Abstract
Caenorhabditis elegans GLD-1, a maxi-KH motif containing RNA-binding protein, has various functions mainly during female germ cell development, suggesting that it likely controls the expression of a selective group of maternal mRNAs. To gain an insight into how GLD-1 specifically recognizes these mRNA targets, we identified 38 biochemically proven GLD-1 binding regions from multiple mRNA targets that are among over 100 putative targets co-immunoprecipitated with GLD-1. The sequence information of these regions revealed three over-represented and phylogenetically conserved sequence motifs. We found that two of the motifs, one of which is novel, are important for GLD-1 binding in several GLD-1 binding regions but not in other regions. Further analyses indicate that the importance of one of the sequence motifs is dependent on two aspects: (1) surrounding sequence information, likely acting as an accessory feature for GLD-1 to efficiently select the sequence motif and (2) RNA secondary structural environment where the sequence motif resides, which likely provides “binding-site accessibility” for GLD-1 to effectively recognize its targets. Our data suggest some mRNAs recruit GLD-1 by a distinct mechanism, which involves more than one sequence motif that needs to be embedded in the correct context and structural environment.
Keywords: GLD-1, mRNA targets, sequence motif, context dependency, binding site accessibility
Introduction
Germline and early development of various organisms is highly dependent on the temporal and spatial control of maternal gene products. In the C. elegans germline, most maternal mRNAs that are destined to function in the oocyte and embryo are initially transcribed during mitosis or early stages of meiosis in germ cell nuclei, the same cells that proceed through the germline to later produce oocytes.1-6 Therefore, precise temporal and spatial regulation of these maternal mRNAs is necessary to produce functional oocytes and is essential to allow specific patterns of protein accumulation to direct normal embryo development.7,8 Key components of these post-transcriptional controls are RNA-binding proteins (RBPs) that bind to regulatory regions generally located in the 5′ untranslated regions (UTRs) and/or the 3′UTRs of their mRNA targets.9-16
One of the major regulators of maternal mRNA expression in the C. elegans germline is an RNA-binding protein, GLD-1 (GermLine Development defective). GLD-1 is a member of a family of proteins, including human/mouse Quaking, SAM68, and Drosophila How that share an approximately 200 amino acid conserved region called the GSG or STAR domain.3,17,18 Within this conserved region is a maxi-KH RNA-binding domain. In the C. elegans germline, GLD-1 primarily functions to inhibit mitosis and/or promote germ cell entry into meiotic development, ensure proper pachytene progression, oogenesis and early embryogenesis, prevent germ cell apoptosis in early meiosis, promote spermatogenesis at early larval stages, and maintain germ cell identity.16,19-25 Upon genetic elimination of gld-1, C. elegans exhibits a loss of sperm production and germ cells that are undergoing oogenesis leave meiotic prophase and proliferate ectopically to form a germline tumor or trans-differentiate into somatic cell lineages.20,22,23
Diverse germline functions of GLD-1 predicted that GLD-1 would control multiple maternal mRNAs, and these predictions were experimentally confirmed.11,13,15,16,19,26-31 In these studies, GLD-1 has been demonstrated to repress the translation of its mRNA targets in the distal region of the wild-type adult hermaphrodite germline where GLD-1 is abundant in the cytoplasm. Then as GLD-1 levels decrease around the loop region and become undetectable in the proximal region, proteins of GLD-1 mRNA targets are expressed in developing oocytes.
Many RBPs, including GLD-1, have multiple RNA targets, which they recognize through specific binding sites.11,27,30,32-36 Therefore, understanding how these RBPs can differentiate targets from non-targets in the cell is crucial to comprehending their selectivity and function. To date, studies have shown that many RBPs generally bind to relatively degenerate, short sequence motifs (core recognition site) that often reside either in the 5′ ends and/or 3′ ends of the mRNAs.11,30,37 However, these motifs alone do not contain enough information for RBPs to select their targets effectively. Several reports have revealed that other factors surrounding the core recognition site can influence the selectivity of RBPs.38-42
There are a number of criteria that have been demonstrated to regulate RBP-mRNA interactions. First, sequences surrounding a core recognition site may cause differential binding. For example, this contextual influence on the recognition sites in mRNA targets was shown for the She-complex that regulates cytoplasmic localization of selected mRNAs in Saccharomyces cerevisiae.39 Via a high-throughput selection strategy, Jambhekar et al. revealed that a highly degenerate core recognition motif predicted to lie in single-stranded regions of mRNAs is important for She-complex-dependent transport. In addition, the selectivity of the She-complex also requires specific sequence features adjacent to the core recognition motif, a phenomenon referred to as “context dependency.”
Second, several mRNA targets of RBPs have been shown to contain binding sites that reside in specific secondary and/or tertiary structures.40,41 Most RNA-binding proteins contain domains that bind RNA in a sequence-specific manner (i.e., RNA recognition motif [RRM] and the K homology [KH] domain) and their ability to recognize the specific sequences is often affected by the secondary structures surrounding the sequences.43,44 For example, the mouse Prrp protein binds two motifs that have to be located in single-stranded regions.45 Other studies also reported similar mechanisms of RNA binding.46,47 Furthermore, it is also demonstrated that the sequestration of binding sites to single- or double-stranded regions promoted or abolished protein binding, respectively.48 These studies highlight that secondary structures dictate the physical accessibility of binding motifs.
To date, two studies have investigated the RNA binding specificity of GLD-1 through comprehensive analyses.27,30 Both groups reported that a core GLD-1 binding motif (GBM) is a degenerate sequence that is similar, yet distinct, from the conserved SBE motif first characterized by Ryder et al. In particular, Wright et al. show that one or a combination of this motif is sufficient to impose GLD-1-mediated translational repression. In this study, we independently identified over 100 additional putative mRNA targets of GLD-1 by co-immunoprecipitation with GLD-1 from cytosolic extracts followed by microarray analysis (RIP-chip).30,49 Next, we selected 32 putative targets from our RIP-chip analysis and identified specific regions in these mRNAs that interact with GLD-1 biochemically (herein referred to as GLD-1 binding regions). GLD-1 is able to interact with a total of 38 GLD-1 binding regions in the 5′, 3′, or both ends of each of the 32 mRNA targets examined. Next, by utilizing the 38 GLD-1 binding regions, and thus, excluding regions of GLD-1 mRNA targets that likely have no relevance in GLD-1 binding, we identified three over-represented sequence motifs through computational analyses. Our findings expand on the results of previous studies in that we demonstrate that two motifs, one of which is novel, are important for GLD-1 binding in four GLD-1 mRNA targets. Moreover, we demonstrated the ability of secondary structure to obscure a GLD-1 binding motif and that functionality of a GLD-1 binding motif is dependent on the surrounding sequences.
Results
Identification of GLD-1 associated mRNAs in C. elegans
Previous study by Wright et al. subjected 3′UTRs of GLD-1 mRNA targets to identify over-represented hexamers based on the assumption that GLD-1 would bind to 3′UTRs of all mRNA targets. Since we previously showed that GLD-1 binds to only 5′UTR of one target28 and both 5′ and 3′ends of another target11 we determined to narrow down true GLD-1 binding regions from many mRNA targets biochemically and to utilize these biochemically proven GLD-1 binding regions to define GLD-1 in vivo binding rules. With multiple biochemically proven GLD-1 binding regions, we were able to identify new unknown binding elements and the requirements of the context and structural environment for GLD-1 to recognize its targets.
To expand and potentially have a complete list of GLD-1 mRNA targets, we employed a systematic approach using GLD-1 immunoprecipitation (IP) of protein-RNA complexes followed by microarray detection strategy. A major improvement over the previous method used to identify GLD-1 mRNA targets11 was the use of microarrays to detect enrichment of mRNAs in the GLD-1 IP. Functional GLD-1 for IP was obtained from cytosol extracts of a transgenic line in which the gld-1(q485)-null mutant was rescued by a genomic insertion (ozIs2 [gld-1::gfp::flag]) encoding wild-type GLD-1 GFP fusion with the FLAG epitope placed at the C terminus (GLD-1::GFP::FLAG). After FLAG and control mouse IgG IP, RNAs were extracted and linearly amplified, then subjected to microarray analysis with arrays that have ~17500 genes (about 90% of protein-coding genes in the C. elegans genome).50,51 We performed four microarray experiments with the IgG and the FLAG immunoprecipitated RNAs. Each experiment used RNA obtained from two independent IP experiments. From the analysis of these four sets, we identified 129 genes that are enriched more than 2-fold in the FLAG IP (P < 0.05). Among these, 49 genes were enriched more than 3-fold (Table S1).
Recently, a study by Wright et al. (2011)11 also identified GLD-1 targets using a similar RIP-chip strategy. They reported that 948 mRNAs were detected as > 3-fold enriched in their analysis, as opposed to 48 in this study, which they were able to further validate through subsequent deep sequencing (RNA-seq) analyses.30 109 of the targets Wright et al. identified were among the 129 identified in this study. We suspect that the discrepancy of our results may be due to mainly two factors: a different immunoprecipitation strategy and the utilization of a different microarray chip.
External validation of our RIP-chip experiments arises from our previous identification of GLD-1 targets via the IP/subtraction/cloning/sequencing strategy. We performed two independent subtraction/cloning/sequencing experiments and identified 16 mRNA targets in both experiments.11 Of the 49 putative targets that were enriched more than 3-fold in the RIP-Chip, we previously identified 12. Of the 80 putative targets enriched between 2- to 3-fold by RIP-chip, two were previously identified (Table S1). Thus, we identified 14 of the 16 GLD-1 mRNA targets identified previously and failed to identify two: lin-45 and B0280.5. lin-45 was not on the array while signals for B0280.5 fell well below the baseline, indicating inefficient spotting on the array. Moreover, targets identified by others were also enriched greater than 2-fold in our experiment (e.g. glp-1).13
To validate the microarray data further, we examined 73 putative targets for enrichment in the FLAG IP via real-time RT-PCR analysis. In addition, we examined several mRNAs that are not enriched in the FLAG IP as controls. For 70 of 73 targets examined, we found that the fold enrichment in the real-time RT-PCR was roughly proportional to that in the RIP-chip analysis. Only three targets appear to be false-positives as their fold enrichment in the RIP-chip by the real-time RT-PCR was approximately 1. In addition, the fold enrichment of several controls was approximately 1 (Table S1). Taken together, these data provide strong proof of principle that the majority of the 129 RIP-chip-defined targets likely interact with GLD-1 in vivo. However, we cannot rule out the possibility that our analysis was not able to pick up other true GLD-1 mRNA targets. In addition, several GLD-1 targets identified previously have fold enrichment of a little less than two, including pie-1 and cey-2.19,52 Thus, we expect that the number of GLD-1 mRNA targets is likely more than 129, which is consistent with the various functions that GLD-1 controls during C. elegans germline development.
