Abstract
Schizophrenia (SZ) and autism spectrum disorders (ASD) are highly heritable neuropsychiatric disorders, although environmental factors, such as maternal immune activation (MIA), play a role as well. Cytokines mediate the effects of MIA on neurogenesis and behavior in animal models. However, MIA stimulators can also induce a febrile reaction, which could have independent effects on neurogenesis through heat shock (HS)-regulated cellular stress pathways. However, this has not been well-studied. To help understand the role of fever in MIA, we used a recently described model of human brain development in which induced pluripotent stem cells (iPSCs) differentiate into 3-dimensional neuronal aggregates that resemble a first trimester telencephalon. RNA-seq was carried out on aggregates that were heat shocked at 39°C for 24 hours, along with their control partners maintained at 37°C. 186 genes showed significant differences in expression following HS (p<0.05), including known HS-inducible genes, as expected, as well as those coding for NGFR and a number of SZ and ASD candidates, including SMARCA2, DPP10, ARNT2, AHI1 and ZNF804A. The degree to which the expression of these genes decrease or increase during HS is similar to that found in copy loss and copy gain copy number variants (CNVs), although the effects of HS are likely to be transient. The dramatic effect on the expression of some SZ and ASD genes places HS, and perhaps other cellular stressors, into a common conceptual framework with disease-causing genetic variants. The findings also suggest that some candidate genes that are assumed to have a relatively limited impact on SZ and ASD pathogenesis based on a small number of positive genetic findings, such as SMARCA2 and ARNT2, may in fact have a much more substantial role in these disorders - as targets of common environmental stressors.
Introduction
Schizophrenia (SZ) and autism spectrum disorders (ASD) are among the most heritable neuropsychiatric disorders. An ever-expanding number of susceptibility genes is being identified through genome wide association studies (GWAS), copy number variant (CNV) analysis, and exome sequencing [1]–[15]. However, environmental factors also appear to play a role, especially maternal immune activation (MIA), which may be brought on by exposure to infectious diseases or autoimmune phenomena [16]–[30]. Supporting an infectious disease and/or autoimmune etiology in a subset of SZ patients is the association that has been found to markers in the HLA locus – one of the most robust GWAS findings in SZ genetics research (although non-immune effects of HLA antigens on brain development and neuronal function is a possible explanation for the association) [1], [14], [31]–[33].
The effects of MIA are mediated by a balance between proinflammatory and anti-inflammatory cytokines, such as interleukins 1β, 6, 10 and 13 [27], [34]–[36]. These cytokines are especially interesting in the context of neuropsychiatric disorders because they have well-established effects on neurogenesis and brain development, which could influence behavior in adult offspring. One model that is commonly used to test the effects of MIA is to treat pregnant animals with cytokines or agents that mimic exposure to infectious organisms, such as bacterial endotoxin (lipopolysaccharide; LPS), and polyinosinic: polycytidylic acid (poly I:C), after which the effects on behavior and neuronal function in the offspring are analyzed. For example, when pregnant mice are treated with poly I:C, altered prefrontal GABAergic gene expression occurs in their adult progeny [37]. Similarly, MIA in first trimester rhesus monkeys leads to offspring with increased repetitive behaviors and abnormal social interactions [38]. Prenatal exposure to low concentrations of poly I:C that do not lead to maternal symptoms or fetal death has also been found to cause impaired non-spatial memory and learning tasks in adult offspring, and decreased hippocampal reelin expression [39]. Similarly, prenatal exposure to poly I:C was shown to reduce the density of parvalbumin GABAergic interneurons in the CA1 region of the hippocampus, similar to that seen in the brains of patients with SZ [40]–[44]. Prenatal exposure to LPS has similar effects on brain development and long-term behavioral effects on offspring [42]–[47].
Although most studies examining the effect of MIA revolve around the effect of cytokines on neurogenesis and brain development, there are very few studies that have examined the effect of fever per se. One study that supports an effect is the finding that the risk of ASD in children born to mothers who experienced a febrile episode during pregnancy is attenuated by antipyretic medications [17]. In animal studies, prenatal exposure to LPS that was accompanied by a febrile reaction resulted in offspring with altered intrinsic excitability of CA1 pyramidal neurons [48]. In a large epidemiological study in Denmark, maternal influenza and febrile episodes were found to increase the risk of ASD, although the findings were not significant after multiple testing corrections were applied [16]. Interestingly, and perhaps somewhat paradoxically, there is also some suggestion that children with ASD have a transient improvement in symptoms following a febrile episode [49], [50]. Thus, there have been some interesting observations related to fever in SZ and ASD, but its potential role as a risk factor has not been well-studied.
As a first step towards understanding the potential effect of fever on the developing human brain, we are using a unique culture system developed by Mariani et al. in which iPSCs are manipulated with rostral neuralizing factors to produce 3-dimensional neuronal aggregates that model the developing first trimester telencephalon [51]. A modification of this culture system has recently been used to grow cortical structures and to model microcephaly [52]. We exposed 50 day old aggregates to HS (39°C for 24 hours) and analyzed transcripts genome-wide using RNA-seq. As expected, the expression level of a number of heat shock (HS) genes markedly increased. Interestingly, the expression of NGFR (nerve growth factor receptor) and several genes that have been implicated in the development of SZ, bipolar disorder (BD) and ASD were differentially expressed following HS.
Methods
Subjects and development of iPSCs from skin fibroblasts
All work involving iPSCs was approved by the Albert Einstein College of Medicine committee on clinical investigation. Participants in this study signed consent forms approved by The Albert Einstein College of Medicine Institution Review Board (IRB). Subjects were also recruited at the NIMH, Child Psychiatry Branch as part of an ongoing study on childhood onset schizophrenia directed by Dr. Judith Rapoport. Subjects in that study signed consents approved by the NIMH IRB. Consent was obtained by skilled members of the research teams who had received prior human subjects training. All lines used in this study were derived from healthy subjects who are serving as controls in ongoing studies in which iPSCs are being developed from patients with SZ who have 22q11.2 deletions. RNA-seq studies were carried out on 2 lines, which we designated as control 1 (C1) and control 2 (C2). C1 is an18 year old male and C2 is a 31 year old female. iPSC lines were generated from skin fibroblasts. In addition, we validated several genes of interest by quantitative real time PCR (qPCR; see below) using another control subject designated as C3, a 27 year old male. The reprogramming procedure is described in the Supporting Information file (Text S1).
Neuronal Differentiation
RNA-seq was carried out on neuronal aggregates as described by Mariani et al. with slight modification (see Text S1) [51]. For the HS experiment, a group of 49 day old aggregates was placed in an incubator set at 39°C for 24 hours, while control sets of aggregates were maintained at 37°C. The incubator conditions were otherwise unchanged (ambient O2, 5% CO2, 85% humidity). After detaching the aggregates, total cellular RNA was isolated using the miRNeasy Kit (Qiagen) according to the manufacturer' protocol. An additional DNAse1 digestion step was performed to ensure that the samples were not contaminated with genomic DNA.
RNA-seq
RNA was extracted from control and HS samples from day 50 mini-brain aggregates derived from the two iPSC lines (C1 and C2; HS1 and HS2). Paired end RNA-seq was carried on an Illumina HiSeq 2000. We obtained 101-bp mate-paired reads from DNA fragments of with an average size of 250-bp (standard deviation for the distribution of inner distances between mate pairs is approximately 100 bp). RNA-seq reads were aligned to the human genome (GRCh37/hg19) using the software TopHat (version 2.0.8) [53]. We counted the number of fragments mapped to each gene annotated in the GENCODE database (version 18) [54]. The category of transcripts is described at http://vega.sanger.ac.uk/info/about/gene_and_transcript_types.html. Transcript abundances were measured in Transcripts Per Million (TPM), which is calculated by multiplying the estimated fraction of transcripts made up by a given gene by 106 [55]. The measure is independent of the mean expressed transcript length and is thus more comparable across samples, so it is favored over another popular transcript measure, FPKM [55]. We used DESeq (an R package developed by Anders and Huber) to evaluate differential expression from the count data [56]. Specifically, DESeq models the variance in fragment counts across replicates using the negative binomial distribution and tests whether, for a given gene, the change in expression strength between the two experimental conditions is significantly large as compared to the variation within each replicate group. In the end, only genes with average TPMs greater than 1 across samples were considered for differential expression. RNA-seq data have been deposited at the Gene Expression Omnibus (GEO) repository (accession # GSE53667).
Quantitative real-time PCR (qPCR)
qPCR was carried out on reverse transcribed PCR. A detailed description and the primers used for this analysis can be found in Text S1.
Results
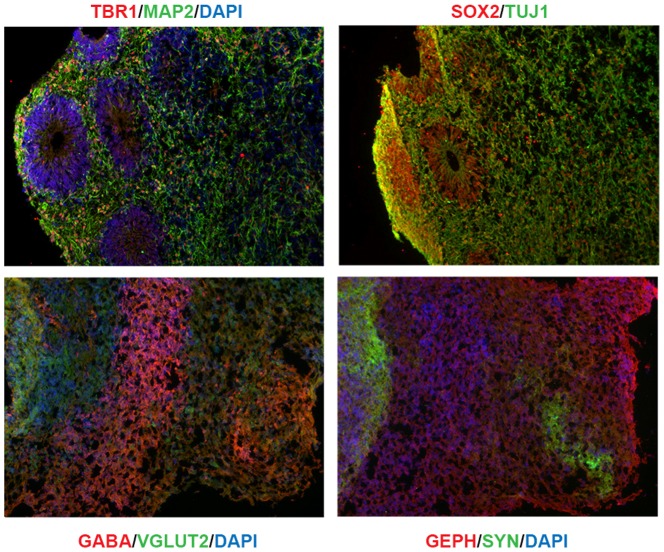
Neuronal aggregates were prepared from two control subjects (C1 and C2); an 18 year old male and 31 year old female, respectively. A representative aggregate is shown in Figure 1. Aggregates are composed of SOX2 positive, radial glial-containing structures surrounded by a field of neurons that are primarily GABAergic and glutamatergic. As seen in Figure 1, the aggregates express pre and postsynaptic markers (synaptophysin and gephryin, respectively). At day 49, while the control aggregates were maintained continuously at 37°C, another set of aggregates from each subject was exposed to HS (39°C for 24 hours; designated HS1 and HS2). RNA was then extracted from the control and HS samples on day 50, and subjected to RNA-seq. The number of RNA-seq reads and fraction of reads that mapped to the genome was similar for all samples (Table S1). In addition, the coefficient of variance was low (0.11 for the two controls; 0.19 for the two HS samples: Pearson correlation coefficients were 0.98 and 0.96, respectively), indicating high reproducibility of our RNA-seq data. The RNA-seq data show that the forebrain transcription factors, FOXP2, GLI2, LHX1, LHX2, POU3F2 and EMX2 are expressed, but not the hindbrain transcription factors HOXA1,HOXA2, HOXB1, HOXB2, and HOXB3 (Table S2). The aggregates express a fairly heterogeneous mix of neurotransmitter receptor genes, although GABAergic and glutamatergic receptors predominate; the glutamate transporter genes, SLC17A6 and SLC17A7, and the GABAergic transporter gene SLC6A1 are also expressed. The aggregates express only a few serotonin and dopamine receptor subtype genes (HTRA1, HTRA2 and HTR5A; DRD2 and DRD4), and several nicotinic cholinergic receptor subtypes (CHRNB1, CHRNA1, CHRNA4, CHRNA7, CHRNB2, CHRNB3 and CHRNB4). The dopamine transporter (SLC6A3), serotonin transporter (SLC6A4) and cholinergic transporter (SLC18A3) genes are not expressed, although TH (tyrosine hydroxylase), a dopaminergic marker is. There were no significant differences in expression of any neurotransmitter receptor or transporter gene in response to HS, with the exception of SLC17A7, as described below.
