Abstract
Two commonly prescribed treatments for opioid addiction are methadone and buprenorphine. While these drugs show some efficacy in treating opioid dependence, treatment response varies among individuals. It is likely that genetic factors play a role in determining treatment outcome. This study analyses the pharmacogenetic association of 6 polymorphisms in OPRD1, the gene encoding the delta-opioid receptor, on treatment outcome in 582 opioid addicted European Americans randomized to either methadone or buprenorphine/naloxone ((Suboxone®) over the course of a 24 week open-label clinical trial. Treatment outcome was assessed as the number of missed or opioid positive urine drug screens over the 24 weeks. In the total sample, no SNPs in OPRD1 were significantly associated with treatment outcome in either treatment arm. However, sex-specific analyses revealed 2 intronic SNPs (rs581111 and rs529520) that predicted treatment outcome in females treated with buprenorphine. Females with the AA or AG genotypes at rs581111 had significantly worse outcomes than those with the GG genotype when treated with buprenorphine (p=0.03, RR=1.67, 95% C.I.[1.06-2.1]). For rs529520, females with the AA genotype had a significantly worse outcome than those with the CC genotype when (p=0.006, RR=2.15, 95%C.I.[1.3-2.29]). No significant associations were detected in males. These findings suggest that rs581111 and rs52920 may be useful when considering treatment options for female opioid addicts, however confirmation in an independent sample is warranted.
Keywords: Opioids, methadone, buprenorphine, OPRD1, pharmacogenetics, females
Introduction
In 2010, 1.17 million people in the United States received treatment for addiction to opioids or illicit use of opioids. The majority of these individuals were abusing or dependent upon prescription opioid analgesics (1). While the number of individuals receiving treatment for alcohol and illicit drug abuse has remained relatively stable since 2002, the number of those receiving treatment for the abuse of opioid analgesics has more than doubled (National Survey on Drug Use and Health, 2010). The two most commonly prescribed FDA-approved treatments for opioid addiction are methadone and buprenorphine, which act by binding to opioid receptors. Methadone acts as an agonist at the μ-opioid receptor (MOR) (2), whereas buprenorphine is a MOR partial agonist and a κ-opioid receptor (KOR) antagonist (Leander, 1987). Opioid signaling via the δ-opioid receptor (DOR) has also been observed in mice chronically treated with methadone (3) and buprenorphine has a high affinity for DOR (4).
The gene encoding DOR (OPRD1) has previously been associated with the risk for opioid addiction. A study of German heroin addicts found a synonymous SNP in OPRD1 to be associated with addiction (5), and a study analyzing OPRD1 in European Americans found a non-synonymous SNP to increase risk for opioid and general substance addiction (6). In a large case-control study of Australian heroin addicts, intronic OPRD1 SNPs, including the SNP rs2236857, were found to associate with addiction (7). A previous study of severe heroin addiction also found rs2236857 to be nominally associated in European Americans (8). Negative findings, that fail to associate OPRD1 polymorphisms with heroin addiction have also been reported (9-11).
Methadone and buprenorphine have substantial efficacy for the treatment of opioid addiction (12); however, a subset of patients relapse, are non-compliant with treatment protocols, and continue to use illicit opioids (12-18). There are a number of factors that influence treatment outcome, including a prior history of drug abuse, medication dosage, polydrug abuse, and ethnic differences (19-22). Patient sex is also known to influence treatment outcome and significant sex differences are reported in the progression from opioid abuse to seeking treatment, the subsequent use of health services, drug metabolism, and the clinical profiles of opioid abusers (23-26).
Sexual dimorphism in the response to opioids is reported in both animals and humans (27). Morphine may be more potent in women (28); however, due to a slower onset of action women require more morphine for analgesic effects (29, 30). Furthermore, women report greater analgesia from KOR agonists compared to males (31-33). Female rats have increased levels of KOR/MOR heterodimers in the spinal cord (34), which affects morphine antinociception, and this has been shown to be dependent on levels of spinal estrogen (35).
Genetic factors are also associated with treatment outcome for opioid addiction (36, 37). A recent pharmacogenetic study of methadone and buprenorphine treatment for opioid addiction analyzed whether polymorphisms in OPRD1 would predict the number of opioid positive urine drug screens in individuals over the course of a 24 week open randomized trial (36). In this study a T/C intronic SNP, rs678849, was found to predict treatment outcome for individuals of African American descent. African Americans carrying a T allele at rs678849 were found to have fewer opioid positive urine samples when treated with methadone; however, individuals with the C/C genotype had better outcomes when treated with buprenorphine.
Despite the growing evidence for sex differences in the response to opioids, little research has been done to identify sex-specific pharmacogenetic effects in treatment for opioid dependence. A recently published paper from our group found SNPs in OPRD1 to be associated with the response to methadone and buprenorphine in African American opioid addicts, however, they found no association in European Americans (36). Using the same sample of European Americans, the present study analyzed the association of SNPs in OPRD1 in males and females separately, in order to identify sex-specific pharmacogenetic effects. The sample utilized was part of an open-label randomized clinical trial designed to assess the effects of methadone and buprenorphine on liver function, which also collected genetic material and data on treatment response (38).
