Abstract
The wilt diseases caused by Verticillium dahliae and Fusarium oxysporum are the major diseases of eggplant (Solanum melongena L.). In order to generate transgenic resistance against the wilt diseases, Agrobacterium-mediated gene transfer was performed to introduce alfalfa glucanase gene encoding an acidic glucanase into eggplant using neomycin phosphotransferase (npt-II) gene as a plant selection marker. The transgene integration into eggplant genome was confirmed by Polymerase chain reaction (PCR) and Southern blot analysis and transgene expression by the glucanase activity and western blot analysis. The selected transgenic lines were challenged with V. dahliae and F. oxysporum under in vitro and in vivo growth conditions, and transgenic lines showed enhanced resistance against the wilt-causing fungi with a delay of 5–7 days in the disease development as compared to wild-type plants.
Keywords: Solanum melongena, Pathogenesis-related (PR) proteins, Glucanase, Plant transformation, Disease resistance
Introduction
Eggplant (Solanum melongena L.) is an important vegetable crop of the temperate and tropical parts of the world. Most of the cultivated varieties are highly susceptible to diseases caused by various pathogens (fungal, bacterial, and viral pathogens) as well as other stresses, and such stresses result in significant loss of crop yield and quality (Collonnier et al. 2001; Kashyap et al. 2003; Wally and Punja 2010; Ceasar and Ignacimuthu 2012). Eggplant wilt caused by a number of fungal genera such as Fusarium, Verticillium, Rhizoctonia, Sclerotium, and Phytophthora cause considerable loss in crop yield annually (Najar et al. 2011). The wild varieties are a source of resistance against the bacterial and fungal diseases (Gousset et al. 2005) and therefore can be used as useful germplasm to develop resistant varieties of eggplant against the pathogens through inter-specific hybridization. However, the inter-specific hybrids between wild and cultivated species have been successful in only a few cases. Therefore, the introgression of desired traits from the wild relatives into the cultivated varieties is not very successful. Thus, there is an urgent need to adopt the transgenic strategies to engineer eggplant for resistance against wilt diseases.
Many complex mechanisms evolved in plants in response to pathogen infection have been identified, and role of multiple genes in response to pathogen infection and various pathways involved therein have been identified (Grover and Gowthaman 2003; Islam 2006). Over-expression of defense response genes in transgenic plants have provided enhanced resistance to a variety of fungal pathogens (Grover and Gowthaman 2003; Wally and Punja 2010). For instance, pathogenesis-related (PR) proteins are induced in plants in response to various pathogens. β-1,3-glucanases (E.C. 3.2.1.39) belong to class II of the PR proteins and are abundant proteins widely distributed among the seed plant species. They are primarily involved in plant defense against pathogens as they catalyze the hydrolysis of β-1,3-glucans which is a major component of the cell wall of most fungi (Rao et al. 1999; Punja 2001; Grover and Gowthaman 2003; Wally and Punja 2010; Ceasar and Ignacimuthu 2012). Enzyme activity is usually low in a healthy plant but increases during fungal infection, and the induction of β-1,3-glucanase in various plants like barley, wheat, rice, and sorghum in response to fungal pathogens clearly demonstrate the involvement of the enzyme in defense response (Cheong et al. 2000; Grover and Gowthaman 2003; Płazek et al. 2009; Cawood et al. 2010; Aggarwal et al. 2011; Salim et al. 2011; Gupta et al. 2013).
The expression of the glucanase gene in transgenic plants has conferred increased resistance against fungal pathogens (Rao et al. 1999; Punja 2001; Maziah et al. 2007; Mondal et al. 2007; Wally and Punja 2010; Sundaresha et al. 2010; O’Kennedy et al. 2011; Ceasar and Ignacimuthu 2012). Increased resistance against Phytophthora infestans due to increase in glucanase activity in transgenic potato plants expressing soybean glucanase gene have been reported (Borkowoska et al. 1998). Reduction in the disease index values was observed in transgenic tobacco plants expressing barley glucanase that correlated well with the levels of glucanase accumulation (Jach et al. 1995). Transgenic tobacco expressing cDNA of soybean β-1,3-glucanase under the control of CaMV 35S promoter was shown to exhibit resistance against fungal diseases (Yoshikawa et al. 1993). Kiwifruit transformed with soybean β-1,3-glucanase showed sixfold increase in the enzyme activity in one of the transformants and decrease in the disease lesion area by the gray mold fungus Botrytis cinerea (Nakamura et al. 1999). Resistance to Peronospora tabacina and Phytophthora parasitica var. nicotianae in the leaves of transgenic tobacco plants was closely associated with high levels of β-1,3-glucanase activity (Lusso and Kuc 1996). Nishizawa et al. (2003) introduced β-1,3-glucanase and β-1,4-glucanse gene (Gns1) of rice, and Akiyama et al. (2004) used another glucanase gene of rice (OsGLN2) to enhance the disease resistance in rice. Bioassays in both these studies showed resistance to blast infection. Expression of β-1,3-glucanase gene (βglu) of potato into flax is found to increase resistance to Fusarium infection (Wróbel-Kwiatkowskaa et al. 2004). Transgenic Indian mustard expressing glucanase gene and resistant against leaf spot disease caused by Alternaria brassicae has been developed (Mondal et al. 2007). Expression of β-1,3-glucanase (gluc78), from the biocontrol fungus Trichoderma atroviride, confers resistance in transgenic pearl millet against Sclerospora graminicola (O’Kennedy et al. 2011). Transgenic groundnut over-expressing a tobacco β-1,3-glucanase has enhanced resistance against two major fungal pathogens, Cercospora arachidicola and Aspergillus flavus (Sundaresha et al. 2010). Transgenic plants expressing glucanase, in combination with other defense-related genes, have also been reported to be resistant against fungal pathogens (Mackintosh et al. 2006; Sridevi et al. 2008; Amian et al. 2011).
