Abstract
Essentially all aboveground plant tissues develop from the stem cells in the primary shoot apical meristem. Proliferation of the stem cell population in the Arabidopsis shoot apical meristem is tightly controlled by a feedback loop formed primarily by the homeodomain transcription factor WUSCHEL (WUS) and the CLAVATA ligand-receptor system. In this study, it is shown that mutation of a translation initiation factor, eIF3h, causes a tendency to develop a strikingly enlarged shoot apical meristem with elevated and ectopic expression of WUS and CLAVATA3 (CLV3). Many of the mRNAs that function in apical meristem maintenance possess upstream open reading frames (uORFs), translational attenuators that render translation partially dependent on eIF3h. Specifically, the mRNA for the receptor kinase, CLV1, is undertranslated in the eif3h mutant as shown by transient and transgenic expression assays. Concordant phenotypic observations include defects in organ polarity and in translation of another uORF-containing mRNA, ASYMMETRIC LEAVES 1 (AS1), in eif3h. In summary, the expression of developmental regulatory mRNAs is attenuated by uORFs, and this attenuation is balanced in part by the translation initiation factor, eIF3h. Thus, translational control plays a key role in Arabidopsis stem cell regulation and organogenesis.
Introduction
In eukaryotic cells, gene expression is highly regulated, often at multiple levels, such as transcription, mRNA structure and stability, translational control, and protein degradation. Translational regulation is arguably least well characterized, and questions concerning the mechanism of translational control abound. In plants, translation is regulated by small metabolites as well as environmental conditions (reviewed in [1]–[3]). In contrast, how translational regulation contributes to plant development remains largely uncharted territory. Mutations that affect specific proteins of the large and small ribosomal subunits, which were recently discovered in genetic interaction screens, suggest a role for translational control in leaf polarity [4]–[7]. Moreover, mutations in RPL24B/SHORTVALVE (STV) cause defects in organ initiation, vascular patterning, and gynoecium structure that could be attributed to undertranslation of mRNAs for transcription factors of the auxin response factor (ARF) class [8].
Among the eukaryotic translation initiation factors, eIF3 is by far the most complex, consisting of 12 subunits in Arabidopsis [9]. eIF3 participates in almost all major steps during initiation, such as tRNA charging of the 40S ribosomal subunit, loading of the charged 40S onto the mRNA’s 5′ Untranslated Region (UTR), mRNA scanning and start codon recognition, and translation reinitiation (reviewed by [10]). The functions of the individual eIF3 subunits remain to be fully characterized. The h subunit of eIF3 is not conserved in budding yeast, but forms part of the functional core of mammalian eIF3 [11], [12]. In Arabidopsis, carboxyl-terminal truncation alleles of eIF3h cause under-translation of specific mRNAs, many of which harbor multiple upstream open reading frames (uORFs) in their 5′ leader [13], [14]. uORFs generally inhibit translation because a ribosome that has translated the uORF must terminate translation, resume scanning and acquire fresh translation initiation factors before it can translate the main ORF downstream. eIF3h ameliorates the inhibitory effect of specific uORFs in part by promoting the reinitiation competence of the translating ribosome [15]. The eif3h mutant shows auxin related phenotypes such as pin-formed inflorescence shoots, misexpression of auxin related genes, and poor translation of ARFs [14], [16], [17]. However, the eif3h mutant displays additional pleiotropic developmental phenotypes, such as growth retardation or growth arrest. It has remained unclear how under-translation of specific mRNAs causes these macroscopic phenotypes.
The plant tissues above ground ultimately develop from the stem cells in the shoot apical meristem (SAM). In Arabidopsis, the stem cell population in the SAM is tightly regulated by the CLAVATA-WUSCHEL (CLV-WUS) circuit (reviewed in [18]). CLV3, an extracellular peptide produced in the outer cell layers in the central zone of the SAM, is the ligand for the receptor kinase CLV1 [19]–[21]. In response to the CLV3 signal, CLV1, the related receptor-like kinase RPK2/TOADSTOOL, and the heterodimer of CLV2 and CORYNE, restrict the spatial expression of the homeodomain transcription factor, WUSCHEL (WUS), to a small cohort of internal cells that form the organizing center of the SAM. Besides other target genes, WUS induces the expression of CLV3, whereby a negative regulatory feedback loop is formed to ensure the stability of the stem cell population [19], [22]–[27].
Here we describe that a mutation in eIF3h causes a variety of defects in SAM maintenance that range from subtle defects in organ positioning and organ polarity to a massively enlarged, yet eventually quiescent, SAM. Translation assays revealed that eIF3h supports the efficient translation of the mRNAs for CLV1 and the leaf transcription factor, AS1, which contain uORFs in their 5′ UTR. Mistranslation of these and other mRNAs in the eif3h mutant may disrupt the otherwise robust feedback circuits that underlie SAM maintenance and organ specification. Thus, the eif3h mutation amounts to a genetic perturbation that unveils a role for translational control in Arabidopsis SAM function and organogenesis.
Results
The eif3h Mutant Plants have Growth Defects in the SAM
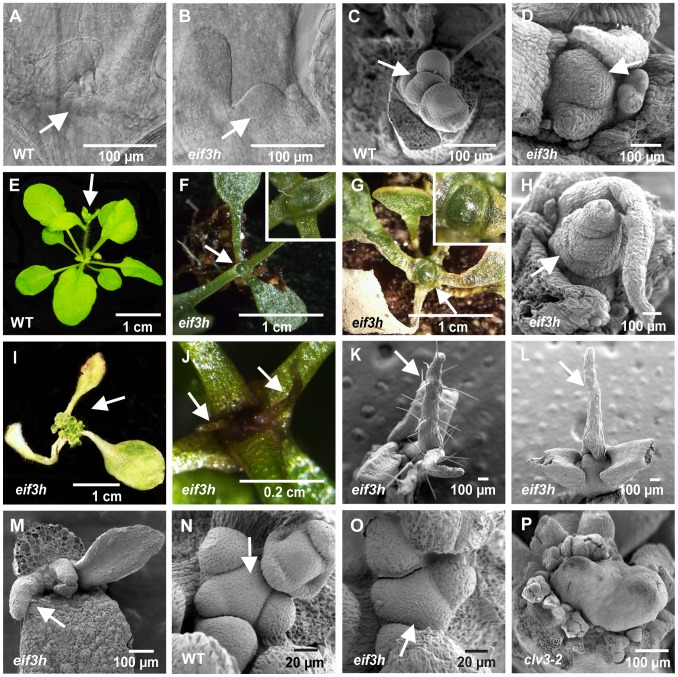
Unlike wild-type Arabidopsis plants, which always initiate a functional inflorescence from the shoot apex under normal growth conditions, a large proportion of eif3h mutant plants never initiated an inflorescence (33%, 45 out of 135). Closer inspection revealed growth defects in the shoot apex. Meristem enlargement could be seen as early as twelve days after germination ( Figure 1A, B ; Table 1 ) but was not reliably detected before that time. At twelve days, the eif3h mutant meristem had a slightly larger diameter than wild type (P = 4.2. 10−7 by two-tailed t-test) and adopted a more dome-like shape. A dome-shape is typical for the inflorescence meristem. Scanning electron microscopy demonstrated that the eif3h mutant apex in a 3-week-old seedling could be significantly enlarged at a time when wild type has begun to produce flowers ( Figure 1C, D and H ). The mutant meristem has enlarged in part through cell proliferation and in part through dramatic cell expansion, a sign that the meristematic cells have adopted a differentiated fate. Cell diameters in our wild-type meristems were 4.2±1.2 micron, close to expectations [28]. For the enlarged meristem in the eif3h mutant ( Figure 1D and H ), the cell diameter in the apex was 14.2±4.0 microns. In contrast, the transverse diameter of differentiated petiole cells was similar between wild type and eif3h, i.e. 13.3±3.2 micron and 15.8±3.7 micron, respectively. In these mutant plants, the shoot apex eventually formed a large dome-shaped structure visible to the naked eye ( Figure 1F , compare wild type in 1E). However, this phenotype was not fully penetrant, as many eif3h plants will produce normal-sized inflorescence meristems ( Figure 1N, O ), in contrast to the clv3 mutant ( Figure 1P ). Plants that formed such a dome-shaped apex typically senesced and died without initiating any additional leaves ( Figure 1G ). Occasionally, plants that had suspended leaf formation would eventually initiate multiple new shoot apices late in development (3 out of 78, 4%) ( Figure 1I ). Radialized leaves were common on the eif3h apex ( Figure 1J–M , Table 1 ). The majority of these leaves were abaxialized as judged by the lack of trichomes at the juvenile stage. An enlarged meristem is characteristic of mutations in the repressors of stem cell proliferation, CLAVATA1 and CLAVATA3. Like clv1 and clv3 mutants, the eif3h mutant occasionally (10–20%) produced fruits with more than the regular two carpels (Figure S1A, B), bifurcated (Figure S1C) or fasciated stems (Figure S1D). Other typical abnormalities in eif3h are shown in Figure S1E–G). In summary, the vegetative phenotypes observed in eif3h mutant plants suggest expansion of SAM size accompanied by a failure to initiate new organs. In plants that managed to flower, SAM enlargement was less pronounced, but could still be deduced from abnormalities, such as fasciated stems, abnormal organ positioning and an enlarged gynoecium.
