Abstract
Most studies of endophytic symbionts have focused on terrestrial plants, neglecting the ecologically and economically important plants present in aquatic ecosystems. We evaluated the diversity, composition, host- and tissue affiliations, and geographic structure of fungal endophytes associated with common aquatic plants in northern Arizona, USA. Endophytes were isolated in culture from roots and photosynthetic tissues during two growing seasons. A total of 226 isolates representing 60 putative species was recovered from 9,600 plant tissue segments. Although isolation frequency was low, endophytes were phylogenetically diverse and species-rich. Comparisons among the most thoroughly sampled species and reservoirs revealed that isolation frequency and diversity did not differ significantly between collection periods, among species, among reservoirs, or as a function of depth. However, community structure differed significantly among reservoirs and tissue types. Phylogenetic analyses of a focal genus (Penicillium) corroborated estimates of species boundaries and informed community analyses, highlighting clade- and genotype-level affiliations of aquatic endophytes with both sediment- and waterborne fungi, and endophytes of proximate terrestrial plants. Together these analyses provide a first quantitative examination of endophytic associations in roots and foliage of aquatic plants and can be used to optimize survey strategies for efficiently capturing fungal biodiversity at local and regional scales.
Keywords: Ascomycota, biodiversity, endophytic fungi, freshwater, lake, symbiosis
Introduction
Fungi are ubiquitous in terrestrial, marine, brackish and freshwater environments, where they play diverse and important ecological roles that often can be tapped for applications in agriculture, medicine, and industry [5, 13, 32, 69]. Despite their abundance and importance, however, the scale of fungal diversity is not well understood: roughly 100,000 species of fungi have been described, yet 1.5–5.1 million species are thought to exist [13, 32]. Many undiscovered fungi likely occur in symbiosis with other organisms, such as plants, and in previously un- or underexplored environments [9, 13, 41–42, 62, 69, 88]. Even though roughly 71% of the Earth’s surface is covered by water [57, 84], relatively few aquatic ecosystems have been examined thoroughly for fungal biodiversity (see [13, 41–42, 46, 69, 87–88]).
Lentic waters (i.e., standing waters) such as lakes and reservoirs, which span a wide array of water quality, seasonality, structural features, and temperature regimes, are home to diverse plants (aquatic plants, aquatic macrophytes, or hydrophytes) that together comprise some of the most productive communities on Earth [84]. Despite their importance to all aspects of human sustainability [52, 55, 84], lentic systems and their plant communities have received particularly little attention in terms of quantitative community sampling of fungi (see [42, 69, 87–88]).
The phylogenetically diverse vascular plants that inhabit lentic waters comprise three general growth forms. Free-floating or floating-leaf plants have leaves and flowers that float on the water surface, and may or may not be rooted in sediment [14]. Emergent plants have foliage that extends above the water surface, as well as submerged stems, roots, and narrow-leaved segments of photosynthetic tissue [14–16, 55, 84]. They grow in shallow waters in littoral zones (i.e., the often plant-rich region near the shoreline; [14, 84, 87–88]). In contrast, all tissues of submergent plants occur beneath the water surface, with occasional floating leaves or flower stalks that protrude only a small distance from the water [14–16, 55, 84]. Many aquatic plants die back in winter in strongly seasonal sites, with new plant growth initiated in spring from overwintering shoots and roots [68, 84].
Although many studies have examined the relationships of terrestrial plants with fungi (e.g., [2, 5–6, 9–10, 17–18, 23, 26, 33, 35, 39, 56, 61, 65–66, 74–76, 80–81]), associations between freshwater aquatic plants and fungal symbionts are not as well characterized (but see [40–43, 48, 71, 73]). Roots of emergent and submergent macrophytes often host arbuscular mycorrhizal fungi (AMF) and dark-septate endophytes (DSE) [12, 20, 40–42], but little is known regarding their culturable fungal symbionts outside of microcosm experiments (see [11, 42–43, 48, 64, 68, 70–71, 73]). In particular, no study to our knowledge has quantitatively evaluated the diversity, distributions, host affiliations, or tissue preferences of root- and shoot endophytes (hereafter, endophytes, sensu [7]) in freshwater plants.
Endophytes live within healthy above- and belowground plant tissues without causing apparent symptoms of disease [7]. In most dicotyledonous plants they are highly diverse, horizontally transmitted, and dominated by Ascomycota [5–7, 10, 33]. Their interactions with hosts can range from defensive mutualism and enhancement of stress tolerance to latent pathogenicity [7, 18, 61]. Endophytes frequently produce diverse secondary metabolites, many of which are important in industry and medicine [30, 67, 72, 77, 90]. In general, however, little is known about the geographic and ecological distributions of endophytes in most plant communities.
Aquatic plants represent an excellent opportunity to address broad questions in endophyte biology. Freshwater plants often inhabit proximate but distinct bodies of water in well-defined watersheds, providing an opportunity to examine geographic structure in a spatially bounded and hierarchical fashion. Because phylogenetically diverse vascular plants in aquatic systems display strong evolutionary convergence in key structural and morphological traits (i.e., thin cuticles, frequently open stomata, aerenchymatous tissues, and often specialized roots [42, 47, 84, 86]), they provide a special opportunity to contrast the role of structural elements vs. host taxonomy in structuring endophyte communities. Because the same individuals may have tissues that are both submerged and exposed to air, they present an opportunity to carefully examine the importance of the surrounding environment in shaping endophyte assemblages. Finally, comparisons with terrestrial plants provide an opportunity to determine whether – like their aquatic plant hosts – these symbionts represent diverse lineages that have colonized water independently, or are instead incidental and potentially transient components of aquatic systems.
Here, we evaluate the diversity, host affiliations, and geographic distributions of endophytes associated with freshwater macrophytes, with a special focus on the most common species of submergent and emergent plants inhabiting lentic waters in northern Arizona, USA. In addition to quantifying the basic ecological traits of a diverse community of symbiotic fungi over two sampling periods, we tested eight main predictions. Because the plants of interest in this study are strictly aquatic and do not occur on land we anticipated marked differences in endophyte communities among lakes (prediction 1), consistent with studies of geographically separated terrestrial plant communities [e.g., 45]. We predicted greater similarity in endophyte communities among lakes within vs. between watersheds (prediction 2), reflecting geographic proximity, shared waters, and similar abiotic features. In parallel with the specialization and convergence of tissue-specific structural features in diverse aquatic plants, we anticipated that their endophytes would show little host specificity (prediction 3), and instead would differ as a function of tissue type (prediction 4). Given the marked difference in environmental conditions for tissues growing in air vs. water, we predicted that endophyte communities would differ significantly between emergent and submerged photosynthetic tissues (prediction 5). Much like their hosts, we anticipated that endophytes of aquatic plants would represent subsets of otherwise terrestrial taxa that have colonized water independently (prediction 6). Finally, we predicted that aquatic endophytes would be largely distinct from communities of endophytes in proximate terrestrial plants (prediction 7), and that they would be readily recovered from lake sediment and water (prediction 8), congruent with the horizontal transmission that characterizes the vast majority of endophyte associations [7].
Materials and Methods
Aquatic macrophytes were collected from each of three microsites along the shore of one natural freshwater lake (Stoneman Lake) and five freshwater reservoirs (Lower Lake Mary, Morton Lake, Mud Lake, Watson Lake, and Willow Creek Reservoir) in northern Arizona, USA, in September-October 2011, and three reservoirs in early June 2012 (Lower Lake Mary, Watson Lake, Willow Creek Reservoir) (Table 1). Microsites were spaced at ca. 30–40 m intervals along accessible shorelines. Average water depth at each sampling site was 33.5 cm in 2011 (range: 4.4–88.4 cm) and 29.0 cm in 2012 (range: 9.4–64.0 cm) (Table 1). Interlake distances ranged from 2 km to 110 km. Water temperatures in Stoneman Lake, Lower Lake Mary, Morton Lake, and Mud Lake (Little Colorado River Watershed, Walnut Creek and Canyon Diablo complexes) typically are cooler than those of Watson Lake and Willow Creek Reservoir (Verde River Watershed) [4].
TABLE 1.
