Abstract
Summary
In Arabidopsis, multisubunit RNA polymerases IV and V orchestrate RNA-directed DNA methylation (RdDM) and transcriptional silencing, but what identifies the loci to be silenced is unclear. We show that heritable silent locus identity at a specific subset of RdDM targets requires HISTONE DEACETYLASE 6 (HDA6) acting upstream of Pol IV recruitment and siRNA biogenesis. At these loci, epigenetic memory conferring silent locus identity is erased in hda6 mutants such that restoration of HDA6 activity cannot restore siRNA biogenesis or silencing. Silent locus identity is similarly lost in mutants for the cytosine maintenance methyltransferase, MET1. By contrast, pol IV or pol V mutants disrupt silencing without erasing silent locus identity, allowing restoration of Pol IV or Pol V function to restore silencing. Collectively, these observations indicate that silent locus specification and silencing are separable steps that together account for epigenetic inheritance of the silenced state.
Introduction
In plants, as in other eukaryotes, transposable elements, repeated sequences and specific genes are silenced in every generation by mechanisms that include cytosine hypermethylation and/or histone post-translational modification (Bonasio et al., 2010; Law and Jacobsen, 2010; Pontvianne et al., 2010). Collectively, these modifications contribute to chromatin states that are refractive to transcription by RNA polymerases I, II or III (Jenuwein and Allis, 2001; Vaillant and Paszkowski, 2007). How genomic loci are identified, or marked, as targets for silencing is unclear. A priori, one might expect silent locus identity to be intrinsic to the locus, perhaps encoded by its nucleic acid sequence. Alternatively, a cell may not so much “know” to silence a locus based on intrinsic features as “remember” to do so based on heritable chromatin marks that confer epigenetic memory.
During transcriptional silencing in Arabidopsis thaliana, previously unmethylated cytosines can be methylated de novo by DRM2 (DOMAINS REARRANGED METHYLTRANSFERASE 2; an ortholog of mammalian DNMT3a and 3b) at sites specified by 24 nt siRNAs (Cao and Jacobsen, 2002). This process, known as RNA-directed DNA methylation (RdDM), can methylate cytosines in any sequence context: CG, CHG or CHH, where H is an A, T or C (Law and Jacobsen, 2010; Matzke et al., 2009; Zhang and Zhu, 2011).
Following de novo cytosine methylation, methylation patterns can be maintained in an RNA-independent manner. At methylated CG motifs, DNA replication generates hemimethylated duplexes that are recognized by VIM proteins (orthologs of mammalian UHRF proteins) that then recruit MET1 (DNA METHYLTRANSFERASE 1; the ortholog of mammalian DNMT1). Resulting CG methylation of the newly synthesized DNA strand (Bostick et al., 2007; Woo et al., 2008) perpetuates the chromatin mark, providing a durable, yet potentially reversible form of epigenetic memory (Becker et al., 2011; Saze et al., 2003; Schmitz et al., 2011). CHG methylation can also be perpetuated in plants, which is accomplished primarily by CMT3 (CHROMOMETHYLASE 3)(Bartee et al., 2001; Lindroth et al., 2001). CMT3 has chromo and bromo adjacent homology (BAH) domains that bind Histone H3 dimethylated on Lysine 9 (H3K9me2). The H3K9 methyltransferase KYP/SUVH4, in turn, has a domain that binds cytosines methylated by CMT3, such that CHG methylation and H3K9me2 specify one another in a feed-forward loop (Du et al., 2012; Johnson et al., 2007; Lindroth et al., 2004).
A recent study suggests that CHH methylation in specific contexts, such as the central regions of long transposable elements, can be maintained via CMT2 (CHROMOMETHYLASE 2) in crosstalk with histone modifications (Zemach et al., 2013). By contrast, DRM2-dependent CHH methylation is not maintained, but requires continuous production of non-coding RNAs that guide RdDM (Haag and Pikaard, 2011). These non-coding RNAs derive from the activities of two multi-subunit RNA polymerases, Pol IV and Pol V (Herr et al., 2005; Kanno et al., 2005; Onodera et al., 2005; Pontier et al., 2005; Ream et al., 2009), that evolved as specialized forms of Pol II (Ream et al., 2009). Genetic and biochemical evidence indicate that Pol IV initiates RdDM by synthesizing RNAs that then serve as templates for RNA-DIRECTED RNA POLYMERASE 2 (RDR2) (Haag et al., 2012; Pontes et al., 2006). RDR2 physically associates with Pol IV (Haag et al., 2012; Law et al., 2011) and may require this association for activity in vitro (Haag et al., 2012). Resulting double-stranded RNAs (dsRNAs) are cleaved by DICER-LIKE 3 (DCL3) (Xie et al., 2004), generating 24-nt siRNA duplexes whose strands are loaded primarily into ARGONAUTE 4 (AGO4) (Qi et al., 2006). AGO4-siRNA complexes find their sites of action by binding to Pol V transcripts generated at target loci (Wierzbicki et al., 2008; Wierzbicki et al., 2009). Through a mechanism that is not well understood, DRM2 is recruited, and cytosine methylation occurs, accompanied by repressive histone modifications that include histone deacetylation and H3K9 and H3K27 methylation.
How Pols IV and V are recruited to specific genomic loci remains unclear. Although 24 nt siRNA biogenesis and RdDM are lost in pol IV or pol V mutants, these processes are typically restored upon transgene complementation or outcrossing to a wild-type plant (Haag et al., 2009; Pontes et al., 2006). Thus, chromosomal information required for Pol IV and Pol V recruitment can persist in their absence. DNA sequences and/or chromatin modification patterns could potentially explain Pol IV recruitment. In support of the latter hypothesis, the SAWADEE domain protein, SHH1/DTF1associates with Pol IV and can bind H3K9me2 (Law et al., 2013; Zhang et al., 2013), consistent with chromatin marks contributing to Pol IV recruitment.
HDA6 is a broad-specificity histone deacetylase with a complex interrelationship with DNA methylation (Earley et al., 2006). Genetic screens identified HDA6 as an activity required for silencing transgenes subjected to RdDM (Aufsatz et al., 2002; Furner et al., 1998; He et al., 2009). However, at 45S ribosomal RNA gene loci, siRNAs that direct CHH hypermethylation by RdDM accumulate in hda6 mutants, indicating that HDA6 is not obligately required for RdDM (Earley et al., 2010). Paradoxically, CG and CHG methylation are lost at rRNA genes in hda6 mutants at the same time that CHH methylation is gained, indicating that HDA6 facilitates maintenance methylation at symmetrical CG and CHG motifs (Earley et al., 2010). Consistent with these findings, HDA6 and MET1 are required for silencing multiple classes of transposable elements (Furner et al., 1998; He et al., 2009; Lippman et al., 2003; Murfett et al., 2001; To et al., 2011), and can physically interact (Liu et al., 2012).
Through investigation of HDA6’s role in RdDM, we identified RdDM targets at which HDA6 and Pol IV display genetic epistasis (Group E loci). At these loci, hda6 mutants are defective for siRNA biogenesis, cytosine methylation, and histone deacetylation, essentially behaving like hda6 pol IV double mutants. Importantly, transgene-mediated restoration of HDA6 activity does not restore siRNA biogenesis, cytosine methylation or histone deacetylation at Group E loci, indicating that the epigenetic memory required for silent locus identity is lost in the absence of HDA6 and is not easily regained. By contrast, loss of silencing in pol IV or pol V mutants is readily restored by complementing transgenes, indicating that silent locus identity persists in the absence of Pol IV or Pol IV. We further show that cytosine methylation by MET1 is required for the heritable epigenetic memory that confers silent locus identity upstream of RdDM. Loss of silent locus identity results in the emergence of expressed epialleles, among them genes known to vary in expression among isogenic populations.