Figure 1. Neuronal aggregates, day 50.
Top panels: SOX2+ structures containing radial glial cells with surrounding field of neurons (MAP+, TUJ1+ cells). Bottom left panel: Neurons contain layers of predominantly GABAergic and glutamatergic neurons (GABA+ and VGLUT2+, respectively). Bottom right panel: The neurons express pre and post synaptic proteins (synaptophysin/SYN, gephyrin/GEPH, respectively).
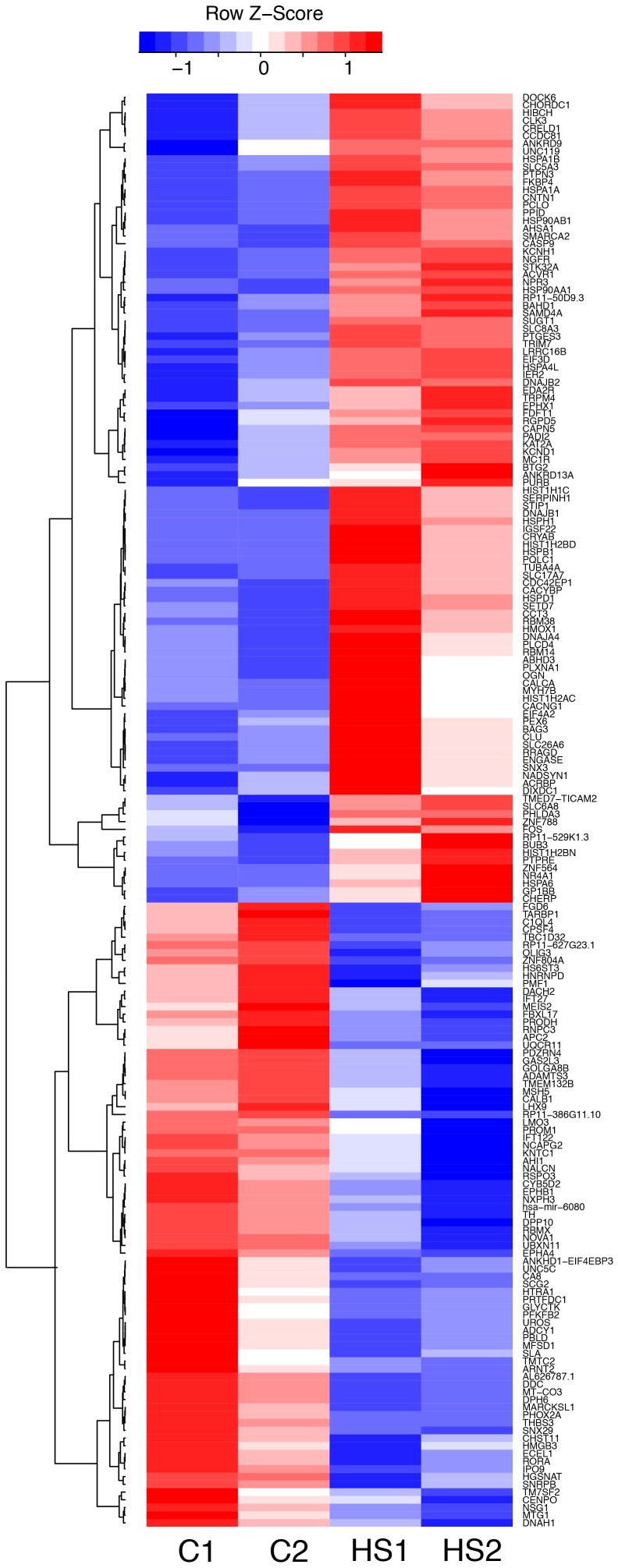
The expression level of 186 genes showed a nominally significant difference in expression following HS (p<0.05: 105 increased; 81 decreased), of which 12 achieved genome wide significance (q<0.05: all increased with HS) (Figure 2; Table S2). Among the 12 genes that increased most dramatically were 8 members of the HS gene family (HSPA1A, HSPA1B, HSP90AA1, HSP90AB1, HSPH1, HSPA6, HSPA4L, DNAJB1), and the HS protein chaperones CRYAB and FKBP4. In addition, several genes of interest with respect to neurogenesis and neuronal function were among the most HS-inducible genes, including SLC5A3 (sodium/myo-inositol co-transporter), which regulates brain inositol levels, and NGFR, which codes for the nerve growth factor (NGF) receptor.
Figure 2. Heat map showing relative expression of 186 genes that exhibited significant change in gene expression following heat shock at a nominally significant level (p<0.05: 105 increased in expression; 81 decreased).
Among the most down-regulated genes were several that code for proteins involved in dopamine transmission, including DDC (dopamine decarboxylase), and PHOX2A (paired mesoderm homeobox protein 2A), a transcription factor that regulates TH and DBH (dopamine beta hydroxylase) gene expression [57]. In addition, other dopaminergic genes - DBH, TH, and ADCY1 - were also down-regulated by HS. This is consistent with the finding that heat stress reduces TH immunoreactivity in striatal dopamine neurons in mice and impairs nigrostriatal dopaminergic neurons and motor function [58].
To identify which of the 186 genes could be the direct transcriptional targets of HS, we reanalyzed the ChIP-seq data that was collected by the ENCODE consortium for HSF1 in the HepG2 cell line [54], [59], since no ChIP-seq data for HSFs are available for human brains or neurons. We found that 28 of our 186 DE genes had at least one HSF1 ChIP-seq peak within 50 kb (P = 3.3e-5, hypergeometric test) (24 up-regulated by HS: ABHD3, AHSA1, CACYBP, CCT3, CHORDC1, CLU, DNAJB1, DNAJB2, FKBP4, HIST1H2BN, HSP90AA1, HSP90AB1, HSPA1A, HSPA1B, HSPA6, HSPD1, HSPH1, KAT2A, PTGES3, SLC5A3, STIP1, SUGT1, TH, TUBA4A; 4 down-regulated: KNTC1, MSH5, SNX29, hsa-mir-6080).
qPCR was used to validate some differentially expressed genes. Because of a lack of RNA remaining after RNA-seq, we were only able to validate a small number of genes. A significant 4-fold increase in HSP90AB1 (Student's t-test, P = 0.006) and a 1.8-fold decrease in ZNF804A were found (P = 0.005), which confirmed the RNA-seq findings.
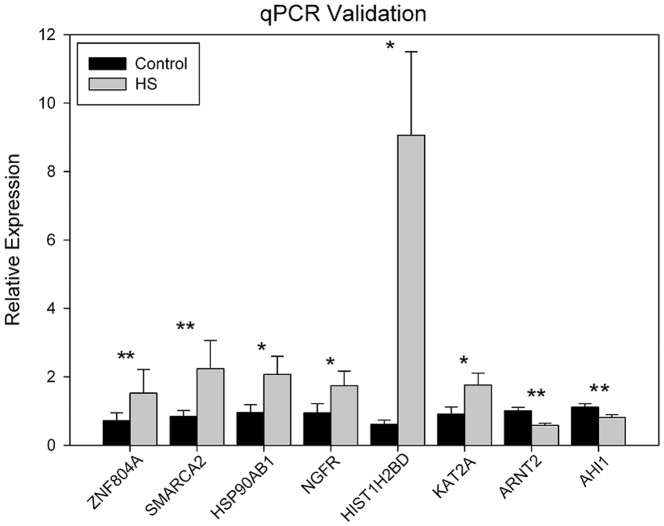
For further qPCR validation, we prepared multiple aggregates from a third control sample and carried out another HS experiment (C3 and HS3). As seen in Figure 3, significant differences in gene expression following HS were observed for every gene analyzed, which all changed in the same direction as in the RNA-seq findings, with the exception of ZNF804A, which increased in C3 upon HS, but decreased in C1 and C2.
Figure 3. qPCR validation for C3 and HS3.
Between 4–8 aggregates were analyzed individually in triplicate using the 2−ΔΔCt relative expression method with β2-microglobulin (β2M) as a control gene. Each sample was normalized against a common control RNA. Mean values +/− standard deviation are shown and a student's t-test was performed. A single asterisk indicates a p<0.05; two asterisks indicate a p<0.01. The actual p-values are ZNF804A (0.0002), SMARCA2 (0.0001), HSP90AB1 (0.03), NGFR (0.04), HIST1H2BD (0.02), KAT2A (0.05), ARNT2 (0.002), AHI1 (0.006).
Pathway Analysis
All differentially expressed genes that were significant at a p<0.05 level were subjected to Ingenuity Pathway Analysis (IPA). The top GO (Gene Ontology) terms for up and down-regulated genes are shown in Table 1. However, only up-regulated genes involved in response to unfolded protein, response to protein stimulus, protein folding and response to organic substance achieved genome-wide significance (a complete list, including the genes that contributed to each GO category, are shown in Table S3).