Locations and characteristics of study sites, and summary of collection data for surveys of endophytic fungi in aquatic plants in northern Arizona, USA. Asterisk denotes one natural lake (Stoneman Lake); all others are reservoirs. Latitude, longitude, and elevation indicate central sampling location among three microsites per reservoir. Minor changes between years reflect changes in shorelines due to fluctuation in water levels. Surface area data are published values ([4, 53]; marked with superscript 1), but fluctuate annually (ranges included in table). Letters used as superscripts indicate that the lake or reservoir was sampled only in 2011 (a), or that a particular host species was collected only in that year (b).
| Lake or Reservoir | County | Area1 (ha) | Elevation (m) | Latitude (2011; 2012) | Longitude (2011; 2012) | Depth (cm) (2011; 2012) | Plant species |
|---|---|---|---|---|---|---|---|
| Morton Lakea | Coconino | 4.1 – 11.4 | 2148 | N 34° 53.577′ | W 111° 17.784′ | 46.6 | P. amphibia; E. bifoliata |
| Mud Lakea | Coconino | 2.8 | 2180 | N 34° 55.530′ | W 111° 21.038′ | 66.7 | P. amphibia, S. pectinata, E. bifoliata |
| Stoneman Lake*a | Coconino | 48.6 | 2045 | N 34° 46.873′ | W 111° 31.227′ | 5.1 | P. amphibia, S. pectinata, E. bifoliata, M. sibiricum |
| Lower Lake Mary | Coconino | 0 – 303.5 | 2068 | N 35° 06.611′; N 35° 06.713′ | W 111° 34.779′ ′; W 111° 34.906′ | 9.2; 16.2 | P. amphibia, S. pectinata, E. bifoliatab |
| Willow Creek Reservoir | Yavapai | 138.4 | 1564 | N 34° 36.126′; N 34° 36.124′ | W 112° 26.319′; W 112° 26.277′ | 42.8; 21.6 | P. amphibia, S. pectinata, E. bifoliatab |
| Watson Lake | Yavapai | 80.9 | 1585 | N 34° 35.125′; N 34° 35.143′ | W 112° 25.374′; W 112° 25.367′ | 30.8; 49.3 | P. amphibia, S. pectinata |
Of the three reservoirs sampled in both years (Lower Lake Mary, first filled in 1905 [4]; Willow Creek Reservoir, first filled in 1936 [4]; and Watson Lake, first filled in 1915 [4]), Lower Lake Mary varies most in water volume: it often dries completely during the summer dry season (Table 1). Both Willow Creek Reservoir and Watson Lake maintain some water volume each year, but shorelines fluctuate according to rainfall and snowmelt. All are stocked regularly with fish [4].
Plant collections
We focused our sampling on three of the most abundant, native species of freshwater plants in the study region (see [27, 60]) (Table 1): Persicaria amphibia (L.) A. Gray (syn. Polygonum amphibium L.; Polygonaceae; emergent), Stuckenia pectinata (L.) Böerner (syn. Potamogeton pectinatus L.; Potamogetonaceae; submergent), and Elodea bifoliata H. St. John (Hydrocharitaceae; submergent). We also examined an additional species common to Arizona [see 27, 60] when it was encountered in our study sites (Myriophyllum sibiricum Komarov; Haloragaceae; submergent). All four species were sampled in 2011 (Table 1). Sampling in 2012 focused only on P. amphibia and S. pectinata, which were especially abundant at all sites (Table 1).
Submergent plants were fully uprooted and partitioned into shoots (photosynthetic stems and leaves) and roots, which were placed immediately into sealable bags with a small amount of water from the collection site. Emergent plants were removed in pieces based on tissue type (roots, submerged stems and leaves, and emergent stems and leaves) and were stored with water (submerged tissues) or without water (emergent tissues) in sealable plastic bags. All plant material was transported to the lab in a cooler for processing within 72 hours of collection. Vouchers of all plants were deposited at the University of Arizona Herbarium (ARIZ: collections DCS001-DCS022, accessions 407794-407797, 411472-411475, 411477-411486, and 411928-411930).
Tissue processing
Samples of each tissue type were rinsed for 30 seconds in running tap water, dried gently with paper towels, and cut into 2 mm2 segments. Segments were then surface-sterilized by sequential immersion in 95% ethanol (10 seconds), 10% Clorox (0.5% sodium hypochlorite; 2 minutes), and 70% ethanol (2 minutes) [8]. After surface-drying under sterile conditions, segments were placed individually into 1.5 mL microcentrifuge tubes containing ca. 0.7 mL of 2% malt extract agar (MEA) (48 pieces/tissue type/species/microsite in 2011; 96 pieces/tissue type/species/microsite in 2012). Isolates were archived at the Robert L. Gilbertson Mycological Herbarium (DM0001-DM0242).
Molecular methods
Most isolates lacked reproductive structures in pure culture and could not be identified beyond the level of phylum based on morphology. Therefore, total genomic DNA was extracted directly from fresh mycelium of each isolate using a phenol:chloroform method [9] or a modified protocol from the Extract-N-Amp tissue PCR kit (Sigma-Aldrich, St. Louis, MO).
We PCR-amplified and sequenced the 600–800 basepair nuclear ribosomal internal transcribed spacers and 5.8s gene (ITS rDNA), and the first 400–600 base pairs of the adjacent portion of the nuclear ribosomal large subunit (LSU rDNA), as a single fragment using primers ITS1F or ITS5 and LR3 [28, 82, 85]. Together these loci span fast- and slow-evolving regions that are widely used in fungal systematics [80]. PCR mixtures for samples extracted using phenol:chloroform consisted of 12.5 μL Sigma REDTaq (Sigma-Aldrich), 8.5 μL PCR-quality water, 1 μL of each primer (10 M), 1 μL dimethyl sulfoxide (DMSO), and 1 μL DNA template. For samples treated with Extract-N-Amp, 20 μL PCR mixtures consisted of 10 μL Extract-N-Amp, 4.4 μL PCR-quality water, 0.8 μL of each primer, and 4 μL DNA template (Sigma Aldrich Extract-N-Amp PCR Tissue Protocol). PCR cycling followed ref. [35]. Products were evaluated by electrophoresis on 1% agarose gels with SYBR Green I (Molecular Probes, Invitrogen; Carlsbad, CA). Positive amplicons were cleaned, quantified, normalized and sequenced bidirectionally at the University of Arizona Genetics Core facility (UAGC) using an Applied Biosystems 3730xl DNA Analyzer (Foster City, CA).
The software programs phred and phrap [21–22] were used to call bases and assemble bidirectional reads into consensus sequences using the ChromaSeq package in Mesquite v. 1.06 [50–51]. All contigs were edited manually in Sequencher version 4.5 (Gene Codes, Ann Arbor, MI) to verify base calls. Edited sequences were submitted to GenBank (accessions KF673551 - KF67377).
ITS rDNA-partial LSU rDNA sequences were used to delimit operational taxonomic units (OTU) using 95% sequence similarity, which approximate species boundaries in several major groups of Ascomycota that are common as endophytes [79]. OTU were assembled in Sequencher v. 4.5 (Gene Codes, Ann Arbor, MI) with a minimum overlap of 40 bases [79]. Sequences also were assembled into groups at 100% and 99% sequence similarity; general conclusions did not differ when operational taxa were delimited at these stringent levels, but the number of singletons increased to the point of prohibiting community analyses (data not shown).
Species accumulation curves and bootstrap estimates of total richness were inferred in EstimateS v. 8.2.0 (http://viceroy.eeb.uconn.edu/EstimateS) using 50 randomizations of sample order. Diversity was measured by Fisher’s alpha, a parameter of the log series model that is robust to variation in sample sizes [see 10]. Hosts that yielded fewer than two isolates were included in whole-community diversity measures, but were not included in diversity assessments for individual host plant species. Fisher’s alpha values over 100, characteristic of exceptionally small sample sizes with high species richness, were excluded from statistical analyses.
Statistical analyses comparing isolation frequency, richness, and diversity were performed in JMP v. 10.0.0 (SAS Institute, Cary, NC). Similarity indices were calculated in EstimateS and in PAST v. 1.88 [31] after removing all singleton OTU. Analyses of similarity (ANOSIM; [19]) were conducted in PAST using nonsingleton OTU only. Results were visualized using hierarchical cluster analysis coupled with z-tests of mean branch-point similarities, and non-metric multidimensional scaling (NMDS), an ordination method that uses rank-order information in a dissimilarity matrix [29, 81]. ANOSIM uses distance measures to test the null hypothesis that there are no differences in species composition between two or more groups [19, 81, 83]. Significance was computed by 10,000 permutations of group membership.