Materials and Methods
Participants and Procedures
The current data were obtained from a National Drug Abuse Treatment Clinical Trials Network (CTN) study. The main outcome measures and study design for this clinical trial have been described in detail previously (38). In summary, individuals seeking treatment for opioid addiction were recruited at federally licensed opioid treatment programs in the United States between May 2006 and October 2009. Institutional review boards at participating sites approved the study, and the NIDA Clinical Trials Network Data Safety and Monitoring Board provided oversight. All patients were at least 18 years of age and met DSM-IV-TR criteria for opioid dependence. Exclusion criteria for the trial included: cardiomyopathy, liver disease, acute psychosis, blood levels of alanine amino transferase or aspartate amino transferase greater than 5 times the maximum normal level, or poor venous access. Patients were randomly assigned to 24 weeks of open-label buprenorphine/naloxone ((Suboxone®) or methadone treatment. Patients were defined as European American if they primarily self-identified as ‘white’ in the study. This also includes a number of patients who identified ‘Latino’ as a secondary identifier (N males=22, N females= 13).
A flexible dosing approach was used, with a wide range allowed in both induction dosing and subsequent maintenance dosing. First day buprenorphine dose began at 2 to 8mg, which could be increased to 16mg in the case of persistent withdrawal. Buprenorphine could be further increased in subsequent days to a maximum dose of 32 mg; the mean maximum daily dose for the trial completers analyzed in this study was 24.5 ± 8.3 mg. The initial maximum dose of methadone was limited to 30 mg. For persistent withdrawal an additional dose was allowed up to a maximum total first day dose of 40 mg as stipulated by US statute. Methadone dose could be increased in subsequent days by 10 mg increments with no maximum. The mean maximum daily methadone dose for the trial completers analyzed in this study was 97.3 ± 45.0 mg. Patients came to the clinic daily for observed dosing except on Sundays and holidays or if local regulations permitted take-home medications. Weekly urine drug samples were taken and tested for opioids. Samples testing positive for methadone were counted as positive for individuals in the buprenorphine group, but not for individuals in the methadone group.
SNP selection and genotyping
SNPs with a minor allele frequency >10% were selected for genotyping using the Tagger algorithm implemented in the Haploview software package (http://www.broadinstitute.org/haploview) (39). 6 SNPs were selected for genotyping (rs1042114, rs678849, rs10753331, rs529520, rs581111, and rs2234918) and these were found to tag 71% of SNPs in OPRD1 with an r2 of 0.8 and a MAF cut-off of 10%, using the International HapMap project CEU population data (HapMap data release 28 phase II & III, August 2010, www.hapmap.org). Using a MAF cut-off of 5%, 62% of SNPs in OPRD1 were captured with an r2 of 0.8. rs2234918 was not genotyped in the HapMap population and is not included in the linkage disequilibrium (LD) calculation.
All SNPs were genotyped using Taqman® SNP Genotyping Assays (Applied Biosystems Inc. (ABI); Foster City, CA, USA) according to the standard Applied Biosystems protocol. Quality control was maintained by genotyping 10% duplicates, which were checked for genotype concordance across the population. The duplicate concordance rate was 100%.
Statistical Analysis
Initial comparisons of average outcome in males compared to females were analyzed by student’s t-test. For each SNP, deviation from Hardy-Weinberg was analyzed and all SNPs were in Hardy-Weinberg Equilibrium (p≥0.05). Gene × environment analyses were performed in the software package PLINK v1.07 (40) for the male and female groups separately, using the percentage of missed or opioid positive urine drug screens over the 24 weeks as the phenotype. Treatment group (buprenorphine or methadone) was used as a covariate. P-values for the gene-environment analyses are reported as significant if they remain significant after correction for multiple testing using the (FDR) procedure with the cut-off for statistical significance after correction set to p≤0.05 (41). As rs581111 and rs529520 were found to be significant in the gene × environment analyses in females, the average percentage of missed or positive urine tests by rs581111 and rs529520 genotype were analyzed by one-way ANOVA with Tukey HSD post hoc analysis. Due to the low frequency of A homozygotes in the population for rs581111, the AA and AG genotypes were combined for the ANOVA analysis. We used Generalized Estimating Equation (GEE) to investigate the associations of genotype and longitudinal urine drug screen outcomes from week 1 to week 24, adjusting for the effects of age, time (week), gender, and treatment group. GEE is a quasi-likelihood based method which produces population averaged estimates for longitudinal binary outcomes (42). As the GEE provides weighted estimates for missing data, the GEE was run with missing tests coded as positive, and again with missing tests coded as missing. We report our estimates as relative risks, and bootstrapped 95% confidence intervals based on 1000 replicate samples. We analyzed urine drug screen outcomes for both treatment groups separately and for the entire sample as a whole, examining the main effects of treatment and rs581111 or rs529520 genotype, as well as the interaction effect of treatment × genotype.
