Abstract
Background
Campylobacter species cause a high proportion of bacterial gastroenteritis cases and are a significant burden on health care systems and economies worldwide; however, the relative contributions of the various possible sources of infection in humans are unclear.
Methods
National-scale genotyping of Campylobacter species was used to quantify the relative importance of various possible sources of human infection. Multilocus sequence types were determined for 5674 isolates obtained from cases of human campylobacteriosis in Scotland from July 2005 through September 2006 and from 999 Campylobacter species isolates from 3417 contemporaneous samples from potential human infection sources. These data were supplemented with 2420 sequence types from other studies, representing isolates from a variety of sources. The clinical isolates were attributed to possible sources on the basis of their sequence types with use of 2 population genetic models, STRUCTURE and an asymmetric island model.
Results
The STRUCTURE and the asymmetric island models attributed most clinical isolates to chicken meat (58% and 78% of Campylobacter jejuni and 40% and 56% of Campylobacter coli isolates, respectively), identifying it as the principal source of Campylobacter infection in humans. Both models attributed the majority of the remaining isolates to ruminant sources, with relatively few isolates attributed to wild bird, environment, swine, and turkey sources.
Conclusions
National-scale genotyping was a practical and efficient methodology for the quantification of the contributions of different sources to human Campylobacter infection. Combined with the knowledge that retail chicken is routinely contaminated with Campylobacter, these results are consistent with the view that the largest reductions in human campylobacteriosis in industrialized countries will come from interventions that focus on the poultry industry.
Campylobacteriosis, caused principally by Campylobacter jejuni and Campylobacter coli, is among the main causes of bacterial gastroenteritis worldwide. In developing countries, campylobacteriosis is primarily a disease that occurs during infancy, because of high levels of early exposure and acquired immunity [1], but in industrialized countries, the epidemiology is characterized by sporadic infection throughout the population at all ages [2]. Campylobacter infection accounts for an estimated 2.5 million cases in the United States and >340,000 cases in the United Kingdom each year [3, 4], which is >3 times the number of cases caused by Salmonella, Escherichia coli O157:H7, and Listeria monocytogenes combined [5]. The estimated annual economic burden of Campylobacter infection is £500 million in the United Kingdom [6] and $8 billion in the United States [7], but despite its significance as a public health problem, the relative contributions of different sources of infection to the human disease burden remain uncertain.
Many species of wild and farm animals, particularly birds, carry Campylobacter species as part of their gut microbiota, and contamination of human food can occur at any point from the farm to the consumer. Potential sources of human infection include contaminated meat, poultry, water, and milk and contact with animals [8]. Analytical epidemiology methods, including risk assessment and case-control studies, provide some indirect evidence for the origin of disease, but because the majority of human Campylobacter infections are sporadic with very few recognized outbreaks that indicate a common infection source [9, 10], these approaches are incomplete. Because interventions for controlling Campylobacter transmission are costly and implementation requires consideration of cost-effectiveness, the uncertainty regarding the sources of human infection has inhibited effective public health intervention by government agencies and industry.
Molecular typing has enhanced many epidemiological studies, including the identification of food-borne outbreaks of infection due to E. coli O157:H7 [11], Salmonella enterica [12], Campylobacter [13], and L. monocytogenes [14]; early identification of an outbreak source enables effective disease containment [14, 15]. Recent advances in bacterial genetic typing and analysis provide the opportunity to determine the origin of Campylobacter isolates obtained from patients on the basis of their genotypes, because there is sufficient genetic variation within the bacterial population to define host or source-associated genotypes [16]. In this study, we used multilocus sequence typing (MLST) [17], which has several advantages over other microbial typing schemes such as serotyping, PFGE, and flaA typing [18-20]. Because it is based on nucleotide sequence, MLST is inherently reproducible, scalable, and portable between laboratories, with data readily shared via the Internet [21].
We addressed the sources of campylobacteriosis in industrialized countries as part of the national Campylobacter MLST Project in Scotland (CaMPS), which included a comprehensive survey of isolates from all confirmed cases of human campylobacteriosis in Scotland during a 15-month period (from mid-July 2005 through mid-October 2006). To attribute isolates to a source, 4 important resources were exploited: (1) a standardized MLST genotyping protocol, (2) a national-scale contemporaneous comparison data set of MLST genotypes from potential disease sources in Scotland, (3) a substantial data archive of MLST genotypes from other sources and locations, and (4) model-based statistical methods that allow quantitative estimation of the genetic attribution of disease to source.