Zhu et al. (1994) expressed alfalfa acidic glucanase gene singly and in conjunction with chitinase gene in tobacco for resistance against Cercospora nicotianae. Masoud et al. (1996) expressed alfalfa acidic glucanase in alfalfa plants for resistance against Phytophthora megasperma f. sp.medicaginis (Pmm). Recently, alfalfa glucanase gene has been coexpressed with rice chitinase in rice and transgenic plants conferred resistance to fungal sheath blight (caused by Rhizoctonia solani) and blast (caused by Magnaporthe oryzae) (Mao et al. 2013). In this work, we report on the production of eggplant constitutively expressing alfalfa acidic glucanase gene, which is similar to the genes that were tested earlier, and the evaluation of these transgenics for tolerance to wilt-causing fungal pathogens, Verticillium dahliae and Fusarium oxysporum. The over-expression of glucanase gene in eggplants significantly enhanced their tolerance to wilt-causing fungal pathogens, with no effect on the phenotype.
Materials and methods
Eggplant transformation
Cotyledon and leaf explants from Pusa Purple Long (PPL) variety were collected from about 15-day-old and 1-month-old axenic seedlings, respectively, and used as explants for Agrobacterium-mediated transformation with binary vector pM42 (Lamb et al. 1996), harboring class II acidic alfalfa glucanase. Transformation and regeneration procedure as well as the selection of transformants on kanamycin were carried out as described earlier (Prabhavathi et al. 2002).
Molecular analyses of transformants
Genomic DNA was isolated from the putative transgenic plants and untransformed control using CTAB method (Doyle and Doyle 1990). Genomic DNA was amplified by PCR using neomycin phosphotransferase (npt-II) gene specific primers (forward - 5′ACGTTGTCA CTGAAGCGGGAAGG 3′ and reverse - 5′ GTAAAGCACGAGGAAGCGGTCAGC 3′) and glucanase gene specific primers, designed for the internal coding region (forward 5′ GATCTCTACAAAGCAAACAACATT 3′ and reverse 5′ AGGGCCAACCACTCTCAG 3′) to check for the integration of npt-II and β-1-3-glucanase gene, respectively. The PCR conditions employed for npt-II and glucanase gene amplification were initial denaturation at 94 °C for 5 min followed by 35 cycles of denaturation at 94 °C for 1 min, primers annealing at 54 °C for 1 min, and synthesis at 72 °C for 2 min, with a final extension step of 72 °C for 10 min. The amplification cycles were carried out in the Techne (UK) PCR machine, and the PCR products were analyzed on 1 % agarose gel along with 1 kb DNA ladder (MBI, Fermentas). Transgene integration was also analyzed by Southern analysis. Ten micrograms of genomic DNA from PCR positive transgenic lines as well as untransformed control were digested with EcoRI, and blots were prepared as per the standard protocols (Sambrook et al. 1989). The glucanase gene fragment was radiolabelled using the nick translation kit (Bangalore Genei, India) following the manufacturer’s instructions and hybridized to the membrane (Hybond-N, Nylon 0.45 μm, Amersham). Southern hybridization and washing was carried out at high stringency (Sambrook et al. 1989). Autoradiographs were obtained by exposing the X-ray film to the membrane.
Western blot analysis
Total proteins were isolated from T1 seedlings using standard protocol (Laemmli 1970). The total protein was quantified using Bradford’s reagent (Bradford 1976), and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as mentioned before (Laemmli 1970). Equal amounts (20 μg) of total protein extract from the transgenic and control were loaded on to 12 % SDS-PAGE and electroblotted overnight onto the PVDF membrane (Winston et al. 1987) in tris-glycine-methanol buffer. The membrane was blocked overnight and probed with anti-barley β-1,3-glucanase antibody at 1:1,000 dilution, and goat anti-rabbit IgG horse radish peroxidase (HRP) was used as the secondary antibody at a dilution of 1:2,000. Blots were developed by ECL following the manufacturer’s instructions (Amersham, USA).
Glucanase enzyme activity
Glucanase enzyme assay was performed to check the levels of β-1,3-glucanase enzyme produced in the T1 progeny plants in comparison to the untransformed control. The enzyme activity of β-1,3-glucanase was colorimetrically assayed by measuring the rate of reducing sugar production by the laminarin-dinitrosalicylate method (Abeles and Forrence 1970) with slight modifications. Crude enzyme extract, 62.5 μl (containing 100 μg of protein), was added to a 62.5 μl laminarin (1 mg ml−1 in 0.1 M sodium acetate buffer, pH 5.2) and then incubated at 40 °C for 10 min. The reaction was stopped by adding 125 μl of dinitrosalicylic reagent and boiled for 5 min in a water bath. The resulting colored solution was diluted with 4.5 ml of water, vortexed and its absorbance at 540 nm was measured. The crude enzyme preparation mixed with laminarin at zero time was kept as control. One unit of enzyme activity was defined as the amount of enzyme that produced reducing sugar equivalent to 1 μmol of glucose equivalent per min under the assay conditions.
Segregation analysis
The T1 seeds obtained from transgenic plants were analyzed for segregation of the transgene. Seeds collected from the selfed T0 plants were screened for kanamycin resistance. The surface-sterilized seeds were inoculated on to basal medium supplemented with 30 mg l−1 kanamycin and incubated at 28 ± 1 °C and 16-h photoperiod. After 15 days, the seeds were scored for kanamycin resistance and the data was validated using χ2 test. The seeds were also inoculated on the MS basal medium without kanamycin to check for the germination frequency (i.e., seed viability).
