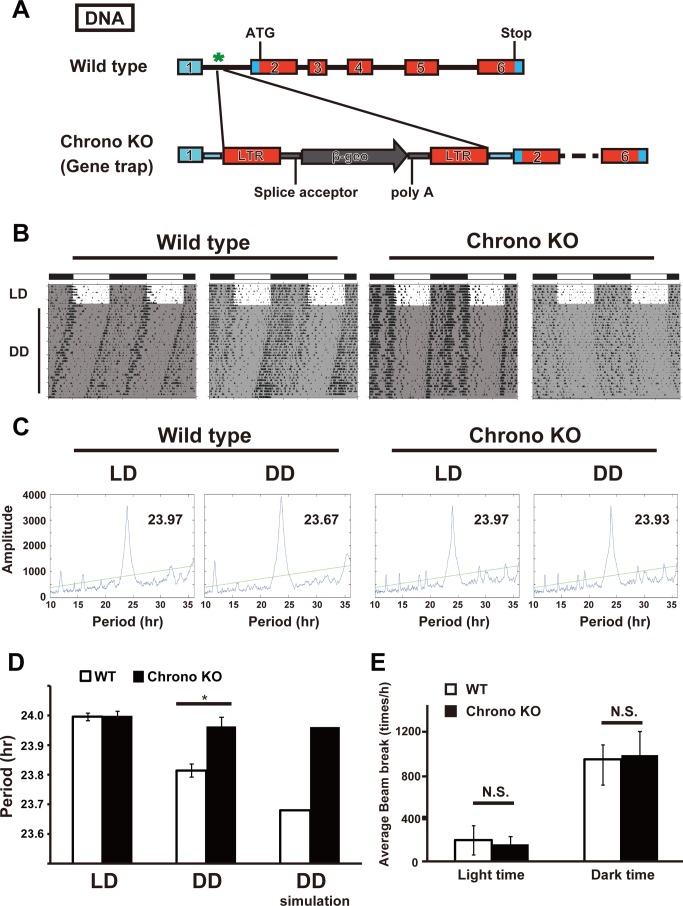
To evaluate the physiological and circadian clock function of Chrono in vivo, we generated Chrono KO mice by using a gene trap method (Figures 4A and S4). After entrainment in the 12–12 h LD condition, the locomotor activity rhythm under DD was recorded. In DD the average circadian period length of wild-type (WT) mice was 23.81±0.08 h (mean ± standard deviation, n = 13), whereas that of Chrono-deficient mice was significantly longer (23.96±0.11 h, n = 12) (*p<0.001; Student's t test) (Figure 4B–D). To logically confirm that the behavioral Chrono KO phenotype is an outcome of the observed biochemical and in vitro characteristics of Chrono (Figure 2), we adopt a mathematical modeling approach. We used a recently developed mathematical model of mammalian circadian clock (Kim–Forger model) because this model successfully reproduced and predicted the circadian period change in response to mutations of clock genes (e.g., Per1/2, Cry1/2, Bmal1, Clock, and etc.) and pharmacological inhibition of kinase (e.g., CK1δ/ε) [20],[21]. The model is extended to include biochemical mechanisms of Chrono, such as binding with other clock components (Figure 2A and B), transcriptional repression by Chrono (Figure 2D–G), and rhythms of Chrono (Figure 1B) (see details in Text S1). Although CHRONO acts similarly to CRY2, we also incorporate key differences, including their very different mRNA time profiles (Figure S5) and the fact that CHRONO does not bind PER1. When the transcription of Chrono is inhibited in the model, the model predicts that Chrono KO lengthens the period (Figure 4D, right), which indicates that the biochemical mechanisms we have identified for Chrono-mediated repression of BMAL1–CLOCK (Figure 2D) are expected to cause the Chrono KO phenotypes. There was no difference of basal locomotor activity between WT and Chrono KO mice during the light phase and dark phase after 1 wk in LD (p>0.5; Student's t test) (Figure 4E). We also examined how Chrono KO mice responded to shifts in the LD cycle. When the lighting cycle was advanced 6 h, both WT and Chrono KO mice re-entrained progressively over 10 d, and there were no difference between the genotypes (Figure S6A and B). Moreover, no induction of Chrono mRNA in the SCN was observed after light stimulation for 30 min (Figure S6C).
Figure 4. Circadian phenotypes of Chrono KO mice.
(A) Genomic structure of a portion of the mouse Chrono gene. Exons are indicated by heavy lines. Gene trapped (KO) mice expressed different transcripts and translated products from WT mice. (B) Representative actograms of WT and Chrono KO mice. Animals were maintained under 12–12 h LD cycles initially and transferred to DD as indicated. Shaded area indicates dark phase. (C) Representative chi-squared periodograms of WT and Chrono KO locomotor activities for 1 wk at LD and DD. (D) Comparison of free-running period estimated for WT and Chrono KO littermates. Pairwise comparisons found no significant differences of circadian periods between WT and KO under LD (left). However, a significantly longer free-running period was observed in Chrono KO mice compared to WT under DD (middle). The computer simulation based on our biochemical data reproduces these phenotypes under DD (right). Free-running periods under DD are 23.96±0.11 h for Chrono KO mice (n = 12, male) and 23.81±0.08 h for WT siblings (n = 13, male) (mean period ± S.D.; *p<0.001, Student's t test). (E) Basal locomotor activity. Amounts of activities during light phase (left) and dark phase (right) for 1 wk in LD show no significant differences. N = 13 for WT and n = 12 for Chrono KO.
Chrono Acts as a Transcriptional Repression Independent of Cry2
Because Chrono is a putative repressor in the circadian clock mechanism, we next asked whether the expression of clock genes is altered after the deletion of Chrono in mice. In mouse embryonic fibroblasts (MEFs) derived from Chrono KO mice (Figure S7A), Per2 expression was increased at 48 h and 52 h after dexamethasone stimulation (Figure S7B), compared with WT MEFs. In the liver derived from Chrono KO mice (Figure S7C), Per3, Cry1, Dbp, and Rev–erbs were increased at CT12 (*p<0.05, **p<0.01, Student's t test), consistent with the idea that Chrono is involved with negative feedback regulation of the core clock component via E-box. The similar trend was observed in the Chrono-deficient SCN (Figure S7D). Moreover, Acetyl-Histone H3 occupancy on Per2 and Dbp promoters was enhanced in Chrono KO MEFs compared to WT (Figure S7E and F; **p<0.01, Student's t test). This result suggests that Chrono changed the epigenetic modification of cells with the expression status.