Taxonomic and phylogenetic analyses
ITS rDNA-partial LSU rDNA data were used to estimate class-level taxonomy based on careful evaluation of the top 50 BLASTn matches in GenBank [1, 81]. Evaluation of the most common OTU groups indicated that the most commonly isolated genus was Penicillium (Trichocomaceae, Eurotiales, Eurotiomycetes, Ascomycota; at least 65 isolates representing multiple OTU). We inferred the phylogenetic relationships of endophytic Penicillium obtained here in the context of (1) 71 strains isolated on 2% MEA in thorough surveys of water and sediment at Lower Lake Mary, Watson Lake, and Willow Creek Reservoir in 2012, concurrent with endophyte surveys [63], (2) 62 strains isolated as endophytes from terrestrial plants in proximate riparian and montane communities [45, Arnold et al. unpubl. data]; and (3) the subset of published Penicillium sequences to which isolates had highest affinity in BLAST searches of GenBank (15 strains). An outgroup sequence representing Aspergillus was chosen based on the literature [37].
The resulting data set was trimmed to consistent start- and end points, and for maximum taxonomic richness was confined only to the ITS rDNA region (final length, 541 base pairs). Redundant sequences were removed and the resulting data set of 124 terminals was aligned using MUSCLE [89] with manual editing in MacClade [51]. The alignment was analyzed using maximum likelihood analyses in GARLI [91] and Bayesian analyses in MrBayes 3.1.2 [38], with the GTR+I+G model of evolution implemented in each case based on analysis in jModelTest [58]. The Bayesian analysis consisted of 3 million generations, sampling every 1000th tree, with four chains and a random starting tree. Completion was assessed by examining –ln li values and the standard deviation of split frequencies. A majority rule consensus tree was inferred from the posterior, with support given by Bayesian posterior probability values. Complementary support values were provided by 100 maximum likelihood bootstrap replicates conducted as above in GARLI.
Results
Surveys of common aquatic plants in six lakes and reservoirs in Arizona yielded a total of 226 isolates of endophytic fungi from 9,600 plant tissue segments (2011, 112 isolates from 5,280 segments; 2012, 114 isolates from 4,320 segments). The overall isolation frequency (i.e., percent of tissue segments bearing culturable fungi) was 2.4% and did not differ significantly as a function of collection period, reservoir, host species, tissue type, or water depth at the sampling point (Table 2; Table S1; Table S2).
TABLE 2.
Statistical analyses reveal that isolation frequency and diversity of endophytes from aquatic macrophytes did not differ significantly as a function of sampling period, reservoir (Lower Lake Mary, Watson Lake, Willow Creek Reservoir), host species (Persicaria amphibia, Stuckenia pectinata), tissue type (roots vs. submerged photosynthetic tissues), or water depth at the sampling site. Only reservoirs and hosts sampled in both years were considered. Because no interannual differences in isolation frequency or diversity were observed, data were combined for both years prior to analysis by reservoir. Thereafter, data were iteratively combined in the order shown below for each subsequent analysis. No interaction terms were significant. Asterisks indicate moderately supported but nonsignificant trends suggesting (a) higher isolation frequency in shallower water, and (b) higher diversity in roots vs. submerged photosynthetic tissues. Survey data and means and standard deviations following appropriate transformations for each analysis are shown in Table S1 and Table S2, respectively.
| Explanatory variable | Isolation frequency | Diversity |
|---|---|---|
| Sampling period | t = −0.39, p = 0.0755 | t = −0.49, p = 0.6435 |
| Reservoir | F = 0.29, p = 0.7477 | F = 0.43, p = 0.6654 |
| Host species | t = −0.08, p = 0.9399 | t = −1.62, p = 0.1441 |
| Tissue | t = −0.47, p = 0.6386 | t = −2.46, p = 0.0577* |
| Water depth | R2 = 0.08, p = 0.0609* | R2 = 0.01, p = 0.7321 |
High-quality sequence data from 225 of 226 isolates yielded 60 OTU based on 95% ITS rDNA-partial LSU rDNA similarity (Fisher’s alpha = 27.8) and 107 genotypes based on 100% similarity (Fisher’s alpha = 83.2). More than half of the 60 OTU (53.3%) were singletons (i.e., found only once). Diversity of endophytes (Fisher’s alpha) did not differ significantly as a function of collection period, reservoir, host species, tissue type, or water depth at the sampling point (Table 2; Table S1; Table S2).
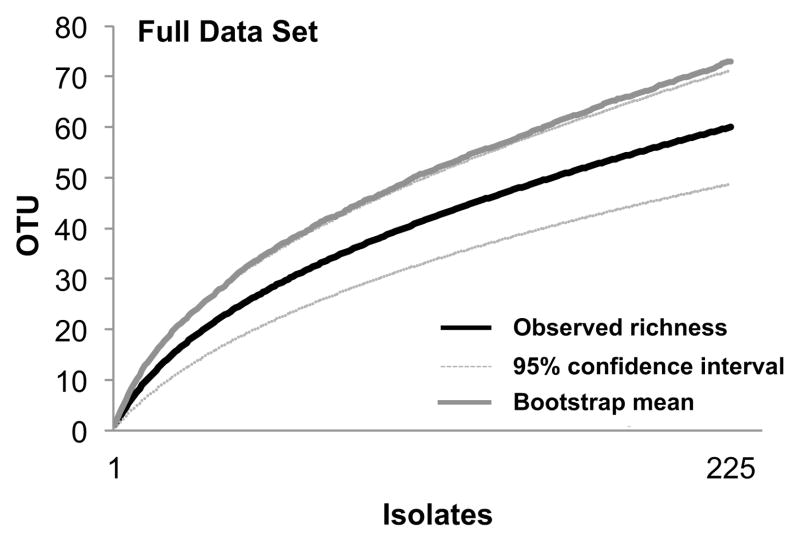
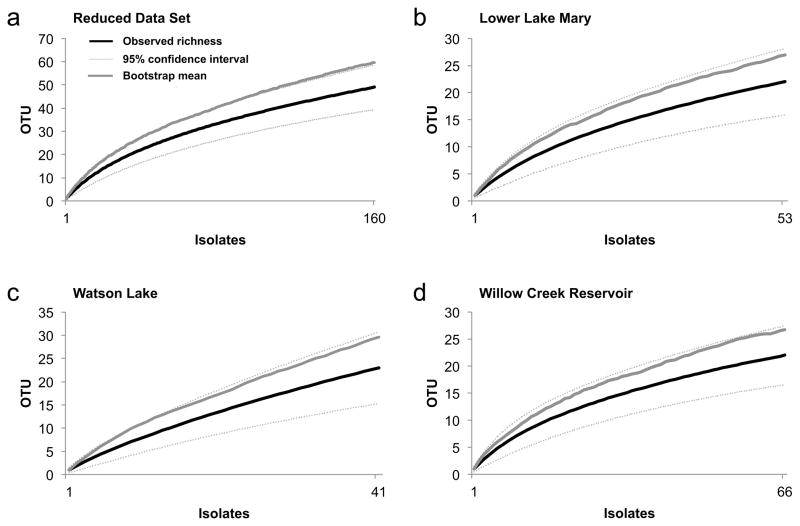
Species accumulation curves for the full data set (one lake and five reservoirs, and all focal species across the two collection periods; Figure 1) and the reduced data set (three reservoirs and two focal species across the two collection periods; Figure 2a) approached statistical completion. Bootstrap estimates of total richness for the two data sets suggested that approximately 80% of expected species richness was found by our surveys (Figures 1 and 2a). Sampling in each reservoir in the reduced data set was statistically complete or nearly complete (Lower Lake Mary = 81.5% of expected richness, Watson Lake = 77.7%, Willow Creek Reservoir = 82.4%; Figures 2b–d), providing a robust basis for the community analyses described below.
Fig. 1.
Species accumulation curve for the full data set: culturable endophytic fungi obtained from photosynthetic tissues and roots of four species of aquatic plants in one lake and five reservoirs in northern Arizona, USA. Figure shows the number of endophyte species (OTU) observed (Mao Tau; black lines), lower and upper 95% confidence intervals (light gray lines), and bootstrap estimate of richness (dark gray lines).
Fig. 2.
Species accumulation curve for (a) the reduced data set (only those species and reservoirs sampled in two collection periods: Persicaria amphibia and Stuckenia pectinata; Lower Lake Mary, Watson Lake, and Willow Creek Reservoir; n = 160 isolates), and each reservoir in the reduced data set analyzed independently: (b) Lower Lake Mary (n = 53 isolates), (c) Watson Lake (n = 41 isolates), and (d) Willow Creek Reservoir (n = 66 isolates). Figures show the number of species (OTU) observed (Mao Tau; black lines), lower and upper 95% confidence intervals (light gray lines), and bootstrap estimate of richness (dark gray lines).
Community composition
ANOSIM indicated that endophyte communities did not differ significantly in composition between the two collection periods (Figure 3a), such that data for the two collections were combined for subsequent analyses.
