SUMMARY
The Arg/N-end rule pathway targets for degradation proteins that bear specific unacetylated N-terminal residues while the Ac/N-end rule pathway targets proteins through their Nα-terminally acetylated (Nt-acetylated) residues. Here we show that Ubr1, the ubiquitin ligase of the Arg/N-end rule pathway, recognizes unacetylated N-terminal methionine if it is followed by a hydrophobic residue. This capability of Ubr1 expands the range of substrates that can be targeted for degradation by the Arg/N-end rule pathway, because virtually all nascent cellular proteins bear N-terminal methionine. We identified Msn4, Sry1, Arl3, and Pre5 as examples of normal or misfolded proteins that can be destroyed through the recognition of their unacetylated N-terminal methionine. Inasmuch as proteins bearing the Nt-acetylated N-terminal methionine residue are substrates of the Ac/N-end rule pathway, the resulting complementarity of the Arg/N-end rule and Ac/N-end rule pathways enables the elimination of protein substrates regardless of acetylation state of N-terminal methionine in these substrates.
Keywords: acetylation, N-end rule, degron, ubiquitin, proteolysis, Ubr1
INTRODUCTION
The N-end rule pathway recognizes proteins containing N-terminal degradation signals called N-degrons, polyubiquitylates these proteins and thereby causes their degradation by the proteasome (Bachmair et al., 1986; Varshavsky, 2011). The main determinant of an N-degron is a destabilizing N-terminal residue of a protein. Recognition components of the N-end rule pathway are called N-recognins. In eukaryotes, N-recognins are E3 ubiquitin (Ub) ligases that can target N-degrons (Figure S1).
Regulated degradation of proteins or their fragments by the N-end rule pathway mediates a strikingly broad range of biological functions, including the sensing of heme, nitric oxide, and oxygen; the control, through degradation, of subunit stoichiometries in multisubunit proteins; the elimination of misfolded proteins; the repression of apoptosis and neurodegeneration; the regulation of chromosome repair, transcription, replication, and cohesion/segregation; the regulation of G proteins, autophagy, peptide import, meiosis, immunity, fat metabolism, cell migration, actin filaments, cardiovascular development, spermatogenesis, neurogenesis, and memory; and the regulation of many processes in plants (Figure S1 and references therein) (Dougan et al., 2011; Finley et al., 2012; Tasaki et al., 2012; Varshavsky, 2008, 2011).
In eukaryotes, the N-end rule pathway consists of two branches. One of them, called the Arg/N-end rule pathway, targets specific unacetylated N-terminal residues (Figure S1B) (Bachmair et al., 1986; Brower et al., 2013; Piatkov et al., 2012). N-terminal Arg, Lys, His, Leu, Phe, Tyr, Trp, and Ile are directly recognized by N-recognins. In contrast, N-terminal Asn, Gln, Asp, and Glu (as well as Cys, under some metabolic conditions) are destabilizing owing to their preliminary enzymatic modifications, which include N-terminal deamidation (Nt-deamidation) and Nt-arginylation (Figure S1B, C). In the yeast S. cerevisiae, the Arg/N-end rule pathway is mediated by the Ubr1 N-recognin, a 225 kDa RING-type E3 Ub ligase and a part of the targeting complex containing the Ubr1-Rad6 and Ufd4-Ubc4/5 holoenzymes (Hwang et al., 2010a).
The other branch, called the Ac/N-end rule pathway, recognizes proteins through their Nα-terminally acetylated (Nt-acetylated) residues (Figure S1A) (Hwang et al., 2010b; Shemorry et al., 2013). The corresponding degradation signals and E3 Ub ligases are called Ac/N-degrons and Ac/N-recognins, respectively. Nt-acetylation of cellular proteins is apparently irreversible, in contrast to acetylation-deacetylation of internal Lys residues. The bulk of Nt-acetylation is cotranslational, being mediated by ribosome-associated Nt-acetylases. Approximately 90% of human proteins are Nt-acetylated (Arnesen et al., 2009; Mischerikow and Heck, 2011). Many, possibly most, Nt-acetylated proteins contain Ac/N-degrons (Figure S1A) (Hwang et al., 2010b; Shemorry et al., 2013).
Natural Ac/N-degrons are regulated through their steric shielding. A protein containing an Ac/N-degron is short-lived unless it can repress (shield) its Ac/N-degron through intramolecular folding or interactions with other proteins (Figure 7A). The resulting hiatus from being vulnerable to degradation can be either long-lasting or transient, depending on the in vivo dynamics (dissociation-reconstitution) of a complex that sequesters the protein’s Ac/N-degron (Shemorry et al., 2013). The cotranslational creation of Ac/N-degrons, their exceptional prevalence, and their conditionality underlie the regulation, by the Ac/N-end rule pathway, of the input stoichiometries of subunits in multisubunit complexes, as well as the elimination of misfolded or otherwise abnormal proteins that cannot shield their Ac/N-degrons (Shemorry et al., 2013).
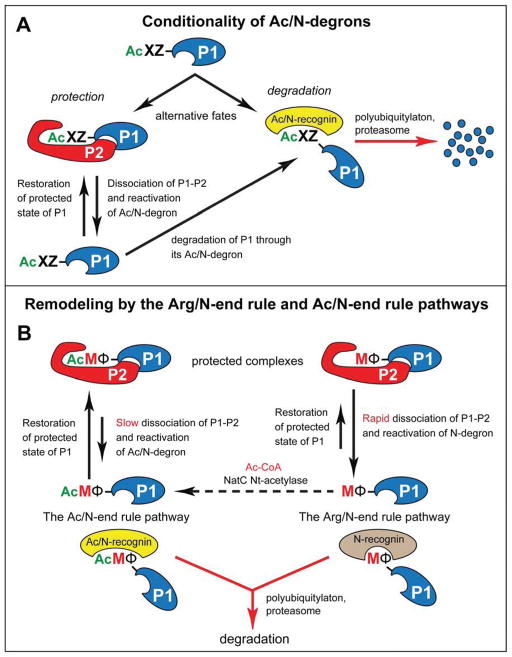
Figure 7. Conditionality of Ac/N-degrons and Protein Remodeling by the N-End Rule Pathway.
(A) Conditionality of Ac/N-degrons. This diagram summarizes the previously attained understanding of the dynamics of Nt-acetylated proteins vis-à-vis the Ac/N-end rule pathway (see Discussion), in conjunction with the initial discovery of the Ac/N-end rule pathway (Hwang et al., 2010b; Shemorry et al., 2013).
(B) The recognition of unacetylated Met-Φ proteins by the Arg/N-end rule pathway and of Nt-acetylated AcMet-Φ proteins by the Ac/N-end rule pathway underlies the proposed remodeling of protein complexes. Owing to different rates of dissociation of Nt-acetylated (AcMet-Φ) vs. unacetylated (Met-Φ) complexes (see Discussion), the subunit-selective degradation of the unacetylated P1 (Met-Φ) subunit of a P1–P2 complex upon its dissociation would allow the replacement-mediated conversion of P1–P2 into a similar but more stable complex containing the Nt-acetylated (AcMet-Φ) counterpart of the Met-Φ P1 subunit. To maximize generality of this description, the Nt-acetylation state of the P2 protein subunit was left unspecified, in contrast to the P1 subunit. Nt-acetylation of cellular proteins is largely cotranslational. The dashed arrow signifies the current uncertainty about rates of posttranslational Nt-acetylation. See Discussion for specific ramifications of this model. See also Figure S1.
This understanding left open the question of what happens when the Nt-acetylation of a specific protein is incomplete. Many cellular proteins are partially Nt-acetylated, i.e., a protein can exist in vivo as a mix of Nt-acetylated and non-Nt-acetylated species (Arnesen et al., 2009). The unacetylated fraction of an incompletely Nt-acetylated protein would be invisible to the Ac/N-end rule pathway. Might there be a system that can gauge and control, through conditional proteolysis, the homeostasis of both Nt-acetylated proteins and their unacetylated counterparts?
Nascent polypeptides bear N-terminal methionine (Met), encoded by the AUG initiation codon. Ribosome-associated Met-aminopeptidases cotranslationally cleave off the N-terminal Met if a residue at position 2, to be made N-terminal by the cleavage, is sufficiently small, i.e., if it is Gly, Ala, Ser, Thr, Cys, Pro, or Val (Xiao et al., 2010). The resulting N-terminal Ala, Ser and Thr, as well as the retained N-terminal Met residue are often Nt-acetylated (Starheim et al., 2012). Other N-terminal residues are Nt-acetylated either less frequently or almost never (Figure S2A).
Until the present work, the unacetylated N-terminal Met was classed as a “stabilizing” (i.e., non-destabilizing) residue (Varshavsky, 2011). We show here that the S. cerevisiae Ubr1 N-recognin and its mouse counterparts Ubr1 and Ubr2 have the previously unknown ability to recognize proteins bearing the unacetylated N-terminal Met if the residue at position 2 is Leu, Phe, Tyr, Trp, Ile, Val or Ala, i.e., a non-Met hydrophobic (Φ) residue. Because Ala2 and Val2 allow the removal of N-terminal Met by Met-aminopeptidases (Xiao et al., 2010), the retention of Met requires a large second-position Φ residue, i.e., Leu, Phe, Tyr, Trp or Ile. Proteins containing this motif, termed Met-Φ proteins, are shown here to be short-lived substrates of both the Arg/N-end rule and Ac/N-end rule pathways.