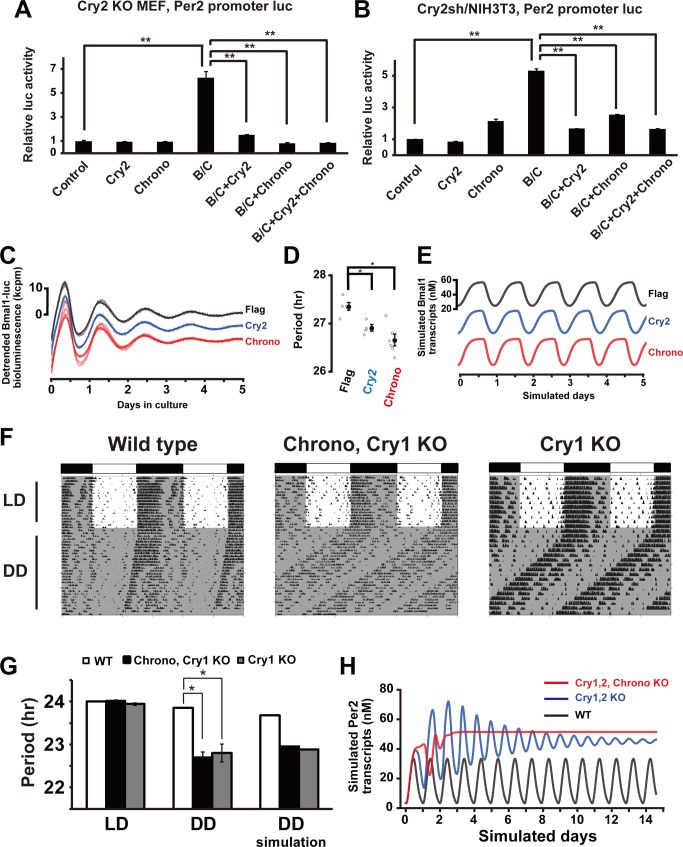
Chrono's role in circadian timekeeping seems to mimic that of Cry2 both in vivo and in cells. Because CHRONO interacts with CRY2 and DEC2, we hypothesized that CHRONO represses BMAL1–CLOCK activity only when partnered with CRY2 or DEC2 to form a complex. To confirm this hypothesis, we performed a luciferase reporter assay using Cry2 KO MEFs (Figure 5A) or Cry2 knockdown (Cry2sh) NIH3T3 cells (Figures 5B and S8) and Dec2 KO MEFs (Figure S3E). The BMAL1–CLOCK complex induced Per2 transcription in all cells and Chrono repressed the BMAL1–CLOCK activity in all cells, including those without Cry2 and Dec2, indicating that CHRONO acts as a transcriptional repressor via an independent pathway from CRY2 and DEC2. Moreover, we established a stable NIH3T3 cell line with Bmal1 promoter luciferase and Cry2 knockdown. The rhythm of fibroblasts with overexpression of Flag as a control showed a robust circadian oscillation with a lengthened period (27.29±0.06 h; mean ± S.E.M.; n = 4) due to Cry2 knockdown. As expected, constitutive overexpression of Cry2 driven by the CMV promoter partially restored the period to a shorter value (26.91±0.07 h; n = 4; *p<0.01, Welch's t test) in the Cry2-sh/NIH3T3 cells. Consistent with the idea that Chrono mimics the Cry2 phenotype, overexpression of Chrono similarly shortened the period (26.66±0.15 h; n = 5; *p<0.01, Welch's t test) of oscillation in the Cry2-sh/NIH3T3 cells (Figure 5C and D). These relative modulations in periods are predicted in parallel by the extended Kim–Forger model (Figure 5E), under the simulated experimental conditions of Cry2 knockdown to 30% of its original value and/or constitutive overexpression of either Cry2 or Chrono. These results indicate that the circadian phenotypes of Chrono are similar to Cry2, although the repression mechanisms operate via different pathways.
Figure 5. CHRONO acts as a transcriptional repressor through an independent pathway.
(A and B) Cry2 KO MEFs (A) and Cry2sh/NIH3T3 cells (B) were transfected with indicated combinations of expression vectors. Chrono inhibited BMAL1–CLOCK (B/C) complex-induced transcriptional activity in the mPer2 promoter. The effects of overexpression of BMAL1, CLOCK, CRY2, and CHRONO proteins on Per2 transcription were evaluated by using luciferase assays. The basal transcription activity of Per2 promoter was set to 1. Data are means ± S.E.M. of triplicate samples. **p<0.001, Student's t test. (C and D) Bioluminescence rhythms (C) and periods (D) of Cry2sh–Bmal1 promoter luc/NIH3T3 cells transfected with indicated expression vectors. All detrended traces (6–7 samples) from each set of experiments (Flag, Cry2, and Chrono overexpression) are superposed. The baselines are intentionally separated for each case for ease of view. The abscissa indicates the day in culture, and the ordinate the relative bioluminescence intensity (kcpm, 1,000 photon counts per minute). Cry2sh/NIH3T3 cells prolonged the period, and Chrono recovered the period length similar to Cry2. *p<0.005. (E) Model prediction of Bmal1 transcript expression in the Cry2 knockdown (Flag, black), under additional constitutive overexpression of Cry2 (blue), and Chrono (red). The baselines are separated intentionally for ease of view. (F) Raw actograms of WT, Chrono, Cry1 double KO and Cry1 KO mice and comparison of free-running period estimates. A significantly shorter free-running period was observed in Chrono, Cry1 double KO and Cry1 KO mice compared to WT in DD (G, middle), in line with the prediction through in silico simulation (G, right). Chrono, Cry1 double KO mice did not show arrhythmicity in DD. *p<0.05, Student's t test. (H) Model prediction of Per2 mRNA time courses in Cry1, Cry2 double KO (blue) and Cry1, Cry2, Chrono triple KO (red) compared to WT (black). The amounts of KO transcripts are set to 0 from the beginning of the simulated day 0, and dynamics before reaching steady state is illustrated.
Given the phenotypic similarities between Chrono and Cry2, we postulated that Chrono could overtake the role of Cry2 under KO conditions. To test this idea, we generated Chrono, Cry1 double KO mice by mating Chrono KO with Cry1 KO mice. The Cry1, Cry2 double KO mouse shows a circadian arrhythmicity in DD [28], and we expected a Chrono, Cry1 double KO would exhibit similar arrhythmicity. However, the circadian locomotor activity persisted under DD. In DD condition, Cry1 KO mice showed a shorter period than WT, similar to Cry1, Chrono double KO mice (Figure 5F). There was no significant difference of period between Cry1 KO and Cry1, Chrono double KO mice (Figure 5G). These results are predicted by our mathematical model, which shows short period rhythms under the double Chrono, Cry1 KO (22.95 h) and Cry1 single KO (22.89 h), respectively (Figure 5G, right). The model also predicted that the Chrono KO does not compromise the amplitude of Per2 mRNA rhythms (Figure 5H). Interestingly, the model predicts initially oscillating but damping Per2 rhythms in Cry1, Cry2 double KO (Figure 5H, blue). However, the triple, Chrono, Cry1, Cry2 KO completely removed any residual oscillations (Figure 5H, red). These results led us to conclude that, although CHRONO acts as a repressor of the BMAL1–CLOCK complex just as CRY2, the mechanism of action of CHRONO is independent of CRY2 and that CHRONO may be required for rhythmicity in certain mutant backgrounds.
Chrono Mediates Clock Repression in the Glucocorticoid Response
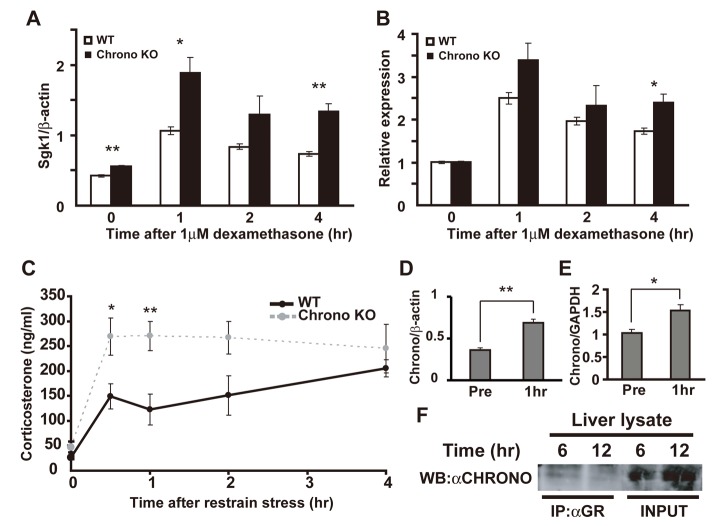
The Cryptochromes (Cry1 and Cry2) have been reported to mediate rhythmic repression of the GR [29]. Because Chrono behaves as a transcriptional repressor with an effect similar to Cry2, we next verified the physiological role of the Chrono KO against stress responses. We first examined the expression of serum/glucocorticoid-regulated kinase 1 (Sgk1) after dexamethasone stimulation. We found that Sgk1 mRNA was up-regulated in Chrono KO MEFs when compared to WT MEFs after 1-h and 4-h exposures to dexamethasone (Figure 6A, *p<0.05, **p<0.01, Student's t test). The relative expression of Sgk1 was also activated by 4-h exposure to dexamethasone (Figure 6B; *p<0.05, Student's t test). Serum corticosterone levels in vivo were also robustly increased in Chrono KO when compared with WT mice after 0.5 h and 1 h of restraint stress (Figure 6C; *p<0.05; one-way ANOVA followed by Tukey–Kramer's multiple comparisons test, as follows, *p<0.05 versus 0.5 h time after restraint stress; **p<0.01 versus 1 h time after restraint stress). In a WT background, Chrono mRNA was up-regulated in MEFs (Figure 6D; **p<0.01, Student's t test) and hypothalamus (Figure 6E; *p<0.05, Student's t test) after 1 h of exposure to dexamethasone and restraint stress, respectively, whereas Cry2 mRNA was not significantly increased under restraint stress in both WT and Chrono KO hypothalamus as well as MEFs (Figure S9). Furthermore, the IP-Western experiment indicated CHRONO endogenously interacted with GR in vivo (Figure 6F). These results suggest that Chrono is up-regulated by stress-dependent behavior and contributes to repressive function in the glucocorticoid response.
