SUMMARY
NLR (nucleotide-binding domain [NBD]- and leucine-rich repeat [LRR]-containing) proteins mediate innate immune sensing of pathogens in mammals and plants. How NLRs detect their cognate stimuli remains poorly understood. Here, we analyzed ligand recognition by NAIP (NLR Apoptosis Inhibitory Protein) inflammasomes. Mice express multiple highly related NAIP paralogs that recognize distinct bacterial proteins. We analyzed a panel of 43 chimeric NAIPs, allowing us to map the NAIP domain responsible for specific ligand detection. Surprisingly, ligand specificity was mediated not by the LRR domain, but by an internal region encompassing several NBD-associated α-helical domains. Interestingly, we find that the ligand specificity domain has evolved under positive selection in both rodents and primates. We further show that ligand binding is required for the subsequent co-oligomerization of NAIPs with the downstream signaling adaptor NLRC4 (NLR family, CARD-containing 4). These data provide a molecular basis for how NLRs detect ligands and assemble into inflammasomes.
INTRODUCTION
Many nucleotide-binding domain (NBD)- and leucine-rich repeat (LRR)-containing proteins (NLRs) function as innate immune sensors that monitor the cytosol for the presence of microbial products and other infection-associated stimuli (Jones and Dangl, 2006; Schroder and Tschopp, 2010; Takeuchi and Akira, 2010; von Moltke et al., 2013). Once activated, some NLRs assemble into high molecular weight complexes termed inflammasomes (Martinon et al., 2002; Schroder and Tschopp, 2010) that recruit and activate pro-inflammatory proteases such as CASPASE-1 (CASP1). CASP1 cleaves the pro-inflammatory cytokines IL-1β and IL-18 into their signaling-competent forms. CASP1 also initiates pyroptosis (Bergsbaken et al., 2009), a rapid, lytic form of cell death that releases pro-inflammatory molecules to trigger rapid and potent immune responses (von Moltke et al., 2012; Yang et al., 2013a).
The NBD-LRR architecture is found in pathogen-sensing proteins in both mammals and plants (Bonardi et al., 2012; Chisholm et al., 2006), but remarkably little is known about how NLRs detect infectious stimuli and initiate signaling. The NBD of NLRs is classified as an AAA+ ATPase (Leipe et al., 2004), a domain found in diverse proteins known to form homo- and hetero-oligomeric complexes (Danot et al., 2009). The NBD is presumed to mediate assembly of NLR protomers into the active oligomerized inflammasome, analogous to the function of the NBD in assembly of the apoptosome (Qi et al., 2010).
The other domain that defines membership in the NLR superfamily, the LRR domain, is believed to have two distinct roles. The first is to function as an autoinhibitory domain, as truncation of this domain generally results in constitutive NLR activation (Chavarria-Smith and Vance, 2013; Kofoed and Vance, 2011; Poyet et al., 2001; Tanabe et al., 2004). The autoinhibitory function of the LRR domain is supported by the recently determined crystal structure of the monomeric/inactive form of NLRC4, in which the LRR domain curves back to occlude the NBD in cis (Hu et al., 2013). In addition to its role in autoinhibition, the LRR domain has also been proposed to act as a ‘sensor’ that directly or indirectly detects ligands (Danot et al., 2009). The ligand-binding function of the LRR domain is supported primarily by analogy to the well-established ligand-binding function of the LRRs in pathogen-sensing Toll-like receptors (TLRs) (Song and Lee, 2012). Association of ligands with the LRR is believed to disrupt autoinhibitory cis interactions between the LRR and the NBD, resulting in NBD-mediated oligomerization and inflammasome assembly (Danot et al., 2009; Faustin et al., 2007; Hu et al., 2013). However, direct evidence for ligand association with the LRR domain, or indeed any other domain, of mammalian NLRs is lacking.
In order to address the fundamental issue of how NLRs detect their specific ligands, we analyzed the ligand specificity of NAIP/NLRC4 inflammasomes. Mice express multiple NAIP paralogs, each of which recognizes a distinct bacterial ligand. Both NAIP5 and NAIP6 detect bacterial flagellin, whereas NAIP2 detects inner rod proteins of type III secretion systems (Kofoed and Vance, 2011; Zhao et al., 2011). Mouse NAIP1 and human NAIP respond to needle proteins of type III secretion systems (Rayamajhi et al., 2013; Yang et al., 2013b; Zhao et al., 2011). Upon recognition of their ligands, NAIPs assemble with NLRC4 into an oligomerized inflammasome that contains both NLRs and the ligand (Kofoed and Vance, 2011). The assembled inflammasome can then directly recruit and activate CASP1 via the NLRC4 CARD domain (von Moltke et al., 2013).
At present, the molecular basis for ligand recognition by the NAIP/NLRC4 inflammasome, or indeed by any mammalian NLR, remains unclear. It has not yet been possible to map the ligand recognition domain of mammalian NLRs by mutagenesis because mutations that disrupt NLR function may not specifically affect ligand binding but may instead disrupt the overall NLR fold or oligomerization competence (Tanabe et al., 2004). We circumvented this difficulty by taking advantage of the fact that, although they recognize distinct bacterial ligands, the mouse NAIP paralogs share a high degree of amino acid identity and the same basic architecture (Figure 1A). Reasoning that chimeric NAIP proteins might retain their overall fold and function, we generated and analyzed a panel of 43 NAIP chimeras to identify the ligand recognition domain. To our surprise, we found that ligand specificity of NAIPs does not require the LRR domain, but instead depends on several largely alpha helical domains associated with the NBD. Furthermore, we observed an evolutionary signature of positive (diversifying) selection that maps to the ligand specificity domain, consistent with the direct association of these domains with rapidly evolving bacterial ligands (Daugherty and Malik, 2012; Mitchell et al., 2012). We further demonstrate that ligand binding is required for NAIPs to co-assemble with NLRC4. These results suggest a new model for innate immune sensing by the NLR superfamily, and provide evidence for an ongoing molecular arms race between NAIPs and the pathogens they sense.