METHODS
Clinical isolates
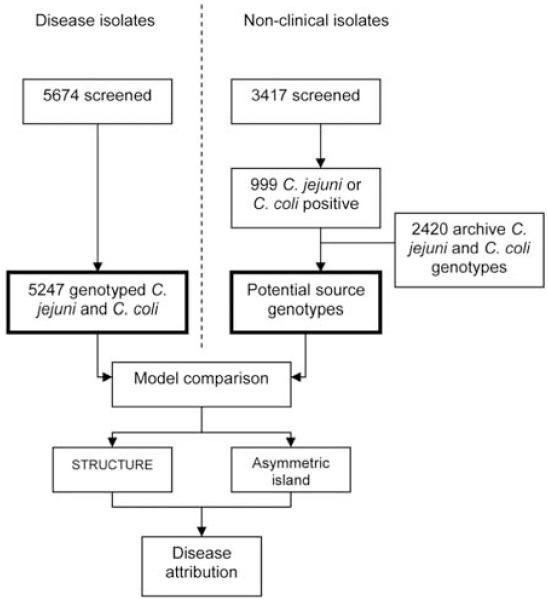
Clinical samples were collected from July 2005 through September 2006 (figure 1), comprising a comprehensive survey of clinical isolates from all 28 diagnostic laboratories in the 15 health board regions in Scotland. Campylobacter isolates were cultured from human stool samples on selective media following microaerophilic (5% O2) incubation, and colony sweeps were submitted to the University of Aberdeen (Aberdeen, United Kingdom) on charcoal transport swabs. Isolates were given a unique identifier code, and sample information was entered into an Access (Microsoft) database. Bacterial isolates were incubated at 37°C, subcultured on mCCDA media (CM0739; Oxoid), and divided for DNA extraction and archiving at −70°C.
Figure 1. Experimental design.
Non-clinical isolates were from food, host animal, and environmental sources.
Environmental isolates
Structured environmental sampling was conducted throughout the study period at 3 urban (Aberdeen, Edinburgh, and Glasgow) and 2 rural (northeast and southwest) locations. Rural farms with livestock were chosen randomly from each postal sector, and urban sampling focused on areas where animal feces coincided with human activity (e.g., parks). Sampled food (i.e., chicken, pate, and liver) was obtained from retail outlets. Approximately 100-g fecal samples were collected for each animal group, and moistened sterile swabs were used to collect smaller fecal specimens (e.g., from wild birds). Meat specimens were incubated in enrichment broth for 1 h before subculture. Sample information, including source species—the only information used in this study—was recorded. Fecal (10 g) and swab samples were homogenized (1:10) in Campylobacter enrichment broth [22] and incubated at 37°C for 5 days in 5% horse blood plus growth supplement (Mast Selectavial SV61), amphotericin (2 μg/mL), cefoperazone (15 μg/mL), and trimethoprim (10 μg/mL). Polymixin B (2500 IU/L) and rifampicin (5 μg/mL) were added to the broth after 7 h. Samples (0.1 mL) of enrichment broth were plated on CCDA (CM0739; Oxoid) and incubated in microaerobic conditions at 37°C. Colonies were enumerated and presumptively identified to be Campylobacter with microscopy (Gram stain) and agglutination (Microscreen latex; Microgen), and single colonies were stored at −80°C in nutrient broth with 15% glycerol.
Genotyping
DNA was prepared from a heat-killed cell suspension with use of the Chelex resin method (Bio-Rad), according to the manufacturer’s instructions. Template DNA was arrayed in 96-well polypropylene plates (Abgene), and 7-locus MLST was performed using standard methods and primers published elsewhere [22, 23]. Sequence extension reaction products were separated and detected with an ABI Prism 3730 automated DNA sequencer (Applied Biosystems). Forward and reverse sequences were assembled from the resultant chromatograms with use of the STADEN suite of computer programs [24]. Consensus sequences for each gene conferred a 7-locus sequence type (ST) and were concatenated (a total of 3309 bp) to give a genotype. A study-specific isolate database was developed using mlstdbNet software [25].
