Abstract
The direct physiological effects that promote nicotine dependence (ND) are mediated by nicotinic acetylcholine receptors (nAChRs). In line with the genetic and pharmacological basis of addiction, many previous studies have revealed significant associations between variants in the nAChR subunit genes and various measures of ND in different ethnic samples. In this study, we first examined the association of variants in nAChR subunits α2 (CHRNA2) and α6 (CHRNA6) genes on chromosome 8 with ND using a family sample consisting of 1,730 European Americans (EAs) from 495 families and 1,892 African Americans (AAs) from 424 families (defined as the discovery family sample). ND was assessed by two standard quantitative measures: Smoking Quantity (SQ) and the Fagerström Test for ND (FTND). We found nominal associations for all seven tested SNPs of the genes with at least one ND measure in the EA sample and for two SNPs in CHRNA2 in the AA sample. Of these, associations of SNPs rs3735757 with FTND (P = 0.0068) and rs2472553 with both ND measures (with a P value of 0.0043 and 0.00086 for SQ and FTND, respectively) continued to be significant in the EA sample even after correction for multiple tests. Further, we found several haplotypes that were significantly associated with ND in the EA sample in CHRNA6 and in the both EA and AA samples in CHRNA2. To confirm the associations of the two genes with ND, we conducted a replication study with an independent case-control sample from the SAGE study, which showed a significant association of the two genes with ND, although the significantly associated SNPs were not always the same in the two samples. Together, these findings indicate that both CHRNA2 and CHRNA6 play a significant role in the etiology of ND in AA and EA smokers. Further replication in additional independent samples is warranted.
Keywords: CHRNA2, CHRNA6, smoking, tobacco dependence association, meta-analysis
Introduction
Tobacco use continues to be an important worldwide health concern. According to World Health Organization, there were 1.3 billion tobacco users world-wide in 2004 (World Health Organization 2012). In the United States, 46.0 million adults were cigarette smokers in 2008, and the number of deaths annually from smoking-related illnesses accounts for 30% of deaths from cancer and nearly 80% of deaths from chronic obstructive pulmonary disease (CDC 2008; Mokdad et al. 2004). The annual economic burden of smoking is also substantial, with a staggering $193 billion in medical costs and productivity losses (CDC 2008; Mokdad et al. 2004).
Cigarette smoking is a complex behavior, with both genetic and environmental components (Al Koudsi and Tyndale 2005; Sullivan and Kendler 1999). Many family, adoption, and twin studies of smoking addiction have indicated a heritability of 11%–78%, with an average heritability of 57% for both male and female smokers (Kendler et al. 1999; Li et al. 2003; Maes et al. 2004; Vink et al. 2005).
Nicotine, the primary psychoactive ingredient in cigarette smoke, exerts its effects by readily crossing the blood–brain barrier and binding to nicotinic acetylcholine receptors (nAChRs) in various brain structures (Wonnacott 1997). Activation of nAChRs on dopaminergic terminals induces dopamine release in the mesolimbic brain reward system (Kleijn et al. 2011; Wonnacott et al. 2000). To date, 17 nAChR subunits have been identified, which are divided into muscle and neuronal types (Kalamida et al. 2007). Neuronal nAChRs are widely expressed in the nervous system in peripheral ganglia and certain areas of the brain, as well as in nonexcitable cells, such as epithelium and cells of the immune system (Cui and Li 2010). Of the neuronal nAChRs, genes for nine α (α2–α10) and three β (β2–β4) subunits have been cloned. The α7–α10 subunits are found either as homopentamers (of five α7, α8, or α9 subunits) or as heteropentamers (of α7/α8 and α9/α10) (Plazas et al. 2005). By contrast, the α2–α6 and β2–β4 subunits form heteropentameric receptors, usually with a (αx)2(βy)3 stoichiometry.
Whereas several human neuronal nAChR subunit genes have been investigated for association with ND and other smoking-related behaviors in human subjects [for reviews, see (Berrettini and Doyle 2012; Greenbaum and Lerer 2009; Li and Burmeister 2009)], CHRNA2 has received less attention. Early linkage analysis of the Collaborative Studies on Genetics of Alcoholism (COGA) data showed modest evidence of linkage to 8p22–23, near CHRNA2, using two smoking phenotypes (ever-smoked and average number of packs per year) (Bergen et al. 1999). The association of CHRNA2 with smoking was reported in the schizophrenia families through linkage analysis and the candidate gene approach (Faraone et al. 2004). Although there is a reported association of CHRNA2 with ND, measured by DSM-IV and FTND score, in the Iowa Adoption Studies, the results were not corrected for multiple comparisons (Philibert et al. 2009; Yates et al. 1998). In a smoking cessation trial, SNP rs2565065 in CHRNA2 appeared to have pharmacogenetic relevance (Heitjan et al. 2008). In contrast, several other studies have failed to reveal a significant association of this gene with ND or other smoking-related phenotypes (Keskitalo-Vuokko et al. 2011).
The CHRNA6 and CHRNB3 genes are located contiguously in a tail-to-tail configuration on chromosome 8. Both α6- and β3-nAChRs are found in various brain regions, including the substantia nigra, ventral tegmental area, striatum, and locus coeruleus (Gotti et al. 2006a; Gotti et al. 2006b), which have significant roles in dopaminergic neurotransmission, thus contributing to reward and reinforcement of behavior (Cui et al. 2003). The α6β2β3- as well as α6α4β2β3-containing receptors in the striatum mediate α-conotoxin MII-sensitive dopamine release. In contrast, α6β2-containing receptors in the superior colliculus seem to be involved in GABA release (Champtiaux et al. 2003; Gotti et al. 2006a; Gotti et al. 2006b; Salminen et al. 2004). A recent meta-GWAS study indicated that rs2304297 in CHRNA6 is significantly associated with ND in the European sample, but the finding did not reach genome-wide significance (Thorgeirsson et al. 2010). Candidate gene-based association studies indicated that SNPs rs2304297 in the 3 -UTR of CHRNA6 was associated with ND in the European sample (Hoft et al. 2009b; Saccone et al. 2007), as was rs1072003 in intron 2 of CHRNA6 with ND in an Israeli female sample (Greenbaum et al. 2006).
Considering that nearly all subjects used in these GWAS or candidate gene-based association studies were of European origin, it would be interesting to know whether CHRNA6 and CHRNA2 genes are also associated with ND in smokers of other ethnicities. Thus, the primary objective of this study was to determine whether significant association of variants in CHRNA6 and CHRNA2 with ND can be detected in independent samples, especially in African American smokers.
Materials and Methods
Subjects and ND measures
Discovery Family Sample
Subjects of this sample include persons of both AA and EA origin who were recruited primarily from the states of Tennessee, Mississippi, Arkansas, and Michigan. Proband smokers were required to be at least 21 years old, to have smoked for at least the last 5 years, and to have smoked at least 20 cigarettes per day during the last 12 months. Once proband smokers were identified, their biological parents and siblings were invited to participate whenever possible. Table 1 provides the detailed characteristics of the two ethnic groups. All participants provided written informed consent, and the Institutional Review Boards of each participating institution approved the study.