Results
Demographics
DNA samples were available from 582 Europeans Americans who received either methadone or buprenorphine for the treatment of opioid dependence. The genetic analysis was restricted to European Americans in order to minimize the effects of genetic population sub-structure between different ethnicities. The number of males and females randomized to each group, the mean age, the average outcome for each treatment group and the average number of missed tests over 24 weeks are summarized in Table 1. The average percentage of positive urine tests over 24 weeks was not significantly different between males (51%) and females (43%, p=0.1) in the buprenorphine group or between males (43%) and females (45%, p=0.63) in the methadone group.
Table 1.
Demographic information and treatment outcomes for participants treated with methadone or buprenorphine for opioid dependence.
| Treatment Group | Methadone | Methadone | Buprenorphine | Buprenorphine |
|---|---|---|---|---|
| Sex | Female | Male | Female | Male |
| Number | 104 | 179 | 81 | 218 |
| Mean Age ± SD | 36.4 ± 10.1 | 35.2 ± 10.9 | 37.0 ± 11.5 | 35.6 ± 11.0 |
| Mean % Positive Urinalysis ± SD | 45.3 ± 32.7% | 43.4 ± 31.6% | 43.0 ± 36.2% | 50.6 ± 35.7% |
| Mean % Missing Tests ± SD | 12.1 ± 17.53% | 13.01 ± 20.15% | 23.1 ± 29.15% | 25.73 ± 27.78% |
SD = standard deviation.
Gene × Environment Analysis
In order to determine if any of the OPRD1 genetic variants were associated with treatment outcome in either males or females, gene × environment analyses were run with the average number of missing or positive urine drug tests over the course of the 24 week trial used as the outcome variable. The results of these analyses are shown in Tables 2 and 3. No significant interactions were observed in males (Table 2); however, in females, rs581111 (p=0.002) and rs529520 (p=0.005) were associated with the number of missing or positive urine drug tests (Table 3). Two additional SNPs, rs1042114 and rs10753331, were nominally associated with treatment outcome in females but these associations did not withstand correction for multiple testing. The GxE analysis was also run with the individuals who self-identified as ‘White/Latino’ removed. This was not found to affect the results, as rs529520 and rs581111 were still significantly associated with treatment outcome in females (data not shown).
Table 2.
The effects of OPRD1 genetic variants on treatment outcome in males.
| SNP ID | Minor Allele | Position | Beta 1 - Methadone | Beta 2 - Buprenorphine | P-value |
|---|---|---|---|---|---|
| rs1042114 | G | 29138975 | -0.08723 | -0.03386 | 0.45 |
| rs678849 | C | 29145188 | 0.03693 | 0.001732 | 0.48 |
| rs10753331 | A | 29164582 | 0.02592 | 0.01808 | 0.88 |
| rs529520 | T | 29174946 | -0.02846 | -0.00568 | 0.64 |
| rs581111 | T | 29175373 | -0.00934 | -0.00478 | 0.94 |
| rs2234918 | C | 29189597 | 0.005875 | -0.02054 | 0.60 |
P-values were generated by gene × environment analyses in PLINK with treatment group as a covariate. Beta 1 and Beta 2 indicate the regression coefficients for methadone and buprenorphine, respectively.
Table 3.
The effects of OPRD1 genetic variants on treatment outcome in females.
| SNP ID | Minor Allele | Position | Beta 1 - Methadone | Beta 2 - Buprenorphine | P-value |
|---|---|---|---|---|---|
| rs1042114 | G | 29138975 | -0.07486 | 0.2055 | 0.015 |
| rs678849 | C | 29145188 | -0.04264 | 0.08234 | 0.11 |
| rs10753331 | A | 29164582 | -0.07338 | 0.08108 | 0.044 |
| rs529520 | T | 29174946 | -0.06821 | 0.1381 | 0.0048 |
| rs581111 | T | 29175373 | -0.07301 | 0.1792 | 0.0020 |
| rs2234918 | C | 29189597 | 0.02903 | 0.01693 | 0.87 |
P-values were generated by gene × environment analyses in PLINK with treatment group as a covariate. Beta 1 and Beta 2 indicate the regression coefficients for methadone and buprenorphine, respectively.
Across the 24 weeks of the trial, female carriers of the rs581111 GG genotype had a fewer missing or positive opioid drug screens when treated with buprenorphine (31.5% ± 30.1%) compared to those patients with an AA or AG genotype (56.8% ±37.7%) (p<0.01). This association was not seen in the methadone treated group, as the percentage of missing or positive drug screens amongst the GG carriers (47.5% ±32.2%) compared to AA or AG carriers (40.4% ±32.0%) was not significantly different. No significant difference in outcome was observed, in either the methadone or buprenorphine treatment groups based on rs529520 genotype.