Functional annotation of GLD-1 targets
To explore functional themes of the putative GLD-1 targets, we subjected the 129 targets that were enriched more than 2-fold in our RIP-chip analysis to enriched Gene Ontology (GO) analysis. As putative GLD-1 mRNA targets, we expected that their encoded functions should play a role in the germline and early embryo. GLD-1 mRNA targets are significantly enriched in 32 functional groups (Table S2), 10 of which correspond to the GO terms enriched in the Wright et al. (2011) study (cytokinesis, cell division, embryonic development, reproductive processes, cell cycle, DNA replication, DNA metabolic process, and cell fate commitment). This was not surprising since 109 of the 129 putative GLD-1 targets presented here were also detected by Wright et al.30 Table 1 represents 20 of the most significantly enriched functional groups. Among them, at least 14 represent the germline and early embryo processes that GLD-1 controls.
Table 1. Functional representation of putative GLD-1 mRNA targets.
| Function* | Gene size† | GLD-1 targets‡ | Significance |
|---|---|---|---|
| embryonic development ending in birth or egg hatching (GO:0009792) | 2338 | 46 | 1.52E-10 |
| cytokinesis (GO:0000910) | 105 | 9 | 8.78E-08 |
| protein binding (GO:0005515) | 2098 | 38 | 1.04E-07 |
| embryonic cleavage (GO:0040016) | 181 | 10 | 1.02E-06 |
| DNA replication (GO:0006260) | 47 | 6 | 1.27E-06 |
| chitin metabolic process (GO:0006030) | 13 | 3 | 1.09E-04 |
| DNA repair (GO:0006281) | 104 | 6 | 1.27E-04 |
| transferase activity (GO:0016740) | 594 | 14 | 1.39E-04 |
| nuclease activity (GO:0004518) | 36 | 4 | 1.44E-04 |
| chitin binding (GO:0008061) | 16 | 3 | 2.11E-04 |
| nucleotidyltransferase activity (GO:0016779) | 43 | 4 | 2.90E-04 |
| glutamate-ammonia ligase activity (GO:0004356) | 5 | 2 | 5.43E-04 |
| glutamine biosynthetic process (GO:0006542) | 5 | 2 | 5.43E-04 |
| centrosome (GO:0005813) | 23 | 3 | 6.42E-04 |
| P granule (GO:0043186) | 24 | 3 | 7.29E-04 |
| oocyte maturation (GO:0001556) | 9 | 2 | 1.92E-03 |
| cleavage furrow (GO:0032154) | 9 | 2 | 1.92E-03 |
| nitrogen compound metabolic process (GO:0006807) | 11 | 2 | 2.90E-03 |
| cell cycle (GO:0007049) | 83 | 4 | 3.43E-03 |
| protein domain specific binding (GO:0019904) | 12 | 2 | 3.46E-03 |
* Function represents similar functional groups of genes classified into the same category according to GO identifier (http://www.geneontology.org); †Gene size represents the total number of genes in each functional group in the entire C. elegans genome; ‡GLD-1 targets indicate the number of GLD-1 targets observed in the same functional group.
First, they include oogenesis, reproduction, and embryogenesis as expected for GLD-1 targets. Second, they also include DNA replication, mitosis, cell division, and cell cycle, again as expected since GLD-1 inhibits mitosis and promotes entry into meiosis.21,22 Third, they also include a few nucleases that likely function during apoptosis as GLD-1 inhibits apoptosis in early meiosis.16 These results are consistent with previous studies of GLD-1 protein.11,16,19,28
GLD-1 binds 5′ and/or 3′ends of its mRNA targets
The mechanism(s) by which GLD-1 specifically recognizes and binds its mRNA targets among thousands of mRNAs expressed in the germline is still not fully understood. To date, two studies have comprehensively explored the RNA binding specificity of GLD-1.27,30 Both groups reported that a core GLD-1 binding motif (GBM) is a degenerate sequence similar to the conserved SBE motif first characterized by Ryder et al. (2003), where Wright et al. (2011)11 showed that one or a combination of these motifs was both necessary and sufficient to confer GLD-1 binding. However, in our analysis (explained below), we found several GLD-1 binding regions do not contain this degenerate GBM or SBE, while others contain only one GBM that is weakly conserved. This led us to hypothesize that GLD-1 recognition and binding cannot be limited to just one universal sequence motif, but perhaps involves more than one or a combination of distinct motifs and/or mechanisms.
To more completely define the rules through which GLD-1 distinguishes mRNA targets from non-targets and represses their translation, we utilized a different approach from previous studies. This systematic approach was based on the hypothesis that the GLD-1 binding motifs should be over-represented in the biochemically proven GLD-1 binding regions (GBRs). We first narrowed down regions from multiple GLD-1 mRNA targets identified through our GLD-1 RIP-chip analysis. Then, we examined whether these regions could bind GLD-1 through GLD-1/Biotin RNA pull-down assays.11 Finally, after identifying a significant number of GBRs, we performed two independent computational analyses to identify over-represented sequence motifs embedded in the GBRs.
We used wild-type cytosol extracts to identify GBRs on multiple targets successfully in earlier studies.11,16,19,28 Because we use cytosol extracts, we could identify binding regions, and therefore binding motifs, of not only GLD-1 but possible in vivo GLD-1-containing complexes. Previously, we examined entire mRNAs, including 5′UTR, ORF, and 3′UTR of six mRNA targets of GLD-1 (gna-2, rme-2, oma-1, oma-2. lin-45, and cep-1) to narrow down GBRs.11,16,28 From these studies, we demonstrated that GLD-1 bound the 5′UTR and/or 3′UTR. In addition, GLD-1 bound to the ORF of a few targets, but only near the 5′ or 3′ ends of the ORF. Thus, to increase the number of GBRs, we focused on the untranslated regions (UTRs) and 150 bases at the beginnings and ends of the open reading frames of additional mRNA targets (see Materials and Methods). We selected 26 additional GLD-1 targets identified through our RIP-chip experiments that have diverse functions throughout the germline development (Table 2).
Table 2. Thirty-two validated GLD-1 targets have 38 GLD-1 binding regions.
| GLD-1 Targets* | Microarray | GLD-1 binding¶ | ||||
|---|---|---|---|---|---|---|
| Sequence name | Gene name | FLAG/IgG† | P-value‡ | DescriptionΠ | 5′ end | 3′ end |
| ZC513.6 | oma-2 | 13.47 | 2.00E-05 | Zinc finger protein of the TIS11 finger type | Y | Y |
| T05G5.7 | rmd-1 | 8.75 | 1.10E-03 | TPR domain | N/D | Y |
| T11F8.3 | rme-2 | 8.36 | 8.00E-05 | Yolk receptor | Y | Y |
| Y75B12B.1 | N/D | 8.01 | 8.00E-04 | Probable transposase | Y | N |
| C50B6.2 | nasp-2 | 7.66 | 6.00E-04 | Histone binding protein | Y | N |
| C07G2.1 | cpg-1 | 7.40 | 2.00E-04 | Chitin binding peritrophin | N | Y |
| F32B6.5 | sss-1 | 6.10 | 1.01E-02 | Sperm-specific protein family, S class | Y | Y |
| F44D12.4 | N/D | 5.11 | 5.90E-03 | PDZ domain | N/D | N |
| H02I12.1 | cbd-1 | 4.87 | 2.00E-04 | Chitin binding peritrophin | Y | N |
| R09B3.1 | exo-3 | 4.83 | 1.10E-03 | Exonuclease | Y | N |
| T01H8.1 | rskn-1 | 4.69 | 1.00E-03 | Protein kinase C | N | Y |
| C09G9.6 | oma-1 | 4.17 | 0.00E+00 | Zinc finger protein of the TIS11 finger type | Y | Y |
| ZK829.5 | tbx-36 | 4.11 | 5.50E-03 | T-box transcription factor | N | Y |
| F14B4.2 | N/D | 4.04 | 2.62E-02 | Hexokinase | N | Y |
| T23G11.2 | gna-2 | 3.99 | 3.00E-04 | Glucosamine phosphate N-Acetyl transferase | Y | N |
| W09C5.2 | unc-59 | 3.90 | 4.20E-03 | Septin required for axon and DTC migration | N | Y |
| T07A9.6 | daf-18 | 3.84 | 7.00E-04 | PTEN lipid phosphatase | N/D | N |
| F43G6.1 | dna-2 | 3.68 | 5.10E-03 | Endonuclease/helicase | N | Y |
| T01C3.2 | N/D | 3.38 | 1.46E-02 | unknown | N/D | Y |
| F26D10.10 | gln-5 | 3.32 | 1.00E-04 | Glutamine sythetase | N | Y |
| T27E9.3 | cdk-5 | 3.13 | 8.30E-03 | Cyclin-dependent kinase | N/D | Y |
| Y49E10.14 | pie-1 | 2.90 | 6.30E-03 | CCCH zinc finger protein | N | Y |
| F19B6.2 | ufd-1 | 2.55 | 3.60E-03 | Ubiquitin fusion degradation protein | N | Y |
| Y47G6A.8 | crn-1 | 2.55 | 3.00E-03 | Cell death-related nuclease | N | Y |
| T13F2.9 | N/D | 2.53 | 2.06E-02 | unknown | N/D | Y |
| M04F3.1 | rpa-2 | 2.37 | 1.00E-03 | unknown | Y | N |
| T27F2.3 | bir-1 | 2.21 | 2.70E-03 | Chromosome segregation | N/D | Y |
| F35B12.5 | sas-5 | 2.16 | 1.00E-04 | Centriole formation | Y | Y |
| T06E6.2 | cyb-3 | 1.86 | 7.10E-03 | A member of the cyclin B family | N | Y |
| F58A4.3 | hcp-3 | 1.80 | 5.60E-03 | Centromere protein (CENP)-A homolog | N | Y |
| F52B5.5 | cep-1 | 1.64 | 1.94E-02 | Ortholog of the human tumor suppressor p52 | N | Y |
| Y73B6A.5§ | lin-45 | N/D | N/D | Ortholog of the vertebrate protein RAF | N | Y |
Sequence and gene names of 32 targets examined; †FLAG/IgG ratios in decreasing order in microarray analysis. With the exception of T06E6.2, F58A4.3, and F52B5.5, all targets have an enrichment of > 2.0 in the GLD-1-FLAG IP compared with the IgG IP; ‡P values of microarray analysis; §Y73B6.A.5 was not spotted in the microarray analysis but was identified previously (Lee and Schedl, 2001); ΠShort descriptions of the functions of targets if known; ¶(Y) and (N) indicate that GLD-1 was detectable (binding) and undetectable (not binding) via western blot analysis after biotin-RNA pull down assays. (N/D) indicate that binding assays were not performed.
After cloning and thorough binding analysis of a combined 32 GLD-1 mRNA targets, we concluded that GLD-1 binds to either the 5′, 3′, or both ends of essentially all mRNA targets examined, validating that our GLD-1 RIP-chip data identified authentic targets. In brief, we were able to identify 38 GLD-1 binding regions from 32 mRNA targets. GLD-1 bound to only 5′ ends of 8, to only 3′ ends of 19, and to both the 5′ and 3′ ends of five mRNA targets (Table 2 and Fig. 1A and B). These findings indicate that GLD-1 translational regulation can be mediated through the 5′ end or 3′ end.