Table 1. GO (Gene Ontology) Analysis of Differentially Expressed Genes.
| Top GO Terms: up-regulated genes | p-value | FDR |
| GO:0006986∼response to unfolded protein | 1.01E-11 | 1.60E-08 |
| GO:0051789∼response to protein stimulus | 2.98E-11 | 4.71E-08 |
| GO:0006457∼protein folding | 4.82E-10 | 7.60E-07 |
| GO:0010033∼response to organic substance | 3.94E-06 | 0.006216 |
| GO:0043066∼negative regulation of apoptosis | 2.35E-04 | 0.369557 |
| GO:0043069∼negative regulation of programmed cell death | 2.60E-04 | 0.409938 |
| GO:0060548∼negative regulation of cell death | 2.66E-04 | 0.418438 |
| GO:0034622∼cellular macromolecular complex assembly | 5.69E-04 | 0.894464 |
| GO:0006916∼anti-apoptosis | 0.001366182 | 2.134406 |
| GO:0009628∼response to abiotic stimulus | 0.001462122 | 2.282681 |
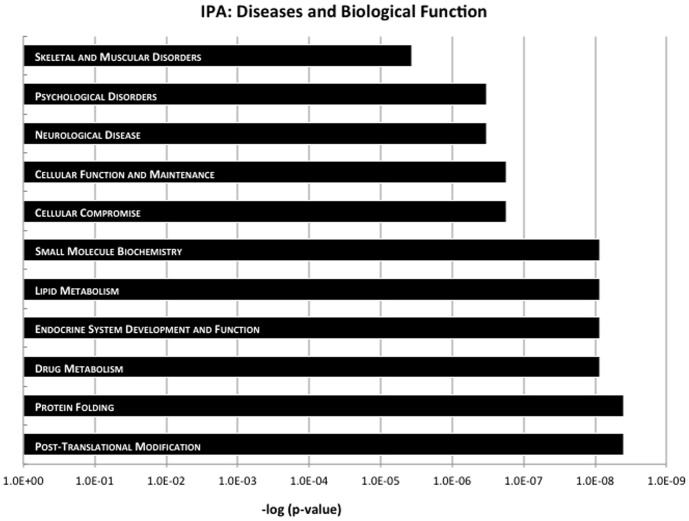
The top diseases and biological functions from the IPA analysis are shown in Figure 4 (see Table S4 for entire set). This included, as expected, genes involved in protein folding. Interestingly, the top disease processes were neurological and psychological disorders, caused by differential expression of a number of genes implicated in SZ, BD and ASD, including ZNF804A, which decreased 1.7-fold, SMARCA2, which increased 1.7-fold, as well as BAG3, KAT2A, HIST1H2BD, SLC6A8, and SLC17A7, which were induced by HS, and PRODH, ARNT2, DPP10, AHI1, IFITM1 and RORA, which decreased (described in detail in the discussion section).
Figure 4. Ingenuity Pathway Analysis showing top diseases and biological functions of differentially expressed genes.
All genes that were significant at a p<0.05 level were subjected to pathway analysis (Ingenuity Pathway Analysis; IPA). The top diseases and biological functions are shown.
The differentially expressed gene list was also examined using IPA's Upstream Regulator Analysis, which predicts factors that affect gene expression. As expected, genes responsive to HS factors (HSF) were the 1st and 3rd most significant upstream regulators (Table 2; Table S5). Interesting, the second most significant upstream regulator hit was RET, a member of the receptor tyrosine kinase family, which has well-established effects on cell growth and oncogenic transformation, as well the differentiation and survival of midbrain dopaminergic neurons [60]. RET CNVs have been found in a subgroup of patients with SZ [11].
Table 2. Upstream Regulator Analysis.
| Up. Regulator | Mol. Type | z-score | p-value | Target molecules in dataset |
| HSF1 | TR | 0.875 | 5.50E-14 | BAG3, CCT3, CLU, CRYAB, DNAJB1, EIF4A2, HMOX1, HSP90AA1, HSP90AB1, HSPA1A/HSPA1B, HSPA4L, HSPB1, HSPD1, HSPH1, KNTC1, SERPINH1, SLC5A3 |
| RET | kinase | 2.778 | 3.08E-09 | CALB1, CLU, DNAJB2, FKBP4, FOS, HSP90AA1, HSPA1A/HSPA1B, HSPD1, HSPH1, STIP1, TH |
| HSF2 | TR | 3.02E-06 | CCT3, CLU, HSPA1A/HSPA1B, HSPB1, HSPH1 | |
| HTT | TR | 4.57E-06 | CLK3, CRYAB, DNAJB1, FDFT1, FKBP4, FOS, HSP90AB1, HSPA1A/HSPA1B, HSPD1, KAT2A, MEIS2, MT-CO3, NGFR, NR4A1, PROM1, PURB, SERPINH1, TH, TUBA4A, UQCR11 | |
| CD437 | CD | 0 | 1.53E-05 | APC2, CCT3, EIF3D, EIF4A2, FOS, HSP90AA1, NR4A1, PURB, RBMX, SNRPB |
| MMTP | CT | 0.749 | 2.47E-05 | CALB1, CASP9, FOS, HSPB1, NR4A1, TH |
| β-estradiol | CEM | −0.14 | 2.66E-05 | ADCY1, ARNT2, BTG2, BUB3, CALB1, CALCA, CASP9, CLU, EIF3D, FDFT1, FGD6, FOS, HMOX1, HNRNPD, HSP90AB1, HSPA1A/HSPA1B, HSPB1, HSPD1, HSPH1, IER2, IFT122, MEIS2, NPR3, NR4A1, OGN, PROM1, SCG2, SETD7, SLA, STIP1, TH |
| NGF | GF | 1.703 | 2.96E-05 | BTG2, CALCA, ECEL1, FOS, HMOX1, NGFR, NR4A1, SCG2, TH |
| PDE | group | 5.63E-05 | FOS, HMOX1, NR4A1 | |
| KCl | CD | 0.571 | 9.25E-05 | BTG2, CALB1, FOS, NR4A1, SLC8A3, TH |
| liquiritigenin | CENM | 1.09E-04 | EPHX1, FOS, HMOX1 | |
| bisphenol A | CENM | 1.941 | 1.86E-04 | ARNT2, CLU, FOS, HSP90AB1, HSPA1A/HSPA1B, NR4A1 |
| phencyclidine | CD | 1.87E-04 | DDC, FOS, SCG2 |
Abbreviations: Up. Regulator (Upstream Regulator); Mol. Type (Molecular Type P); TR (transcription regulator); z-score (activation z-score); p-value (p-value of overlap); GF (growth factor); MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine); CT (chemical toxicant); CD (chemical drug); CEM (chemical - endogenous mammalian); CENM (chemical - endogenous non-mammalian); KCl (potassium chloride)
Upstream Regulator Analysis using IPA predicts factors that affect gene expression.
Another upstream regulator connected to dopaminergic transmission in this analysis is MPTP, a neurotoxin that induces rapid nigrostriatal dopamine neuron degeneration, which is used to generate animal models of Parkinson Disease and can cause the disorder in humans [61]. Several other upstream regulator predictors are relevant to neuropsychiatric disorders, including genes whose expression patterns are affected by phencyclidine (PCP), a drug that mimics the symptoms of SZ in humans and in animal models, and bisphenol A, an endocrine disruptor used in the manufacture of polycarbonate plastics that has been implicated in ASD, and which down-regulates the ASD candidate gene ARNT2 [8], [62]–[65]. Also among the top upstream regulators were CD437 (6-[3-(1-adamantyl)-4-hydroxyphenyl]-2-naphthalene carboxylic acid), a novel synthetic retinoic acid derivative, and NGF.
lncRNAs
An emerging area of interest is the participation of long non-coding RNAs (lncRNAs) in the cellular response to HS and other cellular stresses [66], [67]. Three lncRNAs were affected by HS; RP11-386G11.10 and RP11-627G23.1, which decreased in expression, and RP11-50D9.3, which increased (Table S2). RP11-386G11.10 overlaps and is antisense to the tubulin A encoding genes TUBA1A and TUBA1B. RP11-627G23.1 is ∼25 Kb 3′ to B3GAT1 (beta-1,3-glucuronyltransferase 1), which is highly expressed in the brain and is a key enzyme involved in the biosynthesis of the carbohydrate epitope HNK-1 that is present on a number of cell adhesion molecules important in neurodevelopment and hippocampal long-term potentiation [68]. B3GAT1 is near a balanced translocation that segregates in a family with psychosis and depression, and an association to B3GAT2 has been found in SZ [68], [69]. Finally, RP11-50D9.3 is an antisense transcript that is ∼10 Kb 3′ to the HSP gene, HSPA4L, which is significantly induced in our study. Whether the induction of RP11-50D9.3 by HS is simply a byproduct of HSPA4L transcriptional activation, or is involved in regulating other genes involved in the stress response remains to be determined.
Discussion
MIA can be viewed from two broad perspectives, which may have independent and overlapping effects on neurogenesis through the direct effects of immune cytokines that bind to their specific receptors on neurons, and by fever, which activates cellular stress pathways. In addition, it is likely that the response to MIA is influenced by genetic background; both fetal and maternal. So far, most biologically relevant studies have been carried out in animal models, with studies in humans restricted, with a few exceptions, to retrospective epidemiological studies. However, with the recent advent of iPSC technology there is now an opportunity to study some of the molecular consequences of MIA in vitro using human neurons. The technology also offers the opportunity to study patient-specific neurons to assess potential gene by environment (G × E) interactions.
In this study, we applied iPSC technology to study the effects of HS using a differentiation protocol that generates neuronal aggregates with characteristics of a developing first trimester telencephalon, a period of gestation that has been implicated in both SZ and ASD in some studies [38], [70]–[72]. A relatively brief exposure to HS resulted in a burst of gene expression changes, most notably in members of the HS family, including HSP70 and HSP90, and HSP binding partners. HSPs are ATP-dependent molecular chaperones that play a critical role in maintaining cellular homeostasis following HS and other stressors, such as nutritional deficiency, hypoxia, toxins, heavy metals, infections and inflammation; factors that have each been implicated as risk factors for SZ and ASD [73]–[82]. HSPs target misfolded proteins that accumulate in response to cellular stress, facilitating protein refolding and targeting damaged proteins for degradation in proteasomes. Neurodegenerative disorders, such as Alzheimer Disease, Parkinson Disease, and Huntington Disease, are caused by misfolded proteins, and activation of HSPs is being tested as a novel therapeutic strategy [83]–[86]. The aggregates used in these experiments and perhaps other neuronal induction methods derived from iPSCs, would be ideal systems to test the effects of a host of cellular stressors and the therapeutic effect of drugs on human neurons.
In addition to cellular stress, HSPs also play a role in the response to behavioral stress. HSP70 and HSP90, for example, act as glucocorticoid receptor chaperones aiding in their transport to the nucleus [87]–[89]. The adverse effects of chronic behavioral stress mediated in part by glucocorticoid-inducible genes, play an important role in depression and psychotic disorders [90]–[94]. Thus, the activation of HSP70 and HSP90 gene expression in response to HS and other cellular stressors may overlap with the brain's response to behavioral stress.
In addition to the marked induction of HS related genes - an expected finding - there were a number of genes of interest with respect to neurodevelopmental and neuropsychiatric disorders that showed substantial differences in expression, most notably NGFR, which exhibited the second most significant increase. Although NGF, the ligand for NGFR, was first discovered as a peripheral nervous system growth factor, it does have effects in the brain as well, especially as a trophic factor for cholinergic neurons [95]–[98]. NGF enhances neurite outgrowth in PC12 cells exposed to HS, a response that is affected by the atypical antipsychotic aripiprazole and HSP90α expression [99], [100]. It should be noted that NGF is also induced by IL-1, suggesting an overlap between the effects of HS and cytokines on NGF signaling [101].