Fungal resistance assay of transgenics
One-month-old seedlings of wild-type and T1 segregated transgenic eggplants were tested for resistance against fungal wilts using root-dip and soil mix methods as described before (Prabhavathi and Rajam 2007; Hazarika and Rajam 2011). Each assay was performed three times with six-to-eight replicates of each transgenic line and control. Spores from both fungi were harvested from fungal cultures freshly grown on potato dextrose agar (PDA, HiMedia) plates for about 15 days. Three plants (about 15-days-old) of each of the independent transgenic lines were assayed against V. dahlia and F. oxysporum. In vitro fungal resistance assay was done by inoculating spores (108 spores ml−1) near the root system of the transgenic seedlings growing on 1/10 strength solid MS basal medium. After inoculation, the jam bottles were sealed and kept under the controlled growth conditions with a 16-h photoperiod and temperature of 26 ± 1 °C and the extent of disease resistance was recorded, based on wilting symptoms. For the in vivo root dip method, the plants were kept turgid by spraying with sterile water and covering with a polypropylene bag for 8 h before inoculation. The T1 seedlings from different transgenic lines as well as from untransformed control were inoculated by dipping their root system for 15 min in a freshly prepared spore suspension (108 spores ml−1) of V. dahliae or F. oxysporum (Lin and Xiao 1995). These seedlings were then transplanted into small plastic egg trays containing 1:1 mixture of soil and vermiculite. The seedlings were kept under optimum humidity and temperature conditions, and the degree of resistance to the fungal infection was recorded. For in vivo soil mix assay, 1-month-old seedlings of transgenics and control were inserted into egg trays containing soil:vermiculite (1:1) and mixed with fungal spores (108 spores ml−1). The plants were grown under suitable conditions, and the degree of disease resistance was recorded at regular intervals. Disease severity index (DSI) was scored on a standard scale by the time wild-type plant had developed severe wilting and necrosis (about 10 days post-inoculation). The DSI was calculated by the formula, ∑ PXP0 − 5/∑Pn × 5; where X is the number of plants per phenotypic class; P is the phenotypic class 0: plants showing no aerial symptoms, 1: only bottom leaf necrotic or curled, 2: three leaves developed wilt symptoms, 3: only newest leaves remained healthy, older ones being necrotic and curled, 4: all leaves fallen, plant having only newly formed leaves, 5: plant death; and n is the total number of plants tested. A highly susceptible plant scored 1 on the DSI, while the resistant plants had DSI below 1 and approaching 0 (Lin and Xiao 1995).
Results
Molecular analysis of the transgenic plants
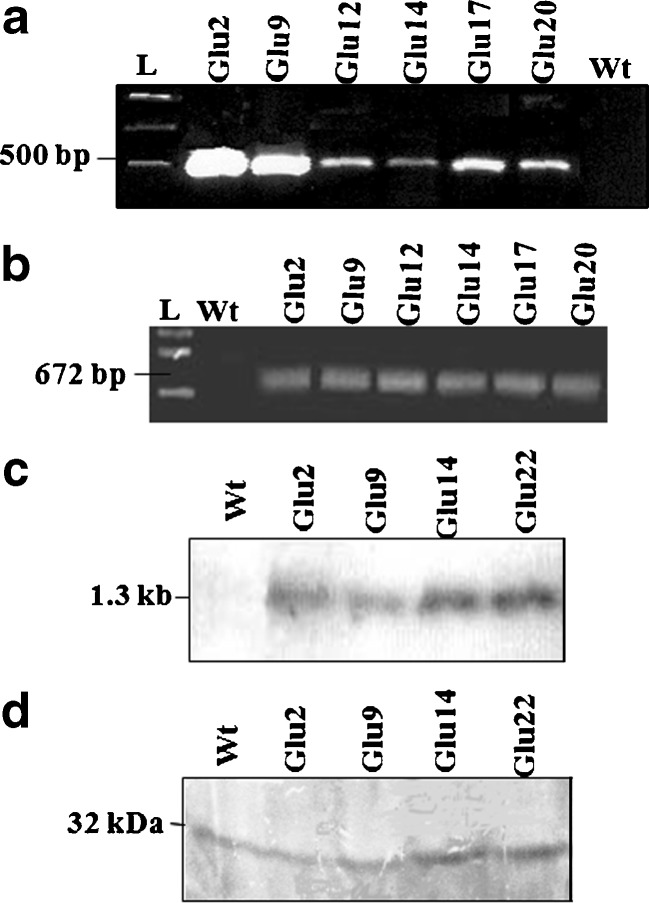
Eggplant cotyledons and leaf explants were subjected to a 2-day pre-culture before Agrobacterium-mediated transformation with the binary vector pM42, harboring class II acidic alfalfa glucanase and npt-II marker genes (Lamb et al. 1996) (Fig. 1). The cocultivated cotyledon and leaf explants regenerated well after transformation on the selection medium. The putative transgenic plants showed normal phenotype and were analyzed by PCR for the presence npt-II and gluacanase gene. The transgenics showing an expected amplification product of 487 bp with the primers specific to npt-II gene (Fig. 2a) and 672 bp with glucanase gene specific primers (Fig. 2b), confirming the successful introduction of alfalfa glucanase gene. To further confirm the integration of glucanase gene, PCR positive transgenic lines were analyzed by Southern hybridization. Upon hybridization, all the transgenic lines analyzed showed the expected band of corresponding to the alfalfa glucanase gene, whereas the untransformed control plants were negative (Fig. 2c). We have utilized some of the transgenic lines for various analyses, but two lines Glu2 and Glu14 were consistently used for all the analyses.
Fig. 1.
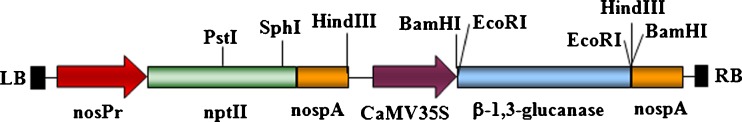
Transfer-DNA map of the binary vector harboring glucanase gene
Fig. 2.
a PCR analysis with primers specific to npt-II gene: Lane 1 indicates 1 kb ladder; lanes 2–7 indicate DNA from different transgenic lines; and lane 8 indicates DNA from wild-type plant. b PCR with primers specific to glucanase gene: Lane 1 indicates 1 kb ladder; lane 2 indicates DNA from untransformed control plant; and lanes 3–8 indicate DNA from different transgenic lines. c Southern blot analysis of the PCR positive transgenics for transgene integration using glucanase gene as probe: Lane 1 indicates DNA from untransformed control plant, and lanes 2–5 indicate DNA from different transgenic lines. d Western blot analysis of the glucanase transgenic lines: Lane 1 indicates protein from untransformed plant and lanes 2–5 indicate protein from different transgenic lines
Glucanase protein was analyzed in PCR and Southern positive transgenic lines by western analysis using barley anti-glucanase polyclonal antibodies. The transgenic lines, viz. Glu2, Glu9, Glu14, and Glu22, showed the presence of the expected 32 kDa protein with varying levels of protein expression (Fig. 2d). The 32 kDa band corresponding to the native glucanase gene was also observed in the control, but its signal was low in comparison to the transgenic lines constitutively expressing the transgene, indicating higher expression of glucanase protein in the transgenic lines.