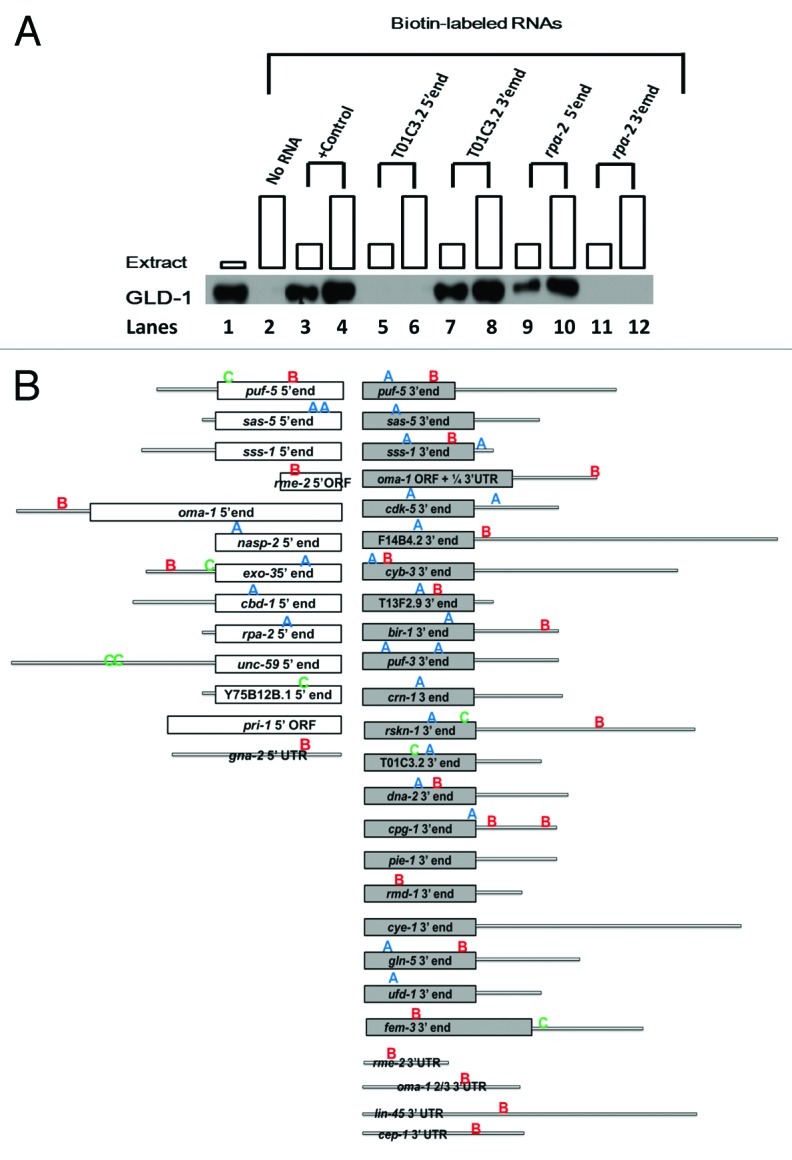
Figure 1. GLD-1 binds to the 5′, 3′ or both ends of 32 mRNA targets. (A) GLD-1 binds to the 3′ end of T01C3.2 and the 5′end of MO4F3.1 RNAs. Western blot analysis of GLD-1/ Biotin-RNA pull-down assays of the 5′ and 3′ ends T01C3.2 and M04F3.1 mRNAs. All biotin-labeled RNAs (400 ng) were incubated with increasing amounts of wild-type cytosol extracts (50–150 ug) and then the RNA-protein complexes were isolated with Streptavidin-magnetic-beads. A western blot analysis was performed to detect the presence of GLD-1 in the complex. GLD-1 was probed in the cytosolic extract in the 1st lane, and no RNA was added to the binding reaction in the second lane. tra-2 3′UTR was used as a positive (+) control with increasing amount of cytosolic extract in the third and fourth lanes. Subsequent lanes represent binding reactions with 5′ and 3′ ends of T01C3.2 and M04F3.1 biotin-labeled RNAs. Boxes at each lane are proportional to the amount of extract used. (B) GLD-1 binds to 38 regions in 32 mRNA targets. 32 GLD-1 targets identified in the RIP-chip analysis were employed to identify GLD-1 binding regions. GLD-1 was able to bind to the 5′, 3′, or both ends of all targets. For each region, the GLD-1/biotin RNA binding assays and western blot analyses were performed at least twice to validate the consistency of the results. White and gray bars indicate the 5′ and 3′ coding regions, respectively, and the gray lines indicate the untranslated regions (UTRs). A Patser scan was performed to locate the over-represented motif A, B, and C identified through PhyloCon and PhyloNet within the 38 GLD-1 binding regions, which are marked at approximate locations. Blue “A,” red “B,” and green “C” represent the Motif A, Motif B, and Motif C, respectively.
Conserved sequence motifs in GLD-1 binding regions
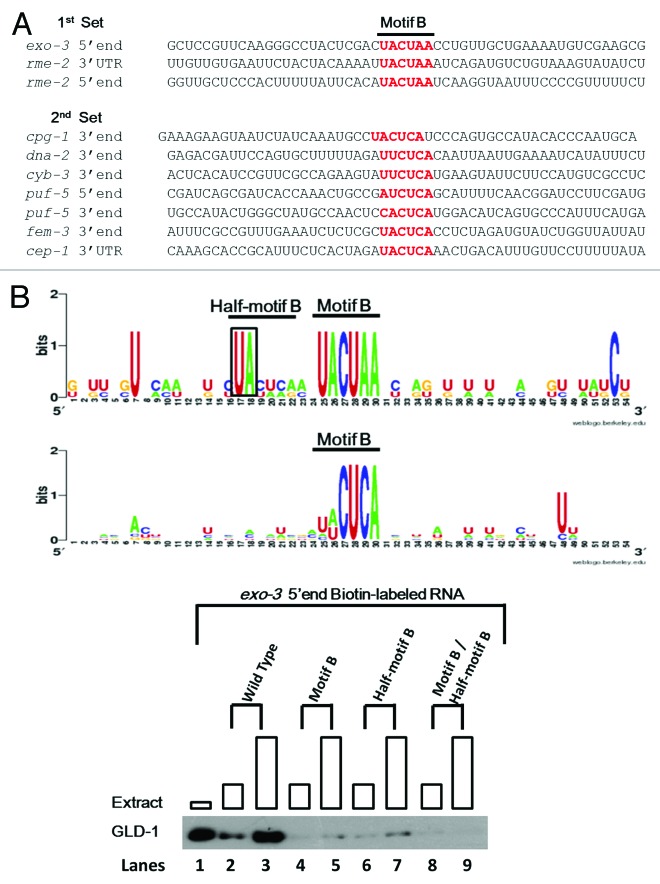
In order to further increase our probability of finding over-represented and conserved sequence motifs, we included orthologous regions to the 38 GBRs from four closely related nematode species: C. briggsae C. remanei, C. japonica, and C. brenneri. The C. elegans GLD-1 and GLD-1 from other nematode species are likely to have a similar set of mRNA targets and, thus, the binding motifs therein.11,53 Two computational analyses were performed: PhyloCon54 and PhyloNet.55 PhyloCon identified two significantly over-represented sequence motifs: “AGAAGC” (Motif A) and “CUACUAAC” (Motif B) (Fig. 2A). Motif B is essentially identical to the previously identified sequence motif, the SBE that is essential for GLD-1 binding to tra-2 and cye-1 mRNA,19,52 and is also important for Quaking (mouse ortholog of GLD-1) binding to its targets.56 Furthermore, it is very similar to the prominent GBM identified previously, encompassing the “strong GBMs” reported by Wright et al. (2011).27,30 PhyloNet also identified Motif A and B as well as a third motif: “GAACGA” (Motif C), which is less prevalent than Motif A and B, yet potentially important for GLD-1 binding (Fig. 2A). More interestingly, both Motif A and Motif C are novel motifs.
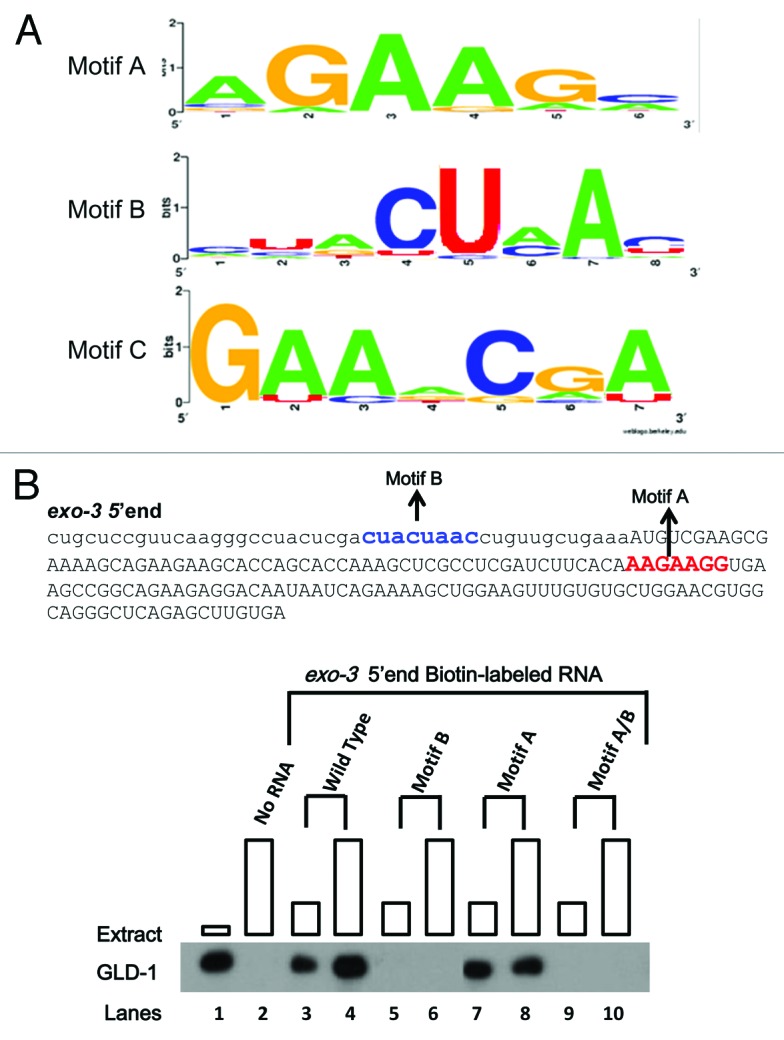
Figure 2. Over-represented sequence Motifs A, B, and C were identified and examined for their importance in GLD-1 binding. (A) The sequences of 38 GBRs from 32 mRNA targets and their corresponding orthologous regions from four closely related nematodes were subjected to PhyloCon and PhyloNet analyses, and sequence Motif A and B (PhyloCon and PhyloNet) and C (PhyloNet) were identified as over-represented. Each motif is represented as a logo that shows a graphical representation of a nucleic acid multiple sequence alignment. The overall height of the stacked nucleotides within each motif indicates the degree of sequence conservation at the position, while the height of symbols within the stack indicates the relative frequency of each nucleic acid at the position. (B) Motif B is important for GLD-1 binding in R09B3.1 5′ end. Single or double mutations containing mutations in Motif B significantly reduces GLD-1 binding to the R09B3.1 3′ end. The RNA sequence of the R09B3.1 3′ end is shown (top). Sequence in lower-case represents the 5′UTR and in upper-case the first 150 nucleotides of the ORF. The Motif A and B are depicted in the sequence. The four most conserved nucleotides were mutated, where the Motif A was mutated to “AUCUUGG” while the Motif B is mutated “UAGAUU”. Biotin-RNA pull-down assays were performed with the wild-type and mutated R09B3.1 3′ end as described previously. Boxes at each lane are proportional to the amount of extract used.
