Abstract
Background: Human monocyte inflammatory responses differ between virulent and attenuated Francisella infection.
Results: A mixed infection model showed that the virulent F. tularensis Schu S4 can attenuate inflammatory cytokine responses to the less virulent F. novicida in human monocytes.
Conclusion: F. tularensis dampens inflammatory response by an active process.
Significance: This suppression may contribute to enhanced pathogenicity of F. tularensis. Francisella tularensis is a Gram-negative facultative bacterium that can cause the disease tularemia, even upon exposure to low numbers of bacteria. One critical characteristic of Francisella is its ability to dampen or subvert the host immune response. Previous work has shown that monocytes infected with highly virulent F. tularensis subsp. tularensis strain Schu S4 responded with a general pattern of quantitatively reduced pro-inflammatory signaling pathway genes and cytokine production in comparison to those infected with the less virulent related F. novicida. However, it has been unclear whether the virulent Schu S4 was merely evading or actively suppressing monocyte responses. By using mixed infection assays with F. tularensis and F. novicida, we show that F. tularensis actively suppresses monocyte pro-inflammatory responses. Additional experiments show that this suppression occurs in a dose-dependent manner and is dependent upon the viability of F. tularensis. Importantly, F. tularensis was able to suppress pro-inflammatory responses to earlier infections with F. novicida. These results lend support that F. tularensis actively dampens human monocyte responses and this likely contributes to its enhanced pathogenicity.
Keywords: Francisella, monocytes, cytokines, signal transduction, innate immunity
Introduction
Francisella tularensis is the highly infectious Gram-negative causative agent of the disease tularemia (Sjostedt, 2007). F. tularensis has further been classified into distinct subspecies including F. tularensis subsp. tularensis (F. tularensis; Type A), F. tularensis subsp. holarctica (F. holarctica; Type B), and F. tularensis subsp. novicida (F. novicida). Francisella is especially recognized for its low infectious dose and ability to cause severe illness and death, which endorses its categorization as a Category A select agent by the CDC (Sharma et al., 2011). Of note, the most life-threatening forms of tularemia are particularly associated with Type A infections regardless of host species (Mohapatra et al., 2013). Although known to infect a range of host organisms and cell types (Rick and Wu, 2007; Hall et al., 2008), F. tularensis has evolved to successfully infect human monocytes/macrophages where it escapes the phagosome, replicates within the cytosol, and then lyses the cell before beginning a new reinfection cycle (Gavrilin et al., 2006; Clemens and Horwitz, 2007; Jones et al., 2012; Celli and Zahrt, 2013).
One critical characteristic of F. tularensis is its ability to attenuate the host inflammatory immune response. Indeed, early studies in humans showed that Francisella-infected individuals exhibited diminished cytokine responses to endotoxin (Greisman et al., 1963). In the murine model, F. tularensis infection does not lead to a classic pro-inflammatory cytokine response, which in turn results in insufficient numbers of immune cells recruited to infection sites (Bosio et al., 2007). Rarther, murine studies have corroborated the findings of Griesman et al. (Greisman et al., 1963), where challenge with LPS after infection does not lead to the production of pro-inflammatory cytokines such as TNF-α in mouse cell lines or in vivo (Telepnev et al., 2003, 2005; Bosio et al., 2007). Similar findings have also been observed in F. tularensis infected murine bone marrow and alveolar macrophages following Pam3CSK administration (Crane et al., 2013a).
It has been shown at the cellular level that dendritic cells infected with F. tularensis respond poorly, exhibiting decreased cytokine production (Bauler et al., 2011). Francisella does not replicate within endothelial cells, nonetheless during their brief interactions (Forestal et al., 2007), Francisella suppresses CCL2 and CXCL8 production thus limiting pro-inflammatory activation of effector immune cells (Bublitz et al., 2010). Multiple investigations, including studies from our group, document that F. tularensis infected cells have a stunted and/or delayed pro-inflammatory cytokine response in contrast to other immune stimulating agents (Telepnev et al., 2003; Sjostedt, 2006; Butchar et al., 2008; Mares et al., 2008; Bosio, 2011; Dai et al., 2013). Francisella's ability to dampen immune response is not only limited to single cell populations, but is also evident in multiple cell environments (Kim et al., 2008). It has been shown in murine models that F. tularensis Schu S4 infections are associated with a weak induction of immune related genes and an overall lack of cell infiltration within the lung, which is in contrast to what is observed in F. tularensis LVS, L. pneumophila, P. aeruginosa or Y. pestis infection (Walters et al., 2013). Concurrently, the respiratory model of tularemia is characterized by the development of tolerogenic dendritic cells, release of anti-inflammatory cytokines in the lungs and stimulation of Treg and Th17 cells (Woolard et al., 2008; Periasamy et al., 2011).