As for a potential role for NGF signaling in neuropsychiatric disorders, a quantitative trait locus (QTL) in an NGF intron was recently found to be associated with nonverbal communication in ASD subjects [102]. Also, NGF levels are reduced in an animal model of Rett Syndrome and in the serum of medication-naïve patients with SZ [103], [104]. In addition, NGF-induced neurite extension is enhanced by DISC1 [105], [106].
In addition to NGFR, the expression of a number of SZ, BD and ASD candidate was significantly affected by HS, including SMARCA2, HIST1H2BD, DPP10, SLC6A8, SLC17A7, ARNT2, AHI1 and ZNF804A. SMARCA2, which increased 1.7 fold with HS, encodes a REST-regulated, SWI/SNF chromatin-remodeling complex that has been implicated in SZ in a low density GWAS and CNV screening, and by molecular analysis following REST knockdown [46], [107], [108]. Point mutations in the gene have also been found in patients with Coffin–Siris syndrome and Nicolaides–Baraitser syndrome, which are characterized by severe developmental delay [109], [110]. Although we show a robust induction of SMARCA2 expression, disease-associated SMARCA2 mutations are primarily loss of function variants. However, it is well-established in psychiatric genetics that both copy gain and copy loss affecting the same locus can cause neurodevelopmental problems (15q11.2, 16p13.1 and 22q11.2, for example) [9], [111]–[113]. As for HIST1H2BD, it was one of 5 differentially expressed genes coding for histone variants found in a large study using lymphoblastoid cell lines derived from patients with SZ and controls [114]. Besides HIST1H2BD, there were 3 other histone variants (HIST1H2BN, HIST1H1C and HIST1H2AC) induced by HS that map to the same region on chromosome 6, near a GWAS signal in SZ, within a cluster of histone variants [115]. And, DPP10, SLC6A8, SLC17A7, ARNT2, and AHI1 have been implicated in SZ, BD and ASD in GWAS, CNV analyses, exome sequencing and molecular studies [8], [10], [116]–[122].
Finally, the effect of HS on ZNF804A requires some discussion. The gene codes for a Zn-finger transcription factor that has been implicated in SZ and BD in replicated GWAS studies and molecular analysis [123]-[125]. In addition, rare copy gain and copy loss CNVs affecting the gene have been found in ASD, psychosis and anxiety disorder [10], [126]–[128]. A significant decrease in ZNF804A expression was found when C1 and C2 were exposed to HS, but an increase was detected in C3. Whether this variability is due to genetic variation within the gene and how this might relate to the GWAS findings remains to be determined; a much larger sample size will be needed.
Another gene implicated in SZ on a molecular level that was significantly affected by HS was BAG3, which codes for an HSP70 co-chaperone that mediates adaptive responses to stressful stimuli [75], [129], [130]. It has been found to be differentially expressed in the prefrontal cortex of patients with SZ, and in neurons derived from SZ-specific iPSCs [131], [132]. In addition to BAG3, SZ-specific neurons reported by Brennand et al. showed differential expression of 12 other genes that were also affected by HS in our experiment: HSPA1A, HSPA1B, KAT2A, SAMD4A, PTPRE, UNC5C, GAS2L3, FGD6, CENPO, NALCN, ECEL1, and TM7SF2 [131]. The finding that genes involved in the cellular stress response are differentially expressed in SZ-specific neurons and in the brains of patients supports a role for these pathways in disease pathogenesis. Interestingly, HSPA1B was one of top four candidate genes, along with DISC1, TCF4, and MBP, identified in a comprehensive functional genomics analysis that combined genetic and molecular findings in SZ [2].
Other differentially expressed genes of interest in our HS experiment with respect to neuropsychiatric disorders were PRODH (proline dehyrdrogenase), IFITM1 (Interferon Induced Transmembrane Protein), and several genes involved in dopamine transmission. PRODH maps to the 22q11.2 region deleted in velocardiofacial syndrome (VCFS), a haploinsufficiency disorder that leads to a variety of physical and psychiatric problems, including SZ and ASD, and IFITM1 has been found to be differentially expressed in the brains of patients with SZ and ASD [133]–[141]. In addition, another member of the interferon-inducible family, IFITM3, along with the HS genes HSPA6, HSPB8 and SERPINH1, has been found to be differentially expressed in the brains of patients with ASD [29], [75].
Among the genes involved in dopaminergic function affected by HS were DDC, PHOX2A, TH, DBH and ADCY1. In addition, two potassium channel encoding genes, KCNH1 and KCND1, increased with HS, while the ASD and BD candidate gene DPP10 decreased; DPP10 codes for a dipeptidyl peptidase that regulates potassium channel function [116], [117], [142]. Potassium channel function, which can affect dopaminergic tone, caused by a variety of genetic, autoimmune and molecular phenomena, is increasingly being recognized as a mechanism underlying the development of SZ, BD and ASD in subgroups of patients [143]–[147].
It is also important to note that in this study we identified genes that were differentially expressed in response to HS by combining the male and female samples. However, an analysis of gender differences in response to HS and other cellular stressors would be of great interest considering the 4-fold higher prevalence of ASD in males compared to females. Addressing this important issue, though, will require a larger sample size.
In summary, the findings reported here show that HS can induce changes in the expression of a number of candidate genes and pathways implicated in SZ, BD and ASD. In fact, the changes in expression that occur for some of these genes is equivalent to the effects of a CNV; ∼2-fold decrease as in a copy loss (DPP10, and ARNT2 for example) or 50% increases and more, as in copy gains (SMARCA2, NGFR). The duration of the HS effect on neurogenesis, however, is likely to be transient, but that remains to be tested. Long term effects on gene expression could potentially occur as a result of the altered expression of chromatin regulators, such as SMARCA2, and KAT2A, and HS-inducible histone variants. However, even if the effects of HS are relatively brief, there may be subsets of neurons and neural progenitor cells that are severely impacted, especially in the context of other cellular stressors and genetic risk variants that can induce effects on gene expression that overlap with those affected by HS. Indeed, some cellular stressors known to operate through, or be influenced by HS pathways can be chronic, such as inflammation, exposure to heavy metals, and endocrine disruptors, as well as emotional stress; these might have a more protracted effect on gene expression and neurogenesis, truly mimicking the effects of CNVs; quantitatively and temporally.
Finally, in addition to the notion that a common environmental stressor like HS can have an effect on the expression of SZ, BD and ASD candidate genes similar in amplitude, if not duration, to a CNV, another important concept emerges from this study. That is, the role that some candidate genes have on disease pathogenesis may be underestimated if based solely on genetic findings. Although genes like SMARCA2 and ARNT2 might be viewed as relatively minor factors based on the small number of positive molecular and genetic studies that have been reported so far, their altered expression in response to HS suggests that they, and similarly affected candidate genes, could play a much more substantial role in SZ and ASD - as targets of common environmental stressors.
Supporting Information
Text S1. Supplementary methods
Table S1. RNA-seq statistics. C1 and C2 refer to controls 1 and 2: HS1 and HS2 are the heat shocked counterparts.
Table S2. RNA-seq reads in TPM (Transcripts Per Million) for all genes arranged by p-value (lowest to highest). The 186 genes that showed nominally significant differences in the mean log2 fold-change (HS/control) are in bold type.
Table S3: Top Gene Ontology (GO) Terms for genes up and down-regulated by HS.
Table S4. Ingenuity Pathway Analysis showing disease and biological functions of differentially expressed genes
Table S5: Upstream Regulator Analysis, all genes
(ZIP)
Acknowledgments
We wish to thank Dr. Judith L. Rapoport, Chief of the Child Psychiatry Branch at the NIMH, and her staff, for providing fibroblasts on control subjects.