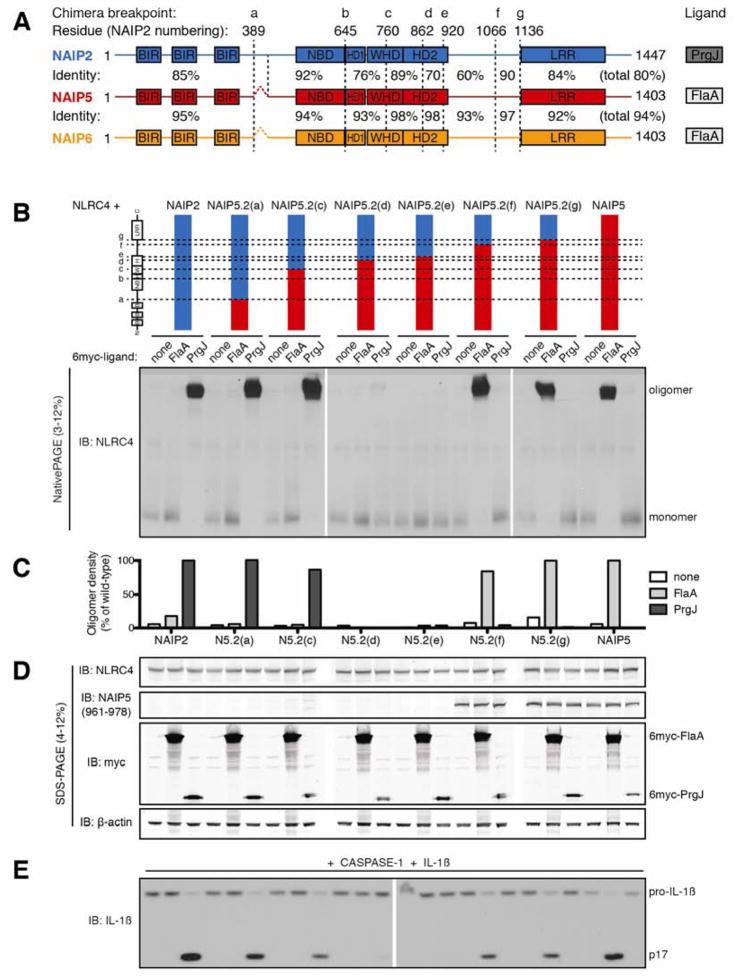
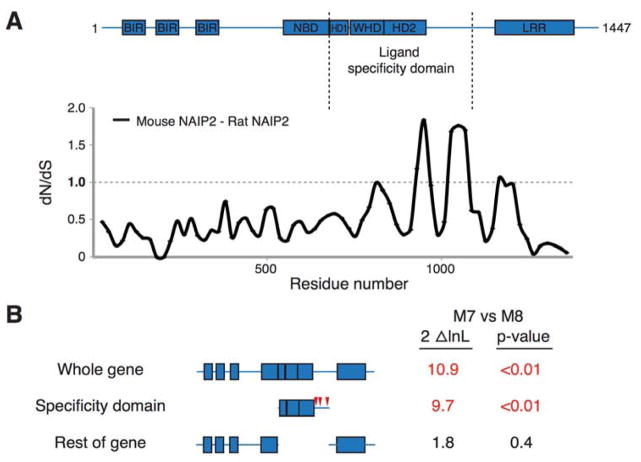
Figure 1. NAIP5 LRR is dispensable for recognition of flagellin.
(A) Schematic of NAIP predicted domains drawn to scale. Baculovirus inhibitor of apoptosis repeat (BIR) and leucine-rich repeat (LRR) domains were identified by amino acid query against the NCBI Conserved Domain Database. The nucleotide binding domain (NBD), helix domain 1 (HD1), winged helix domain (WHD), and helix domain 2 (HD2) were annotated by homology modeling to NLRC4 (PDB 4KXF). Chimera breakpoints are identified by lowercase letters (a–g) and by dashed lines. Amino acid identity between NAIP2 and NAIP5 or NAIP5 and NAIP6 is indicated for each segment and across total protein length.
(B) Oligomerization assay to test the specificity of NAIP5.2 chimeras. HEK293T cells were transfected with NLRC4 and the indicated NAIP chimera and 6myc-tagged ligand. After 48 hours, cell lysates were harvested, normalized for total protein, subjected to blue native PAGE, and immunoblotted (IB) for NLRC4, as previously described (Kofoed and Vance, 2011). Results shown are representative of at least 3 independent trials. See also Figure S1.
(C) NAIP chimera responses to each ligand in (B) were quantified by densitometry of the oligomer species and normalized to wild-type NAIP2 or NAIP5 response to PrgJ or FlaA, respectively.
(D) Lysates from (B) were subjected to denaturing SDS-PAGE and immunoblotted for NLRC4, NAIP5, myc, and β-actin to control for equal transfection and loading.
(E) NAIP2.5 inflammasomes are functional and induce IL-1β cleavage. HEK293T cells were transfected as in (B) but with the addition of CASP-1 and pro-IL-1β. Cell lysates were harvested at 24 hours and immunoblotted for IL-1β; full-length (pro) and cleaved (p17) forms are indicated. Results shown are representative of 2 independent trials.
RESULTS
In order to map the region(s) of NAIPs required for specific recognition of ligands, we generated chimeras between murine NAIP paralogs that respond to distinct ligands. We swapped homologous sequences between NAIP2 and either NAIP5 or NAIP6 at seven breakpoints (a–g) along the length of the protein (Figure 1A). These breakpoints were chosen because they are located within stretches of high sequence identity among the NAIP paralogs. We then assayed the ability of chimeric NAIPs to induce NLRC4 inflammasome assembly in response to Legionella pneumophila flagellin (FlaA) or Salmonella typhimurium type III secretion system inner rod protein (PrgJ). Assembly of the high molecular weight (~1MDa) inflammasome was monitored by native gel electrophoresis using an established reconstituted inflammasome assay (Kofoed and Vance, 2011). Because this is the most sensitive assay for NAIP function thus far described, we reasoned it might allow us to detect NAIP regions that only weakly contribute to ligand recognition.
As previously observed (Kofoed and Vance, 2011), transient transfection of HEK293T cells with NLRC4 and NAIP2 yields an oligomerized inflammasome only in response to PrgJ, while NAIP5 and NAIP6 induce NLRC4 oligomerization only in response to FlaA (Figure 1 and S1). Thus, the parental NAIPs exhibit a high degree of selectivity for their ligands.
BIRs, NBD and LRR of NAIPs do not affect ligand specificity
We first analyzed a series of chimeras in which the N-terminal domains of NAIP5 or NAIP6 were fused to the C-terminal domains of NAIP2 (termed NAIP5.2 or NAIP6.2 chimeras; Figure 1B-E, S1). The NAIP5.2(a) chimera, containing the NAIP5 BIR (baculovirus inhibitor of apoptosis repeat) domains, still oligomerized normally in response to the NAIP2 ligand PrgJ (Figure 1B, C). The NAIP5.2(c) chimera, containing the BIRs, NBD, HD1 (helical domain 1), and part of the WHD (winged helix domain) of NAIP5, exhibited a slightly diminished response to PrgJ. The reduced response to PrgJ was more pronounced in the corresponding NAIP6.2(c) chimera (Figure S1), suggesting that a portion of the PrgJ-detecting element had been replaced in these chimeras. The NAIP5.2(d) and NAIP5.2(e) chimeras, with breakpoints in HD2 (helical domain 2), did not respond to either ligand (Figure 1B, C). Importantly, however, the NAIP5.2(f) and NAIP5.2(g) chimeras, which contained the NAIP5 N-terminal domains but the LRR domain of NAIP2, assembled inflammasomes only in response to the NAIP5 ligand FlaA. Both chimeras were also able to activate CASP-1 to initiate cleavage of IL-1β (Figure 1E). In addition, the corresponding NAIP6.2(f) and NAIP6.2(g) chimeras recognized FlaA as well as wild-type NAIP6 (Figure S1), despite containing the LRR domain from NAIP2. These results demonstrate that the LRR domains of NAIP5 and NAIP6 are dispensable for the specific response to FlaA, while the BIR domains of NAIP2 are dispensable for detection of PrgJ.