Table 1.
Description of discovery and replication samples
| Discovery Family Samples | Replication Case-Control Samples | |||
|---|---|---|---|---|
| Ethnicity | AA | EA | AA | EA |
| No. of nuclear families | 424 | 495 | - | - |
| Avg. members/family (SD) | 4.46 (0.88) | 3.49 (0.80) | - | - |
| No. of subjects | 1,892 | 1,730 | 1,136 | 2,428 |
| Female (%) | 57 | 57 | 52 | 56 |
| Age; years (SD) | 40.32 (14.60) | 45.39 (15.77) | 39.68 (6.71) | 38.37 (9.65) |
| No. of smokers | 1,013 | 1,088 | 626 | 1,048 |
| CPD (SD) | 21.55 (12.07) | 22.07 (12.60) | 24.33 (17.46) | 26.16 (19.71) |
| FTND score (SD) | 5.49 (3.60) | 4.62 (3.55) | 3.90 (2.88) | 2.96 (3.27) |
The ND of each smoker was assessed with the two commonly used measures of smoking quantity (SQ; the number of cigarettes smoked per day) and the Fagerström test for ND (FTND; 0–10 scale) (Fagerstrom 1978). Because of the overlap of the contents of the two measures, a fairly robust correlation exists among them in both populations (r = 0.88 for AAs and 0.89 for EAs).
Replication Case-Control Sample
All subjects included in this sample were participants in the Study of Addiction: Genetics and Environment (SAGE) (Bierut et al. 2010) through the NCBI dbGaP database (dbGaP study accession phs000092.v1.p1). Quantitative measurements of severity of addiction to various substances, including nicotine, are provided in this dataset. For a detailed description, please see http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1.
Genotyping and imputation
For the discovery family samples, genomic DNA was either extracted from cells in the peripheral venous blood of each participant using a Maxi kit (Qiagen Inc, Valencia, CA) or obtained from the NIDA Genetics Repository at Rutgers University. Seven SNPs in CHRNA2 (rs2292976, rs3735757, rs891398, and rs2472553) and CHRNA6 (rs9298628, rs892413, and rs2217732) were selected based on results reported by other researchers (Heitjan et al. 2008; Hoft et al. 2009b; Philibert et al. 2009), the location of each SNP and a uniform coverage of the gene of interest, and allele frequency in samples with European and African origins from NCBI SNP database. All SNPs were genotyped using TaqMan assays in the 384-well microplate format (Applied Biosystems Inc., Foster City, CA) as reported previously (Beuten et al. 2005; Li et al. 2005; Ma et al. 2005). Briefly, 15 ng of DNA was amplified in a total volume of 7 μl containing an MGB probe and 2.5 μl of TaqMan universal PCR master mix. Allelic discrimination analyses were performed on the ABI Prism 7900HT. To ensure the quality of genotyping, four no-template negative controls and four positive controls were added to each 384-well plate.
The SAGE samples were genotyped on commercially available platforms, including Illumina (San Diego, CA, USA). Quality control was performed in each group separately, with the goal of excluding those samples with sex or chromosomal anomalies, a low call rate, or first- or second-degree relatedness. Imputations of non-genotyped SNPs in the 1000 Genome CEU v2 (2010–11 release) and the HapMap Phase II CHB+JPT were carried out for the SAGE data using MaCH (Li et al. 2009; Li et al. 2010) and IMPUTE v2 (Howie et al. 2009; Marchini et al. 2007), respectively.
Statistical analysis
To test for genotyping quality in the discovery family sample, we assessed Mendelian inconsistencies and departure from Hardy-Weinberg equilibrium (HWE) using Haploview (v. 4.0) software (Barrett et al. 2005). Subjects with any inconsistent SNP data for a given genetic variant were excluded from analysis.
Individual and haplotype-based association analysis for the discovery family sample
Associations between the seven SNPs in CHRNA2 and CHRNA6 and the two ND measures were determined using the pedigree-based association test (PBAT v. 3.5) based on the generalized estimating equation approach (Lange et al. 2004). Pair-wise linkage disequilibrium (LD) and haplotype blocks for the four SNPs in CHRNA2 and three SNPs in CHRNA6 were assessed by Haploview (v. 4.0) software (Barrett et al. 2005; Gabriel et al. 2002). Association analysis for haplotypes located in each LD block with the two ND measures was performed using the family-based association test (FBAT v.1.7.3) (Horvath et al. 2004). Three genetic models (additive, dominant, and recessive) were tested for all association analyses, with sex and age as covariates in the AA and EA samples. Statistically significant results (P < 0.05) for individual SNPs and major haplotypes (frequency ≥ 5%) were adjusted for multiple testing using Bonferroni correction.
Individual and haplotype-based association analysis for the replication case-control SAGE sample
The association analysis was performed using a linear regression model by regressing two ND measures in PLINK (Purcell et al. 2007) on age, sex, SNP allele dosage, and other drug dependences (alcohol, cocaine, marijuana, opiates, and other drugs) covariates. Non-smokers were excluded.
Meta-analysis of individual SNP association for both the discovery and replication samples
Prior to conducting meta-analysis, we measured the heterogeneity for the SAGE case-control AA and EA samples using the program METAL (Willer et al. 2010). To combine the association analysis results from the discovery family samples and replication case-control samples, we conducted our meta-analysis of each SNP under the same genetic model used for analyzing each individual sample by using Fisher’s combining P value method (Fisher 1932). Considering that the PBAT approach used in the discovery family sample provides only Z-score and P value, we used an equal weight for each studied sample.
Results
Individual SNP-based association analysis for discovery and replication samples
Results from the individual SNP-based association analyses of the discovery family sample are shown in Table 2. Of the CHRNA2 polymorphisms, SNP rs891398 showed strong associations with FTND in the AAs (p = 0.0079) under an additive model and weak association in the EAs (p = 0.0338) under a dominant model. Also, rs373575 (p = 0.00782 and 0.00675) and rs2472553 (p = 0.00429 and 0.000863) showed strong association with SQ and FTND in the EAs under a dominant model. For the CHRNA6 polymorphisms, the only variant significantly associated with SQ in the EAs was rs892413, with a P value of 0.00769 under the additive model. All these associations remained significant after Bonferroni correction for multiple testing.
Table 2.
