Abstract
We recently conducted a study that aimed to describe the differentiation mechanisms used to generate O2 and CO2 sensing neurons in C. elegans. We identified egl-13/Sox5 to be required for the differentiation of both O2 and CO2 sensing neurons. We found that egl-13 functions cell autonomously to drive O2 and CO2 sensing neuron fate and is therefore essential for O2 and CO2 sensing-induced behaviors. Through systematic dissection of the egl-13 promoter we identified upstream regulators of egl-13 and proposed a model of how differentiation of O2 and CO2 sensing neurons is regulated. In this commentary we discuss our findings and open questions we wish to address in future studies.
Keywords: neuronal specification, neuronal diversity, EGL-13, Sox5, oxygen and carbon dioxide sensing, BAG neurons, URX neurons
Introduction
How the human brain, which contains ~86 billion neurons, generates the correct complement of diverse neuron types is a long-standing question.1-3 In order for a sensory neuron to correctly differentiate to its terminal neuronal fate, and thus, be able to respond to specific sensory cues, it needs to obtain a specific morphological, spatial, and synaptic repertoire. Such differentiation events are tightly regulated during development by both intrinsic and extrinsic signals.3 It has been proposed that terminal differentiation of specific neurons utilizes unique combinations of transcription factors to achieve neuronal diversity.4-7 One example is the function of the conserved transcription factors AST-1 (ETS factor) and CEH-43 (homeobox factor), which act in a combinatorial manner to specify dopaminergic neurons in C. elegans.8,9 The mechanism controlling dopaminergic specification seems to be conserved, at least for AST-1, since knockout mice for the AST-1 ortholog Etv1 fail to differentiate dopaminergic neurons in the olfactory bulb.9 Another example showing mechanistic conservation across species is the specification of cholinergic neurons. This event requires the COE-type transcription factor UNC-3 in C. elegans.10 The role of unc-3 was further investigated in the chordate C. intestinalis where the ortholog of UNC-3, COE, also regulates its own expression in cholinergic motor neurons.10 These studies are good examples of how neuronal differentiation and diversity utilizes conserved mechanisms across the phylogeny and how studies in model organisms can provide crucial information on questions that are difficult to address in higher organisms.
In their terminal differentiated state, oxygen (O2) and carbon dioxide (CO2) sensing neurons express specific gene batteries that enable them to sense and initiate a behavioral response to changes in these respiratory gases. In humans, O2 and CO2 are sensed by the carotid body.11,12 CO2 is also sensed in its hydrated form through changes in pH by neurons in the brainstem.11-15 The carotid body comprises two major cell types, glomus cells (type I) and sustentacular cells (type II). Glomus cells are neurosecretory cells that release neurotransmitters in a Ca2+-dependent manner, when the pressure of O2 or CO2 (pO2/pCO2) changes in the blood.16 The signal is projected to the medulla oblongata before it is transmitted to motor neurons in the diaphragm that responds with increased or decreased contractions.17 In C. elegans, behavioral responses to changes in the environmental concentrations of O2 and CO2 are controlled by six neurons: BAGL/R, URXL/R, AQR, and PQR. The BAG neurons respond to downsteps in O2 and upsteps in CO2, whereas the URX, AQR, and PQR neurons respond to upsteps in O2.18-21 O2 sensing is mediated by soluble guanylate cyclases (sGC), GCY-31/33 in BAG, and GCY-35/36 in URX, which bind O2 through a heme group.20,22 The binding of O2 to a sGC induces the conversion of GTP to cGMP that in turn allows the cation channels TAX-2/4 to open and depolarize the neurons.22,23 A response to upsteps in CO2 is initiated by binding of CO2 or a CO2 metabolite to the receptor guanylate cyclase GCY-9 expressed by the BAG neurons.24-26 The CO2 response, like the O2 response, utilizes cGMP and TAX-2/4 to transmit the signal.23 Since the molecular basis of O2 and CO2 sensing in humans are not well understood, studies in model organisms may provide important insights. Humans do not have a direct ortholog of the guanylate cyclase GC-D used by rodents or GCY-9 used by C. elegans to detect CO2. However, it is proposed that the human CO2 response is activated, like GC-D in rodents, by a change in pH.13,14 The conserved link is further strengthened by the direct regulation of gcy-9 expression by ETS-5 in the BAG neurons of C. elegans25,26 because the mouse ortholog Pet1b is expressed and required for differentiation of CO2 responsive neurons.27 Defining direct targets of Pet1b might therefore identify CO2 responding receptor candidates.