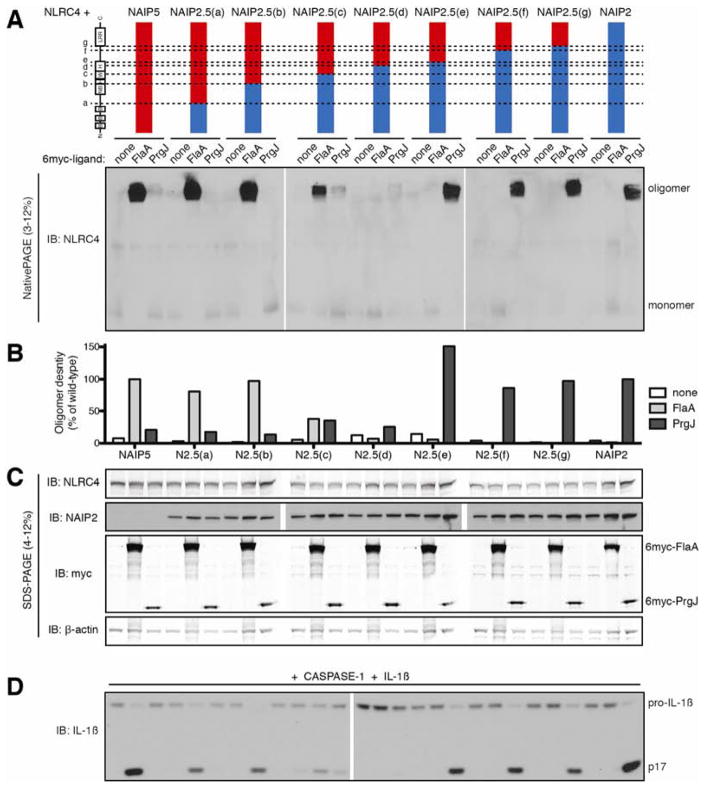
We next generated a set of reciprocal NAIP2.5 chimeras, in which the N-terminal domains of NAIP2 were fused to the C-terminal domains of NAIP5 (Figure 2). Replacement of the NAIP5 BIRs and NBD with those of NAIP2 (NAIP2.5(b)) did not alter the extent or specificity of oligomerization in response to FlaA, indicating that these domains were dispensable for detection of FlaA. However, the NAIP2.5(c) chimera, which additionally contains HD1 and the N-terminal half of the WHD from NAIP2, responded only weakly to FlaA, and notably, also responded weakly to PrgJ. This result suggests a contribution of HD1 and the WHD to ligand recognition. The partial response to each ligand could not be explained by decreased expression of NAIP2.5(c) (Figure 2C). Whereas NAIP2.5(d) did not respond to either ligand, the NAIP2.5(e) chimera detected PrgJ at least as well as wild-type NAIP2 (Figure 2B, C) and was capable of inducing IL-1β cleavage specifically in response to PrgJ (Figure 2D). These data indicate that NAIP2 domains C-terminal to HD2 are not required for the response of NAIP2 to PrgJ. The analogous set of NAIP2.6 chimeras confirmed that the LRR domain does not dictate the ligand specificity of NAIP2 (Figure S2).
Figure 2. NAIP2 LRR is dispensable for recognition of PrgJ.
(A) Oligomerization assay to test the specificity of NAIP2.5 chimeras. HEK293T cells were transfected with NLRC4 and the indicated NAIP chimera and 6myc-tagged ligand. Cell lysates were subjected to blue native PAGE as in Figure 1. Results shown are representative of at least 3 independent trials. See also Figure S2.
(B) NAIP chimera responses to each ligand in (A) were quantified by densitometry of the oligomer species and normalized to wild-type NAIP2 or NAIP5 response to PrgJ or FlaA, respectively.
(C) Lysates from (A) were subjected to denaturing SDS-PAGE and immunoblotted for NLRC4, NAIP2, myc, and β-actin.
(D) NAIP5.2 inflammasomes are functional. HEK293T cells were transfected as in (A) but with the addition of CASP-1 and pro-IL-1β. Cell lysates were analyzed for IL-1β cleavage after 24 hours. Results shown are representative of 2 independent trials.
Central NBD-associated domains dictate ligand specificity
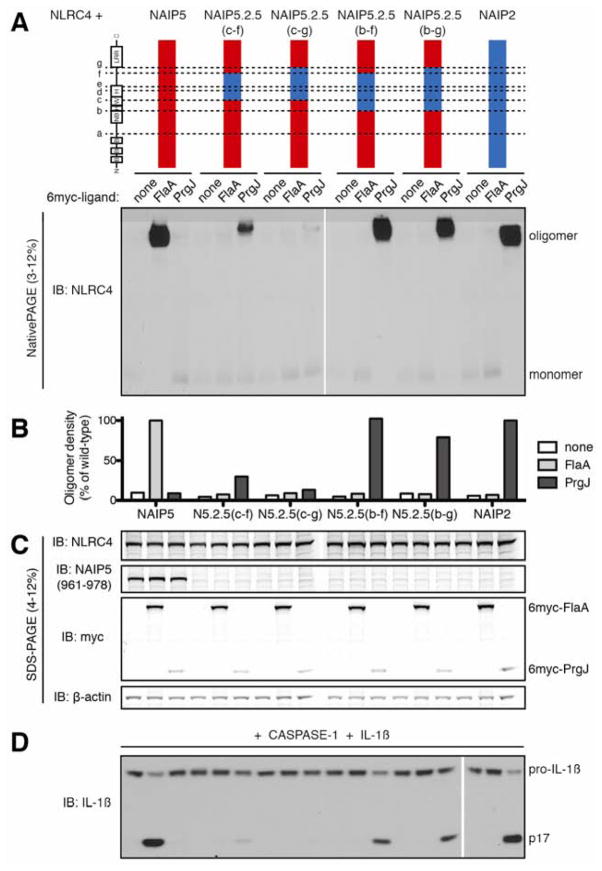
The above results suggested that the central NBD-associated helical domains of NAIPs dictate ligand specificity. To confirm this, we analyzed ‘double’ chimeric NAIPs in which only the central domains were exchanged between paralogs. NAIP5.2.5(c–f) or NAIP6.2.6(c–f) chimeras, containing the C-terminal half of the WHD, HD2, and most of the subsequent unannotated domain from NAIP2, responded weakly or not at all to PrgJ (Figures 3, S3). However, additional inclusion of the HD1 and full WHD from NAIP2 allowed the NAIP5.2.5(b–f) chimera to oligomerize in response to PrgJ as well as wild-type NAIP2 (Figure 3). The same domains of NAIP2 within a NAIP6 context also conferred a specific, albeit reduced, response to PrgJ (Figure S3, NAIP6.2.6(b–f)). These results demonstrate that HD1, WHD, HD2 and part of the unannotated domain of NAIP2 dictate specific recognition of PrgJ.
Figure 3. Central NBD-associated domains of NAIP2, including HD1, WHD, and HD2, are sufficient for recognition of PrgJ.
(A) Oligomerization assay to test the specificity of NAIP5.2.5 chimeras. HEK293T cells were transfected with NLRC4 and the indicated NAIP chimera and 6myc-tagged ligand. Cell lysates were subjected to blue native PAGE as in Figure 1. Results shown are representative of at least 3 independent trials. See also Figure S3.
(B) NAIP chimera responses to each ligand in (A) were quantified by densitometry of the oligomer species and normalized to wild-type NAIP2 or NAIP5 response to PrgJ or FlaA, respectively.
(C) Lysates from (A) were subjected to denaturing SDS-PAGE and immunoblotted for NLRC4, NAIP5, myc, and β-actin.
(D) NAIP5.2.5 inflammasomes are functional. HEK293T cells were transfected as in (A) but with the addition of CASP-1 and pro-IL-1β. Cell lysates were analyzed for IL-1β cleavage after 24 hours. Results shown are representative of 2 independent trials.