We chose to examine human peripheral blood monocytes, because a higher percentage of monocytes are infected by F. tularensis than either F. holarctica or F. novicida during the course of infection (Hall et al., 2008). It is well documented that avirulent F. novicida is capable of inducing a potent inflammatory program (Rick and Wu, 2007; Sjostedt, 2007; Cremer et al., 2009; Dai et al., 2013). In human monocytes, the focus of this study, we have previously shown that infection with F. tularensis, leads to diminished responses of cytokines such as TNF-α, IL-6, IL-8, and IL-12 among others (Butchar et al., 2008). Infection with F. tularensis also leads to the downregulation of critical host response pathway members such as TLR2, MyD88, the PI3K regulatory subunit, Type I/Type II Interferon pathway components, and factors related to autophagy (Butchar et al., 2008; Cremer et al., 2011).
The precise mechanism(s) by which the virulent F. tularensis can elicit dampened immune responses upon infection is still not completely understood (Oyston et al., 2004; Bosio, 2011). There is strong evidence suggesting that this bacterium can evade many of the host detection mechanisms, which leads to suboptimal immune responses and permits bacterial growth. In addition, some studies have suggested that active mechanisms are also at play, wherein Francisella not only escapes detection but also preemptively dampens host cell responses. For example, it has recently been shown that the lipids of F. tularensis but not those of F. novicida were capable of dampening responses to subsequent innate immune stimuli both in vitro and in vivo (Crane et al., 2013b; Ireland et al., 2013) and that interaction between C3-opsonized F. tularensis and Complement Receptor 3 led to host cell immunosuppression (Dai et al., 2013).
However, differentiating between active suppression by Francisella and the more general phenomenon of endotoxin tolerance/cross-tolerance (Greisman and Hornick, 1975; West and Heagy, 2002; Morris and Li, 2012) has not been straightforward. Tolerance consists of both an early and late phase, depends on mediators such as the inositol phosphatase SHIP (Sly et al., 2004), and can be abrogated via molecules such as IFNγ (Chen and Ivashkiv, 2010). Interestingly, IFNγ has been shown to be important for cellular resistance against F. tularensis in both human and mouse macrophages (Edwards et al., 2010).
Using a mixed infection model, we show that the virulent F. tularensis Schu S4 can attenuate pro-inflammatory cytokine responses to the less virulent F. novicida in human monocytes. This process is dose-dependent and requires that F. tularensis is viable. Importantly, our results show that F. tularensis can dampen monocyte responses that are already in progress, suggesting that it is bacterially-driven suppression rather than host-cell-mediated tolerance. These results indicate that although F. tularensis may evade detection by host innate immune sensors, it also actively antagonizes host cell responses.
Materials and methods
Human peripheral blood monocyte isolation
Human Peripheral blood monocytes (PBM) were isolated as previously described (Butchar et al., 2008) using centrifugation through a Ficoll gradient followed by CD14-positive selection by Magnet-Assisted Cell Sorting (MACS, Miltenyi Biotec, Auburn, CA). This results in a ≥98% pure CD14-positive population of monocytes that has been confirmed by flow cytometry. Cells were incubated at 37°C with 5% CO2 supplementation.
Bacterial strains and monocyte infections
All infections were performed with PBM in antibiotic-free RPMI-1640 media (Gibco-BRL, Rockville, MD) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Hyclone, Logan, UT) and 1% L-glutamine (Invitrogen, Carlsbad, CA). F. novicida - U112 (JSG1819) and F. tularensis (Schu S4) were generously provided by Dr. John Gunn (The Ohio State University, Columbus, OH) and grown on Chocolate II agar plates (Becton, Dickinson and Company, Sparks, MD) at 37°C. All experiments involving F. tularensis were performed in a BSL3 facility at The Ohio State University as previously described (Butchar et al., 2007). Bacteria were resuspended in RPMI-1640 and then quantified by a spectrometer at 600 nm wave-length to calculate Multiplicity of Infection (MOI). Heat-killed F. tularensis (t°) was prepared by heating at 95°C for 10 min. Paraformaldehyde (pf) -killed F. tularensis was prepared by treating with 4% paraformaldehyde for 40 min, followed by five washes in PBS and two washes in RPMI to quench residual aldehydes as previously described (Gavrilin et al., 2006; Cremer et al., 2012). Treated bacterial suspensions were plated on chocolate II agar plates to ensure effective killing.
For infection, monocytes were resuspended in polypropylene tubes (Fisher Scientific) in RPMI medium 1640 supplemented with 10% FBS (endotoxin-free; HyClone), at 1 or 2 × 106 cells per tube. Live or killed bacteria were added to cells individually or together at various multiplicities of infection (MOI), specified for every experiment. Cells were harvested 2, 4, 16-18, and 24 h after infection; separated from bacteria by low-speed centrifugation at 1000 g for 5 min; and lysed in TRIzol® (Invitrogen, Carlsbad, CA) or hypotonic lysis buffer for RNA or protein isolation, respectively. After low-speed centrifugation, cell culture media was cleared from bacteria by high speed centrifugation at 16,000 g for 5 min, filtered and used for cytokine determination.