Many diseases are characterized by an inability to sense O2 and CO2 or progress due to altered pO2. These diseases include solid tumors where altered gene expression within hypoxic regions lead to resistance to radio- and chemotherapy28 resulting in bad prognosis for patients.29 In addition, the inability to sense increases in CO2 concentration by infants is believed to be the major cause of sudden infant death syndrome (SIDS).30 There is also an interest in understanding the CO2-sensing mechanisms used by mosquitoes carrying malaria and how parasites infecting farm animals locate their hosts.31,32 We believe that obtaining a deeper insight into the mechanisms used to sense O2 and CO2 can yield a better understanding of such pathophysiological and ethological situations. In our study, we aimed to describe how neurons required to sense environmental changes in O2 and CO2 are specified (Fig. 1).
Figure 1. Specification of distinct sensory neuron fates. As in other organisms, C. elegans has specialized neurons that are able to detect changes in O2 and CO2. However, the molecular mechanisms that drive these specializations are poorly understood.
egl-13 is Required for Differentiation of O2- and CO2-Sensing Neurons
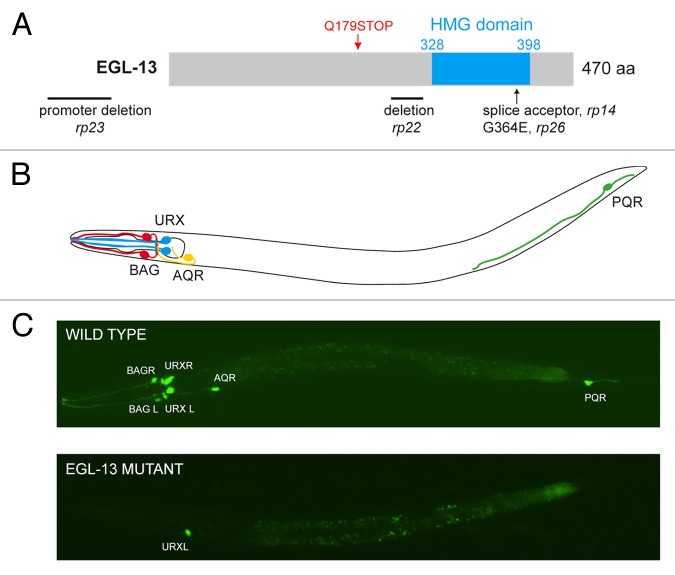
To identify factors required for the differentiation of O2- and CO2-sensing neurons, we conducted a forward genetic screen using a fluorescent marker for the gas-sensing neurons. From this screen, we isolated four independent mutant alleles of egl-13 (rp13, rp22, rp23, and rp26)33 that all phenocopied the previously published null allele egl-13(ku194)34 (Fig. 2A). We found that egl-13 is required for the correct expression of the terminal gene battery in both the BAG and URX neurons. Previous studies in vertebrates have described roles of the egl-13 ortholog Sox5 in chondrogenesis and cell cycle progression of neuronal progenitors in the spinal cord in mice and chickens, respectively.35-37 Data presented below indicate that, in the nervous system of C. elegans, egl-13 specifically acts to determine O2 and CO2 sensing neuron specification (Fig. 2B and C). When we analyzed the expression of a transcriptional reporter of egl-13, we found that expression is confined to the O2- and CO2-sensing neurons, muscle and vulval cells. Further, we observed that egl-13 expression is initiated during embryogenesis where the BAG and URX neurons are generated.38 In addition, we found that egl-13 is not required for expression of terminal differentiation markers of the sister cells of BAG and URX (SMDV L/R and CEPD L/R). These data indicate that in the nervous system EGL-13 functions specifically to regulate terminal differentiation of BAG and URX neurons.