Results
HDA6 and Pol IV/V collaborate to silence transposable elements and genes
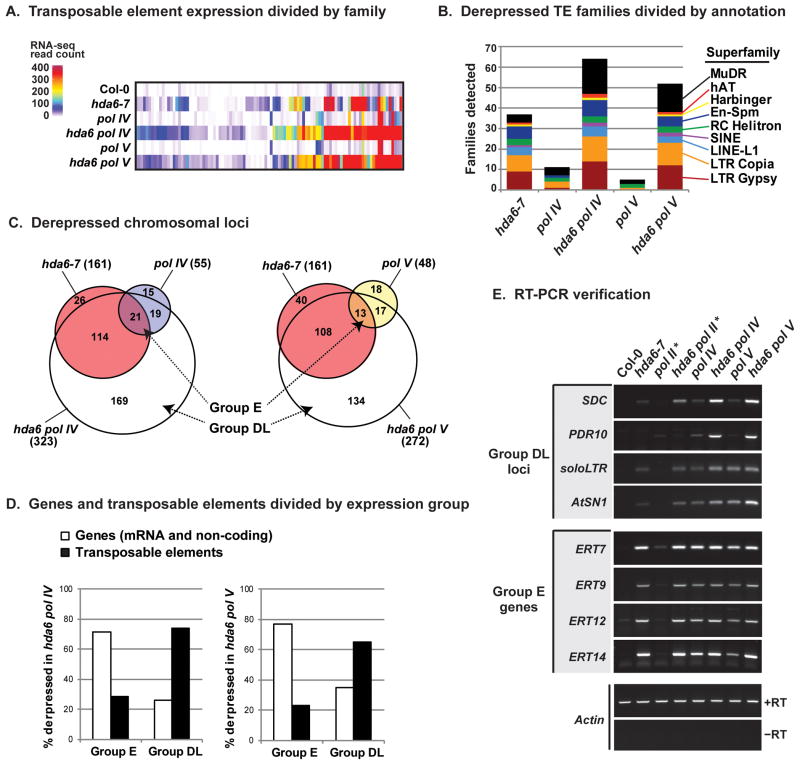
Widespread derepression of transposable element families occurs in A. thaliana hda6 null mutants, as shown by deep sequencing of polyA+ RNAs, comparing hda6 mutants to wild type (ecotype Col-0) plants (Figure 1A). Fewer transposable element families are derepressed in null mutants lacking the largest subunits of Pol IV (nrpd1-3) or Pol V (nrpe1-11), hereafter referred to as pol IV or pol V mutants (Figure 1A). In hda6 pol IV or hda6 pol V double mutants, additive or synergistic derepression of transposable elements occurs (Figure 1A), with long-terminal repeat (LTR) retrotransposons and MuDR transposons being the most frequently derepressed (Figure 1B).
Figure 1. Functional relationships among HDA6, Pol IV and Pol V in transcriptional silencing.
(A) Hierarchical clustering of 99 transposable element families based on polyA+ RNA-seq read counts. The single mutants hda6-7, nrpd1-3 (pol IV) or nrpe1-11 (pol V), and the double mutants hda6 pol IV and hda6 pol V are compared to wild-type (Col-0). Expression levels are normalized relative to the total number of mapped reads per genotype.
(B) Derepressed transposable element (TE) Superfamilies. Families were considered “derepressed” if at least 40 polyA+ RNA-seq reads mapped to a family in the given mutant and fewer than 20 reads were detected in wild-type Col-0.
(C) Venn diagrams summarizing locus derepression in single and double mutants of hda6, pol IV and pol V. Derepressed loci were determined using NOISeq (Tarazona et al., 2012) by comparing polyA+ RNA-seq read counts in the mutant relative to Col-0. A minimum 4-fold increase in the mutant relative to wild-type, and p value < 0.01 were set as thresholds.
(D) Relative abundance of genes versus transposons among Group E and Group DL loci detected in hda6 pol IV (left) or hda6 pol V (right) double mutants.
(E) RT-PCR analyses of Group DL and Group E loci in wild-type (Col-0) and the indicated mutant backgrounds. Actin serves as a loading control. Pol II* denotes nrpb2-3, a mutant allele of the Pol II second-largest subunit that affects RdDM at some loci (Zheng et al., 2009). Controls omitting reverse transcriptase (−RT) were performed for all loci; no products were obtained, as shown for the Actin control (See also Figure S1).
Approximately 330–370 genetic loci are derepressed more than 4-fold (log2-ratio > 2.0; p < 0.01) in hda6-7, pol IV or pol V single mutants, or in hda6 pol IV or hda6 pol V double mutants (Figure 1C). Of these, 55 and 48 loci are derepressed in pol IV or pol V single mutants, respectively, whereas three times as many (161) are derepressed in hda6-7. In hda6 pol IV double mutants, 323 total loci are upregulated at least 4-fold, including most loci derepressed in each single mutant plus an additional 169 loci for which the effects of hda6 and pol IV null mutations are additive or synergistic (Figure 1C, white area).
We define loci that are derepressed more strongly in hda6 pol IV double mutants than in either single mutant as Group DL (double-locked) loci, because HDA6 and Pol IV contribute independently to locking down their silencing. By contrast, loci that are similarly derepressed in hda6-7 or pol IV single mutants, and are not further derepressed in the double mutant, we define as Group E loci because hda6-7 and pol IV display genetic epistasis. Group DL and Group E loci are also apparent when the pol V mutant is considered in combination with hda6-7 (Figure 1C). Hierarchical clustering reveals a high degree of overlap among pol IV and pol V-affected loci (Figure 1A).
Group E and Group DL loci are extensively methylated on cytosines in CG, CHG and CHH contexts, unlike housekeeping genes (Figure S1A). A notable difference between Groups E and DL is a higher proportion of genes in Group E; 71% of Group E loci are genes, whereas only 29% are transposable elements. Conversely, only 26% of Group DL loci are genes whereas 74% are transposable elements (Figure 1D, left graph). This bias is observed for both pol IV or pol V mutants in combination with hda6 (Figure 1D, compare left and right panels; see also Figures S1C, S1D).
Group DL loci include well-studied targets of RdDM including the SDC gene, soloLTR and AtSN1 transposable elements (Henderson and Jacobsen, 2008; Herr et al., 2005; Huettel et al., 2006; Onodera et al., 2005). These loci are silenced in wild-type Col-0 plants, thus no RT-PCR signals are detected (Figure 1E, first column). Their transcripts are detectable in hda6-7, pol IV or pol V single mutants but are most highly expressed in hda6 pol IV or hda6 pol V double mutants, in which the double-lock is lost (Figure 1E). In contrast, Group E genes such as ERT7, ERT9, ERT12 or ERT14 (Epistatic HDA6-RdDM Targets) are derepressed in hda6-7, pol IV or pol V single mutants and are not further derepressed in hda6 pol IV or hda6 pol V double mutants (Figure 1E, Figure S1D). These results were confirmed for three independent mutant alleles of HDA6 (hda6-5, hda6-6 and hda6-7) (Figure S1E).