Data Analysis
To assess evidence for the presence or absence of genetic structuring by geography, the MLST genotype composition across the health board regions was assessed using the exact test of population differentiation in the ARLEQUIN software package [26]; the test was run with 100,000 Markov chain steps and 100,000 dememorization steps. The 3 island health boards were excluded because of the low number of clinical cases of campylobacteriosis in these regions. Specifically, the number of cases in Orkney (19), Shetland (24), and the Western Isles (9) was lower than the regional mean number of cases (379) and lower than the number of cases in the region with the lowest incidence, Argyll and Clyde (132). Therefore, estimates for regional differentiation in ST composition were more accurate if the island regions were excluded.
Different Campylobacter sources harbored different STs; for example, ST-257 and ST-61 were particularly common in chicken and cattle, respectively. Although there is overlap, the association of different STs with particular host sources [16] makes it possible to quantitatively estimate the source of human infection. Two genetic attribution models were used for formal probabilistic assignment of human isolates to the putative origin populations. The first, the no admixture model in STRUCTURE [27], determined population structure from genetic data and assigned individual isolates in the test set of human isolates independently to a source with use of the training set. The second, the asymmetric island model [28], combined information regarding population structure across all of the isolates in the test set to estimate the overall proportion of isolates attributable to each source.
The attribution methods were validated to determine the limitations of the attribution discrimination achievable with a 7-locus genetic profile. This was performed by calculating the probability of a randomly selected subset of each host species being assigned to the correct population of origin. In each case, 50% of the genotypes from each host source were removed and compared with all the remaining genotypes, and the assignment probability to each source was calculated. Probabilistic assignment in STRUCTURE was performed using the no admixture model with 10,000 burn-in iterations and with 10,000 subsequent iterations. For each analysis, assignment of isolate sources was performed on the basis of a training set of isolates. In STRUCTURE, these were distinguished from the test data with use of the USEPOPINFO flag. After calibration of the models, clinical C. jejuni and C. coli isolates for which complete 7-locus genotype data were available were assigned to putative sources by comparison to data sets comprising all the contemporaneous host and environmental and food isolate genotype data augmented with archive data sets of 2420 isolates from published sources [16, 29-31]. This provided a representative attribution data set for analysis that included C. jejuni isolates from 586 cattle, 1288 chickens, 170 wild birds, 91 environmental samples, and 249 sheep and C. coli isolates from 86 cattle, 459 chickens, 57 sheep, 322 swine, and 111 turkeys.
RESULTS
From mid-July 2005 through mid-October 2006, 5674 isolates were obtained from confirmed clinical cases of campylobacteriosis from diagnostic bacteriology laboratories in all hospitals reporting Campylobacter cases in the 15 National Health Service Scotland Health Boards. Of these, 427 isolates were excluded because they were mixed cultures, gave incomplete typing results, were successive samples from the same patient, or represented species other than C. coli or C. jejuni. Most (90.4%) of the clinical isolates were C. jejuni, and the remaining 9.6% were C. coli. A total of 999 environmental isolates from potential sources of human infection (i.e., cattle, chickens, wild birds, environment, sheep, and swine) were successfully typed with use of MLST. These were combined with data from other studies to give a total comparison data set including 3419 environmental isolates. With regard to MLST results, the clinical isolates of both Campylobacter species were diverse, with 501 STs found among C. jejuni isolates and 108 STs found among C. coli isolates. Most (81%) of the C. jejuni isolates belonged to the 10 most common clonal complexes (ST-21, ST-257, ST-45, ST-48, ST-206, ST-443, ST-574, ST-354, ST-353, and ST-61) and most (94%) of the C. coli isolates belonged to the ST-828 clonal complex. Pair-wise comparisons among groups of isolates from the 12 mainland health boards yielded no evidence for differentiation in ST composition (overall, P = .284, by ARLEQUIN exact test of population differentiation) [26].
The source attribution models were tested with isolates that had a known source. Self-assignment was carried out on random subsets of the comparison data sets for all animal source populations. Assignment of 50% of C. jejuni isolates to a host source based on a reduced training set with use of STRUCTURE gave self-assignment probabilities of 70% for chicken, 84% for ruminant, 54% for wild bird, and 38% for environmental isolates. The asymmetric island model consistently provided a greater probability of assignment to the correct source with self-assignment probabilities of 97% for chicken, 98% for ruminant, 77% for wild bird, and 62% for environmental isolates.