Next, we performed a Patser scan to determine the location of these motifs within the 38 GBRs. We found that 27 Motif As are present within 23 GBRs, 22 Motif Bs are present within 22 GBRs, and eight Motif Cs are present within seven GBRs. Among these, Motif A, B, and C are exclusively present in 10 GBRs, seven GBRs, and two GBRs, respectively. Moreover, Motif A and B, Motif A and C, and Motif B and C are present collectively in 10, one, and two GBRs, respectively. Lastly, two GBRs harbor all three motifs, while three GBRs contain none (Fig. 1B). These findings support our initial hypothesis in that GLD-1 binding sites are likely not confined to a universal motif, i.e., the GBM/SBE;30,52 instead, GLD-1 may recognize more than one distinct motif that may dictate GLD-1 binding to different sets of mRNA targets.
Motifs B and C are important for GLD-1 binding to a small number of targets
We examined the importance of the Motifs A, B, and C for GLD-1 binding in vitro. To accomplish this, we mutated the motifs within several GBRs at their four most phylogenetically conserved nucleotides and assessed GLD-1 binding to the mutated compared with wild-type GBRs using the GLD-1/RNA pull-down assay. Any reduction in binding to the mutated GBRs compared with its wild-type counterpart indicates that the motif is important for GLD-1 binding in vitro.
We examined a total of 34 over-represented sequence motifs within 28 GBRs: 18 Motif As in 13 GBRs; 10 Motif Bs in 10 GBRs; and six Motif Cs in five GBRs. Consistent with previous reports on motifs that are similar to Motif B, i.e. the SBE and GBM, mutations of three independent Motif Bs significantly reduced GLD-1 binding in three GBRs (Fig. 2B and Table 3). Strikingly, a mutation in Motif C significantly reduced GLD-1 binding in puf-5 5′end, whereas a mutation in a Motif B within the same region did not (Table 3 and Fig. S1). Also, the puf-5 5′end does not have other motifs in addition to Motif C and B, which suggests that the Motif C is sufficient to promote strong GLD-1 binding to this region. These findings support the possibility that some mRNAs recruit GLD-1 by a distinct mechanism. Lastly, none of the 18 mutations of Motif A reduced GLD-1 binding, suggesting that Motif A is not important even though it is most significantly enriched in GBRs. It appears that Motif B, Motif C, and possibly even Motif A, represent only part of a larger context required for GLD-1 binding. Other factors or information, i.e. sequences or structures, surrounding these motifs may dictate their importance in GLD-1 binding. Furthermore, these findings suggest that translational regulation via GLD-1 may require other binding partners, i.e. proteins or RNAs, which may require yet unknown and distinct mRNA binding specificity.
Table 3. Motif B and Motif C are important for GLD-1 binding in several GBRs.
| Motif mutations¶ | ||||
|---|---|---|---|---|
| GLD-1 Binding region* | Motif A | Motif B | Motif C | Doubles |
| exo-3 (R09B3.1) 5′end† | N/S | S | S | |
| cep-1 F52B5.5 3′UTR‡ | N/S | |||
| rme-2 (T11F8.3) 3′UTR§ | S | |||
| rme-2 (T11F8.3) 5′end | S | |||
| cpg-1 (C07G2.1a) 3′end | N/S (2)a | N/S | N/S | |
| sas-5 (F35B12.5) 5′end | N/S (2) | N/S | ||
| sas-5 (F35B12.5) 3′end | N/S (2) | N/S | ||
| Y45F10A.2.1 3′end | N/S (2) | N/S | ||
| cdk-5 (T27E9.3) 3′end | N/S (2) | N/S | ||
| sss-1 (F32B6.5) 3′end | N/S | |||
| dna-2 (F43G6.1) 3′end | N/S | N/S | N/S | |
| cyb-3 (T06E6.2) 3′end | N/S | N/S | N/S | |
| puf-5 (F54C9.8) 5′endΠ | N/S | S | S | |
| puf-5 (F54C9.8) 3′end | N/S | N/S | N/S | |
| fem-3 (C01F6.4) 3′end | N/S | N/S | N/S | |
| rskn-1 (T01H8.1) 3′end | N/S | N/S | N/S | |
| T01C3.2 3′end | N/S | N/S | N/S | |
| unc-59 (W09C5.2) 5′end | N/S | N/S (2) | N/S | |
* Gene names and regions of GLD-1 mRNA targets. Total 34 over-represented motifs were biochemically examined in 18 GBRs; †,‡,§ Motif B is important for GLD-1 binding in exo-3 5′end, rme-2 3′UTR, and rme-2 5′end; ΠMotif C is important for GLD-1 binding in puf-5 5′end; ¶(S) and (N/S) denotes those Motifs that are significant or not significant for GLD-1 binding, respectively. Motifs are designated as “significant” if mutation causes significant reduction in GLD-1 binding compared with the wild-type counterpart, whereas motifs were designated as “not significant” if mutation does not cause reduction in GLD-1 binding; a(2) denotes that two of the same motif exist and were examined independently within a single GBR.
We also examined whether GLD-1 binds directly to these sequence motifs. Since we use cytosol extract in our binding studies, we cannot ignore the possibility that GLD-1 may complex with other proteins or RNAs within the extract which alters GLD-1 binding specificity. A previous study by Ryder et al. (2003) showed that GLD-1 directly binds the 28 nucleotide-long recognition element within the 3′ UTR of tra-2 mRNA.52 To examine if this is consistent with other GLD-1 mRNA targets, we performed electrophoresis mobility shift assays (EMSA)57 using 32P-labeled RNAs and recombinant GLD-1 to address whether GLD-1 can directly bind to the Motif Bs in two GBRs, rme-2 3′UTR and exo-3 5′end, in the absence of other RNAs and/or proteins. We found that recombinant GLD-1 alone can bind effectively to two wild-type GBRs while this binding was reduced to the mutated GBRs (Fig. S2), suggesting that GLD-1 likely binds directly to Motif B in these regions in vitro.
Sequences surrounding Motif B are important for GLD-1 binding
Even though we were able to demonstrate that the Motif B and C are important for GLD-1 binding in several GBRs, mutations of Motif B and C in other GBRs did not affect GLD-1 binding. More specifically, we found that three independent mutations of the Motif B affected GLD-1/mRNA interaction while the seven others did not. Thus, we explored the possibility that the sequences surrounding Motif B may contribute to GLD-1 binding in a context-dependent manner. We separated the 10 GBRs containing the examined Motif Bs into two sets. The first set contained the three GBRs where mutations of the Motif B reduced or eliminated GLD-1 binding, which indicates that Motif Bs in these GBR are important for GLD-1 binding. The second set contained the seven GBRs to which GLD-1 bound well even though the Motif Bs were mutated, indicating that the Motif Bs do not have biochemical importance for this interaction. Next, these two data sets were aligned so that Motif B was centered within the alignment with additional 25 nucleotides flanking both sides (Fig. 3A). We examined the sequences flanking the Motif B to identify any conserved nucleotides in the first data set but not in the second. Interestingly, while no other sequences are highly conserved in the second data set, a UA site just upstream of Motif B is highly conserved in the first data set. Furthermore, sequences that immediately follow this conserved UA site, although not fully conserved, represent a weakly conserved Motif B as shown by the matrix in Figure 3B; this putative site (UA C/U U/A C/A G/A) was renamed Half-motif B.
Figure 3. The identification of the Half-motif B and its importance in GLD-1 binding. (A) The possibility that sequences surrounding Motif B may contribute in a context-dependent manner to GLD-1 binding was examined. Two sets of sequences were compiled. The first set contains three GBRs where mutations of the Motif B reduced or eliminated GLD-1 binding, indicating that the Motif Bs in these GBR are important for GLD-1 binding. The second set contains the seven GBRs where GLD-1 still binds well even though the Motif Bs are mutated, indicating that the Motif Bs in these regions have no biochemical importance. Next, these two data sets were aligned so that Motif B was centered within the alignment with additional ~25 nucleotides flanking either side of the Motif Bs. (B) With the hypothesis that adjacent sequences important for GLD-1 recognition should be conserved, two independent sequence alignments were performed to see if any nucleotides were conserved in the first but not in the second set. Sequence alignment matrixes of the two sets are shown. Motif B is highly conserved in both data sets. Also, a Uridine-Adenine (UA) site just upstream of Motif B is highly conserved in the sequence alignment of the first set, which is not detected in the sequence alignment of the second set. Sequences that immediately follow the conserved UA site in the first set represent a weakly conserved Motif B, (UA C/U U/A C/A G/A) and is therefore designated as a Half-motif B. (C) The importance of Half-motif B for GLD-1/RNA interaction was examined. A western blot of the GLD-1/ Biotin-RNA pull-down assay with the wild-type R09B3.1 5′end and corresponding mutations is shown. The first lane shows GLD-1 in cytosolic extract. Lanes 2 through 9 represent binding reactions with wild-type and mutated R09B3.1 5′end biotin-labeled RNAs (400 ng) with increasing amount of cytosolic extract (50 and 150 ug). The conserved “UA” dinucleotide of the Half-motif B was mutated to “AU” and the Motif B (“UACUAA”) was mutated “UAGAUU” within R09B3.1 5′end (see Fig. 3B). The mutation of the Half-motif B reduces GLD-1 binding as significantly as the mutation of the Motif B compared with the wild-type. A double mutation of both motifs appears to reduce GLD-1 binding further, where the residual GLD-1 bands are no longer detected. Boxes are each lane are proportional to the amount of extract used.
To examine whether the Half-motif B is an important component for GLD-1/mRNA interactions, we mutated the highly conserved UA site in the Half-motif B within two GBRs (exo-3 5′end and rme-2 3′UTR) of the first data set and performed the Biotin-RNA pull-down assays. Not surprisingly, these significantly reduced GLD-1 binding to exo-3 5′end and rme-2 3′UTR, a reduction in binding that is comparable to that observed for mutations in Motif B alone (Fig. 3C). In addition, when both the Motif Bs and Half-motif Bs were mutated in these regions, GLD-1 binding was further reduced to undetectable levels (Fig. 3C; data not shown but both exo-3 5′end and rme-2 3′UTR show essentially identical results). These results suggest that the Motif B alone does not confer specificity to GLD-1 binding in exo-3 5′end and rme-2 3′UTR. Rather, GLD-1 binding may be context dependent, requiring nearby sequences, i.e., the Half-motif B, as an accessory feature for efficient GLD-1/RNA interactions.