Real-time RT-qPCR
Quantitative Reverse-Transcription PCR was performed in detail as described previously (Gavrilin et al., 2006). In short, RNA was extracted from human PBM using TRIzol® reagent (Invitrogen, Carlsbad, CA), reverse transcribed to cDNA, and then amplified using SYBR Green PCR master mix (Eurogentec North America, San Diego, Ca). Real-time PCR was performed on an Applied Biosystems StepOne Plus system, with automatically-calculated thresholds. Primer sequences used to amplify cDNA are as follows: IL6 (forward, 5′-CACAGACAGCCACTCACCTC-3′; reverse, 5′-TTTTCTGCC AGTGCCTCTTT-3′), IL8 (forward, 5′-AGTTTTTGAAGAGGGCTGAGAAT-3′; reverse, 5′-CAACAGACCCACACAATACATGA-3′), and TNF (forward, 5′-GCTTGTTCCTCAGCC TCTTCT-3′; reverse, 5′-GGTTTGCTACAACATGGGCTA-3′). The housekeeping gene sequences are CAP1 (forward, 5′-ATTCCCTGGATTGTGAAATAGTC-3′; reverse, 5′-ATT AAAGTCACCGCCTTCTGTAG-3′) and GAPDH (forward, 5′-ACTTTGGTATCGTGGAAG GAC T-3′; reverse, 5′-GTAGAGGCAGGGATGATGTTC T-3′). Relative copy numbers (RCN) were calculated as 2−ΔCt, with ΔCt calculated by subtracting the average Ct of two housekeeping controls (CAP1 and GAPDH) from the experimental sample Ct (Gavrilin et al., 2006; Butchar et al., 2008).
ELISA
Cell-free supernatants were collected from resting and infected PBM and analyzed using sandwich ELISA kits specific for human TNF-α, IL-6, and IL-8 (R&D Systems). Each sample was tested in biological triplicates and instructions were followed according to manufacturer protocols.
Immunostaining
Infected cells were fixed in 3.7% paraformaldehyde supplemented with 0.2% FBS for 30 min. Next, cells were washed two times with 1X PBS to remove residual paraformaldehyde. Cells were placed on poly-Lysine coated slides and allowed to adhere prior to blocking with a 5% FBS and 1% BSA solution in 1X PBS for 30 min. F. novicida-infected cells were incubated with primary anti F. tularensis subsp. novicida monoclonal antibody Fn8.2 (Immuno-Precise Antibodies Ltd; Victoria, British Columbia, Canada) at a 1:100 dilution for 4 h followed by the addition of Alexa Fluor 488 rabbit anti-goat antibody (Invitrogen, Carlsbad, CA). F. tularensis infected cells were incubated with primary mouse antibody raised against F. tularensis subsp tularensis LPS (Abcam, Cambridge, MA) at a 1:1000 dilution for 4 h followed by the addition of Alexa Fluor 594 goat anti-mouse antibody (Invitrogen, Carlsbad, CA). Cover slips were mounted onto the slides using VECTASHIELD mounting media. Images were captured using an Olympus BX41 fluorescent microscope equipped with a DP20 digital camera (Olympus) at 100X magnification. A minimum of 50 cells were analyzed per test group.
Lactate dehydrogenase (LDH) cytotoxicity assay
LDH release from the cell was used as an indicator of cell death using an NAD+ reduction assay (Roche Applied Science). Supernatants from treated cells were collected, clarified by centrifugation at 400 g for 5 min, filtered and used for the assay. For a positive control, total LDH content in untreated monocytes was obtained by lysing cells with 1% Triton X-100. RPMI-1640 media was used as a blank and OD values were subtracted from readings of samples and positive control. LDH concentration in the medium was measured at 490 nm. Cell death was calculated by the formula: cytotoxicity (%) = [(sample-blank)/(positive control-blank) × 100], as described earlier (Gavrilin et al., 2012).
Statistics
Student's t-test was used for comparison between two groups, and One-Way ANOVA was used for multiple group comparisons with a Tukey's Multiple Comparison post-hoc test to analyze significant differences. p ≤ 0.05 was considered to be significant. All experiments were performed a minimum of 4 independent times (n = 4) and results are expressed as mean values ± s.e.m.