Generalized Estimating Equation (GEE) analysis
GEE were used to calculate the effect of the gene × environment interaction when the 24 weeks of urinalysis data were taken as repeated measures. A significant interaction between genotype at rs581111 and treatment group was observed (p=0.03, RR=1.67, 95%C.I.[1.06-2.1]), confirming the original gene × environment finding. When treatment groups were analyzed separately, females with the AA or AG genotypes at rs581111 had significantly worse outcomes than those with the GG genotype when treated with buprenorphine (p=0.031, RR=1.72, 95%C.I.[1.25-1.97]) (Figure 1). However, no significant interaction was found in the methadone treated group (data not shown). For rs529520, a significant interaction of treatment and genotype was observed as females with the AA genotype had a worse outcome than those with the CC genotype when treated with buprenorphine (p=0.006, RR=2.15, 95% C.I.[1.3-2.29]) (Figure 2A). The number of missing or positive urine tests was not significantly different between the AC and AA genotypes (p=0.072, RR=1.51, 95% C.I.[0.99-2.06] (Figure 2B). When the buprenorphine treated cohort were analyzed separately the association of AA genotype with treatment outcome was still significant (p=0.05, RR=1.8, 95% C.I.[1.04-2.02]). No association was observed in the methadone treated individuals (data not shown).
Figure 1.
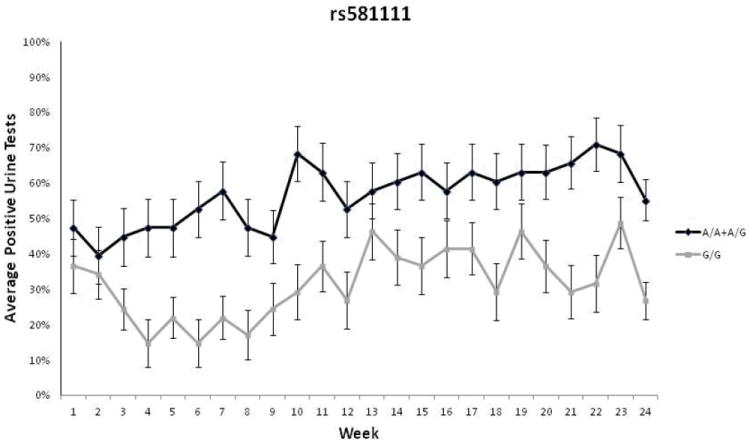
Longitudinal urinalysis data for females treated with buprenorphine for 24 weeks based on rs581111 genotype. Weekly urine drug screens were administered for the presence of opioids other than the one prescribed. The average percentage of missing or opioid positive urine tests during each week is provided for individuals with each genotype. Due to the low minor allele frequency of rs581111, A/A and A/G carriers are grouped together. Patients with the AA/AG genotypes were more likely to have opioid-positive urine drug screens(p=0.031, RR=1.72, 95% C.I.[1.25-1.97]). Error bars represent standard error of the mean.
Figure 2.
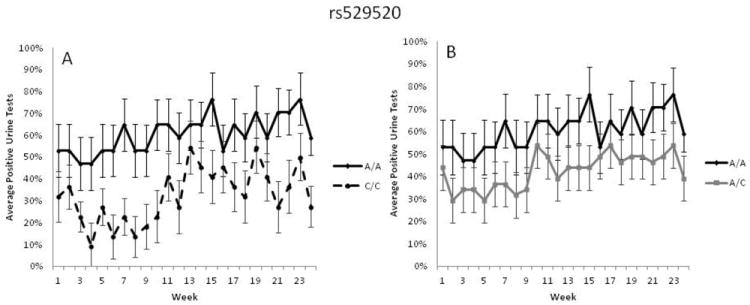
Longitudinal urinalysis data for females treated with buprenorphine for 24 weeks based on rs529520 genotype. Weekly urine drug screens were administered for the presence of opioids other than the one prescribed. The average percentage of missing or opioid positive urine tests during each week is provided for individuals with each genotype. A) Patients with the AA genotype were more likely to have missing or opioid-positive drug screens compared to patients with the CC genotype (p=0.025, RR=1.65). B) Patients with the AA genotype were not significantly different from patients with the AC genotype (p=0.072, RR=1.51, 95% C.I.[0.99-2.06]). Error bars represent standard error of the mean.
The GEE was re-run with missing urine drug screens coded as ‘missing’ rather than ‘positive’. Significant interactions of treatment and genotype remained for females treated with buprenorphine. For rs529520, carriers of the AA genotype did significantly worse than those with the CC genotype (p=0.025, RR=1.65, 95% C.I. [1.5-2.06]). For rs581111, the interaction of treatment and genotype was also significant, with carriers of the A allele having significantly worse outcomes than those with the GG genotype (p=0.009, RR=1.56, 95% C.I. [1.41-1.78]).
Discussion
OPRD1 has been previously associated with heroin addiction and treatment outcome (5-8, 36). The present study demonstrates that 2 SNPs located in intron 1, rs581111 and rs529520, predict treatment outcome in females treated with buprenorphine. Female opioid addicts with the GG genotype at rs581111 were found to have significantly better outcomes when treated with buprenorphine, compared to patients with the AG or AA genotype. Females with the CC genotype at rs529520 had significantly fewer missing or opioid positive drug screens over 24 weeks compared to those with the AA genotype. Levran et al. found 2 SNPs in intron 1 of OPRD1, rs2236857 and rs2236861, to be associated with heroin addiction and these SNPs are in LD (D’=1) with rs529520 and rs581111 respectively, however the r2 between these SNPs in Europeans is modest (0.02-0.3). Furthermore, Nelson et al. demonstrated a haplotype of rs2236857 and rs581111 to be associated with heroin addiction. As we find rs581111 to also be associated with treatment outcome in females, these data lend further support to the importance of OPRD1 genotypes for opioid addiction.