The substrate range of the Ac/N-end rule pathway is exceptionally broad, as ~90% of human proteins are Nt-acetylated and many Nt-acetylated proteins contain Ac/N-degrons (Figure S1A) (Hwang et al., 2010b; Shemorry et al., 2013). The substrate range of the Arg/N-end rule pathway was thought to be much narrower, because the exposure of the previously known unacetylated destabilizing N-terminal residues in substrates of this pathway (Figure S1B) requires preliminary cleavages of proteins by nonprocessive proteases that include calpains, caspases, separases and secretases. The discovery that this proteolytic system can target unacetylated Met-Φ proteins greatly expands the substrate range of the Arg/N-end rule pathway.
We found that the natural Met-Φ proteins Msn4, Sry1, Arl3, and Pre5 bear unacetylated Met-based N-degrons. We also found that the previously reported degradation of misfolded proteins by the Arg/N-end rule pathway (Eisele and Wolf, 2008; Heck et al., 2010) can involve the Ubr1-mediated recognition of these abnormal proteins through their Met-based N-degrons. The cited proteins are a part of an apparently much larger set of normal or misfolded proteins that can be destroyed through the recognition of their unacetylated N-terminal Met.
In either yeast or mammals, approximately 15% of genes encode Met-Φ proteins. As described below, many, possibly most, unacetylated Met-Φ proteins contain Met-based N-degrons. The resulting functional complementarity between the two branches of the N-end rule pathway makes possible the degradation-mediated control of Met-Φ proteins irrespective of the extent of their Nt-acetylation. Specifically, it is shown here that an Nt-acetylated Met-Φ protein can be destroyed by the Ac/N-end rule pathway while the otherwise identical unacetylated protein can be eliminated, independently, by the Arg/N-end rule pathway (Figures 6, 7 and S1).
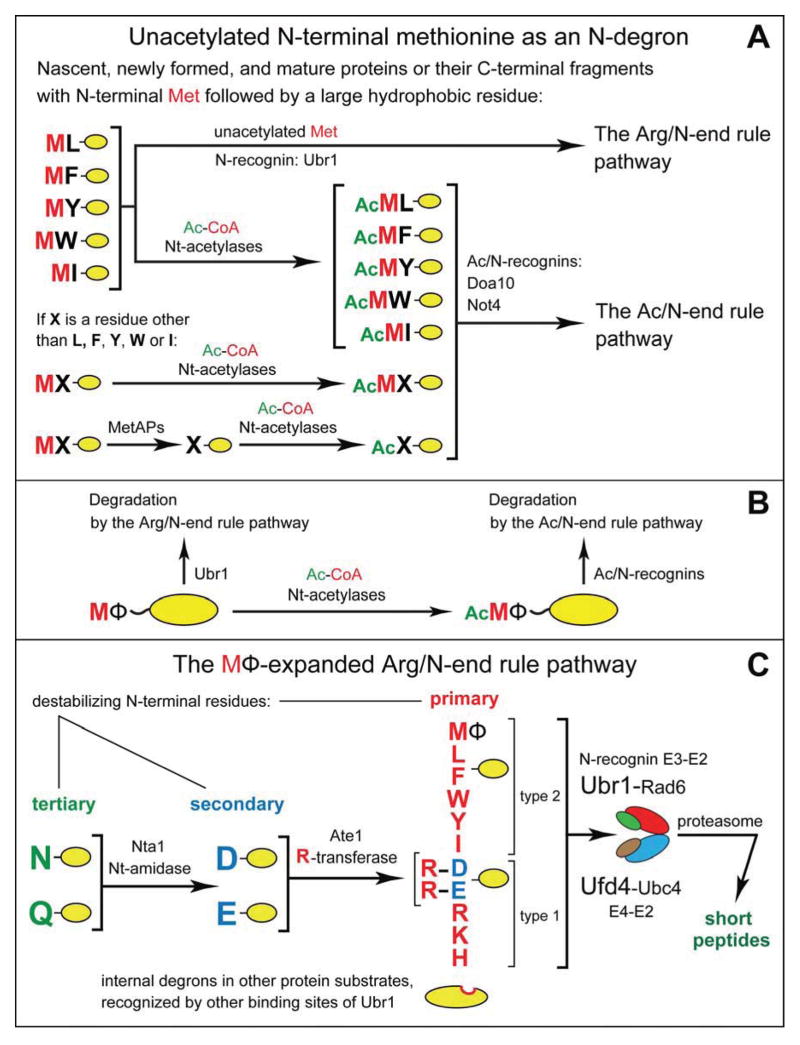
Figure 6. Complementary Specificities of the Arg/N-End Rule Pathway and the Ac/N-End Rule Pathway.
(A) This diagram summarizes the main discovery of the present work, a functional complementarity between the Arg/N-end rule and Ac/N-end rule pathways. This complementarity stems from the recognition of the previously unknown MetΦ/N-degrons in Met-Φ proteins vs. the recognition of the previously characterized AcMetΦ/N-degrons in AcMet-Φ proteins. AcMetΦ/N-degrons are a subset of Ac/N-degrons in Nt-acetylated cellular proteins (Hwang et al., 2010b; Shemorry et al., 2013). Met-Φ proteins are defined, in this study, as those that bear N-terminal Met followed by a large hydrophobic (Φ) non-Met residue.
(B) Condensed summary of the dual-pathway circuit shown in A.
(C) The MΦ-based expansion of the Arg/N-end rule pathway in the present work, through the addition of a large set of new substrates, Met-Φ proteins. See also Figure S1.
RESULTS
Binding of N-Recognins to Met-Φ Peptides
First indications that S. cerevisiae Ubr1 has a broader than previously known recognition specificity were provided by peptide arrays on membrane support (SPOT). In these assays, XZ-eK(3–10) peptides were C-terminally linked to a membrane in equal molar amounts and probed for binding to purified, flag-tagged S. cerevisiae Ubr1 (ScfUbr1) or to its mouse counterparts MmfUbr1 and MmfUbr2 (Figure 1A, B). The notation eK(3–10) (extension (e) containing lysine (K)) denotes 8 residues (after the varying residues X and Z) of the previously characterized ~40-residue eK sequence upstream of engineered N-end rule reporters (Hwang et al., 2010b; Varshavsky, 2011).
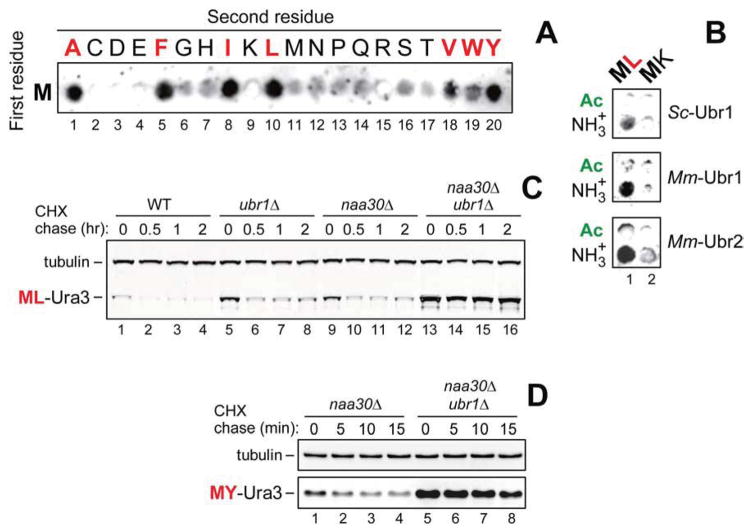
Figure 1. Specific Binding of Ubr1 to Unacetylated N-Terminal Methionine Followed by a Hydrophobic Residue.
(A) SPOT assay with S. cerevisiae fUbr1 and Met-Z-eK(3–10) peptides MZGSGAWLLP (Z = Ala, Cys, Asp, Glu, Phe, Gly, His, Ile, Lys, Leu, Met, Asn, Pro, Gln, Arg, Ser, Thr, Val, Trp, Tyr).
(B) Same as in A but with S. cerevisiae fUbr1 (ScfUbr1), mouse fUbr1 (MmfUbr1) and mouse fUbr2 (MmfUbr2) vs. Met-Z-eK(3–10) (Z = Leu, Lys) peptides and their Nt-acetylated counterparts.
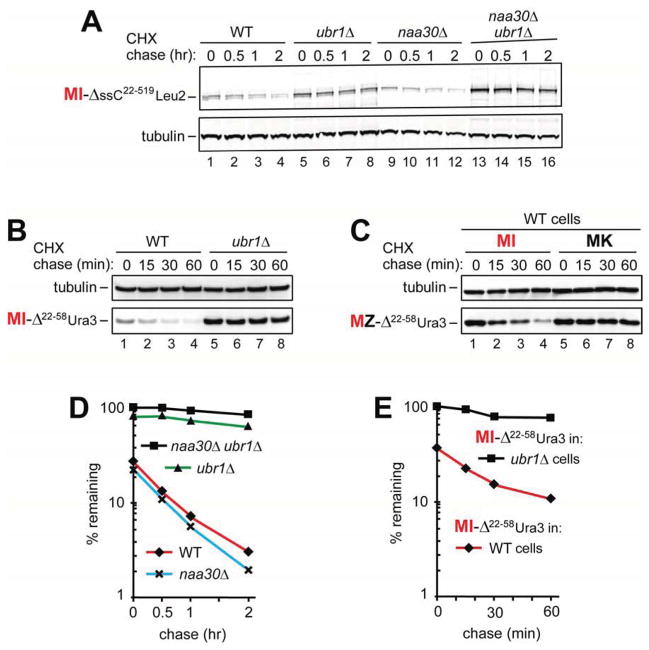
(C) Cycloheximide (CHX) chases with ML-Ura3 in WT (lanes 1–4), ubr1Δ (lanes 5–8), naa30Δ (lanes 9–12), and naa30Δ ubr1Δ cells (lanes 13–16).