Figure 1. Defects of eif3h mutant Arabidopsis in shoot apical meristem maintenance.
(A–B) 12 day old SAM imaged by differential interference contrast after clearing with chloral hydrate; arrows point to SAM. (A) wild type. (B) eif3h mutant. (C–D, H) Scanning electron micrographs of 3 week old wild-type inflorescence meristem (C) and equivalent in eif3h (D and H). Note enlargement of cells and of the entire SAM in eif3h. Arrow points to the SAM. The meristem in (D) is fasciated, i.e. branched into two, and both (D) and (H) have formed a radialized leaf. (E–G) Enlarged shoot apex in eif3h. White arrows point to the inflorescence (in wild type) or the shoot apex (in eif3h). (E) Wild type. (F) The enlarged quiescent eif3h SAM (inset shows a close-up of the apex). (G) Further enlarged dome-shaped eif3h SAM prior to senescence (inset shows a close-up). (I) Reactivated eif3h SAM with multiple apices initiating. (J–M) Filamentous organs emerging from the eif3h apex (arrows). (J) 3 week old eif3h mutant. (K) Filamentous organ with trichomes on 3 week old eif3h apex. (L) Filamentous organ without trichomes on 3 week old eif3h apex. (M) A filamentous leaf on a 1 week old eif3h apex. (N–P) Inflorescence apices. Arrows point to the SAM. (N) Wild type. (O) eif3h. (P) 3-week old clv3-2.
Table 1. Meristem abnormalities in the eif3h mutant.
| eif3h | Wild type | |
| Meristem diameter1) | 93±14 µm | 71±10 µm |
| Meristem morphology2) | ||
| Openly visible and enlarged | 29/49 | 0/50 |
| with radialized leaves | 17/29 | none |
| Not openly visible | 20/49 | 50/50 |
| with radialized leaves | 8/20 | 0/50 |
Determined in situ by differential interference microscopy from 22 (eif3h) and 52 (wild type) 12 day old seedlings.
Determined as in Figure 1J under a stereomicroscope.
eIF3h Boosts Translation of CLV1 mRNAs
eIF3h counteracts the translational repression by uORFs [13]–[15]. Among genes involved in SAM maintenance, the receptor kinase CLV1 harbors five uAUGs, suggesting that CLV1 is a potential client of eIF3h. A protoplast transformation assay based on in vitro transcribed mRNA was adopted to observe the translation efficiency of specific mRNA 5′ leaders in the eif3h mutant. The translational efficiency on the uORF-containing CLV1 leader was lower in eif3h than wild type, whereas the translation on the uORF-free WUS leader was not altered ( Figure 2B, C ). The ribosomal occupancy of the native Arabidopsis CLV1 mRNAs was also reduced in the eif3h mutant compared to wild-type ( Table 2 ).
Figure 2. The 5′ leader of the CLV1 mRNA renders translation dependent on eIF3h.
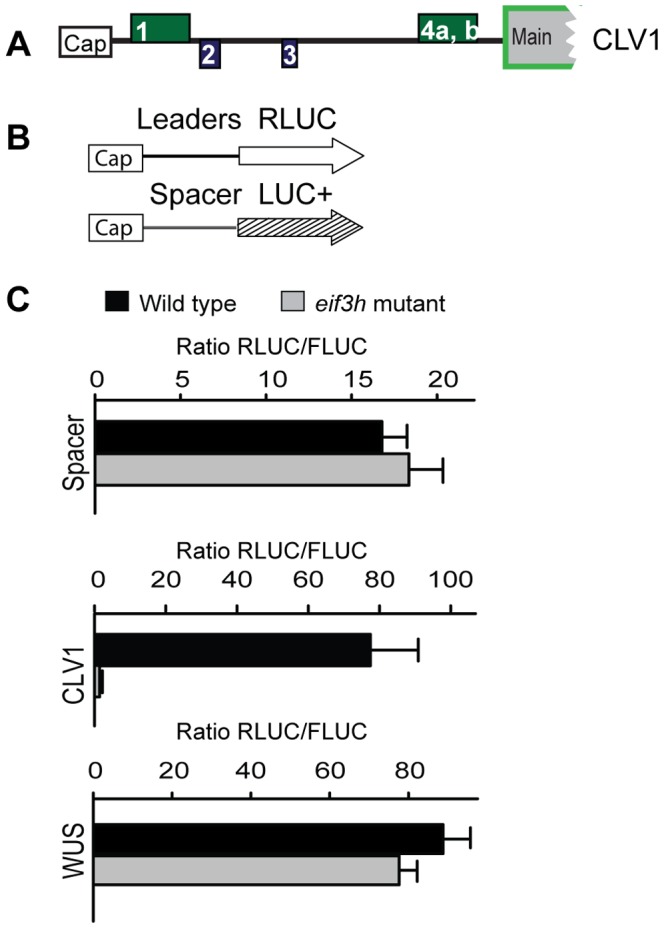
(A) The 5′ leader of CLV1 harbors multiple uORFs. The boxes stand for uORFs that are in the –1 frame (green), or in the +1 frame (blue) with the main ORF. The cDNA sequence corresponds to the longest known gene model: CLV1 (At1g75820.1). (B) Schematic view of the mRNAs for protoplast transformation. mRNAs were prepared by in vitro transcription with SP6 RNA polymerase. An equal amount of internal control (Spacer-LUC+) mRNA was added to the 5′ leader-RLUC mRNA to be tested as an internal control for transformation efficiency. (C) Translational efficiency on the CLV1 and WUS 5′ leader is expressed as the mean RLUC/FLUC ratio with standard errors from three replicate transformations.
Table 2. Polysome loading state of selected mRNAs in eif3h mutant versus wild type.
| Gene | AGI | TL eif3h/WT | uORFs |
| CLV1 | At1g75820 | −0.89 | 4 |
| AS1 | At2g37630 | −1.44 | 3 |
| TOADSTOOL | At3g02130 | −1.65 | NA |
| CORYNE | At5g13290 | −0.36 | 4 |
| CLV2 | At1g65380 | −0.46 | 0 |
| CORONA | At1g52150 | −0.59 | 8 |
| SPLAYED | At2g28290 | −0.20 | 2 |
| KNAT1/BP | At4g08150 | −0.40 | 2 |
| ARF4 | At5g60450 | −0.59 | 3 |
| ARF5 | At1g19850 | −0.48 | 6 |
| ARF6 | At1g30330 | −0.20 | 6 |
| ARF7 | At5g20730 | −0.61 | 4 |
| ARF11 | At2g46530 | −0.47 | 6 |
| ARF18 | At3g61830 | −0.76 | 2 |
| HY5 | At5g11260 | +0.17 | 1 |
| ATBZIP11 | At4g34590 | −0.65 | 4 |
| TIR1 | At3g62980 | +0.18 | 1 |
| AUX1 | At2g38120 | +0.31 | 1 |
Note: TL eif3h/WT. Translation state (TL) is the ratio of mRNA in polysomal and non-polysomal RNA fractions; it is log-transformed and unitless. When compared between eif3h mutant and wild-type seedlings, negative numbers indicate that the mRNA is less polysomal, i.e. undertranslated in the eif3h mutant. The values are means from duplicate polysome microarrays [13]. TOADSTOOL and the heterodimer of CLV2 and CORYNE are receptor like kinases that can function as CLV3 receptors in parallel to CLV1. Data for ARFs, TIR1 and AUX1 are included for the purpose of calibration, given that ARFs are also undertranslated in eif3h while TIR1 and AUX1 are not [16]. The median value from 8832 genes was −0.10±0.21. TOADSTOOL and AS1 and CLV1 rank 19th and 55th and 495th, respectively, among cytosolic mRNAs. uORF numbers are from TAIR10. NA, not available.