Sex differences that distinguish male and female opioid addicts have been previously reported (24, 43, 44). Female opioid addicts exhibit different drug abuse profiles and present with different medical and psychiatric problems compared to their male counterparts (23, 25). While some of the differences in the clinical profiles of male and female opioid addicts may be societally or environmentally influenced, it is likely that some of this variance has a biological basis.
The opioid binding capacity of men and women in the brain has been shown to differ, as a study found women to have higher MOR binding as determined by positron emission tomography (PET) scanning (45). Furthermore, sex differences in the analgesic response to opioids have been reported for KOR agonists/antagonists, with KOR-acting analgesics producing greater analgesia in females compared to men (31-33). Furthermore, the pharmacokinetics of buprenorphine have been shown to differ between males and females and this is influenced by sex differences in body composition (26). Sexual dimorphism in DOR expression and function has also been observed. Female rats show an increase level of DOR associated with the plasma membrane in the nucleus accumbens core, following withdrawal from cocaine (46) and stress-induced analgesia was found to be decreased in female Oprm1-/-/Oprd1-/- mice when compared to males (47).
Given the inherent differences observed in the opioid system between males and females it is plausible that a pharmacogenetic effect involving OPRD1 polymorphisms and the response to buprenorphine would be sex-specific. Interestingly, a bioinformatic analysis of intron 1 of OPRD1 revealed a perfect estrogen response element (ERE) located just 77 base pairs away from rs581111. Furthermore, an ERE element is located in the promoter of OPRD1 (48), suggesting that OPRD1 gene expression may be regulated by estrogen. Levels of DOR are also affected by buprenorphine, as treatment in mice leads to an upregulation of DOR in the forebrain (49, 50). Furthermore, norbuprenorphine, a metabolite of buprenorphine, acts as a DOR agonist in vitro (51). These data, suggest that the OPRD1 locus is worthy of further study when understanding treatment response to buprenorphine, specifically in females.
There are a number of limitations to this study. rs581111 and rs529520 are located 427 base pairs apart in intron 1 of OPRD1. From the genotyping data and statistical analysis alone, it is not possible to determine whether both of these SNPs are relevant for treatment outcome or whether the association at one SNP is affecting the result at the other. There is moderate LD between these 2 SNPs in the European population (D’=1, r2=0.46), and it is possible that these SNPs together tag another locus which is the causal variant for the phenotype observed in this study. Further work on the role of rs581111 and rs529520 in the context of OPRD1 gene expression and DOR protein levels and re-sequencing of the OPRD1 locus is required to understand how these SNPs affect the response to treatment for opioid dependence in females. Another limitation to this study pertains to the handling of missing data. During the START trial, a higher rate of dropouts in the buprenorphine group was observed (38) compared to those treated with methadone. This may have influenced our analyses as our data show an association of genotype with treatment response in the buprenorphine arm of the trial. However, as the participants enrolled in this trial were opioid dependent, it is a reasonable assumption that if they were not present to provide a urine sample and receive agonist replacement therapy, then it is likely that they were abusing opioids.
Finally, the number of women enrolled in the START trial is relatively low, with 104 patients in the methadone arm of the trial and 81 in the buprenorphine arm. Therefore, in order to validate these findings, replication in an independent study is warranted. Confirming these associations will be an important finding in the field of opioid addiction treatment as these genotypes have the potential to determine the appropriate treatment regime for female opioid addicts.
Acknowledgments
Main START study funding came from the National Institute on Drug Abuse through the Clinical Trials Network (CTN) through a series of grants provided to each participating node: the Pacific Northwest Node (U10 DA01714), the Oregon Hawaii Node (U10 DA013036), the California/Arizona Node (U10 DA015815), the New England Node (U10 DA13038), the Delaware Valley Node (U10 DA13043), the Pacific Region Node (U10 DA13045), and the New York Node (U10 DA013046). Dr. Berrettini was also supported by the Delaware Valley Node (U10 DA13043). This work was supported by the Center for Neurobiology and Behavior, Department of Psychiatry, University of Pennsylvania, Training Program in Neuropsychopharmacology (T32MH014654, P.I.: I. Lucki), NIDA grant P20DA025995 (P.I.: W. Berrettini), the Veterans Administration Mental Illness Research Education and Clinical Center MIRECC) at the Philadelphia VAMC (David Oslin, MD, PI) and NIDA grant P60DA05186 (P.I.: Charles O’Brien).