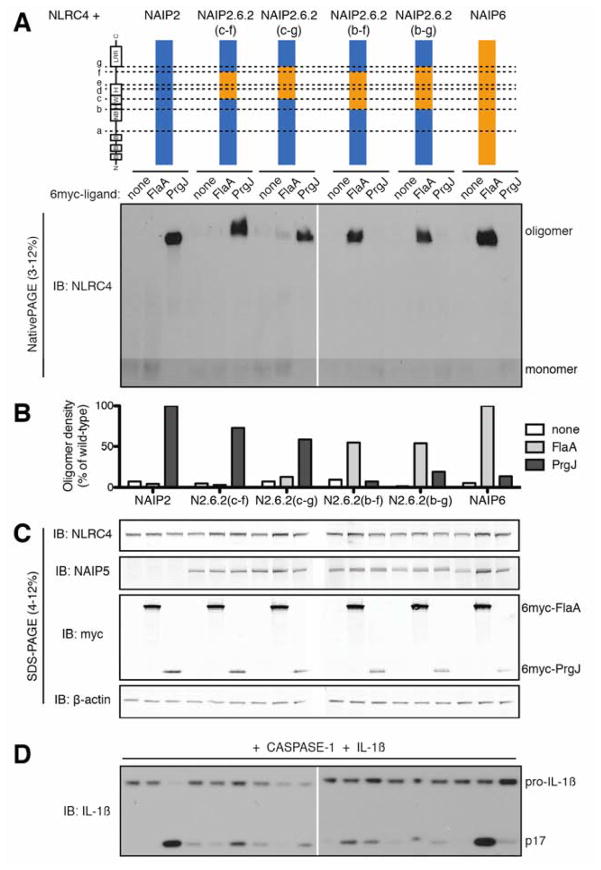
We next tested whether the same domains also control the specificity of NAIP6 for FlaA (Figure 4). Analogous to NAIP2, the C-terminal half of the WHD, HD2 and unannotated domain of NAIP6 (as in chimeras NAIP2.6.2(c–f) and NAIP2.6.2(c–g)) were not sufficient to confer a robust response to FlaA. Surprisingly, these chimeras still responded partially to PrgJ, further emphasizing the importance of HD1 and the N-terminal half of the WHD from NAIP2 for PrgJ recognition. Extension of the NAIP6 region to include HD1 and the full WHD abrogated the response to PrgJ and conferred substantial response to FlaA (i.e., NAIP2.6.2(b–f) and NAIP2.6.2(b–g)). However, these chimeras supported only partial oligomerization in response to FlaA (Figure 4B) and exhibited background IL-1β processing even in the absence of ligand (Figure 4D). Furthermore, although NAIP5 chimeras with a single breakpoint were functional (Figure 1 and 2), we were unable to generate ‘double’ chimeras containing the NAIP5 central domains that were capable of responding to FlaA (Figure S4).
Figure 4. Central NBD-associated domains of NAIP6, including HD1, WHD and HD2, are sufficient for recognition of flagellin.
(A) Oligomerization assay to test the specificity of NAIP2.6.2 chimeras. HEK293T cells were transfected with NLRC4 and the indicated NAIP chimera and 6myc-tagged ligand. Cell lysates were subjected to blue native PAGE as in Figure 1. See also Figure S4.
(B) NAIP chimera responses to each ligand in (A) were quantified by densitometry of the oligomer species and normalized to wild-type NAIP2 or NAIP6 response to PrgJ or FlaA, respectively.
(C) Lysates from (A) were subjected to denaturing SDS-PAGE and immunoblotted for NLRC4, NAIP5, myc, and β-actin. The NAIP5 antibody also detects NAIP6.
(D) NAIP2.6.2 chimeras exhibit basal IL-1β cleavage. HEK293T cells were transfected as in (A) but with the addition of CASP-1 and pro-IL-1β. Cell lysates were analyzed for IL-1β cleavage after 24 hours. All results shown are representative of 3 independent trials.
The ligand specificity and degree of response for the 43 chimeras tested are summarized in Figure S7. Collectively, our results demonstrate that the NBD-associated HD1, WHD, HD2, and unannotated domain of NAIPs are both necessary and sufficient to dictate ligand specificity. In contrast to our expectations and those of others in the field (e.g., (Danot et al., 2009)), our extensive analysis revealed no role for the annotated LRR domain in specific ligand recognition.
The NAIP ligand specificity domain has evolved under positive selection
We hypothesized that the NAIP ligand specificity domain might be subject to diversifying evolutionary selection, driven by the need to recognize relatively rapidly evolving bacterial ligands. We found evidence for extensive recombination between rodent Naip paralogs, with several recombination breakpoints occurring within the ligand specificity domain. While it is difficult to assess whether these recombinants have been adaptively selected for, recombinants that yield chimeric specificity domains could be a rapid means to evolve novel NAIP specificities. Indeed, recombination has contributed to differing specificities of at least one allelic series of plant NLRs (Ellis et al., 1999). Nonetheless, the rampant recombination precludes codon-based analyses of positive selection. Instead, we performed a pairwise sliding window analysis comparing the rate of synonymous changes (dS) to the rate of nonsynonymous changes (dN) between the mouse and rat Naip2 genes to investigate whether the ligand specificity domain displays a signature of positive selection. This sliding-window approach is less susceptible to artifacts arising due to recombination. We found that much of Naip2 is evolving with a dN/dS ratio of less than 1, indicative of purifying selection (Figure 5A). Strikingly, however, we found two peaks in which the dN/dS ratio was much greater than 1. Both of these peaks lie in the domains functionally implicated in ligand recognition, although the second peak falls within the unannotated domain that has no apparent role in PrgJ recognition by mouse NAIP2. The unannotated domain does appear to play a role in flagellin recognition by mouse NAIP5, and may also be important for recognition of ligand by rat NAIP2, thereby providing a possible explanation for the peak of elevated dN/dS ratio in this region. Overall, these data are consistent with the ligand specificity domain evolving under positive selection.
Figure 5. The ligand specificity domain has evolved under positive selection.
(A) Sliding window comparison of the dN/dS ratio between mouse and rat Naip2 genes. dN/dS ratios were calculated every 20 codons with a window size of 50 codons. Shown above is the domain structure of mouse NAIP2.
(B) Results of PAML analyses on the entire primate NAIP gene, the ligand specificity domain alone, and the entire gene outside the ligand specificity domain. The left column shows two times the log likelihood difference between a model that allows (M8) or disallows (M7) positive selection. The right column shows the statistical significance of support for the gene, or domain, having evolved under positive selection. Values in red indicate strong support for positive selection. Red triangles indicate primate NAIP codons (corresponding to mouse NAIP2 residues 941, 965 and 1049) identified as having evolved under recurrent positive selection with a posterior probability of >0.95. See also Figure S5.
We next extended our evolutionary analysis to primate NAIP genes. Primate genomes encode only one intact NAIP ortholog, and unlike in rodents, we found no evidence for recombination. We therefore analyzed these genes by maximum likelihood methods for evidence of recurrent positive selection (Figure 5B). We found a statistically significant signature (p < 0.01) of positive selection, consistent with NAIP evolving adaptively in primates. Moreover, when we analyzed the ligand specificity domain alone, we found strong evidence for positive selection, as well as three individual codons that have evolved under recurrent positive selection. Importantly, removal of the specificity domain from our analysis of primate NAIP genes resulted in a loss of the signature of positive selection. Taken together, these data suggest that NAIP genes in both primate and rodent genomes have evolved under positive selection, specifically in the region of the protein that confers ligand specificity.
Localized positive selection is consistent with direct association of these domains with rapidly evolving bacterial ligands. Indeed, co-immunoprecipitation of chimeric NAIPs with either FlaA or PrgJ required the full ligand specificity domain from either NAIP6 or NAIP2, respectively (Figure S5). In contrast, the C-terminal LRR domain and N-terminal BIRs and NBD of NAIP6 were dispensable for association with FlaA, and the NAIP2 LRR domain was dispensable for binding PrgJ. Thus, the ligand specificity domain also dictates the physical association of NAIPs with their cognate ligands.