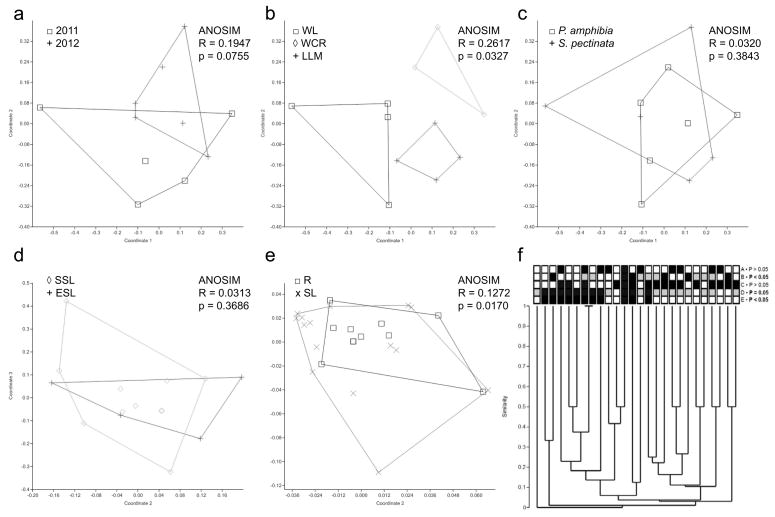
Fig. 3.
Community analyses of fungal endophyte communities. Panels indicate results of non-metric multidimensional scaling using Jaccard’s index, and relevant ANOSIM results (panels a–e), and results of cluster analyses (panel f). Only taxa and reservoirs sampled in both collection periods are included, and all singleton OTU were excluded. Endophyte communities did not differ significantly as a function of collection period (a), but did differ significantly among reservoirs (b) (WL, Watson Lake; WCR, Willow Creek Reservoir; LLM, Lower Lake Mary). Communities did not differ significantly between host species (c), nor between emergent (ESL) and submerged (SSL) photosynthetic tissues (d). However, communities differed significantly overall between roots (R) and shoots (SL) (panel e). Panel (f) summarizes these results in a hierarchical manner through cluster analysis with terminals coded as follows: row A, sampling year (black, 2011; white, 2012); row B, reservoir (black, WCR; grey, WL; white, LLM); row C, host species (black, Persicaria; white, Stuckenia); row D, tissue type (black, roots; grey, submerged photosynthetic tissue; white, emergent photosynthetic tissue); row E, roots (black) vs. photosynthetic tissue (white). P-values for each row indicate results of z-test evaluating relative similarity of terminals in the same category (e.g., same year, same reservoir, same host species, same tissue type) vs. different categories. Significant values (P<0.05) reveal strong effects of tissue type and reservoir on endophyte community structure, complementing NMDS and ANOSIM results (panels b and e).
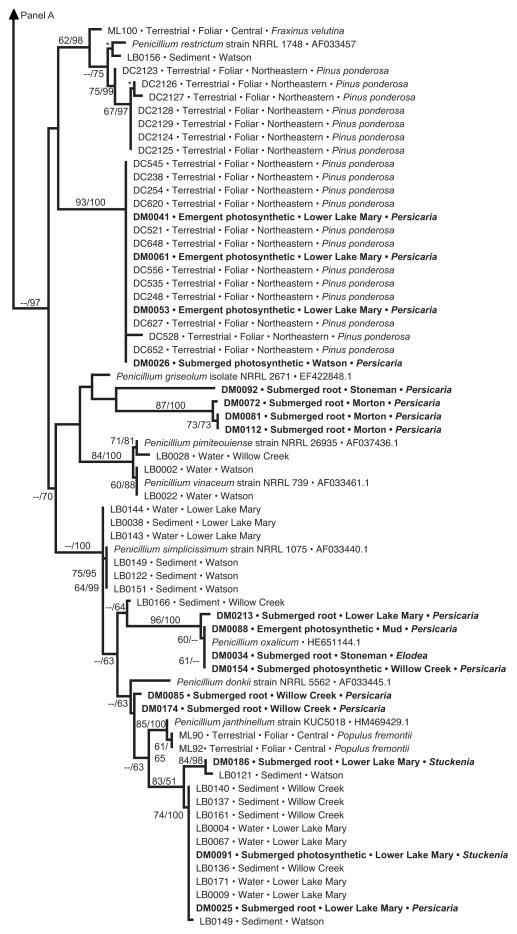
Consistent with prediction 1, we found strong evidence for differences in endophyte community structure among reservoirs (Figure 3b). In contrast to prediction 2, however, we did not find evidence for greater similarity among communities within vs. between watersheds: Watson Lake and Willow Creek Reservoir, both part of the Verde River watershed and separated by only 2 km, had significantly different endophyte communities (Figure 3b). Phylogenetic analyses of Penicillium did not strongly corroborate community-level inferences regarding differences among reservoirs (Figure 4), suggesting that Penicillium species may be particularly widespread at a regional scale, and that differences among reservoirs reflect the presence/absence of other taxa.
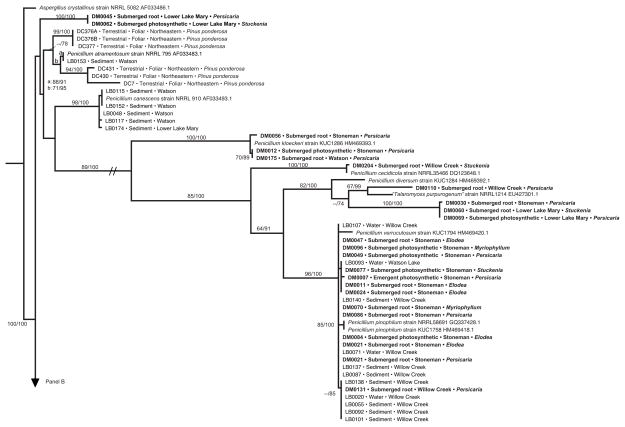
Fig. 4.
Phylogenetic analysis of Penicillium endophytes of aquatic plants, spread over two panels (a, b). Topology reflects the most likely tree based on analyses of ITS rDNA, with support values indicating maximum likelihood bootstrap (before slash; ≥60% shown) and Bayesian posterior probabilities (after slash; ≥60% shown). Strains in bold were obtained from aquatic plants in this study; terminals indicate isolate number (cf. Table S2), tissue type, lake or reservoir, and genus of the host species. These are placed into the larger context of (1) closely related species of Penicillium obtained from GenBank; (2) endophytes from terrestrial plants in north-central (‘central’) or northeastern Arizona, with terminal labels indicating isolate number, status as foliar endophytes of terrestrial plants, and host species [45 and Arnold et al., unpublished data]; and (3) samples of Pencillium spp. collected directly from water or sediment of the same reservoirs, with terminal labels indicating isolate number, substrate, and site [63]. One terminal is labeled ‘Talaromyces purpurogenus’; quotation marks are used because the species is considered equivalent to Pencillium purpurogenum, but is labeled as Talaromyces in GenBank. Hatch marks (panel a) indicate long branch shortened 25% for illustration purposes. Asterisks indicate high support in cases in which too little space was available to depict support values
Consistent with prediction 3, we found no evidence for host specificity among the aquatic-plant endophytes considered here: endophyte assemblages in members of two families of aquatic plants did not differ significantly (Figure 3c). This result was further corroborated by analyses within a focal genus (Pencillium), which revealed that relatively few clades showed clear structure as a function of host taxonomy (Figure 4).
As expected (prediction 4), endophyte assemblages differed as a function of tissue type, but not as a function of the air- vs. water environment (prediction 5). Communities did not differ significantly between emergent and submerged photosynthetic tissues (Figure 3d), such that these data were combined as ‘shoot-associated endophytes’ and contrasted with endophyte communities in roots. In turn, root- and shoot communities differed significantly (Figure 3e). Phylogenetic analyses of Penicillium do not strongly corroborate this result (Figure 4), suggesting tissue-generalism in that genus and indicating that root- and shoot communities likely differ on the basis of other taxa. Together these findings were placed into a hierarchical context through cluster analysis (Figure 3f), which summarizes the significant association metrics for endophyte communities with regard to geographical structure (row B: Lower Lake Mary, Watson Lake, Willow Creek Reservoir) and tissue type (row E: root vs. shoot).