Footnotes
Conflicts of Interest:
Andrew Saxon: Paid consultant to Reckitt Benckiser Pharmaceuticals; Walter Ling: Paid consultant to Reckitt Benckiser Pharmaceuticals; R. Douglas Bruce: Research grant support from Gilead Sciences, Inc., Merck & Co., Bristol Myers Squibb, Boehringer Ingelheim, Reckitt Benckiser Pharmaceuticals, Abbott Laboratories, Pfizer, Inc., and honorarium from Reckitt Benckiser Pharmaceuticals; All other authors report no financial or other possible conflicts of interest.
References
- 1.Administration SAaMHS. Results from the 2011 National Survey on Drug Use and Health: Summary of National Findings. NSDUH Series H-44. 2010 HHS Publication No. (SMA) 12-4713. [Google Scholar]
- 2.Nicholls L, Bragaw L, Ruetsch C. Opioid dependence treatment and guidelines. J Manag Care Pharm. 2010 Feb;16(1 Suppl B):S14–21. doi: 10.18553/jmcp.2010.16.S1-B.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rady JJ, Portoghese PS, Fujimo JM. Methadone and heroin antinociception: predominant delta-opioid-receptor responses in methadone-tolerant mice. Japanese journal of pharmacology. 2002 Mar;88(3):319–31. doi: 10.1254/jjp.88.319. [DOI] [PubMed] [Google Scholar]
- 4.Negus SS, Bidlack JM, Mello NK, Furness MS, Rice KC, Brandt MR. Delta opioid antagonist effects of buprenorphine in rhesus monkeys. Behavioural pharmacology. 2002 Nov;13(7):557–70. doi: 10.1097/00008877-200211000-00005. [DOI] [PubMed] [Google Scholar]
- 5.Mayer P, Rochlitz H, Rauch E, Rommelspacher H, Hasse HE, Schmidt S, et al. Association between a delta opioid receptor gene polymorphism and heroin dependence in man. Neuroreport. 1997 Jul 28;8(11):2547–50. doi: 10.1097/00001756-199707280-00025. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Molecular psychiatry. 2008 May;13(5):531–43. doi: 10.1038/sj.mp.4002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand FL, et al. Association of OPRD1 polymorphisms with heroin dependence in a large case-control series. Addiction biology. 2012 Apr 13; doi: 10.1111/j.1369-1600.2012.00445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levran O, Londono D, O’Hara K, Nielsen DA, Peles E, Rotrosen J, et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes, brain, and behavior. 2008 Oct;7(7):720–9. doi: 10.1111/j.1601-183X.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franke P, Nothen MM, Wang T, Neidt H, Knapp M, Lichtermann D, et al. Human delta-opioid receptor gene and susceptibility to heroin and alcohol dependence. American journal of medical genetics. 1999 Oct 15;88(5):462–4. doi: 10.1002/(sici)1096-8628(19991015)88:5<462::aid-ajmg4>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 10.Xu K, Liu XH, Nagarajan S, Gu XY, Goldman D. Relationship of the delta-opioid receptor gene to heroin abuse in a large Chinese case/control sample. American journal of medical genetics. 2002 Jun 1;110(1):45–50. doi: 10.1002/ajmg.10374. [DOI] [PubMed] [Google Scholar]
- 11.Xuei X, Flury-Wetherill L, Bierut L, Dick D, Nurnberger J, Jr, Foroud T, et al. The opioid system in alcohol and drug dependence: family-based association study. Am J Med Genet B Neuropsychiatr Genet. 2007 Oct 5;144B(7):877–84. doi: 10.1002/ajmg.b.30531. [DOI] [PubMed] [Google Scholar]
- 12.Mattick RP, Kimber J, Breen C, Davoli M. Buprenorphine maintenance versus placebo or methadone maintenance for opioid dependence. Cochrane database of systematic reviews. 2008(2) doi: 10.1002/14651858.CD002207.pub3. (Online) CD002207. [DOI] [PubMed] [Google Scholar]
- 13.Fiellin DA, O’Connor PG, Chawarski M, Pakes JP, Pantalon MV, Schottenfeld RS. Methadone maintenance in primary care: a randomized controlled trial. Jama. 2001 Oct 10;286(14):1724–31. doi: 10.1001/jama.286.14.1724. [DOI] [PubMed] [Google Scholar]
- 14.Johnson RE, Fudala PJ, Payne R. Buprenorphine: considerations for pain management. Journal of pain and symptom management. 2005 Mar;29(3):297–326. doi: 10.1016/j.jpainsymman.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Ling W, Wesson DR, Charuvastra C, Klett CJ. A controlled trial comparing buprenorphine and methadone maintenance in opioid dependence. Archives of general psychiatry. 1996 May;53(5):401–7. doi: 10.1001/archpsyc.1996.01830050035005. [DOI] [PubMed] [Google Scholar]
- 16.Pani PP, Maremmani I, Pirastu R, Tagliamonte A, Gessa GL. Buprenorphine: a controlled clinical trial in the treatment of opioid dependence. Drug and alcohol dependence. 2000 Jul 1;60(1):39–50. doi: 10.1016/s0376-8716(99)00140-4. [DOI] [PubMed] [Google Scholar]