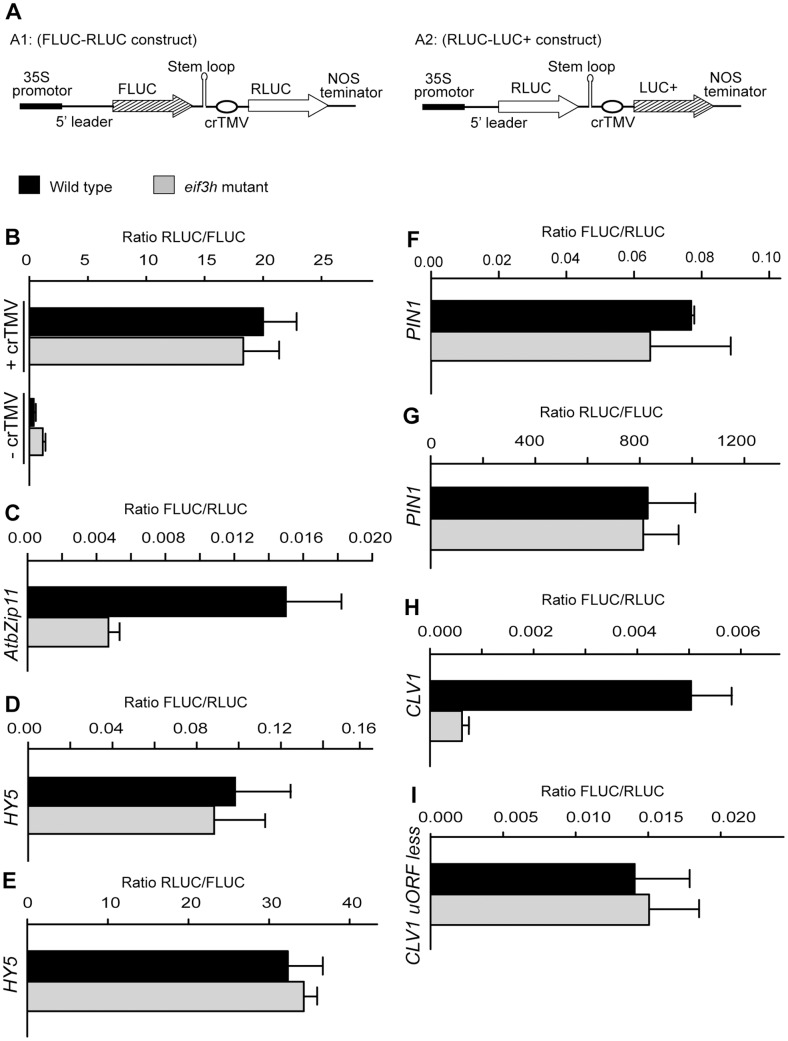
The translation efficiency under the control of the CLV1 5′ leader was further examined using a novel DNA-based, single-plasmid, assay, in which the translational reporter (upstream) and the transformation control (downstream) were fused via a sequence from the crucifer strain of tobacco mosaic virus (crTMV), which was reported to act as an internal ribosome entry site [29] (IRES; Figure 3A ). The crTMV element had minimal IRES activity in our hands, but had fortuitous promoter activity when the plasmids were transformed into Arabidopsis protoplasts (Figure S2). However, since the expression from the crTMV element was not affected by the eif3h mutation ( Figure 3B ), the constructs were deemed adequate for determining the translation potential of specific mRNA leader sequences. For additional validation, we confirmed that the leader of AtbZip11 was eif3h-dependent in this assay, while HY5 was not, as expected [14] ( Figure 3C–E ). The uORF-less PIN1 leader was also not affected by the eif3h mutation ( Figure 3F, G ). However, the CLV1 leader again showed clear eIF3h-dependence ( Figure 3H ).
Figure 3. Translation assays with reference gene driven by the crTMV intergenic sequence.
(A) Structure of the expression plasmids. In A1, the experimental FLUC reporter gene is transcribed by the 35S promoter and harbors the 5′ leader to be tested for translational efficiency. The RLUC reference gene is located further downstream on the same plasmid and is expressed due to transcriptional promoter activity of the crTMV sequence element. In A2, the experimental reporter is RLUC and the reference is LUC+. (B–I) Plasmids were transiently transformed into 10 day old wild-type or eif3h-1 seedlings. The expression is given as the ratio of the reporter luciferase activity divided by the reference luciferase from three replicate transformations with standard error. (B) The A1 construct with or without the crTMV sequence was used to confirm that the crTMV element is not eIF3h dependent. In this exceptional case, data are shown as downstream : upstream activity. (C–H) Tests of four plant 5′ leaders. (C) AtbZip11; (D) HY5; (E) HY5 leader in the RLUC-LUC+ construct (A2). AtbZip11 and HY5 leaders served to evaluate the translation assay system. (F, G) PIN1; (H) CLV1; (I) CLV1 uORF-less. HY5 has only one very short uORF and PIN1 has none.
Multiple uORFs in the CLV1 Leader Contribute to its eIF3h Dependence
The four uORFs present in the CLV1 5′ leader have 16, four, one, and twelve codons respectively; the fourth one contains an internal AUG codon. Translation assays using in vitro transcribed mRNAs indicated that the dependence on eIF3h was significantly reduced when all five uAUGs were removed ( Figure 4A ). uORFs1 and 2 contributed most strongly to eIF3h dependence (3rd and 4th constructs), while uORF3 or uORF4 alone (5th, 8th and 9th) had less of an effect. uORFs 1 and 2 also caused the largest absolute reduction in FLUC activity (not shown). We note that, even on the uORF-less CLV1 leader, expression was lower in eif3h compared to wild type ( Figure 4A ). Among other possible reasons, this might be due to the length of the mRNA, which is a factor in its ribosome occupancy in eif3h [13], or there might be fortuitous initiation at non-AUG codons in the 5′ UTR. Nonetheless, eIF3h mitigates the cumulative inhibition of translation caused by multiple uORFs of different length and position.
Figure 4. Removal of uORFs from the CLV1 5′ leader reduces its eIF3h dependence.
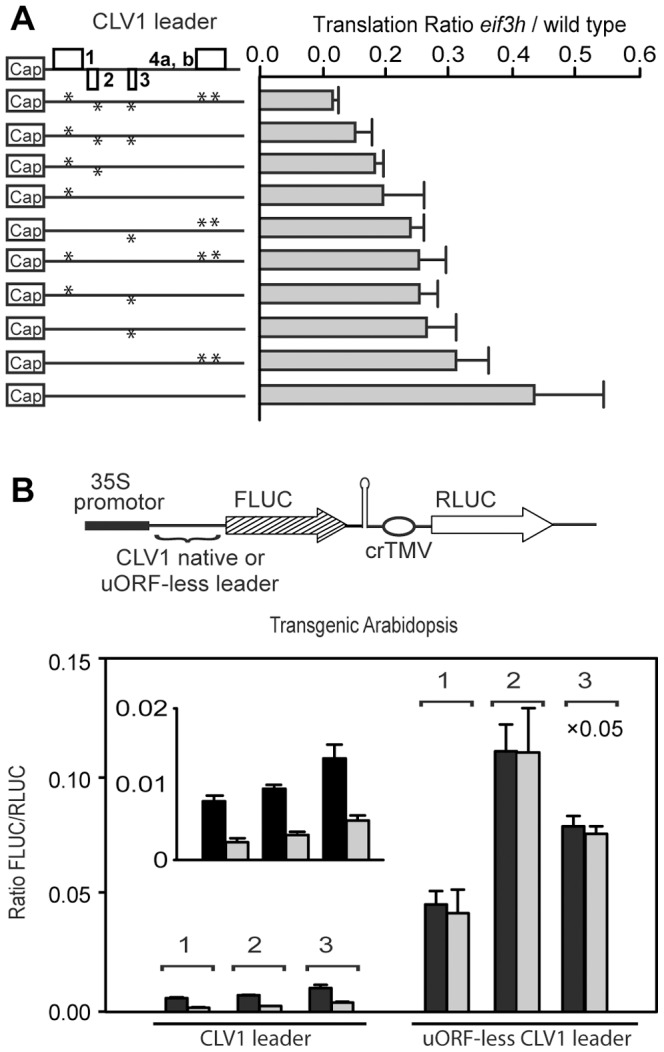
(A) Transient dual luciferase assays were performed in protoplasts as in Figure 2 but data are presented as the relative expression in the eif3h mutant as compared to wild type with standard deviations. Asterisks represent up to five uAUGs and open boxes represent uORFs. (B) Stable transgenic plants harboring the dual-luciferase construct illustrated at the top. The CLV1 native leader or uORF-less leader is linked to FLUC, and the RLUC ORF driven by the crTMV-element serves as a reference. Translational efficiency of the 5′ leader was established via dual luciferase assays in 10-day-old seedlings from three independent transgenic lines. Statistical significance was determined by t-test (***for p-value <0.001 and **for P-value<0.05). x0.05: FLUC activities of one particularly highly expressing line were multiplied by 0.05 for convenience of display.