(D) CHX chases with MY-Ura3 in naa30Δ (lanes 1–4) and naa30Δ ubr1Δ cells (lanes 5–8). See also Figures S1–S3.
ScfUbr1 was found to bind to unacetylated Met-Z-eK(3–10) peptides in which Z = Ala, Val, Leu, Ile, Phe, Trp, Tyr (Figure 1A, spots 1, 5, 8, 10, 18–20), but did not significantly bind to the otherwise identical Met-Z-eK(3–10) peptides in which Z = Cys, Asp, Glu, Gly, His, Lys, Met, Asn, Pro, Gln, Arg, Ser, Thr (Figure 1A, spots 2–4, 6, 7, 9, 11–17). In agreement with these data, a SPOT assay with ScfUbr1 as well as mouse MmfUbr1 and MmfUbr2 showed that yeast Ubr1 could bind to Met-Leu-eK(3–10) but not to Met-Lys-eK(3–10) (with Lys at position 2), and that the binding patterns of mouse Ubr1 and Ubr2 were similar to those of yeast Ubr1, including the absence of binding to Nt-acetylated counterparts of the unacetylated peptides (Figure 1B).
Given the pattern of retention of N-terminal Met in nascent proteins (see Introduction), the SPOT results suggested that Ubr1 can target proteins in vivo through their retained unacetylated N-terminal Met if it is followed by one of the large Φ residues Leu, Phe, Tyr, Trp, or Ile. We show, below, that this is indeed the case. The corresponding degrons of Met-Φ and AcMet-Φ proteins were termed MetΦ/N-degrons and AcMetΦ/N-degrons, respectively.
Degradation of Met-Φ Proteins by the Arg/N-End rule Pathway
The 35 kDa Met-Z-eK-ha-Ura3 (MZ-Ura3) reporters comprised N-terminal Met; a varying residue Z at position 2; the eK extension (see above); the ha epitope; and S. cerevisiae Ura3. MZ-Ura3 proteins were produced through the cotranslational deubiquitylation of Ub-MZ-Ura3, expressed in yeast using low copy plasmids and the PCUP1 promoter (Hwang et al., 2010b).
We showed previously that ML-Ura3 in wild-type (WT) cells was at least partially Nt-acetylated in vivo by the NatC Nt-acetylase and that the resulting AcML-Ura3 was targeted for degradation by the Ac/N-end rule pathway (Figures S1A and S2A) (Hwang et al., 2010b). Cycloheximide (CHX) chases indicated that the short-lived ML-Ura3 was longer-lived in ubr1Δ cells (lacking the Arg/N-end rule pathway) and in naa30Δ (m ak3Δ) cells (lacking the NatC Nt-acetylase) (Figures 1C and S2B). Moreover, ML-Ura3 was synergistically and nearly completely stabilized, in addition to a further increase of its pre-chase level, in naa30Δ ubr1Δ cells (Figure 1C). In contrast to pulse-chase assays, CHX-chases do not distinguish between “young” and “old” protein molecules. 35S-pulse-chases with ML-Ura3 (expressed from Ub-ML-Ura3 or directly as ML-Ura3) yielded results similar to those with CHX-chases (Figure 1C), including higher pre-chase levels of 35S-pulse-labeled ML-Ura3 in naa30Δ ubr1Δ cells vs. naa30Δ cells (Figure S3A–D).
The CHX-chase patterns with MI-Ura3 and MY-Ura3, in which Leu2 was replaced by other Φ residues, Ile or Tyr, were similar to those with ML-Ura3. Specifically, MI-Ura3 and MY-Ura3 were short-lived in naa30Δ cells but became nearly completely stable in naa30Δ ubr1Δ cells, with striking increases in their pre-chase levels (Figures 1D and 2A, C).
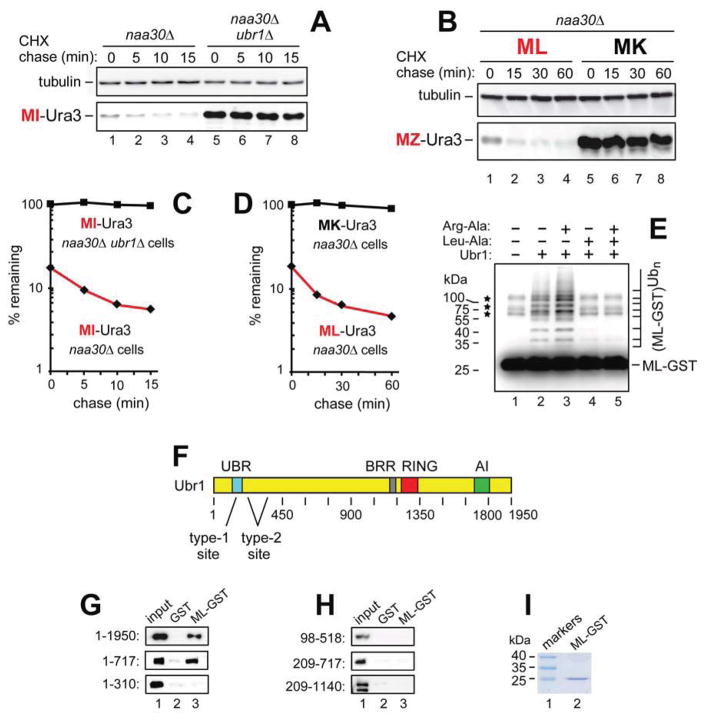
Figure 2. Unacetylated N-Terminal Methionine as an N-Degron of the Arg/N-End Rule Pathway.
(A) CHX chases with MI-Ura3 in naa30Δ (lanes 1–4) and naa30Δ ubr1Δ cells (lanes 5–8).
(B) CHX chases with ML-Ura3 (lanes 1–4) and MK-Ura3 (lanes 5–8) in naa30Δ cells.
(C) Quantification of data in A.
(D) Quantification of data in B. In A–D, the corresponding CHX-chase assays were carried out at least three times and yielded results within 10% of the data shown.
(E) In vitro polyubiquitylation of purified ML-GST by the purified Ubr1-Rad6 Ub ligase. Lane 1, complete assay but without Ubr1 (the asterisks indicate three minor contaminants in purified ML-GST, whose band is indicated on the right). Lane 2, same as lane 1 but with Ubr1. Lane 3, same as lane 2 but with Arg-Ala (1 mM). Lane 4, same as lane 2 but with Leu-Ala (1 mM). Lane 5, same as lane 2 but with Arg-Ala and Leu-Ala.
(F) The UBR, BRR, RING, and AI regions of the S. cerevisiae Ubr1 N-recognin (Varshavsky, 2011).
(G) GST-pulldown assays with ML-GST versus GST and either full-length fUbr11–1950 or its fragments fUBR1–717 and fUbr11–310.
(H) Same as in G but with fUbr198–518, fUbr1209–717 and Ubr1209–1140f.
(I) SDS-PAGE of purified ML-GST (lane 2). See also Figures S1–S4.
The unacetylated state of ML-Ura3 in naa30Δ cells (Hwang et al., 2010b), the binding of Ubr1 to unacetylated N-terminal Met-Φ sequences (Figure 1A, B), and the synergistic stabilization of ML-Ura3, MI-Ura3 and MY-Ura3 by the ablation of both Ubr1 and NatC in naa30Δ ubr1Δ cells (Figures 1C, D, 2A, C and S3A–D) indicated that these reporters were destroyed through their MetΦ/N-degrons in naa30Δ cells. In an independent support of this conclusion, and in striking contrast to the instability of ML-Ura3, MI-Ura3 and MY-Ura3 in naa30Δ cells, the otherwise identical MK-Ura3, containing Lys2 (instead of Leu2 in ML-Ura3), was long-lived in naa30Δ cells under the same conditions (Figure 2B, D). Moreover, the pre-chase level of MK-Ura3 was ~5-fold higher than that of ML-Ura3 (Figure 2B, D). These in vivo results were predicted by in vitro data, as there was no significant binding of Ubr1 to N-terminal Met if position 2 was occupied by a basic residue such as Lys (Figure 1A, B).
RT-PCR of mRNA encoding ML-Ura3 indicated no significant changes in the level of this mRNA between naa30Δ cells and naa30Δ ubr1Δ cells (Figure S4C), in striking contrast to the considerably higher levels of the ML-Ura3 protein (and of the analogous MI-Ura3 and MY-Ura3) both before and during CHX-chases in naa30Δ ubr1Δ cells (Figures 1C, D and 2A, C). Thus, the observed increases of protein levels stemmed either largely or entirely from changes in the rate of degradation of these proteins.