Taxonomic and phylogenetic analyses
Endophytes of these aquatic plants were phylogenetically diverse. The majority were members of the Pezizomycotina (Ascomycota; n = 219 sequences with defined taxonomy; Table 3; Table S3). Pezizomycotina isolates spanned five classes and at least 13 orders, 19 families, and 37 genera (Table 3). The remaining six isolates consisted of one isolate of Basidiomycota (Agaricomycetes) with BLAST affinity for Ceratobasidium, and five Mucoromycotina (Mucorales) representing 1 OTU with BLAST affinity for Rhizopus.
TABLE 3.
Taxonomic composition of endophytes representing the Pezizomycotina (Ascomycota) (219 isolates; Table S3). The data shown here include two isolates that were not identifiable to the class level (incertae sedis) and are referred to as “mitosporic Ascomycota” with BLAST affinity for Ochrocornis, a genus of uncertain placement. Two Pezizomycetes isolates were from an unknown family and genus.
| Class | Total Isolates (%) | Root Isolates (%) | Shoot Isolates (%) | Families | Genera | Total OTU (%) | Root OTU (%) | Shoot OTU (%) |
|---|---|---|---|---|---|---|---|---|
| Eurotiomycetes | 90 (41.1%) | 50 (40.7%) | 40 (41.7%) | 3 | 7 | 19 (32.8%) | 13 (35.1%) | 14 (37.8%) |
| Dothideomycetes | 82 (37.5%) | 42 (34.1%) | 40 (41.7%) | 8 | 16 | 20 (34.5%) | 12 (32.4%) | 12 (32.4%) |
| Sordariomycetes | 23 (10.5%) | 13 (10.6%) | 10 (10.4%) | 7 | 9 | 12 (20.7%) | 6 (16.2%) | 8 (21.6%) |
| Leotiomycetes | 20 (9.1%) | 16 (13.0%) | 4 (4.2%) | 1 | 4 | 5 (8.6%) | 5 (13.5%) | 2 (5.4%) |
| Pezizomycetes | 2 (0.9%) | 0 (0%) | 2 (2.1%) | Unk | Unk | 1 (1.7%) | N/A | 1 (2.7%) |
| Incertae sedis | 2 (0.9%) | 2 (1.6%) | 0 (0%) | N/A | 1 | 1 (1.7%) | 1 (2.7%) | N/A |
Overall, the taxonomic groups recovered here are known primarily from terrestrial systems, consistent with prediction 6. Major taxonomic groups were found with similar frequency in root vs. shoot tissues (except Leotiomycetes), suggesting that tissue preferences reflect differences at lower taxonomic levels than that of fungal class (Table 3).
Phylogenetic analyses of the most common genus recovered here (Penicillium) corroborated OTU designations (Figure 4, Supplementary Table 1); reveal the phylogenetic diversity of the OTU recovered from that genus; highlight the novelty of several clades relative to known and sequenced species of Penicillium available through GenBank; and highlight the relationships of endophytes from aquatic plants to fungi from water- and sediment samples of the same water bodies, and to fungi from proximate terrestrial plants (Figure 4). The topology provided only mixed support with regard to our final two predictions. Some endophytes from aquatic plants clustered with strong support in clades consisting primarily of terrestrial endophytes, whereas others were distinct from those in terrestrial systems (prediction 7). Similarly, some aquatic endophytes were nested within clades containing water-borne and sediment fungi, consistent with prediction 8 – but others did not show clear affinities to these communities. There was no preferential association between endophytes of roots and fungi in sediment, suggesting mixing of these fungi among substrates within each body of water. Similarity of some strains from aquatic plants with endophytes in terrestrial plants in northeastern Arizona (e.g., DM0041, DM0061, DM0053, and DM0026; Figure 4b), and the presence of particular clades across water bodies in multiple watersheds (Figure 4a, b) suggest that some of Penicillium clades are widespread at a regional scale.
Discussion
Ecological associations between fungi and terrestrial plants have received substantive attention over the last four decades (e.g., [2, 5–6, 9–10, 17–18, 23, 26, 33, 35, 39, 56, 61, 65–66, 74–76, 80–81]). In contrast, community-level sampling of fungi from marine, brackish, and fresh waters have been relatively neglected (reviewed in [44, 69, 87–88]; see also [42]). In part due to increasing recognition of the frailty and importance of aquatic ecosystems and the plants that inhabit them, recent studies have begun to focus on fungi from plants in diverse aquatic environments (e.g., [11–12, 20, 24, 40–43, 48, 59, 64, 68, 70–71, 73]). However, relatively few studies have adopted a replicated or quantitative approach to community sampling in the context of regional-scale sampling.
We examined the abundance, diversity, geographic distributions, and taxonomic composition of endophytic symbionts from the roots and photosynthetic tissues of locally abundant aquatic plants in lentic waters in northern Arizona. Using both surveys of endophyte communities (Figures 1–3) and phylogenetic analyses of the most prevalent genus (Penicillium, Figure 4), we tested eight predictions. Overall, our results reveal a low isolation frequency but high species- and phylogenetic diversity of endophytes in these aquatic systems, with strong evidence for structure at the community level as a function of tissue type and water body. We found no evidence for interannual variation, or differences in communities immediately above and below the air/water interface. Endophytes from these aquatic plants included genotypes known from proximate terrestrial plants, and others that were represented in water and sediment samples. Our results suggest that tissue specificity reflects distributions of taxa below the class level; that tissue- and geographic structure is not consistently evident in the most common genus (Penicillium) and instead reflects differences in the occurrence of other taxa; and that several Penicillium clades appear to have wide distributions over the geographic areas studied here and in related studies [45, 63, Arnold, unpubl. data]. Overall, the results presented here not only inform our understanding of endophytic symbioses in the diverse environments in which plants occur, but also suggest strategies optimizing strategies to efficiently capture distinctive endophytes for studies of fungal biodiversity.
Isolation frequency
The isolation frequency we observed (ca. 2.4% of tissue segments yielding a fungus in culture) was low relative to that observed in previous studies of aquatic plants. Kohout et al. [43] recorded an isolation frequency of 8.8% from roots of five submergent isoetids from freshwater oligotrophic lakes in southern and central Norway, and Li et al. [48] reported an isolation frequency of 42.8% from stem and leaf tissues from five riparian plants in China (three submergent; two in the vicinity of a stream with stem pieces in water). In contrast to our study, both focused on riparian areas that were not man-made and were located in regions with a high density of natural riparian systems.
The isolation frequency we observed also was lower than in many terrestrial plants, consistent with Li et al. [48], who found a lower isolation frequency in truly aquatic samples (18–30%) relative to semi- and non-aquatic tissues in proximity to water (41–63%). Isolation frequency from leaves of terrestrial riparian species such as Fraxinus velutina, Quercus emoryi, and Populus fremontii in riparian areas of north-central Arizona ranged from 0–16% in a recent study, but were greater overall than those observed here (mean = 6.5%; [45]). Notably Lau et al. [45] showed that isolation frequency in terrestrial plants was positively associated with rainfall. Our study shows that immersion in water does not ensure a high frequency of infection with culturable fungi.
One important difference among studies of endophytes is the approach for surface sterilization (see [5, 23, 25, 43, 49, 66]). We used a method that was developed and tested with terrestrial plants [8]. When photosynthetic tissues of aquatic plants were placed in 10% bleach, they began to lose their pigment after ca. 30 seconds. As observed by Kandalepas [42], such tissue pieces typically turned yellow or light brown, raising the possibility that our sterilization process may have infiltrated and damaged endophytes in these delicate structures. Similar concerns were raised by Kohout et al. [43], who suggested that bleach may infiltrate large root cavities. In that study, 100% household bleach was used (vs. 10% bleach in the present work). However, Li et al. [48] recovered a relatively high isolation frequency following surface-sterilization by an approach similar to ours.
Notably, both Li et al. [48] and Kohout et al. [43] used a different medium (a modified recipe based on potato dextrose agar), suggesting its use in future studies. Different media can lead to differences in isolation frequency and apparent diversity of fungi (Sandberg and Arnold, in prep) and should be explored further for studies in aquatic systems. Notably, 2% MEA is mildly acidic, but Willow Creek Reservoir, Watson Lake, and Lower Lake Mary are slightly alkaline (www.azdeq.gov, accessed 4/2013; Sandberg, pers. obs.; [3, 78]). Here, 2% malt extract agar was used to enhance comparability with endophytes of terrestrial plants in the same biogeographic region ([45, 81]).