The eIF3h dependence of translation on the native and uORF-less CLV1 leaders was further examined after stable transformation of eif3h-heterozygous plants. Dual-luciferase assays were performed after Mendelian segregation of wild-type and eif3h mutant seedlings in the progeny ( Figure 4B ). The uORFs clearly attenuated translation in this assay. While the native CLV1 leader yielded less expression in the eif3h mutant than wild type for all three transgenic lines tested, the uORF-less CLV1 leader drove equal expression levels in both genotypes ( Figure 4B ). The reduced FLUC expression from the CLV1 mRNA in eif3h could not be attributed to reduced transcript stability (Figure S3). Notwithstanding the quantitative disparity in the eIF3h defect between transient and stable expression, one may conclude that the uORF-studded CLV1 leader requires eIF3h for maximal expression under both conditions.
WUS and CLV3 Gene Expression in the eif3h Mutant
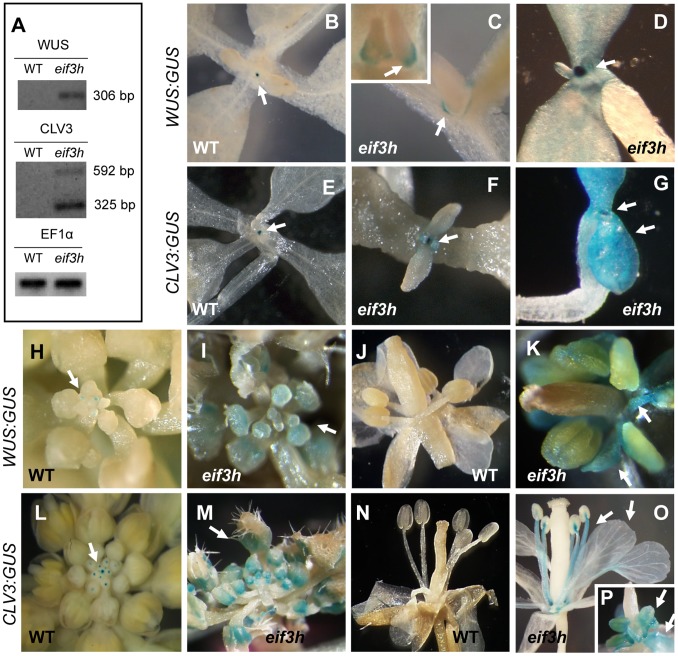
Because CLV1 suppresses expression of the stem cell regulator WUSCHEL, any reduction in translation of CLV1 in the eif3h mutant would be expected to increase WUS transcription, which will in turn promote CLV3 transcription according to the canonical CLV-WUS feedback regulation. Indeed, RT-PCR results indicated that WUS mRNA and two CLV3 mRNAs were overexpressed in the eif3h mutant ( Figure 5A ). Moreover, while expression of WUS:GUS and CLV3:GUS promoter:reporter transgenes were restricted to a small domain in the shoot apex of wild-type seedlings ( Figure 5B, E ), in the apex of eif3h mutant seedlings they tended to be expressed more highly and also ectopically around the base of young leaves ( Figure 5C, F ) and in the cotyledons ( Figure 5D, G ). In keeping with the incomplete penetrance of the meristem overgrowth defect in eif3h-1, the size of the meristematic expression domain was still near normal in this experiment. WUS:GUS expression was also elevated in the inflorescence tip in the eif3h mutant ( Figure 5H–I ), and could often be seen ectopically in floral organs, such as stamens, petals ( Figure 5J–K ), and ovules (data not shown). Consistent with elevated WUS expression, the CLV3:GUS was also expressed more highly ( Figure 5L–M ) in the eif3h inflorescence and ectopically in floral organs ( Figure 5N–P ). Although ectopic CLV3:GUS was observed in the eif3h mutant embryo, its level in the embryonic SAM was in the normal range (data not shown). Together, these results are consistent with the notion that translational defects in the eif3h mutant cause activation of WUS, which in turn contributes to the meristem expansion observed in the eif3h mutant.
Figure 5. WUS and CLV3 expression in the eif3h mutant.
(A) Reverse transcription (RT) PCR results for WUS and CLV3 mRNAs from 2 week old plants along with translation elongation factor 1α as a control. In eif3h, two CLV3 transcripts corresponding to gene models At2g27250.1 and At2g27250.3 were detected. To detect the transcripts in wild type, more amplification cycles are needed. (B–D) WUS:GUS expression [57] in wild type (B) and eif3h (C, D) seedlings. (E–G) CLV3:GUS expression in wild type (E) and eif3h (F, G) seedlings. (E) is at half the magnification of F and G. (H, I) WUS:GUS expression in wild type (H) and eif3h (I) inflorescences. (J, K) WUS:GUS expression in wild type (J) and eif3h (K) flowers. (L, M) CLV3:GUS expression in the wild type (L) and eif3h (M) inflorescences. (N, O) CLV3:GUS expression in wild type (N) and eif3h (O) flowers. (P) shows elevated CLV3:GUS expression in stamens and petals of a developing eif3h flower bud.
Relationship between eIF3h and ASYMMETRIC LEAVES1
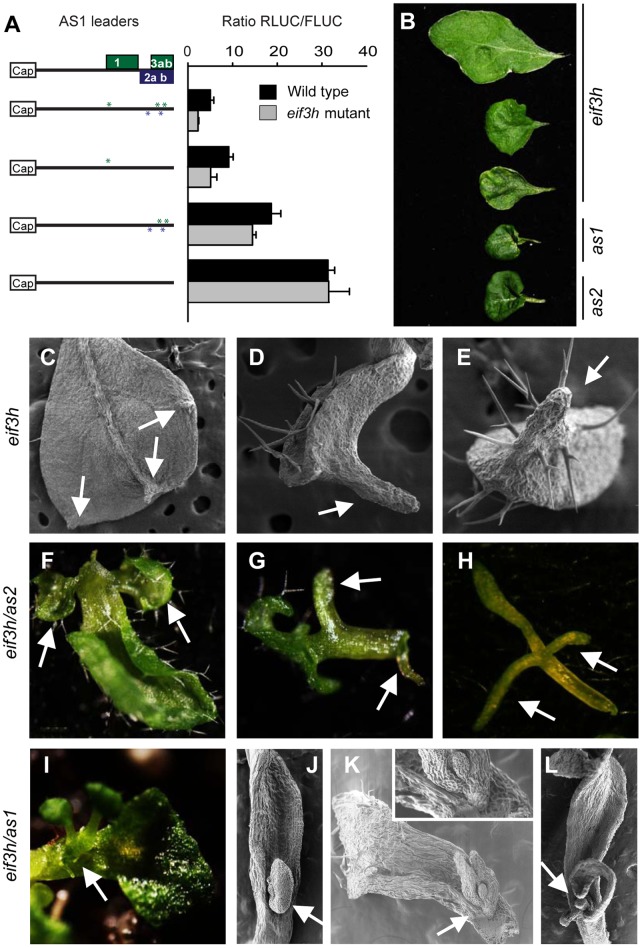
The radialized leaves that are often seen in eif3h mutant plants toward the end of the growth period may be due to defects in leaf polarity. ASYMMETRIC LEAVES1 (AS1) and AS2 code for transcription factors that cooperate as adaxializing factors in leaf polarity. In snapdragon, a mutation in the AS1 homolog, PHANTASTICA, alone causes radialized and abaxialized leaves [30], [31], and AS1/PHAN possesses a cluster of evolutionarily conserved uORFs [32]. eif3h mutant leaves were crinkly, similar to those of as1 and as2 mutants ( Figure 6B ). The AS1 uORFs were indeed inhibitory to translation and caused a slight dependence on eIF3h ( Figure 6A ). In addition, the AS1 mRNA has reduced ribosome occupancy in the eif3h mutant ( Table 2 ). Together, these results suggest that crinkly and radialized leaves in eif3h may be due in part to poor translation of AS1.