ND-associated p values under the three genetic models for the first given allele of each SNP in CHRNA2 and CHRNA6 in the discovery family sample
| Gene | dbSNP ID (Gene Location) | Alleles | African American | European American | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Allele Freq | SQ | FTND | Allele Freq | SQ | FTND | |||
| CHRNA2 | rs2292976 (Exon 8) | A/G | 0.13/0.87 | 0.120a | 0.0866a | 0.14/0.86 | −0.902a | −0.664a |
| 0.0578d | 0.0461d | 0.502d | 0.631d | |||||
| −0.444r | −0.700r | −0.0378r | −0.0221r | |||||
|
| ||||||||
| rs3735757 (Intron 5) | G/C | 0.22/0.78 | −0.378a | −0.613a | 0.14/0.86 | −0.589a | −0.291a | |
| −0.635d | −0.810d | 0.750d | −0.891d | |||||
| −0.239r | −0.464r | −0.00782r | −0.00675r | |||||
|
| ||||||||
| rs891398 (Exon 5) | T/C (T125A) | 0.25/0.75 | 0.0201a | 0.00790a | 0.52/0.48 | −0.511a | −0.450a | |
| 0.026d | 0.0287d | 0.228d | 0.294d | |||||
| 0.302r | 0.0814r | −0.0357r | −0.0338r | |||||
|
| ||||||||
| rs2472553 (Exon 2) | T/C (T22I) | 0.16/0.84 | 0.481a | 0.534a | 0.13/0.87 | −0.595a | −0.192a | |
| 0.324d | 0.387d | 0.776d | −0.724d | |||||
| −0.538r | −0.649r | −0.00429r | −0.000863r | |||||
|
| ||||||||
| CHRNA6 | rs9298628 (3′-flanking) | C/T | 0.25/0.75 | −0.422a | −0.375a | 0.81/0.19 | −0.0455a | −0.216a |
| −0.266d | −0.232d | −0.336d | −0.547d | |||||
| −0.913r | −0.996r | −0.0625r | −0.251r | |||||
|
| ||||||||
| rs892413 (Intron 2) | C/A | 0.25/0.75 | −0.612a | −0.397a | 0.80/0.20 | −0.00769a | −0.152a | |
| −0.566d | −0.340d | −0.117d | −0.189d | |||||
| −0.704r | −0.732r | −0.0195r | −0.304r | |||||
|
| ||||||||
| rs2217732 (Intron 2) | A/G | 0.26/0.74 | −0.423a | −0.405a | 0.81/0.19 | −0.0195a | −0.126a | |
| −0.247d | −0.215d | −0.335d | −0.433d | |||||
| −0.972r | 0.864r | −0.0235r | −0.163r | |||||
Notes: (1) Superscripts indicate genetic model used for analysis: a = additive; d = dominant; and r = recessive. (2) For each ethnic-specific sample, age and sex were used as covariates. (3) Negative signs indicate protective effect with the model specified in superscript letters.
The results from the replication case-control SAGE sample showed that SNP rs2292976 in CHRNA2 had a significant association with FTND in the AAs under both an additive (p = 0.00533) and a dominant (p = 0.0079) model (Table 3). Of the CHRNA6 polymorphisms, rs9298628, rs892416, and rs2217732 showed a significant association with FTND in the EAs, with P values of 0.000889, 0.00153, and 0.000865, respectively, under the additive model and 0.000218, 0.00053, and 0.000282 under the recessive model. Again, all these associations remained significant after Bonferroni correction.
Table 3.
ND-associated p values under the three genetic models for the first given allele of each SNP in CHRNA2 and CHRNA6 with ND in the replication case-control sample
| Gene | dbSNP ID (Gene Location) | Alleles | African American | European American | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Allele Freq | SQ | FTND | Allele Freq | SQ | FTND | |||
| CHRNA2 | rs2292976 (Exon 8) | A/G | 0.10/0.90 | −0.041a | −0.00533a | 0.13/0.87 | 0.684a | 0.0487a |
| −0.058d | −0.00790d | 0.619d | 0.0767d | |||||
| −0.209r | −0.147r | −0.952r | 0.157r | |||||
|
| ||||||||
| rs3735757 (Intron 5) | G/C | 0.20/0.80 | −0.152a | −0.267a | 0.13/0.87 | 0.982a | 0.114a | |
| −0.209d | −0.380d | 0.975d | 0.167d | |||||
| −0.275r | −0.290r | −0.876r | 0.217r | |||||
|
| ||||||||
| rs891398 (Exon 5) | T/C (T125A) | 0.25/0.75 | −0.463a | 0.343a | 0.51/0.49 | −0.172a | −0.297a | |
| −0.462d | 0.398d | 0.043d | −0.124d | |||||
| −0.724r | 0.506r | −0.825r | −0.864r | |||||
|
| ||||||||
| rs2472553 (Exon 2) | T/C (T22I) | 0.16/0.84 | 0.185a | 0.102a | 0.13/0.87 | −0.942a | −0.0785a | |
| 0.192d | 0.122d | 0.812d | −0.0953d | |||||
| 0.518r | 0.340r | −0.635r | −0.326r | |||||
|
| ||||||||
| CHRNA6 | rs9298628 (3′-flanking) | C/T | 0.29/0.71 | 0.0323a | 0.056a | 0.79/0.21 | 0.104a | 0.000889a |
| 0.0502d | 0.013d | 0.589d | 0.680d | |||||
| 0.143r | 0.941r | 0.036r | 0.000218r | |||||
|
| ||||||||
| rs892413 (Intron 2) | C/A | 0.31/0.69 | −0.119a | 0.208a | 0.80/0.20 | −0.189a | 0.00153a | |
| −0.0799d | −0.486d | −0.083d | 0.688d | |||||
| −0.586r | 0.035r | −0.515r | 0.000530r | |||||
|
| ||||||||
| rs2217732 (Intron 2) | A/G | 0.29/0.71 | 0.105a | 0.098a | 0.80/0.20 | 0.080a | 0.000865a | |
| 0.125d | 0.026d | 0.828d | 0.513d | |||||
| 0.304r | −0.938r | 0.034r | 0.000282r | |||||
Notes: (1) Superscripts indicate genetic model used for analysis: a = additive; d = dominant; and r = recessive. (2) For each ethnic-specific sample, age, sex, and other non-nicotine drug dependences were used as covariates. (3) Negative signs indicate protective effect with the model specified in superscript letters.
Meta-analysis associations for both discovery and replication samples
Meta-analysis was performed for the seven SNPs by combining the results from the discovery and replication samples, which included the AA sample only, EA sample only, and the AA and EA samples together (Table 4). The reason to perform meta-analysis on the AA and EA samples together was that the heterogeneity test for SAGE AA and EA case-control samples revealed no heterogeneity between the two ethnic samples on these SNPs. Of the meta-analyzed SNPs, rs2292976 in CHRNA2 (P = 0.0053) and rs892413 in CHRNA6 (P = 0.00311) showed the strongest association with FTND.
Table 4.
Meta-analysis results of SNPs in CHRNA2 and CHRNA6 with ND in both the discovery and replication samples
| Gene | dbSNP ID | Allele | African American | European American | AA + EA Samples | I2 |
|---|---|---|---|---|---|---|
| CHRNA2 | rs2292976 | A | 0.00325d | 0.195d | 0.00530d | 0 |
| rs3735757 | G | 0.404r | 0.011r | 0.0252r | 0 | |
| rs891398 | T | 0.0187a | 0.402a | 0.0443a | 0 | |
| rs2472553 | T | 0.554r | 0.00258r | 0.0108r | 0 | |
| CHRNA6 | rs9298628 | C | 0.998r | 0.000592r | 0.00498r | 0 |
| rs892413 | C | 0.120r | 0.00290r | 0.00311r | 0 | |
| rs2217732 | A | 0.981r | 0.000505r | 0.00426r | 0 |
Notes: 1) For each SNP, meta-analysis was performed on only one genetic model, which was selected on the basis of the association analysis result for both the discovery and the replication samples; 2) I2 was calculated only for the replication case-control samples with the METAL program.