Ligand binding is strictly required for NAIP oligomerization
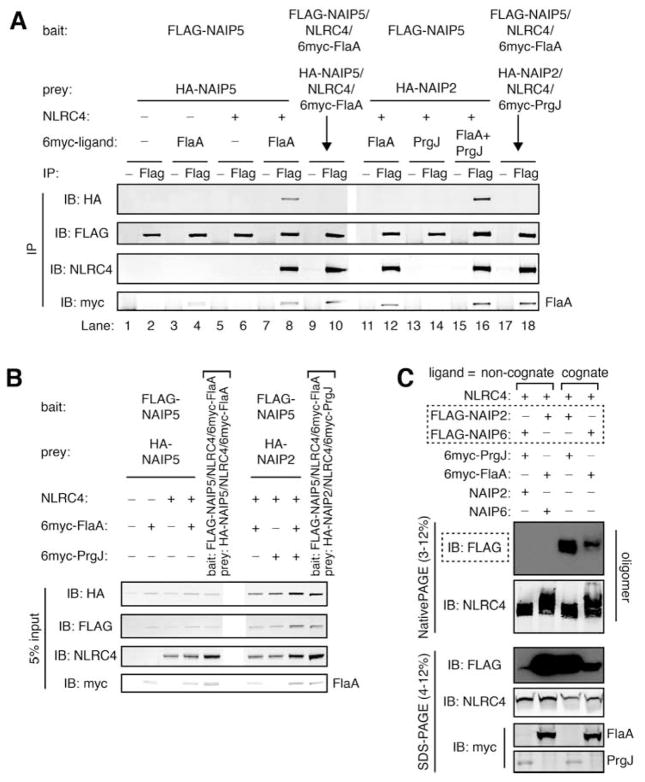
Because our analyses mapped NAIP ligand recognition to NBD-associated domains, we investigated the effects of ligand binding on NAIP oligomerization. Consistent with a recent study (Halff et al., 2012), we found that NAIPs are monomeric in the absence of ligand, as evidenced by the inability of FLAG-tagged NAIP5 to co-immunoprecipitate HA-tagged NAIP5 when co-transfected into HEK293T cells (Figure 6A, lane 2). In the absence of NLRC4, FLAG-NAIP5 bound FlaA as previously reported (Zhao et al., 2011) but remained unable to associate with HA-NAIP5 (Figure 6A, lane 4). However, binding FlaA did permit FLAG-NAIP5 to associate with co-expressed NLRC4, and within this assembled inflammasome, FLAG-NAIP5 was able to co-immunoprecipitate HA-NAIP5 (Figure 6A, lane 8). This association was not detected upon mixing pre-assembled FLAG-NAIP5 and HA-NAIP5 inflammasomes (Figure 6A, lane 10), suggesting that HA-NAIP5 and FLAG-NAIP5 co-immunoprecipitated within a single assembled inflammasome rather than through non-specific inter-oligomer association.
Figure 6. NAIPs require cognate ligand to assemble into an inflammasome.
(A) HA-NAIP2 associates with FLAG-NAIP5 only when both cognate ligands are present. HEK293T cells were co-transfected with multiple NAIP constructs, distinguishable by FLAG or HA tags, and NLRC4 and 6myc-tagged ligands as indicated. After 48 hours, cell lysates were divided equally, and assembled inflammasomes were immunoprecipitated (IP) with anti-FLAG antibody (Flag) or normal mouse IgG (−) as a negative control. To control for non-specific inter-oligomer association, separately assembled inflammasomes were mixed in lysate (denoted by arrows).
Immunoprecipitates were separated by SDS-PAGE and immunoblotted for HA, FLAG, NLRC4 and myc. Results are representative of at least 4 independent trials.
(B) Prior to immunoprecipitations in (A), 5% of lysate volume was removed as input, separated by SDS-PAGE, and immunoblotted in parallel with IP samples.
(C) FLAG-NAIPs are detectable in assembled inflammasomes only when provided with their cognate ligand. HEK293T cells were co-transfected with NLRC4 and the indicated 6myc-tagged ligand and NAIP construct(s). After 48 hours, cell lysates were harvested, separated in parallel by blue native PAGE and SDS-PAGE, and immunoblotted for FLAG followed by NLRC4 and myc. See also Figure S6.
The above results imply that more than one NAIP protomer can be incorporated into a single inflammasome oligomer. To confirm this, we assessed the stoichiometry of NAIP and NLRC4 constituents within assembled inflammasomes. We labeled both NLRs with the same FLAG epitope, immunoprecipitated the complex via the 6myc-tagged ligand, and determined the ratio of NLRs by densitometry of an anti-FLAG immunoblot. We analyzed the stoichiometry of the NAIP2/NLRC4/PrgJ inflammasome because under our experimental conditions 6myc-PrgJ did not associate with either unassembled NAIP2 or NLRC4 (Figure S6A). Therefore, the observed ratio of 5 NLRC4 to 2 NAIP2 (Figure S6B, ratio = 2.5) reflects the average composition of assembled inflammasomes free of PrgJ-associated unassembled components. Inflammasome stoichiometry was insensitive to the relative expression levels of each NLR (Figure S6C). However, it remains possible that assembled inflammasomes are heterogeneous in composition, and thus the stoichiometry of individual oligomers could vary considerably from the observed 5:2 average.
We took advantage of the fact that inflammasomes can contain multiple NAIP protomers to determine under what conditions NAIP2 and NAIP5 could co-oligomerize. We found that a FLAG-NAIP5/NLRC4/FlaA inflammasome did not associate with co-expressed HA-NAIP2 in the absence of PrgJ, nor did FLAG-NAIP5 pull down an HA-NAIP2/NLRC4/PrgJ inflammasome in the absence of FlaA (Figure 6A, lanes 11–14). However, HA-NAIP2 was incorporated into a FLAG-NAIP5 inflammasome, at roughly the same efficiency as HA-NAIP5, when the NAIP2 cognate ligand PrgJ was present in addition to the NAIP5 ligand FlaA (Figure 6A, lane 16). Again, this association was not detected upon mixing pre-assembled FLAG-NAIP5 and HA-NAIP2 inflammasomes (Figure 6A, lane 18), suggesting that it was not due to non-specific inter-oligomer association. These data indicate that ligand binding is strictly required for assembly of NAIPs into inflammasomes. We confirmed this finding using the oligomerization assay to monitor inflammasome assembly (Figure 6C). Again, FLAG-NAIP2 and FLAG-NAIP6 were detectable in the oligomer only when provided with their cognate ligands, i.e., PrgJ and FlaA, respectively.
The model that ligand binding is required for assembly of each NAIP protomer into inflammasomes predicts a 1:1 ligand:NAIP stoichiometry. We therefore analyzed the ratio of GFP-NAIP and GFP-ligand in inflammasomes immunoprecipitated by FLAG-NLRC4, which does not associate with either NAIP or ligand individually (Figure S6E). Within assembled inflammasomes, both NAIP5 and NAIP2 were present at a 1:1 stoichiometry with their respective ligands (Figure S6F). These results are consistent with the model that cognate ligand is required for NAIP incorporation into NLRC4-containing inflammasomes.