Figure 6. Reduced translation behind the ASYMMETRIC LEAVES 1 leader in the eif3h mutant, and defects on leaf polarity in the eif3h mutant and eif3h/as2 or eif3h/as1 double mutants.
(A) Translation assay results for the AS1 (At2g37630.1) leader and its uORF-removed variants. uAUGs in the AS1 leader were mutated as described in material and methods for removing uAUGs from the CLV1 leader. The translation assay was performed as in Figure 2 and data analyzed as in Figure 4 . Statistical significance was determined by t-test (**for p-value <0.05). (B) Rosette leaves of eif3h, as1 and as2 (At1g65620) mutants. (C–E) Scanning electron microscopic images for eif3h rosette leaves with outgrowth on the abaxial side. Arrows point to outgrowths without (C and D) or with (E) trichomes. (F–G) Rosette leaves and needle-like leaf of the eif3h/as2 double mutant with additional leaflets. Arrows point to an expanded leaflet (F), needle-like leaflets (G) and needle-like leaflets on a needle-like leaf. (I–L) Ectopic structures growing on the eif3h/as1 double mutant rosette leaves. Arrows point to an ectopic shoot growing (I) and SEM images of ectopic ovule like structures (J, K, L).
The eif3h mutant occasionally had ectopic outgrowths on its leaves ( Figure 6C–E ), but these did not normally reveal any pluripotential stem cell character. However, the eif3h leaf phenotype was strongly enhanced by both as1 and as2 mutations to similar degrees. Double mutants formed elaborate outgrowths on their leaves, which suggested that the leaf tissue can adopt meristematic potential ( Figure 6F–L ). The enhancement of the as2 phenotype in particular suggests that eIF3h may be rate limiting for AS1 expression.
Discussion
Maintenance of the stem cell population in the SAM is critically important for the continuous initiation of lateral organs including leaves, branches, and flowers. The underlying regulatory feedback loop composed of the WUS and CLV genes has been studied intensively from different angles, including its interface with auxin and cytokinin [18], [33], [34]. This study revealed another previously underappreciated aspect of SAM maintenance, an interplay between uORF containing mRNAs and the machinery supporting their efficient translation. The striking formation of an enlarged meristematic dome ( Figure 1 ) or of a pinformed stem in the eif3h mutant [16] suggests that Arabidopsis eIF3h supports stem cell homeostasis, organ initiation, and morphogenesis. We propose that eIF3h does so in part by overcoming the inherent inhibition of translation by clusters of uORFs that reside in mRNAs encoding several regulators of meristem activity.
Clients of eIF3h Maintain Stem Cell Homeostasis
We identified two new uORF containing clients of eIF3h, CLV1 and AS1, and demonstrated their translational defects in the eif3h mutant. The CLV1 mRNA has reduced polysome loading in the eif3h mutant ( Table 2 ). The full length CLV1 mRNA harbors four uORFs in its 5′ leader, which are responsible in part for the eIF3h-dependence of translation of a fused reporter gene ( Figure 2 , 3 , 4 ). Underexpression of CLV1 should result in overexpression of WUS and in turn CLV3 [19], [26]. WUS:GUS and CLV3:GUS overexpression was indeed observed ( Figure 5 ), along with enlargement of the vegetative SAM ( Figure 1 ).
Full length AS1 mRNA harbors three uORFs. Again, eIF3h maintains ribosome occupancy on the AS1 mRNA and partially alleviates the translational suppression by its inhibitory uORFs ( Figure 6 ). Interestingly, the position of the AS1 uORFs is highly conserved throughout the dicotyledonous plants, suggesting their regulatory significance [32], although their peptide sequence is not conserved. Underexpression of AS1 will cause wrinkled leaves, which were observed in the eif3h mutant. Underexpression of AS1 would also sensitize plants to other mutations affecting leaf polarity, such as as2, which was also observed. For comparison, the conservation status of uORFs in the 5′UTRs of CLV1 orthologs will require better cDNA sequence information. However, among nine putative CLV1 orthologs identified from public genomic DNA sequences of various eudicots (Vitis, Citrus, three Brassicaceae, four legumes), all had upstream AUGs within 120 nucleotides of the main AUG, albeit with a pattern different from Arabidopsis (not shown). In summary, CLV1 and AS1 thus join the ranks of several other uORF containing mRNAs that are translated poorly in the eif3h mutant, most notably auxin response factors [16], [17].
The phenotypic defects of the eif3h mutants are consistent with the simultaneous disruption of multiple translation units, not just CLV1 and AS1. The WUS-CLV circuit is generally robust and not easily perturbed by external circumstances such as nitrogen starvation, or herbicide, conditions that reduce translation. Broader changes, presumably affecting multiple transcripts, will be necessary to disrupt the circuit. The translational attenuation of CLV1 mRNA in the eif3h mutant would certainly not be sufficient to cause the loss of control over the meristem in the eif3h mutant plants. Even complete loss of CLV1 translation should produce only a relatively mild phenotype, i.e. an enlarged SAM and an increased number of flowers initiated on the shoot apex [20], [23]. However, in eif3h mutants, the shoot apex often enlarges continuously and stops producing lateral organs, before eventually arresting its growth. Similar enlarged leafless apical domes arise as a consequence of a variety of genetic perturbations, for example upon excessive or ectopic WUS expression [35]. Likewise, excessive silencing of Homeodomain-Zipper class III transcription factors, which are adaxial leaf determinants, in conjunction with a mutation of the ERECTA receptor kinase also causes meristem overproliferation [36]. Finally a similar defect arises upon misregulation of the auxin equilibrium in the shoot apex, such as in pin1 arf5/mp double mutants, in yucca aux1 double mutants, as well as in arf5/mp mutants treated with the auxin efflux inhibitor 1-N-naphthylphthalamic acid [37], [38]. Again, we surmise that it is the mistranslation of multiple regulatory mRNAs that causes similarly severe phenotypes in the eif3h mutant.
Notably, CLV1 functions in concert with several other receptor kinase like genes, CORYNE, which also contains uORFs, TOADSTOOL, and CLAVATA2. Moreover, other genes that regulate WUS, such as CORONA and SPLAYED [39]–[41], harbor multiple uORFs in their 5′ leaders. Not unlike CLV1, several of these mRNAs have reduced ribosome occupancy in the eif3h mutant, which may well add to the misregulation in the eif3h SAM. We suggest that the misregulation of the meristem in the eif3h mutant is caused by the combined undertranslation of several if not many meristem regulators. The full range of mRNAs affected by the eif3h mutation is unknown, but certainly includes mRNAs with functions beyond meristem maintenance. Aside from translational suppression, some mRNAs are translationally stimulated, for example ribosomal protein mRNAs [13], [16], [17], [42].
A complex system such as the SAM is maintained by multiple positive and negative feedback loops. Induction of CLV3 by WUS maintains the SAM. Besides the well known, stabilizing, negative feedback from CLV3 via the receptor kinases to WUS, WUS overexpression also causes further repression of CLV1 expression [43], and overexpression of CLV3 can downregulate CLV1 protein posttranslationally [44]. If such a system is perturbed simultaneously at multiple steps, as may well be the case when eIF3h activity is defective, the eventual outcome could be of two types. Either the system manages to rebalance itself, or it collapses. The eif3h mutant may teeter on this verge, and this may be the reason why only a fraction of eif3h mutant plants lose control over the meristem.