Haplotype-based association analysis
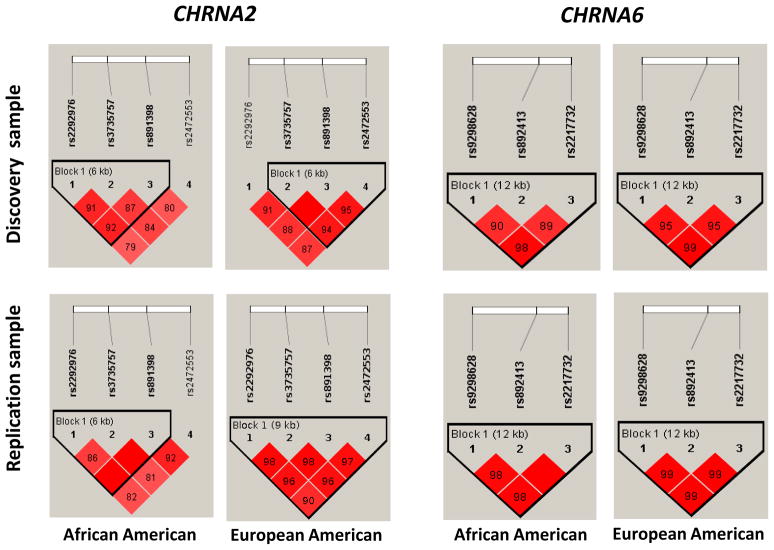
According to the haplotype block criteria defined by Gabriel et al. (2002), only one block was identified within each ethic sample in the CHRNA2 as well as in CHRNA6 (Figure 1). We employed the FBAT program to perform haplotype-based association analysis for all major (defined as >5%) haplotypes in each of the above-mentioned LD blocks with the two ND measures in CHRNA2 (Table 5) and CHRNA6 (Table 6) from the discovery family sample.
Figure 1.
LD structures for CHRNA2 (left) and CHRNA6 (right) SNPs in AAs and EAs from discovery and replication samples. Haploview (Barrett et al. 2005) was used to calculate all D values, and haplotype blocks were defined according to Gabriel et al. (2002). The number in each box represents the D value for each SNP pair surrounding that box.
Table 5.
Association analysis results for haplotypes in CHRNA2 and CHRNA6 with ND in the discovery family sample
|
A) CHRNA2
| |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2292976 | rs3735757 | rs891398 | rs2472553 | Freq | SQ | FTND | |||||||
|
| |||||||||||||
| P haplotype | Z-score | P global | Family # | P haplotype | Z-score | P global | Family # | ||||||
| African American | G | C | C | 54.9 | 0.36d | −0.92 | 0.13 | 130 | 0.097a | −1.66 | 0.023 | 221 | |
| G | C | T | 22.3 | 0.058a | 1.90 | 175 | 0.011a | 2.54 | 175 | ||||
| A | G | C | 10.6 | 0.44d | 0.77 | 105 | 0.28d | 1.09 | 109 | ||||
| G | G | C | 9.6 | 0.0099d | −2.58 | 101 | 0.0063d | −2.74 | 102 | ||||
|
| |||||||||||||
| European American | C | C | T | 34.4 | 0.020d | 2.32 | 0.012 | 193 | 0.0043d | 2.85 | 0.0039 | 193 | |
| C | T | T | 51.9 | 0.014d | −2.46 | 152 | 0.0093d | −2.60 | 152 | ||||
| G | C | C | 11.7 | 0.049d | −1.97 | 10 | 0.015d | −2.42 | 10 | ||||
|
B) CHRNA6
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs9298628 | rs892413 | rs2217732 | Freq | SQ | FTND | |||||||
|
| ||||||||||||
| P haplotype | Z-score | P global | Family # | P haplotype | Z-score | P global | Family # | |||||
| African American | C | C | A | 22.1 | 0.66a | −0.44 | 0.54 | 176 | 0.66d | −0.44 | 0.32 | 157 |
| T | A | G | 72.9 | 0.26d | 1.12 | 163 | 0.13d | 1.50 | 163 | |||
|
| ||||||||||||
| European American | C | C | A | 79.1 | 0.0067a | −2.71 | 0.014 | 176 | 0.12a | −1.54 | 0.18 | 179 |
| T | A | G | 18.3 | 0.011a | 2.55 | 171 | 0.094a | 1.67 | 170 | |||
Notes: (1) Superscripts indicate genetic model used for analysis: a = additive; d = dominant; and r = recessive; (2) Corrected P value at 0.05 is 0.0125 in AAs and 0.0167 in EAs for CHRNA2 and 0.025 in both EAs and AAs for CHRNA6.
Table 6.
Association analysis results for SNPs in CHRNA2 and CHRNA6 with ND in the replication case-control sample
| A) CHRNA2
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| rs2292976 | rs3735757 | rs891398 | rs2472553 | Freq | SQ | FTND | |||
|
| |||||||||
| P haplotype | P global | P haplotype | P global | ||||||
| African American | G | C | C | 54.0 | 0.0713a | 0.220 | 0.744a | 0.099 | |
| G | C | T | 24.8 | 0.139a | 0.379a | ||||
| G | G | C | 10.9 | 0.823a | 0.324a | ||||
| A | G | C | 9.16 | 0.0275a | 0.00649a | ||||
|
| |||||||||
| European American | A | G | C | T | 11.4 | 0.769a | 0.533 | 0.0686a | 0.408 |
| G | C | C | C | 35.7 | 0.178a | 0.826a | |||
| G | C | T | C | 50.1 | 0.158a | 0.240a | |||
|
B) CHRNA6
| ||||||||
|---|---|---|---|---|---|---|---|---|
| rs9298628 | rs892413 | rs2217732 | Freq | SQ | FTND | |||
|
| ||||||||
| P haplotype | P global | P haplotype | P global | |||||
| African American | T | A | G | 68.4 | 0.0584a | 0.238 | 0.145a | 0.243 |
| C | C | A | 28.9 | 0.105a | 0.0885a | |||
|
| ||||||||
| European American | T | A | G | 20.0 | 0.178a | 0.228 | 0.000947a | 0.00792 |
| C | C | A | 79.2 | 0.122a | 0.00121a | |||
Notes: (1) Superscripts indicate genetic model used for analysis: a = additive; (2) Corrected P value at 0.05 is 0.0125 in AAs and 0.0167 in EAs for CHRNA2 and 0.025 in both EAs and AAs for CHRNA6.