DISCUSSION
Despite their shared domain architecture, members of the NLR family respond to a surprisingly diverse set of ligands and agonists (Schroder and Tschopp, 2010; Takeuchi and Akira, 2010; von Moltke et al., 2013). To gain insight into how the NBD-LRR platform can evolve such divergent specificities, we investigated the mechanism of ligand recognition by NAIP/NLRC4 inflammasomes. We took advantage of the existence of several highly related murine NAIPs with distinct ligand specificities. We mapped the NAIP specificity domain using a panel of 43 chimeric NAIPs, of which, remarkably, 31 (72%) retained at least some function.
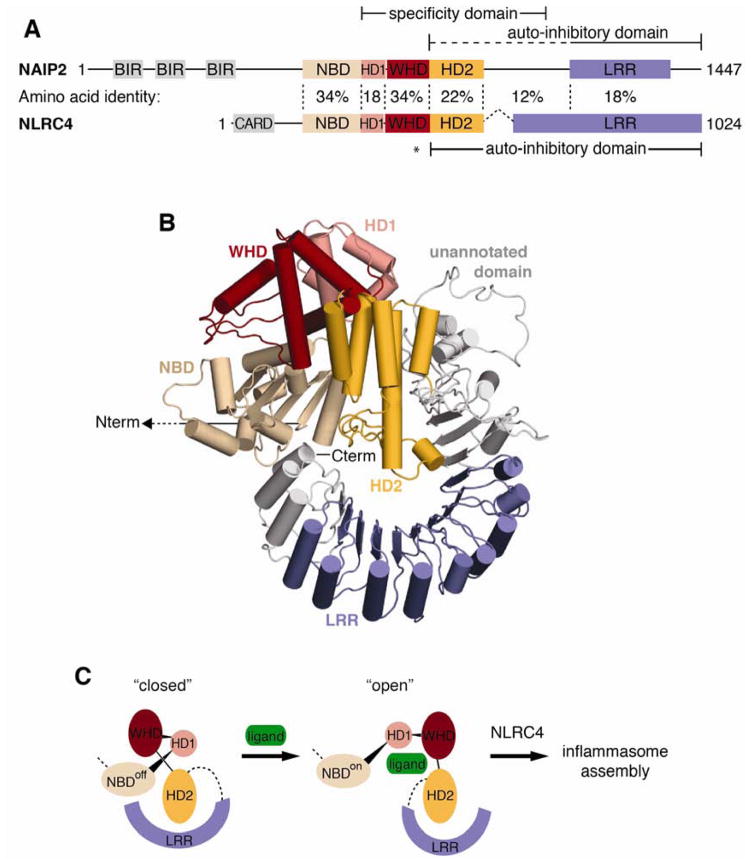
The LRR domain has been identified as the specificity determinant in several plant NLR immune sensors (Dodds et al., 2001; Ellis et al., 1999; Jia et al., 2000; Krasileva et al., 2010). Mammalian NLR specificity domains have not previously been conclusively mapped, although some evidence implicates the LRR in NOD2 and NLRP3 (Girardin et al., 2005; Iyer et al., 2013). However, our analysis of NAIP chimeras indicated that the annotated NAIP LRR domain was dispensable for ligand specificity. The SwissModel repository (Kiefer et al., 2009) predicts that the LRR domain may extend N-terminally to NAIP2 residue 1069, but even this extended region had no effect on specificity (Figure 1, NAIP5.2(f); Figure 2, NAIP2.5(f)). It is possible that the unannotated region between HD2 and breakpoint “f” might adopt an LRR-like tertiary structure, although structural modeling predicts near-total alpha helicity (Figure 7B). However, recognition of PrgJ by NAIP2 does not require any domains C-terminal to HD2 (Figure 2), ruling out a role for LRRs in PrgJ detection. Although we did not uncover a role for the NAIP LRR domain in dictating ligand specificity, our data do not undermine the previously established role of the LRR domain in NAIP autoinhibition (Kofoed and Vance, 2011). Thus, taken together, our data demonstrate that ligand recognition and autoinhibition are mediated by separable domains.
Figure 7. Model of NAIP autoinhibition relief by ligand binding.
(A) Comparison of NLRC4 (PDB 4KXF) and predicted NAIP2 domain architecture. Solid lines indicate the NAIP ligand specificity domain identified in this study and autoinhibitory domains previously identified for NAIP (Kofoed and Vance, 2011) and NLRC4 (Hu et al., 2013). The dashed line denotes a potential extension of the autoinhibitory domain based on comparison with NLRC4; the asterisk marks the NLRC4 autoinhibitory residue H443 that is not conserved in NAIPs. Amino acid identity between NAIP2 and NLRC4 is indicated for each segment. See also Figure S7.
(B) Homology model of NAIP2 by multiple template threading using the Phyre2 server. NAIP2 predicted domains are colored as follows: NBD (tan), HD1 (pink), WHD (red), HD2 (orange), and LRR (blue). The N-terminal BIR domains of NAIP2 are not shown. Regions outside of the predicted domains are depicted in gray; predicted structure of the unannotated domain (especially residues 987–1040) is based on low-confidence ab initio modeling.
(C) Model for NAIP activation by ligand binding. Regions of low-confidence structural modeling have been replaced with dashed lines. Ligand, depicted as a green oval, is predicted to bind primarily within the NAIP ligand specificity domain. Ligand binding may sterically occlude autoinhibitory interactions and/or allosterically induce rotation of autoinhibitory domains away from the NBD in a manner similar to apoptosome assembly (Reubold et al., 2011). Exposure of NBD oligomerization surfaces triggers assembly of the NAIP/NLRC4 inflammasome.
Our data indicate that the LRR domains of NLRs are not necessarily pathogen-detection domains. Consistent with this possibility, the NLR family member NLRP1B can be activated upon direct cleavage by Bacillus anthracis lethal factor protease (Chavarria-Smith and Vance, 2013; Levinsohn et al., 2012). Importantly, cleavage of NLRP1B is sufficient for its activation, even by a heterologous protease, arguing against a requirement for specific recognition of lethal factor by the NLRP1B LRR domain (Chavarria-Smith and Vance, 2013). Instead, the NLRP1B LRR is essential only for mediating NLR autoinhibition, analogous to the LRR of NAIPs. The mechanism of recognition by other NLRs remains undetermined and may involve the LRR domain. However, we propose that the total surface area of NLRs available for evolvable ligand recognition includes the NBD-associated domains in addition to the LRR. Recombination or mutation anywhere within this expanded region could result in novel ligand specificities. Indeed, our data suggest that the ligand specificity domain of NAIP5 is shifted C-terminally relative to the corresponding ligand specificity domain of NAIP2 (compare Figure 1, NAIP5.2(f) and Figure 2, NAIP2.5(e)). The existence of a large and evolvable ligand specificity domain may be critical for NAIP proteins to keep pace with rapidly evolving bacterial ligands.
Based on both functional and evolutionary analysis, we define the central NBD-associated domains as the NAIP ligand specificity domain (Figure 7A). However, these domains from NAIP5 were unable to mediate a response to FlaA (Figure S4), despite ‘single’ chimera analysis mapping NAIP5 recognition of FlaA to this region (Figures 1, 2, S7). The most likely explanation for the failure of NAIP2.5.2 chimeras to respond to FlaA is that chimerism at these breakpoints disturbed the tertiary structure enough to disrupt function without dramatically reducing expression levels (Figure S4B). The partial oligomerization and background IL-1β processing of NAIP2.6.2 chimeras (Figure 4) support this interpretation. The ligand recognition domains are thus unlikely to be entirely modular, but instead require placement within a compatible structural framework. Indeed, homology modeling of NAIP2 (Figure 7B) suggests that these largely alpha helical domains adopt a compact, closed structure with substantial inter-domain contacts. Chimerism at breakpoints that interfere with critical inter-domain contacts could therefore disrupt NAIP structure and function. This may explain why NAIP5.2.5(c–f) responds partially to PrgJ, whereas NAIP5.2.5(c–g), containing a larger NAIP2 fragment, does not (Figure 3).