- 17.Petitjean S, Stohler R, Deglon JJ, Livoti S, Waldvogel D, Uehlinger C, et al. Double-blind randomized trial of buprenorphine and methadone in opiate dependence. Drug and alcohol dependence. 2001 Mar 1;62(1):97–104. doi: 10.1016/s0376-8716(00)00163-0. [DOI] [PubMed] [Google Scholar]
- 18.Strain EC, Bigelow GE, Liebson IA, Stitzer ML. Moderate- vs high-dose methadone in the treatment of opioid dependence: a randomized trial. Jama. 1999 Mar 17;281(11):1000–5. doi: 10.1001/jama.281.11.1000. [DOI] [PubMed] [Google Scholar]
- 19.del Rio M, Mino A, Perneger TV. Predictors of patient retention in a newly established methadone maintenance treatment programme. Addiction (Abingdon, England) 1997 Oct;92(10):1353–60. [PubMed] [Google Scholar]
- 20.Farrell M, Ward J, Mattick R, Hall W, Stimson GV, des Jarlais D, et al. Methadone maintenance treatment in opiate dependence: a review. BMJ (Clinical research ed) 1994 Oct 15;309(6960):997–1001. doi: 10.1136/bmj.309.6960.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamal F, Flavin S, Campbell F, Behan C, Fagan J, Smyth R. Factors affecting the outcome of methadone maintenance treatment in opiate dependence. Irish medical journal. 2007 Mar;100(3):393–7. [PubMed] [Google Scholar]
- 22.Wu LT, Pan JJ, Blazer DG, Tai B, Stitzer ML, Woody GE. Using a latent variable approach to inform gender and racial/ethnic differences in cocaine dependence: a National Drug Abuse Treatment Clinical Trials Network study. Journal of substance abuse treatment. 2010 Jun;38(Suppl 1):S70–9. doi: 10.1016/j.jsat.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Back SE, Payne RL, Wahlquist AH, Carter RE, Stroud Z, Haynes L, et al. Comparative profiles of men and women with opioid dependence: results from a national multisite effectiveness trial. The American journal of drug and alcohol abuse. 2011 Sep;37(5):313–23. doi: 10.3109/00952990.2011.596982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez-Avila CA, Rounsaville BJ, Kranzler HR. Opioid-, cannabis- and alcohol-dependent women show more rapid progression to substance abuse treatment. Drug and alcohol dependence. 2004 Jun 11;74(3):265–72. doi: 10.1016/j.drugalcdep.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Kosten TR, Rounsaville BJ, Kleber HD. Ethnic and gender differences among opiate addicts. The International journal of the addictions. 1985 Aug;20(8):1143–62. doi: 10.3109/10826088509056356. [DOI] [PubMed] [Google Scholar]
- 26.Moody DE, Fang WB, Morrison J, McCance-Katz E. Gender differences in pharmacokinetics of maintenance dosed buprenorphine. Drug and alcohol dependence. 2011 Nov 1;118(2-3):479–83. doi: 10.1016/j.drugalcdep.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dahan A, Kest B, Waxman AR, Sarton E. Sex-specific responses to opiates: animal and human studies. Anesthesia and analgesia. 2008 Jul;107(1):83–95. doi: 10.1213/ane.0b013e31816a66a4. [DOI] [PubMed] [Google Scholar]
- 28.Sarton E, Olofsen E, Romberg R, den Hartigh J, Kest B, Nieuwenhuijs D, et al. Sex differences in morphine analgesia: an experimental study in healthy volunteers. Anesthesiology. 2000 Nov;93(5):1245–54. doi: 10.1097/00000542-200011000-00018. discussion 6A. [DOI] [PubMed] [Google Scholar]
- 29.Aubrun F, Salvi N, Coriat P, Riou B. Sex- and age-related differences in morphine requirements for postoperative pain relief. Anesthesiology. 2005 Jul;103(1):156–60. doi: 10.1097/00000542-200507000-00023. [DOI] [PubMed] [Google Scholar]
- 30.Cepeda MS, Carr DB. Women experience more pain and require more morphine than men to achieve a similar degree of analgesia. Anesthesia and analgesia. 2003 Nov;97(5):1464–8. doi: 10.1213/01.ANE.0000080153.36643.83. [DOI] [PubMed] [Google Scholar]
- 31.Gear RW, Gordon NC, Heller PH, Paul S, Miaskowski C, Levine JD. Gender difference in analgesic response to the kappa-opioid pentazocine. Neuroscience letters. 1996 Mar 1;205(3):207–9. doi: 10.1016/0304-3940(96)12402-2. [DOI] [PubMed] [Google Scholar]
- 32.Gear RW, Miaskowski C, Gordon NC, Paul SM, Heller PH, Levine JD. Kappa-opioids produce significantly greater analgesia in women than in men. Nature medicine. 1996 Nov;2(11):1248–50. doi: 10.1038/nm1196-1248. [DOI] [PubMed] [Google Scholar]
- 33.Gear RW, Miaskowski C, Gordon NC, Paul SM, Heller PH, Levine JD. The kappa opioid nalbuphine produces gender- and dose-dependent analgesia and antianalgesia in patients with postoperative pain. Pain. 1999 Nov;83(2):339–45. doi: 10.1016/s0304-3959(99)00119-0. [DOI] [PubMed] [Google Scholar]