In CHRNA2, significant haplotypes in the AAs were: (1) G-C-T, formed by SNPs rs2292976, rs3735757, and rs891398 (Figure 1), with a frequency of 22.3%, which was associated significantly with FTND (Z = 2.54, P = 0.011); and (2) G-G-C, formed by the same SNPs, with a frequency of 9.6%, which was associated significantly with FTND (Z = −2.74; P = 0.0063) under a dominant model. The identified haplotypes in the EAs were: (1) C-T-T, formed by SNPs rs3735757, rs891398, and rs2472553, with a frequency of 51.9%, which was significantly associated with FTND (Z = −2.60; P = 0.0093); and (2) C-C-T, formed by SNPs rs3735757, rs891398, and rs2472553, with a frequency of 34.4%, which was significantly associated with FTND (Z = 2.85; P = 0.0043).
In CHRNA6, for the AA sample, we found no haplotypes showing significant association with ND. In the EA sample (Table 5), we found one haplotype, C-C-A, formed by SNPs rs9298628, rs892413, and rs2217732, with a frequency of 79.1%, significantly associated with SQ (Z = −2.71; P = 0.0067). Several haplotype-based associations remained significant after Bonferroni correction for each LD block.
In the replication case-control sample, only one haplotype in CHRNA2, A-G-C, formed by SNPs rs2292976, rs3735757, and rs891398, was significantly associated with FTND in the AA sample (P = 0.00649) (Table 6). Two haplotypes formed by SNPs rs9298628, rs892413, and rs2217732 in CHRNA6 (Figure 1) showed significant associations in the EA sample: (1) T-A-G, with a frequency of 20.0%, was significantly associated with FTND (P = 0.000947); and (2) C-C-A, with a frequency of 79.2%, was significantly associated with FTND (P = 0.00121).
Discussion
Nicotinic acetylcholine receptors α2 and α6 play vital roles in the nervous system. To test for their association with ND, seven SNPs in CHRNA2 and CHRNA6 were investigated in two independent samples of either African or European origin. Association analysis revealed that multiple SNPs and haplotypes are significantly associated with ND in the both discovery and replication samples. In the discovery sample, individual SNP analysis for CHRNA2 revealed a significant association of two SNPs in the EA sample and one SNP in the AA sample with FTND and/or SQ. In particular, we found that associations of SNPs rs3735757 and rs2472553 in CHRNA2 with SQ and FTND remained significant after correction for multiple testing. However, such associations were not exactly the same at the SNP level in the replication sample, where only SNP rs2292976 showed significant association with FTND in the AA population. Although SNP rs892413 showed significant association with SQ in the EA discovery samples, all three CHRNA6 SNPs exhibited significant association with FTND in the EA replication sample, even after correction for multiple testing. Further, we found a significant association between several haplotypes of CHRNA2/CHRNA6 and ND in both the discovery and the replication samples, although haplotypes formed by particular SNPs were sometimes different in the two samples. Meta-analysis of the discovery and the replication samples added further support for the association of CHRNA2 and CHRNA6 with ND.
Compared with other nAChR subunit genes such as CHRNA5-A3-B4, CHRNA4, CHRNB2, and CHRNB3 (Bierut 2010; Cui et al. 2013; Li and Burmeister 2009; Saccone et al. 2009; Saccone et al. 2010; Thorgeirsson et al. 2010; Wang et al. 2012), CHRNA2 has not received much attention in nicotine research. Although CHRNA2 was one of the first nAChRs investigated as an ND candidate gene in several GWAS and candidate studies, no significant associations have been reported after correction for multiple testing or replicated in independent studies. In the present study, we demonstrated that CHRNA2 shows a strong association with FTND after correction for multiple testing. Importantly, the SNP rs2472553, which evinced the strongest association with ND, appears to encode a functional variant in the signal peptide, causing an amino acid change from threonine to isoleucine at the 22nd residue. Our functional study with oocyte electrophysiology indicates that this mutation changes the sensitivity of functional receptors (Dash et al, in preparation).
CHRNA2 plays a vital role in other neurologic disorders such as epilepsy, with an estimated prevalence in Europeans that ranges from 3–8 per 1,000 individuals (Forsgren et al. 2005). The mutation I279N in the α2 subunit (i.e., rs104894063) was the first identified functional variant associated with epilepsy in CHRNA2; electrophysiological investigation of the I279N mutation in HEK 293 cells indicates that the α2I279N/β4 receptor has a significantly higher sensitivity to the natural agonist than does the wild-type α2/β4 receptor (Aridon et al. 2006). Another oocyte electrophysiology study found that α2I279N, co-expressed with the β2 subunit, causes a gain-of-function effect whose distinct biopharmacological profile includes reduced inhibition by carbamazepine and greater nicotine sensitivity (Hoda et al. 2009).
Several GWAS and candidate gene studies have revealed strong associations between CHRNA6 and ND. However, most studies tested CHRNB3-CHRNA6 associations as a cluster, in which most of the significant association was attributable to variants in CHRNB3; moreover, the association of CHRNA6 SNPs typically did not survive correction for multiple testing (Hoft et al. 2009b; Saccone et al. 2009; Zeiger et al. 2008). In the present study, we demonstrated that CHRNA6 is still significantly associated with FTND after correction for multiple testing. In a study by Hoft and colleagues (Hoft et al. 2009a), two SNPs from the CHRNA6-CHRNB3 cluster were found to be associated with smoking quit-attempts also: SNP rs2304297 (P = 0.0044) from CHRNA6 and rs7004381 (P = 0.0024) from CHRNB3. Complementing our association study, animal self-administration studies suggest that β2*nAChRs assembled with α6 subunits would be useful pharmacological targets for smoking cessation products (Brunzell 2012): nicotine self-administration is absent in α6-knockout mice, and targeted re-expression of the α6 subunit in the ventral tegmental area (VTA) of α6-KO mice promptly restores nicotine self-administration (Pons et al. 2008). Further, several in vitro electrophysiological, synaptosome-release-assay, and cyclic-voltammetry studies have demonstrated that nicotine-mediated elevation of dopamine release is blocked following antagonism of α6β2*nAChRs with α-CTX MII (Champtiaux et al. 2003; Drenan et al. 2008; Perez et al. 2010; Perez et al. 2009; Salminen et al. 2007; Zhao-Shea et al. 2011). Finally, an in vivo function study of α6*nAChRs in mesolimbic DA neurons has shown that the elevation of DA release caused by nicotine can be inhibited by intra-VTA infusion of α-CTX MII, implicating α6β2*nAChRs in the regulation of this effect (Gotti et al. 2010). Collectively, the association and functional studies of CHRNA6 suggest that α6*nAChRs are strong candidates for drug-development research on smoking cessation.
Recently, CHRNA6 has been found to be associated, not only with ND, but also with alcohol dependence; three SNPs (rs1072003, P = 0.015; rs892413, P = 0.0033; and rs2304297, P = 0.012) were associated with alcohol dependence in the National Youth Survey Family Study in a sample that was mostly EAs (Hoft et al. 2009a). Another study showed that two haplotypes of the CHRNA6, CCCC and TCGA, formed by SNPs rs10087172, rs10109429, rs2196129, and rs16891604, were associated with heavy alcohol consumption (P = 0.004 and P = 0.035, respectively) and with increased alcohol intake (P = 0.004) for the CCCC haplotype in a Spanish population (Landgren et al. 2009).