Double-locked silencing at transposable elements and the Group DL gene, SDC
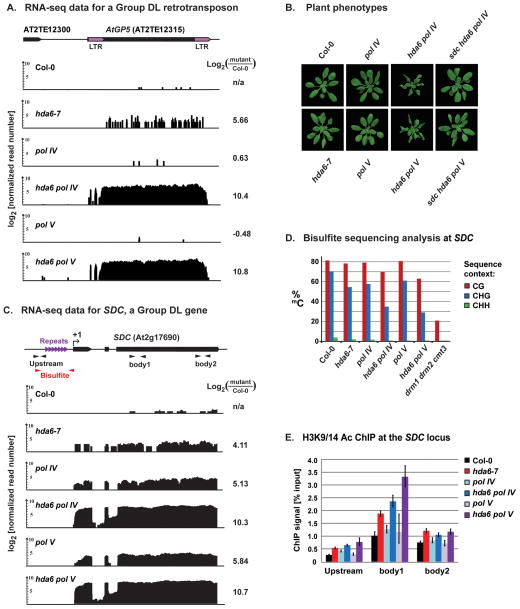
A representative Group DL locus is AT2TE12315, a member of the AtGP5 transposable element family. RNA-seq data reveal partial derepression of this element in hda6-7 and an additional 26 to 36-fold derepression (4.7–5.2 log2 units) in hda6 pol IV or hda6 pol V double mutants (Figure 2A). Interestingly, the AtGP5 element is not significantly derepressed in either pol IV or pol V single mutants, suggesting that RdDM is not essential for silencing so long as HDA6-dependent silencing is intact. However, a synergistic loss of silencing occurs when both pathways are disrupted (Figure 2A).
Figure 2. Molecular and phenotypic consequences of “double-locked” silencing involving HDA6 and Pol IV/V.
(A) AtGP5 retrotransposon RNA-seq data. Long terminal repeats (LTRs) flank the transposable element body (see diagram). Vertical black bars denote polyA+ RNA read numbers (log2 units), normalized to total mapped reads, in wild-type Col-0, homozygous hda6-7, pol IV and pol V mutants and the double mutants, hda6 pol IV and hda6 pol V. Numbers at right are RPKM (reads per kilobase per million) log-ratios, comparing expression in each mutant to Col-0.
(B) Phenotypes of 21-day-old plants: wild-type Col-0, or the indicated mutants (see also Figure S2).
(C) Diagram of the SDC locus (At2g17690), showing upstream tandem repeats (purple arrows), the Pol II transcription start site (+1), and PolyA+ RNA-seq data plotted on a log2 scale (black). PCR amplicons for chromatin immunoprecipitation (ChIP) and bisulfite sequencing (black and red arrows, respectively) are indicated.
(D) Bisulfite sequencing analysis of DNA methylation upstream of +1 (red amplicon, panel C). Data from triple mutant drm1 drm2 cmt3 serves as a hypomethylated control (Henderson and Jacobsen, 2008).
(E) ChIP analysis of Histone 3 Lysine 9/14 acetylation at three positions in the SDC locus. Error bars show the propagated standard error of the mean from three replicates.
Gross morphological defects are not apparent in single hda6-7, pol IV or pol V null mutants (Aufsatz et al., 2002; Murfett et al., 2001; Onodera et al., 2005; Pontier et al., 2005). However, hda6 pol IV or hda6 pol V double mutants are dwarfed and have curled leaves (Figures 2B, S2). In fact, any double mutant pairing of hda6-7 with a mutation disrupting RdDM yields this phenotype (Figure S2 and data not shown). Similar phenotypes were previously described for plants that were doubly mutant for the CHG maintenance methyltransferase system (cmt3 or kyp/suvh4) in combination with the de novo cytosine methyltransferase system (drm2, or other RdDM pathway mutants) due to derepression of SDC, a gene encoding an F-box protein (Chan et al., 2006; Henderson and Jacobsen, 2008). Triple mutant sdc hda6 nrpd1 or sdc hda6 nrpe1 plants resemble wild-type Col-0, confirming that SDC is necessary for the hda6 pol IV/V double mutant phenotypes (Figure 2B).
Relative to wild type, SDC is derepressed up to 57-fold (~4–6 log2 units) in hda6, pol IV or pol V single mutants, but over 1000-fold (10 log2 units) in hda6 pol IV or hda6 pol V double mutants (Figure 2C). Repeats adjacent to the gene promoter influence SDC silencing (see diagram in Figure 2C)(Henderson and Jacobsen, 2008). Bisulfite sequencing of this region revealed minor reductions in DNA methylation in the hda6-7 single mutant, but additional hypomethylation in hda6 pol IV or hda6 pol V double mutants, predominantly at CHG and CHH sites (Figure 2D). By comparison, a drm1 drm2 cmt3 triple mutant control shows a four-fold reduction in CG methylation and an essentially complete loss of CHH and CHG methylation (Figure 2D), as reported previously (Henderson and Jacobsen, 2008).
Chromatin immunoprecipitation (ChIP) performed using an antibody specific for Histone H3 diacetylated on lysines 9 and 14 (H3K9/14), a mark of active chromatin, reveals elevated H3K9/14 acetylation in hda6-7 relative to wild type at three intervals of the SDC locus (Figure 2E), with the highest enrichment in the 5′ half of the gene (body 1 region; see diagram in panel C). Neither pol IV nor pol V single mutants show this increased H3K9/14 acetylation. However, the double mutants hda6 pol IV and hda6 pol V display additive effects greater than those observed in hda6-7 alone. Collectively, the data indicate a partial functional overlap between HDA6 and RdDM for cytosine methylation and histone modification at SDC.
Epistatic HDA6 and Pol IV/V functions at Group E loci
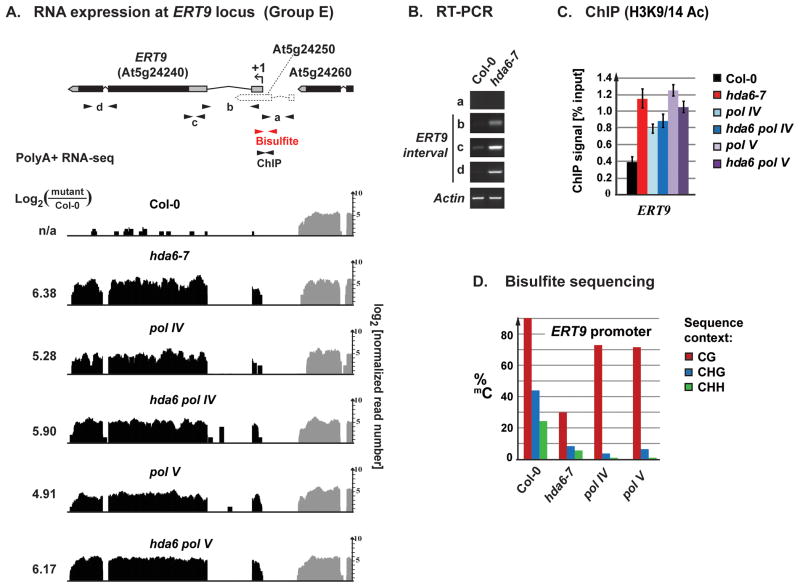
ERT9 (At5g24240) encodes a putative phosphatidylinositol 3-/4-kinase (Figure 3A). Its first exon overlaps a predicted, but unsupported, gene model, At5g24250 for which no RNA-seq (Figure 3A) or RT-PCR confirmation was obtained (Figure 3B). However, the adjacent gene, At5g24260, is expressed at similar levels in wild-type Col-0 or hda6-7, pol IV or pol V mutants (grey vertical bars in Figure 3A). ERT9 transcripts (black vertical bars) are detected at low levels in wild-type plants, but are 30–84 fold (4.9–6.4 log2 units) more abundant in hda6-7, pol IV or pol V mutants (Figure 3A). Unlike Group DL loci (see Figure 2), ERT9 is derepressed in hda6 pol IV or hda6 pol V double mutants to the same extent as in hda6-7, the defining characteristic of a Group E locus. Chromatin immunoprecipitation experiments reveal increased H3K9/14 acetylation at the ERT9 promoter in hda6, pol IV, pol V, hda6 pol IV or hda6 pol V mutants compared to wild-type plants, with no apparent synergy or additivity in the double mutants versus single mutants (Figure 3C).