Funding Statement
This work was supported by the National Institute of Mental Health (MH073164, MH097893 and MH087840). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. The Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium, Ripke S, Sanders AR, Kendler KS, Levinson DF, et al. (2011) Genome-wide association study identifies five new schizophrenia loci. Nat Genet 43: 969–976 10.1038/ng.940;10.1038/ng.940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, et al. (2012) Convergent functional genomics of schizophrenia: From comprehensive understanding to genetic risk prediction. Mol Psychiatry 17: 887–905 10.1038/mp.2012.37 ; 10.1038/mp.2012.37 10.1038/mp.2012.37; 10.1038/mp.2012.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O'Donovan MC, Craddock NJ, Owen MJ (2009) Genetics of psychosis; insights from views across the genome. Hum Genet 126: 3–12 10.1007/s00439-009-0703-0 [DOI] [PubMed] [Google Scholar]
- 4. Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, et al. (2011) Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet 43: 864–868 10.1038/ng.902;10.1038/ng.902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corvin A, Morris DW. (2013) Genome-wide association studies: Findings at the major histocompatibility complex locus in psychosis. Biol Psychiatry. 10.1016/j.biopsych.2013.09.018 ; 10.1016/j.biopsych.2013.09.018 10.1016/j.biopsych.2013.09.018; 10.1016/j.biopsych.2013.09.018 [DOI] [PubMed] [Google Scholar]
- 6. Giusti-Rodriguez P, Sullivan PF (2013) The genomics of schizophrenia: Update and implications. J Clin Invest 123: 4557–4563 10.1172/JCI66031;10.1172/JCI66031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gejman PV, Sanders AR, Duan J (2010) The role of genetics in the etiology of schizophrenia. Psychiatr Clin North Am 33: 35–66 10.1016/j.psc.2009.12.003 ; 10.1016/j.psc.2009.12.003 10.1016/j.psc.2009.12.003; 10.1016/j.psc.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, et al. (2012) Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485: 242–245 10.1038/nature11011 ; 10.1038/nature11011 10.1038/nature11011; 10.1038/nature11011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ahn K, Gotay N, Andersen TM, Anvari AA, Gochman P, et al. (2013) High rate of disease-related copy number variations in childhood onset schizophrenia. Mol Psychiatry. 10.1038/mp.2013.59 ; 10.1038/mp.2013.59 10.1038/mp.2013.59; 10.1038/mp.2013.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Griswold AJ, Ma D, Cukier HN, Nations LD, Schmidt MA, et al. (2012) Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet. 10.1093/hmg/dds164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Glessner JT, Reilly MP, Kim CE, Takahashi N, Albano A, et al. (2010) Strong synaptic transmission impact by copy number variations in schizophrenia. Proc Natl Acad Sci U S A 107: 10584–10589 10.1073/pnas.1000274107 ; 10.1073/pnas.1000274107 10.1073/pnas.1000274107; 10.1073/pnas.1000274107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Need AC, Ge D, Weale ME, Maia J, Feng S, et al. (2009) A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet 5: e1000373 10.1371/journal.pgen.1000373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, et al. (2009) Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459: 569–573 10.1038/nature07953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, et al. (2009) Common variants conferring risk of schizophrenia. Nature. 10.1038/nature08186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Genetic Risk and Outcome in Psychosis (GROUP) Consortium, et al. (2008) Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet 83: 504–510 10.1016/j.ajhg.2008.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Atladottir HO, Henriksen TB, Schendel DE, Parner ET, (2012) Autism after infection, febrile episodes, and antibiotic use during pregnancy: An exploratory study. Pediatrics. – 10.1542/peds.20121107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zerbo O, Iosif AM, Walker C, Ozonoff S, Hansen RL, et al. (2012) Is maternal influenza or fever during pregnancy associated with autism or developmental delays? results from the CHARGE (CHildhood autism risks from genetics and environment) study. J Autism Dev Disord. 10.1007/s10803-012-1540-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parker-Athill EC, Tan J (2010) Maternal immune activation and autism spectrum disorder: Interleukin-6 signaling as a key mechanistic pathway. Neurosignals 18: 113–128 10.1159/000319828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsiao EY, Patterson PH (2011) Activation of the maternal immune system induces endocrine changes in the placenta via IL-6. Brain Behav Immun 25: 604–615 10.1016/j.bbi.2010.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ghanizadeh A (2011) Could fever and neuroinflammation play a role in the neurobiology of autism? A subject worthy of more research. Int J Hyperthermia 27: 737–738 10.3109/02656736.2011.604665 [DOI] [PubMed] [Google Scholar]
- 21. Canetta SE, Brown AS (2012) Prenatal infection, maternal immune activation, and risk for schizophrenia. Transl Neurosci 3: 320–327 10.2478/s13380-012-0045-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fatjo-Vilas M, Pomarol-Clotet E, Salvador R, Monte GC, Gomar JJ, et al. (2012) Effect of the interleukin-1beta gene on dorsolateral prefrontal cortex function in schizophrenia: A genetic neuroimaging study. Biol Psychiatry 72: 758–765 10.1016/j.biopsych.2012.04.035; 10.1073/pnas.100027410710.1016/j.biopsych.2012.04.035; 10.1016/j.biopsych.2012.04.035 [DOI] [PubMed] [Google Scholar]
- 23. Roumier A, Pascual O, Bechade C, Wakselman S, Poncer JC, et al. (2008) Prenatal activation of microglia induces delayed impairment of glutamatergic synaptic function. PLoS One 3: e2595 10.1371/journal.pone.0002595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goshen I, Yirmiya R (2009) Interleukin-1 (IL-1): A central regulator of stress responses. Front Neuroendocrinol 30: 30–45 10.1016/j.yfrne.2008.10.001 ; 10.1016/j.yfrne.2008.10.001 10.1016/j.yfrne.2008.10.001; 10.1016/j.yfrne.2008.10.001 [DOI] [PubMed] [Google Scholar]
- 25. Koo JW, Duman RS (2009) Interleukin-1 receptor null mutant mice show decreased anxiety-like behavior and enhanced fear memory. Neurosci Lett 456: 39–43 10.1016/j.neulet.2009.03.068 ; 10.1016/j.neulet.2009.03.068 10.1016/j.neulet.2009.03.068; 10.1016/j.neulet.2009.03.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nilsberth C, Elander L, Hamzic N, Norell M, Lonn J, et al. (2009) The role of interleukin-6 in lipopolysaccharide-induced fever by mechanisms independent of prostaglandin E2. Endocrinology 150: 1850–1860 – 10.1210/en.20080806 [DOI] [PubMed] [Google Scholar]
- 27. Wei H, Alberts I, Li X (2013) Brain IL-6 and autism. Neuroscience 252: 320–325 10.1016/j.neuroscience.2013.08.025 ; 10.1016/j.neuroscience.2013.08.025 10.1016/j.neuroscience.2013.08.025; 10.1016/j.neuroscience.2013.08.025 [DOI] [PubMed] [Google Scholar]
- 28. Wei H, Chadman KK, McCloskey DP, Sheikh AM, Malik M, et al. (2012) Brain IL-6 elevation causes neuronal circuitry imbalances and mediates autism-like behaviors. Biochim Biophys Acta 1822: 831–842 10.1016/j.bbadis.2012.01.011 [DOI] [PubMed] [Google Scholar]
- 29. Garbett KA, Hsiao EY, Kalman S, Patterson PH, Mirnics K (2012) Effects of maternal immune activation on gene expression patterns in the fetal brain. Transl Psychiatry 2: e98 10.1038/tp.2012.24 ; 10.1038/tp.2012.24 10.1038/tp.2012.24; 10.1038/tp.2012.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li X, Chauhan A, Sheikh AM, Patil S, Chauhan V, et al. (2009) Elevated immune response in the brain of autistic patients. J Neuroimmunol 207: 111–116 10.1016/j.jneuroim.2008.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McAllister AK (2013) Major histocompatibility complex I in brain development and schizophrenia. Biol Psychiatry. 10.1016/j.biopsych.2013.10.003 ; 10.1016/j.biopsych.2013.10.003 10.1016/j.biopsych.2013.10.003; 10.1016/j.biopsych.2013.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boulanger LM (2004) MHC class I in activity-dependent structural and functional plasticity. Neuron Glia Biol 1: 283–289 10.1017/S1740925X05000128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, et al. (2000) Functional requirement for class I MHC in CNS development and plasticity. Science 290: 2155–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fineberg AM, Ellman LM (2013) Inflammatory cytokines and neurological and neurocognitive alterations in the course of schizophrenia. Biol Psychiatry 73: 951–966 10.1016/j.biopsych.2013.01.001 ; 10.1016/j.biopsych.2013.01.001 10.1016/j.biopsych.2013.01.001; 10.1016/j.biopsych.2013.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Arrode-Bruses G, Bruses JL (2012) Maternal immune activation by poly I:C induces expression of cytokines IL-1beta and IL-13, chemokine MCP-1 and colony stimulating factor VEGF in fetal mouse brain. J Neuroinflammation 9: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meyer U, Feldon J, Fatemi SH (2009) In-vivo rodent models for the experimental investigation of prenatal immune activation effects in neurodevelopmental brain disorders. Neurosci Biobehav Rev 33: 1061–1079 10.1016/j.neubiorev.2009.05.001 ; 10.1016/j.neubiorev.2009.05.001 10.1016/j.neubiorev.2009.05.001; 10.1016/j.neubiorev.2009.05.001 [DOI] [PubMed] [Google Scholar]
- 37. Richetto J, Calabrese F, Riva MA, Meyer U (2013) Prenatal immune activation induces maturation-dependent alterations in the prefrontal GABAergic transcriptome. Schizophr Bull. 10.1093/schbul/sbs195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bauman MD, Iosif AM, Smith SE, Bregere C, Amaral DG, et al. (2013) Activation of the maternal immune system during pregnancy alters behavioral development of rhesus monkey offspring. Biol Psychiatry. 10.1016/j.biopsych.2013.06.025 ; 10.1016/j.biopsych.2013.06.025 10.1016/j.biopsych.2013.06.025; 10.1016/j.biopsych.2013.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ratnayake U, Quinn TA, Castillo-Melendez M, Dickinson H, Walker DW (2012) Behaviour and hippocampus-specific changes in spiny mouse neonates after treatment of the mother with the viral-mimetic poly I:C at mid-pregnancy. Brain Behav Immun 26: 1288–1299 10.1016/j.bbi.2012.08.011 ; 10.1016/j.bbi.2012.08.011 10.1016/j.bbi.2012.08.011; 10.1016/j.bbi.2012.08.011 [DOI] [PubMed] [Google Scholar]