Our results indicate that both CHRNA2 and CHRNA6 are significantly associated with ND. Such association with ND at both the individual SNP and haplotype level makes these genes good subjects for research on molecular mechanisms of dependence. A better understanding of the role of these genetic variants—especially the functional variants—may provide key insights for pharmacologic targeting to reduce or possibly eliminate some of the addictive properties of nicotine in susceptible individuals.
Acknowledgments
We acknowledge the invaluable contributions of personal information and blood samples by all participants in the study. This project was supported by National Institutes of Health grant R01 DA012844 to MDL. We are thankful to the NIH GWAS data repository for providing us access to their dataset through project 771 to Ming D. Li under the title “Genome-wide association analysis for addiction and type 2 diabetes.”
We also thank NIH GWAS Data Repository, the investigators who contributed the phenotype and genotype data from original studies, and the primary funding organization that supported the contributing study. Funding support for SAGE was provided through the NIH Genes, Environment and Health Initiative (GEI) Grant U01 HG004422; the GENEVA Coordinating Center (U01 HG004446); the National Institute on Alcohol Abuse and Alcoholism (U10 AA008401); the National Institute on Drug Abuse (R01 DA013423); the National Cancer Institute (P01 CA089392); and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). Assistance with data cleansing was provided by the National Center for Biotechnology Information. Genotyping was performed at the Johns Hopkins University Center for Inherited Disease Research or at deCODE. Funding support for the GWAS of Lung Cancer and Smoking was provided through the NIH Genes, Environment and Health 7 Initiative [GEI] (Z01 CP 010200). The human subjects participating in the GWAS derive from The Environment and Genetics in Lung Cancer Etiology (EAGLE) case-control study and the Prostate, Lung Colon and Ovary Screening Trial, and these studies are supported by intramural resources of the National Cancer Institute. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, by the Gene Environment Association Studies, GENEVA Coordinating Center (U01HG004446). Assistance with data cleansing was provided by the National Center for Biotechnology Information. Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000093.
Footnotes
Contributors
SW, AV, CS, and QX performed the genotyping and statistical analysis and/or wrote the first draft of the manuscript; JZM performed statistical analysis and database management; OP, CP, and TJP undertook subject recruitment and clinical data collection; MDL designed the study and wrote the manuscript. All authors contributed to and have approved the final manuscript.
Conflict of Interest
MDL has served as a consultant and board member of ADial Pharmaceuticals, LLC. All other authors declare that they have no conflicts of interest.
References
- Al Koudsi N, Tyndale RF. Genetic influences on smoking: a brief review. Ther Drug Monit. 2005;27:704–9. doi: 10.1097/01.ftd.0000179842.63515.c6. [DOI] [PubMed] [Google Scholar]
- Aridon P, Marini C, Di Resta C, Brilli E, De Fusco M, Politi F, Parrini E, Manfredi I, Pisano T, Pruna D, Curia G, Cianchetti C, Pasqualetti M, Becchetti A, Guerrini R, Casari G. Increased sensitivity of the neuronal nicotinic receptor alpha 2 subunit causes familial epilepsy with nocturnal wandering and ictal fear. Am J Hum Genet. 2006;79:342–50. doi: 10.1086/506459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bergen AW, Korczak JF, Weissbecker KA, Goldstein AM. A genome-wide search for loci contributing to smoking and alcoholism. Genet Epidemiol. 1999;17:S55–S60. doi: 10.1002/gepi.1370170710. [DOI] [PubMed] [Google Scholar]
- Berrettini WH, Doyle GA. The CHRNA5-A3-B4 gene cluster in nicotine addiction. Mol Psychiatry. 2012;17:856–66. doi: 10.1038/mp.2011.122. [DOI] [PubMed] [Google Scholar]
- Beuten J, Ma JZ, Payne TJ, Dupont RT, Crews KM, Somes G, Williams NJ, Elston RC, Li MD. Single- and Multilocus Allelic Variants within the GABAB Receptor Subunit 2 (GABAB2) Gene Are Significantly Associated with Nicotine Dependence. Am J Hum Genet. 2005;76:859–64. doi: 10.1086/429839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ. Convergence of genetic findings for nicotine dependence and smoking related diseases with chromosome 15q24–25. Trends Pharmacol Sci. 2010;31:46–51. doi: 10.1016/j.tips.2009.10.004. S0165-6147(09)00168-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI, Jr, Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A. 2010;107:5082–7. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunzell DH. Preclinical evidence that activation of mesolimbic alpha 6 subunit containing nicotinic acetylcholine receptors supports nicotine addiction phenotype. Nicotine Tob Res. 2012;14:1258–69. doi: 10.1093/ntr/nts089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Smoking-attributable mortality, years of potential life lost, and productivity losses--United States, 2000–2004. MMWR Morb Mortal Wkly Rep. 2008;57:1226–8. mm5745a3 [pii] [PubMed] [Google Scholar]
- Champtiaux N, Gotti C, Cordero-Erausquin M, David DJ, Przybylski C, Lena C, Clementi F, Moretti M, Rossi FM, Le Novere N, McIntosh JM, Gardier AM, Changeux JP. Subunit composition of functional nicotinic receptors in dopaminergic neurons investigated with knock-out mice. J Neurosci. 2003;23:7820–9. doi: 10.1523/JNEUROSCI.23-21-07820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Booker TK, Allen RS, Grady SR, Whiteaker P, Marks MJ, Salminen O, Tritto T, Butt CM, Allen WR, Stitzel JA, McIntosh JM, Boulter J, Collins AC, Heinemann SF. The beta3 nicotinic receptor subunit: a component of alpha-conotoxin MII-binding nicotinic acetylcholine receptors that modulate dopamine release and related behaviors. J Neurosci. 2003;23:11045–53. doi: 10.1523/JNEUROSCI.23-35-11045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui WY, Li MD. Nicotinic Modulation of Innate Immune Pathways Via alpha7 Nicotinic Acetylcholine Receptor. J Neuroimmune Pharmacol. 2010:479–488. doi: 10.1007/s11481-010-9210-2. [DOI] [PubMed] [Google Scholar]
- Cui WY, Wang S, Yang J, Yi SG, Yoon D, Kim YJ, Payne TJ, Ma JZ, Park T, Li MD. Significant association of CHRNB3 variants with nicotine dependence in multiple ethnic populations. Mol Psychiatry. 2013 doi: 10.1038/mp.2012.190. [DOI] [PubMed] [Google Scholar]