A Binding Site of Ubr1 That Recognizes MetΦ/N-Degrons
The ~80-residue UBR domain of the 1,950-residue Ubr1 contains its type-1 binding site, which recognizes the N-terminal basic residues Arg, Lys or His (Choi et al., 2010; Matta-Camacho et al., 2010). The nearby type-2 binding site of Ubr1 recognizes the large N-terminal Φ residues Leu, Phe, Tyr, Trp or Ile (Figure 2F) (Xia et al., 2008). To map a site that recognizes the unacetylated N-terminal Met of Met-Φ proteins, we performed glutathione-S-transferase (GST)-pulldowns with purified ML-eK-GST (ML-GST) and extracts from S. cerevisiae that expressed the flag-tagged full-length Ubr11–1950 or its fragments fUBR1–717, Ubr1209–1140f, fUbr11–310, fUbr198–518, and fUbr1209–717. The fUBR1–717 fragment, which contained the type-1 and type-2 binding sites, could bind to ML-GST, but shorter N-terminal fragments of Ubr1 or its C-terminal fragments did not bind to ML-GST (Figure 2F–I).
Dipeptides with type-1 or type-2 destabilizing N-terminal residues (Figure S1B) can inhibit, through competition, the binding of Ubr1 to proteins bearing these N-terminal residues (Varshavsky, 2011). We used the type-1 Arg-Ala and/or type-2 Leu-Ala dipeptides with the previously characterized, completely defined in vitro ubiquitylation system (Hwang et al., 2010a). It comprised purified Ub, the Uba1 E1 enzyme, the Rad6 E2 enzyme, the Ubr1 E3 N-recognin, and the unacetylated ML-GST reporter. Leu-Ala completely inhibited the Ubr1-dependent polyubiquitylation of ML-GST (Figure 2E, lanes 4 vs. 2), whereas Arg-Ala enhanced its polyubiquitylation, without negating the inhibitory effect of Leu-Ala if the two dipeptides were added together (Figure 2E, lanes 3 vs. 5). The positive allosteric effect of Arg-Ala was previously observed with other type-2 N-end rule substrates, whose binding to the type-2 site of Ubr1 was shown to be allosterically enhanced by the occupancy of its type-1 site (Varshavsky, 2011). Together, the ubiquitylation data and the pattern of ML-GST binding to Ubr1 and its fragments (Figure 2E–I) strongly suggested that the recognition of N-terminal Met in Met-Φ proteins by Ubr1 is mediated by its previously characterized (Xia et al., 2008) type-2 binding site.
Degradation of a Misfolded Protein Through Its MetΦ/N-Degron
Studies by Wolf, Hampton and colleagues showed that a variety of misfolded proteins can be destroyed by the Ubr1-dependent Arg/N-end rule pathway (Eisele and Wolf, 2008; Fredrickson and Gardner, 2012; Heck et al., 2010; Prasad et al., 2010; Summers et al., 2013; Theodoraki et al., 2012). One Ubr1 substrate of this class is the short-lived 110 kDa ΔssC22–519Leu2myc, comprising a misfolded PRC1-derived moiety, the Leu2 moiety, and the myc13 tag (Eisele and Wolf, 2008). Because ΔssC22–519Leu2myc starts with Met-Ile (a Met-Φ sequence), we asked whether this protein, denoted as MI-ΔssC22–519Leu2myc, was targeted through its N-terminal Met.
CHX-chases of MI-ΔssC22–519Leu2myc showed it to be short-lived in WT cells and significantly stabilized in ubr1Δ cells (Figure 3A, D), in agreement with earlier findings (Eisele and Wolf, 2008). MI-ΔssC22–519Leu2myc was stabilized in double-mutant naa30Δ ubr1Δ cells even stronger than in ubr1Δ cells, with a further increase of its pre-chase level (Figure 3A, D). MI-ΔssC22–58Ura3ha, containing the first 37 residues of the ΔssC22–519 moiety, the Ura3 moiety, and the ha tag, was also short-lived in WT cells, and was strikingly stabilized in ubr1Δ cells (Figure 3B, E).
Figure 3. Misfolded Proteins Containing Met-Based N-Degrons.
(A) CHX chases with MI-ΔssC22–519Leu2myc in WT (lanes 1–4), ubr1Δ (lanes 5–8), naa30Δ (lanes 9–12), and naa30Δ ubr1Δ cells (lanes 13–16).
(B) CHX-chases with MI-ΔssC22–58Ura3ha in WT (lanes 1–4) and ubr1Δ cells (lanes 5–8).
(C) CHX-chases with MI-ΔssC22–58 Ura3ha (lanes 1–4) and MK-ΔssC22–58 Ura3ha (lanes 5–8) in WT cells.
(D) Quantification of data in A.
(E) Quantification of data in B. In A–E, the corresponding CHX-chase assays were carried out at least three times and yielded results within 10% of the data shown. See also Figures S1 and S2.
Given the presence of Ubr1-dependent MetΦ/N-degrons in MZ-Ura3 proteins (Figures 1C, D and 2A–D), the results with MI-ΔssC22–519Leu2myc and MI-ΔssC22–58Ura3ha (Figure 3A, B, D) indicated that these proteins were destroyed largely through their MetΦ/N-degrons. If so, the Ile2→Lys2 mutation should abrogate this degradation in WT cells, because Ubr1 does not bind to N-terminal Met-Lys (Figure 1A, B). Indeed, MK-ΔssC22–58Ura3ha was completely stable in WT cells, in striking contrast to the degradation of the otherwise identical MI-ΔssC22–58Ura3ha (Figure 3C). In sum, at least some misfolded Met-Φ proteins, including MI-ΔssC22–519Leu2myc and MI-ΔssC22–58Ura3ha, are destroyed by the Arg/N-end rule pathway through their MetΦ/N-degrons.
MetΦ/N-Degrons and AcMetΦ/N-Degrons in Natural Proteins
The findings so far indicated the following mode of degradation of a Met-Φ protein (Figures 1–3):
The unacetylated N-terminal Met of a Met-Φ protein can act as a MetΦ/N-degron, leading to the degradation of this protein by the Ubr1-dependent Arg/N-end rule pathway.
Nt-acetylation converts a MetΦ/N-degron into AcMetΦ/N-degron and thereby shifts the targeting of the resulting AcMet-Φ protein to the Ac/N-end rule pathway.
The dual-pathway circuit that is revealed by this understanding (Figures 6 and 7) comprises the Nt-acetylated AcMet-Φ protein, the at least transient presence of its unacetylated Met-Φ counterpart, and the targeting of these otherwise identical proteins by two mechanistically distinct branches of the N-end rule pathway. An unacetylated Met-Φ protein is vulnerable to the Arg/N-end rule pathway. However, at least some molecules of this protein would be irreversibly Nt-acetylated before their encounters with Ubr1. Nt-acetylation of these Met-Φ molecules would preclude their capture by Ubr1 while making them vulnerable to the Ac/N-end rule pathway (Figures 6 and 7). To begin exploring this deeper understanding, we chose the S. cerevisiae Met-Φ proteins Msn4, Sry1, Arl3, and Pre5 from hundreds of S. cerevisiae Met-Φ proteins. These partly random choices were determined largely by the fact that the cited proteins are Nt-acetylated in WT yeast (Arnesen et al., 2009). (Met-Φ proteins are expected to be at least partially Nt-acetylated by the NatC Nt-acetylase (Figure S2A).) The proteins were C-terminally ha-tagged and expressed from the PCUP1 promoter on a low copy plasmid.
Msn4
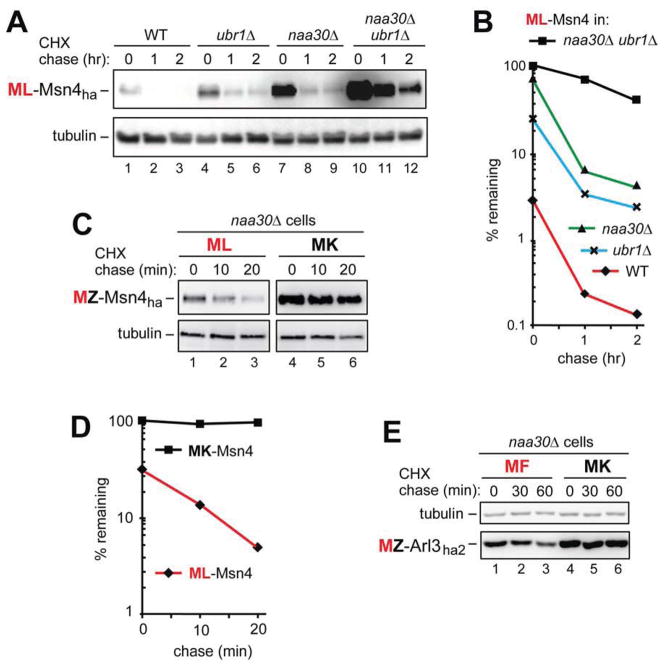
The 70 kDa Msn4 (N-terminus: MLV-), denoted as ML-Msn4ha, is a short-lived transcriptional activator that induces specific genes in response to stresses (Takatsume et al., 2010). ML-Msn4ha was a highly unstable protein (t1/2 ≪ 1 hr) in WT cells (Figure 4A, B). ML-Msn4ha was partially stabilized in ubr1Δ cells, indicating the presence of MetΦ/N-degron in some molecules of ML-Msn4ha in WT cells (Figure 4A, B). ML-Msn4ha was more strongly but still partially stabilized in naa30Δ cells, indicating the presence of AcMetΦ/N-degron in other (Nt-acetylated) molecules of ML-Msn4ha in WT cells (Figure 4A, B).
Figure 4. The Natural ML-Msn4 and MF-Arl3 Proteins Contain Met-Based N-Degrons.
(A) CHX chases with ML-Msn4ha in WT (lanes 1–3), ubr1Δ (lanes 4–6), naa30Δ (lanes 7–9), and naa30Δ ubr1Δ cells (lanes 10–12).