- 40. Ducharme G, Lowe GC, Goutagny R, Williams S (2012) Early alterations in hippocampal circuitry and theta rhythm generation in a mouse model of prenatal infection: Implications for schizophrenia. PLoS One 7: e29754 10.1371/journal.pone.0029754 ; 10.1371/journal.pone.0029754 10.1371/journal.pone.0029754; 10.1371/journal.pone.0029754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arnold SE, Franz BR, Gur RC, Gur RE, Shapiro RM, et al. (1995) Smaller neuron size in schizophrenia in hippocampal subfields that mediate cortical-hippocampal interactions. Am J Psychiatry 152: 738–748. [DOI] [PubMed] [Google Scholar]
- 42. Benes FM (2011) Regulation of cell cycle and DNA repair in post-mitotic GABA neurons in psychotic disorders. Neuropharmacology 60: 1232–1242 10.1016/j.neuropharm.2010.12.011 [DOI] [PubMed] [Google Scholar]
- 43. Benes FM, Lim B, Matzilevich D, Subburaju S, Walsh JP (2008) Circuitry-based gene expression profiles in GABA cells of the trisynaptic pathway in schizophrenics versus bipolars. Proc Natl Acad Sci U S A 105: 20935–20940 10.1073/pnas.0810153105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, et al. (2007) Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci U S A 104: 10164–10169 10.1073/pnas.0703806104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fatemi SH, Folsom TD (2009) The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr Bull 35: 528–548 10.1093/schbul/sbn187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, et al. (2008) Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 10.1126/science.1155174 [DOI] [PubMed] [Google Scholar]
- 47. Lang UE, Puls I, Muller DJ, Strutz-Seebohm N, Gallinat J (2007) Molecular mechanisms of schizophrenia. Cell Physiol Biochem 20: 687–702 10.1159/000110430 [DOI] [PubMed] [Google Scholar]
- 48. Lowe GC, Luheshi GN, Williams S (2008) Maternal infection and fever during late gestation are associated with altered synaptic transmission in the hippocampus of juvenile offspring rats. Am J Physiol Regul Integr Comp Physiol 295: R1563–71 10.1152/ajpregu.90350.2008 ; 10.1152/ajpregu.90350.2008 10.1152/ajpregu.90350.2008; 10.1152/ajpregu.90350.2008 [DOI] [PubMed] [Google Scholar]
- 49. Mehler MF, Purpura DP (2009) Autism, fever, epigenetics and the locus coeruleus. Brain Res Rev 59: 388–392 10.1016/j.brainresrev.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Curran LK, Newschaffer CJ, Lee LC, Crawford SO, Johnston MV, et al. (2007) Behaviors associated with fever in children with autism spectrum disorders. Pediatrics 120: e1386–92 – 10.1542/peds.20070360 [DOI] [PubMed] [Google Scholar]
- 51. Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, et al. (2012) Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci U S A 109: 12770–12775 10.1073/pnas.1202944109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, et al. (2013) Cerebral organoids model human brain development and microcephaly. Nature 501: 373–379 10.1038/nature12517 ; 10.1038/nature12517 10.1038/nature12517; 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, et al. (2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, et al. (2012) GENCODE: The reference human genome annotation for the ENCODE project. Genome Res 22: 1760–1774 10.1101/gr.135350.111 ; 10.1101/gr.135350.111 10.1101/gr.135350.111; 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN (2010) RNA-seq gene expression estimation with read mapping uncertainty. Bioinformatics 26: 493–500 10.1093/bioinformatics/btp692 ; 10.1093/bioinformatics/btp692 10.1093/bioinformatics/btp692; 10.1093/bioinformatics/btp692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106-2010-11-10-r106. Epub 2010 Oct 27. 10.1186/gb-2010-11-10-r106 ; 10.1186/gb-2010-11-10-r106 10.1186/gb-2010-11-10-r106; 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fan Y, Huang J, Duffourc M, Kao RL, Ordway GA, et al. (2011) Transcription factor Phox2 upregulates expression of norepinephrine transporter and dopamine beta-hydroxylase in adult rat brains. Neuroscience 192: 37–53 10.1016/j.neuroscience.2011.07.005 ; 10.1016/j.neuroscience.2011.07.005 10.1016/j.neuroscience.2011.07.005; 10.1016/j.neuroscience.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim HG, Kim TM, Park G, Lee TH, Oh MS (2013) Repeated heat exposure impairs nigrostriatal dopaminergic neurons in mice. Biol Pharm Bull 36: 1556–1561. [DOI] [PubMed] [Google Scholar]
- 59. ENCODE Project Consortium, Bernstein BE, Birney E, Dunham I, Green ED, et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 10.1038/nature11247 ; 10.1038/nature11247 10.1038/nature11247; 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kowsky S, Poppelmeyer C, Kramer ER, Falkenburger BH, Kruse A, et al. (2007) RET signaling does not modulate MPTP toxicity but is required for regeneration of dopaminergic axon terminals. Proc Natl Acad Sci U S A 104: 20049–20054 10.1073/pnas.0706177104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Iderberg H, Francardo V, Pioli EY (2012) Animal models of L-DOPA-induced dyskinesia: An update on the current options. Neuroscience 211: 13–27 10.1016/j.neuroscience.2012.03.023 ; 10.1016/j.neuroscience.2012.03.023 10.1016/j.neuroscience.2012.03.023; 10.1016/j.neuroscience.2012.03.023 [DOI] [PubMed] [Google Scholar]
- 62. Wolstenholme JT, Goldsby JA, Rissman EF (2013) Transgenerational effects of prenatal bisphenol A on social recognition. Horm Behav 64: 833–839 10.1016/j.yhbeh.2013.09.007 ; 10.1016/j.yhbeh.2013.09.007 10.1016/j.yhbeh.2013.09.007; 10.1016/j.yhbeh.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wolstenholme JT, Edwards M, Shetty SR, Gatewood JD, Taylor JA, et al. (2012) Gestational exposure to bisphenol a produces transgenerational changes in behaviors and gene expression. Endocrinology 153: 3828–3838 – 10.1210/en.2012–1195 ; – 10.1210/en.20121195 10.1210/en.20121195; – 10.1210/en.2012–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Qin XY, Zaha H, Nagano R, Yoshinaga J, Yonemoto J, et al. (2011) Xenoestrogens down-regulate aryl-hydrocarbon receptor nuclear translocator 2 mRNA expression in human breast cancer cells via an estrogen receptor alpha-dependent mechanism. Toxicol Lett 206: 152–157 10.1016/j.toxlet.2011.07.007 ; 10.1016/j.toxlet.2011.07.007 10.1016/j.toxlet.2011.07.007; 10.1016/j.toxlet.2011.07.007 [DOI] [PubMed] [Google Scholar]
- 65. Jodo E (2013) The role of the hippocampo-prefrontal cortex system in phencyclidine-induced psychosis: A model for schizophrenia. J Physiol Paris 107: 434–440 10.1016/j.jphysparis.2013.06.002 ; 10.1016/j.jphysparis.2013.06.002 10.1016/j.jphysparis.2013.06.002; 10.1016/j.jphysparis.2013.06.002 [DOI] [PubMed] [Google Scholar]
- 66. Lakhotia SC (2012) Long non-coding RNAs coordinate cellular responses to stress. Wiley Interdiscip Rev RNA 3: 779–796 10.1002/wrna.1135 ; 10.1002/wrna.1135 10.1002/wrna.1135; 10.1002/wrna.1135 [DOI] [PubMed] [Google Scholar]
- 67. Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, et al. (2013) NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 10.1091/mbc.E13-09-0558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jeffries AR, Mungall AJ, Dawson E, Halls K, Langford CF, et al. (2003) Beta-1,3-glucuronyltransferase-1 gene implicated as a candidate for a schizophrenia-like psychosis through molecular analysis of a balanced translocation. Mol Psychiatry 8: 654–663 10.1038/sj.mp.4001382 [DOI] [PubMed] [Google Scholar]
- 69. Kahler AK, Djurovic S, Rimol LM, Brown AA, Athanasiu L, et al. (2011) Candidate gene analysis of the human natural killer-1 carbohydrate pathway and perineuronal nets in schizophrenia: B3GAT2 is associated with disease risk and cortical surface area. Biol Psychiatry 69: 90–96 10.1016/j.biopsych.2010.07.035 ; 10.1016/j.biopsych.2010.07.035 10.1016/j.biopsych.2010.07.035; 10.1016/j.biopsych.2010.07.035 [DOI] [PubMed] [Google Scholar]
- 70. Sorensen HJ, Mortensen EL, Reinisch JM, Mednick SA (2009) Association between prenatal exposure to bacterial infection and risk of schizophrenia. Schizophr Bull 35: 631–637 10.1093/schbul/sbn121 ; 10.1093/schbul/sbn121 10.1093/schbul/sbn121; 10.1093/schbul/sbn121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brown AS, Begg MD, Gravenstein S, Schaefer CA, Wyatt RJ, et al. (2004) Serologic evidence of prenatal influenza in the etiology of schizophrenia. Arch Gen Psychiatry 61: 774–780 10.1001/archpsyc.61.8.774 [DOI] [PubMed] [Google Scholar]
- 72. Malaspina D, Corcoran C, Kleinhaus KR, Perrin MC, Fennig S, et al. (2008) Acute maternal stress in pregnancy and schizophrenia in offspring: A cohort prospective study. BMC Psychiatry 8: 71–244X-8-71 10.1186/1471-244X-8-71 ; 10.1186/1471-244X-8-71 10.1186/1471-244X-8-71; 10.1186/1471-244X-8-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. El-Ansary A, Al-Ayadhi L (2012) Neuroinflammation in autism spectrum disorders. J Neuroinflammation 9: 265–2094-9-265 10.1186/1742-2094-9-265 ; 10.1186/1742-2094-9-265 10.1186/1742-2094-9-265; 10.1186/1742-2094-9-265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Deng MY, Lam S, Meyer U, Feldon J, Li Q, et al. (2011) Frontal-subcortical protein expression following prenatal exposure to maternal inflammation. PLoS One 6: e16638 10.1371/journal.pone.0016638 ; 10.1371/journal.pone.0016638 10.1371/journal.pone.0016638; 10.1371/journal.pone.0016638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Garbett K, Ebert PJ, Mitchell A, Lintas C, Manzi B, et al. (2008) Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol Dis 30: 303–311 10.1016/j.nbd.2008.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Evers M, Cunningham-Rundles C, Hollander E (2002) Heat shock protein 90 antibodies in autism. Mol Psychiatry 7 Suppl 2S26–8 10.1038/sj.mp.4001171 [DOI] [PubMed] [Google Scholar]
- 77. Pongrac JL, Middleton FA, Peng L, Lewis DA, Levitt P, et al. (2004) Heat shock protein 12A shows reduced expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry 56: 943–950 10.1016/j.biopsych.2004.09.005 [DOI] [PubMed] [Google Scholar]
- 78. Kakiuchi C, Ishiwata M, Nanko S, Kunugi H, Minabe Y, et al. (2005) Functional polymorphisms of HSPA5: Possible association with bipolar disorder. Biochem Biophys Res Commun 336: 1136–1143 10.1016/j.bbrc.2005.08.248 [DOI] [PubMed] [Google Scholar]
- 79. Uenishi R, Gong P, Suzuki K, Koizumi S (2006) Cross talk of heat shock and heavy metal regulatory pathways. Biochem Biophys Res Commun 341: 1072–1077 10.1016/j.bbrc.2006.01.066 [DOI] [PubMed] [Google Scholar]