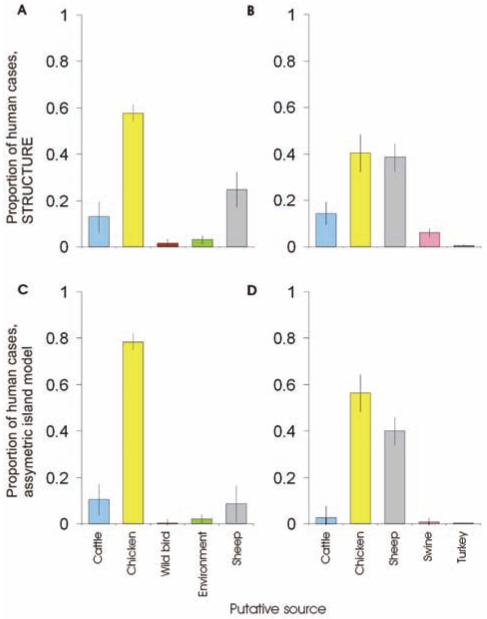
The assignment probability for each potential source was calculated for each clinical isolate individually (figure 2), and the percentage of all clinical isolates attributed to each source was calculated as the sum of these probabilities (figure 3). The clinical C. jejuni isolates were attributed to sources according to the STRUCTURE analysis as follows: chickens, 58%; ruminants, 38%; and wild birds and environment, 4%. The sources of isolates were similar according to the asymmetric island model: chickens, 78%; ruminants, 18%; and wild birds and environment, 4%. The clinical C. coli isolates were attributed to sources as follows: chickens, 40%; sheep, 40%; cattle, 14%; pigs, 6%; and turkey, <1%. These isolates were attributed to sources according to the asymmetric island model as follows: chickens, 56%; sheep, 40%; cattle, 2%; pigs, <1%; and turkeys, <1% (figure 3).
Figure 2. Assignment of Human clinical cases of campylobacteriosis to source using the Bayesian clustering algorithm STRUCTURE (A and B) and the asymmetric island model (C and D).
Each isolate is represented by a vertical bar, showing the estimated probability that it comes from each of the putative sources. Sources for Campylobacter jejuni were cattle (blue), chickens (yellow), wild birds (brown), the environment (green), and sheep (light gray). Sources for Campylobacter coli were cattle (blue), chickens (yellow), sheep (gray), swine (pink), and turkeys (black). Isolates are ordered by attributed source.
Figure 3. The origin of human campylobacteriosis in Scotland (2005–2006).
Probabilistic assignment of the source of human infection with Campylobacter jejuni (A and C) and Campylobacter coli (B and D) was determined using STRUCTURE and asymmetric island attribution models. Sequence types of disease-causing C. jejuni and C. coli were compared with data sets with isolates from cattle, chicken, and sheep. In addition, C. jejuni was compared with wild bird and environmental data sets and C. coli was compared with swine and turkey data sets. In each diagram, 5 equal-sized columns would be expected in the absence of any genetic differentiation by host species.
DISCUSSION
A national-scale high-throughput molecular typing approach was employed to quantify the contributions of different sources of human Campylobacter infection in Scotland. Attributing clinical isolates to sources of infection is efficient when different sources harbor strains with characteristic genotypes. Partially clonal bacteria such as Campylobacter species have genetic diversity in the housekeeping genes that form the basis of MLST systems [17, 32]. The different categories of hosts and sources had genetic coherence sufficient to outweigh temporal or spatial variation in sample type, which allowed the attribution of isolates to specific sources [16].
Molecular typing has previously been used to infer the source of campylobacteriosis, and overlap in the genotypes of Campylobacter species present in humans and chickens has been demonstrated [33-35]. To progress beyond this qualitative observation, formal statistical attribution was employed with use of 2 models, to ensure that conclusions were independent of methodological bias. In self-attribution model-validation tests, STRUCTURE consistently gave a lower probability and the asymmetric island model provided a greater probability of correct assignment of an isolate to origin. There were 2 differences in the models that account for the improved estimation of source by the asymmetric island model. The first was that the asymmetric island model allows loci to be linked by modelling bacterial recombination explicitly, compared with STRUCTURE, which models linkage disequilibrium caused by admixture and assumes that loci are unlinked in the source populations. The second difference involved a hyper-parameter, F, that was used to share information about source across all cases. By directly parameterizing the quantity of interest (the proportion of cases attributable to each source), we exploited information more efficiently and source estimation became more accurate. Although the attribution statistics are different between the 2 models, the results are consistent, which enhances the robustness of the conclusion that chicken is the principal source of human disease.