Not all the Half-motif B and Motif B combinations are important in GLD-1 binding
The Half-motif B/Motif B appears to be an important feature required for GLD-1 binding in three GBRs. To extend our understanding of the interactions between GLD-1 and the Half-motif B/Motif B, we searched other GBRs in order to find additional neighboring Motif Bs and Half-motif Bs. For this search, we used a stringent Motif B UACU(C/A)A since only these stringent sequences are previously shown to be important in GLD-1 binding biochemically52,30 and the UA dinucleotide since it appears to be the most conserved and significant portion of the Half-motif B. To note, we omitted the importance of phylogenetic conservation of these motifs in this search. We found that 27 out of 38 GBRs contain a stringent Motif. Among the 27 GBRs, 22 contain one or more UA dinucleotide(s) within 11 nucleotides of the stringent Motif B (Table S3). To verify the importance of the stringent Motif B and its nearby Half-motif B for GLD-1 binding, we mutated these motifs within additional GBRs and assessed GLD-1 binding in vitro. We had examined the stringent Motif B and the Half-motif B in five GBRs, where two out of the five (exo-3 5′end and rme-2 3′UTR) were shown to be important for GLD-1 binding (Fig. 3 and data not shown). We extended our analysis by examining four other GBRs: rskn-1 3′ end, dna-2 3′end, nasp-2 5′end, and fem-3 3′end (Table S3), all of which contain the stringent Motif Bs and the UA dinucleotides within 10 nucleotides of each other (Table S3). However, mutagenesis and in vitro binding experiments revealed no importance of these motifs for GLD-1 binding; neither the single nor double mutations resulted in a reduction in GLD-1 binding compared with the wild-type (data not shown). These experiments suggest that the mere presence of the Half-motif B and/or the stringent Motif B is not sufficient for GLD-1 recognition.
In the studies by Galarneau and Richard (2005 and 2009) and Ryder et al. (2004),52,56 it was suggested that Quaking and GLD-1 recognize a shorter “half-site,” in addition to their core RNA binding motif. However, the Wright et al. study (2011)30 did not find any evidence for this. This may be due to the fact that many of the RNAs in which the “half-site” was reported contain multiple GBMs, suggesting that at least in some cases, the “half-site” is part of a degenerate full-length binding motif. Moreover, most Motif Bs were not functional within the majority of the targets examined in this study. This suggests (1) that there may be other GLD-1 binding determinants or (2) the GBRs examined may contain GBMs elsewhere that were not tested for their importance in GLD-1 binding. These possibilities could explain the residual GLD-1 binding when the Motif B or the Half-motif B was mutated in many of the GBRs. To address this, we scanned the 38 GLD-1 binding regions to see if they contained any of the 38 possible GBMs reported by Wright et al. (2011).30 We found that six out of 38 GLD-1 binding regions (sss-1 5‘end, sss-1 3′end, nasp-2 5‘end, ufd-1 3′end, sas-5 5‘end, and cdk-5 3′end) do not contain any of the 38 possible GBMs. Although these mRNAs were identified by Wright et al., they were not sufficiently enriched in their GLD-1 IPs (> 6.5) and, therefore, were not included in their computational analysis. In addition, three GBRs contain one GBR that were categorized to have weak predicted GLD-1 binding scores (rmd-1 3′end, Y75B12B.1 5′end, and puf-5 3′end).30 This indicates that, for at least six GBRs identified in this study, other GLD-1 binding determinants exist.
Target site accessibility within GBRs may provide specificity for GLD-1 binding
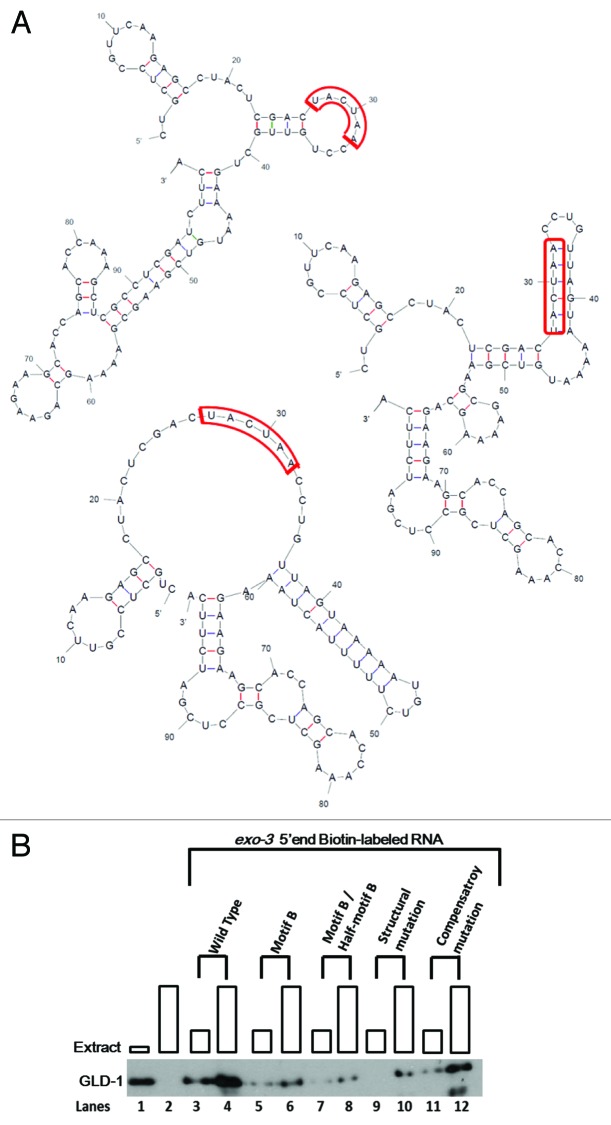
Thus far, we examined the importance of the Motif B and/or the Half-motif B for GLD-1 binding in a total of 12 GBRs. We found only three were important: in exo-3 5′end, rme-2 3′UTR, and rme-2 5′ end. Since significant sequence difference does not exist between biochemically important and unimportant motifs, we hypothesized that the difference may be the accessibility of these motifs dictated by RNA secondary structures. More specifically, if the sequence motifs reside in single-stranded regions of the RNA, this should provide accessibility for GLD-1 binding. This hypothesis is supported in that (1) KH-domain RBPs are generally known to bind ssRNA and (2) a similar phenomenon has been reported both in vitro and in vivo for GLD-1 and the Drosophila homolog How in previous studies.30,58 Nevertheless, we sought to extend the study of the target RNA secondary structure in GLD-1 binding by utilizing the GBRs in which the Motif B and the Half-motif B were examined biochemically. We considered a maximum of three most stable structures as predicted by MFold67 for each GBR since a few have only one or two dominantly stable and others have three most stable structures (Fig. 4A, wild-type).
Figure 4. GLD-1 binding to the Motif B within R09B3.1 5′end is dependent on the RNA secondary structure. (A) The most dominant structures of exo-3 5′end of the wild-type, structural mutant, and compensatory mutant are shown as predicted by MFold. The Motif Bs within each structure are outlined in red. The Motif B in the wild-type R09B3.1 5′end predicted to be completely unstructured, likely rendering the Motif B highly accessible for GLD-1 binding. A structural mutation forces the Motif B to become structured while maintain the wild-type Motif B sequence. A compensatory mutation places the Motif B back into unstructured conformation, likely bringing back Motif B’s accessibility to GLD-1 (right). (B) Western blot analysis of biotin/RNA pull-down assays of wild-type R09B3.1 5′end and corresponding mutations. Binding reactions were performed as identical to the previous assays. GLD-1 was probed in the cytosolic extract in the first lane, and no RNA was subjected as a negative control in the second lane. GLD-1 binding is significantly reduced when the Motif B and/or Half-motif B are mutated within R09B3.1 5′end as shown previously. GLD-1 binding is also significantly reduced with the structural mutation of the R09B3.1 5′end, whereas the GLD-1 binding is rescued with the compensatory mutation, comparable to the wild-type.
When we located the Motif B and the Half-motif B in each predicted secondary structure, we found that the majority of biochemically important sites (78%) were predicted to be in single-stranded regions, as opposed to only 31% that were confirmed to be not important motifs (Table 4). This significant difference between the two groups suggests a correlation between GLD-1’s ability to recognize a motif and the accessibility of the motif within the secondary structure. To experimentally test if secondary structure can interfere with binding, we introduced mutations to exo-3 5′end to alter predicted structures and assayed GLD-1 binding. Within the three most stable structures of exo-3 5′end predicted by MFold, three Motif Bs, and two Half-motif Bs were single stranded, whereas only one Half-motif B was structured. It suggests that Motif B/Half-motif B is accessible for GLD-1 binding (Table 4).
Table 4. The secondary structures surrounding the Half-motif B and Motif B may dictate binding-site accessibility for GLD-1.