While the severe phenotypic defects indicate a major shift away from regular stem cell homeostasis, the eif3h mutant evidently retains some ability to translate genes with uORF-containing mRNAs, given that eif3h does not consistently phenocopy, for example, severe arf5/mp or arf3/ettin alleles. To explain this, we invoke that many of the implicated client genes are transcribed from multiple transcription start sites, which may result in shorter 5′ leaders with fewer uORFs [45]. Moreover, one should also expect significant leaky ribosome scanning across those uAUGs that are in a weak initiation context.
Translational Control in Arabidopsis Development
The concept that translational control of regulatory mRNAs adds a novel layer of gene regulation in the meristem merits further exploration ( Figure 7 ). In keeping with this notion, several groups reported that mutations in genes for ribosomal proteins enhance mutations in leaf polarity factors, such as AS1 and AS2 [5]–[7]. For example, a double mutant of rpl4d and as2 forms trumpet-like and needle-like leaves [4] similar to those observed in eif3h as1 double mutants. Certain phenotypes of the rpl4d mutant can be partially rescued with specific auxin response factor genes once their uORFs have been removed from their 5′ leaders [46]. Similar to these ribosomal protein mutations, eif3h strongly enhanced leaf polarity defects of as2 and as1 mutations and often generated radialized leaves on its own. These results indicate that the developmental clients of eIF3h are not restricted to the meristem, but include organogenesis factors. Several ribosomal mutations, including rpl4d, cause a pin-like phenotype when combined with as2 [4]–[7]. A mutation in rpl4d can be pin-like when CLV3 expression is altered, rpl5a has pin-like shoots in the Landsberg background [46], and rpl24b/stv1 enhances arf3 to form pins [8]. The spectrum of the eif3h mutation is clearly reminiscent of these phenotypes seen in ribosomal protein mutants. However, no meristem overgrowth phenotype like the one presented here for eif3h has been described for these ribosomal mutants on their own (e.g. see [47]).
Figure 7. A concept map for the role of eIF3h in Arabidopsis SAM maintenance and auxin response.
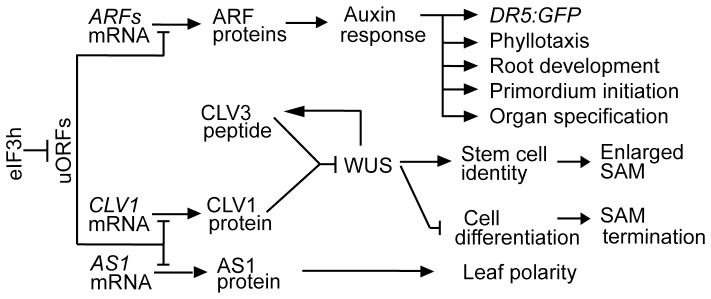
By overcoming the translational repression by uORFs, eIF3h promotes the translation of ARFs [16] and CLV1 and AS1 (this work), and therefore plays an important role in SAM maintenance and organogenesis.
We propose that the phenotypic spectrum of the eif3h mutant points to a group of mRNAs that are particularly sensitive to defects in the translation apparatus. Evidently, the phenotype of mutants in the translation apparatus will be driven by those mRNAs that have special requirements during translation. mRNAs with uORFs pose special requirements for translation, because, either, the translation machinery must perform two successive initiation events on the same mRNA. Or the translation machinery must bypass uORFs by leaky scanning. In the absence of either reinitiation or leaky scanning, the main open reading frame will be loaded skimpily with ribosomes, which may trigger other forms of posttranscriptional inhibition such as nonsense-mediated decay [48]. Here, we demonstrated, using the eif3h mutant as an example, that developmental regulatory genes such as CLV1 possess uORFs that render them sensitive to mutations in the translation apparatus.
The value of this study probably lies less in laying bare specific new mechanisms in stem cell regulation. Rather, it opens the window onto a previously underappreciated RNA sequence element that is present in a multitude of mRNAs that function in stem cell maintenance. What aspect of translational control in the meristem should be attributed to uORFs? uORFs may fine-tune the expression of many developmental regulator genes. In addition, weak transcriptional activity is believed to be noisy and stochastic in living cells [49], [50]. Because uORFs generally reduce translation they may serve to permit a high, by inference less noisy, transcription rate while keeping the rate of protein synthesis at the low level expected for a regulatory protein. One role of the eIF3h protein appears to be to maintain a moderate efficiency of translation on such uORF containing mRNAs.
Materials and Methods
Plant Growth Conditions and Transformation
Growth conditions for wild type (Wassilewskija ecotype) and the eif3h-1 allele, which harbors a T-DNA insertion in the 10th of 12 exons and results in a truncated protein, have been described [14]. Transgenes were transformed into heterozygous eif3h mutant plants using floral dip of Agrobacterium. Transgenic plants were selected on 10 mg/l Basta and verified by PCR.
Molecular Cloning
The plasmids for in vitro transcription were made in the TA-cloning vector pKRX [51] and contained the SP6 phage promoter, the translational leader from tobacco etch virus (TL) and the coding region of firefly luciferase (FLUC) or LUC+ (from pGL3-basic, Promega, Madison, WI) or Renilla luciferase (RLUC; [52]) followed by a 70 nucleotide long poly-A tail. The TL 5′ leader was replaced with the respective leader sequence to be assayed. The 5′ leader called Spacer is the multiple cloning site of pGL3-basic.
The crTMV based dual-luciferase expression cassettes were assembled in pBluescript II using the cauliflower mosaic virus 35S promoter, tobacco etch virus leader (TL), and RLUC, the crTMV internal ribosome entry site sequence from plasmid pYY376 [29], [53], FLUC or LUC+, and the nopaline synthase terminator. A truncated crTMV element retaining only 7 bp on the left and 20 bp on the right end was generated as a negative control. Stop codons were introduced between the upstream luciferase gene and the IRES with the oligonucleotides (LOOPADP-for: 5′ GATCTATCTAGTCTAGATAGCGTAGCCTAGGGGTG ACCACTAGTACCGGTGACGTCGG 3′ and LOOPADP-rev 5′ CGCGCCGACGTCACCGGT ACTAGTGGTCACCCCTAGGCTACGCTATCTAGACTAGATA-3′. A stem loop (ΔG = −41.7 kcal/mol) was introduced into the SpeI site in the loop adaptors by annealing the two oligos LOOP-for: 5′ CTAGAGCCACCACGGCCCCCAAGCTTGGGCCGTGGTGGCT 3′ and LOOP-rev: 5′ CTAGAGCCACCACGGCCCAAGCTTGGGGGCCGTGGTGGCT 3′ (A1 in Figure 3A) [54].
Site Directed Mutagenesis
The primers for removing uAUGs from the CLV1 leader are listed in Table S1. Briefly, wild type CLV1 leader was amplified with primers AT1G75820-FOR1 and REV1, and cloned between the SP6 promoter and RLUC in a pKRX vector. The CLV1 leader was re-amplified with primers AT1G75820-FOR2, REV1 and cloned between SP6 and RLUC as before to remove the first uAUG. To further remove uAUG 2, two short PCR products made with M13-FOR, AT1G75820-REV3 and AT1G75820 FOR3, RLUC-REV were fused by a double template PCR with primers M13-FOR and RLUC-REV. Using the double template PCR product as template, a similar approach was applied to further remove uORF 3, and 4a, 4b with primers AT1G75820-FOR4, REV4 and primers AT1G75820-FOR5, REV5 respectively. Equivalent procedures were followed for mutagenesis of the AS1 5′ leader.
Microscopy
For scanning electron microscopy plant material was dissected and placed into 0.1M sodium cacodylate buffered 3% glutaraldehyde for 60 minutes. Samples were then washed in buffer over 30 minutes before being post-fixed in buffered 2% osmium tetroxide for 60 minutes. Samples were washed in water, dehydrated in a graded ethanol series then critical point dried in CO2. Once dried, samples were mounted, coated with gold in a SPI sputter coater and examined with a Zeiss 1525 scanning electron microscope.