Figure 3. Epistasis of HDA6 and Pol IV/V at a Group E locus.
(A) Diagram of the ERT9 gene (At5g24240) and adjacent annotated loci, At5g24250 and At5g24260. Arrows mark PCR amplicons used for RT-PCR, bisulfite sequencing or ChIP analyses. Data tracks show polyA+ RNA read counts (log2 units) in wild-type Col-0, hda6-7, pol IV, pol V and the double mutants hda6 pol IV and hda6 pol V. Numbers to the left are log-ratios, comparing expression in each mutant to Col-0. Grey shading represents RNA-seq reads mapped to the 3′-terminal exons of At5g24260.
(B) RT-PCR verification of ERT9 gene derepression in the mutant hda6-7 (intervals b, c, d), and lack of detectable At5g24250 expression (interval a).
(C) ChIP analysis of Histone 3 Lysine 9/14 acetylation at the ERT9 promoter region. Error bars show the propagated standard error of the mean from three replicates.
(D) Bisulfite sequencing analysis of DNA methylation in the interval −131 to +190 relative to the ERT9 transcription start site (+1) (see also Figure S3).
Bisulfite sequencing of an interval flanking the ERT9 transcription start site and promoter shows that 90% of CG motifs are methylated in wild-type Col-0. CG methylation drops to 30% in hda6-7 but is ~70% in pol IV or pol V mutants (Figure 3D). 44% of CHG sites are methylated in Col-0, but CHG methylation drops to 8% in hda6-7, 4% in pol IV and 6% in pol V. Similarly, 24% of CHH sites are methylated in Col-0, but CHH methylation falls to 5% in hda6-7, and less than 1% in pol IV or pol V. Analogous results are observed at the Group E locus, ERT7 (Figure S3A). Collectively, these data indicate that HDA6 is important for CG, CHG and CHH methylation at Group E loci, consistent with roles in both maintenance methylation, involving MET1 and CMT3 (Figure S3B) and de novo RdDM, involving DRM2. In contrast, Pols IV and V are important for RdDM at CHG and CHH sites, but less important for CG methylation.
HDA6 facilitates Pol IV-dependent siRNA biogenesis at Group E loci
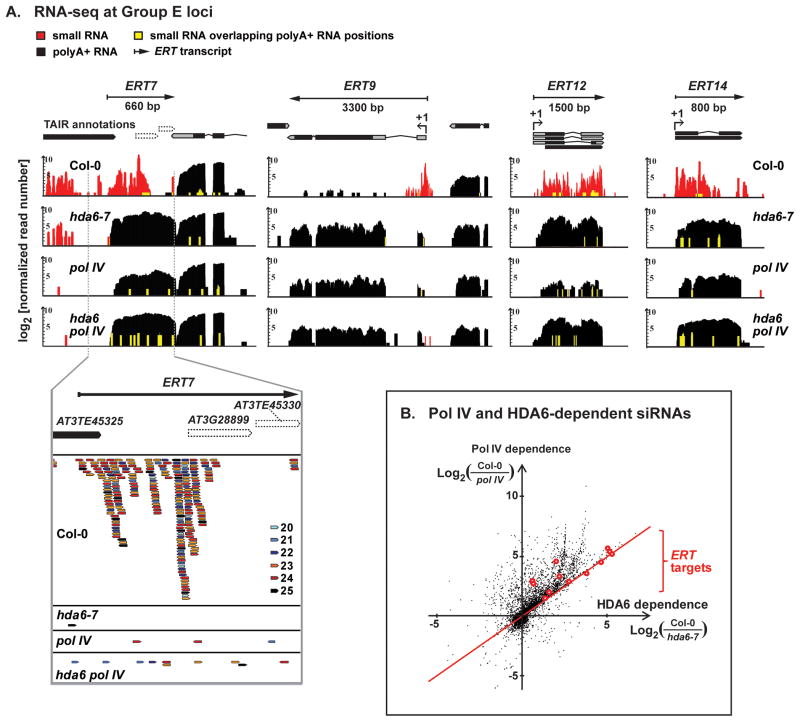
Key insight into the role of HDA6 in RdDM came from small RNA deep sequencing. In wild-type Col-0, abundant siRNAs are detected at Group E loci, such as ERT7, ERT9, ERT12 or ERT14 (Figure 4A, red signals), and expression of polyA+ RNA from these loci is correspondingly low (black signals). The siRNAs are predominantly 23–24 nt, the size of DCL3 products that guide RdDM (see Figure 4A detail, at bottom). Surprisingly, these siRNAs are absent in hda6-7, as in pol IV mutants, indicating that HDA6 is required for Pol IV-dependent siRNA biogenesis at Group E loci. A plot of the requirement for Pol IV versus HDA6 in siRNA biogenesis, genome-wide (Figure 4B), shows that most ERT loci (red circles) fall along the red line that denotes a 1:1 correspondence between Pol IV and HDA6 dependence.
Figure 4. HDA6 facilitates Pol IV-dependent siRNA biogenesis.
(A) RNA-seq profiles at three Group E targets. Data tracks show polyA+ RNA reads (black vertical bars) and small RNA reads (red and yellow vertical bars; yellow denotes positions where siRNA and poly A+ reads overlap) in wild-type (Col-0), hda6-7, nrpd1-3 (pol IV), and the double mutant, hda6 pol IV. Read counts (log2 units) were normalized by total mapped reads. Analogous data for Group DL loci are in Figure S4.
(B) Scatter plot showing the relative dependence of 24 nt siRNA production on Pol IV and HDA6, genome-wide. Each data point represents a TAIR10 gene locus, with the x-axis indicating the log-ratio of small RNA read density in Col-0 relative to hda6-7, and the y-axis indicating the log-ratio of small RNA read density in Col-0 relative to pol IV. Group E loci are marked in red.
Abundant Pol IV-dependent siRNAs are also detected at Group DL loci, such as SDC, soloLTR and the ROMANIAT5 retrotransposon (Figure S4, red signals), and correlate with silencing of partially overlapping polyA+ RNA transcription units (Figure S4, black signals). However, unlike Group E loci, siRNA levels at Group DL loci are not significantly impacted in hda6-7, consistent with Pol IV-dependent RdDM acting in parallel with HDA6-mediated chromatin modification to achieve double-locked silencing at these loci.