Chickens were the dominant source of campylobacteriosis in Scotland in 2005–2006. Using the asymmetric island model, which was more accurate in the self-assignment tests, genotypes associated with human disease were most similar to those from chicken sources in 78% of cases of C. jejuni and 56% of C. coli infection. Thus, 76% of the cases of campylobacteriosis in Scotland could be attributed to consumption of contaminated chicken. Cattle and sheep, representing a single C. jejuni gene pool [16], contributed far fewer cases of C. jejuni infection (<20%), and the contribution from wild bird and environment sources was low (<4% of cases).
The data set that was used to represent sources of human infection did not include all possible sources of Campylobacter infection; however, it did represent a diverse range of sources, with the main types of known food animal sources included alongside isolates from wild birds and the environment. Unless an omitted source harbored genotypes that were very similar to those genotypes found in chickens, the impact of any excluded sources on the main finding of the proportion of infection attributable to a chicken origin is likely to have been very small. Because the study included isolates from ruminants, which were more similar to isolates from chickens than were those that have been typed from other sources [36], this was unlikely. One possible disease source for which subtype distribution is poorly characterized is companion animals, especially cats and dogs. Although these have been implicated as a risk factor in Campylobacter infection, their contribution appears to be small [8], they do not represent a reservoir, and their sources of infection may overlap those of humans. Thus, even if companion animals sometimes act as part of the chain of transmission to humans, their infection may have originated in a reservoir source, and the absence of companion animal reference populations in this study will have resulted in the attribution of such infections to the more distant reservoir source.
The high attribution of human disease isolates to a chicken source was consistent with previous studies. Contaminated poultry meat has been associated with disease by risk assessment [37], outbreak investigation [9, 10], and analytical epidemiology studies of sporadic cases [8]. In a review of analytical studies, chicken consumption was the most commonly identified risk factor for campylobacteriosis [8]; however, all of these approaches have substantial difficulties in obtaining an unbiased estimate of the proportion of human disease that is transmitted from chicken sources [38]. A unique opportunity to assess the contribution of chicken quantitatively was presented by the “dioxin crisis” in Belgium in 1999, which led the authorities to remove all domestically produced poultry products from sale. This comprised approximately one-half of the poultry available at retail, with imported meat remaining available. There was a 40% reduction in the incidence of human campylobacteriosis after this withdrawal [39], consistent with the estimate reported here. The exploitation of this event was valuable in providing an estimate of the contribution of chicken meat to disease; however, ongoing monitoring of the contribution of chicken sources to human disease in the countries where campylobacteriosis remains a problem is central to the development and evaluation of effective control measures, which cannot rely on data that is only opportunistically available.
The molecular typing and statistical genetic approaches employed here provide a means of routine monitoring of the transmission of Campylobacter species and other bacteria from chicken meat and other sources to humans. The model-based molecular attribution of campylobacteriosis in Scotland is, therefore, of widespread applicability. It provides a means of evaluating interventions aimed at the prevention of disease transmission through the food chain from farms to retail outlets to final consumption. This capacity to monitor the key intended outcome of interventions, such as reduction of infection on the farm, changes to slaughtering procedures, and population education [40], will support progress to evidence-based disease control programs.
Acknowledgments
We thank the National Health Service of Scotland (NHSS) diagnostic laboratories and Consultants in Public Health Medicine, the staff of Aberdeen University and NHSS, local farmers, Local Authorities Coordinators of Regulatory Services, the veterinary laboratories of the Scottish Agricultural College and the University of Glasgow, animal rescue centers and wildlife reserves, postgraduate research students, and visiting research students for their participation in the collection of bacterial isolates. This study used the C. jejuni Multilocus Sequence Typing Web site (http://pubmlst.org/campylobacter/), developed by Keith Jolley and Man-Suen Chan at the University of Oxford.