DNA Based Expression Assay after Transient or Stable Transformation
Wild-type and eif3h mutant plants were grown for ten days on MS agar plates with 1% sucrose. Plasmids carrying dual-luciferase constructs were introduced by particle bombardment as previously described [14]. Transformed seedlings were incubated at 22°C in a lighted growth chamber for 8 hours before assaying for luciferase activity. Activities of the experimental luciferase and the reference luciferase were measured in a single protein extract using the Dual Luciferase system (Promega, Madison, WI) in the TD-20/20 luminometer (Turnerdesigns, Sunnyvale, CA). Mean ratios of experimental and reference luciferase from 3 or 4 biological replicates were used to compare the translation efficiency between wild type and eif3h mutant.
Protoplast Preparation and PEG Mediated mRNA Transformation
Protoplasts were prepared from shoots of wild type or mutant 7-day-old Arabidopsis seedlings [55] and were transformed with 200 ng mRNA using the polyethyleneglycol method [56] as described [15]. The protoplasts were incubated in a 24 well plate for 3 hours in the dark at room temperature, then harvested by centrifugation for luciferase assays.
GUS Staining
Arabidopsis seedlings or inflorescences were prefixed in 90% acetone for 20 min, rinsed briefly in staining buffer without X-Gluc and infiltrated in staining buffer (0.05M phosphate buffer, pH 7.2; 0.2% Triton X-100; 2 mM potassium ferrocyanide and 2 mM potassium ferricyanide; 2 mM X-Gluc) in vacuum for 30 min, followed by incubation at room temperature for 6 hours. After dehydration with an ethanol series (20%, 35% and 50%), tissue was fixed in FAA (50% ethanol, 10% acetic acid and 5% formaldehyde) for 30 min at room temperature, then dehydrated completely with 70%, 85% and 100% ethanol.
Reverse Transcription PCR
First strand cDNA were synthesized with M-MLV reverse transcriptase (Promega) and oligo (dT) primers using RNAs prepared from 7 day old Arabidopsis seedlings. PCR for WUS and CLV3 was for 40 cycles and for eEF1α was 28 cycles. The gene specific primers for each gene are: WUS (forward: 5′-CCCAGCTTCAATAACGGGAAT-3′, reverse: 5′-ACCGTGCATAGGGAAGAGAG-3′), CLV3 (forward: 5′-cacctcgagCACTCAGTCACTTTCTCTCTAA-3, reverse: 5′-TCAAGGGAGCTGAAAGTTGT-3′) and eEF1α (forward: 5′-GATGAGACTTTCGTTATGA TCGAC-3′; reverse: 5′-ATTGAAAACCATAATAAAAAGTCTCAGA-3′). To measure RNA stability, 2-week-old transgenic seedlings were transferred to incubation buffer (1 mM Pipes, pH 6.25, 1 mM sodium citrate, 1 mM KCl, 15 mM sucrose) for 30 minutes, followed by addition of transcriptional inhibitor (100 µg/mL cordycepin; Sigma, St. Louis, MO). Samples were harvested at specific time points and analyzed by RT-PCR using primers listed in Table S1.
Supporting Information
eif3h inflorescence phenotypes indicative of defects in meristem regulation. (A) 3-carpel phenotype of eif3h. (B) Siliques of clv1-1 and clv3-2 showing 3 to 5 carpels are shown for comparison. (C) The eif3h inflorescence may be bifurcated. (D) eif3h inflorescence with fasciated shoots. (E) eif3h lateral branches that are not subtended by cauline leaves. (F) eif3h inflorescence showing spontaneous arrest of internode elongation in the main apex (arrows) that fails to exert apical dominance. (G) Reactivated eif3h inflorescence after premature termination.
(TIF)
The crTMV IRES element has promoter activity. DNA fragments harboring the elements outlined on the left were isolated from plasmids by gel purification and transformed into tobacco or Arabidopsis seedlings using the particle gun. Luciferase activity was measured in triplicate using a dual luciferase assay (Promega). Means and standard deviations are shown. Orange bar, 35S promoter. Black bar, SP6 promoter. Green arrow, FLUC coding sequence. Blue arrow, RLUC coding sequence. Three red circles, stop codons in all three reading frames. Stem-loop, Forms hairpin-loop when single stranded. Red oval, crTMV IRES element. Hatched red oval, truncated crTMV element.
(TIF)
Transcript stability of transcripts harboring the CLV1 5′ leader. The decay of mRNA levels for transgenic FLUC and RLUC mRNAs as well as for the highly stable, endogenous, translation elongation factor 1 alpha (EF1α) mRNA was monitored by RT-PCR after blocking transcription with cordycepin. Control amplifications with higher PCR cycle numbers (sat.) were performed for representative samples to confirm that the experimental amplifications had not reached saturation. RNA was isolated from transgenic seedlings used in Figure 4 . The gene expression cassettes are CLV1-FLUC transgenes used in Figure 3H and 3I .
(TIF)
Primers for removing uAUGs from the CLV1 leader and for RT-PCR.
(DOC)
Acknowledgments
We are grateful for transgenic lines contributed by Rüdiger Simon, Jennifer Fletcher, a plasmid construct by Yoshiharu Yamamoto, and cDNAs clones distributed by the Arabidopsis stock center. We appreciate access to microscopes granted by Mariano Labrador, Elena Shpak, and Andreas Nebenführ and discussions with John Golz, Jennifer Fletcher, Elliot Meyerowitz, and Elena Shpak.
Funding Statement
This work was supported by a grant from the DOE Energy Biosciences Program (DEFG02- 96ER20223). Partial stipend support for FZ was provided by BARD grant IS3901-06C. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Roy B, von Arnim AG (2013) Translational regulation of cytoplasmic mRNAs. In: Torii K, Chang C, editors. The Arabidopsis Book. Rockville: American Society of Plant Biologists. pp. e0165. [DOI] [PMC free article] [PubMed]
- 2. Echevarria-Zomeno S, Yanguez E, Fernandez-Bautista N, Castro-Sanz AB, Ferrando A, et al. (2013) Regulation of Translation Initiation under Biotic and Abiotic Stresses. Int J Mol Sci 14: 4670–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Muench DG, Zhang C, Dahodwala M (2012) Control of cytoplasmic translation in plants. Wiley Int Rev RNA 3: 178–194. [DOI] [PubMed] [Google Scholar]
- 4. Horiguchi G, Molla-Morales A, Perez-Perez JM, Kojima K, Robles P, et al. (2011) Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J 65: 724–736. [DOI] [PubMed] [Google Scholar]
- 5. Horiguchi G, Van Lijsebettens M, Candela H, Micol JL, Tsukaya H (2012) Ribosomes and translation in plant developmental control. Plant Sci 191–192: 24–34. [DOI] [PubMed] [Google Scholar]
- 6. Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, et al. (2008) Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135: 1315–1324. [DOI] [PubMed] [Google Scholar]
- 7. Yao Y, Ling Q, Wang H, Huang H (2008) Ribosomal proteins promote leaf adaxial identity. Development 135: 1325–1334. [DOI] [PubMed] [Google Scholar]
- 8. Nishimura T, Wada T, Yamamoto KT, Okada K (2005) The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17: 2940–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burks EA, Bezerra PP, Le H, Gallie DR, Browning KS (2001) Plant initiation factor 3 subunit composition resembles mammalian initiation factor 3 and has a novel subunit. J Biol Chem 276: 2122–2131. [DOI] [PubMed] [Google Scholar]
- 10. Valasek LS (2012) ‘Ribozoomin’–translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr Prot & Pep Sci 13: 305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Masutani M, Sonenberg N, Yokoyama S, Imataka H (2007) Reconstitution reveals the functional core of mammalian eIF3. EMBO J 26: 3373–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun C, Todorovic A, Querol-Audi J, Bai Y, Villa N, et al. (2011) Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). Proc Natl Acad Sci U S A 108: 20473–20478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim BH, Cai X, Vaughn JN, von Arnim AG (2007) On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol 8: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim TH, Kim BH, Yahalom A, Chamovitz DA, von Arnim AG (2004) Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell 16: 3341–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roy B, Vaughn JN, Kim BH, Zhou F, Gilchrist MA, et al. (2010) The h subunit of eIF3 promotes reinitiation competence during translation of mRNAs harboring upstream open reading frames. RNA 16: 748–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou F, Roy B, von Arnim AG (2010) Translation reinitiation and development are compromised in similar ways by mutations in translation initiation factor eIF3h and the ribosomal protein RPL24. BMC Plant Biol 10: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schepetilnikov M, Dimitrova M, Mancera-Martinez E, Geldreich A, Keller M, et al. (2013) TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J 32: 1087–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aichinger E, Kornet N, Friedrich T, Laux T (2012) Plant stem cell niches. Annu Rev Plant Biol 63: 615–636. [DOI] [PubMed] [Google Scholar]