Figure 6. Chrono contributes to the repressive function in glucocorticoid response.
(A) Expression of Sgk1 in MEFs from WT and Chrono KO mice after treatment with dexamethasone for 1–4 h. The relative level of Sgk1 mRNA was normalized to the corresponding β-actin mRNA. Data are means ± S.E.M. of triplicate samples (*p<0.05, **p<0.01, Student's t test). (B) The relative Sgk1 expression level was normalized to the pretreatment (0 h) level. (C) Serum corticosterone level at 0, 0.5, 1, 2, and 4 h after restraint stress. Results are the means ± S.E.M. Asterisks indicate the existence of significant differences between the time points means within each group by one-way ANOVA followed by Tukey–Kramer's multiple comparisons test, as follows: *p<0.05 versus 0.5 h time after restraint stress; **p<0.01 versus 1 h time after restraint stress. (D) Expression of Chrono in WT MEFs between pre– and post–1-h treatment with dexamethasone. **p<0.01, Student's t test. (E) Expression of Chrono in hypothalamus between pre– and post–1-h restraint stress. *p<0.05, Student's t test. (D and E) The relative level of Chrono mRNA was normalized to the corresponding β-actin and GAPDH mRNA. (F) Endogenous CHRONO and GR interaction in the WT mouse liver at ZT 6 and 12.
Chrono in the SCN Plays a Central Role in Behavioral Rhythms
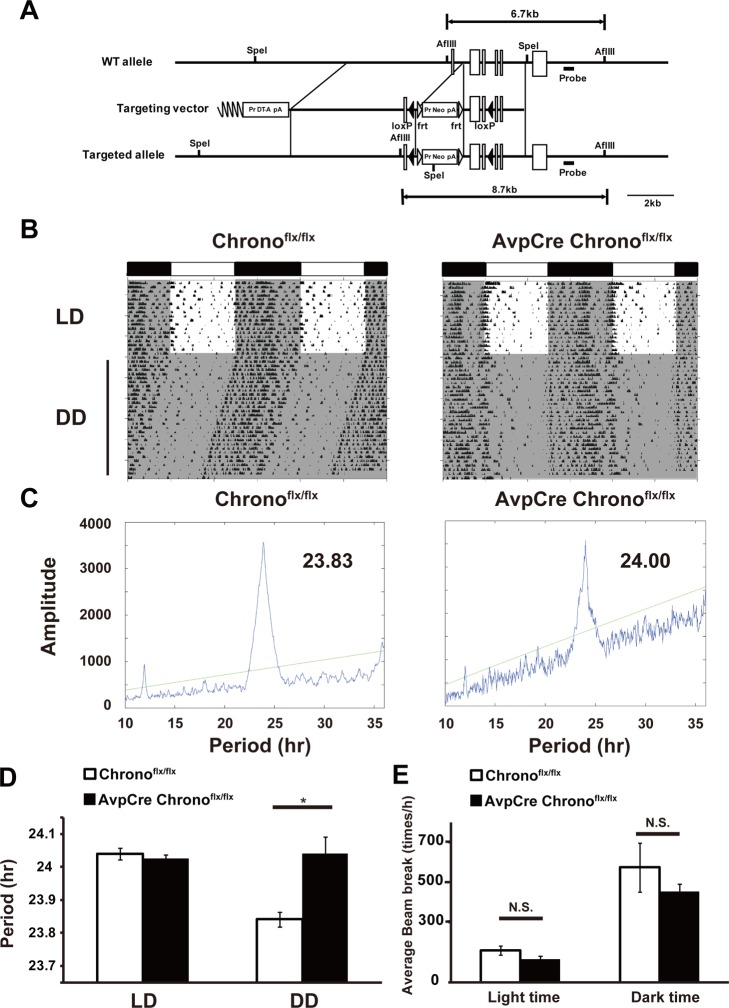
To identify the role of Chrono in the SCN, we generated mice carrying a conditional Chrono KO allele using the Cre-loxP system (Figures 7A and S10). Avp-Cre mice express Cre specifically in arginine vasopressin (AVP) neurons (GENSAT project) [30]. AVP is the most abundant neuropeptide in the hypothalamus and specifically in the SCN [31],[32]; therefore, Avp-Cre Chrono flx/flx mice can be considered as SCN-targeted Chrono KO mice. The average locomotor activity rhythm of Avp-Cre Chrono flx/flx male mice (24.04±0.12 h, n = 6) was significantly longer than that of Chrono flx/flx littermates (23.84±0.06 h, n = 8) (*p<0.01, Student's t test) (Figure 7B–D). The basal locomotor activity was measured during the light phase and the dark phase after 1 wk in LD by the infrared beam breaking. The activity amount of the Avp-Cre Chrono flx/flx mouse was not altered compared with Chrono flx/flx mouse (Figure 7E). These results demonstrate that Chrono expression in the Avp neurons plays a central role in behavioral rhythms.
Figure 7. Chrono in the Avp neurons plays a central role in behavioral rhythms.
(A) The targeting strategy is illustrated with the structure of the WT Chrono allele, the targeting vector, the floxed allele Chrono flx. Chrono exons are shown as open boxes; 5′ genomic DNA, intronic sequences, and 3′ genomic DNA as solid lines. The Pr Neo pA and the Pr DT-A pA cassettes are indicated as open boxes. The loxP sequences are shown as triangles. (B) Representative actograms of Chrono flx/flx and AvpCre Chrono flx/flx mice. Animals were maintained on 12–12 h LD cycles initially and transferred to DD. Shaded area indicates dark phase. (C) Representative chi-squared periodograms of Chrono flx/flx and AvpCre Chrono flx/flx locomotor activities for 1 wk under DD. (D) Comparison of free-running periods estimated for Chrono flx/flx and AvpCre Chrono flx/flx mice. Pairwise comparisons indicated that there were no significant differences in circadian periods under LD compared with the Chrono flx/flx littermates. However, a significantly longer free-running period was observed in AvpCre Chrono flx/flx mice compared with the Chrono flx/flx littermates in DD. AvpCre Chrono flx/flx mice (24.04±0.12 h, n = 6, male) and Chrono flx/flx siblings (23.84±0.06 h, n = 8, male) in DD (mean period ± S.D.; *p<0.01, Student's t test). (E) Basal locomotor activity. Amounts of activities during the light phase (left) and the dark phase (right) for 1 wk in LD show no significant differences. n = 8 for Chrono flx/flx and n = 6 for AvpCre Chrono flx/flx.
Discussion
In this study, we characterized a gene called Chrono that serves as a component in the negative arm of the core mammalian circadian clock. Indeed, based on our genomic analysis of circadian promoters, CHRONO appears to be one of the last remaining components of the clock. The most interesting feature of CHRONO is its regulation by epigenetic control and behavioral stress. These findings place CHRONO in a central position to couple stress metabolism to clock regulation.