Loss of HDA6 erases epigenetic memory required for Group E silent locus identity
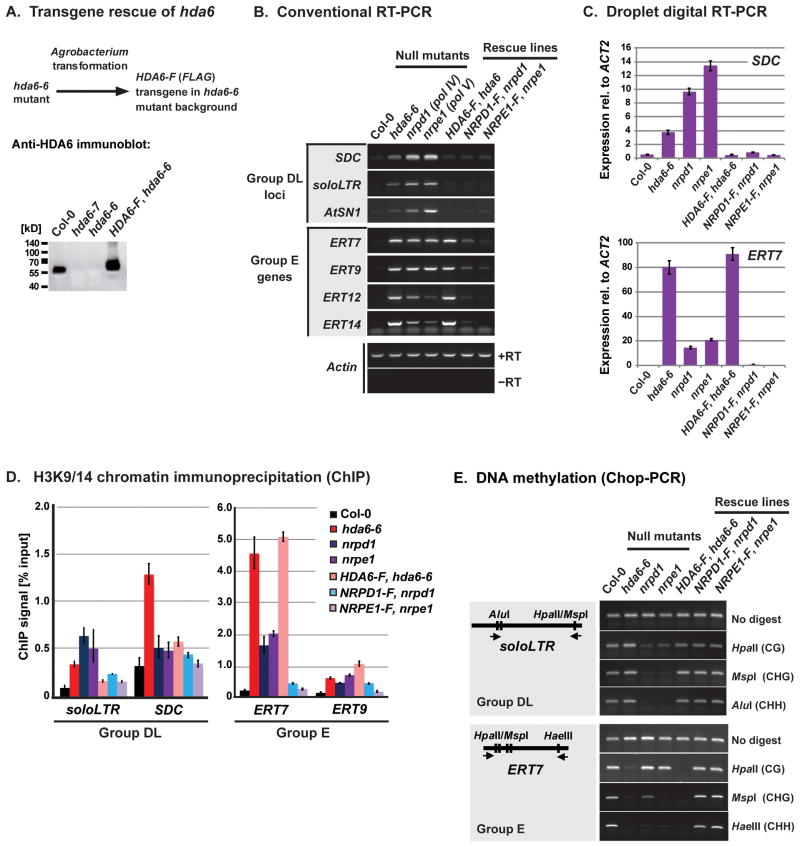
Transforming the hda6-6 null mutant with a transgene encoding FLAG-tagged HDA6 (HDA6-F) (Earley et al., 2010) restores HDA6 protein expression (Figure 5A) and silencing at Group DL loci such as SDC and soloLTR (Figures 5B, 5C, S5A), showing that the HDA6-F protein is functional. Versions of HDA6-F transgenes bearing alanine substitutions of amino acids critical for HDA6 activity (Earley et al., 2010) fail to restore silencing at Group DL loci, indicating that histone deacetylase activity is required for rescue (Figure S5B).
Figure 5. HDA6 rescue fails to restore silent locus identity at Group E loci.
(A) Agrobacterium-mediated transformation of the hda6-6 null mutant with a transgene expressing HDA6 fused to a FLAG epitope (HDA6-F). HDA6 was detected by immunoblotting using an antibody raised against a C-terminal peptide.
(B) RT-PCR assays for Group DL and Group E locus expression in Col-0 or hda6-6, nrpd1 (pol IV) or nrpe1 (pol V) mutants, or in lines in which these mutants were rescued by corresponding full-length, FLAG-tagged transgenes (HDA6-F, NRPD1-F, or NRPE1-F). Actin serves as a loading control. Controls omitting reverse transcriptase (−RT) produced no amplification products, as shown for Actin (see also Figure S5).
(C) Quantitative droplet digital RT-PCR assays of SDC (Group DL) and ERT7 (Group E) expression. Transcript levels are reported relative to ACT2. Controls omitting RT yielded almost no positive droplets.
(D) H3K9/14 acetylation measured by chromatin immunoprecipitation (ChIP). Percentage enrichment was determined by real-time PCR analysis of immunoprecipitated DNA compared to input chromatin. Error bars show the propagated standard error of the mean for three replicates.
(E) Chop-PCR assay assessing DNA methylation by digesting genomic DNA with methylation-sensitive restriction endonucleases prior to PCR amplification using flanking primers. The endonucleases report on cytosine methylation in different sequence contexts: HpaII (CG), MspI (CHG), HaeIII (CHH) or AluI (CHH), where H is any base other than G. See also Figures S5 and S6.
At both Group DL and Group E loci, silencing that is lost in pol IV (nrpd1) or pol V (nrpe1) null mutants can be restored by transgenes expressing FLAG-tagged NRPD1 or NRPE1 (Figures 5B, 5C). Restoration of silencing correlates with the resetting of histone acetylation (Figure 5D) and cytosine methylation (Figure 5E) to levels similar to wild-type. Importantly, a key distinction between Group DL and Group E loci is that transgene-mediated restoration of HDA6 activity fails to restore silencing at Group E loci such as ERT7, ERT9, ERT12 or ERT14 (Figures 5B, 5C). Consistent with this failure to restore Group E silencing, Histone H3 hyperacetylation (Figure 5D) and CG, CHG and CHH hypomethylation persist at hda6 mutant levels (Figures 5E, S6). These results suggest that HDA6 potentiates an epigenetic memory at Group E loci that once lost is not readily regained simply by restoring HDA6 activity.
siRNA production is not restored in hda6 mutants upon restoration of HDA6
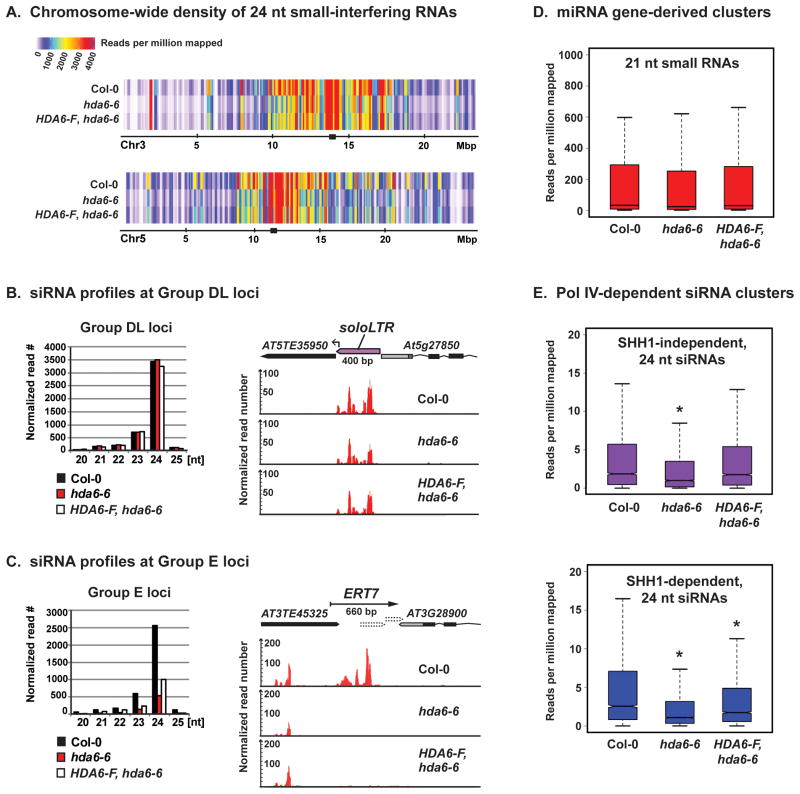
Chromosome-wide plots reveal a modest but widespread reduction in Pol IV-dependent 24 nt siRNA density in hda6-6 relative to wild-type (Figure 6A). At many intervals, 24 nt siRNA levels return to near wild-type levels upon HDA6 rescue, but localized siRNA deficiencies persist. At Group DL loci, the abundance of 24 nt siRNAs is unchanged in hda6-6 relative to wild-type Col-0 (Figures 6B, S7A, B). By contrast, at Group E loci, 24 nt siRNA levels are reduced by nearly 80% in hda6-6 and do not recover upon restoration of HDA6 activity (Figure 6C, S7A, C). Other small RNAs, such as 21 nt miRNAs, are unaffected in hda6 mutants (Figure 6D).