Financial Support. Food Standards Agency of Scotland and a Wellcome Trust Senior Research Fellowship (to M.C.J.M.).
Footnotes
Potential conflicts of interest. All authors: no conflicts.
References
- 1.Oberhelman RA, Taylor DN. Campylobacter infections in developing countries. In: Nachamkin I, Blaser MJ, editors. Campylobacter. 2nd ed. ASM Press; Washington, DC: 2000. pp. 139–53. [Google Scholar]
- 2.Olson CK, Ethelberg S, van Pelt W, Tauxe RV. Epidemiology of Campylobacter jejuni infections in industrialized nations. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. 3rd ed. ASM Press; Washington, DC: 2008. pp. 163–91. [Google Scholar]
- 3.Allos B. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis. 2001;32:1201–6. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 4.Kessel AS, Gillespie IA, O’Brien SJ, Adak GK, Humphrey TJ, Ward LR. General outbreaks of infectious intestinal disease linked with poultry, England and Wales, 1992-1999. Commun Dis Public Health. 2001;4:171–7. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention (CDC) Division of Food-borne, Bacterial and Mycotic Diseases (DFBMD) listing. CDC; United States: 2008. [Google Scholar]
- 6.Humphrey T, O’Brien S, Madsen M. Campylobacters as zoonotic pathogens: a food production perspective. Int J Food Microbiol. 2007;117:237–57. doi: 10.1016/j.ijfoodmicro.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Buzby JC, Roberts T. Economic costs and trade impacts of microbial foodbourne illness. World Health Stat Q. 1997;50:57–66. [PubMed] [Google Scholar]
- 8.Neimann J, Engberg J, Molbak K, Wegener HC. A case-control study of risk factors for sporadic Campylobacter infections in Denmark. Epidemiol Infect. 2003;130:353–66. doi: 10.1017/s0950268803008355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pebody RG, Ryan MJ, Wall PG. Outbreaks of Campylobacter infection: rare events for a common pathogen. Commun Dis Rep CDR Rev. 1997;7:R33–7. [PubMed] [Google Scholar]
- 10.Frost JA, Gillespie IA, O’Brien SJ. Public health implications of Campylobacter outbreaks in England and Wales, 1995-9: epidemiological and microbiological investigations. Epidemiol Infect. 2002;128:111–8. doi: 10.1017/s0950268802006799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bender JB, Hedberg CW, Besser JM, Boxrud DJ, MacDonald KL, Osterholm MT. Surveillance by molecular subtype for Escherichia coli O157:H7 infections in Minnesota by molecular subtyping. N Engl J Med. 1997;337:388–94. doi: 10.1056/NEJM199708073370604. [DOI] [PubMed] [Google Scholar]
- 12.Bender JB, Hedberg CW, Boxrud DJ, et al. Use of molecular subtyping in surveillance for Salmonella enterica serotype typhimurium. N Engl J Med. 2001;344:189–95. doi: 10.1056/NEJM200101183440305. [DOI] [PubMed] [Google Scholar]
- 13.Sails AD, Swaminathan B, Fields PI. Utility of multilocus sequence typing as an epidemiological tool for investigation of outbreaks of gastroenteritis caused by Campylobacter jejuni. J Clin Microbiol. 2003;41:4733–9. doi: 10.1128/JCM.41.10.4733-4739.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsen SJ, Patrick M, Hunter SB, et al. Multistate outbreak of Listeria monocytogenes infection linked to delicatessen turkey meat. Clin Infect Dis. 2005;40:962–7. doi: 10.1086/428575. [DOI] [PubMed] [Google Scholar]
- 15.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg Infect Dis. 2005;11:603–9. doi: 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarthy ND, Colles FM, Dingle KE, et al. Host-associated genetic import in Campylobacter jejuni. Emerg Infect Dis. 2007;13:267–72. doi: 10.3201/eid1302.060620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dingle KE, Colles FM, Wareing DRA, et al. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrington CS, Thomson Carter FM, Carter PE. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J Clin Microbiol. 1997;35:2386–92. doi: 10.1128/jcm.35.9.2386-2392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinbrueckner B, Ruberg F, Kist M. Bacterial genetic fingerprint: a reliable factor in the study of the epidemiology of human Campylobacter enteritis? J Clin Microbiol. 2001;39:4155–9. doi: 10.1128/JCM.39.11.4155-4159.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wassenaar TM, Geilhausen B, Newell DG. Evidence for genome instability in Campylobacter jejuni isolated from poultry. Appl Environ Microbiol. 