Figure 2. Identification of EGL-13, a factor that specifies both O2- and CO2-sensing neurons. (A) Schematic presentation of egl-13 genetic lesions showing the resultant effects on the EGL-13 protein. (B) In C. elegans, the O2/CO2-sensing neural system comprises of six neurons called BAGL/R, URXL/R, AQR, and PQR (only single BAG and URX cells are shown). This neural system senses and transmits information regarding O2/CO2 status in the external environment and in the body fluid to the nerve ring. (C) Wild-type animals have six functional O2/CO2-sensing (top picture). egl-13 mutant animals (bottom picture) fail to correctly express an O2/CO2-sensing cell fate marker and are unable to sense O2 and CO2.
In our rescue analysis we asked three questions that further characterized the role of EGL-13 during differentiation of the BAG and URX neurons: (1) Does EGL-13 act autonomously? (2) Is EGL-13 needed to initiate and maintain a correct cellular fate? (3) Since egl-13 has four different splice variants (A to D) distinguished by the length of the N-terminal tail, we decided to test if one of the EGL-13 isoforms has a predominant role in specifying BAG and URX neuronal fate?
To answer the first question, we used BAG and URX cell-specific promoters that we developed during the study. Using these promoters to drive EGL-13, we rescued the expression of the terminal differentiation markers flp-19::GFP and gcy-33::GFP in BAG and URX neurons, respectively. These data showed that EGL-13 functions cell autonomously in BAG and URX neurons.
To answer the second question, we used a heat shock (HS) inducible promoter to drive the expression of egl-13. Using this technology to express egl-13 at the L3 stage (URX and BAG neurons generated during embryogenesis), we were able to answer the fundamental question of whether the BAG and URX neurons are generated at all or remain in an undifferentiated state in egl-13 mutant animals. We concluded the latter was the case since we were able to induce expression of a URX terminal fate marker post-developmentally. Further, we showed that the URX neurons lost expression of the marker, and thus, their O2-sensing fate, when the HS promoter was returned to an inactive state by lowering the incubation temperature. Together, these data demonstrated that EGL-13 is sufficient to drive terminal differentiation of both BAG and URX neurons, and EGL-13 is continuously needed to maintain the O2-sensing fate in URX neurons.
The third question was addressed by evaluating the rescuing potency of the EGL-13 long isoform A and short isoform D. Previous studies have shown that Sox proteins often work in complexes with other proteins, via their long N-terminal tails,39 and we speculated this could also be the case for EGL-13 during differentiation of O2- and CO2-sensing neurons. However, we found that the short isoform D of EGL-13 without a long N-terminal tail rescued equally well as the long isoform A, indicating no requirement of the N-terminal tail in this context. Although we did not further investigate EGL-13 protein interactions in our manuscript, we have not discarded the possibility of interaction domains in the remainder of the protein. To identify proteins that interact with EGL-13 to drive the differentiation of the BAG and URX neurons, we will express affinity-tagged EGL-13 specifically in BAG and URX neurons and use this to pull down potential interaction partners, which will be identified by mass spectrometry analysis. These experiments would add an additional layer of mechanistic insight on how transcription in BAG and URX is regulated at the protein level but also show how Sox proteins act in transcriptional complexes.