- 19. Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619. [DOI] [PubMed] [Google Scholar]
- 20. Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585. [DOI] [PubMed] [Google Scholar]
- 21. Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y (2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- 22. Guo Y, Han L, Hymes M, Denver R, Clark SE (2010) CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J 63: 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, et al. (2010) RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920. [DOI] [PubMed] [Google Scholar]
- 24. Mayer KF, Schoof H, Häcker A, Lenhard M, Jürgens G, et al. (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815. [DOI] [PubMed] [Google Scholar]
- 25. Müller R, Bleckmann A, Simon R (2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schoof H, Lenhard M, Häcker A, Mayer KF, Jürgens G, et al. (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644. [DOI] [PubMed] [Google Scholar]
- 27. Yadav RK, Perales M, Gruel J, Girke T, Jonsson H, et al. (2011) WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev 25: 2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laufs P, Grandjean O, Jonak C, Kieu K, Traas J (1998) Cellular parameters of the shoot apical meristem in Arabidopsis. Plant Cell 10: 1375–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ivanov PA, Karpova OV, Skulachev MV, Tomashevskaya OL, Rodionova NP, et al. (1997) A tobamovirus genome that contains an internal ribosome entry site functional in vitro. Virology 232: 32–43. [DOI] [PubMed] [Google Scholar]
- 30. Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, et al. (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971. [DOI] [PubMed] [Google Scholar]
- 31. Waites R, Selvadurai HR, Oliver IR, Hudson A (1998) The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93: 779–789. [DOI] [PubMed] [Google Scholar]
- 32. Vaughn JN, Ellingson SR, Mignone F, Arnim A (2012) Known and novel post-transcriptional regulatory sequences are conserved across plant families. RNA 18: 368–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM (2009) Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci U S A 106: 16529–16534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, et al. (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260. [DOI] [PubMed] [Google Scholar]
- 35. Yadav RK, Tavakkoli M, Reddy GV (2010) WUSCHEL mediates stem cell homeostasis by regulating stem cell number and patterns of cell division and differentiation of stem cell progenitors. Development 137: 3581–3589. [DOI] [PubMed] [Google Scholar]
- 36. Mandel T, Moreau F, Kutsher Y, Fletcher JC, Carles CC, et al. (2014) The ERECTA receptor kinase regulates Arabidopsis shoot apical meristem size, phyllotaxy and floral meristem identity. Development 141: 830–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schütz M, Berleth T, Mattsson J (2008) Multiple MONOPTEROS-dependent pathways are involved in leaf initiation. Plant Physiol 148: 870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Green KA, Prigge MJ, Katzman RB, Clark SE (2005) CORONA, a member of the class III homeodomain leucine zipper gene family in Arabidopsis, regulates stem cell specification and organogenesis. Plant Cell 17: 691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kwon CS, Chen C, Wagner D (2005) WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev 19: 992–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lenhard M, Bohnert A, Jürgens G, Laux T (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814. [DOI] [PubMed] [Google Scholar]
- 42. Tiruneh BS, Kim BH, Gallie DR, Roy B, von Arnim AG (2013) The global translation profile in a ribosomal protein mutant resembles that of an eIF3 mutant. BMC Biol 11: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, et al. (2010) Transcriptional control of a plant stem cell niche. Dev Cell 18: 849–861. [DOI] [PubMed] [Google Scholar]
- 44. Nimchuk ZL, Tarr PT, Ohno C, Qu X, Meyerowitz EM (2011) Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr Biol 21: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yamamoto YY, Yoshitsugu T, Sakurai T, Seki M, Shinozaki K, et al. (2009) Heterogeneity of Arabidopsis core promoters revealed by high-density TSS analysis. Plant J 60: 350–362. [DOI] [PubMed] [Google Scholar]
- 46. Rosado A, Li R, van de Ven W, Hsu E, Raikhel NV (2012) Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc Natl Acad Sci U S A 109: 19537–19544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Szakonyi D, Byrne ME (2011) Involvement of ribosomal protein RPL27a in meristem activity and organ development. Plant Sign Behav 6: 712–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rayson S, Arciga-Reyes L, Wootton L, De Torres Zabala M, Truman W, et al. (2012) A role for nonsense-mediated mRNA decay in plants: pathogen responses are induced in Arabidopsis thaliana NMD mutants. PloS One 7: e31917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blake WJ, M KA, Cantor CR, Collins JJ (2003) Noise in eukaryotic gene expression. Nature 422: 633–637. [DOI] [PubMed] [Google Scholar]
- 50. Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A (2002) Regulation of noise in the expression of a single gene. Nat Genet 31: 69–73. [DOI] [PubMed] [Google Scholar]
- 51.Schutte BC, Ranade K, Pruessner J, Dracopoli N (1997) Optimized conditions for cloning PCR products into an XcmI T-vector. Biotechniques 22: 40–42, 44. [DOI] [PubMed]
- 52. Subramanian C, Woo J, Cai X, Xu X, Servick S, et al. (2006) A suite of tools and application notes for in vivo protein interaction assays using bioluminescence resonance energy transfer (BRET). Plant J 48: 138–152. [DOI] [PubMed] [Google Scholar]
- 53. Yamamoto YY, Tsuhara Y, Gohda K, Suzuki K, Matsui M (2003) Gene trapping of the Arabidopsis genome with a firefly luciferase reporter. Plant J 35: 273–283. [DOI] [PubMed] [Google Scholar]
- 54. Kozak M (1986) Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci U S A 83: 2850–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- 56. Gallie DR (1993) Introduction of mRNA to plant protoplasts using polyethylene glycol. Plant Cell Reports 13: 119–122. [DOI] [PubMed] [Google Scholar]
- 57. Brand U, Grunewald M, Hobe M, Simon R (2002) Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol 129: 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eif3h inflorescence phenotypes indicative of defects in meristem regulation. (A) 3-carpel phenotype of eif3h. (B) Siliques of clv1-1 and clv3-2 showing 3 to 5 carpels are shown for comparison. (C) The eif3h inflorescence may be bifurcated. (D) eif3h inflorescence with fasciated shoots. (E) eif3h lateral branches that are not subtended by cauline leaves. (F) eif3h inflorescence showing spontaneous arrest of internode elongation in the main apex (arrows) that fails to exert apical dominance. (G) Reactivated eif3h inflorescence after premature termination.
(TIF)
The crTMV IRES element has promoter activity. DNA fragments harboring the elements outlined on the left were isolated from plasmids by gel purification and transformed into tobacco or Arabidopsis seedlings using the particle gun. Luciferase activity was measured in triplicate using a dual luciferase assay (Promega). Means and standard deviations are shown. Orange bar, 35S promoter. Black bar, SP6 promoter. Green arrow, FLUC coding sequence. Blue arrow, RLUC coding sequence. Three red circles, stop codons in all three reading frames. Stem-loop, Forms hairpin-loop when single stranded. Red oval, crTMV IRES element. Hatched red oval, truncated crTMV element.
(TIF)
Transcript stability of transcripts harboring the CLV1 5′ leader. The decay of mRNA levels for transgenic FLUC and RLUC mRNAs as well as for the highly stable, endogenous, translation elongation factor 1 alpha (EF1α) mRNA was monitored by RT-PCR after blocking transcription with cordycepin. Control amplifications with higher PCR cycle numbers (sat.) were performed for representative samples to confirm that the experimental amplifications had not reached saturation. RNA was isolated from transgenic seedlings used in Figure 4 . The gene expression cassettes are CLV1-FLUC transgenes used in Figure 3H and 3I .
(TIF)
Primers for removing uAUGs from the CLV1 leader and for RT-PCR.
(DOC)