Diversity
In contrast to isolation frequency, endophytes from aquatic plants in northern Arizona are highly diverse. The diversity of fungi observed here (Fisher’s alpha = 27.8) and the prevalence of singleton OTU (53.3%) resemble Fisher’s alpha values of ‘hyperdiverse’ endophytes in species-rich systems such as tropical forests (e.g., Fisher’s alpha = 30.9 with 51.6% singletons, [9]; Fisher’s alpha = 25.9 with 62.8% singletons, [34]). The diversity we discovered was much greater than that observed in roots of aquatic plants in Norway by Kohout et al. [43] (Fisher’s alpha = 3.9), and was greater than that observed in leaves from two freshwater plant species collected from Louisiana wetlands by Kandalepas [42] (Fisher’s alpha = 11.06).
Because this study spanned multiple reservoirs, some of which are quite distant from one another (>100 km) and host different fungal communities (see below), our overall diversity values are inflated by regional comparisons relative to studies conducted within single sites. Interestingly, Lau et al. [45] recorded a total Fisher’s alpha of 14.3 (ca. twofold less than in the present study) among endophytes of three species of terrestrial plants in six riparian areas sampled at a similar spatial scale in northern Arizona. In our work, the overall diversity per host species in each lake or reservoir was 10.2 ± 6.5 (range per species, Fisher’s alpha = 0.9–20.3), which was 1.4 times greater than the values found by U’Ren et al. [81] in terrestrial plants from a similar bioclimatic region (Fisher’s alpha = 7.2 ± 6.7; range per species, Fisher’s alpha = 0.8–10.9). Importantly, each of those studies included only foliage; here, we sampled foliage, stems, and roots, and the distinctive communities in each tissue type thus increased overall diversity. When we examine only endophytes from photosynthetic tissues, diversity again exceeds values in terrestrial plants of the region (see [45, 81]). Thus in general, our study points to a high diversity of endophytes in aquatic plants despite a low isolation frequency.
Although our data reveal a high diversity of endophytes, it is likely that additional taxa could be recovered using culture-free methods. Kohout et al. [43] showed that a culture-independent approach yielded a higher diversity of fungi in Norwegian aquatic plants (culture-free method, Fisher’s alpha = 8.5) than did a culture-based approach with the same plants (Fisher’s alpha = 3.9). Similarly, Neubert et al. [54] found a high diversity of endo- and ectophytic fungi (>600 OTU) associated with all tissue types of the opportunistically aquatic plant Phragmites australis in dry and flooded sites in Lake Constance, Germany. Kandalepas et al. [41] observed a high frequency of root colonization by AMF and DSE in wetland plants in Louisiana, suggesting that unculturable fungi may be common in aquatic plant tissues more generally. Together, these and studies of terrestrial plants argue strongly for using culture-independent methods to evaluate endophyte diversity in future work.
Taxonomic composition of communities
We recovered a very high degree of phylogenetic richness among endophytes of focal aquatic plants, with members of at least 37 genera, 19 families, 13 orders, seven classes, and three phyla present among 225 sequenced isolates. Clearly a high diversity of lineages and species are capable of forming endophytic symbioses in these freshwater systems.
At higher taxonomic levels, fungal assemblages in these aquatic plants were distinctive relative to those in terrestrial plants of the region, but resembled other surveys of aquatic macrophytes. Terrestrial angiosperms in the region typically harbor endophyte communities that are dominated by Sordariomycetes, Dothideomycetes, and Pezizomycetes, with Leotiomycetes also very common in some conifers (see [35–36, 45, 80–81]). Our results resemble those of Li et al. [48], who found high numbers of Eurotiomycetes and Dothideomycetes in aquatic macrophytes (especially Cladosporium, Penicillium, Alternaria, and Aspergillus). Many of these fungi are highly cosmopolitan, occurring opportunistically in many different terrestrial and aquatic environments [e.g., 87]. In contrast, Kandalepas [42] observed a high number of Sordariomycetes and low numbers of Dothideomycetes and Eurotiomycetes in wetland plants in Louisiana. Although not all of the isolates were identified in that study, taxa such as Cladosporium, Penicillium, Alternaria, and Aspergillus were common [42]. In general, a review of the available evidence suggests that culture-based surveys of plants in lentic waters may yield many previously known fungal genera from terrestrial systems, which are represented by distinctive species or genotypes in aquatic plants. Notably we found no evidence for morphology or sequence data resembling Ingoldian or aeroaquatic fungi (Table S3), nor the distinctive fungi described from submerged wood (e.g., [24, 59]), suggesting that endophytes of aquatic plants may be an important but previously overlooked group of fungi in freshwater systems.
In addition to differences at higher taxonomic levels, the presence of distinctive communities of endophytes in aquatic vs. terrestrial plants would argue for sampling aquatic systems for adequately capturing regional fungal diversity. Aquatic plants differ from terrestrial plants not only in their habit of growing in water, but also in a suite of morphological characteristics: thinner cuticles, feathery roots, mucilaginous surfaces, the presence of aerenchyma, and frequently open stomata [84, 86]. Our results suggest that communities differ in taxa other than Penicillium (Figure 4) and that such differences may only be detectable in broad community surveys. To further test the prediction that endophytes of aquatic plants differ from those in proximate terrestrial species, we compared the full data set obtained in the present study (225 isolates) with those found in terrestrial plants in northern Arizona riparian zones (111 isolates; [45]). Species-accumulation curves in both studies approached saturation, suggesting that each study captured the majority of available species richness. Using 95% sequence similarity, the overall pool of 336 isolates represented 93 OTU (Fisher’s alpha = 43.7), of which 56 were found only once (60.2%). Among the 37 OTU found more than once, 33 were found in only aquatic or only terrestrial plants (89.2%), whereas only four were found in both aquatic and terrestrial plants. Thus aquatic plants appear to represent an important complement to surveys of terrestrial plants in studying endophyte biodiversity.
Together these analyses provide a first quantitative estimation of endophytic fungal distributions in the aquatic plants and lentic waters of the southwestern USA. Our work reveals that despite a low isolation frequency, endophytes associated with roots and photosynthetic tissues of aquatic plants in northern Arizona are highly diverse and distinctive at low- and high taxonomic levels relative to those in proximate terrestrial communities. These data suggest that efficient biodiversity- or bioprospecting surveys seeking species- or genotype-level diversity could be achieved by including aquatic plants in regional surveys, and by examining multiple lakes/reservoirs and tissue types, with less emphasis on multiple aquatic plant taxa, sampling depths, or sampling years. More generally, our study provides a basis for evaluating general trends in endophyte biology that have been based mostly on plants in terrestrial ecosystems, and uncovers the compelling features of fungal communities in aquatic systems.
Supplementary Material
Acknowledgments
We thank the School of Plant Sciences and the College of Agriculture and Life Sciences at The University of Arizona for supporting this work. DCS was supported in part by a Pierson Fellowship through the Plant Pathology graduate major at The University of Arizona. Additional support was provided by the National Institutes of Health (R01 to A.A.L. Gunatilaka and AEA) and the National Science Foundation (NSF DEB-1045766 to AEA). We thank Kayla Arendt, Mariana del Olmo-Ruiz, Nicholas Massimo, Jakob Riddle, Justin Shaffer, and especially Jana U’Ren for lab assistance and helpful discussion; and Lauren Dominick, Chan Jung, Thaddeus Metz, Jamie Moy, Brittany Peña, Ethan Posey, Adrian Ramirez, Cole Steen, and Brittany Wohl for assistance in the field. We are especially grateful to Anthony Robinson and the Arizona Game and Fish Department, Jacob Butler, Kevin Fitzsimmons, and William Matter for helpful discussion and sharing their knowledge regarding limnology and aquatic biology, Marc J. Orbach and Barry M. Pryor for helpful guidance, and two anonymous reviewers for improving the manuscript. This paper represents a portion of the MS research of DCS in the Plant Pathology major within the School of Plant Sciences at The University of Arizona.
References
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arenal F, Platas G, Pelaez F. A new endophytic species of Preussia (Sporormiaceae) inferred from morphological observations and molecular phylogenetic analysis. Fungal Divers. 2007;25:1–17. [Google Scholar]
- 3.Anonymous. Arizona Department of Environmental Quality. Lake Mary regional TMDL for mercury in fish tissue. Arizona Department of Environmental Quality open file report. 2010 http://www.azdeq.gov/environ/water/assessment/download/Lake_Mary_Region_Draft-6-16-2010.pdf.