- Drenan RM, Grady SR, Whiteaker P, McClure-Begley T, McKinney S, Miwa JM, Bupp S, Heintz N, McIntosh JM, Bencherif M, Marks MJ, Lester HA. In vivo activation of midbrain dopamine neurons via sensitized, high-affinity alpha 6 nicotinic acetylcholine receptors. Neuron. 2008;60:123–36. doi: 10.1016/j.neuron.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerstrom KO. Measuring degree of physical dependence to tobacco smoking with reference to individualization of treatment. Addict Behav. 1978;3:235–41. doi: 10.1016/0306-4603(78)90024-2. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Su J, Taylor L, Wilcox M, Van Eerdewegh P, Tsuang MT. A novel permutation testing method implicates sixteen nicotinic acetylcholine receptor genes as risk factors for smoking in schizophrenia families. Hum Hered. 2004;57:59–68. doi: 10.1159/000077543. [DOI] [PubMed] [Google Scholar]
- Forsgren L, Beghi E, Oun A, Sillanpaa M. The epidemiology of epilepsy in Europe - a systematic review. Eur J Neurol. 2005;12:245–53. doi: 10.1111/j.1468-1331.2004.00992.x. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gotti C, Guiducci S, Tedesco V, Corbioli S, Zanetti L, Moretti M, Zanardi A, Rimondini R, Mugnaini M, Clementi F, Chiamulera C, Zoli M. Nicotinic acetylcholine receptors in the mesolimbic pathway: primary role of ventral tegmental area alpha6beta2* receptors in mediating systemic nicotine effects on dopamine release, locomotion, and reinforcement. J Neurosci. 2010;30:5311–25. doi: 10.1523/JNEUROSCI.5095-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotti C, Riganti L, Vailati S, Clementi F. Brain neuronal nicotinic receptors as new targets for drug discovery. Curr Pharm Des. 2006a;12:407–28. doi: 10.2174/138161206775474486. [DOI] [PubMed] [Google Scholar]
- Gotti C, Zoli M, Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol Sci. 2006b;27:482–91. doi: 10.1016/j.tips.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Greenbaum L, Kanyas K, Karni O, Merbl Y, Olender T, Horowitz A, Yakir A, Lancet D, Ben-Asher E, Lerer B. Why do young women smoke? I. Direct and interactive effects of environment, psychological characteristics and nicotinic cholinergic receptor genes. Mol Psychiatry. 2006;11:312–22. 223. doi: 10.1038/sj.mp.4001774. [DOI] [PubMed] [Google Scholar]
- Greenbaum L, Lerer B. Differential contribution of genetic variation in multiple brain nicotinic cholinergic receptors to nicotine dependence: recent progress and emerging open questions. Mol Psychiatry. 2009;14:912–45. doi: 10.1038/mp.2009.59. [DOI] [PubMed] [Google Scholar]
- Heitjan DF, Guo M, Ray R, Wileyto EP, Epstein LH, Lerman C. Identification of pharmacogenetic markers in smoking cessation therapy. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:712–9. doi: 10.1002/ajmg.b.30669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoda JC, Wanischeck M, Bertrand D, Steinlein OK. Pleiotropic functional effects of the first epilepsy-associated mutation in the human CHRNA2 gene. FEBS Lett. 2009;583:1599–604. doi: 10.1016/j.febslet.2009.04.024. [DOI] [PubMed] [Google Scholar]
- Hoft NR, Corley RP, McQueen MB, Huizinga D, Menard S, Ehringer MA. SNPs in CHRNA6 and CHRNB3 are associated with alcohol consumption in a nationally representative sample. Genes Brain Behav. 2009a;8:631–7. doi: 10.1111/j.1601-183X.2009.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoft NR, Corley RP, McQueen MB, Schlaepfer IR, Huizinga D, Ehringer MA. Genetic association of the CHRNA6 and CHRNB3 genes with tobacco dependence in a nationally representative sample. Neuropsychopharmacology. 2009b;34:698–706. doi: 10.1038/npp.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–9. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamida D, Poulas K, Avramopoulou V, Fostieri E, Lagoumintzis G, Lazaridis K, Sideri A, Zouridakis M, Tzartos SJ. Muscle and neuronal nicotinic acetylcholine receptors. Structure, function and pathogenicity. FEBS J. 2007;274:3799–845. doi: 10.1111/j.1742-4658.2007.05935.x. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Sullivan P, Corey LA, Gardner CO, Prescott CA. A population-based twin study in women of smoking initiation and nicotine dependence. Psychol Med. 1999;29:299–308. doi: 10.1017/s0033291798008022. [DOI] [PubMed] [Google Scholar]
- Keskitalo-Vuokko K, Pitkaniemi J, Broms U, Heliovaara M, Aromaa A, Perola M, Ripatti S, Salminen O, Salomaa V, Loukola A, Kaprio J. Associations of nicotine intake measures with CHRN genes in Finnish smokers. Nicotine Tob Res. 2011;13:686–90. doi: 10.1093/ntr/ntr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijn J, Folgering JH, van der Hart MC, Rollema H, Cremers TI, Westerink BH. Direct effect of nicotine on mesolimbic dopamine release in rat nucleus accumbens shell. Neurosci Lett. 2011;493:55–8. doi: 10.1016/j.neulet.2011.02.035. [DOI] [PubMed] [Google Scholar]
- Landgren S, Engel JA, Andersson ME, Gonzalez-Quintela A, Campos J, Nilsson S, Zetterberg H, Blennow K, Jerlhag E. Association of nAChR gene haplotypes with heavy alcohol use and body mass. Brain Res. 2009;1305(Suppl):S72–9. doi: 10.1016/j.brainres.2009.08.026. [DOI] [PubMed] [Google Scholar]
- Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: Tools for Family-Based Association Studies. Am J Hum Genet. 2004;74:367–9. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Beuten J, Ma JZ, Payne TJ, Lou XY, Garcia V, Duenes AS, Crews KM, Elston RC. Ethnic- and gender-specific association of the nicotinic acetylcholine receptor alpha4 subunit gene (CHRNA4) with nicotine dependence. Hum Mol Genet. 2005;14:1211–9. doi: 10.1093/hmg/ddi132. [DOI] [PubMed] [Google Scholar]
- Li MD, Burmeister M. New insights into the genetics of addiction. Nat Rev Genet. 2009;10:225–31. doi: 10.1038/nrg2536. nrg2536 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Cheng R, Ma JZ, Swan GE. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98:23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JZ, Beuten J, Payne TJ, Dupont RT, Elston RC, Li MD. Haplotype analysis indicates an association between the DOPA decarboxylase (DDC) gene and nicotine dependence. Hum Mol Genet. 2005;14:1691–8. doi: 10.1093/hmg/ddi177. [DOI] [PubMed] [Google Scholar]