Glucanase activity in transgenic lines
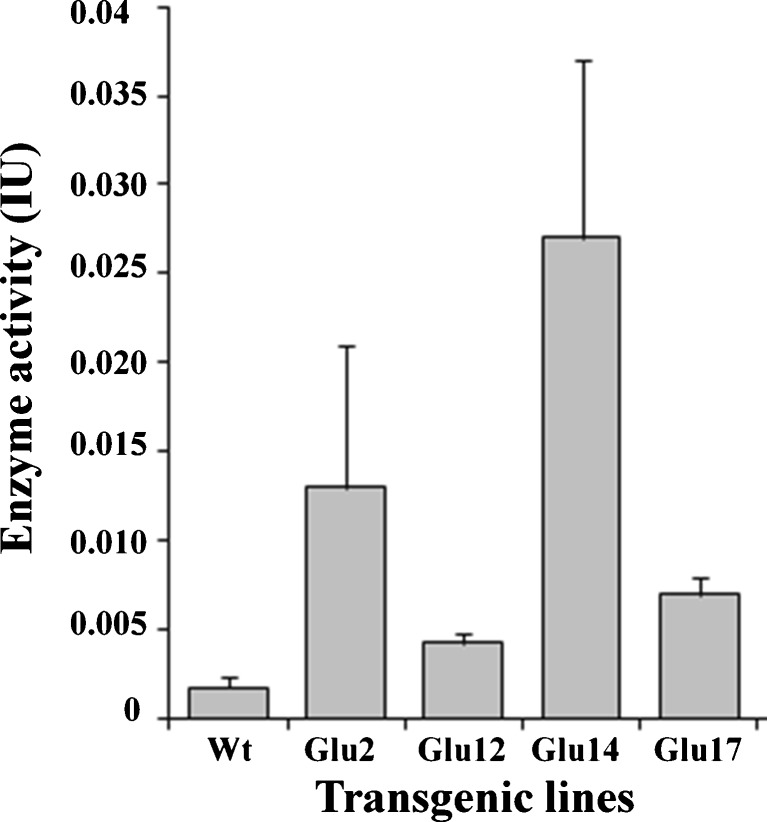
Glucanase activity was assayed on positive transgenic lines (Fig. 3). The seedlings of transgenic lines (Glu2, Glu12, Glu14, Glu17) showed higher glucanase enzyme activity as compared to the untransformed control (Fig. 3). The Glu14 line exhibited approximately eightfold increase in glucanase activity, while the transgenic lines Glu2, Glu12, and Glu22 showed two to fourfold increase in the enzyme activity as compared to the enzyme activity in control seedlings.
Fig. 3.
Analysis of β-1,3-glucanase activity. β-1,3-glucanase activity in total protein extracts from leaves of untransformed and T1 transgenic lines. Standard error of mean for all the lines are indicated at the top of the bars
Segregation analysis of the marker gene
In order to determine the segregation of the marker gene (npt-II), T1 selfed seeds were germinated on MS basal medium fortified with 30 mg l−1 kanamycin. The transgenic seeds started germinating after 1 week of inoculation and the seedlings grew well on antibiotic-amended medium, while there was no germination in control seeds. Six out of the nine transgenic lines tested, viz Glu9, Glu14, Glu22, Glu24, Glu26, and Glu34, showed 3:1 transgene segregation, suggesting single copy transgene integration, while Glu2, Glu12, and Glu38 lines deviated from 3:1 ratio (Table 1).
Table 1.
Segregation analysis of the transgene in the T1 glucanase transgenics of eggplant based on seed germination on kanamycin (30 mg l−1) amended MS basal medium
| Transgenic line | Total no. of seeds inoculated | KanR | KanS | Χ 2 |
|---|---|---|---|---|
| Wt | 87 | 0 | 87 | – |
| Glu2 | 45 | 40 | 5 | 4.63 |
| Glu9 | 50 | 34 | 16 | 7.31 |
| Glul2 | 50 | 44 | 6 | 4.51 |
| Glu14 | 50 | 35 | 15 | 0.67 |
| Glu22 | 50 | 41 | 9 | 4.31 |
| Glu24 | 45 | 38 | 7 | 2.14 |
| Glu26 | 50 | 33 | 17 | 2.16 |
| Glu34 | 50 | 36 | 14 | 0.24 |
| Glu38 | 83 | 70 | 13 | 3.86 |
Chi-square test was performed at P > 0.05 for 3:1 ratio
Fungal resistance assay of the transgenics
The T1 seedlings (positive segregants) of transgenic lines Glu2, Glu12, Glu14, and Glu17 were evaluated for resistance against the wilt disease caused by V. dahliae and F. oxysporum by in vitro (data not shown) and in vivo (Fig. 4) infection assays. In the in vitro infection assay, the seedlings of T1 transgenics and untransformed control were transferred to jam bottles containing 1/10th strength MS medium and then fungal spores were inoculated near the root system. The germination of the fungal spores and mycelial growth were observed within 1 week of inoculation in both untransformed control and transgenic seedlings. The transgenic lines Glu2, Glu12, Glu14, and Glu17 showed enhanced resistance to fungal pathogens with a delay of the appearance of wilt symptoms. There were no wilt symptoms in the transgenic lines within 10 days of inoculation, while the untransformed control seedlings showed wilting of lower leaves within 5–7 days of the inoculation. However, the seedlings of transgenic lines showed wilting and slight necrosis in the lower and second leaves after about 15 days of inoculation, while the untransformed control seedlings showed severe wilting and necrosis and died eventually. On the other hand, the transgenic seedlings could survive even after 1 month of inoculation and also showed the development of new leaves. The degree of fungal resistance varied between different transgenic lines against the two fungal pathogens.