Our results suggest that CHRONO operates as a repressor of the core circadian feedback loop through the recruitment of HDAC. The repressive effects of CHRONO were also seen in Cry2 KO MEFs and Dec2 KO MEFs, which shows that CHRONO repression does not require cooperative interactions with CRY2 or DEC2 [33],[34]. It has been reported that the recruitment of histone-modifying enzymes is regulated in a circadian manner [35]–[39] and some complexes are observed in transcriptional repression as in PER and SIN3–HDAC [14]. HDAC1 rhythmically bound to the Per2 promoter (Figure S3F), and this result was consistent with a recent report [40]. Acetyl-Histone H3 occupancy around E-boxes was enhanced in Chrono KO MEFs compared to WT (Figure S7E and F), suggesting that Chrono also has the potential to form complexes with other histone-modifying enzymes. However, HAT (histone acetyltransferase) activity by CLOCK was not affected by CHRONO in vitro experiments (Figure S11). Further mechanistic study is required. CHRONO endogenously binds to the E-boxes of circadian genes with circadian rhythmicity, suggesting that Chrono is a core circadian repressor that operates as an auto-regulated component of the clock.
The Chrono KO mouse showed a lengthened circadian period similar to the Cry2 KO mouse [28]. Overexpression of Chrono restored the period of Bmal1-luc oscillations in Cry2 knockdown cells in a manner comparable to Cry2 overexpression. On the other hand, the Chrono, Cry1 double KO mouse did not show arrhythmic behavior as seen in the Cry1, Cry2 double KO mouse [28]. Together, these results suggest that Chrono plays a similar but independent role from Cry2. A conditional Chrono KO mouse driven by the Avp promoter (AvpCre Chrono flx/flx) also showed a lengthened circadian period. This points to Chrono's central role in the core clock and that its expression in Avp neurons is critical for the determination of behavioral circadian period. It also demonstrates that the AvpCre system (GENSAT line number QZ20_CRE) can potentially prove useful in dissecting the output pathway of the SCN that may perform coding by internally distributed periods [41].
The role of Chrono was also tested in silico by modeling mechanisms of Chrono-mediated repression of BMAL1–CLOCK (Figure 2D) within the most comprehensive and realistic mathematical model of the mammalian circadian clock [20]. The extended model successfully predicted that the reduced (or enhanced) Chrono expression results in a lengthened (or shortened) period (Figures 4D and 5E), matching the phenotypes of Chrono KO or overexpression (Figures 4D and 5D). This indicates that the proposed biochemical mechanisms of Chrono-mediated repression of BMAL1–CLOCK (Figure 2D) are consistent with the overall phenotypes observed when Chrono levels are changed.
Chrono is likely to be a physiological response-dependent regulator. In response to dexamethasone application, Sgk1 was up-regulated in Chrono KO MEFs when compared with WT MEFs. In vivo serum corticosterone levels were also increased in the Chrono KO mouse when compared with the WT mouse under restraint stress and CHRONO itself interacted with the GR. Along with the observation that Chrono mRNA expression was induced by the stress response in MEFs and hypothalamus, we conclude that Chrono is a potential repressor activated by behavioral stress and can couple the clock with the hypothalamic–pituitary–adrenal (HPA) axis. Our previous results showed acute physical stress also elevated Per1 mRNA through a glucocorticoid-responsive element [42]. Further experimental work is needed to reveal the detailed molecular interactions between the circadian clock and the HPA axis.
Our discovery of the role of Chrono opens up many possibilities for future work. Our mathematical modeling raises the interesting possibility that the Per2 oscillations from Cry1, Cry2 double KO neonatal SCN [25] is mediated by Chrono. In further studies, it would be interesting to see if the Cry1, Cry2, Chrono triple KO eliminates the ability to oscillate in neonatal SCNs as predicted by our model (Figure 5H). Further work should also examine the role of Chrono in linking the HPA to the circadian clock. Taken together, we conclude that Chrono is a novel circadian clock gene that acts as a repressor in the circadian system and modulates physiology.
Materials and Methods
Ethics Statement
All protocols of animal experiments followed in the present study were approved by the Animal Research Committee of Hiroshima University and Animal Care and Use Committees of the RIKEN Brain Science Institute.
Cell Culture
NIH3T3, COS7 cells and MEFs were maintained in Dulbecco's Modified Eagle Medium (DMEM; Nacalai Tesque, Kyoto, Japan) supplemented with 10% fetal bovine serum (FBS) and penicillin–streptomycin antibiotics at 37°C and 5% CO2.
Chromatin IP
NIH3T3 and MEF cells were stimulated with 100 nM dexamethasone containing medium at each time point. Cells were fixed in 1× PBS containing 0.5% formaldehyde. Glycine was added to a final concentration of 0.125 M, and the incubation was continued for an additional 15 min. After washing the samples with ice-cold phosphate-buffered saline, the samples were homogenized in 1 mL of ice-cold homogenize buffer (5 mM PIPES [pH 8.0], 85 mM KCl, 0.5% NP-40, and protease inhibitors cocktail) and centrifuged (15,000× g, 4°C, 5 min). The pellets were suspended in nucleus lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1% SDS, protease inhibitors) and sonicated 20 times for 30 s each time at intervals of 60 s with a Bioruptor (Diagenode, Inc.) or sonicated 10 times for 10 s each time at intervals of 50 s with a MICROSON (Misonix, Inc.) for brain samples. The samples were centrifuged at 15,000 rpm at 4°C for 5 min. Supernatants were diluted 10-fold in ChIP dilution buffer (50 mM Tris-HCl [pH 8.0], 167 mM NaCl, 1.1% Triton X-100, 0.11% sodium deoxycholate, protease inhibitor).
Chromatin IP Sequencing (ChIP-seq)
Whole brain samples from mice (C57BL/6J) were used for ChIP-seq. BMAL1-bound DNA was purified by SDS-PAGE to obtain 150–200 bp fragments and sequenced on an Illumina GA sequencer at the Research Center of Research Institute for Radiation Biology and Medicine (RiRBM), Hiroshima University. We generated 15,000–20,000 clusters per “tile,” and 26 cycles of the sequencing reactions were performed according to the manufacturer's instructions. The identification of each DNA fragments was performed using Genome Studio software (Illumina Inc.).
ChIP-PCR
CHRONO, BMAL1, HDAC1 (ab7028, Abcam), Acetyl-Histone H3 (06-599, Millipore), and IgG-bound DNA were used for quantitative real-time reverse-transcription PCR (RT-PCR). The primers were designed for amplifying the E-box-like regions in Per2 and Dbp promoters.
In Vitro Real-Time Oscillation Monitoring System (IV-ROMS)
NIH3T3 cultures at the concentration of 1×105 cells in Opti-MEM (Gibco) supplemented with 10% FBS in a 35 mm dish were transfected with the desired plasmids by using the Lipofectamine reagent (Invitrogen). The medium was exchanged 24 h after transfection with 100 nM dexamethasone containing medium, and 2 h later this medium was replaced with Opti-MEM supplemented with 1% FBS and 0.1 mM luciferin–10 mM HEPES (pH 7.2). Bioluminescence was measured by using the IV-ROMS (Hamamatsu Photonics) as described previously [26],[43].
Luciferase Assay
NIH3T3 cells and MEFs were cultured and transfected with the desired plasmids by using Lipofectamine 2000 (Invitrogen) or Nucleofection (Lonza). Cells were harvested 24 h after transfection, and cell lysates were prepared and then used in the dual luciferase assay system (Promega). For exogenous expression, we transfected cells with pcDNA3 driven by ubiquitous cytomegalovirus (CMV) promoter.
Western Blotting and IP
Rabbit antibody against HA-tag and mouse antibodies against Flag-tag and Myc-tag were subjected to Western blot according to the manufacturer's protocol. For IP, COS7 cells transfected with the desired plasmids by using Lipofectamine 2000 (Invitrogen) were lysed in TNE buffer with protease inhibitor. Following the standard protocol, lysates were precleared with Dynabeads Protein G (Invitrogen) and then immunoprecipitated with rabbit anti-HA antibodies (Cell signaling), mouse anti-Flag antibodies (Sigma), or mouse anti–c-Myc antibodies (Sigma). After washing three times, the precipitates were resuspended in the 2× SDS-PAGE sample buffer, boiled for 5 min, and run on a 10% SDS-PAGE gel followed by Western blot analysis. Immunoreactive bands were detected by ODYSSEY Infrared Imaging System (LI-COR). These experiments were independently performed three times. The intensity of the band (BMAL1 and CHRONO rpar; was calculated by using ODYSSEY Infrared Imaging System.