- 4.Anonymous. U.S. Fish and Wildlife Service. Wildlife and Sport Fish Restoration Program. U.S. Fish and Wildlife Service; Albuquerque, NM: 2011. Biological assessment of the Arizona Game and Fish Department’s Statewide and Urban Fisheries Stocking Program for the years 2011–2012. [Google Scholar]
- 5.Arnold AE, Maynard Z, Gilbert GS, Coley PD, Kursar TA. Are tropical fungal endophytes hyperdiverse? Ecol Lett. 2000;3:267–274. [Google Scholar]
- 6.Arnold AE, Maynard Z, Gilbert G. Fungal endophytes in dicotyledonous neotropical trees: Patterns of abundance and diversity. Mycol Res. 2001;105:1502–1507. [Google Scholar]
- 7.Arnold AE. Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol Rev. 2007;21:51–66. [Google Scholar]
- 8.Arnold AE, Henk DA, Eells R, Lutzoni F, Vilgalys R. Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia. 2007;99:185–206. doi: 10.3852/mycologia.99.2.185. [DOI] [PubMed] [Google Scholar]
- 9.Arnold AE, Lutzoni F. Diversity and host range of foliar fungal endophytes: Are tropical leaves biodiversity hotspots? Ecology. 2007;88:541–549. doi: 10.1890/05-1459. [DOI] [PubMed] [Google Scholar]
- 10.Arnold AE, Miadlikowska J, Higgins KL, Sarvate SD, Gugger P, Way A, Hofstetter V, Kauff F, Lutzoni F. A phylogenetic estimation of trophic transition networks for ascomycetous fungi: Are lichens cradles of symbiotrophic fungal diversification? Syst Biol. 2009;58:283–297. doi: 10.1093/sysbio/syp001. [DOI] [PubMed] [Google Scholar]
- 11.Arya P, Sati SC. Evaluation of endophytic aquatic hyphomycetes for their antagonistic activity against pathogenic bacteria. Int Res J Microbiol. 2011;2:343–7. [Google Scholar]
- 12.Beck-Nielsen D, Madsen TV. Occurrence of vesicular arbuscular mycorrhiza in aquatic macrophytes from lakes and streams. Aquat Bot. 2001;71:141–148. [Google Scholar]
- 13.Blackwell M. The fungi: 1, 2, 3 … 5.1 million species? Am J Bot. 2011;98:426–38. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- 14.Borman S, Korth R, Temte J, Watkins C. Through the looking glass: A field guide to aquatic plants. Stevens Point, Wis: Wisconsin Lakes Partnership 1997 [Google Scholar]
- 15.Brix H, Schierup HH. The use of aquatic macrophytes in water-pollution control. Ambio. 1989;18:100–107. [Google Scholar]
- 16.Brönmark C, Hansson LA. The biology of lakes and ponds. Oxford: Oxford University Press; 2005. [Google Scholar]
- 17.Carroll GC, Carroll FE. Studies on the incidence of coniferous needle endophytes in the Pacific Northwest. Can J Bot. 1978;56:3034–3043. [Google Scholar]
- 18.Carroll G. Fungal endophytes in stems and leaves: From latent pathogen to mutualistic symbiont. Ecology. 1988;69:2–9. [Google Scholar]
- 19.Clarke KR. Non-parametric multivariate analysis of changes in community structure. Austral J Ecol. 1993;18:117–143. [Google Scholar]
- 20.de Marins JF, Carrenho R, Thomaz SM. Occurrence and coexistence of arbuscular mycorrhizal fungi and dark septate fungi in aquatic macrophytes in a tropical river flood plain system. Aquat Bot. 2009;91:13–19. [Google Scholar]
- 21.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 22.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 23.Faeth SH, Hammon KE. Fungal endophytes in oak trees: Experimental analyses of interactions with leafminers. Ecology. 1997;78:820–827. [Google Scholar]
- 24.Ferrer A, Miller AN, Sarmiento C, Shearer CA. Three new genera representing novel lineages of Sordariomycetidae (Sordariomycetes, Ascomycota) from tropical freshwater habitats in Costa Rica. Mycologia. 2012;104:865–879. doi: 10.3852/11-111. [DOI] [PubMed] [Google Scholar]
- 25.Fisher PJ, Anson AE, Petrini DO. Fungal endophytes in Ulex europaeus and Ulex gallii. T Brit Mycol Soc. 1986;86:153–156. [Google Scholar]
- 26.Fisher PJ. Survival and spread of the endophyte Stagonospora pteridiicola in Pteridium aquilinum, other ferns and some flowering plants. New Phytol. 1996;132:119–122. doi: 10.1111/j.1469-8137.1996.tb04516.x. [DOI] [PubMed] [Google Scholar]
- 27.Fulmer JE, Robinson AT. Aquatic plant species distributions and associations in Arizona’s reservoirs. J Aquat Plant Manage. 2008;46:100–106. [Google Scholar]
- 28.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes: Application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 29.Gauch HG., JR . Multivariate analysis in community structure. Cambridge University Press; Cambridge, UK: 1982. [Google Scholar]
- 30.Gunatilaka AAL. Natural products from plant-associated microorganisms: Distribution, structural diversity, bioactivity, and implications of their occurrence. J Nat Prod. 2006;69:509–526. doi: 10.1021/np058128n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammer Ø, Harper DAT, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:9. [Google Scholar]
- 32.Hawksworth DL. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol Res. 2001;105:1422–1432. [Google Scholar]
- 33.Higgins KL, Arnold AE, Miadlikowska J, Sarvate SD, Lutzoni F. Phylogenetic relationships, host affinity, and geographic structure of boreal and arctic endophytes from three major plant lineages. Mol Phylogenet Evol. 2007;42:543–555. doi: 10.1016/j.ympev.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 34.Higgins KL, Coley PD, Kursar TA, Arnold AE. Culturing and direct PCR suggest prevalent host generalism among diverse fungal endophytes of tropical forest grasses. Mycologia. 2011;103:247–260. doi: 10.3852/09-158. [DOI] [PubMed] [Google Scholar]
- 35.Hoffman M, Arnold AE. Geography and host identity interact to shape communities of endophytic fungi in cupressaceous trees. Mycol Res. 2008;112:331–344. doi: 10.1016/j.mycres.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 36.Hoffman M, Arnold AE. Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Apple Environ Microb. 2010;76:4063–4075. doi: 10.1128/AEM.02928-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Houbraken J, Samson RA. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol. 2011;70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 39.Janos DP. Mycorrhizae influence tropical succession. Biotropica. 1980;12:56–64. [Google Scholar]
- 40.Jayachandran K, Shetty KG. Growth response and phosphorus uptake by arbuscular mycorrhizae of wet prairie sawgrass. Aquat Bot. 2003;76:281–290. [Google Scholar]
- 41.Kandalepas D, Stevens KJ, Shaffer GP, Platt WJ. How abundant are root-colonizing fungi in southeastern Louisiana’s degraded marshes? Wetlands. 2010;30:189–199. [Google Scholar]
- 42.Kandalepas D. Dissertation. Louisiana State University; 2012. Effects of coastal dynamics on colonization of Louisiana wetland plants by fungal endophytes. [Google Scholar]
- 43.Surprising spectra of root-associated fungi in submerged aquatic plants. FEMS Microbiol Ecol. 2012;80:216–35. doi: 10.1111/j.1574-6941.2011.01291.x. [DOI] [PubMed] [Google Scholar]
- 44.Krauss G-J, Sole M, Krauss G, Schlosser D, Wesenberg D, Baerlocher F. Fungi in freshwaters: ecology, physiology and biochemical potential. FEMS Microbiol Rev. 2011;35:620–651. doi: 10.1111/j.1574-6976.2011.00266.x. [DOI] [PubMed] [Google Scholar]
- 45.Lau MK, Arnold AE, Johnson NC. Factors influencing communities of foliar fungal endophytes in riparian woody plants. Fungal Ecol. 2013;6:365–378. [Google Scholar]
- 46.Le Calvez T, Burgaud G, Mahe S, Barbier G, Vandenkoornhuyse P. Fungal diversity in deep-sea hydrothermal ecosystems. Apple Environ Microb. 2009;75:6415–6421. doi: 10.1128/AEM.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Les DH, Garvin DH, Wimpee CF. Molecular evolutionary history of ancient aquatic angiosperms. Proc Natl Acad Sci USA. 1991;88:10119–1023. doi: 10.1073/pnas.88.22.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li HY, Zhao CA, Liu CJ, Xu XF. Endophytic fungi diversity of aquatic/riparian plants and their antifungal activity in vitro. J Microbiol. 2010;48:1–6. doi: 10.1007/s12275-009-0163-1. [DOI] [PubMed] [Google Scholar]
- 49.Lodge DJ, Fisher PJ, Sutton BC. Endophytic fungi of Manilkara bidentata leaves in Puerto Rico. Mycologia. 1996;88:733–738. [Google Scholar]
- 50.Maddison DR, Maddison WP. ChromaSeq module. Mesquite: a modular system for evolutionary analysis. Version 1.06. 2005 http://mesquiteproject.org/
- 51.Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.6. 2009 http://mesquiteproject.org/
- 52.Maier RM, Pepper IL, Gerba CP. Environmental microbiology. Burlington, MA: Academic Press; 2009. [Google Scholar]
- 53.Malcolm Pirnie and Arizona Department of Environmental Quality. Stoneman Lake TMDL. Arizona Department of Environmental Quality open file report. 2000 http://www.epa.gov/waters/tmdldocs/11720_stonemanlaketmdl.pdf.