Fig. 4.
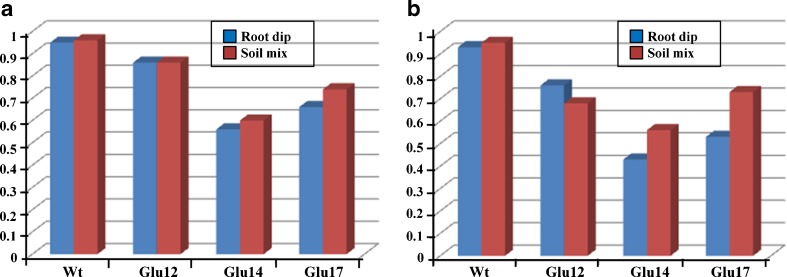
Biotic stress assay of T1 progenies of glucanase transgenic lines and untransformed control in root dip and soil mix fungal resistance assays against infection by F. oxysporum and V. dahliae. a In vivo root dip assay using V. dahliae. b In vivo root dip assay using F. oxysporum. c In vivo soil mix assay using V. dahliae. d In vivo soil mix assay using F. oxysporum. Wt indicates untransformed control. Glu2, Glu12, Glu14, and Glu17 indicate transgenic lines. Data were recorded after 1 month of inoculation. 6–8 T1 progeny seedlings per transgenic line tested for all the experiments. The experiments were repeated thrice with 6–8 T1 progeny seedlings per transgenic lines tested
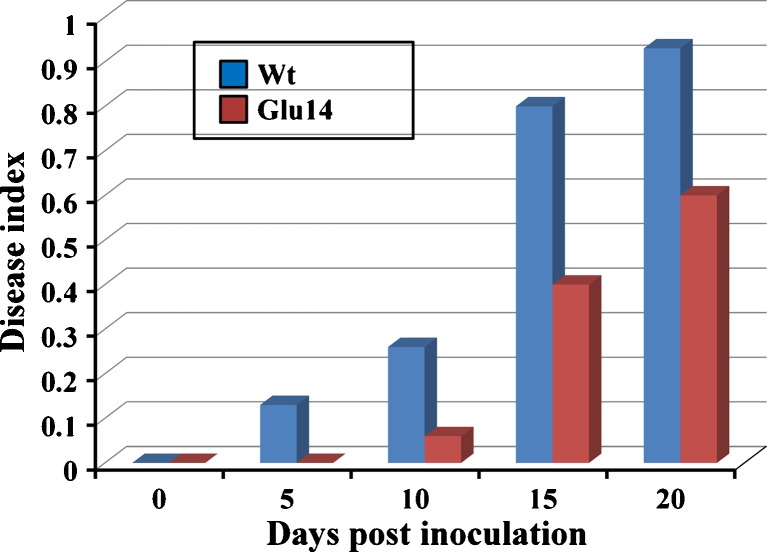
In the in vivo assay, 1-month-old seedlings of untransformed control and segregants of T1 progeny of transgenic lines were transferred to the soil mixed with fungal spores; whereas in the root dip assay, roots of the seedlings were dipped in the fungal spores, transferred to sterile soil, and scored for the development and severity of wilt symptoms. The transgenic lines Glu2, Glu12, Glu14, and Glu17 showed enhanced resistance to fungal pathogens marked by delayed onset of disease symptoms as compared to the wild-type control. While no wilting symptoms were seen in the transgenic seedlings up to 10 days of inoculation, while the untransformed control showed wilting of lower leaves and upper leaves with necrotic stem after 7 and 10 days of inoculation, respectively (Fig. 4). Transgenic seedlings showed slight wilting and necrosis of the lower and second leaves were observed after 15 days of inoculation, whereas the control seedlings were heavily infected and eventually died, while the transgenic seedlings developed new leaves even after 1 month of inoculation. Similar degree of resistance was observed in transgenic seedlings against V. dahliae and F. oxysporum. The response of different transgenic lines toward the fungal infection was variable (Fig. 4). Among all the tested transgenic lines, Glu14 line showed high resistance to fungal pathogens as compared to other lines, based on DI (Fig. 5). The fungal resistance assays were done on the segregated T1 seedlings. The assays were repeated thrice, and 6-8 seedlings of each transgenic line along with the untransformed control were used. The T1 seedlings for every independent transgenic line showed enhanced resistance against the fungal pathogens. The progression of disease symptoms were carefully observed and plotted as disease severity index (Fig. 6). Even after 5 days of inoculation, wilt disease progression was delayed in the transgenic lines as compared to untransformed control that had started showing wilting and necrosis in the lower leaves. After 15 days of inoculation, the untransformed control was severely infected and eventually died, whereas the transgenic lines showed wilting and necrosis only on the lower leaves (Fig. 6). There was no increase in the disease index values of Glu14 even after 20 days of inoculation, indicating enhanced resistance in the transgenics expressing glucanase gene.
Fig. 5.
Disease resistance index of glucanase transgenic lines and untransformed control in root dip and soil mix assays. a V. dahliae. b F. oxysporum
Fig. 6.
Kinetics of the disease index of Glu14 transgenic line and untransformed control in root dip infection assay with F. oxysporum
Discussion
Genetic engineering of disease resistance by transfer of plant defense-related genes into crops is valuable in terms of cost, efficacy, and reduction of pesticide usage (Lin et al. 2004; Wally and Punja 2010; Ceasar and Ignacimuthu 2012). In this study, we explored the possibility of the use of a glucanase gene from alfalfa against two important fungal pathogens of eggplant, viz. V. dahliae and F. oxysporum.