The simplest interpretation of our data is that the ‘specificity’ domain we identify is also the site of ligand binding. This domain is both necessary and sufficient to dictate ligand specificity and confers the ability for chimeric NAIPs to physically associate with ligand (Figure S5). Moreover, we find evidence for positive selection acting on this domain but not on the rest of the protein (Figure 5). This signature of positive selection is consistent with direct contact between this domain and rapidly evolving pathogen ligands, as has been observed in other host-pathogen interactions (Daugherty and Malik, 2012; Mitchell et al., 2012) and even some plant NLRs (McDowell et al., 1998).
We therefore propose a model in which NAIP activation is triggered by ligand binding to the NBD-associated helical domains (Figure 7C). Intriguingly, the NBD-associated HD2 of NLRC4 was recently shown to participate in autoinhibition (Hu et al., 2013). If the HD2 of NAIPs plays a similar role, then ligand binding at HD2 could sterically preclude autoinhibition. Alternatively, ligand binding may allosterically activate NAIPs through rigid body rotation of the autoinhibitory LRR domain away from the NBD, reminiscent of the mechanism by which cytochrome C activates APAF-1 to trigger apoptosome assembly (Reubold et al., 2011). Finally, we cannot rule out the possibility that the LRR contributes to ligand binding, even though the LRR domain does not define PrgJ vs. FlaA specificity. For example, the LRR might recognize structural features common to both ligands.
Our data indicate that NAIPs only assemble into inflammasomes in the presence of their cognate ligands (Figure 6). Future structural studies will be required to determine how ligand-bound NAIPs are licensed to co-oligomerize with NLRC4 but remain unable to self-associate (Figure 6A). The lack of NAIP self-association is particularly interesting in light of our observation that an inflammasome can contain more than one NAIP protomer (Figure S6). One possible model is that in an inflammasome containing multiple NAIP protomers, the NAIP protomers do not occupy adjacent positions within the oligomeric structure. We observed an average ratio of 2.5 NLRC4 protomers per NAIP within assembled inflammasomes. This may reflect an inflammasome composition of 5 NLRC4 and 2 NAIP protomers, consistent with the observed ~1 MDa molecular weight (Kofoed and Vance, 2011) and the tendency of AAA+ proteins to form oligomers of 5–7 protomers (Danot et al., 2009). However, these data could also be consistent with oligomers of 8 NLRC4 and 3 NAIP protomers (ratio = 2.67). Indeed, NAIP/NLRC4 disks of 11-fold symmetry have been reported (Halff et al., 2012). It is intriguing to speculate that inflammasomes not only contain 2 or 3 NAIP protomers but may even require the activation of multiple NAIPs to trigger assembly. This heightened threshold could help to prevent spontaneous activation of the NAIP/NLRC4 inflammasome due to rare population of the active NAIP conformation, circumventing autoinflammatory complications linked to inappropriate inflammasome activation (Schroder and Tschopp, 2010). Regardless, taken together, our data identify the NBD-associated helical domains as a highly evolvable surface that mediates pathogen recognition by a class of NLR innate immune sensors.
EXPERIMENTAL PROCEDURES
Expression constructs
All constructs except IL-1β (pSPORT, CMV promoter) were cloned into the MSCV2.2 retroviral vector, and expression was driven by the viral LTR. Murine NLRC4, NAIP2, NAIP5, NAIP6, CASPASE-1, pro-IL-1β, and N-terminally 6myc-tagged FlaA (L. pneumophila) and PrgJ (S. typhimurium) have been described (Kofoed and Vance, 2011), except that NAIP5 was modified to replace a C-terminal codon missing relative to the reference sequence (NCBI refseq. NP_035000.2). NAIP chimeras were generated by splicing-by-overlap extension PCR. N-terminal epitope tags were added by conventional PCR using forward primers containing either a 1xFLAG or 1xHA tag immediately after the start codon. See Supplemental Experimental Procedures for details.
Cell culture and transient transfection
HEK293T cells were grown in complete medium (DMEM, 10% FBS, 2mM L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin). Cells were seeded into 6-well plates at a density of 8 × 105 cells per well or into 24-well plates at 1.5 × 105 cells per well, and transfected the following day using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instruction.
Reconstituted inflammasome and native PAGE
Inflammasome reconstitution was performed as previously described (Kofoed and Vance, 2011, 2013). In brief, HEK293T cells were transfected with NLRC4, a NAIP construct(s), and the appropriate ligand(s). Cells were lysed with 1% digitonin, and oligomerization was assessed by blue native PAGE (polyacrylamide gel electrophoresis) followed by immunoblotting. For additional details, see Supplemental Experimental Procedures and (Kofoed and Vance, 2013).
Immunoprecipitation
Immunoprecipitation protocol was essentially as previously described (Zhao et al., 2011). In brief, transfected HEK293T cells were lysed with 1% Triton X-100. Lysates were divided equally and subjected to immunoprecipitation with specific antibody or isotype controls. Where indicated, separately transfected HEK293T cells were mixed prior to lysis. For additional details, see Supplemental Experimental Procedures.
IL-1β processing
HEK293T cells were transfected in 24-well plates with 50 ng of pro-IL-1β and 160 ng each of the remaining plasmids. After 24 hours, media was removed and cells were lysed in plate in RIPA buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1mM PMSF, 1x Roche protease inhibitor tablet [no EDTA], pH 8.0) for 20 min at 4°C. Lysates were clarified by centrifugation at 16,000 g for 30 min at 4°C, and supernatants (18% of total) were separated by 12% SDS-PAGE in MES buffer (Invitrogen) and immunoblotted for IL-1β.
Immunoblotting and densitometry
Proteins separated by either native or denaturing PAGE were transferred to Immobilon-FL PVDF membranes (Millipore). Membranes were blocked with Li-Cor Odyssey blocking buffer (denaturing gels) or 5% milk (native gels). See Supplemental Experimental Procedures for antibody details. Immunoblots were imaged using a Li-Cor fluorimeter, followed by conventional chemiluminescent immunoblotting for native gels. Native gel images shown are chemiluminescent immunoblots. Densitometry was performed on Li-Cor immunoblots using ImageJ. In at least one experiment, a FLAG-NLRC4 dilution series was used as a standard curve to ensure quantified bands of anti-FLAG immunoblots were in the linear range of signal.
Domain annotation and homology modeling
NAIP domains were identified by amino acid query using the NCBI Conserved Domain Database or by homology modeling NAIPs to the NLRC4 crystal structure (Hu et al., 2013) using the Phyre2 one-to-one threading tool (Kelley and Sternberg, 2009). A homology model of full-length NAIP2 was generated using the Phyre2 intensive modeling mode to allow multiple template modeling with the following templates: PDB 1I3O, RKXF, 1TFQ, 1PGV, 2A5Y, 3T6P.