- 80. Kakiuchi C, Ishiwata M, Nanko S, Kunugi H, Minabe Y, et al. (2007) Association analysis of HSP90B1 with bipolar disorder. J Hum Genet 52: 794–803 10.1007/s10038-007-0188-4 [DOI] [PubMed] [Google Scholar]
- 81. Koizumi S, Suzuki K, Yamaguchi S (2013) Heavy metal response of the heat shock protein 70 gene is mediated by duplicated heat shock elements and heat shock factor 1. Gene 522: 184–191 10.1016/j.gene.2013.03.090 ; 10.1016/j.gene.2013.03.090 10.1016/j.gene.2013.03.090; 10.1016/j.gene.2013.03.090 [DOI] [PubMed] [Google Scholar]
- 82. Muralidharan S, Mandrekar P, (2013) Cellular stress response and innate immune signaling: Integrating pathways in host defense and inflammation. J Leukoc Biol. 10.1189/jlb.0313153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chaari A, Hoarau-Vechot J, Ladjimi M (2013) Applying chaperones to protein-misfolding disorders: Molecular chaperones against alpha-synuclein in parkinson's disease. Int J Biol Macromol 60: 196–205 10.1016/j.ijbiomac.2013.05.032 ; 10.1016/j.ijbiomac.2013.05.032 10.1016/j.ijbiomac.2013.05.032; 10.1016/j.ijbiomac.2013.05.032 [DOI] [PubMed] [Google Scholar]
- 84. Bobkova NV, Garbuz DG, Nesterova I, Medvinskaya N, Samokhin A, et al. (2014) Therapeutic effect of exogenous hsp70 in mouse models of alzheimer's disease. J Alzheimers Dis 38: 425–435 10.3233/JAD-130779 ; 10.3233/JAD-130779 10.3233/JAD-130779; 10.3233/JAD-130779 [DOI] [PubMed] [Google Scholar]
- 85. Pierce A, Podlutskaya N, Halloran JJ, Hussong SA, Lin PY, et al. (2013) Over-expression of heat shock factor 1 phenocopies the effect of chronic inhibition of TOR by rapamycin and is sufficient to ameliorate alzheimer's-like deficits in mice modeling the disease. J Neurochem 124: 880–893 10.1111/jnc.12080 ; 10.1111/jnc.12080 10.1111/jnc.12080; 10.1111/jnc.12080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhao H, Michaelis ML, Blagg BS (2012) Hsp90 modulation for the treatment of alzheimer's disease. Adv Pharmacol 64: 1–25 10.1016/B978-0-12-394816-8.00001-5 ; 10.1016/B978-0-12-394816-8.00001-5 10.1016/B978-0-12-394816-8.00001-5; 10.1016/B978-0-12-394816-8.00001-5 [DOI] [PubMed] [Google Scholar]
- 87. Grad I, Picard D (2007) The glucocorticoid responses are shaped by molecular chaperones. Mol Cell Endocrinol 275: 2–12 10.1016/j.mce.2007.05.018 [DOI] [PubMed] [Google Scholar]
- 88. Beck IM, Drebert ZJ, Hoya-Arias R, Bahar AA, Devos M, et al. (2013) Compound A, a selective glucocorticoid receptor modulator, enhances heat shock protein Hsp70 gene promoter activation. PLoS One 8: e69115 10.1371/journal.pone.0069115 ; 10.1371/journal.pone.0069115 10.1371/journal.pone.0069115; 10.1371/journal.pone.0069115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Numakawa T, Adachi N, Richards M, Chiba S, Kunugi H (2013) Brain-derived neurotrophic factor and glucocorticoids: Reciprocal influence on the central nervous system. Neuroscience 239: 157–172 10.1016/j.neuroscience.2012.09.073 ; 10.1073/pnas.1000274107; 10.1016/j.neuroscience.2012.09.073 [DOI] [PubMed] [Google Scholar]
- 90. McEwen BS (2012) Brain on stress: How the social environment gets under the skin. Proc Natl Acad Sci U S A 109 Suppl 217180–17185 10.1073/pnas.1121254109 ; 10.1073/pnas.1121254109 10.1073/pnas.1121254109; 10.1073/pnas.1121254109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Datson NA, van den Oever JM, Korobko OB, Magarinos AM, de Kloet ER, et al. (2013) Previous history of chronic stress changes the transcriptional response to glucocorticoid challenge in the dentate gyrus region of the male rat hippocampus. Endocrinology 154: 3261–3272 – 10.1210/en.2012–2233 ; – 10.1210/en.20122233 10.1210/en.20122233; – 10.1210/en.2012–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Struber N, Struber D, Roth G (2013) Impact of early adversity on glucocorticoid regulation and later mental disorders. Neurosci Biobehav Rev 38C: 17–37 10.1016/j.neubiorev.2013.10.015 ; 10.1016/j.neubiorev.2013.10.015 10.1016/j.neubiorev.2013.10.015; 10.1016/j.neubiorev.2013.10.015 [DOI] [PubMed] [Google Scholar]
- 93. Silverman MN, Sternberg EM (2012) Glucocorticoid regulation of inflammation and its functional correlates: From HPA axis to glucocorticoid receptor dysfunction. Ann N Y Acad Sci 1261: 55–63 10.1111/j.1749-6632.2012.06633.x ; 10.1111/j.1749-6632.2012.06633.x 10.1111/j.1749-6632.2012.06633.x; 10.1111/j.1749-6632.2012.06633.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wyrwoll CS, Holmes MC (2012) Prenatal excess glucocorticoid exposure and adult affective disorders: A role for serotonergic and catecholamine pathways. Neuroendocrinology 95: 47–55 10.1159/000331345 ; 10.1159/000331345 10.1159/000331345; 10.1159/000331345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Morcuende S, Munoz-Hernandez R, Benitez-Temino B, Pastor AM, de la Cruz RR (2013) Neuroprotective effects of NGF, BDNF, NT-3 and GDNF on axotomized extraocular motoneurons in neonatal rats. Neuroscience 250: 31–48 10.1016/j.neuroscience.2013.06.050 ; 10.1016/j.neuroscience.2013.06.050 10.1016/j.neuroscience.2013.06.050; 10.1016/j.neuroscience.2013.06.050 [DOI] [PubMed] [Google Scholar]
- 96. Hotta H, Kagitani F, Kondo M, Uchida S (2009) Basal forebrain stimulation induces NGF secretion in ipsilateral parietal cortex via nicotinic receptor activation in adult, but not aged rats. Neurosci Res 63: 122–128 10.1016/j.neures.2008.11.004 ; 10.1016/j.neures.2008.11.004 10.1016/j.neures.2008.11.004; 10.1016/j.neures.2008.11.004 [DOI] [PubMed] [Google Scholar]
- 97. Houeland G, Romani A, Marchetti C, Amato G, Capsoni S, et al. (2010) Transgenic mice with chronic NGF deprivation and alzheimer's disease-like pathology display hippocampal region-specific impairments in short- and long-term plasticities. J Neurosci 30: 13089–13094 10.1523/JNEUROSCI.0457-10.2010 ; – 10.1523/JNEUROSCI.0457-10.2010; – 10.1523/JNEUROSCI.0457–10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Madziar B, Shah S, Brock M, Burke R, Lopez-Coviella I, et al. (2008) Nerve growth factor regulates the expression of the cholinergic locus and the high-affinity choline transporter via the Akt/PKB signaling pathway. J Neurochem 107: 1284–1293 – 10.1111/j.1471–4159.2008.05681.x ; – 10.1111/j.14714159.2008.05681.x 10.1111/j.14714159.2008.05681.x; – 10.1111/j.1471–4159.2008.05681.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ishima T, Iyo M, Hashimoto K (2012) Neurite outgrowth mediated by the heat shock protein Hsp90alpha: A novel target for the antipsychotic drug aripiprazole. Transl Psychiatry 2: e170 10.1038/tp.2012.97 ; 10.1038/tp.2012.97 10.1038/tp.2012.97; 10.1038/tp.2012.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Read DE, Reed Herbert K, Gorman AM (2008) Heat shock enhances NGF-induced neurite elongation which is not mediated by Hsp25 in PC12 cells. Brain Res 1221: 14–23 10.1016/j.brainres.2008.05.028 ; 10.1016/j.brainres.2008.05.028 10.1016/j.brainres.2008.05.028; 10.1016/j.brainres.2008.05.028 [DOI] [PubMed] [Google Scholar]
- 101. Gruber HE, Hoelscher GL, Bethea S, Hanley EN (2012) Jr (2012) Interleukin 1-beta upregulates brain-derived neurotrophic factor, neurotrophin 3 and neuropilin 2 gene expression and NGF production in annulus cells. Biotech Histochem 87: 506–511 10.3109/10520295.2012.703692 ; 10.3109/10520295.2012.703692 10.3109/10520295.2012.703692; 10.3109/10520295.2012.703692 [DOI] [PubMed] [Google Scholar]
- 102. Lu AT, Yoon J, Geschwind DH, Cantor RM (2013) QTL replication and targeted association highlight the nerve growth factor gene for nonverbal communication deficits in autism spectrum disorders. Mol Psychiatry 18: 226–235 10.1038/mp.2011.155 ; 10.1038/mp.2011.155 10.1038/mp.2011.155; 10.1038/mp.2011.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Martinotti G, Di Iorio G, Marini S, Ricci V, De Berardis D, et al. (2012) Nerve growth factor and brain-derived neurotrophic factor concentrations in schizophrenia: A review. J Biol Regul Homeost Agents 26: 347–356. [PubMed] [Google Scholar]
- 104. Schaevitz LR, Moriuchi JM, Nag N, Mellot TJ, Berger-Sweeney J (2010) Cognitive and social functions and growth factors in a mouse model of rett syndrome. Physiol Behav 100: 255–263 10.1016/j.physbeh.2009.12.025 ; 10.1016/j.physbeh.2009.12.025 10.1016/j.physbeh.2009.12.025; 10.1016/j.physbeh.2009.12.025 [DOI] [PubMed] [Google Scholar]
- 105. Nihonmatsu-Kikuchi N, Hashimoto R, Hattori S, Matsuzaki S, Shinozaki T, et al. (2011) Reduced rate of neural differentiation in the dentate gyrus of adult dysbindin null (sandy) mouse. PLoS One 6: e15886 10.1371/journal.pone.0015886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Hattori T, Shimizu S, Koyama Y, Yamada K, Kuwahara R, et al. (2010) DISC1 regulates cell-cell adhesion, cell-matrix adhesion and neurite outgrowth. Mol Psychiatry 15: 778, 798–809. 10.1038/mp.2010.60 [DOI] [PubMed] [Google Scholar]
- 107. Loe-Mie Y, Lepagnol-Bestel AM, Maussion G, Doron-Faigenboim A, Imbeaud S, et al. (2010) SMARCA2 and other genome-wide supported schizophrenia-associated genes: Regulation by REST/NRSF, network organization and primate-specific evolution. Hum Mol Genet 19: 2841–2857 10.1093/hmg/ddq184 [DOI] [PubMed] [Google Scholar]
- 108. Koga M, Ishiguro H, Yazaki S, Horiuchi Y, Arai M, et al. (2009) Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum Mol Genet 18: 2483–2494 10.1093/hmg/ddp166 [DOI] [PubMed] [Google Scholar]
- 109. Kosho T, Okamoto N, Ohashi H, Tsurusaki Y, Imai Y, et al. (2013) Clinical correlations of mutations affecting six components of the SWI/SNF complex: Detailed description of 21 patients and a review of the literature. Am J Med Genet A 161A: 1221–1237 10.1002/ajmg.a.35933 ; 10.1002/ajmg.a.35933 10.1002/ajmg.a.35933; 10.1002/ajmg.a.35933 [DOI] [PubMed] [Google Scholar]
- 110. Gana S, Panizzon M, Fongaro D, Selicorni A, Memo L, et al. (2011) Nicolaides-baraitser syndrome: Two new cases with autism spectrum disorder. Clin Dysmorphol 20: 38–41 10.1097/MCD.0b013e32833edaa9 ; 10.1097/MCD.0b013e32833edaa9 10.1097/MCD.0b013e32833edaa9; 10.1097/MCD.0b013e32833edaa9 [DOI] [PubMed] [Google Scholar]
- 111. Derks EM, Ayub M, Chambert K, Del Favero J, Johnstone M, et al. (2013) A genome wide survey supports the involvement of large copy number variants in schizophrenia with and without intellectual disability. Am J Med Genet B Neuropsychiatr Genet. 10.1002/ajmg.b.32189 ; 10.1002/ajmg.b.32189 10.1002/ajmg.b.32189; 10.1002/ajmg.b.32189 [DOI] [PubMed] [Google Scholar]