- 54.Neubert K, Mendgen K, Brinkmann H, Wirsel SGR. Only a few fungal species dominate highly diverse mycofloras associated with the common reed. Appl Environ Microb. 2006;72:1118–1128. doi: 10.1128/AEM.72.2.1118-1128.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niereg WA National Audubon Society. Wetlands. New York: Knopf; 1985. [Google Scholar]
- 56.Petrini O. Fungal endophytes of tree leaves. In: Andrews JH, Hirano SS, editors. Microbial ecology of leaves. Springer; New York: 1991. [Google Scholar]
- 57.Polunin N. Aquatic ecosystems: Trends and global prospects. Cambridge, UK: Cambridge University Press; 2008. [Google Scholar]
- 58.Posada D. jModelTest: Phylogenetic model averaging. Molec Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 59.Raja HA, Hirayama K, Tanaka K, Miller AN, Shearer CA. Freshwater Ascomycetes: two new species of Lindgomyces (Lindgomycetaceae, Pleosporales, Dothideomycetes) from Japan and USA. Mycologia. 2011;103:1421–1432. doi: 10.3852/11-077. [DOI] [PubMed] [Google Scholar]
- 60.Robinson AT, Fulmer JE, Avenetti LD. Aquatic plant surveys and evaluation of aquatic plant harvesting in Arizona reservoirs. Arizona Game and Fish Department, Research Branch, Technical Guidance Bulletin No. 9, Phoenix. 2007:39. [Google Scholar]
- 61.Rodriguez R, White J, Arnold AE, Redman R. Fungal endophytes: diversity and ecological roles. New Phytol. 2009;182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- 62.Rosling A, Cox F, Cruz-Martinez K, Ihrmark K, Grelet G-A, Lindahl BD, Menkis A, James TY. Archaeorhizomycetes: Unearthing an ancient class of ubiquitous soil fungi. Science. 2011;333:876–879. doi: 10.1126/science.1206958. [DOI] [PubMed] [Google Scholar]
- 63.Sandberg DC. MS thesis. University of Arizona; 2013. Host affiliations and geographic distributions of fungal endophytes inhabiting aquatic plants in northern Arizona, USA. [Google Scholar]
- 64.Sati SC, Belwal M. Aquatic hyphomycetes as endophytes of riparian plant roots. Mycologia. 2005;97:45–49. doi: 10.3852/mycologia.97.1.45. [DOI] [PubMed] [Google Scholar]
- 65.Schulthess FM, Faeth SH. Distribution, abundances, and associations of the endophytic fungal community of Arizona fescue (Festuca arizonica) Mycologia. 1998;90:569–578. [Google Scholar]
- 66.Schulz B, Wanke U, Draeger S, Aust HJ. Endophytes from herbaceous plants and shrubs – effectiveness of surface sterilization methods. Mycol Res. 1993;97:1447–1450. [Google Scholar]
- 67.Schulz B, Boyle C, Draeger S, Römmert A-K, Krohn K. Endophytic fungi: A source of novel biological active secondary metabolites. Mycol Res. 2002;106:996–1004. [Google Scholar]
- 68.Shearer JF. Recovery of endophytic fungi from Myriophyllum spicatum. APCRP Technical Notes Collection. Vicksburg, MS: U.S. Army Engineer Research and Development Center; 2001. ERDC TN-APCRP-BC-03. [Google Scholar]
- 69.Shearer CA, Descals E, Kohlmeyer B, Kohlmeyer J, Marvanova L, Padgett D, Porter D, Raja HA, Schmit JP, Thorton HA, Voglymayr H. Fungal biodiversity in aquatic habitats. Biodiv Conserv. 2007;16:49–67. [Google Scholar]
- 70.Shearer JF. APCRP Technical Notes Collection. Vicksburg, MS: U.S. Army Engineer Research and Development Center; 2010. Relationship between Eurasian watermilfoil phenology and endophyte presence. ERDC/TN APCRP-BC-20. [Google Scholar]
- 71.Sridhar KR, Bärlocher F. Endophytic aquatic hyphomycetes of roots from spruce, birch and maple. Mycol Res. 1992;96:305–308. [Google Scholar]
- 72.Strobel G, Daisey B. Bioprospecting for microbial endophytes and their natural products. Microbiol Molec Biol Rev. 2003;67:491–502. doi: 10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suryanarayanan TS, Kumaresan V. Endophytic fungi of some halophytes from an estuarine mangrove forest. Mycol Res. 2000;104:1465–1467. [Google Scholar]
- 74.Suryanarayanan TS, Murali TS, Venkatesan G. Occurrence and distribution of fungal endophytes in tropical forests across a rainfall gradient. Can J Bot. 2002;80:818–826. [Google Scholar]
- 75.Suryanarayanan TS, Wittlinger SK, Faeth SH. Endophytic fungi associated with cacti in Arizona. Mycol Res. 2005;109:635–639. doi: 10.1017/s0953756205002753. [DOI] [PubMed] [Google Scholar]
- 76.Suryanarayanan TS, Murali TS, Thirunavukkarasu N, Rajulu MBG, Venkatesan G, Sukumar R. Endophytic fungal communities in woody perennials of three tropical forest types of the Western Ghats, southern India. Biodiv Conserv. 2011;20:913–928. [Google Scholar]
- 77.Tan RX, Zou WX. Endophytes: A rich source of functional metabolites. Nat Prod Rep. 2001;18:448–459. doi: 10.1039/b100918o. [DOI] [PubMed] [Google Scholar]
- 78.Upper Granite Creek Watershed Management Plan. Improvement plan for the Upper Granite Creek Watershed, Arizona, Version 1.0. Prescott Creeks and the Granite Creek Watershed Improvement Council open report. 2011 http://www.prescottcreeks.org/sites/prescottcreeks.org/files/.wysiwyg/WIP-full_wm.pdf.
- 79.U’Ren JM, Dalling JW, Gallery RE, Maddison DR, Davis EC, Gibson CM, Arnold AE. Diversity and evolutionary origins of fungi associated with seeds of a neotropical pioneer tree: A case study for analyzing fungal environmental samples. Mycol Res. 2009;113:432–449. doi: 10.1016/j.mycres.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 80.U’Ren JM, Lutzoni F, Miadlikowska J, Arnold AE. Community analysis reveals close affinities between endophytic and endolichenic fungi in mosses and lichens. Microb Ecol. 2010;60:340–353. doi: 10.1007/s00248-010-9698-2. [DOI] [PubMed] [Google Scholar]
- 81.U’Ren JM, Lutzoni F, Miadlikowska J, Laetsch AD, Arnold AE. Host and geographic structure of endophytic and endolichenic fungi at a continental scale. Am J Bot. 2012;99:898–914. doi: 10.3732/ajb.1100459. [DOI] [PubMed] [Google Scholar]
- 82.Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Warwick RM, Clarke KR, Suharsono A statistical analysis of coral community responses to the 1982–83 El Niño in the Thousand Islands, Indonesia. Coral Reefs. 1990;8:171–179. [Google Scholar]
- 84.Wetzel RG. Limnology: Lake and river ecosystems. 3. Academic Press; 2001. [Google Scholar]
- 85.White TJ, Bruns T, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A guide to methods and applications. Academic Press; San Diego, CA: 1990. [Google Scholar]
- 86.Willoughby LG. Freshwater biology. New York: Pica Press; 1977. [Google Scholar]
- 87.Wurzbacher C, Brlocher M, Grossart HP. Fungi in lake ecosystems. Aquat Microb Ecol. 2010;59:125–149. [Google Scholar]
- 88.Wurzbacher C, Williams J, Grossart HP. Aquatic fungi. In: Grillo, editor. Biodiversity, Book 2. Intech; 2011. [DOI] [Google Scholar]
- 89.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comp Biol. 2000;7:203–14. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 90.Zhang HW, Song YC, Tan RX. Biology and chemistry of endophytes. Nat Prod Rep. 2006;23:753–771. doi: 10.1039/b609472b. [DOI] [PubMed] [Google Scholar]
- 91.Zwickl DJ. Dissertation. The University of Texas; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.