- Maes HH, Sullivan PF, Bulik CM, Neale MC, Prescott CA, Eaves LJ, Kendler KS. A twin study of genetic and environmental influences on tobacco initiation, regular tobacco use and nicotine dependence. Psychol Med. 2004;34:1251–61. doi: 10.1017/s0033291704002405. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. JAMA. 2004;291:1238–45. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- Perez XA, Bordia T, McIntosh JM, Quik M. alpha6ss2* and alpha4ss2* nicotinic receptors both regulate dopamine signaling with increased nigrostriatal damage: relevance to Parkinson’s disease. Mol Pharmacol. 2010;78:971–80. doi: 10.1124/mol.110.067561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez XA, O’Leary KT, Parameswaran N, McIntosh JM, Quik M. Prominent role of alpha3/alpha6beta2* nAChRs in regulating evoked dopamine release in primate putamen: effect of long-term nicotine treatment. Mol Pharmacol. 2009;75:938–46. doi: 10.1124/mol.108.053801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Todorov A, Andersen A, Hollenbeck N, Gunter T, Heath A, Madden P. Examination of the nicotine dependence (NICSNP) consortium findings in the Iowa adoption studies population. Nicotine Tob Res. 2009;11:286–92. doi: 10.1093/ntr/ntn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plazas PV, Katz E, Gomez-Casati ME, Bouzat C, Elgoyhen AB. Stoichiometry of the alpha9alpha10 nicotinic cholinergic receptor. J Neurosci. 2005;25:10905–12. doi: 10.1523/JNEUROSCI.3805-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons S, Fattore L, Cossu G, Tolu S, Porcu E, McIntosh JM, Changeux JP, Maskos U, Fratta W. Crucial role of alpha4 and alpha6 nicotinic acetylcholine receptor subunits from ventral tegmental area in systemic nicotine self-administration. J Neurosci. 2008;28:12318–27. doi: 10.1523/JNEUROSCI.3918-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. S0002-9297(07)61352-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, Pergadia ML, Agrawal A, Breslau N, Grucza RA, Hatsukami D, Johnson EO, Madden PA, Swan GE, Wang JC, Goate AM, Rice JP, Bierut LJ. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:453–66. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Schwantes-An TH, Wang JC, Grucza RA, Breslau N, Hatsukami D, Johnson EO, Rice JP, Goate AM, Bierut LJ. Multiple cholinergic nicotinic receptor genes affect nicotine dependence risk in African and European Americans. Genes Brain Behav. 2010;9:741–50. doi: 10.1111/j.1601-183X.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, Swan GE, Goate AM, Rutter J, Bertelsen S, Fox L, Fugman D, Martin NG, Montgomery GW, Wang JC, Ballinger DG, Rice JP, Bierut LJ. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen O, Drapeau JA, McIntosh JM, Collins AC, Marks MJ, Grady SR. Pharmacology of alpha-conotoxin MII-sensitive subtypes of nicotinic acetylcholine receptors isolated by breeding of null mutant mice. Mol Pharmacol. 2007;71:1563–71. doi: 10.1124/mol.106.031492. [DOI] [PubMed] [Google Scholar]
- Salminen O, Murphy KL, McIntosh JM, Drago J, Marks MJ, Collins AC, Grady SR. Subunit composition and pharmacology of two classes of striatal presynaptic nicotinic acetylcholine receptors mediating dopamine release in mice. Mol Pharmacol. 2004;65:1526–35. doi: 10.1124/mol.65.6.1526. 10.1124/mol.65.6.1526 65/6/1526. [pii] [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS. The genetic epidemiology of smoking. Nicotine Tob Res. 1999;1(Suppl 2):S51–7. doi: 10.1080/14622299050011811. discussion S69–70. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, Gieger C, Rawal R, Mangino M, Prokopenko I, Magi R, Keskitalo K, Gudjonsdottir IH, Gretarsdottir S, Stefansson H, Thompson JR, Aulchenko YS, Nelis M, Aben KK, den Heijer M, Dirksen A, Ashraf H, Soranzo N, Valdes AM, Steves C, Uitterlinden AG, Hofman A, Tonjes A, Kovacs P, Hottenga JJ, Willemsen G, Vogelzangs N, Doring A, Dahmen N, Nitz B, Pergadia ML, Saez B, De Diego V, Lezcano V, Garcia-Prats MD, Ripatti S, Perola M, Kettunen J, Hartikainen AL, Pouta A, Laitinen J, Isohanni M, Huei-Yi S, Allen M, Krestyaninova M, Hall AS, Jones GT, van Rij AM, Mueller T, Dieplinger B, Haltmayer M, Jonsson S, Matthiasson SE, Oskarsson H, Tyrfingsson T, Kiemeney LA, Mayordomo JI, Lindholt JS, Pedersen JH, Franklin WA, Wolf H, Montgomery GW, Heath AC, Martin NG, Madden PA, Giegling I, Rujescu D, Jarvelin MR, Salomaa V, Stumvoll M, Spector TD, Wichmann HE, Metspalu A, Samani NJ, Penninx BW, Oostra BA, Boomsma DI, Tiemeier H, van Duijn CM, Kaprio J, Gulcher JR, McCarthy MI, Peltonen L, Thorsteinsdottir U, Stefansson K. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–53. doi: 10.1038/ng.573. ng.573 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Willemsen G, Boomsma DI. Heritability of smoking initiation and nicotine dependence. Behav Genet. 2005;35:397–406. doi: 10.1007/s10519-004-1327-8. [DOI] [PubMed] [Google Scholar]
- Wang JC, Kapoor M, Goate AM. The genetics of substance dependence. Annu Rev Genomics Hum Genet. 2012;13:241–61. doi: 10.1146/annurev-genom-090711-163844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonnacott S. Presynaptic nicotinic ACh receptors. Trends Neurosci. 1997;20:92–8. doi: 10.1016/s0166-2236(96)10073-4. [DOI] [PubMed] [Google Scholar]
- Wonnacott S, Kaiser S, Mogg A, Soliakov L, Jones IW. Presynaptic nicotinic receptors modulating dopamine release in the rat striatum. Eur J Pharmacol. 2000;393:51–8. doi: 10.1016/s0014-2999(00)00005-4. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Drug Report 2012. World Health Organization; 2012. [Google Scholar]
- Yates W, Cadoret R, Troughton E. The Iowa adoption studies methods and results. In: LaBuda M, Grigorenko E, editors. On the Way to Individuality: Methodological Issues in Behavioral Genetics. Nova Science Publishers; Hauppauge, NY: 1998. pp. 95–125. [Google Scholar]
- Zeiger JS, Haberstick BC, Schlaepfer I, Collins AC, Corley RP, Crowley TJ, Hewitt JK, Hopfer CJ, Lessem J, McQueen MB, Rhee SH, Ehringer MA. The neuronal nicotinic receptor subunit genes (CHRNA6 and CHRNB3) are associated with subjective responses to tobacco. Hum Mol Genet. 2008;17:724–34. doi: 10.1093/hmg/ddm344. [DOI] [PubMed] [Google Scholar]
- Zhao-Shea R, Liu L, Soll LG, Improgo MR, Meyers EE, McIntosh JM, Grady SR, Marks MJ, Gardner PD, Tapper AR. Nicotine-mediated activation of dopaminergic neurons in distinct regions of the ventral tegmental area. Neuropsychopharmacology. 2011;36:1021–32. doi: 10.1038/npp.2010.240. [DOI] [PMC free article] [PubMed] [Google Scholar]