| GBR* | Sequence† |
|---|---|
| rme-2 5′end | CCAUGAGAACCAUGCGCCUUGCUUGGUUGCUCCCACUUUUUAUUCACAUACUAAUCAAGAACACAGCUCAAGCUCCGGCUGUCAACAAACUCGACAUGCG |
| rme-2 3′UTR | GAGAACCAUGCGCCUUGCUUGGUUGCUCCCACUUUUUAUUCACAAUUCUACUACAAAAUUACUAAAUCAGAUGUCUGUAAAGUAUAUCUAUUUUUGCCUA |
| exo-3 5′end | CUGCUCCGUUCAAGAGCCUACUCGACUACUAACCUGUUGCUGAAAAUGUCGAAGCGAAAAGCAGAAGAAGCACCAGCACCAAAGCUCGCCUCGAUCUUCA |
| fem-3 3′end | UCAAUUUCCCGUCUACAGUUACAAAUUUCGCCGUUUGAAAUCUCUCGCUACUCACCUCUAGAUGUAUCUGGUUAUUAUGAAACAAUUAAAAGAAAAAAAG |
| dna-2 3′end | CAGUGCUUUUUAGAUUCUCACAAUUAAUUGAAAAUCAUAUUUCUAUUCCUACUCAACUGUAAAAUUCCCGCCUUUUUUACUCAUUUAUUAGAUUUCUCAC |
| nasp-2 5′end | ACUACGUAACUUCAUAAUGAGCGCUUUUGCUGAUUCUACUAACAUUAUCGCUCAUGAUAAGGAAACGAUCGCGAAGAAUGUCGACUCAACGGCGAAGAGA |
| rskn-1 3′end | GAUUCAUUUUUACGCACGGAUUUUACUUUUUUCUUUUUUAAAUACUACUAACAUUUAUCCAAUUGUUUCAACAAAUGCUCCUCAUUGAAGCCCAAUCACC |
| cpg-1 3′end | GUUCAGUACUACUGAAAGAAGUAAUCUAUCAAAUGCCUACUCAUCCCAGUGCCAUACACCCAAUGCAUAUUACUUACCCAUAUCACAACUAACCUGUAAU |
| cep-1 3′UTR | AAUAUUCAUCCUUUUCAAAGCACCGCAUUUCUCACUAGAUACUCAAACUGACAUUUGUUCCUUUUUAUAAUCGCAUUUUUUCGUAAUAAAGUUUGUUCGU |
| C09G9.6.1 3′end | ACAACUUCUCUACAUUUGUCAAAUUUUUAAACACUUUCAAUAUUCGUGUCCCCCAUACUAACUAAUAAUGCGUGCAUUUGUGAAUAUAAUAAGUUUUAUG |
| C09G9.6.1 5′end | UCAAACUAAUCCGAAUGCCCAGAUUGGAGAUUUGGUUACUCAAACUGCUAACUUGAUUGCUAUCAAAAAGCAGUUGCUUGAAGAUAUUGCAUUCAACCAA |
| T01C3.2 3′end | AGAAGCUCUACGAUCAAGUUCUCGAAUUCGUCACAACUACUAACAAGAAAUGAUUCGCUUUUACGAGGAAAGAAAUCAACUGUGAUCUCAUUAUAUUUUG |
| Three most stable structures§ | ||||
|---|---|---|---|---|
| GLD-1 binding region‡ | First stable Second stable |
Third stable | % AccessibilityΠ | |
| rme-2 5′end | Half (SS) | B (SS) Half (SS) | B (SS) |
Half (SS) | B (SS) | 100 | Accessibility of functional Motifs:78%¶ |
| rme-2 3′UTR | Half (SS) | B (DS) Half (DS) | B (DS) |
Half (SS) | B (SS) | 50 | |
| exo-3 5′end | Half (SS) | B (SS) Half (SS) | B (SS) |
Half (SS) | B (DS) | 83 | |
| fem-3 3′end | Half (DS) | B (DS) Half (DS) | B (DS) |
N/A | 0 | Accessibility of non-functional Motifs: 31%a |
| dna-2 3′end | Half (DS) | B (DS) Half (DS) | B (SS) |
Half (SS) | B (DS) | 33 | |
| nasp-2 5′end | Half (DS) | B (SS) Half (SS) | B (DS) |
Half (DS) | B (DS) | 67 | |
| rskn-1 3′end | Half (SS) | B (SS) Half (DS) | B (SS) |
Half (SS) | B (SS) | 83 | |
| cpg-1 3′end | B (DS) B (SS) |
B (DS) | 33 | |
| cep-1 3′UTR | B (DS) B (DS) |
B (DS) | 0 | |
| C09G9.6.1 3′end | B (DS) B (SS) |
B (DS) | 33 | |
| C09G9.6.1 5′endb | B (DS) N/A |
N/A | 0 | |
| T01C3.2 3′end | B (SS) B (DS) |
B (DS) | 33 | |
* GLD-1 binding regions containing the Motif Bs and/or Half-motif Bs that were examined biochemically, where exo-3 5′end, rme-2 3′ UTR, and rme-2 5′end (top 3) harbor motifs that are important for GLD-1 binding; †Sequences subjected to secondary structure predictions by MFold. The UA sites of the Half-motif B and the Motif Bs are in bold; ‡GLD-1 binding regions assessed for their target site accessibility. The motifs within were considered accessible for GLD-1 binding if they are in single-stranded conformation, whereas the motifs were considered inaccessible if they are in double-stranded conformation of the secondary structures; §The three most thermodynamically stable secondary structures for each GBR were evaluated for their binding-site accessibility. “Half” and “B” represents the Half-motif B and Motif B, respectively. (SS) and (DS) denote whether the motifs are within single-stranded or double-stranded regions, respectively; ΠThe percentage of motifs that are single-stranded from the top three stable structures of a given GBR; ¶78% of the biochemically functional GLD-1 binding motifs (Motif Bs and/or the Half-motif Bs) were predicted to be single-stranded; a31% of the biochemically non-functional GLD-1 binding motifs (Motif Bs and/or the Half-motif Bs) were predicted to be single-stranded; bMFold predicted only 2 and 1 stable structures from the C01F6.4 3′end and C09G9.6.1 5′end GBRs, respectively, and therefore the second and/or third stable structures of these GBRs were denoted as N/A (not applicable).
We mutated six nucleotides downstream to the Motif B to be completely complementary to the Motif B. This forced Motif B to become structured in the MFold prediction (Fig. 4A, structural mutation). Then, we examined whether this structural mutation reduced GLD-1 binding to the exo-3 5′end compared with its wild-type counterpart (Fig. 4B). Strikingly, the structural mutation significantly reduced GLD-1 binding compared with the wild-type, reducing GLD-1 binding at least as much as the single mutation of the Motif B or the Half-motif B (Fig. 4B). These results suggest that GLD-1 binding to exo-3 5′end RNA is largely dependent upon the Motif B/Half-motif B being accessible in the single-stranded region.
To further demonstrate the importance of the accessibility of Motif B/Half-motif B, we introduced another mutation to re-establish these sites in a single-stranded region. A mutation that introduced complementarity to the structural mutation was predicted by Mfold to restore single-strandedness of the Motif B (Fig. 4A, compensatory mutation) by making a double-stranded stem structure between the structural and compensatory mutations. As shown in Figure 4B, the compensatory mutation restored GLD-1 binding to a similar level as to the wild-type RNA.
We explored the possibility that the biochemically unimportant Motif Bs and Cs (Table 3) may be inaccessible for GLD-1 binding due to their secondary structures. We found that among the top three most thermodynamically stable structures predicted by Mfold, 91% of the non-functional motifs were at least partly structured, suggesting a role of secondary structure in inhibiting GLD-1 binding. Next, we ask if the Half-motif B (UA mutation) affects the secondary structure of the Motif B and if this could explain the effect on binding to rme-2 3′UTR, rme-2 5′end, and exo-3 5′end. However, mutations to the UA of Half-motif B within these regions have no apparent effect on Motif B accessibility, indicating that the functionality of the Half-motif in rme-2 3′UTR, rme-2 5′end, and exo-3 5′end is independent of RNA structure. Taken together, these data and observations strengthen previous knowledge that the presence of GLD-1 binding sites (i.e., the Half-motif B and Motif B) must reside in single-stranded RNA for GLD-1 to recognize and bind efficiently.
It would be interesting to examine whether predicted structures of GBRs with non-binding Motif Bs are what prevent those motifs from binding, because we cannot rule out that other possibilities are plausible for the non-binding motifs. First, the spacing of specific key nucleotides within the motif(s) may not be optimal for GLD-1 contact. Second, specific nucleotides may poison GLD-1 contact nucleotides. Third, a variety of unusual folds, kinks, and twists can be present within or surrounding Motif B, which likely inhibit GLD-1 from recognizing Motif B. Lastly, GLD-1 binding may require a local, non-canonical fold or geometry, which could be prevented by specific nucleotides within or neighboring the motif.
The requirement of the Motif B for GLD-1-dependent regulation in vivo
To examine the functional importance of the Motif B and the Half-motif B, we determined their ability to achieve GLD-1-dependent translational repression in vivo. We hypothesized that a mutation in the Motif B and/or the Half-motif B that disrupted GLD-1 binding in vitro would relieve GLD-1-mediated translational repression in vivo. To address this, we performed transgenic assays in which the wild-type and variously mutated rme-2 3′UTRs were independently cloned downstream of green fluorescent protein (GFP) fused to Histone H2B and used the pie-1 promoter to drive expression in all germ cell types. Transgenic constructs were then randomly integrated in the genome by a biolistic transformation procedure as described,59 and at least two independent lines were analyzed per construct. A similar transgenic assay was conducted by Wright et al. (2011),30 but on several more GLD-1 targets, including rme-2 3′UTR. We recapitulated their results in that the Motif B (same as the GBM within rme-2 3′UTR) was required for GLD-1-mediated translation repression in vivo (Fig. S3A). Moreover, we saw that in the fusion-containing mutations in both Motif B and Half-motif B in otherwise the same transgenic mRNA, GFP:H2B expression expanded into the distal meiotic regions where GLD-1 is abundant (Fig. S3C and D). To further examine whether these transgenic transcripts are regulated by GLD-1, we depleted GLD-1 by RNA-mediated interference. Depletion of GLD-1 protein caused severe germline tumor as expected, and GFP::H2B expression was ubiquitous and at comparable levels throughout the germline (Fig. S3B). Taken together, we confirmed that GLD-1 represses translation through the Motif B and Half-motif B in rme-2 3′UTR.
Discussion
Protein-RNA recognition plays an important role in regulating cell function, including the assembly and function of ribonucleoprotein particles (RNPs) and the post-transcriptional regulation of gene expression. However, many studies have focused primarily on how an RBP interacts with a single or limited number of RNA targets, despite the growing evidence that most RBPs have many targets.11,27,30,32-36 Here, we used our knowledge that GLD-1 regulates numerous targets to explore GLD-1’s binding specificity. Any comprehensive understanding of how an RNA binding protein recognizes and regulates its RNA targets requires the identification of most RNA targets, and our identification of over 100 such targets has provided an entrée for this investigation (Table S1). Our analysis of these putative targets suggests that they are functionally inter-related to act during germline development.60,61
In this study, we identified 38 GBRs from 32 GLD-1 targets identified in our GLD-1 RIP-chip analysis (Fig. 1). We were able to detect three over-represented sequence motifs and demonstrated that Motif B and C are important in GLD-1 binding to several GBRs. More importantly, not only is Motif C a novel motif that promotes GLD-1 binding, further analysis showed that at least six GBRs do not contain the prominent SBE or GBM previously reported to be necessary and sufficient for GLD-1 binding. Our data clearly suggest that other factors dictate GLD-1 and mRNA associations and that some mRNAs can recruit GLD-1 by distinct mechanisms.
Complexity of GLD-1 recognition of mRNA targets
Our data indicate strong plasticity in the sequence requirements for GLD-1 binding: three out of 10 Motif Bs examined were important for GLD-1 binding. Similarly, one out of six Motif Cs examined was important for GLD-1 binding. What accounted for such variability? First, we found that three GLD-1 binding sites carry an important accessory sequence feature, a Half-motif B upstream to the Motif B, indicating that context dependency exists. Yet, the presence of the Half-motif B/Motif B alone was not sufficient for GLD-1 binding in other GBRs. For example, mutations in the Half-motif B and/or Motif B did not reduce GLD-1 binding to rskn-1 3′ end, dna-2 3′end, nasp-2 5′end, and fem-3 3′end (Table S3).
What makes the Half-motif Bs and Motif Bs within rme-2 3′UTR, rme-2 5′end, and exo-3 5′end important in GLD-1 binding while the same motifs in other GBRs are not? It has been shown that intrinsic mRNA secondary structure that forms prior to trans-factor binding can constrain subsequent binding events at all levels of post-transcriptional regulation.62-64 Moreover, it was also demonstrated that target-site accessibility has a significant impact on mRNA target selection with the previously determined sequence-binding preferences.65 In light of these findings, RNA secondary structures may limit GLD-1-RNA interactions if the sequence motifs are embedded in double-stranded regions. Consistent with this hypothesis, we have shown that 78% of the biochemically confirmed important GLD-1 binding sites (the Half-motif Bs and/or Motif Bs) are predicted to be in single-stranded regions, whereas only 31% of the motifs that were confirmed to be not important GLD-1 binding sites are predicted to be within single-stranded regions (Table 4). More importantly, we were able to show experimentally that the binding site accessibility influences GLD-1 binding to the Motif B within exo-3 5′end (Fig. 4). GLD-1 binding assays with structural and compensatory mutations confirmed that at least one core binding site (the Motif B) was required to be within single-stranded or unstructured regions to allow efficient GLD-1 binding. This outcome explains why some Motif Bs and Half-motif Bs are not functional in GLD-1 binding.