Figure 6. Epigenetic memory loss results in failure to produce Pol IV-dependent siRNAs.
(A) Abundance of 24 nt siRNAs mapping to chromosomes 3 and 5 in wild-type (Col-0), hda6-6 and HDA6-FLAG rescued hda6-6 plants. Small RNA clusters are plotted using a 200-kb sliding window stepped in 100-kb increments. Black squares mark the estimated positions of centromeres.
(B) Relative abundance of siRNAs of different size classes mapping to Group DL loci (left panel) and a browser view of siRNA abundance and distribution at the soloLTR locus (right panel).
(C) Relative abundance of siRNAs of different size classes mapping to Group E loci (left panel) and a browser view of siRNA profiles at ERT7 (right panel). Supporting polyA+ RNA-seq data and gene annotations are in Figures 4A and S4.
(D) Boxplot analysis of 21 nt miRNAs, revealing no significant overall change in miRNA abundance in hda6 or hda6 HDA6-F rescue lines relative to wild-type Col-0.
(E) Boxplot analysis of the effects of HDA6 mutation, and HDA6 rescue, on 24 nt siRNAs that are either SHH1-independent or SHH1-dependent (as defined in Law et. al. 2013). All read counts are normalized to total mapped read numbers. Asterisks above boxplots in panel E indicate significant reduction relative to Col-0 (p < 0.002, Wilcoxon rank-sum test). See also Figure S7.
The loss of Pol IV-dependent 24 nt siRNAs in hda6 mutants suggests that HDA6 establishes a chromatin state required for Pol IV recruitment. We hypothesized that this chromatin state might facilitate recruitment of the Pol IV-interacting protein, SHH1, whose mutation affects approximately one half, but not all, Pol IV-dependent siRNA clusters (Law et al., 2013). Whereas SHH1-independent siRNA clusters show reduced abundance in hda6-6 but full restoration upon HDA6 rescue (Figure 6E, top panel), SHH1-dependent siRNA clusters are strongly reduced in hda6-6 and do not fully recover upon restoration of HDA6 activity (Figure 6E, bottom panel). These data indicate that HDA6 confers silent locus identity at loci where SHH1 helps recruit Pol IV.
Maintenance cytosine methylation confers epigenetic memory at Group E targets
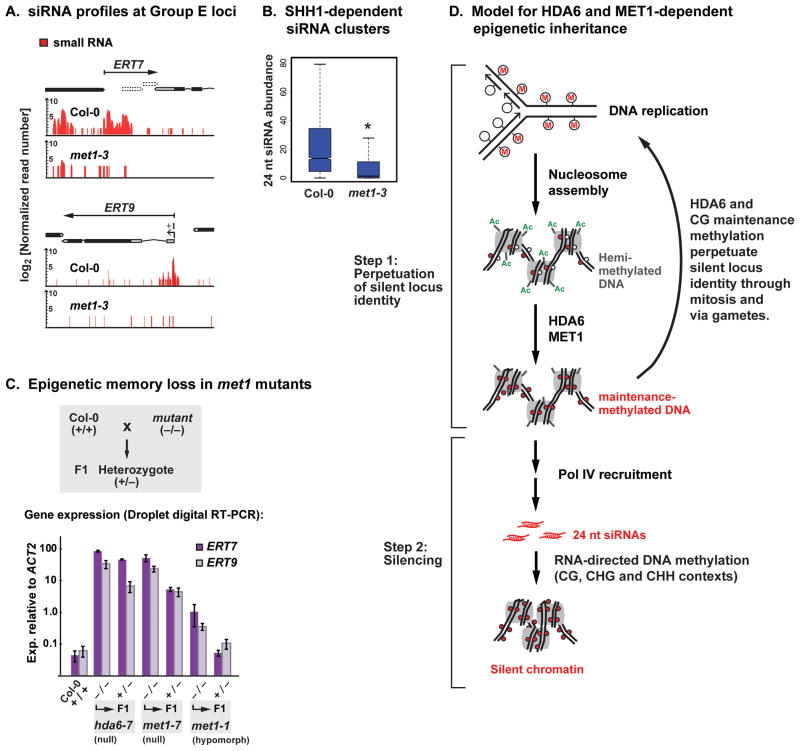
HDA6 influences maintenance methylation at CG motifs of transposable elements and 45S ribosomal RNA genes (Earley et al., 2010; Lippman et al., 2003; Liu et al., 2012; To et al., 2011), suggesting that HDA6-dependent epigenetic memory might involve maintenance methylation by MET1. Consistent with this hypothesis, Pol IV-dependent siRNAs at Group E loci are reduced in met1 mutants (Figure 7A), as are SHH1-dependent siRNAs (Figure 7B). Moreover, Group E loci are strongly derepressed in met1 mutants, as in hda6 mutants (Figure S3B).
Figure 7. HDA6, MET1, SHH1 and Pol IV collaborate in gene silencing.
(A) 24 nt siRNAs at Group E loci are reduced in met1 mutants. Red vertical bars depict the abundance of siRNA reads (log2 units) detected in Col-0 or met1-3 null mutants in the datasets of Lister et al. 2008, normalized relative to the number of mapped reads.
(B) Boxplot analysis of the MET1-dependence of SHH1-dependent siRNA clusters (*indicates significant reduction; p < 0.002, Wilcoxon rank-sum test).
(C) Erasure of silent locus identity occurs in met1 mutants, as in hda6 mutants. The histograms show the expression levels of the Group E loci, ERT7 and ERT9 in wild-type Col-0 (+/+), homozygous mutants (−/−) for hda6-7 (a null mutant), met1-7 (a null mutant) or met1-1 (a partial loss-of-function, or hypomorphic, mutant), or in F1 heterozygous progeny (+/−) that result from outcrossing the homozygous mutants to a wild-type plant. Quantitative droplet digital RT-PCR was used to quantify ERT7 and ERT9 expression levels (on a Log10 scale) relative to an ACT2 control.
(D) Two-step model for epigenetic inheritance in which silent locus identity at Group E loci is heritable and is a prerequisite for, but separable from, silencing by RNA-directed DNA methylation. Following replication of fully CG-methylated DNA, hemi-methylated DNA is assembled into nucleosomes by chromatin assembly factors that use acetylated histones as substrates. De-acetylation of these histones by HDA6 may be a prerequisite for CG methylation maintenance by MET1. HDA6-dependent CG maintenance methylation perpetuates the heritable epigenetic memory accounting for silent locus identity. Subsequent recruitment of Pol IV, assisted by SHH1 or other chromatin binding partners, brings about silencing of the loci by RdDM in each new generation.
To test whether met1 mutants display heritable loss of silent locus identity at Group E loci, like hda6 mutants, we crossed homozygous met1-7 or hda6-7 null mutants to wild-type Col-0. In resulting F1 heterozygous progeny, neither ERT7 nor ERT9 silencing was restored to wild-type levels (Figure 7C). Repeating this experiment using met1-1, a hypomorphic allele that causes only a partial loss of CG methylation, and partial loss of ERT7 and ERT9 silencing, allowed for restoration of ERT7 and ERT9 silencing to wild-type levels in F1 progeny. These results suggest that partial CG methylation is sufficient for silent locus identity to persist, thus permitting Pol IV-dependent silencing in subsequent generations.