- 112. Lo-Castro A, Galasso C, Cerminara C, El-Malhany N, Benedetti S, et al. (2009) Association of syndromic mental retardation and autism with 22q11.2 duplication. Neuropediatrics 40: 137–140 10.1055/s-0029-1237724 ; 10.1055/s-0029-1237724 10.1055/s-0029-1237724; 10.1055/s-0029-1237724 [DOI] [PubMed] [Google Scholar]
- 113. Ingason A, Kirov G, Giegling I, Hansen T, Isles AR, et al. (2011) Maternally derived microduplications at 15q11-q13: Implication of imprinted genes in psychotic illness. Am J Psychiatry 168: 408–417 10.1176/appi.ajp.2010.09111660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Sanders AR, Goring HH, Duan J, Drigalenko EI, Moy W, et al. (2013) Transcriptome study of differential expression in schizophrenia. Hum Mol Genet. 10.1093/hmg/ddt350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, et al. (2009) Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 460: 753–757 10.1038/nature08192 ; 10.1038/nature08192 10.1038/nature08192; 10.1038/nature08192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Djurovic S, Gustafsson O, Mattingsdal M, Athanasiu L, Bjella T, et al. (2010) A genome-wide association study of bipolar disorder in norwegian individuals, followed by replication in icelandic sample. J Affect Disord 126: 312–316 10.1016/j.jad.2010.04.007 ; 10.1016/j.jad.2010.04.007 10.1016/j.jad.2010.04.007; 10.1016/j.jad.2010.04.007 [DOI] [PubMed] [Google Scholar]
- 117. Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, et al. (2008) Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 82: 477–488 10.1016/j.ajhg.2007.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Chakrabarti B, Dudbridge F, Kent L, Wheelwright S, Hill-Cawthorne G, et al. (2009) Genes related to sex steroids, neural growth, and social-emotional behavior are associated with autistic traits, empathy, and asperger syndrome. Autism Res 2: 157–177 10.1002/aur.80 [DOI] [PubMed] [Google Scholar]
- 119. Dreha-Kulaczewski S, Kalscheuer V, Tzschach A, Hu H, Helms G, et al. (2013) A novel SLC6A8 mutation in a large family with X-linked intellectual disability: Clinical and proton magnetic resonance spectroscopy data of both hemizygous males and heterozygous females. JIMD Rep. 10.1007/8904_2013_261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lotan A, Lifschytz T, Slonimsky A, Broner EC, Greenbaum L, et al. (2013) Neural mechanisms underlying stress resilience in Ahi1 knockout mice: Relevance to neuropsychiatric disorders. Mol Psychiatry. 10.1038/mp.2013.123 ; 10.1038/mp.2013.123 10.1038/mp.2013.123; 10.1038/mp.2013.123 [DOI] [PubMed] [Google Scholar]
- 121. Kato T (2007) Molecular genetics of bipolar disorder and depression. Psychiatry Clin Neurosci 61: 3–19 10.1111/j.1440-1819.2007.01604.x [DOI] [PubMed] [Google Scholar]
- 122. Rivero O, Reif A, Sanjuan J, Molto MD, Kittel-Schneider S, et al. (2010) Impact of the AHI1 gene on the vulnerability to schizophrenia: A case-control association study. PLoS One 5: e12254 10.1371/journal.pone.0012254 ; 10.1371/journal.pone.0012254 10.1371/journal.pone.0012254; 10.1371/journal.pone.0012254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Riley B, Thiselton D, Maher BS, Bigdeli T, Wormley B, et al. (2010) Replication of association between schizophrenia and ZNF804A in the irish case-control study of schizophrenia sample. Mol Psychiatry 15: 29–37 10.1038/mp.2009.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, et al. (2008) Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet 40: 1053–1055 10.1038/ng.201 [DOI] [PubMed] [Google Scholar]
- 125. Riley B, Thiselton D, Maher BS, Bigdeli T, Wormley B, et al. (2010) Replication of association between schizophrenia and ZNF804A in the irish case-control study of schizophrenia sample. Mol Psychiatry 15: 29–37 10.1038/mp.2009.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, et al. (2012) Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell 149: 525–537 10.1016/j.cell.2012.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Steinberg S, Mors O, Borglum AD, Gustafsson O, Werge T, et al. (2010) Expanding the range of ZNF804A variants conferring risk of psychosis. Mol Psychiatry. 10.1038/mp.2009.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Steinberg S, Mors O, Borglum AD, Gustafsson O, Werge T, et al. (2011) Expanding the range of ZNF804A variants conferring risk of psychosis. Mol Psychiatry 16: 59–66 10.1038/mp.2009.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Rosati A, Graziano V, De Laurenzi V, Pascale M, Turco MC (2011) BAG3: A multifaceted protein that regulates major cell pathways. Cell Death Dis 2: e141 10.1038/cddis.2011.24 ; 10.1038/cddis.2011.24 10.1038/cddis.2011.24; 10.1038/cddis.2011.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Yunoki T, Kariya A, Kondo T, Hayashi A, Tabuchi Y (2013) The combination of silencing BAG3 and inhibition of the JNK pathway enhances hyperthermia sensitivity in human oral squamous cell carcinoma cells. Cancer Lett 335: 52–57 10.1016/j.canlet.2013.01.049 ; 10.1016/j.canlet.2013.01.049 10.1016/j.canlet.2013.01.049; 10.1016/j.canlet.2013.01.049 [DOI] [PubMed] [Google Scholar]
- 131. Brennand KJ, Gage FH (2012) Modeling psychiatric disorders through reprogramming. Dis Model Mech 5: 26–32 10.1242/dmm.008268 ; 10.1242/dmm.008268 10.1242/dmm.008268; 10.1242/dmm.008268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Perez-Santiago J, Diez-Alarcia R, Callado LF, Zhang JX, Chana G, et al. (2012) A combined analysis of microarray gene expression studies of the human prefrontal cortex identifies genes implicated in schizophrenia. J Psychiatr Res. 10.1016/j.jpsychires.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 133. Chinnadurai S, Goudy S (2012) Understanding velocardiofacial syndrome: How recent discoveries can help you improve your patient outcomes. Curr Opin Otolaryngol Head Neck Surg 20: 502–506 10.1097/MOO.0b013e328359b476 ; 10.1097/MOO.0b013e328359b476 10.1097/MOO.0b013e328359b476; 10.1097/MOO.0b013e328359b476 [DOI] [PubMed] [Google Scholar]
- 134. Shprintzen RJ, Goldberg RB, Lewin ML, Sidoti EJ, Berkman MD, et al. (1978) A new syndrome involving cleft palate, cardiac anomalies, typical facies, and learning disabilities: Velo-cardio-facial syndrome. Cleft Palate J 15: 56–62. [PubMed] [Google Scholar]
- 135. Scambler PJ, Kelly D, Lindsay E, Williamson R, Goldberg R, et al. (1992) Velo-cardio-facial syndrome associated with chromosome 22 deletions encompassing the DiGeorge locus. Lancet 339: 1138–1139. [DOI] [PubMed] [Google Scholar]
- 136. Driscoll DA, Spinner NB, Budarf ML, McDonald-McGinn DM, Zackai EH, et al. (1992) Deletions and microdeletions of 22q11.2 in velo-cardio-facial syndrome. Am J Med Genet 44: 261–268 10.1002/ajmg.1320440237 [DOI] [PubMed] [Google Scholar]
- 137. Murphy KC, Owen MJ (2001) Velo-cardio-facial syndrome: A model for understanding the genetics and pathogenesis of schizophrenia. Br J Psychiatry 179: 397–402. [DOI] [PubMed] [Google Scholar]
- 138. Kates WR, Antshel KM, Fremont WP, Shprintzen RJ, Strunge LA, et al. (2007) Comparing phenotypes in patients with idiopathic autism to patients with velocardiofacial syndrome (22q11 DS) with and without autism. Am J Med Genet A 143A: 2642–2650 10.1002/ajmg.a.32012 [DOI] [PubMed] [Google Scholar]
- 139. Papolos DF, Veit S, Faedda GL, Saito T, Lachman HM (1998) Ultra-ultra rapid cycling bipolar disorder is associated with the low activity catecholamine-O-methyltransferase allele. Mol Psychiatry 3: 346–349. [DOI] [PubMed] [Google Scholar]
- 140. Shprintzen RJ, Goldberg R, Golding-Kushner KJ, Marion RW (1992) Late-onset psychosis in the velo-cardio-facial syndrome. Am J Med Genet 42: 141–142 10.1002/ajmg.1320420131 [DOI] [PubMed] [Google Scholar]
- 141. Pulver AE, Nestadt G, Goldberg R, Shprintzen RJ, Lamacz M, et al. (1994) Psychotic illness in patients diagnosed with velo-cardio-facial syndrome and their relatives. J Nerv Ment Dis 182: 476–478. [DOI] [PubMed] [Google Scholar]
- 142. Zagha E, Ozaita A, Chang SY, Nadal MS, Lin U, et al. (2005) DPP10 modulates Kv4-mediated A-type potassium channels. J Biol Chem 280: 18853–18861 10.1074/jbc.M410613200 [DOI] [PubMed] [Google Scholar]
- 143. Schmunk G, Gargus JJ (2013) Channelopathy pathogenesis in autism spectrum disorders. Front Genet 4: 222 10.3389/fgene.2013.00222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Martinez-Martinez P, Molenaar PC, Losen M, Stevens J, Baets MH, et al. (2013) Autoantibodies to neurotransmitter receptors and ion channels: From neuromuscular to neuropsychiatric disorders. Front Genet 4: 181 10.3389/fgene.2013.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Yao JJ, Sun J, Zhao QR, Wang CY, Mei YA (2013) Neuregulin-1/ErbB4 signaling regulates Kv4.2-mediated transient outward K+ current through the Akt/mTOR pathway. Am J Physiol Cell Physiol 305: C197–206 10.1152/ajpcell.00041.2013 ; 10.1152/ajpcell.00041.2013 10.1152/ajpcell.00041.2013; 10.1152/ajpcell.00041.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Georgiev D, Arion D, Enwright JF, Kikuchi M, Minabe Y, et al. (2013) Lower gene expression for KCNS3 potassium channel subunit in parvalbumin-containing neurons in the prefrontal cortex in schizophrenia. Am J Psychiatry. 10.1176/appi.ajp.2013.13040468 ; 10.1176/appi.ajp.2013.13040468 10.1176/appi.ajp.2013.13040468; 10.1176/appi.ajp.2013.13040468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Lee YH, Kim JH, Song GG (2013) Pathway analysis of a genome-wide association study in schizophrenia. Gene 525: 107–115 10.1016/j.gene.2013.04.014 ; 10.1016/j.gene.2013.04.014 10.1016/j.gene.2013.04.014; 10.1016/j.gene.2013.04.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Text S1. Supplementary methods
Table S1. RNA-seq statistics. C1 and C2 refer to controls 1 and 2: HS1 and HS2 are the heat shocked counterparts.
Table S2. RNA-seq reads in TPM (Transcripts Per Million) for all genes arranged by p-value (lowest to highest). The 186 genes that showed nominally significant differences in the mean log2 fold-change (HS/control) are in bold type.
Table S3: Top Gene Ontology (GO) Terms for genes up and down-regulated by HS.
Table S4. Ingenuity Pathway Analysis showing disease and biological functions of differentially expressed genes
Table S5: Upstream Regulator Analysis, all genes
(ZIP)