How does GLD-1 select its targets?
Our data show that GLD-1 recognizes its targets using at least two different sequence motifs, Motif B and C. Motif B is essentially the same motif as previously identified in various studies as a GLD-1 or Quaking binding motif, suggesting it is an important sequence determinant employed by GLD-1 to select its targets.27,30,52,56 However, it cannot be a sole sequence determinant since several GBRs identified in this study do not contain the Motif B. This finding indicates that GLD-1 recognizes these GBRs using other sequences. In addition, even though the Motif B is present in the majority of GBRs, we showed that the same Motif B is important in GLD-1 binding in several but not in all GBRs. Thus, in GBRs where the Motif B was not important in GLD-1 binding, the Motif B is either a minor determinant of GLD-1 binding or other equally important motif(s) are present.
Our analysis of two sets of GBRs, one where Motif B is important for GLD-1 binding and one where it is not, showed that the ability of GLD-1 to utilize the sequence motif depends on two features. First, the Half-motif B with a strong UA requirement acts as an accessory feature for GLD-1 to efficiently select the Motif B. Second, the sequence motif resides in the single-stranded region, which provides accessibility for GLD-1 to effectively recognize the motif. Taken together, GLD-1 selects its targets that contain the sequence motif embedded in the correct context and structural environment.
GLD-1 targets and the GLD binding motif identified from previous studies
Several studies have identified numerous GLD-1 targets and provided extensive analyses of RNA determinant(s) that mediate GLD-1 binding.27,29,30 Jungkamp et al. utilized the iPAR-CLIP (in vivo PAR-CLIP) strategy to identify transcriptome-wide binding sites and ~440 targets of GLD-1. Wright et al. also identified GLD-1 targets using the RIP-chip strategy similar to the one presented here; however, they report 948 mRNA were 3-fold enriched in their RIP-chip analysis as opposed to 49 in this study. This disparity may have resulted from (1) the difference in the way the IPs were performed and/or (2) the utilization of different microarray chips. Furthermore, both the Jumpkamp and Wright studies relied on a computational program, MEME motif finder,66 to identify a GLD-1 binding site similar to the one initially published by Ryder et al. (2004). An advantage of our study was that we combined biochemically proven data sets (38 GBRs) with two computational programs (PhyloCon and PhyloNet) to identify GLD-1 binding sites. In doing so, we identified a similar site to the GLD-1 binding motif (GBM) found by previous groups, but we also found a novel site (Motif C) that was important for GLD-1 to bind puf-5 5′end in vitro. Additionally, while previous studies demonstrated that the GBM is a degenerate motif of ~38 possibilities that is necessary and sufficient for GLD-1 binding, careful analysis of our 38 biochemically proven GBRs revealed that 6 regions do not contain a GBM, and 3 more contain only one GBM that was categorized to be a weak GLD-1 binder.27,30 Taking our findings together, it is evident that the GBM is not the sole GLD-1 binding determinant.
The difficulty of obtaining a comprehensive understanding of target selection could be explained by a recent finding that GLD-1 associates with other RNA binding proteins such as CGH-1 and CAR-1.29 While for some GBRs, the binding motifs of GLD-1 (e.g., Motif B/GBM or C) could be a major determinant of binding to GLD-1 associated with other RNA binding proteins, in other GBRs, binding motifs of other RNA binding proteins could be the major binding determinant. It will require further studies to understand how target selection occurs when two or more RNA binding proteins directly associate with each other.
Finally, our current study adds to our understanding of GLD1 binding sites by (1) definitively demonstrating the existence and importance of the Half motif B site, (2) demonstrating the existence and importance of the novel Motif C site, (3) demonstrating that secondary structure inhibits GLD-1 binding to otherwise active sites, thereby explaining at least in part the failure of some appropriate sequences to bind GLD-1, and (4) proving that not all apparent Motif B/Half motif B sites and Motif C sites present in single-stranded regions are active in vitro and in vivo, indicating that our understanding of the specificity of GLD-1 binding is still incomplete. We believe that this complexity of GLD-1 interaction with RNA is not unique but rather may well represent the complexity of protein–RNA interactions is general. This complexity is required for RNA protein complexes to regulate many genes in many cases in slightly different ways.
Materials and Methods
Nematode culture and strains
Standard procedures for nematode culture and genetic manipulation were followed with growth at 20 °C. The strains used in this study were: (1) wild-type N2, (2) gld-1(q485), (3) gld-1(q485); ozIs2[gld-1::gfp::flag], and (4) unc-119 (ed3). unc-119 (ed3) was obtained from Caenorhabditis Genetics Center (CGC).
Preparation of cytosolic extracts, immmunoprecipitation, linear RNA amplification, and microarray and GO analysis
Cytosol extracts of the rescued gld-1(q485); ozIs2[gld-1::gfp::flag] strain was prepared and subjected to immuprecipitation (IP) with anti-FLAG Ab and mouse IgG as described previously.11 RNAs that were co-immunoprecipitated were extracted with Trizol (Invitrogen) and RNA was prepared following the manufacturer’s protocol. Then two rounds linear RNA amplification was performed using 200 ng RNA from two IPs employing the ExpressArt mRNA Amplification kit (Artus-Biotech). Microarray analysis was performed as described (Reinke et al., 2004) to measure enrichment of FLAG compared IgG IPed RNAs. Microarrays representing 17744 genes were probed with four independent sets of FLAG and IgG IPed RNA. The 129 significantly enriched genes were annotated with gene symbols and Entrez IDs (http://ncbi.nlm.nih.gov). A total of 17319 Entrez-based genes were matched into 2692 functional gene sets labeled by GO identifier (http://geneontology.org/). For convenience, “Celegans.na26.annot.csv” were downloaded from Affymetrix (http://www.affymetrix.com/) and were referenced for annotation and matching. The significance of enrichment was calculated by hypergeometric distribution using PERL script.
Cloning 5′ and 3′ ends of GLD-1 mRNA targets
The first strand cDNA of adult wild-type hermaphrodites28 was used as templates for PCR. The 5′ end of each target gene was PCR amplified using Splice Leader 1 (SL1) or SL2 primers and the target gene-specific primers, complementary to a region ~150 nucleotide (nt) downstream of the predicted start codon. The 3′ end of each target gene was PCR amplified using a target gene-specific primer ~150 nt upstream of the predicted stop codon and a primer that complements the 3′ end of the oligo-dT-linker primer used in the first strand cDNA synthesis. PCR products were gel-purified, cloned into a TOPO vector (pCRII-TOPO, Invitrogen) and sequenced to confirm the orientation and nucleotide sequences.
Biotin-RNA pull-down assays
All template DNAs for the biotin RNA synthesis were amplified by PCR using reverse or M13 primer and downstream primers. Biotin-RNA synthesis was performed as described.11,28 ~400 ng of biotin-labeled RNA was incubated with cytosol extracts in IP buffer (5 mM Hepes (pH7.6), 1 mM MgCl2, 75 mMKCl, 1 mM DTT, 1% glycerol, 600 ug/mL tRNA, 6 mg/mL heparin, 10 units of RNase Inhibitor) in a final volume of 50 uL for 20 min at room temperature (RT). 100 uL of Streptavidin-magnetic beads (Promega) was first washed in IP buffer with 20 ug/mL tRNA, and then resuspended in 15 uL of the IP buffer containing 20 ug/mL tRNA. The resuspended beads were added to each binding reaction and incubated for 20 min at room temperature (RT). The magnetic beads were then isolated, washed three times in IP buffer with 20 ug/mL tRNA. All binding reactions were mixed with SDS sample buffer at least 1:2 proportions and heated to a 95 °C for 2–4 min. Samples were loaded on to a 10% SDS-PAGE containing 30% Acrylamide/Bis (29:1) and ran at 150 V for ~1.5 h. Gels were transferred onto Immobilon-P transfer membrane at 12 V for 1 h and blocked in blocking solution for at least 30 min. After blocking, the membrane was incubated with primary antibody (GLD-1 antibody) at 4 °C overnight. Incubation with secondary antibody (HRP anti-Mouse IgG antibody) was performed in room temperature for 2.5 h. Protein was detected using Luminol substrate mixture (1mL).
Specificity of GLD-1 association with GBRs were assessed by determining the detectability of GLD-1 western blot analysis as compared with the negative control in which no RNA was subjected to the binding reaction. Generally, negative controls where RNA is omitted in the binding reaction have undetectable levels of GLD-1 in the western blot analysis; thus, RNAs that exhibit detectable GLD-1 in western blots were considered GLD-1 binders.
Computational analyses to identify over-represented motifs
The sequences of 38 confirmed GBRs and their corresponding orthologous regions from C. briggsae, C. remanei, C. japonica, and C. brenneri were subjected to PhyloCon and PhyloNet to search for the over-represented and phylogenetically conserved sequence motifs.54,55 With the over-represented sequence motif information identified through PhyloCon and/or PhyloNet, a Patser scan was conducted to determine the presence and location of the motifs within the 38 GBRs.
The PhyloCon program searches for consensus sequence motifs that are embedded in co-regulated but otherwise unconnected sequences in related species.54 A statistical limitation of PhyloCon is that only the most significant or prevalent motifs are detected, with weak signals lost mainly because they cannot be distinguished from random patterns in complex data sets. Therefore, the PhyloNet program was utilized to identify motifs. PhyloNet and PhyloCon are similar in that they both use phylogenetic data from related species; however, while the PhyloCon’s algorithm considers co-regulation of genes within species, PhyloNet’s algorithm does not. This allows PhyloNet to identify (more) motifs based solely on their occurrence within phylogenetically related data set. In principle, the sequences of the GLD-1 binding regions subjected to the analysis are presumably co-regulated by GLD-1.
As a control, we subjected 25 random germline-expressed but not enriched in the GLD-1 IP to PhyloCon and PhyloNet analyses and we did not identify any significantly enriched motifs.