Discussion
Most genomic loci subjected to RNA-directed DNA methylation (RdDM) are transposable elements at which HDA6 and RdDM act independently, contributing to silencing through a double-lock mechanism (See also, Mirouze et al., 2009; Yokthongwattana et al., 2010). However, at a subset of silenced loci (Group E loci), HDA6 is needed for the initiating step of the RdDM pathway in which Pol IV-dependent 24 nt siRNAs are produced. At these Group E loci, HDA6, Pol IV and Pol V are each required for RdDM and silencing, but HDA6 has the distinction of conferring epigenetic memory needed for Pol IV and Pol V-dependent RdDM. This memory remains intact in pol IV or pol V single mutant lineages such that restoration of Pol IV or Pol V activity is sufficient to restore RdDM and silencing. By contrast, hda6 mutants lose the epigenetic memory that accounts for silent locus identity at Group E loci, such that 24 nt siRNA biogenesis, RdDM and silencing are not restored simply by restoring HDA6 activity.
HDA6 is a broad-specificity histone deacetylase (Earley et al, 2006), thus trans-generational perpetuation of positioned, deacetylated nucleosomes through mitosis and meiosis could conceivably account for epigenetic memory at Group E loci. However, studies from our laboratory (see Figures 3D, S6 and Earley et al., 2010), and others (Lippman et al., 2003; Liu et al., 2012; Stroud et al., 2013; To et al., 2011) have shown that HDA6 facilitates MET1-dependent CG methylation. If CG methylation is lost, it is not consistently regained, as shown in F1 heterozygotes or recombinant inbred populations derived from crossing methylation-deficient and wild-type parents (Kakutani et al., 1999; Lippman et al., 2004; Reinders et al., 2009; Saze et al., 2003; Vongs et al., 1993). Failure to regain CG methylation is especially true for low copy number loci (Teixeira et al., 2009). Collectively, these considerations suggest that HDA6-dependent CG maintenance methylation may account for epigenetic memory. This hypothesis is consistent with met1 null mutants phenocopying hda6 null mutants with respect to loss of Pol IV-dependent siRNAs and the inability to restore Group E locus silencing simply by restoring MET1 activity.
Our results indicate that chromatin dependent silencing of Group E loci reflects two separable processes, depicted in the model of Figure 7D. The first confers silent locus identity through the actions of HDA6 and MET1, perpetuating epigenetic marks that are heritable through mitosis and meiosis, most likely in the form of CG methylation patterns. The model envisions that CG maintenance methylation occurs in the context of nucleosomes, with newly deposited histones, which are acetylated (Avvakumov et al., 2011), needing to be deacetylated by HDA6 prior to methylation of daughter strand cytosines by MET1. HDA6 may facilitate additional chromatin modifications, perhaps not limited to histone deacetylation, independent of DNA methylation. Although HDA6 and MET1-dependent chromatin modifications are not sufficient to silence Group E loci, they are a prerequisite for subsequent Pol IV recruitment and RdDM, bringing about further cytosine methylation, associated chromatin modifications, and transcriptional silencing in step two (Figure 7D).
It is not clear from our results how Pol IV or Pol V are recruited to Group DL loci. Helper proteins recognizing non-CG methylation or specific histone modification patterns may facilitate Pol IV/V recruitment at these loci in an HDA6-independent manner.
Two recent studies examined the stability of cytosine methylation and gene expression in isogenic A. thaliana lines. Bisulfite sequencing showed that cytosine methylation patterns tend to be faithfully maintained over at least 30 generations. However, spontaneous loss of CG methylation occurs at individual loci, resulting in the sporadic emergence of transcriptionally active epialleles (Becker et al., 2011; Schmitz and Ecker, 2012; Schmitz et al., 2011). Two loci shown to be susceptible to such spontaneous losses of CG methylation and silencing are among the Group E loci identified in our study: At5g24240 and At4g14548 (which we call ERT9 and ERT11, respectively). Sporadic erasure of HDA6- and MET1-dependent silent locus identity can presumably account for the emergence of stable, active epialleles at these loci. We speculate that sporadic DNA damage, followed by replacement of methylated cytosines by unmethylated cytosines during the repair process, might be one mechanism by which epigenetic memory loss can occur in plants whose HDA6 and MET1 genes are fully functional.
Experimental Procedures
Plant materials
Arabidopsis thaliana hda6-5 and hda6-6 (a.k.a., axe1-4 and axe1-5), and hda6-7 (a.k.a., rts1-1) mutants were described in Murfett et al. (2001) and Aufsatz et al. (2002). RNA polymerase mutants pol IV (nrpd1-3) and pol V (nrpe1-11) were described in (Onodera et al., 2005) and (Pontes et al., 2006); pol II* (nrpb2-3) was provided by X. Chen (Zheng et al. 2009). Single mutant sdc (SALK_017593, Henderson and Jacobsen, 2008) and triple mutant drm1-2 drm2-2 cmt3-11t (Chan et al. 2006) were obtained from the Arabidopsis Biological Resource Center. drd1-6 was provided by M. Matzke, dms3-4 and drm2-2b (SAIL_70_E12) by A. Wierzbicki and met1-1 by Eric Richards. met1-7 (SALK_076522) was obtained from the Nottingham Arabidopsis Stock Center.
RNA analyses
Total RNA was extracted from two-week-old Arabidopsis plant leaf tissue using TRI-reagent (MRC, Inc.). For semi-quantitative RT-PCR, 1.5 μg of DNase I-treated total RNA was used for random-primed cDNA synthesis by Superscript III reverse transcriptase (Invitrogen). Standard PCR was performed on cDNA aliquots (~100 ng RNA input) using GoTaq Green (Promega) and primers listed in Table S2. Droplet Digital PCR and RNA-sequencing methods are provided in Supplemental Experimental Procedures.
DNA methylation analyses
Chop PCR assays were performed using 100 ng of restriction enzyme-digested (“chopped”) genomic DNA as in (Earley et al. 2010). Bisulfite sequencing was by the procedure of (Foerster et al., 2010). In brief, PCR fragments amplified from bisulfite-treated DNA were cloned into pGEM-T-Easy and sequenced using a T7 primer. At least 30 sequences per amplicon were analyzed in CyMATE (Hetzl et al. 2007). Chop PCR and bisulfite sequencing primers are listed in Table S2.
Protein blot analysis
Approximately 300 mg of 14-day-old leaf tissue was ground in liquid nitrogen and suspended in 1 mL of phosphate buffered saline, pH 7.4,1mM PMSF and 1% Plant Protease Inhibitors (Sigma, St. Louis). After 10 min at room temperature, with mixing by inversion, extracts were subjected to centrifugation 2 min at 800 x g. The supernatant was filtered through Miracloth and 60 μg total protein, determined using a Coomassie blue dye binding assay, then subjected to electrophoresis on a 7.5% SDS-PAGE gel and transferred to nitrocellulose. The blot was incubated overnight with anti-HDA6 antisera (raised in rabbits against the C-terminal peptide CDEMDDDNPEPDVNPPSS), followed by anti-Rabbit-HRP for 1 h and film detection using ECL reagent (Amersham).