It was observed that the eggplant transgenics constitutively expressing alfalfa glucanase gene conferred moderate to high resistance against fungal wilts with a delay in disease symptoms development. Such a delay in disease symptom development has been observed in earlier studies (Mao et al. 2013; Mondal et al. 2007; Broglie et al. 1991), and it was attributed to the antifungal activity. The PR proteins (chitinase and glucanase) that are mainly localized in the chloroplasts before infection, but they were translocated to the vacuole and cell wall upon infection (Mao et al. 2013). The increase in the expression of the glucanase protein proved the fact that the resistance behavior of the transgenic plants was because of the over-expression of the glucanase gene in these transgenic plants (Zhu et al. 1994; Jongedijk et al. 1995; Masoud et al. 1996). Further evidence for the efficacy of the introduced glucanase gene was by the strong correlation seen between the glucanase activity and plant disease index (Jach et al. 1995). The plants that had low disease index exhibited high resistant to both the fungi tested (Lin and Xiao 1995).
The variation in the degree of fungal resistance in different transgenic lines could be due to the position effect mediated variation in transgene expression (Prabhavathi and Rajam 2007). Further, there was a correlation between transgene product (glucanase activity) and the degree of fungal resistance, and similar observations were also made in other studies (Yoshikawa et al. 1993; Jach et al. 1995; Lusso and Kuc 1996).
There are only a few reports of studies employing glucanases to enhance fungal resistance in transgenic plants (Rao et al. 1999; Punja 2001; Wally and Punja 2010). The disease resistance conferred by alfalfa glucanase may be due to its involvement in the solubilization of elicitors from the fungal cell walls which induce production of antifungal phytoalexins (Keen and Yoshikawa 1983) and/or the induction of the transcription of a plant defense gene, phenylalanine ammonialyase in a response to fungal attack to greater extent than control leaves (Yoshikawa et al. 1993). The restricted growth of pathogens leading to reduction in disease symptoms in transgenic plants expressing glucanase has been well documented in different host-pathogen systems (Zhu et al. 1994). The hydrolytic enzyme glucanase is lytic on the cell wall of necrotrophic fungal pathogens whose major cell wall component is glucan. It is well accepted that coordinated expression of many PR proteins may provide requisite levels of tolerance against the pathogen (Mao et al. 2013; Anand et al. 2003; Jach et al. 1995; Jongedijk et al. 1995). On pathogen infection, the defense signaling cascade is upregulated bringing about enhanced expression of PR proteins besides triggering the hypersensitive response. It is likely that in the background of expression of defense responsive proteins, enhanced expression of glucanase might have increased the tolerance levels of the transgenic plants. The transgenic eggplant lines with enhanced fungal resistance would be useful as parents in traditional breeding programs for fungal disease resistance.
Acknowledgments
This work was generously supported by the Indo–French Centre for Promotion of Advanced Research (Project No. 1803-1). We are grateful to Dr. Christopher Lamb, Salk Institute, USA, for providing pM42 plasmid carrying glucanase gene, and Prof. S. Muthukrishnan (Kansas State University, USA) for providing the glucanase antibodies.
Abbreviations
- BAP
Benzylaminopurine
- CaMV 35S
Cauliflower mosaic virus 35S promoter
- Glu
Glucanase
- IAA
Indole-3-acetic acid
- LB
Left border
- MS
Murashige and Skoog
- npt II
Neomycin phosphotransferase
- PR
Pathogenesis-related
- RB
Right border
- SRM
Shoot regeneration medium
References
- Abeles FB, Forrence LE. Temporal and hormonal control of β-1,3-glucanase in Phaseolus vulgaris L. Plant Physiol. 1970;45:395–400. doi: 10.1104/pp.45.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal R, Purwar S, Kharbikar L, Sangeeta Gupta L. Induction of a wheat β-1,3-glucanase gene during the defense response to Bipolaris sorokiniana. Acta Phytopathol Entomol Hung. 2011;46:37–49. doi: 10.1556/APhyt.46.2011.1.5. [DOI] [Google Scholar]
- Akiyama T, Pillai MA, Sentoku N. Cloning, characterization and expression of OsGLN2, a rice endo-1,3-beta-glucanase gene regulated developmentally in flowers and hormonally in germinating seeds. Planta. 2004;220:129–139. doi: 10.1007/s00425-004-1312-8. [DOI] [PubMed] [Google Scholar]
- Amian AA, Papenbrock J, Jacobsen HJ, Hassan F. Enhancing transgenic pea (Pisum sativum L.) resistance against fungal diseases through stacking of two antifungal genes (chitinase and glucanase) GM Crops. 2011;2:104–109. doi: 10.4161/gmcr.2.2.16125. [DOI] [PubMed] [Google Scholar]
- Anand A, Zhou T, Trick HN, Gill BZ, Bockus WW, Muthukrishnan S. Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin like protein, chitinase and glucanase against Fusarium graminearum. J Exp Bot. 2003;54:1101–1111. doi: 10.1093/jxb/erg110. [DOI] [PubMed] [Google Scholar]