(B) Quantification of data in A.
(C) CHX chases with ML-Msn4ha (lanes 1–3) and MK-Msn4ha (lanes 4–6) in naa30Δ cells.
(D) Quantification of data in C.
(E) CHX chases with MF-Arl3ha2 (lanes 1–3) and MK-Arl3ha2 (lanes 4–6) in naa30Δ cells.
In A–E, the corresponding CHX-chase assays were carried out at least three times and yielded results within 10% of the data shown. See also Figures S1 and S2.
In a most telling pattern analogous to but even more striking than the results with engineered MZ-Ura3 proteins (Figures 1C, D and 2A–D), ML-Msn4ha was synergistically stabilized in naa30Δ ubr1Δ cells (Figure 4A, lanes 10–12 vs. 1–3, and Figure 4B). Remarkably, the pre-chase level of ML-Msn4ha in naa30Δ ubr1Δ cells was at least 30-fold higher than in WT cells (Figure 4A, B). Despite this enormous increase, ML-Msn4ha retained a part of its instability in double-mutant cells, suggesting the presence of an internal degron as well (Figure 4A, B).
Independent evidence that the unacetylated ML-Msn4ha was degraded by the Arg/N-end rule pathway in NatC-lacking naa30Δ cells was provided by the Leu2→Lys2 mutation, which stabilized the resulting MK-Msn4ha, in addition to strikingly increasing its pre-chase level (Figure 4C, D). These in vivo results were predicted by the in vitro evidence that Ubr1 does not bind to N-terminal Met-Lys (Figure 1A, B).
Sry1
The 35 kDa Sry1 (N-terminus: MIV-), denoted as MI-Sry1, is a 3-hydroxyaspartate dehydratase. It modifies 3-hydroxyaspartate, a potentially toxic microbial metabolite (Wada et al., 2003). We produced MI-Sry1 in two ways, either as MI-Sry1ha3, expressed from the native PSRY1 promoter and the endogenous (chromosomal) SRY1 locus instead of WT MI-Sry1, or as MI-Sry1ha, moderately overexpressed from the PCUP1 promoter on a low copy plasmid in Sry1+ cells. Analyzed by CHX-chases, the level of exogenously expressed MI-Sry1ha was low in WT cells and even lower in naa30Δ cells, in which MI-Sry1ha was not Nt-acetylated. Tellingly, the levels of MI-Sry1ha were strikingly (more than 25-fold) higher in either ubr1Δ or naa30Δ ubr1Δ cells than in WT or naa30Δ cells (Figure S3E). 35S-pulse-chases of exogenously expressed MI-Sry1ha in naa30Δ cells vs. naa30Δ ubr1Δ cells were in agreement with CHX-chase results in that pulse-labeled MI-Sry1ha was unstable in naa30Δ cells and was stabilized in naa30Δ ubr1Δ cells, including its strongly increased pre-chase level (Figure S3F, G).
In CHX-chases with the endogenous MI-Sry1ha3, which was expressed from the PSRY1 promoter and the SRY1 chromosomal locus, the level of MI-Sry1ha3 was too low for detection in either WT or naa30Δ cells, which lacked the cognate NatC Nt-acetylase (Figure 5D). Strikingly, however, and similarly to the exogenously expressed MI-Sry1ha, the level of endogenous MI-Sry1ha3 became at least 20-fold higher in ubr1Δ cells before the chase, and was even further increased in naa30Δ ubr1Δ cells (Figure 5D).
Figure 5. The Natural MF-Pre5 and MI-Sry1 Proteins Contain Met-Based N-Degrons.
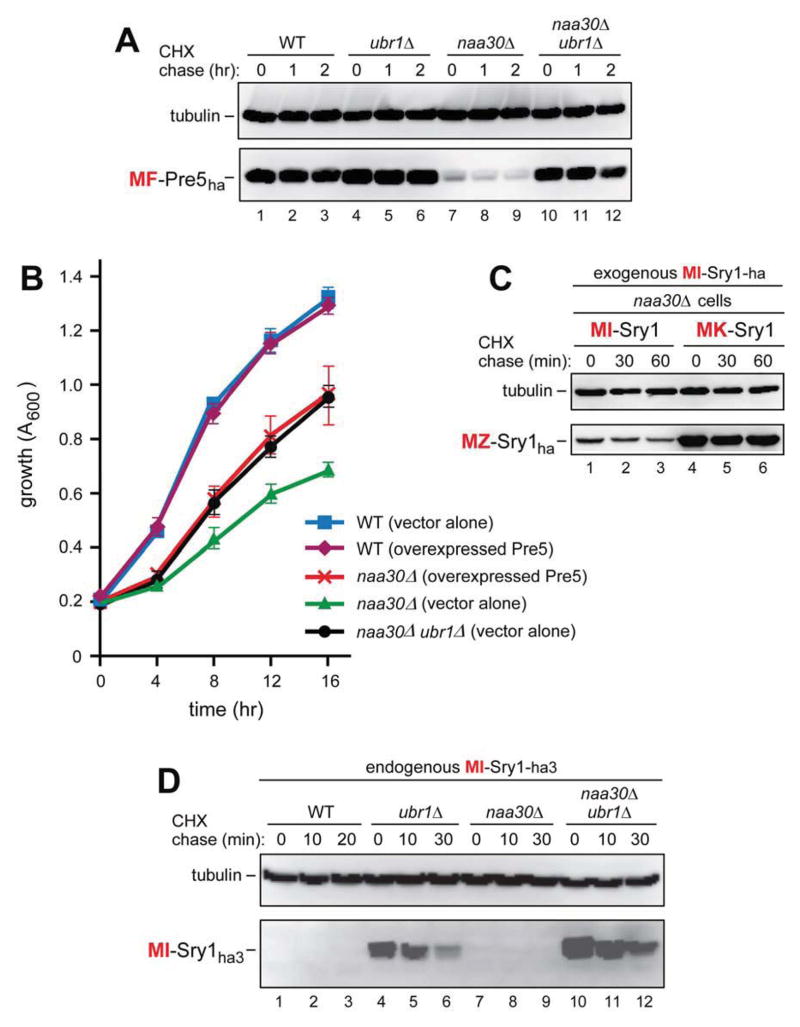
(A) CHX chases with MF-Pre5ha in WT (lanes 1–3), ubr1Δ (lanes 4–6), naa30Δ (lanes 7–9), and naa30Δ ubr1Δ cells (lanes 10–12).
(B) Growth rates of WT, naa30Δ and naa30Δ ubr1Δ S. cerevisiae strains, including WT and naa30Δ strains that overexpressed the MF-Pre5 proteasomal subunit. Standard errors (of triplicate measurements) are shown as well.
(C) CHX chases with MI-Sry1ha (lanes 1–3) and MK-Sry1ha (lanes 4–6) in naa30Δ cells.
(D) CHX chases with endogenously expressed MI-Sry1ha in WT (lanes 1–3), ubr1Δ (lanes 4–6), naa30Δ (lanes 7–9), and naa30Δ ubr1Δ cells (lanes 10–12). See also Figures S1, S2, and S4.
Independent evidence that the unacetylated MI-Sry1ha was degraded by the Arg/N-end rule pathway in NatC-lacking naa30Δ cells was provided by the Ile2→Lys2 mutation. It stabilized the resulting MK-Sry1ha and greatly increased its level before the chase (Figure 5C), in agreement with the in vitro evidence that Ubr1 does not bind to N-terminal Met-Lys (Figure 1A, B).
Arl3
The 22 kDa Arl3 (N-terminus: MFH-), denoted as MF-Arl3ha2, is a Golgi-associated cytosolic GTPase. Nt-acetylation of Arl3 is required for its targeting to Golgi (Behnia et al., 2004; Setty et al., 2004). MF-Arl3ha2 was unstable (t1/2 ≈ 35 min) in naa30Δ cells (Figure 4E). Given the results with other Met-Φ proteins (Figures 1–5), the unacetylated (in naa30Δ cells) N-terminal Met-Phe of MF-Arl3ha2 was expected to be targeted by the Arg/N-end rule pathway (Figure 6). If so, the Phe2→Lys2 mutation in MF-Arl3ha2 should abrogate this degradation. Indeed, MK-Arl3ha2 was completely stable during CHX-chase in naa30Δ cells, in contrast to MF-Arl3ha2 (Figure 4E). Similarly to the findings with MZ-Ura3, MZ-ΔssC22–58Ura3ha, MZ-Msn4ha, and MZ-Sry1ha (Figures 2B, D, 3C, F, 4C, D and 5C), this in vivo result was predicted by the in vitro data about the absence of Ubr1 binding to N-terminal Met-Lys (Figure 1A, B). In sum, the unacetylated MF-Arl3ha2 contains a MetΦ/N-degron.
Pre5
The 25 kDa Pre5 (N-terminus: MFR-), denoted as MF-Pre5ha, is a subunit of the 20S proteasome (Heinemeyer et al., 1994). MF-Pre5ha was a relatively long-lived protein in WT cells (t1/2 > 1 hr), in ubr1Δ cells, and in naa30Δ ubr1Δ cells (Figure 5A, lanes 1–3 vs. lanes 4–6 and 10–12). Remarkably, however, MF-Pre5ha became so short-lived in naa30Δ cells (lacking the NatC Nt-acetylase) that it was barely detectable even before the chase (Figure 5A, lanes 7–9 vs. 1–3). An illuminating explanation of this striking effect is described in Discussion. 35S-pulse-chases of MF-Pre5ha were in agreement with CHX-chase results in that MF-Pre5ha was unstable in naa30Δ cells and was stabilized in naa30Δ ubr1Δ cells, including its strongly increased pre-chase level (Figure S4A, B).