Chromatin Immunoprecipitation (ChIP)
ChIP was performed based on the protocol of (Wierzbicki et al., 2008). Real-time PCR amplification of immunoprecipitated DNA was accomplished using SYBR Green JumpStart Taq ReadyMix (Sigma) on a Stratagene MX3000P instrument. ChIP samples were normalized to input controls using the comparative CT method (Livak and Schmittgen, 2001). Real-time PCR primers are listed in Table S2.
Bioinformatic analyses
Illumina sequences for polyA+ RNA-seq libraries were aligned to the A. thaliana reference with Bowtie and Tophat (Langmead et al., 2009), and filtered for perfect matches. Annotated TAIR10 “mRNA” and “ncRNA” loci were considered to be “Genes”. Transposable elements as annotated in TAIR10 were refined in Cufflinks to restrict the set to expressed regions. For small RNA analyses, trimmed reads were aligned to the reference with Novoalign (Novocraft) and perfectly matched, 20–25 nt inserts were analyzed further. For details, see Supplemental Experimental Procedures.
Supplementary Material
Highlights.
HDA6 and Pols IV/V collaborate in silencing via at least two modes of action.
Silent locus identity can be epigenetically inherited without silencing.
HDA6 and MET1 facilitate epigenetic memory and silent locus identity.
Silent locus specification precedes silencing via RNA-directed DNA methylation.
Acknowledgments
RC performed ChIP assays in Figures 2E, 3C and 5D. CC performed bisulfite sequencing in Figures 2D, 3D, S6. KM directed Illumina library construction and sequencing. TB, RP, SY, CB, DR and HT performed bioinformatic analyses. TB and FP conducted all other experiments. TB and CSP wrote the manuscript. We thank Zach Smith and James Ford of the IU Center for Genomics and Bioinformatics for technical assistance. CSP is an Investigator of the Howard Hughes Medical Institute (HHMI) and Gordon and Betty Moore Foundation (GBMF). This work was supported by National Institutes of Health grant GM077590 and GBMF grant GBMF3036 to CSP. TB was supported by a W.M. Keck Postdoctoral Fellowship, an NIH Ruth L. Kirschstein National Research Service Award, and funding from HHMI.
Footnotes
Accession Numbers
Sequencing data have been deposited in the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP026486.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aufsatz W, Mette MF, Van Der Winden J, Matzke M, Matzke AJ. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. Embo J. 2002;21:6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov N, Nourani A, Cote J. Histone chaperones: modulators of chromatin marks. Mol Cell. 2011;41:502–514. doi: 10.1016/j.molcel.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Bartee L, Malagnac F, Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C, Hagmann J, Muller J, Koenig D, Stegle O, Borgwardt K, Weigel D. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Role of the Arabidopsis DRM Methyltransferases in De Novo DNA Methylation and Gene Silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Zhang X, Shah G, Chien JS, Jacobsen SE. RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis. PLoS Genet. 2006;2:e83. doi: 10.1371/journal.pgen.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151:167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006;20:1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Pontvianne F, Wierzbicki AT, Blevins T, Tucker S, Costa-Nunes P, Pontes O, Pikaard CS. Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev. 2010;24:1119–1132. doi: 10.1101/gad.1914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster AM, Hetzl J, Mullner C, Mittelsten Scheid O. Analysis of bisulfite sequencing data from plant DNA using CyMATE. Methods Mol Biol. 2010;631:13–22. doi: 10.1007/978-1-60761-646-7_2. [DOI] [PubMed] [Google Scholar]
- Furner IJ, Sheikh MA, Collett CE. Gene silencing and homology-dependent gene silencing in Arabidopsis: genetic modifiers and DNA methylation. Genetics. 1998;149:651–662. doi: 10.1093/genetics/149.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nature reviews. Molecular cell biology. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- Haag JR, Pontes O, Pikaard CS. Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing. PLoS ONE. 2009;4:e4110. doi: 10.1371/journal.pone.0004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Ream TS, Marasco M, Nicora CD, Norbeck AD, Pasa-Tolic L, Pikaard CS. In Vitro Transcription Activities of Pol IV, Pol V, and RDR2 Reveal Coupling of Pol IV and RDR2 for dsRNA Synthesis in Plant RNA Silencing. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23:318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev. 2008;22:1597–1606. doi: 10.1101/gad.1667808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJ, Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. Embo J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T, Munakata K, Richards EJ, Hirochika H. Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics. 1999;151:831–838. doi: 10.1093/genetics/151.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Du J, Hale CJ, Feng S, Krajewski K, Palanca AM, Strahl BD, Patel DJ, Jacobsen SE. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498:385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet. 2011;7:e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 2004;23:4286–4296. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yu CW, Duan J, Luo M, Wang K, Tian G, Cui Y, Wu K. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant physiology. 2012;158:119–129. doi: 10.1104/pp.111.184275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Mirouze M, Reinders J, Bucher E, Nishimura T, Schneeberger K, Ossowski S, Cao J, Weigel D, Paszkowski J, Mathieu O. Selective epigenetic control of retrotransposition in Arabidopsis. Nature. 2009;461:427–430. doi: 10.1038/nature08328. [DOI] [PubMed] [Google Scholar]
- Murfett J, Wang XJ, Hagen G, Guilfoyle TJ. Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell. 2001;13:1047–1061. doi: 10.1105/tpc.13.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Pikaard CS. Arabidopsis Histone Lysine Methyltransferases. Advances in botanical research. 2010;53:1–22. doi: 10.1016/S0065-2296(10)53001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu JK, Hagen G, Guilfoyle TJ, Pasa-Tolic L, Pikaard CS. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Wulff BB, Mirouze M, Mari-Ordonez A, Dapp M, Rozhon W, Bucher E, Theiler G, Paszkowski J. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 2009;23:939–950. doi: 10.1101/gad.524609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Mittelsten Scheid O, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Ecker JR. Epigenetic and epigenomic variation in Arabidopsis thaliana. Trends Plant Sci. 2012;17:149–154. doi: 10.1016/j.tplants.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S, Furio-Tari P, Ferrer A, Conesa A. NOISeq: Exploratory analysis and differential expression for RNA-seq data. R package version 1.1.5 2012 [Google Scholar]
- Teixeira FK, Heredia F, Sarazin A, Roudier F, Boccara M, Ciaudo C, Cruaud C, Poulain J, Berdasco M, Fraga MF, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323:1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- To TK, Kim JM, Matsui A, Kurihara Y, Morosawa T, Ishida J, Tanaka M, Endo T, Kakutani T, Toyoda T, et al. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 2011;7:e1002055. doi: 10.1371/journal.pgen.1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant I, Paszkowski J. Role of histone and DNA methylation in gene regulation. Curr Opin Plant Biol. 2007;10:528–533. doi: 10.1016/j.pbi.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Dittmer TA, Richards EJ. Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet. 2008;4:e1000156. doi: 10.1371/journal.pgen.1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokthongwattana C, Bucher E, Caikovski M, Vaillant I, Nicolet J, Mittelsten Scheid O, Paszkowski J. MOM1 and Pol-IV/V interactions regulate the intensity and specificity of transcriptional gene silencing. EMBO J. 2010;29:340–351. doi: 10.1038/emboj.2009.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh P-H, Coleman-Derr D, Eshed Williams L, Thao K, Harmer SL, Zilberman D. The nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013 doi: 10.1016/j.cell.2013.02.033. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ma ZY, Zeng L, Tanaka K, Zhang CJ, Ma J, Bai G, Wang P, Zhang SW, Liu ZW, et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1300585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu JK. RNA-directed DNA methylation. Curr Opin Plant Biol. 2011;14:142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009;23:2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.