Animal Care and Behavioral Analysis
Circadian rhythms of locomotor activity were analyzed as previous described [43]. Each mouse was individually housed for 2 wk in 12–12 h LD cycles, and then for 4 wk in constant darkness (DD). Locomotor activity was monitored with a locomotor activity recording apparatus (Biotex, Kyoto, Japan) that measures events of infrared beam breaking in 1-min bins. The data from the first week under DD were used to estimate the period of circadian locomotor activities of WT and Chrono KO mice. For RNA sampling, dissected tissues were immediately frozen in liquid nitrogen and stored at −80°C until processing.
Methods for Light Treatment
The experiment was performed as described previously [44]. Male ICR mice (SLC, Japan) were exposed to an incandescent light stimulus (1,000 lux, 30 min) at CT16 in the second DD cycle. Animals were sacrificed 60 min after the initiation of the light exposure. Total RNA was prepared from six pooled pairs of SCN at each time point using PicoPure RNA Isolation kit (Applied Biosystems).
In Vitro Translation
CHRONO protein was synthesized using the TNT T7 Coupled Reticulocyte Lysate System (Promega). We added 2 µg of template DNA (Chrono/pcDNA3 or Chrono–Flag/pcDNA3) to an aliquot of the TNT Quick Master Mix and incubated it in a 50 µl reaction volume for 60–90 min at 30°C. Synthesized proteins were detected by 10 mCi/ml (specific activity, 30 TBq/mmol) [35S]methionine, and resolved on SDS-PAGE (10%) gels using one-fifth of each translation reaction product mixed with an equal volume of sample buffer (15% glycerol, 5% β-mercaptoethanol, 4.5% SDS, 100 mM Tris-Cl, pH 6.8, 0.03% bromophenol blue). Gels were fixed, dried, and exposed to Hyper-film (Amersham) for 16 h to 2 d. The molecular masses (in kDa) of the translated proteins were derived using standard curves generated from protein size standards (Bio-Rad).
Quantitative RT-PCR
Each quantitative real-time RT-PCR was performed using the ABI Prism 7900HT sequence detection system as described previously [42],[45]. The PCR primers were designed with the Primer Express software (Applied Biosystems). The reaction was first incubated at 50°C for 2 min and then at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min.
Corticosterone Level Under the Restraint Stress
Blood samples were collected by tail bleeding at time points of 0, 0.5, 1, 2, and 4 h after restraint stress. Corticosterone in mouse serum was measured using YK240 corticosterone enzyme immunoassay (EIA) kit (Yanaihara Institute).
RNAi Experiment
BLOCK-iT Lentiviral RNAi System (Invitrogen) was used for RNAi experiments. NIH3T3 cells at the density of 5×104 were infected with a lentiviral vector and cultured. We selected stably transfected cells with zeocin. The shRNA/NIH3T3 cells were infected with Bmal1 promoter-driven luciferase lentiviral vector and selected for stable expression with blasticidin.
Antibodies
Antibody against BMAL1 was generated as described previously [22]. Purified glutathione S-transferase (GST) –Gm129 N-terminal (amino acids 1 to 187) protein was produced as a recombinant protein in the competent cells BL21 (DE3) (Stratagene). After removing GST by using GSTrap FF and PreScision Protease (GE Healthcare), the produced antigen was used to immunize rabbits. The antiserum was subjected to affinity purification using Affi gel 10 (Bio-Rad) conjugated with the antigen. The anti-CHRONO antibody recognized its target protein in immunochemical analysis (Figure S4D).
Chrono (Gm129) Gene Trap Mice
C57BL/6 gene trap ES cell clone IST11761C7 was an embryonic stem cell provided by the gene trap method [46]. Long terminal repeat (LTR) –splice acceptor–βgeo–polyA–LTR sequence was inserted between exons 1 and 2 in one allele of the Chrono (Gm129) gene (Figure 4A) (TIGM, Texas A&M Institute for Genomic Medicine). In substitution for mRNA producing CHRONO protein, mRNA producing fusion protein of the neomycin-resistant gene product and β-galactosidase (B-geo) was transcribed from this variation allele. We generated chimeric mice from C57BL/6 gene trap ES cell clone IST11761C7, mated the chimeric mice with WT C57BL/6 (+/+), and subsequently obtained heterozygous KO mice (+/−). Ultimately, we obtained homozygous KO (−/−) mice by mating heterozygous KO mice. Absence of Chrono mRNA and CHRONO protein was confirmed by PCR, RT-PCR, and Western blotting (Figure S4B–D).
mRNA Expression of Core Circadian Genes in Chrono KO MEFs and Liver
WT and Chrono KO MEFs were stimulated with 100 nM dexamethasone containing medium. After 32 h, mRNA were extracted by TRI reagent (Molecular Research Center, Inc.) every 4 h. Quantitative real-time RT-PCR was described above. Adult WT and Chrono KO mice were exposed to 2 wk of LD cycles and then kept in complete darkness as a continuation of the dark phase of the last LD cycle. mRNA expression was examined CT12 and next CT0 from the third DD cycle.
Generation of Chrono Conditional KO Mice
The Chrono conditional KO mice (Accession No. CDB0913K; http://www.cdb.riken.jp/arg/mutant%20mice%20list.html) were generated as described (http://www.cdb.riken.jp/arg/Methods.html). To generate a targeting vector, genomic fragments of the Chrono locus were obtained from the RP23-385O12 BAC clone (BACPAC Resources). A 950 bp region containing exons 2 and 3 of the Chrono gene was flanked by loxP sites (Figure 7A). Targeted ES clones were microinjected into ICR eight-cell stage embryos, and injected embryos were transferred into pseudopregnant ICR females. The resulting chimeras were bred with C57BL/6 mice, and heterozygous offspring were identified by PCR. Primers for 5′ loxP were used—forward E1F (5′-CAGACAGTGAAGAAGCTGCATA-3′) and reverse loxR2 (5′-CAGACTGCCTTGGGAAAAGC-3′)—yielding no product for WT allele and 603 bp products for the targeted allele, respectively. Primers for 3′ loxP were used—forward loxF1 (5′-GGCATGGGCTATTCTGTTTG-3′) and reverse loxR1 (5′-TTGAGGGAAACAGCAGAGGT-3′)—yielding 121 bp products for WT allele and 179 bp products for the targeted allele, respectively (Figure S10A and C).
In Silico Evaluation of Chrono in the Oscillator Network
We incorporated Chrono into a recently developed mathematical model of the mammalian circadian clock [20] based on our biochemical findings. We assumed that the mRNA degradation rate of Chrono is the same as Per2, as the Chrono mRNA time course is similar to Per2 mRNA [42]. We further assumed the time course of PER2–CHRONO nuclear entry is similar to the PER–CRY complexes. However, as found in our experiments, the CHRONO protein binds to the PER2 protein but not to PER1 protein (Figure 2A) and that the CHRONO protein interacts with BMAL1–CLOCK to inhibit its E-box activation on Per1, Per2, Cry1, Cry2, and Rev–erbs promoters. In the model, we did not consider binding between CHRONO and CRY2, as BMAL1–CLOCK repression by CHRONO did not depend on the presence of CRY2 (Figure 5). In total, 38 variables were newly added to the Kim–Forger model, which account for all the possible complexes involving CHRONO (details of computer simulation are described in Text S1, Tables S1, S2, S3, and Appendix S1). All the simulations were performed with Mathematica 8.0 (Wolfram Research).
Supporting Information
Sequence conservation of Chrono . The protein sequence alignment of Chrono across species was performed with Homologene database (NCBI). Chrono is highly conserved in mammals.
(TIF)
Characterization of CHRONO. (A–C) CHRONO protein expression showed circadian rhythm antiphasic to BMAL1 in the liver. We prepared liver samples at CT 2, 8, 14, and 20. Each time point has four or five samples, which were dissolved with RIPA buffer. The x-axis represents time in CT and y-axis protein amounts. The relative levels of protein were normalized to the β-ACTIN protein levels. The maximum protein amount was set to 100. (D) A schematic model of CHRONO interaction. CHRONO interacts with BMAL1, PER2, CRY2, and DEC2 (red line), but not PER1, CRY1, and DEC1 (see Figure 2A and B).