RT-PCR of mRNAs encoding ML-Ura3, MI-Sry1, and MF-Pre5 indicated no significant changes in the level of these mRNAs between naa30Δ cells and naa30Δ ubr1Δ cells (Figure S4C–F), in contrast to much higher levels of the corresponding proteins in naa30Δ ubr1Δ cells (Figures 1C, D, 2A, C, 5A, D, and S3E–G). Thus, strong increases in the levels of these proteins stemmed either largely or entirely from changes in the rate of their degradation, particularly the one that occurred either cotranslationally or shortly afterward, in agreement with evidence for a significant degradation of nascent and newly formed proteins (Duttler et al., 2013; Hartl et al., 2011; Turner and Varshavsky, 2000; Wang et al., 2013; Yewdell et al., 2011).
Overexpression of Pre5 Rescues Growth of naa30Δ Cells but Is Unnecessary in naa30Δ ubr1Δ Cells
The instability of (unacetylated) MF-Pre5 in naa30Δ cells suggested that the previously unexplained slow growth phenotype of naa30Δ cells (Starheim et al., 2012) might be caused, in part, by low levels of the normally abundant, in WT cells, but now unacetylated and short-lived MF-Pre5 proteasomal subunit, owing to its degradation by the Arg/N-end rule pathway. We asked, therefore, whether overexpression of MF-Pre5 might partially rescue the slow growth of naa30Δ cells. Indeed, overexpression of MF-Pre5 (at 37°C, to increase the dependence of growth on the proteasome activity (Finley et al., 2012)), was found to accelerate the growth of naa30Δ cells from ~50% to ~85% of the WT growth rate under the same conditions (Figure 5B). Remarkably, we also found that naa30Δ ubr1Δ cells, in the absence of MF-Pre5 overexpression, grew as fast as the “rescued” naa30Δ cells overexpressing MF-Pre5 (Figure 5B) (see Discussion).
Stress Hypersensitivity of Cells Lacking Both Ubr1 and the NatC Nt-Acetylase
Our findings (Figures 1–5 and S3) suggested that cells in which both MetΦ/N-degrons and AcMetΦ/N-degrons are inactive may be hypersensitive to stress, in comparison to the loss of just one of two alternative degradation routes (Figures 6 and 7B). Indeed, naa30Δ ubr1Δ cells, which lacked both the Arg/N-end rule pathway and the NatC Nt-acetylase of Met-Φ proteins (Figures S1B and S2A), were more sensitive to a range of stresses than the corresponding single mutants, let alone WT cells (Figure S4G, H). The examined stressors were amino acid analogs (production of misfolded proteins), 2% ethanol (perturbations of membranes and other structures), and Congo Red or Calcofluor White, whose effects include impairments of cell wall synthesis (Figure S4G, H).
DISCUSSION
The main discovery of this study revealed a link between the Ac/N-end rule pathway and the Arg/N-end rule pathway, two universally present pathways of protein degradation (Figures 6, 7 and S1). We found that the S. cerevisiae Ubr1 N-recognin of the Arg/N-end rule pathway as well as its mouse counterparts Ubr1 and Ubr2 can recognize Met-Φ proteins through their unacetylated N-terminal Met residues (Figures 1–5, S3 and S4). (A Met-Φ protein bears N-terminal Met followed by a large hydrophobic (Φ) non-Met residue, i.e., Leu, Phe, Tyr, Trp or Ile.) The resulting complementarity between the Arg/N-end rule and Ac/N-end rule pathways makes possible the proteolysis-mediated control of Met-Φ proteins irrespective of the extent of their Nt-acetylation. Specifically, the Ac/N-end rule pathway can target an Nt-acetylated AcMet-Φ protein but not the otherwise identical unacetylated Met-Φ protein. The latter, however, can be destroyed as well, because it contains a MetΦ/N-degron. This previously unknown class of N-degrons is recognized by the Ubr1-dependent Arg/N-end rule pathway. The resulting dual-pathway circuit is summarized in Figures 6 and 7B.
In either yeast or mammals, ~15% of genes encode Met-Φ proteins. For this reason alone, the present advance has significant biological ramifications. Msn4 (transcriptional activator), Sry1 (3-hydroxyaspartate dehydratase), Arl3 (GTPase of the Ras superfamily), and Pre5 (proteasomal subunit) (Figures 4, 5, and S3) are the initial examples of natural Met-Φ proteins whose unacetylated N-terminal Met residues are shown here to function as MetΦ/N-degrons. Many other proteins of the Met-Φ class are likely to be similar to Msn4, Sry1, Arl3 and Pre5 in their vulnerability to the Arg/N-end rule pathway (through their unacetylated N-terminal Met) and also, alternatively, to the Ac/N-end rule pathway, through their Nt-acetylated AcMet (Figures 6 and 7).
We also showed here that the misfolded protein ΔssC22–519Leu2myc, a previously identified short-lived substrate of the Arg/N-end rule pathway (Eisele and Wolf, 2008), contains a MetΦ/N-degron and is targeted by Ubr1 through this, previously unknown class of N-degrons (Figures 3, 6 and 7B). Given these results, it is likely that prematurely terminated (truncated) polypeptides of the Met-Φ class can also be targeted by the Arg/N-end rule pathway through their MetΦ/N-degrons or, alternatively, by the Ac/N-end rule pathway through their AcMetΦ/N-degrons. It remains to be determined whether the degradation of other misfolded Met-Φ proteins is mediated by their MetΦ/N-degrons and/or AcMetΦ/N-degrons, or whether the Arg/N-end rule pathway can target some of these proteins through their internal degrons as well.
Natural Ac/N-degrons can be repressed through steric shielding (Shemorry et al., 2013). Specifically, a protein subunit that contacts an Nt-acetylated subunit in an oligomeric complex may sequester that subunit’s Ac/N-degron and thereby preclude, reversibly, its recognition by the Ac/N-end rule pathway (Figures 7A and S1A). For example, S. cerevisiae Cog1, a subunit of the COG complex, was shown to contain an Ac/N-degron. Nevertheless, Cog1 is long-lived if expressed at normal (endogenous) levels. However, an overexpressed Cog1 is short-lived (Shemorry et al., 2013). Moreover, the previously long-lived endogenous Cog1 becomes short-lived if the production of Cog1 goes up in a cell. The cause of Cog1 destabilization was traced to the loss of stoichiometry, as only a minority of overproduced Cog1 molecules could repress their Ac/N-degrons through the binding to other, less abundant (normally expressed) subunits of the COG complex (Shemorry et al., 2013). This understanding has explained, among other things, how the prevalence of Ac/N-degrons (~90% of human proteins are Nt-acetylated, apparently irreversibly) can be consistent with the fact that most Nt-acetylated proteins are at least intermittently long-lived in vivo.
The targeting of MetΦ/N-degrons and AcMetΦ/N-degrons by, respectively, the Arg/N-end rule and Ac/N-end rule pathways (Figures 6 and 7B) may also account for the previously noted destabilization of some chloroplast DNA-encoded proteins upon a partial retention of their N-terminal Met, a finding that had not been understood in mechanistic terms (Giglione et al., 2003). Produced in bacteria-like chloroplasts of plant cells, these proteins bear, initially, the N-terminal formyl-Met (fMet) residue. We suggested that the transient (eventually deformylated) N-terminal fMet of bacterial proteins may act as an fMet-based N-degron (Hwang et al., 2010b). Such a degradation signal would be analogous to the previously identified eukaryotic Ac/N-degrons, but with the transient formyl moiety (instead of the permanent acetyl moiety) at the N-termini of nascent bacterial proteins. This possibility remains to be examined.
MF-Pre5, a subunit of the 20S proteasome, was relatively long-lived in WT, ubr1Δ, and naa30Δ ubr1Δ cells (t1/2 > 1 hr). However, in naa30Δ cells (lacking NatC but containing the Arg/N-end rule pathway), MF-Pre5ha became so short-lived (t1/2 ≪ 30 min) that it was barely detectable even before the chase (Figure 5A). An explanation, below, of the striking destabilization of MF-Pre5ha in the absence of its Nt-acetylation is based on structural studies of Nt-acetylated proteins by Schulman, Barford and colleagues (Monda et al., 2012; Scott et al., 2011; Zhang et al., 2010).
The Nt-acetylation of MF-Pre5ha in WT cells (Arnesen et al., 2009) precludes its targeting by the Arg/N-end rule pathway. The degradation of Nt-acetylated AcMF-Pre5ha is slow in WT cells, in contrast to the rapid destruction of unacetylated MF-Pre5ha in naa30Δ cells (Figure 5A). We suggest this is so because the AcMetΦ/N-degron of Nt-acetylated AcMF-Pre5ha is rapidly repressed (shielded), owing to its interactions with other subunits of the proteasome and/or with proteasomal chaperones (Matias et al., 2010; Tomko and Hochstrasser, 2013), by analogy with the efficacious repression of the previously characterized natural Ac/N-degrons, in proteins such as Cog1 and Hcn1 (Shemorry et al., 2013). However, in naa30Δ cells, the now unacetylated N-terminal Met of MF-Pre5ha acts as an active MetΦ/N-degron of the Arg/N-end rule pathway, resulting in the observed destruction of MF-Pre5ha (Figure 5A). So far, this line of reasoning invoked concepts validated earlier, including the conditionality of Ac/N-degrons (Shemorry et al., 2013). The remaining step, described below, is to explain why the MetΦ/N-degron of unacetylated MF-Pre5ha (in naa30Δ cells) is much more active than the AcMetΦ/N-degron of Nt-acetylated AcMF-Pre5ha (in WT cells) (Figure 5A).