Analysis of positive selection
Publicly available NAIP gene sequences were used for all analyses. Rodent NAIP sequences (mouse NAIP1, 2, 5, 6 and 7; rat NAIP2 and 5; and hamster NAIP) were aligned and trimmed to remove all ambiguities and gaps. Similarly, NAIP orthologs from 8 primate species (human, chimpanzee, bonobo, gorilla, orangutan, rhesus macaque, baboon and African green monkey) were aligned and trimmed. Alignments were analyzed by GARD in the HyPhy package (Kosakovsky Pond et al., 2006) for evidence of recombination. Maximum likelihood analyses were performed on primate NAIP genes using PAML (Yang, 2007) to compare models that disallow (M7) or allow (M8) for codons to evolve under positive selection. Reported p-values compare the log likelihood values for each model using a chi-squared test with two degrees of freedom. Specific codons that evolved under recurrent positive selection with a posterior probability of >0.95 were identified using the Naive Empirical Bayes analysis within PAML. Sliding window analyses were performed using K-estimator (Comeron, 1999).
Supplementary Material
HIGHLIGHTS.
The LRR domain of NAIPs is not utilized for discrimination of bacterial ligands.
The NBD-associated helical domains of NAIPs dictate specific ligand recognition.
The ligand specificity domain has evolved under positive selection.
Ligand binding is required for NAIP assembly into inflammasomes.
Acknowledgments
Work in the Vance Laboratory is supported by NIH grants AI075039, AI063302, AI080749, AI103817, and by an Investigator Award in the Pathogenesis of Infectious Disease from the Burroughs Wellcome Fund. R.E.V. and H.S.M. are Investigators of the Howard Hughes Medical Institute. J.L.T. is supported by an NSF graduate student fellowship. M.D.D. is supported by a Cancer Research Institute Irvington Fellowship. We thank S. Mariathasan and V. Dixit (Genentech) for the gift of anti-NLRC4 antibody; B. Staskawicz and A. Steinbrenner (UC Berkeley) for discussion of plant NLRs; and members of the Barton and Vance labs for discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Cherkis K, Nishimura MT, Dangl JL. A new eye on NLR proteins: focused on clarity or diffused by complexity? Curr Opin Immunol. 2012;24:41–50. doi: 10.1016/j.coi.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarria-Smith J, Vance RE. Direct proteolytic cleavage of NLRP1B is necessary and sufficient for inflammasome activation by anthrax lethal factor. PLoS pathogens. 2013;9:e1003452. doi: 10.1371/journal.ppat.1003452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Comeron JM. K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics. 1999;15:763–764. doi: 10.1093/bioinformatics/15.9.763. [DOI] [PubMed] [Google Scholar]
- Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E. Wheel of Life, Wheel of Death: A Mechanistic Insight into Signaling by STAND Proteins. Structure. 2009;17:172–182. doi: 10.1016/j.str.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annual review of genetics. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Ellis JG. Six amino acid changes confined to the leucine-rich repeat beta-strand/beta-turn motif determine the difference between the P and P2 rust resistance specificities in flax. The Plant cell. 2001;13:163–178. doi: 10.1105/tpc.13.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JG, Lawrence GJ, Luck JE, Dodds PN. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. The Plant cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustin B, Lartigue L, Bruey JM, Luciano F, Sergienko E, Bailly-Maitre B, Volkmann N, Hanein D, Rouiller I, Reed JC. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Molecular cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Jehanno M, Mengin-Lecreulx D, Sansonetti PJ, Alzari PM, Philpott DJ. Identification of the critical residues involved in peptidoglycan detection by Nod1. The Journal of biological chemistry. 2005;280:38648–38656. doi: 10.1074/jbc.M509537200. [DOI] [PubMed] [Google Scholar]
- Halff EF, Diebolder CA, Versteeg M, Schouten A, Brondijk TH, Huizinga EG. Formation and structure of a NAIP5-NLRC4 inflammasome induced by direct interactions with conserved N- and C-terminal regions of flagellin. The Journal of biological chemistry. 2012;287:38460–38472. doi: 10.1074/jbc.M112.393512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, Wang R, Zhang Y, Martinon F, Miao D, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341:172–175. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- Iyer SS, He Q, Janczy JR, Elliott EI, Zhong Z, Olivier AK, Sadler JJ, Knepper-Adrian V, Han R, Qiao L, et al. Mitochondrial cardiolipin is required for nlrp3 inflammasome activation. Immunity. 2013;39:311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. The EMBO journal. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic acids research. 2009;37:D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoed EM, Vance RE. Blue native polyacrylamide gel electrophoresis to monitor inflammasome assembly and composition. Methods Mol Biol. 2013;1040:169–183. doi: 10.1007/978-1-62703-523-1_13. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- Krasileva KV, Dahlbeck D, Staskawicz BJ. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. The Plant cell. 2010;22:2444–2458. doi: 10.1105/tpc.110.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, Aravind L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. Journal of molecular biology. 2004;343:1–28. doi: 10.1016/j.jmb.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Levinsohn JL, Newman ZL, Hellmich KA, Fattah R, Getz MA, Liu S, Sastalla I, Leppla SH, Moayeri M. Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS pathogens. 2012;8:e1002638. doi: 10.1371/journal.ppat.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Molecular cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- McDowell JM, Dhandaydham M, Long TA, Aarts MG, Goff S, Holub EB, Dangl JL. Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. The Plant cell. 1998;10:1861–1874. doi: 10.1105/tpc.10.11.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PS, Patzina C, Emerman M, Haller O, Malik HS, Kochs G. Evolution-guided identification of antiviral specificity determinants in the broadly acting interferon-induced innate immunity factor MxA. Cell host & microbe. 2012;12:598–604. doi: 10.1016/j.chom.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyet JL, Srinivasula SM, Tnani M, Razmara M, Fernandes-Alnemri T, Alnemri ES. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. The Journal of biological chemistry. 2001;276:28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, He T, Liang Q, Liu Y, Yuan X, et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Rayamajhi M, Zak DE, Chavarria-Smith J, Vance RE, Miao EA. Cutting Edge: Mouse NAIP1 Detects the Type III Secretion System Needle Protein. J Immunol. 2013 doi: 10.4049/jimmunol.1301549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold TF, Wohlgemuth S, Eschenburg S. Crystal structure of full-length Apaf-1: how the death signal is relayed in the mitochondrial pathway of apoptosis. Structure. 2011;19:1074–1083. doi: 10.1016/j.str.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Song DH, Lee JO. Sensing of microbial molecular patterns by Toll-like receptors. Immunol Rev. 2012;250:216–229. doi: 10.1111/j.1600-065X.2012.01167.x. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tanabe T, Chamaillard M, Ogura Y, Zhu L, Qiu S, Masumoto J, Ghosh P, Moran A, Predergast MM, Tromp G, et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. The EMBO journal. 2004;23:1587–1597. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J, Ayres JS, Kofoed EM, Chavarria-Smith J, Vance RE. Recognition of bacteria by inflammasomes. Annual review of immunology. 2013;31:73–106. doi: 10.1146/annurev-immunol-032712-095944. [DOI] [PubMed] [Google Scholar]
- von Moltke J, Trinidad NJ, Moayeri M, Kintzer AF, Wang SB, van Rooijen N, Brown CR, Krantz BA, Leppla SH, Gronert K, et al. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature. 2012;490:107–111. doi: 10.1038/nature11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Antoine DJ, Andersson U, Tracey KJ. The many faces of HMGB1: molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J Leukoc Biol. 2013a;93:865–873. doi: 10.1189/jlb.1212662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhao Y, Shi J, Shao F. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proceedings of the National Academy of Sciences of the United States of America; 2013b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular biology and evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, Liu L, Shao F. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.