(TIF)
Chrono represses transcriptional activity. (A) Effects of Chrono expression on the Per2 promoter luciferase activities. Chrono repressed the Per2 transactivation by BMAL1 and CLOCK (B/C) overexpression with the potency similar to Dec2. The bar plots represent means ± S.E.M. of four samples (*p<0.05, Student's t test). (B) Effects of Chrono expression on the Chrono promoter luciferase activity. Overexpressed CHRONO repressed transactivation by exogenous expression of BMAL1 and CLOCK (B/C) in Chrono promoter, same as Cry2. Bars represent means ± S.E.M. of four samples (*p<0.05, **p<0.001, Student's t test). (C) Effects of Chrono and Cry2 expression on the Dbp promoter luciferase activities. Chrono repressed the Dbp promoter activity with the potency similar to Cry2. The abscissa indicates the day in culture, and the ordinate the relative bioluminescence intensity (kcpm, 1,000 photon counts per minute). The first amplitude (D) was significantly decreased with overexpression of Chrono compared to control. The bar plots indicate the mean ± S.E.M (shaded area) of eight samples. **p<0.0001, Student's t test. (E) Dec2 KO MEFs were transfected with combinations of expression vectors as indicated. Chrono inhibited BMAL1–CLOCK complex-induced transcriptional activity on the mPer2 promoter. The effects of overexpression of BMAL1, CLOCK, DEC2, and CHRONO proteins on Per2 transcription were evaluated by measuring bioluminescence from luciferase activities. The basal transcription level of the Per2 promoter was set to 1. The bar plots indicate the mean ± S.E.M of triplicate samples. *p<0.001, Student's t test. (F) ChIP analyses for HDAC1 and IgG (negative control). The HDAC1 occupancies at the endogenous E-box of the Per2 promoter were detected in the WT MEF cells at 28, 36, 44, and 52 h after induction with dexamethasone. The graph showed relative real-time PCR values. The maximum value of WT was set to 100. Data are means ± S.E.M. of three samples. (F) ChIP analysis for BMAL1 and IgG (negative control). The BMAL1 occupancy at the endogenous E-box of Per2 promoter was detected in NIH3T3 cells after 100 nM dexamethasone stimulation. The graph showed relative real-time PCR values. The data were plotted as percentages relative to the input DNA. Data are means ± S.E.M. of 3–4 samples.
(TIF)
Construction of Chrono KO mice. (A) The targeting strategy for PCR genotyping. An LTR–splice acceptor–βgeo–polyA–LTR sequence is inserted between exons 1 and 2 of Chrono allele (TIGM, Texas A&M Institute for Genomic Medicine). Primer locations are schematically displayed in (A). (B) PCR genotyping of DNA extracted from mouse tails of KO (Chrono −/−), heterozygous (Chrono +/−), WT (Chrono +/+) offspring. (C) RT-PCR analysis of Chrono expression in the hypothalamus. (D) Western blot analysis of Chrono expression in the liver of KO (Chrono −/−) and WT (Chrono +/+).
(TIF)
The simulated time courses of Chrono mRNA and Cry2 mRNA in the mathematical model. The amplitude and phase of Chrono mRNA and Cry2 mRNA are very different, matching experimental data [22],[47]. That is, the amplitude of the Chrono mRNA rhythm is much larger than that of Cry2 mRNA, and the phase of the Chrono mRNA rhythm is more advanced than that of Cry2 mRNA.
(TIF)
Characterization of Chrono KO and light response of Chrono in vivo . (A) Representative actograms from WT and Chrono KO mice that were subjected first to LD cycles, followed by a 6-h jet-lag light phase advance. Shaded areas indicate the dark phase. (B) Re-entrainment traces from an average of WT (red), Chrono KO (green), and merged (right). (C) Although a 30 min light pulse (1,000 lux) delivered from CT16.0 to CT16.5 induced Per1 mRNA expression (*p<0.05, Student's t test), Chrono expression was not induced.
(TIF)
Characterization of Chrono KO and Chrono may change the epigenetic modification of cells via Acetyl-Histone H3. Analysis of mRNA of core clock genes in WT and Chrono KO MEFs. Temporal mRNA expression of Chrono (A) and Per2 (B) in WT and Chrono KO MEFs. The abscissa represents time after dexamethasone stimulation and the ordinate the mRNA amounts. (C) mRNA expressions of circadian genes in WT and Chrono KO liver at CT12 and CT0. The relative levels of mRNA were normalized to the corresponding GAPDH mRNA levels. Plots and error bars represent mean ± S.E.M. of four samples. *p<0.05, **p<0.01, Student's t test. (D) Expression patterns of circadian genes in the SCN of WT and Chrono KO mice. The SCN samples from four or five mice were mixed at each time point. Solid lines with white circles and dotted lines with black circles represent WT and Chrono KO, respectively. The relative levels of each mRNA are normalized to the corresponding GAPDH RNA level. (E and F) ChIP analysis for Acetyl-Histone H3. The Acetyl-Histone H3 occupancies at the endogenous E-boxes of Per2 (E) and Dbp (F) promoters were detected in WT and Chrono KO MEFs at 52 h after induction with dexamethasone. The data were plotted as percentages relative to the input DNA. Data are means ± S.E.M. of five samples.
(TIF)
RT-PCR quantification of Cry2 mRNA abundance in NIH3T3 fibroblasts that stably express the Cry2 shRNA. LacZsh was used as a control. The relative levels of mRNA were normalized to the corresponding GAPDH mRNA levels. The maximum mRNA amount was set to 100. *p<0.01, Student's t test.
(TIF)
Expression of Cry2 in hypothalamus under restraint stress. Expression of Cry2 in hypothalamus under restraint stress (pre- and after 1 h) of WT (A) and Chrono KO mice (B). The relative level of Cry2 mRNA was normalized to the corresponding GAPDH mRNA. Expression of Cry2 in WT MEFs after Dex stimulation (C) (pre- and after 1 h).
(TIF)
Construction of Avp -specific KO of Chrono in mice. (A) The targeting strategy for PCR genotyping illustrated by the structures of the WT Chrono allele and the floxed allele Chrono flx. Chrono exons are represented as open boxes and 5′ genomic DNA, intronic sequences, and 3′ genomic DNA as solid lines. The Pr Neo pA and the Pr DT-A pA cassettes are shown as open boxes. The loxP sequences are indicated as solid triangles and the frt sequences as open triangles. (B) Southern blot verification of mouse tail DNA containing the Chrono flx allele. Extracted DNA samples were digested with AflIII and hybridized with the probe (Figure 7A). The WT (6.7 kb) and Chrono flx (8.7 kb) alleles were detected. (C) Mouse tail DNA from mice carrying WT and/or the Chronoflx allele were genotyped by PCR using primer sets E1F/loxR2 or loxF1/loxR1, as shown in (A). E1F/loxR2 amplifies a fragment of 603 bp only from the Chrono flx allele. loxF1/loxR1 amplifies fragments of 121 bp from the WT allele and 179 bp from the Chrono flx allele.
(TIF)
HAT activity in NIH3T3. HAT activity in NIH3T3 cells, which were transfected with the desired plasmids by using Lipofectamine 2000 (Invitrogen). After 24 h from transfection, the nuclear protein was extracted and the Histone acetyltransferase activity was measured by using HAT assay kits (ab65352, Abcam).
(TIF)
Newly added variables for mRNA dynamics of Chrono .
(DOCX)
Extended variables of protein complexes.
(DOCX)
Newly added/modified parameters for Chrono dynamics.
(DOCX)
Supplementary methods.
(DOCX)
Newly added and modified equations.
(DOCX)
Acknowledgments
We thank A. Kanai and all of the technicians of the Takumi laboratory for their technical assistance and C. Yokoyama for comments on the manuscript.