Nt-acetylation of a protein makes its initially charged N-terminus uncharged, more bulky, and more hydrophobic. Schulman and colleagues have shown that the strength of cognate protein interactions that involve the Nt-acetyl group is higher, by approximately 100-fold, in the presence vs. the absence of this group (Monda et al., 2012). Because MF-Pre5ha is not Nt-acetylated in naa30Δ cells, a degron-shielding “protective” complex that contains unacetylated MF-Pre5ha would still be expected to form but would be much less tight in these cells than in WT cells, which contain Nt-acetylated AcMF-Pre5ha. Given the findings with other Nt-acetylated proteins (Monda et al., 2012), a significant difference in the rate of dissociation of protective complexes containing Nt-acetylated versus unacetylated MF-Pre5ha is likely to account for the faster degradation of MF-Pre5ha in naa30Δ cells (Figure 5A). This mechanism, to be verified and explored in future studies, may prove to be general and illuminating (Figure 7B).
The degradation, in naa30Δ cells, of the (unacetylated) MF-Pre5 proteasomal subunit by the Arg/N-end rule pathway (Figure 5A) suggested that the slow growth phenotype of naa30Δ cells might be caused, in part, by low levels of MF-Pre5 in these cells. Indeed, the growth rate of naa30Δ cells (but not of WT cells) was significantly increased by overexpression of MF-Pre5 (Figure 5B). Thus, the slow growth of naa30Δ cells (which cannot Nt-acetylate Met-Φ proteins) is caused, at least in part, by abnormally low levels of unacetylated and therefore short-lived Met-Φ proteins that include MF-Pre5. These proteins are longer-lived in WT cells (owing to efficacious shielding of their Ac/N-degrons) but become vulnerable to the Arg/N-end rule pathway in the absence of Nt-acetylation (Figures 6 and 7B). In a telling sequel to this result, we also found that naa30Δ ubr1Δ cells that did not overexpress Pre5 grew as fast as naa30Δ cells that overexpressed Pre5 (Figure 5B). Thus, not only it is true that a significant part of the growth defect of naa30Δ cells (in which MF-Pre5 is degraded) can be rescued by overexpressing MF-Pre5, but a nearly identical extent of growth rescue can also be produced by deleting UBR1 in the naa30Δ genetic background and thereby stabilizing MF-Pre5 (Figure 5A, B).
As discussed above, the previously characterized conditionality of Ac/N-degrons (Shemorry et al., 2013), including AcMetΦ/N-degrons, is likely to be more pronounced than the conditionality of MetΦ/N-degrons, because protective complexes containing unacetylated Met-Φ subunits would dissociate more rapidly (Monda et al., 2012). If so, one function of the dual-pathway circuit in Figures 6A, B and 7B is the degradation-mediated remodeling of protein complexes. This remodeling, based on the previously discovered subunit selectivity of protein degradation by the Arg/N-end rule pathway (Johnson et al., 1990), can eliminate an unacetylated Met-Φ subunit from a complex, thereby making possible the replacement of that subunit by its Nt-acetylated counterpart.
An incomplete Nt-acetylation of some newly formed proteins in WT cells stems, in part, from substoichiometric levels of ribosome-associated Nt-acetylases vis-à-vis the levels of ribosomes. A complex containing an unacetylated Met-Φ subunit instead of its cognate Nt-acetylated counterpart would be more prone to dissociation (Monda et al., 2012). Although a faster disassembly of the complex may compromise its function, it would also facilitate the repair (remodeling) of this complex through the degradation-mediated replacement of its unacetylated subunits by their Nt-acetylated counterparts. One prediction of this model is that some Met-Φ proteins may be Nt-acetylated in vivo to a lower extent than they appear to be in Nt-acetylation databases (which are based on steady-state measurements), because unacetylated versions of these proteins would be preferentially destroyed by the Arg/N-end rule pathway through their MetΦ/N-degrons.
The conditional and processive protein degradation by the Arg/N-end rule pathway and the Ac/N-end rule pathway encompasses full-length proteins and their protein-sized fragments generated by nonprocessive proteases that include calpains, caspases, separases, secretases, and Met-aminopeptidases (Figures 6 and S1). While distinct mechanistically (they involve different N-degrons and N-recognins), the two branches of the N-end rule pathway have now been shown to interact functionally (Figures 6 and 7). One remarkable conclusion from this – still continuing – expansion of the N-end rule pathway is that nearly all 20 amino acids of the genetic code can act, in specific sequence contexts, as destabilizing N-terminal residues (N-degrons), both in their unacetylated states and after Nt-acetylation. Yet another astonishing realization, brought about by the present study and by the 2010 discovery of the Ac/N-end rule pathway (Hwang et al., 2010b; Shemorry et al., 2013), is that most proteins in a cell are conditionally short-lived N-end rule substrates, either as full-length proteins or as protease-generated fragments. Some physiological ramifications of these insights are described above, and many more remain to be explored.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Genetic Techniques
Tables S1–S3 cite S. cerevisiae strains, plasmids and PCR primers, respectively. Standard techniques were employed for strain construction and transformation. Details are described in Extended Experimental Procedures.
Purification of ScfUbr1, MmfUBR1 and MmfUBR2
The N-terminally flag-tagged S. cerevisiae Ubr1 (ScfUbr1), mouse Ubr1 (MmfUBR1) and mouse Ubr2 (MmfUBR2) were expressed in protease-deficient S. cerevisiae SC295 (Table S1) and purified by affinity chromatography as described in Extended Experimental Procedures.
SPOT Binding Assays
These assays employed a set of synthetic Met-Z-eK(3–10) peptides (Z = Ala, Cys, Asp, Glu, Phe, Gly, His, Ile, Lys, Leu, Met, Asn, Pro, Gln, Arg, Ser, Thr, Val, Trp, Tyr) as well as the independently prepared Met-Leu-eK(3–10) and Met-Lys-eK(3–10) peptides, and their Nt-acetylated counterparts AcMet-Leu-eK(3–10) and AcMet-Lys-eK(3–10). The peptides were C-terminally linked, as “dots”, to a cellulose-PEG membrane in equal molar amounts. Except for varying residues at positions 1 and 2, the sequences of the 10-residue SPOT-arrayed peptides were identical to the N-terminal sequence of the extensively characterized eK extension (see the main text). The peptides were synthesized by GmbH (JPT) (Berlin, Germany) using the JPT Peptide Technology. SPOT assays were carried out as described in Extended Experimental Procedures.
Cycloheximide-Chase and Pulse-Chase Assays
They were performed largely as described (Hwang et al., 2010b; Shemorry et al., 2013). Briefly, S. cerevisiae strains expressing epitope-tagged test proteins were treated with cycloheximide (CHX), and the samples were processed at indicated times for protein extraction, SDS-PAGE and immunoblotting with either anti-ha, anti-myc, or anti-tubulin antibodies. The latter antibodies were used to verify the uniformity of total protein loads. In 35S-pulse-chase assays, S. cerevisiae were labeled with 35S-methionine/cysteine for 2 to 5 min (as indicated), followed by a chase, the processing of a cell extract for immunoprecipitation with anti-ha antibody, SDS-PAGE, autoradiography, and quantification, as described in Extended Experimental Procedures.
GST-Pulldown Assays
ML-eK-GST (ML-GST) (see the main text) was expressed in E. coli and purified by affinity chromatography, using Glutathione HiCap Matrix (Qiagen). The N-terminally flag-tagged full-length S. cerevisiae Ubr1 (fUbr1) or its flag-tagged fUBR1–717, Ubr1209–1140f, fUbr11–310, fUbr198–518, and fUbr1209–717 fragments were expressed in S. cerevisiae SC295 (Table S1) from the PADH1 promoter on a high copy plasmid. GST-pulldown assays with purified ML-GST and S. cerevisiae extracts containing fUbr1 or its fragments were carried out as described in Extended Experimental Procedures.
Ubr1-Mediated Ubiquitylation Assay
This completely defined in vitro ubiquitylation assay comprised purified human Ub, purified S. cerevisiae Uba1 (the E1 enzyme), purified S. cerevisiae Rad6 (the E2 enzyme), purified fUbr1 (the E3 N-recognin), and purified ML-GST (see the preceding section about fUbr1 and ML-GST). Details of purification of these proteins as well as the ubiquitylation assay (Hwang et al., 2010a) are described in Extended Experimental Procedures.
RT-PCR Assays
Total RNAs were extracted using RiboPure-Yeast Kit (Life Technologies) from indicated S. cerevisiae strains that expressed either ML-Ura3 (Ub-ML-eK-ha-Ura3), MF-Pre5ha, MI-Sry1ha, or the endogenous MI-Sry1ha3 and were grown to A600 of ~1 in 15 ml of SC(-Trp) or SC(-His) media containing 20 μM CuSO4. The steps of reverse transcription, PCR amplification of resulting DNAs, and other RT-PCR procedures are described in Extended Experimental Procedures.