- 34.Chakrabarti S, Liu NJ, Gintzler AR. Formation of mu-/kappa-opioid receptor heterodimer is sex-dependent and mediates female-specific opioid analgesia. Proceedings of the National Academy of Sciences of the United States of America. 2010 Nov 16;107(46):20115–9. doi: 10.1073/pnas.1009923107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu NJ, Chakrabarti S, Schnell S, Wessendorf M, Gintzler AR. Spinal synthesis of estrogen and concomitant signaling by membrane estrogen receptors regulate spinal kappa- and mu-opioid receptor heterodimerization and female-specific spinal morphine antinociception. J Neurosci. 2011 Aug 17;31(33):11836–45. doi: 10.1523/JNEUROSCI.1901-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crist R, Clarke T, Ang A, Ambrose-Lanci LM, Lohoff F, Saxon AJ, et al. An intronic variant in OPRD1 predicts treatment outcome for opioid dependence in African-Americans. Neuropsychopharmacology. 2013 doi: 10.1038/npp.2013.99. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oneda B, Crettol S, Bochud M, Besson J, Croquette-Krokar M, Hammig R, et al. beta-Arrestin2 influences the response to methadone in opioid-dependent patients. The pharmacogenomics journal. 2011 Aug;11(4):258–66. doi: 10.1038/tpj.2010.37. [DOI] [PubMed] [Google Scholar]
- 38.Saxon AJ, Ling W, Hillhouse M, Thomas C, Hasson A, Ang A, et al. Buprenorphine/Naloxone and methadone effects on laboratory indices of liver health: A randomized trial. Drug and alcohol dependence. 2012 Aug 22; doi: 10.1016/j.drugalcdep.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (Oxford, England) 2005 Jan 15;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 40.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007 Sep;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behavioural brain research. 2001 Nov 1;125(1-2):279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 42.Liang KYZ. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 43.Anglin MD, Hser YI, McGlothlin WH. Sex differences in addict careers. 2. Becoming addicted. The American journal of drug and alcohol abuse. 1987;13(1-2):59–71. doi: 10.3109/00952998709001500. [DOI] [PubMed] [Google Scholar]
- 44.Longshore D, Hsieh S, Danila B, Anglin MD. Methadone maintenance and needle/syringe sharing. The International journal of the addictions. 1993 Aug;28(10):983–96. doi: 10.3109/10826089309062178. [DOI] [PubMed] [Google Scholar]
- 45.Zubieta JK, Dannals RF, Frost JJ. Gender and age influences on human brain mu-opioid receptor binding measured by PET. The American journal of psychiatry. 1999 Jun;156(6):842–8. doi: 10.1176/ajp.156.6.842. [DOI] [PubMed] [Google Scholar]
- 46.Ambrose-Lanci LM, Peiris NB, Unterwald EM, Van Bockstaele EJ. Cocaine withdrawal-induced trafficking of delta-opioid receptors in rat nucleus accumbens. Brain research. 2008 May 19;1210:92–102. doi: 10.1016/j.brainres.2008.02.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Contet C, Gaveriaux-Ruff C, Matifas A, Caradec C, Champy MF, Kieffer BL. Dissociation of analgesic and hormonal responses to forced swim stress using opioid receptor knockout mice. Neuropsychopharmacology. 2006 Aug;31(8):1733–44. doi: 10.1038/sj.npp.1300934. [DOI] [PubMed] [Google Scholar]
- 48.Bourdeau V, Deschenes J, Metivier R, Nagai Y, Nguyen D, Bretschneider N, et al. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Molecular endocrinology (Baltimore, Md) 2004 Jun;18(6):1411–27. doi: 10.1210/me.2003-0441. [DOI] [PubMed] [Google Scholar]
- 49.Belcheva MM, Barg J, McHale RJ, Dawn S, Ho MT, Ignatova E, et al. Differential down- and up-regulation of rat brain opioid receptor types and subtypes by buprenorphine. Molecular pharmacology. 1993 Jul;44(1):173–9. [PMC free article] [PubMed] [Google Scholar]
- 50.Belcheva MM, Ho MT, Ignatova EG, Jefcoat LB, Barg J, Vogel Z, et al. Buprenorphine differentially alters opioid receptor adaptation in rat brain regions. The Journal of pharmacology and experimental therapeutics. 1996 Jun;277(3):1322–7. [PMC free article] [PubMed] [Google Scholar]
- 51.Kajiwara M, Aoki K, Ishii K, Numata H, Matsumiya T, Oka T. Agonist and antagonist actions of buprenorphine on three types of opioid receptor in isolated preparations. Japanese journal of pharmacology. 1986 Jan;40(1):95–101. doi: 10.1254/jjp.40.95. [DOI] [PubMed] [Google Scholar]