- Borkowoska M, Krzymowska M, Talarczyk A, Awan MF, Yakovleva L, Kleczkowski K, Wielgat B. Transgenic potato plants expressing soybean beta-1,3-endoglucanase gene exhibit an increased resistance to Phytophthora infestans. Z Naturforsch. 1998;53:1012–1016. doi: 10.1515/znc-1998-11-1212. [DOI] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantification of microgram quantities of proteins utilizing the principle of protein dye binding. Ann Biochem. 1976;72:248–250. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Broglie K, Chet I, Holliday M, Cressman R, et al. Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science. 1991;254:1194–1197. doi: 10.1126/science.254.5035.1194. [DOI] [PubMed] [Google Scholar]
- Cawood ME, Pretorius JC, van der Westhuizen AJ, Pretorius ZA. Disease development and PR-protein activity in wheat (Triticum aestivum) seedlings treated with plant extracts prior to leaf rust (Puccinia triticina) infection. Crop Prot. 2010;29:1311–1319. doi: 10.1016/j.cropro.2010.06.017. [DOI] [Google Scholar]
- Ceasar SA, Ignacimuthu S. Genetic engineering of crop plants for fungal resistance: role of antifungal genes. Biotechnol Lett. 2012;34:995–1002. doi: 10.1007/s10529-012-0871-1. [DOI] [PubMed] [Google Scholar]
- Cheong YH, Kim CY, Chun HJ, Moon BC, Park HC, Kim JK, Lee S, Han C, Lee SY, Cho MJ. Molecular cloning of a soybean class III beta-1,3-glucanase gene that is regulated both developmentally and in response to pathogen infection. Plant Sci. 2000;154:71–81. doi: 10.1016/S0168-9452(00)00187-4. [DOI] [PubMed] [Google Scholar]
- Collonnier C, Fock I, Kashyap V, Rotino GL, Daunay MC, Lian Y, Mariska IK, Rajam MV, Servaes A, Ducreux G, Sihachakr D. Applications of biotechnology in eggplant. Plant Cell Tissue Org. 2001;65:91–107. doi: 10.1023/A:1010674425536. [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Gousset C, Collonnier C, Mulya K, Mariska I, Rotino GL, Besse P, Servaes A, Sihachakr D. Solanum torvum, as a useful source of resistance against the bacterial and fungal diseases for improvement of eggplant (S. melongena L.) Plant Sci. 2005;168:319–327. doi: 10.1016/j.plantsci.2004.07.034. [DOI] [Google Scholar]
- Grover A, Gowthaman R. Strategies for development of fungus-resistant transgenic plants. Curr Sci. 2003;84:330–340. [Google Scholar]
- Gupta P, Ravi I, Sharma V. Induction of β-1,3-glucanase and chitinase activity in the defense response of Eruca sativa plants against the fungal pathogen Alternaria brassicicola. J Plant Interact. 2013;8:155–161. doi: 10.1080/17429145.2012.679705. [DOI] [Google Scholar]
- Hazarika P, Rajam MV. Biotic and abiotic stress tolerance in transgenic tomatoes by constitutive expression of S-adenosylmethionine decarboxylase gene. Physiol Mol Biol Plants. 2011;17:115–128. doi: 10.1007/s12298-011-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam A. Fungus resistant transgenic plants: strategies, progress and lessons learnt. Plant Tissue Cult Biotechnol. 2006;16:117–138. [Google Scholar]
- Jach G, Görnhardt B, Mundy J, Logemann J, Pinsdorf E, Leah R, Schell J, Mass C. Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J. 1995;8:97–109. doi: 10.1046/j.1365-313X.1995.08010097.x. [DOI] [PubMed] [Google Scholar]
- Jongedijk E, Tigelaar H, van Roekel JSC, Bres-Vloemans SA, Dekker I, van den Elzen PJM, Cornelissen BJC, Melchers LS. Synergistic activity of chitinases and β-1,3-glucanases enhances fungal resistance in transgenic tomato plants. Euphytica. 1995;85:173–180. doi: 10.1007/BF00023946. [DOI] [Google Scholar]
- Kashyap V, Kumar SV, Collonier C, Fusari F, Haicor R, Rotino GL, Sihachakr D, Rajam MV. Biotechnology of eggplant. Sci Hortic. 2003;97:1–25. doi: 10.1016/S0304-4238(02)00140-1. [DOI] [Google Scholar]
- Keen NT, Yoshikawa M. β-1,3-endoglucanase from soybean releases elicitor-active carbohydrates from fungus cell walls. Plant Physiol. 1983;71:460–465. doi: 10.1104/pp.71.3.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamb CJ, Zhu Q, Maher EA, Dixon RA (1996) Transgenic plants containing multiple disease resistance genes. US patent- 5530187
- Lin B, Xiao Y. Sources of resistance to Verticillium wilt in Solanum melongena and its affinities identified by improved root dip method. Capsicum Eggplant Newsl. 1995;14:81–84. [Google Scholar]
- Lin WC, Lu CF, Wu JW, Cheng ML, Lin YM, Yang NS, Black L, Green SK, Wang JF, Cheng CP. Transgenic tomato plants expressing the Arabidopsis AtNPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 2004;13:567–581. doi: 10.1007/s11248-004-2375-9. [DOI] [PubMed] [Google Scholar]
- Lusso M, Kuc J. The effect of sense and antisense expression of the PR-N gene for β-1,3-glucanase on disease resistance of tobacco to fungi and viruses. Physiol Mol Plant Pathol. 1996;49:267–283. doi: 10.1006/pmpp.1996.0054. [DOI] [Google Scholar]
- Mackintosh CA, Garvin DF, Radmer LE, Heinen SJ, Muehlbauer GJ. A model wheat cultivar for transformation to improve resistance to Fusarium head blight. Plant Cell Rep. 2006;25:313–319. doi: 10.1007/s00299-005-0059-4. [DOI] [PubMed] [Google Scholar]
- Mao B, Liu X, Hu D, Li D (2013) Co-expression of RCH10 and AGLU1 confers rice resistance to fungal sheath blight Rhizoctonia solani and blast Magnaporthe oryzae and reveals impact on seed germination. World J Microbiol Biotechnol. doi:10.1007/s11274-013-1546-3 [DOI] [PubMed]