Cell Growth Assays
To determine whether overexpression of PRE5 influences cell growth in specific genetic backgrounds, S. cerevisiae JD53 (WT), CHY371 (naa30Δ) and CHY372 (naa30Δ ubr1Δ) that carried either pCH690 (p424CUP1) or pCH1658 (p424CUP1-PRE5) were used (Table S2). Cells were grown overnight in SC-Trp medium at 30°C, then re-inoculated, in triplicate, into 10 ml of SC-Trp medium in a 100 ml flask to the final A600 of 0.2 and thereafter incubated, with rotary shaking, at 37°C for 16 hr. A600 of cultures (each of them grown in triplicate) were measured every 4 hr. To examine effects of stressors on cell growth, JD53 (WT), JD83-1A (ubr1Δ), CHY371(naa30), or CHY372 (naa30Δ ubr1Δ) S. cerevisiae were grown in YPD medium in the presence of indicated stressors (Figure S4G, H), and the relative rates of cell growth were assayed as described in Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
N-terminal Met followed by a hydrophobic residue functions as an N-degron.
Unacetylated Met-bearing proteins can be destroyed by the Arg/N-end rule pathway.
The Arg/N-end rule and Ac/N-end rule pathways have complementary specificities.
Both unacetylated and AcMet-proteins can be destroyed by these N-end rule pathways.
Acknowledgments
We thank S. Y. Kim (Korea Research, Institute of Biosciences and Biotechnology) for calculating the relative content of encoded human Met-Φ proteins, and D. H. Wolf (University of Stuttgart, Germany) for providing pFE15. We also thank C. Brower, K. Piatkov, J. Raskatov and B. Wadas (California Institute of Technology), and I. Hwang (Pohang University of Science and Technology) for helpful comments on the manuscript. We are grateful to members of the Hwang and Varshavsky laboratories for their assistance and advice. This work was supported by grants to C.S.H from the National Research Foundation (NRF) of the Korea government (MSIP) (NRF-2011-0021975 and 2012R1A4A1028200), the Korean Healthcare Technology R&D Project of the Ministry of Health & Welfare (HI11C1279), and the T. J. Park Science Fellowship of POSCO T. J. Park Foundation, and also by grants to A.V. from the U.S. National Institutes of Health (DK039520 and GM031530). H.K.K was supported by the Korean Government’s NRF-2013-Global Ph.D. Fellowship Program (NRF-2013H1A2A1033225) and the BK21 PLUS Program.
Footnotes
Supplemental Information includes Extended Experimental Procedures, Figures S1–S4, and Tables S1–S3.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnesen T, Van Damme P, Polevoda B, Helsens K, Evjenth R, Colaert N, Varhaug JE, Vandekerckhove J, Lillehaug JR, Sherman F, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast to humans. Proc Natl Acad Sci USA. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Behnia R, Panic B, Whyte JRC, Munro S. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat Cell Biol. 2004;6:405–413. doi: 10.1038/ncb1120. [DOI] [PubMed] [Google Scholar]
- Brower CS, Piatkov KI, Varshavsky A. Neurodegeneration-associated protein fragments as short-lived substrates of the N-end rule pathway. Mol Cell. 2013;50:161–171. doi: 10.1016/j.molcel.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WS, Jeong BC, Joo YJ, Lee MR, Kim J, Eck MJ, Song HK. Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat Struct Mol Biol. 2010;17:1175–1181. doi: 10.1038/nsmb.1907. [DOI] [PubMed] [Google Scholar]
- Dougan DA, Micevski D, Truscott KN. The N-end rule pathway: from recognition by N-recognins to destruction by AAA+ proteases. Biochim Biophys Acta. 2011;1823:83–91. doi: 10.1016/j.bbamcr.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Duttler S, Pechmann S, Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol Cell. 2013;50:379–393. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisele F, Wolf DH. Degradation of misfolded proteins in the cytoplasm by the ubiquitin ligase Ubr1. FEBS Lett. 2008;582:4143–4146. doi: 10.1016/j.febslet.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Finley D, Ulrich HD, Sommer T, Kaiser P. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics. 2012;192:319–360. doi: 10.1534/genetics.112.140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson EK, Gardner RG. Selective destruction of abnormal proteins by ubiquitin-mediated protein quality control degradation. Semin Cell Dev Biol. 2012;23:530–537. doi: 10.1016/j.semcdb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglione C, Vallon O, Meinnel T. Control of protein life-span by N-terminal methionine excision. EMBO J. 2003;22:13–23. doi: 10.1093/emboj/cdg007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and homeostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci USA. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W, Tröndle N, Albrecht G, Wolf DH. PRE5 and PRE6, the last missing genes encoding 20S proteasome subunits from yeast? Biochemistry. 1994;33:12229–12237. doi: 10.1021/bi00206a028. [DOI] [PubMed] [Google Scholar]
- Hwang CS, Shemorry A, Varshavsky A. The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat Cell Biol. 2010a;12:1177–1185. doi: 10.1038/ncb2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010b;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Gonda DK, Varshavsky A. Cis-trans recognition and subunit-specific degradation of short-lived proteins. Nature. 1990;346:287–291. doi: 10.1038/346287a0. [DOI] [PubMed] [Google Scholar]
- Matias AC, Ramos PC, Dohmen RJ. Chaperone-assisted assembly of the proteasome core particle. Biochem Soc Trans. 2010;38:29–33. doi: 10.1042/BST0380029. [DOI] [PubMed] [Google Scholar]
- Matta-Camacho E, Kozlov G, Li FF, Gehring K. Structural basis of substrate recognition and specificity in the N-end rule pathway. Nat Struct Mol Biol. 2010;17:1182–1188. doi: 10.1038/nsmb.1894. [DOI] [PubMed] [Google Scholar]
- Mischerikow N, Heck AJ. Targeted large-scale analysis of protein acetylation. Proteomics. 2011;11:571–589. doi: 10.1002/pmic.201000397. [DOI] [PubMed] [Google Scholar]
- Monda JK, Scott DC, Miller DJ, Lydeard J, King D, Harper JW, Bennett EJ, Schulman BA. Structural conservation of distinctive N-terminal acetylation-dependent interactions across a family of mammalian NEDD8 ligation enzymes. Structure. 2012;21:1–12. doi: 10.1016/j.str.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkov KI, Brower CS, Varshavsky A. The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc Natl Acad Sci USA. 2012;109:E1839–E1847. doi: 10.1073/pnas.1207786109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Kawaguchi S, Ng DTW. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell. 2010;21:2117–2127. doi: 10.1091/mbc.E10-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SRG, Strochlic TI, Tong AHY, Boone C, Burd CG. Golgi targeting of ARF-like GTPase Arl3p requires its N-alpha-acetylation and the integral membrane protein Sys1p. Nat Cell Biol. 2004;6:414–419. doi: 10.1038/ncb1121. [DOI] [PubMed] [Google Scholar]
- Shemorry A, Hwang CS, Varshavsky A. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol Cell. 2013;50:540–551. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starheim KK, Gevaert K, Arnesen T. Protein N-terminal acettyltransferases: when the start matters. Trends Biochem Sci. 2012;37:152–161. doi: 10.1016/j.tibs.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Summers DW, Wolfe KJ, Ren HY, Cyr DM. The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS One. 2013;8:e52099. doi: 10.1371/journal.pone.0052099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsume Y, Ohdate T, Maeta K, Nomura W, Izawa S, Inoue Y. Calcineurin/Crz1 destabilizes Msn2 and Msn4 in the nucleus in response to Ca(2+) in Saccharomyces cerevisiae. Biochem J. 2010;427:275–287. doi: 10.1042/BJ20091334. [DOI] [PubMed] [Google Scholar]
- Tasaki TS, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu Rev Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoraki MA, Nillegoda NB, Saini J, Caplan AJ. A network of ubiquitin ligases is important for the dynamics of misfolded protein aggregates in yeast. J Biol Chem. 2012;287:23911–23922. doi: 10.1074/jbc.M112.341164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko RJ, Jr, Hochstrasser M. Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem. 2013;82:415–445. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GC, Varshavsky A. Detecting and measuring cotranslational protein degradation in vivo. Science. 2000;289:2117–2120. doi: 10.1126/science.289.5487.2117. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. Discovery of cellular regulation by protein degradation. J Biol Chem. 2008;283:34469–34489. doi: 10.1074/jbc.X800009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule pathway and regulation by proteolysis. Prot Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M, Nakamori S, Takagi H. Serine racemase homologue of Saccharomyces cerevisiae has L-threo-3-hydroxyaspartate dehydratase activity. FEMS Microbiol Lett. 2003;225:189–193. doi: 10.1016/S0378-1097(03)00484-1. [DOI] [PubMed] [Google Scholar]
- Wang F, Durfee LA, Huibregtse JM. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol Cell. 2013;50:368–378. doi: 10.1016/j.molcel.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Webster A, Du F, Piatkov K, Ghislain M, Varshavsky A. Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. J Biol Chem. 2008;283:24011–24028. doi: 10.1074/jbc.M802583200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Zhang F, Nacev BA, Liu JO, Pei D. Protein N-terminal processing: substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry. 2010;49:5588–5599. doi: 10.1021/bi1005464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW, Lascina JR, Rechsteiner MC, Nicchitta CV. Out with the old, in with the new? Comparing methods for measuring protein degradation. Cell Biol Int. 2011;35:457–462. doi: 10.1042/CBI20110055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kulkarni K, Hanrahan SJ, Thompson AJ, Barford D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homodimer interface. EMBO J. 2010;29:3733–3744. doi: 10.1038/emboj.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.