Abbreviations
- AVP
arginine vasopressin
- ChIP
chromatin immunoprecipitation
- CT
circadian time
- DD
dark–dark
- GR
glucocorticoid receptor
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- IP
immunoprecipitation
- KO
knockout
- LD
light–dark
- MEF
mouse embryonic fibroblast
- SCN
suprachiasmatic nucleus
- TSA
trichostatin A
- TTFL
transcription–translation feedback loop
- WT
wild–type
Funding Statement
Japan Society of Promotion of Science and Ministry of Education, Culture, Sports, Science, and Technology KAKENHI, Strategic International Coorperative Program and CREST, Japan Science and Technology Agency, Human Frontier Science Program (HFSP) grant RPG 24/2012, the Takeda Science Foundation, Mitsui Life Social Welfare Foundation, Sony Corporation, and Nippon Boehringer Ingelheim Co. NSF grant DMS-0931642 to the Mathematical Biosciences Institute (JKK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hastings MH, Reddy AB, Maywood ES (2003) A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci 4: 649–661. [DOI] [PubMed] [Google Scholar]
- 2. Eckel-Mahan K, Sassone-Corsi P (2013) Metabolism and the circadian clock converge. Physiol Rev 93: 107–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Albrecht U (2012) Timing to perfection: the biology of central and peripheral circadian clocks. Neuron 74: 246–260. [DOI] [PubMed] [Google Scholar]
- 4. Mohawk JA, Green CB, Takahashi JS (2012) Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35: 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Asher G, Schibler U (2011) Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab 13: 125–137. [DOI] [PubMed] [Google Scholar]
- 6. Bargiello TA, Jackson FR, Young MW (1984) Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature 312: 752–754. [DOI] [PubMed] [Google Scholar]
- 7. Reddy P, Zehring WA, Wheeler DA, Pirrotta V, Hadfield C, et al. (1984) Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell 38: 701–710. [DOI] [PubMed] [Google Scholar]
- 8. Zehring WA, Wheeler DA, Reddy P, Konopka RJ, Kyriacou CP, et al. (1984) P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell 39: 369–376. [DOI] [PubMed] [Google Scholar]
- 9. Zhang EE, Kay SA (2010) Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol 11: 764–776. [DOI] [PubMed] [Google Scholar]
- 10. Zheng X, Sehgal A (2012) Speed control: cogs and gears that drive the circadian clock. Trends Neurosci 35: 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown SA, Kowalska E, Dallmann R (2012) (Re)inventing the circadian feedback loop. Dev Cell 22: 477–487. [DOI] [PubMed] [Google Scholar]
- 12. Ukai-Tadenuma M, Kasukawa T, Ueda HR (2008) Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat Cell Biol 10: 1154–1163. [DOI] [PubMed] [Google Scholar]
- 13. Brown SA, Ripperger J, Kadener S, Fleury-Olela F, Vilbois F, et al. (2005) PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 308: 693–696. [DOI] [PubMed] [Google Scholar]
- 14. Duong HA, Robles MS, Knutti D, Weitz CJ (2011) A molecular mechanism for circadian clock negative feedback. Science 332: 1436–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Padmanabhan K, Robles MS, Westerling T, Weitz CJ (2012) Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science 337: 599–602. [DOI] [PubMed] [Google Scholar]
- 16. Robles MS, Boyault C, Knutti D, Padmanabhan K, Weitz CJ (2010) Identification of RACK1 and protein kinase Calpha as integral components of the mammalian circadian clock. Science 327: 463–466. [DOI] [PubMed] [Google Scholar]
- 17. Forger DB, Peskin CS (2003) A detailed predictive model of the mammalian circadian clock. Proc Natl Acad Sci U S A 100: 14806–14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gallego M, Eide EJ, Woolf MF, Virshup DM, Forger DB (2006) An opposite role for tau in circadian rhythms revealed by mathematical modeling. Proc Natl Acad Sci U S A 103: 10618–10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leloup JC, Goldbeter A (2003) Toward a detailed computational model for the mammalian circadian clock. Proc Natl Acad Sci U S A 100: 7051–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim JK, Forger DB (2012) A mechanism for robust circadian timekeeping via stoichiometric balance. Mol Syst Biol 8: 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim JK, Forger DB, Marconi M, Wood D, Doran A, et al. (2013) Modeling and validating chronic pharmacological manipulation of circadian rhythms. CPT Pharmacometrics Syst Pharmacol 2: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hatanaka F, Matsubara C, Myung J, Yoritaka T, Kamimura N, et al. (2010) Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol Cell Biol 30: 5636–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koike N, Yoo SH, Huang HC, Kumar V, Lee C, et al. (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338: 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, et al. (2011) Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol 9: e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ono D, Honma S, Honma K (2013) Cryptochromes are critical for the development of coherent circadian rhythms in the mouse suprachiasmatic nucleus. Nat Commun 4: 1666. [DOI] [PubMed] [Google Scholar]
- 26. Yamamoto T, Nakahata Y, Soma H, Akashi M, Mamine T, et al. (2004) Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol Biol 5: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bellet MM, Sassone-Corsi P (2010) Mammalian circadian clock and metabolism—the epigenetic link. J Cell Sci 123: 3837–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, et al. (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398: 627–630. [DOI] [PubMed] [Google Scholar]
- 29. Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, et al. (2011) Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480: 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, et al. (2003) A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425: 917–925. [DOI] [PubMed] [Google Scholar]
- 31. Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, et al. (1999) A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96: 57–68. [DOI] [PubMed] [Google Scholar]
- 32. Moore RY, Speh JC, Leak RK (2002) Suprachiasmatic nucleus organization. Cell Tissue Res 309: 89–98. [DOI] [PubMed] [Google Scholar]
- 33. Langmesser S, Tallone T, Bordon A, Rusconi S, Albrecht U (2008) Interaction of circadian clock proteins PER2 and CRY with BMAL1 and CLOCK. BMC Mol Biol 9: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fujimoto K, Hamaguchi H, Hashiba T, Nakamura T, Kawamoto T, et al. (2007) Transcriptional repression by the basic helix-loop-helix protein Dec2: multiple mechanisms through E-box elements. Int J Mol Med 19: 925–932. [PubMed] [Google Scholar]
- 35. Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, et al. (2008) The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134: 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, et al. (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134: 317–328. [DOI] [PubMed] [Google Scholar]
- 37. DiTacchio L, Le HD, Vollmers C, Hatori M, Witcher M, et al. (2011) Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science 333: 1881–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ripperger JA, Schibler U (2006) Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet 38: 369–374. [DOI] [PubMed] [Google Scholar]
- 39. Etchegaray JP, Lee C, Wade PA, Reppert SM (2003) Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421: 177–182. [DOI] [PubMed] [Google Scholar]
- 40. Duong HA, Weitz CJ (2014) Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat Struct Mol Biol 21: 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Myung J, Hong S, Hatanaka F, Nakajima Y, De Schutter E, et al. (2012) Period coding of Bmal1 oscillators in the suprachiasmatic nucleus. J Neurosci 32: 8900–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamamoto T, Nakahata Y, Tanaka M, Yoshida M, Soma H, et al. (2005) Acute physical stress elevates mouse period1 mRNA expression in mouse peripheral tissues via a glucocorticoid-responsive element. J Biol Chem 280: 42036–42043. [DOI] [PubMed] [Google Scholar]
- 43. Akashi M, Takumi T (2005) The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol 12: 441–448. [DOI] [PubMed] [Google Scholar]
- 44. Takumi T, Taguchi K, Miyake S, Sakakida Y, Takashima N, et al. (1998) A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. Embo J 17: 4753–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, et al. (2009) Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell 137: 1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zambrowicz BP, Abuin A, Ramirez-Solis R, Richter LJ, Piggott J, et al. (2003) Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci U S A 100: 14109–14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ueda HR, Hayashi S, Chen W, Sano M, Machida M, et al. (2005) System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet 37: 187–192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence conservation of Chrono . The protein sequence alignment of Chrono across species was performed with Homologene database (NCBI). Chrono is highly conserved in mammals.