Ectopic Expression of egl-13 Induces O2 and CO2 like Fates
We tested whether egl-13 was sufficient to induce O2- or CO2-sensing neuronal fate in other neurons by ectopically expressing egl-13 under the control of the unc-86 promoter that is widely expressed in the nervous system.40 Using this promoter to drive egl-13 expression, we were able to induce expression of multiple O2 and CO2 neuronal markers in neurons that do not normally express these markers. Since the neurons we observed to adopt an O2/CO2 sensing-like fate were not consistent from animal to animal we were unable to experimentally show if they responded to changes in O2 or CO2, indicating a total change of fate, or alternatively, if their fate was pushed toward an O2- or CO2-sensing fate while still having features of their original fate. That ectopic egl-13 expression can induce O2 or CO2 neuronal fate in other neurons further underlines the importance of EGL-13 in BAG and URX differentiation and resembles findings from other studies.9,10 One such example is the adoption of dopaminergic neuronal fate by a selection of non-dopaminergic neurons when AST-1 is ectopically expressed.9 Another study described how UNC-3 is able to induce cholinergic fate in a subset of non-cholinergic neurons.10 The general assumption on cell identity and fate maintenance is that both positive and negative regulators tightly control it.41,42 Negative control that prevent cells from changing fates has been shown to be directed by chromatin modifications that prevent expression of genes from other cell fate programs.43,44 Recently, it was shown that removing LIN-53, a member of the PRC2 complex, thereby altering the chromatin state, made it possible to induce germ cell conversion into specific neurons, depending on the transcription factor used to drive the fate.43,44
egl-13 Mutants Are Unable to Respond to Changes in O2 and CO2
To test the functionality of the BAG and URX neurons in egl-13 mutant animals, we conducted behavioral assays where the locomotory response to changes in either O2 or CO2 was tested. Populations of wild-type animals decrease their average speed in response to changes in either O2 or CO2.20,45,46 This behavior was absent in egl-13 mutants but transgenic expression of egl-13 under its own promoter rescued the sensory defects. These data clearly showed that egl-13 was needed not only for differentiation but also for correct functionality of the BAG and URX neurons.
egl-13 Regulation in BAG and URX Neurons
Since egl-13 is required for terminal differentiation of both BAG and URX neurons, we speculated whether the upstream regulator(s) of egl-13 expression was the same for both neurons. To address that question, we drove expression of fluorescent protein with truncated fragments of the egl-13 promoter and analyzed the expression patterns in BAG and URX neurons. It became apparent that egl-13 expression is regulated through different motifs in the BAG and URX neurons. In the BAG neurons, we found that egl-13 expression is regulated through two ETS-5 binding sites, whereas in URX, egl-13 is regulated through AHR-1 and EGL-13 binding sites. We were able to confirm this regulation by crossing the mutants of ahr-1, egl-13, and ets-5 into the full-length egl-13 reporter, and by crossing egl-13 mutants with reporters of ets-5 and ahr-1. These experiments showed that the regulation is complex and the expression of the factors involved, in both BAG and URX, are dependent on each other for correct expression (Fig. 3). We speculate that egl-13 is involved in the general specification of both O2- and CO2-sensing neurons, but in order to distinguish between the two different fate programs, giving rise to BAG and URX individually, there is a requirement for co-factors ETS-5 and AHR-1, respectively.
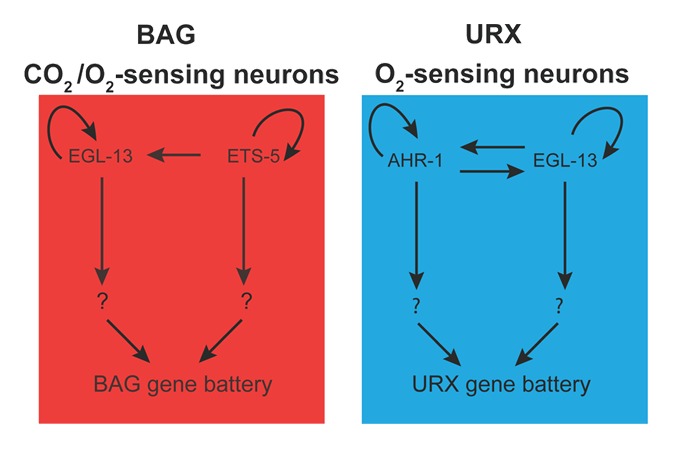
Figure 3. Working model for O2/CO2-sensing neuron specification. So far we have placed three factors (EGL-13, ETS-5, and AHR-1) into the genetic network of O2/CO2-sensing neuron specification. Ongoing work will help us understand how other factors work together to precisely control this important developmental decision.