Biolistic transformation
All transgenic constructs used in this study were generated using the Multisite Gateway Cloning Technology (Invitrogen). The Promoter Entry Clones (pCG142: pie-1 promoter) and the GFP Entry Clones (pCM1.35: GFP::Histone H2B) were kindly provided by Dr Geraldine Seydoux (Johns Hopkins University). 3′ Entry Clones were generated from PCR products flanked by attR2 and attL3 sites and subjected to BP reaction with a pDONR P2R-P3 Entry vector. Promoter Entry Clones, GFP Entry Clones, and 3′ Entry Clones were subjected to an Ligation Reaction (Invitrogen) with the Destination Vector - pCG150 to generate Expression clones.59 Biolistic transformations of the Expression clones into C. elegans unc-119(ed3) hermaphrodites were performed using a BioRad Biolistic PDS-1000/HE. For each bombardment, 7 μg plasmid DNA was coupled to ~5 mg of 1.0 μm microcarrier gold beads, and bombarded onto a layer of 1–2 million unc-119(ed3) L4 and adult hermaphrodites placed on a 70 mm diameter lawn of NA22 on NEP plates. Worms were allowed to recover for 0.5 h after bombardment and were then transferred onto 12 NA22 seeded NEP plates and grown at 25 °C. From each plate containing animals rescued for the unc-119 mutation, homozygous stable lines were identified by the complete absence of unc-119 mutant progeny over several generations. Homozygous lines were examined for GFP expression under a compound microscope at 400X magnification. Dissected gonads from all transgenic lines were prepared as described.11,21,22
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Hua Shi and Richard Zitomer for comments on the manuscript. We thank Mailien Ho, Luis Daniel Diaz, and John Ryoo for their technical assistance. This work was supported by American Cancer Society grant RSG-07-181-01-DDC to MHL.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/worm/article/26548
Footnotes
Previously published online: www.landesbioscience.com/journals/worm/article/26548
References
- 1.Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–3. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 2.D’Agostino I, Merritt C, Chen PL, Seydoux G, Subramaniam K. Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev Biol. 2006;292:244–52. doi: 10.1016/j.ydbio.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 3.Jones AR, Schedl T. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev. 1995;9:1491–504. doi: 10.1101/gad.9.12.1491. [DOI] [PubMed] [Google Scholar]
- 4.Lublin AL, Evans TC. The RNA-binding proteins PUF-5, PUF-6, and PUF-7 reveal multiple systems for maternal mRNA regulation during C. elegans oogenesis. Dev Biol. 2007;303:635–49. doi: 10.1016/j.ydbio.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat Rev Genet. 2003;4:626–37. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 6.Bettegowda A, Smith GW. Mechanisms of maternal mRNA regulation: implications for mammalian early embryonic development. Front Biosci. 2007;12:3713–26. doi: 10.2741/2346. [DOI] [PubMed] [Google Scholar]
- 7.de Moor CH, Meijer H, Lissenden S. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin Cell Dev Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Lasko P. Translational control during early development. Prog Mol Biol Transl Sci. 2009;90:211–54. doi: 10.1016/S1877-1173(09)90006-0. [DOI] [PubMed] [Google Scholar]
- 9.Jadhav S, Rana M, Subramaniam K. Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development. 2008;135:1803–12. doi: 10.1242/dev.013656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kershner AM, Kimble J. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc Natl Acad Sci U S A. 2010;107:3936–41. doi: 10.1073/pnas.1000495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee MH, Schedl T. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 2001;15:2408–20. doi: 10.1101/gad.915901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lublin AL, Evans TC. The RNA-binding proteins PUF-5, PUF-6, and PUF-7 reveal multiple systems for maternal mRNA regulation during C. elegans oogenesis. Dev Biol. 2007;303:635–49. doi: 10.1016/j.ydbio.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Marin VA, Evans TC. Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development. 2003;130:2623–32. doi: 10.1242/dev.00486. [DOI] [PubMed] [Google Scholar]
- 14.Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–82. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mootz D, Ho DM, Hunter CP. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development. 2004;131:3263–72. doi: 10.1242/dev.01196. [DOI] [PubMed] [Google Scholar]
- 16.Schumacher B, Hanazawa M, Lee MH, Nayak S, Volkmann K, Hofmann ER, Hengartner M, Schedl T, Gartner A. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–68. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 17.Di Fruscio M, Chen T, Bonyadi S, Lasko P, Richard S. The identification of two Drosophila K homology domain proteins. Kep1 and SAM are members of the Sam68 family of GSG domain proteins. J Biol Chem. 1998;273:30122–30. doi: 10.1074/jbc.273.46.30122. [DOI] [PubMed] [Google Scholar]
- 18.Vernet C, Artzt K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 1997;13:479–84. doi: 10.1016/S0168-9525(97)01269-9. [DOI] [PubMed] [Google Scholar]
- 19.Biedermann B, Wright J, Senften M, Kalchhauser I, Sarathy G, Lee MH, Ciosk R. Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev Cell. 2009;17:355–64. doi: 10.1016/j.devcel.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Ciosk R, DePalma M, Priess JR. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science. 2006;311:851–3. doi: 10.1126/science.1122491. [DOI] [PubMed] [Google Scholar]
- 21.Francis R, Barton MK, Kimble J, Schedl T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics. 1995;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francis R, Maine E, Schedl T. Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics. 1995;139:607–30. doi: 10.1093/genetics/139.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen D, Hubbard EJ, Schedl T. Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev Biol. 2004;268:342–57. doi: 10.1016/j.ydbio.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 24.Hansen D, Wilson-Berry L, Dang T, Schedl T. Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development. 2004;131:93–104. doi: 10.1242/dev.00916. [DOI] [PubMed] [Google Scholar]
- 25.Kadyk LC, Kimble J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998;125:1803–13. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- 26.Jan E, Motzny CK, Graves LE, Goodwin EB. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 1999;18:258–69. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jungkamp AC, Stoeckius M, Mecenas D, Grün D, Mastrobuoni G, Kempa S, Rajewsky N. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol Cell. 2011;44:828–40. doi: 10.1016/j.molcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee MH, Schedl T. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev. 2004;18:1047–59. doi: 10.1101/gad.1188404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheckel C, Gaidatzis D, Wright JE, Ciosk R. Genome-wide analysis of GLD-1-mediated mRNA regulation suggests a role in mRNA storage. PLoS Genet. 2012;8:e1002742. doi: 10.1371/journal.pgen.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright JE, Gaidatzis D, Senften M, Farley BM, Westhof E, Ryder SP, Ciosk R. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J. 2011;30:533–45. doi: 10.1038/emboj.2010.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu L, Paulsen J, Yoo Y, Goodwin EB, Strome S. Caenorhabditis elegans MES-3 is a target of GLD-1 and functions epigenetically in germline development. Genetics. 2001;159:1007–17. doi: 10.1093/genetics/159.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halbeisen RE, Galgano A, Scherrer T, Gerber AP. Post-transcriptional gene regulation: from genome-wide studies to principles. Cell Mol Life Sci. 2008;65:798–813. doi: 10.1007/s00018-007-7447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–52. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr., Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–39. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Juan L, Lv J, Wang K, Sanford JR, Liu Y. Predicting sequence and structural specificities of RNA binding regions recognized by splicing factor SRSF1. BMC Genomics. 2011;12(Suppl 5):S8. doi: 10.1186/1471-2164-12-S5-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–51. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 39.Jambhekar AK, McDermott K, Sorber KA, Shepard KA, Vale RD, Takizawa PA, DeRisi JL. Unbiased selection of localization elements reveals cis-acting determinants of mRNA bud localization in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2005;102:18005–10. doi: 10.1073/pnas.0509229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allain FH, Howe PW, Neuhaus D, Varani G. Structural basis of the RNA-binding specificity of human U1A protein. EMBO J. 1997;16:5764–72. doi: 10.1093/emboj/16.18.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hentze MW, Kühn LC. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc Natl Acad Sci U S A. 1996;93:8175–82. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kazan H, Ray D, Chan ET, Hughes TR, Morris Q. RNA context: A New Method for Learning the Sequence and Structure Binding Preferences of RNA-Binding Proteins. PLOS Comput Biol. 2010;6:255–449. doi: 10.1371/journal.pcbi.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall KB. RNA-protein interactions. Curr Opin Struct Biol. 2002;12:283–8. doi: 10.1016/S0959-440X(02)00323-8. [DOI] [PubMed] [Google Scholar]
- 44.Messias AC, Sattler M. Structural basis of single-stranded RNA recognition. Acc Chem Res. 2004;37:279–87. doi: 10.1021/ar030034m. [DOI] [PubMed] [Google Scholar]
- 45.Hori T, Taguchi Y, Uesugi S, Kurihara Y. The RNA ligands for mouse proline-rich RNA-binding protein (mouse Prrp) contain two consensus sequences in separate loop structure. Nucleic Acids Res. 2005;33:190–200. doi: 10.1093/nar/gki153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thisted T, Lyakhov DL, Liebhaber SA. Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and alphaCP-2KL, suggest Distinct modes of RNA recognition. J Biol Chem. 2001;276:17484–96. doi: 10.1074/jbc.M010594200. [DOI] [PubMed] [Google Scholar]
- 47.Buckanovich RJ, Darnell RB. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol. 1997;17:3194–201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meisner NC, Hackermüller J, Uhl V, Aszódi A, Jaritz M, Auer M. mRNA openers and closers: modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. Chembiochem. 2004;5:1432–47. doi: 10.1002/cbic.200400219. [DOI] [PubMed] [Google Scholar]
- 49.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci U S A. 2000;97:14085–90. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reinke VJ. Germline genomics. In The C. Elegans Community (Ed.) 2006; WormBook (1-10). doi: 10.1895/wormbook.1.74.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reinke VJ, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–23. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- 52.Ryder SP, Frater LA, Abramovitz DL, Goodwin EB, Williamson JR. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat Struct Mol Biol. 2004;11:20–8. doi: 10.1038/nsmb706. [DOI] [PubMed] [Google Scholar]
- 53.Nayak S, Goree J, Schedl T. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 2005;3:e6. doi: 10.1371/journal.pbio.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang T, Stormo GD. Combining phylogenetic data with co-regulated genes to identify regulatory motifs. Bioinformatics. 2003;19:2369–80. doi: 10.1093/bioinformatics/btg329. [DOI] [PubMed] [Google Scholar]
- 55.Wang T, Stormo GD. Identifying the conserved network of cis-regulatory sites of a eukaryotic genome. Proc Natl Acad Sci U S A. 2005;102:17400–5. doi: 10.1073/pnas.0505147102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galarneau A, Richard S. The STAR RNA binding proteins GLD-1, QKI, SAM68 and SLM-2 bind bipartite RNA motifs. BMC Mol Biol. 2009;10:47. doi: 10.1186/1471-2199-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garner MM, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981;9:3047–60. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Israeli D, Nir R, Volk T. Dissection of the target specificity of the RNA-binding protein HOW reveals dpp mRNA as a novel HOW target. Development. 2007;134:2107–14. doi: 10.1242/dev.001594. [DOI] [PubMed] [Google Scholar]
- 59.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–26. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–43. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 61.Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell. 2002;9:1161–7. doi: 10.1016/S1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- 62.Akerman M, David-Eden H, Pinter RY, Mandel-Gutfreund Y. A computational approach for genome-wide mapping of splicing factor binding sites. Genome Biol. 2009;10:R30. doi: 10.1186/gb-2009-10-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stadler MB, Shomron N, Yeo GW, Schneider A, Xiao X, Burge CB. Inference of splicing regulatory activities by sequence neighborhood analysis. PLoS Genet. 2006;2:e191. doi: 10.1371/journal.pgen.0020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–6. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 65.Morris J, Singh JM, Eberwine JH. Transcriptome analysis of single cells. J Vis Exp. 2011;50:2634. doi: 10.3791/2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.