(TIF)
Characterization of CHRONO. (A–C) CHRONO protein expression showed circadian rhythm antiphasic to BMAL1 in the liver. We prepared liver samples at CT 2, 8, 14, and 20. Each time point has four or five samples, which were dissolved with RIPA buffer. The x-axis represents time in CT and y-axis protein amounts. The relative levels of protein were normalized to the β-ACTIN protein levels. The maximum protein amount was set to 100. (D) A schematic model of CHRONO interaction. CHRONO interacts with BMAL1, PER2, CRY2, and DEC2 (red line), but not PER1, CRY1, and DEC1 (see Figure 2A and B).
(TIF)
Chrono represses transcriptional activity. (A) Effects of Chrono expression on the Per2 promoter luciferase activities. Chrono repressed the Per2 transactivation by BMAL1 and CLOCK (B/C) overexpression with the potency similar to Dec2. The bar plots represent means ± S.E.M. of four samples (*p<0.05, Student's t test). (B) Effects of Chrono expression on the Chrono promoter luciferase activity. Overexpressed CHRONO repressed transactivation by exogenous expression of BMAL1 and CLOCK (B/C) in Chrono promoter, same as Cry2. Bars represent means ± S.E.M. of four samples (*p<0.05, **p<0.001, Student's t test). (C) Effects of Chrono and Cry2 expression on the Dbp promoter luciferase activities. Chrono repressed the Dbp promoter activity with the potency similar to Cry2. The abscissa indicates the day in culture, and the ordinate the relative bioluminescence intensity (kcpm, 1,000 photon counts per minute). The first amplitude (D) was significantly decreased with overexpression of Chrono compared to control. The bar plots indicate the mean ± S.E.M (shaded area) of eight samples. **p<0.0001, Student's t test. (E) Dec2 KO MEFs were transfected with combinations of expression vectors as indicated. Chrono inhibited BMAL1–CLOCK complex-induced transcriptional activity on the mPer2 promoter. The effects of overexpression of BMAL1, CLOCK, DEC2, and CHRONO proteins on Per2 transcription were evaluated by measuring bioluminescence from luciferase activities. The basal transcription level of the Per2 promoter was set to 1. The bar plots indicate the mean ± S.E.M of triplicate samples. *p<0.001, Student's t test. (F) ChIP analyses for HDAC1 and IgG (negative control). The HDAC1 occupancies at the endogenous E-box of the Per2 promoter were detected in the WT MEF cells at 28, 36, 44, and 52 h after induction with dexamethasone. The graph showed relative real-time PCR values. The maximum value of WT was set to 100. Data are means ± S.E.M. of three samples. (F) ChIP analysis for BMAL1 and IgG (negative control). The BMAL1 occupancy at the endogenous E-box of Per2 promoter was detected in NIH3T3 cells after 100 nM dexamethasone stimulation. The graph showed relative real-time PCR values. The data were plotted as percentages relative to the input DNA. Data are means ± S.E.M. of 3–4 samples.
(TIF)
Construction of Chrono KO mice. (A) The targeting strategy for PCR genotyping. An LTR–splice acceptor–βgeo–polyA–LTR sequence is inserted between exons 1 and 2 of Chrono allele (TIGM, Texas A&M Institute for Genomic Medicine). Primer locations are schematically displayed in (A). (B) PCR genotyping of DNA extracted from mouse tails of KO (Chrono −/−), heterozygous (Chrono +/−), WT (Chrono +/+) offspring. (C) RT-PCR analysis of Chrono expression in the hypothalamus. (D) Western blot analysis of Chrono expression in the liver of KO (Chrono −/−) and WT (Chrono +/+).
(TIF)
The simulated time courses of Chrono mRNA and Cry2 mRNA in the mathematical model. The amplitude and phase of Chrono mRNA and Cry2 mRNA are very different, matching experimental data [22],[47]. That is, the amplitude of the Chrono mRNA rhythm is much larger than that of Cry2 mRNA, and the phase of the Chrono mRNA rhythm is more advanced than that of Cry2 mRNA.
(TIF)
Characterization of Chrono KO and light response of Chrono in vivo . (A) Representative actograms from WT and Chrono KO mice that were subjected first to LD cycles, followed by a 6-h jet-lag light phase advance. Shaded areas indicate the dark phase. (B) Re-entrainment traces from an average of WT (red), Chrono KO (green), and merged (right). (C) Although a 30 min light pulse (1,000 lux) delivered from CT16.0 to CT16.5 induced Per1 mRNA expression (*p<0.05, Student's t test), Chrono expression was not induced.
(TIF)
Characterization of Chrono KO and Chrono may change the epigenetic modification of cells via Acetyl-Histone H3. Analysis of mRNA of core clock genes in WT and Chrono KO MEFs. Temporal mRNA expression of Chrono (A) and Per2 (B) in WT and Chrono KO MEFs. The abscissa represents time after dexamethasone stimulation and the ordinate the mRNA amounts. (C) mRNA expressions of circadian genes in WT and Chrono KO liver at CT12 and CT0. The relative levels of mRNA were normalized to the corresponding GAPDH mRNA levels. Plots and error bars represent mean ± S.E.M. of four samples. *p<0.05, **p<0.01, Student's t test. (D) Expression patterns of circadian genes in the SCN of WT and Chrono KO mice. The SCN samples from four or five mice were mixed at each time point. Solid lines with white circles and dotted lines with black circles represent WT and Chrono KO, respectively. The relative levels of each mRNA are normalized to the corresponding GAPDH RNA level. (E and F) ChIP analysis for Acetyl-Histone H3. The Acetyl-Histone H3 occupancies at the endogenous E-boxes of Per2 (E) and Dbp (F) promoters were detected in WT and Chrono KO MEFs at 52 h after induction with dexamethasone. The data were plotted as percentages relative to the input DNA. Data are means ± S.E.M. of five samples.
(TIF)
RT-PCR quantification of Cry2 mRNA abundance in NIH3T3 fibroblasts that stably express the Cry2 shRNA. LacZsh was used as a control. The relative levels of mRNA were normalized to the corresponding GAPDH mRNA levels. The maximum mRNA amount was set to 100. *p<0.01, Student's t test.
(TIF)
Expression of Cry2 in hypothalamus under restraint stress. Expression of Cry2 in hypothalamus under restraint stress (pre- and after 1 h) of WT (A) and Chrono KO mice (B). The relative level of Cry2 mRNA was normalized to the corresponding GAPDH mRNA. Expression of Cry2 in WT MEFs after Dex stimulation (C) (pre- and after 1 h).
(TIF)
Construction of Avp -specific KO of Chrono in mice. (A) The targeting strategy for PCR genotyping illustrated by the structures of the WT Chrono allele and the floxed allele Chrono flx. Chrono exons are represented as open boxes and 5′ genomic DNA, intronic sequences, and 3′ genomic DNA as solid lines. The Pr Neo pA and the Pr DT-A pA cassettes are shown as open boxes. The loxP sequences are indicated as solid triangles and the frt sequences as open triangles. (B) Southern blot verification of mouse tail DNA containing the Chrono flx allele. Extracted DNA samples were digested with AflIII and hybridized with the probe (Figure 7A). The WT (6.7 kb) and Chrono flx (8.7 kb) alleles were detected. (C) Mouse tail DNA from mice carrying WT and/or the Chronoflx allele were genotyped by PCR using primer sets E1F/loxR2 or loxF1/loxR1, as shown in (A). E1F/loxR2 amplifies a fragment of 603 bp only from the Chrono flx allele. loxF1/loxR1 amplifies fragments of 121 bp from the WT allele and 179 bp from the Chrono flx allele.
(TIF)
HAT activity in NIH3T3. HAT activity in NIH3T3 cells, which were transfected with the desired plasmids by using Lipofectamine 2000 (Invitrogen). After 24 h from transfection, the nuclear protein was extracted and the Histone acetyltransferase activity was measured by using HAT assay kits (ab65352, Abcam).
(TIF)
Newly added variables for mRNA dynamics of Chrono .
(DOCX)
Extended variables of protein complexes.
(DOCX)
Newly added/modified parameters for Chrono dynamics.
(DOCX)
Supplementary methods.
(DOCX)
Newly added and modified equations.
(DOCX)