The question marks in our model illustrate that we are still questioning the directness of regulation of the terminal gene battery in the BAG and URX neurons (Fig. 3). However, ETS-5 was shown to bind directly to the promoter of gcy-9 in the BAG neurons26 and our preliminary data show that both AHR-1 and EGL-13 bind the promoter of the URX terminal differentiation gene flp-8. To complete the picture we are currently analyzing the direct regulation with the rest of the BAG and URX promoters.
Concluding Remarks
Collective data from other neuronal cell fate studies have been used to create a paradigm on how neuronal diversity is obtained by the selective expression of one or few key transcription factors referred to as terminal selectors.8,10,47 In the definition of terminal selectors, four characteristics are listed: (1) Essential for terminal neuronal differentiation but not for general neuronal features. (2) Directly regulates the expression of the terminal gene battery through a specific motif. (3) Regulates its own expression to maintain differentiated features of the neuron. (4) One neuron might have more that one terminal selector and the correct fate is only obtained when co-expressed in the same cellular context.4,5,48
Reviewing the data we obtained during our studies, and in addition to our preliminary data showing that ahr-1 and egl-13 bind directly to the flp-8 promoter in URX neurons, strongly suggests that EGL-13 is a terminal selector. However, since it has a dual role in both BAG and URX neurons, the need of co-factors is obvious. We believe that ETS-5 and AHR-1 are strong EGL-13 interaction candidates in the BAG and URX neurons respectively and might prove to work with EGL-13 in a terminal selector complex. However, we do not exclude the possibility that other factors are needed for the correct differentiation of the O2- and CO2-sensing neurons and additional genetic screens would enable the identification of such genes.
The information we have provided on the differentiation mechanism of O2- and CO2-sensing neurons deepens and solidifies the understanding of how specific neurons differentiate and neuronal diversity is obtained. Further work pursuing the role(s) of egl-13/Sox5 in neuronal differentiation in higher organisms may potentially provide a better understanding of aspects of complex diseases.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Financial Disclosure
This work was supported by a grant from the Lundbeck Foundation (Grant number R93-A8391) and by an ERC Starting Grant (260807 - HYPOXICMICRORNAS).
Footnotes
Previously published online: www.landesbioscience.com/journals/worm/article/27284
References
- 1.Azevedo FA, Carvalho LR, Grinberg LT, Farfel JM, Ferretti RE, Leite RE, Jacob Filho W, Lent R, Herculano-Houzel S. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J Comp Neurol. 2009;513:532–41. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- 2.Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134:3771–80. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- 3.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–9. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 4.Hobert O. Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc Natl Acad Sci U S A. 2008;105:20067–71. doi: 10.1073/pnas.0806070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hobert O, Carrera I, Stefanakis N. The molecular and gene regulatory signature of a neuron. Trends Neurosci. 2010;33:435–45. doi: 10.1016/j.tins.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budry L, Balsalobre A, Gauthier Y, Khetchoumian K, L’honoré A, Vallette S, Brue T, Figarella-Branger D, Meij B, Drouin J. The selector gene Pax7 dictates alternate pituitary cell fates through its pioneer action on chromatin remodeling. Genes Dev. 2012;26:2299–310. doi: 10.1101/gad.200436.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doitsidou M, Flames N, Topalidou I, Abe N, Felton T, Remesal L, Popovitchenko T, Mann R, Chalfie M, Hobert O. A combinatorial regulatory signature controls terminal differentiation of the dopaminergic nervous system in C. elegans. Genes Dev. 2013;27:1391–405. doi: 10.1101/gad.217224.