1998;64:1816–21. doi: 10.1128/aem.64.5.1816-1821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maiden MC. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561–88. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 22.Gormley FJ, Macrae M, Forbes KJ, Ogden ID, Dallas JF, Strachan NJ. Has retail chicken played a role in the decline of human campylobacteriosis? Appl Environ Microbiol. 2008;74:383–90. doi: 10.1128/AEM.01455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller WG, On SL, Wang G, Fontanoz S, Lastovica AJ, Mandrell RE. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J Clin Microbiol. 2005;43:2315–29. doi: 10.1128/JCM.43.5.2315-2329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5:233–41. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 25.Jolley KA, Chan MS, Maiden MC. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics. 2004;5:86. doi: 10.1186/1471-2105-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Excoffer L, Laval G, Schneider S. ARLEQUIN (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2007;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 27.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson DJ, Gabriel E, Leatherbarrow AJ, et al. Tracing the source of campylobacteriosis. PLoS Genet. 2008;4:e1000203. doi: 10.1371/journal.pgen.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller WG, Englen MD, Kathariou S, et al. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology. 2006;152:245–55. doi: 10.1099/mic.0.28348-0. [DOI] [PubMed] [Google Scholar]
- 30.Dingle KE, Colles FM, Falush D, Maiden MC. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J Clin Microbiol. 2005;43:340–7. doi: 10.1128/JCM.43.1.340-347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thakur S, Gebreyes WA. Campylobacter coli in swine production: antimicrobial resistance mechanisms and molecular epidemiology. J Clin Microbiol. 2005;43:5705–14. doi: 10.1128/JCM.43.11.5705-5714.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maiden MC. Population genomics: diversity and virulence in the Neisseria. Curr Opin Microbiol. 2008;11:467–471. doi: 10.1016/j.mib.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kramer JM, Frost JA, Bolton FJ, Wareing DR. Campylobacter contamination of raw meat and poultry at retail sale: identification of multiple types and comparison with isolates from human infection. J Food Prot. 2000;63:1654–9. doi: 10.4315/0362-028x-63.12.1654. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen EM, Nielsen NL. Serotypes and typability of Campylobacter jejuni and Campylobacter coli isolated from poultry products. Int J Food Microbiol. 1999;46:199–205. doi: 10.1016/s0168-1605(98)00194-9. [DOI] [PubMed] [Google Scholar]
- 35.Saito S, Yatsuyanagi J, Harata S, et al. Campylobacter jejuni isolated from retail poultry meat, bovine feces and bile, and human diarrhealsamples in Japan: comparison of serotypes and genotypes. FEMS Immunol Med Microbiol. 2005;45:311–9. doi: 10.1016/j.femsim.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Dingle KE, Colles FM, Ure R, et al. Molecular characterisation of Campylobacter jejuni clones: a rational basis for epidemiological investigations. Emerg Infect Dis. 2002;8:949–955. doi: 10.3201/eid0809.02-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nauta MJ, Jacobs-Reitsma WF, Havelaar AH. A risk assessment model for Campylobacter in broiler meat. Risk Anal. 2007;27:845–61. doi: 10.1111/j.1539-6924.2006.00834.x. [DOI] [PubMed] [Google Scholar]
- 38.Evers EG, Van der Fels-Klerx HJ, Nauta MJ, Schijven JF, Havelaar AH. Campylobacter source attribution by exposure assessment. Int J Risk Assess Manage. 2008;8:174–90. [Google Scholar]
- 39.Vellinga A, Van Loock F. The dioxin crisis as experiment to determine poultry-related Campylobacter enteritis. Emerg Infect Dis. 2002;8:19–22. doi: 10.3201/eid0801.010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nauta MJ, Fischer AR, van Asselt ED, de Jong AE, Frewer LJ, de Jonge R. Food safety in the domestic environment: the effect of consumer risk information on human disease risks. Risk Anal. 2008;28:179–92. doi: 10.1111/j.1539-6924.2008.01012.x. [DOI] [PubMed] [Google Scholar]