- Masoud SA, Zhu Q, Lamb C, Dixon RA. Constitutive expression of an inducible β-1,3-glucanase in alfalfa reduces disease severity caused by the oomycete pathogen Phytophthora megasperma f. spmedicaginis, but does not reduce disease severity of chitin-containing fungi. Transgenic Res. 1996;5:313–323. doi: 10.1007/BF01968941. [DOI] [Google Scholar]
- Maziah M, Saraih M, Sreeramanan S. Transgenic banana Rastali (AAB) with β-1,3-glucanase gene for tolerance to Fusarium wilt race 1 disease via Agrobacterium-mediated ransformation system. Plant Pathol J. 2007;6:271–282. doi: 10.3923/ppj.2007.271.282. [DOI] [Google Scholar]
- Mondal KR, Bhattacharya RC, Koundal KR, Chatterjee SC. Transgenic Indian mustard (Brassica juncea) expressing tomato glucanase leads to arrested growth of Alternaria brassicae. Plant Cell Rep. 2007;26:247–252. doi: 10.1007/s00299-006-0241-3. [DOI] [PubMed] [Google Scholar]
- Najar AG, Anwar A, Masoodi L, Khar MS. Evaluation of native biocontrol agents against Fusarium solani f. sp. melongenae causing wilt disease of brinjal in Kashmir. J Phytology. 2011;3:31–34. [Google Scholar]
- Nakamura Y, Sawada H, Kobayashi S, Nakajima I, Yoshikawa M. Expression of soybean β-1,3-glucanase cDNA and effect on disease tolerance in kiwifruit plants. Plant Cell Rep. 1999;18:527–532. doi: 10.1007/s002990050616. [DOI] [Google Scholar]
- Nishizawa Y, Saruta M, Nakazono K, Nishio Z, Soma M, Yoshida T, Nakajima E, Hibi T. Characterization of transgenic rice plants over-expressing the stress-inducible beta-glucanase gene Gns1. Plant Mol Biol. 2003;51:143–152. doi: 10.1023/A:1020714426540. [DOI] [PubMed] [Google Scholar]
- O’Kennedy MM, Crampton BG, Lorito M, Chakauya E, Breese WA, Burger JT, Botha FC. Expression of a β-1,3-glucanase from a biocontrol fungus in transgenic pearl millet. S Afr J Bot. 2011;77:335–345. doi: 10.1016/j.sajb.2010.09.016. [DOI] [Google Scholar]
- Płazek A, Skoczowski A, Hura K, Libik M, Barna B. Accumulation of H2O2 and changes in activities of antioxidative enzymes and β-1,3-glucanase in barley and meadow fescue leaves attacked by Bipolaris sorokiniana. Cereal Res Commun. 2009;37:399–408. doi: 10.1556/CRC.37.2009.3.9. [DOI] [Google Scholar]
- Prabhavathi V, Rajam MV. Mannitol-accumulating transgenic eggplants exhibit enhanced resistance to fungal wilts. Plant Sci. 2007;173:50–54. doi: 10.1016/j.plantsci.2007.04.004. [DOI] [Google Scholar]
- Prabhavathi V, Yadav JS, Kumar PA, Rajam MV. Abiotic stress tolerance in transgenic eggplant (Solanum melongena L.) by introduction of bacterial mannitol phosphodehydrogenase gene. Mol Breeding. 2002;9:137–147. doi: 10.1023/A:1026765026493. [DOI] [Google Scholar]
- Punja ZK. Genetic engineering of plants to enhance resistance to fungal pathogens—a review of progress and future prospects. Can J Plant Pathol. 2001;23:216–235. doi: 10.1080/07060660109506935. [DOI] [Google Scholar]
- Rao GU, Kaur M, Verma A, Sihachakr D, Rajam MV. Genetic engineering of crop plants for resistance to fungal pathogens. J Plant Biol. 1999;26:31–42. [Google Scholar]
- Salim AP, Saminaidu K, Marimuthu M, Perumal Y, Rethinasamy V, Palanisami JR, Vadivel K. Defense responses in tomato landrace and wild genotypes to early blight pathogen Alternaria solani infection and accumulation of pathogenesis-related proteins. Arch Phytopathol Plant Protect. 2011;44:1147–1164. doi: 10.1080/03235408.2010.482763. [DOI] [Google Scholar]
- Sambrook J, Fristsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring harbour: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sridevi G, Parameswari C, Sabapathi N, Raghupathy V, Veluthambi K. Combined expression of chitinase and β-1,3-glucanase genes in indica rice (Oryza sativa L.) enhances resistance against Rhizoctonia solani. Plant Sci. 2008;175:283–290. doi: 10.1016/j.plantsci.2008.04.011. [DOI] [Google Scholar]
- Sundaresha S, Manoj Kumar A, Rohini S, Math S, Keshamma E, Chandrashekar S, Udayakumar M. Enhanced protection against two major fungal pathogens of groundnut, Cercospora arachidicola and Aspergillus flavus in transgenic groundnut over-expressing a tobacco β-1,3-glucanase. Eur J Plant Pathol. 2010;126:497–508. doi: 10.1007/s10658-009-9556-6. [DOI] [Google Scholar]
- Wally O, Punja ZK. Genetic engineering for increasing fungal and bacterial disease resistance in crop plants. GM Crops. 2010;1:199–206. doi: 10.4161/gmcr.1.4.13225. [DOI] [PubMed] [Google Scholar]
- Winston S, Fuller S, Hurrul J. Western blotting. In: Ausubel FM, editor. Curr Protoc in Mol Biol. New York: Wiley; 1987. pp. 10.8.1–10.8.6. [Google Scholar]
- Wróbel-Kwiatkowskaa M, Lorenc-Kukulaa K, Starzyckib M, Oszmiańskic J, Kepczyńskad E, Szopaa J. Expression of β-1,3-glucanase in flax causes increased resistance to fungi. Physiol Mol Plant Pathol. 2004;65:245–256. doi: 10.1016/j.pmpp.2005.02.008. [DOI] [Google Scholar]
- Yoshikawa M, Tsuda M, Takeuchi Y. Resistance to fungal disease in transgenic tobacco plants expressing the phytoalexin elicitor-releasing factor, β-1,3-glucanase from soybean. Naturwissenschaften. 1993;80:417–420. doi: 10.1007/BF01168337. [DOI] [Google Scholar]
- Zhu Q, Maher EA, Masoud S, Dixon RA, Lamb CJ. Enhanced protection against fungal attack by constitutive co-expression of chitinase and glucanase genes in transgenic tobacco. Nat Biotechnol. 1994;12:807–812. doi: 10.1038/nbt0894-807. [DOI] [Google Scholar]