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flames N, Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature. 2009;458:885–9. doi: 10.1038/nature07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kratsios P, Stolfi A, Levine M, Hobert O. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat Neurosci. 2012;15:205–14. doi: 10.1038/nn.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lahiri S, Roy A, Baby SM, Hoshi T, Semenza GL, Prabhakar NR. Oxygen sensing in the body. Prog Biophys Mol Biol. 2006;91:249–86. doi: 10.1016/j.pbiomolbio.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Ma DK, Ringstad N. The neurobiology of sensing respiratory gases for the control of animal behavior. Front Biol (Beijing) 2012;7:246–53. doi: 10.1007/s11515-012-1219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richerson GB. Serotonergic neurons as carbon dioxide sensors that maintain pH homeostasis. Nat Rev Neurosci. 2004;5:449–61. doi: 10.1038/nrn1409. [DOI] [PubMed] [Google Scholar]
- 14.Spyer KM. To breathe or not to breathe? That is the question. Exp Physiol. 2009;94:1–10. doi: 10.1113/expphysiol.2008.043109. [DOI] [PubMed] [Google Scholar]
- 15.Buchanan GF, Richerson GB. Central serotonin neurons are required for arousal to CO2. Proc Natl Acad Sci U S A. 2010;107:16354–9. doi: 10.1073/pnas.1004587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng YJ, Nanduri J, Raghuraman G, Souvannakitti D, Gadalla MM, Kumar GK, Snyder SH, Prabhakar NR. H2S mediates O2 sensing in the carotid body. Proc Natl Acad Sci U S A. 2010;107:10719–24. doi: 10.1073/pnas.1005866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teppema LJ, Dahan A. The ventilatory response to hypoxia in mammals: mechanisms, measurement, and analysis. Physiol Rev. 2010;90:675–754. doi: 10.1152/physrev.00012.2009. [DOI] [PubMed] [Google Scholar]
- 18.Chang AJ, Chronis N, Karow DS, Marletta MA, Bargmann CI. A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol. 2006;4:e274. doi: 10.1371/journal.pbio.0040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang AJ, Bargmann CI. Hypoxia and the HIF-1 transcriptional pathway reorganize a neuronal circuit for oxygen-dependent behavior in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2008;105:7321–6. doi: 10.1073/pnas.0802164105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmer M, Gray JM, Pokala N, Chang AJ, Karow DS, Marletta MA, Hudson ML, Morton DB, Chronis N, Bargmann CI. Neurons detect increases and decreases in oxygen levels using distinct guanylate cyclases. Neuron. 2009;61:865–79. doi: 10.1016/j.neuron.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogers C, Persson A, Cheung B, de Bono M. Behavioral motifs and neural pathways coordinating O2 responses and aggregation in C. elegans. Curr Biol. 2006;16:649–59. doi: 10.1016/j.cub.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 22.Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marletta MA, Bargmann CI. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–22. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 23.Bretscher AJ, Kodama-Namba E, Busch KE, Murphy RJ, Soltesz Z, Laurent P, de Bono M. Temperature, oxygen, and salt-sensing neurons in C. elegans are carbon dioxide sensors that control avoidance behavior. Neuron. 2011;69:1099–113. doi: 10.1016/j.neuron.2011.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallem EA, Sternberg PW. Acute carbon dioxide avoidance in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2008;105:8038–43. doi: 10.1073/pnas.0707469105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guillermin ML, Castelletto ML, Hallem EA. Differentiation of carbon dioxide-sensing neurons in Caenorhabditis elegans requires the ETS-5 transcription factor. Genetics. 2011;189:1327–39. doi: 10.1534/genetics.111.133835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandt JP, Aziz-Zaman S, Juozaityte V, Martinez-Velazquez LA, Petersen JG, Pocock R, Ringstad N. A single gene target of an ETS-family transcription factor determines neuronal CO2-chemosensitivity. PLoS One. 2012;7:e34014. doi: 10.1371/journal.pone.0034014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, Deneris ES. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron. 2003;37:233–47. doi: 10.1016/S0896-6273(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 28.Giaccia AJ, Simon MC, Johnson R. The biology of hypoxia: the role of oxygen sensing in development, normal function, and disease. Genes Dev. 2004;18:2183–94. doi: 10.1101/gad.1243304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melillo G. Inhibiting hypoxia-inducible factor 1 for cancer therapy. Mol Cancer Res. 2006;4:601–5. doi: 10.1158/1541-7786.MCR-06-0235. [DOI] [PubMed] [Google Scholar]
- 30.Panigrahy A, Filiano J, Sleeper LA, Mandell F, Valdes-Dapena M, Krous HF, Rava LA, Foley E, White WF, Kinney HC. Decreased serotonergic receptor binding in rhombic lip-derived regions of the medulla oblongata in the sudden infant death syndrome. J Neuropathol Exp Neurol. 2000;59:377–84. doi: 10.1093/jnen/59.5.377. [DOI] [PubMed] [Google Scholar]
- 31.Guerenstein PG, Hildebrand JG. Roles and effects of environmental carbon dioxide in insect life. Annu Rev Entomol. 2008;53:161–78. doi: 10.1146/annurev.ento.53.103106.093402. [DOI] [PubMed] [Google Scholar]
- 32.Haas W. Parasitic worms: strategies of host finding, recognition and invasion. Zoology (Jena) 2003;106:349–64. doi: 10.1078/0944-2006-00125. [DOI] [PubMed] [Google Scholar]
- 33.Gramstrup Petersen J, Rojo Romanos T, Juozaityte V, Redo Riveiro A, Hums I, Traunmüller L, Zimmer M, Pocock R. EGL-13/SoxD specifies distinct O2 and CO2 sensory neuron fates in Caenorhabditis elegans. PLoS Genet. 2013;9:e1003511. doi: 10.1371/journal.pgen.1003511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanna-Rose W, Han M. COG-2, a sox domain protein necessary for establishing a functional vulval-uterine connection in Caenorhabditis elegans. Development. 1999;126:169–79. doi: 10.1242/dev.126.1.169. [DOI] [PubMed] [Google Scholar]
- 35.Kwan KY, Lam MM, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci U S A. 2008;105:16021–6. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–47. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 37.Perez-Alcala S, Nieto MA, Barbas JA. LSox5 regulates RhoB expression in the neural tube and promotes generation of the neural crest. Development. 2004;131:4455–65. doi: 10.1242/dev.01329. [DOI] [PubMed] [Google Scholar]
- 38.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 39.Lefebvre V, Dumitriu B, Penzo-Méndez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39:2195–214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-X. [DOI] [PubMed] [Google Scholar]
- 41.Blau HM, Baltimore D. Differentiation requires continuous regulation. J Cell Biol. 1991;112:781–3. doi: 10.1083/jcb.112.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tursun B. Cellular reprogramming processes in Drosophila and C. elegans. Curr Opin Genet Dev. 2012;22:475–84. doi: 10.1016/j.gde.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Patel T, Tursun B, Rahe DP, Hobert O. Removal of Polycomb repressive complex 2 makes C. elegans germ cells susceptible to direct conversion into specific somatic cell types. Cell Rep. 2012;2:1178–86. doi: 10.1016/j.celrep.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science. 2011;331:304–8. doi: 10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Couto A, Oda S, Nikolaev VO, Soltesz Z, de Bono M. In vivo genetic dissection of O2-evoked cGMP dynamics in a Caenorhabditis elegans gas sensor. Proc Natl Acad Sci U S A. 2013;110:E3301–10. doi: 10.1073/pnas.1217428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Busch KE, Laurent P, Soltesz Z, Murphy RJ, Faivre O, Hedwig B, Thomas M, Smith HL, de Bono M. Tonic signaling from O₂ sensors sets neural circuit activity and behavioral state. Nat Neurosci. 2012;15:581–91. doi: 10.1038/nn.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Etchberger JF, Flowers EB, Poole RJ, Bashllari E, Hobert O. Cis-regulatory mechanisms of left/right asymmetric neuron-subtype specification in C. elegans. Development. 2009;136:147–60. doi: 10.1242/dev.030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hobert O. Regulation of terminal differentiation programs in the nervous system. Annu Rev Cell Dev Biol. 2011;27:681–96. doi: 10.1146/annurev-cellbio-092910-154226. [DOI] [PubMed] [Google Scholar]