Results
Virulent F. tularensis elicits a dampened cytokine response in human monocytes and suppresses responses to F. novicida
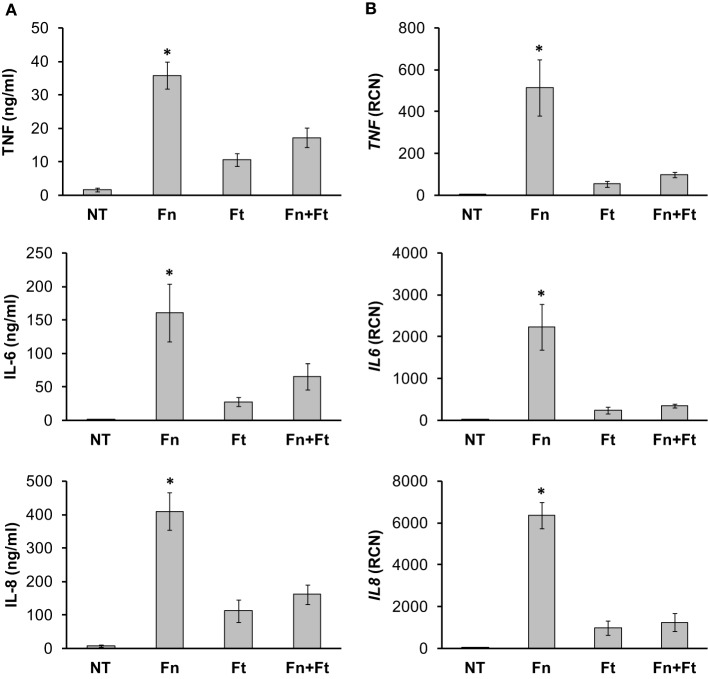
It has previously been shown that F. tularensis-infected monocytes generate a limited pro-inflammatory cytokine response in contrast to those infected with F. novicida (Gavrilin et al., 2006, 2009; Butchar et al., 2008; Cremer et al., 2009), and that F. tularensis could lead to weaker responses to subsequent stimuli such as LPS (Bosio et al., 2007). Here, we aimed to determine the effects of F. tularensis on monocyte responses to F. novicida infection, with the expectation that active suppression (as opposed to evasion of detection) by F. tularensis would significantly dampen F. novicida-induced cytokine production. As a first step, we infected monocytes overnight with F. novicida (Fn) or F. tularensis SchuS4 (Ft) independently and measured cytokine responses by ELISAs and RT-qPCR. As expected, monocytes infected with F. novicida showed high production of TNF-α, IL-6 and IL-8 compared to those infected with F. tularensis SchuS4 (Ft) (Figure 1). After establishing the effects of these bacteria as single agents we proceeded to examine the effects of virulent Francisella on monocyte responses to the more pro-inflammatory F. novicida. To explore the possibility that active mechanisms are facilitating the observed immune suppression, we performed overnight infections of human monocytes with F. novicida, F. tularensis or both. Cell-free supernatants were collected, filtered, and assayed by ELISA for cytokine/chemokine production. Monocytes infected concurrently with F. novicida and F. tularensis displayed a dampened response similar to that seen with F. tularensis infections (Figure 1A), suggesting that F. tularensis was attenuating the response to F. novicida. We also assessed the cytokine mRNA levels induced after infection. Our results showed that F. tularensis infection led to significantly lower transcript levels both in single and mixed infections (Figure 1B). These results indicate that F. tularensis has a dominant immunosuppressive effect, as it was able to blunt the monocyte responses to the less virulent F. novicida.
Figure 1.
Virulent strains of F. tularensis elicit a dampened cytokine response in monocytes. Primary human monocytes were left untreated (NT) or infected in triplicate for 16 h with either F. novicida (Fn), F. tularensis Schu S4 (Ft), or both at an MOI of 50 for each bacteria. Cell-free supernatants from infected monocytes were collected and assayed by (A) sandwich ELISAs and (B) RT-qPCR for TNF-α, IL-6 and IL-8. “RCN” represents Relative Copy Number for the Y-axis. Graphs represent the mean ± s.e.m. from 4 independent donors. Data were analyzed by ANOVA. *p < 0.01 (Fn vs. all groups). There was no significant difference between Ft and Fn + Ft.
Bacterial interactions with monocytes do not differ between F. novicida and F. tularensis
Since monocyte responses differ dramatically between F. novicida and F. tularensis, we aimed to determine if the effects of F. tularensis on monocyte responses compared to F. novicida infection were due to a difference in the number of bacteria associating with monocytes. To test this, we first infected monocytes for 5 h with F. novicida or F. tularensis at an MOI of 50. Following this, cells were washed two times, fixed in paraformaldehyde and stained with antibodies generated toward each specific bacterium as seen in representative images (Figure 2A). Our results show that although monocytes are associated with slightly lower numbers of F. novicida compared to F. tularensis, there is no statistically significant difference in the number of bacteria that associate with each cell (Figure 2B). The total percentage of cells associated with either F. novicida or F. tularensis is comparable, i.e., both bacteria associate with about 70% of the cells (Figure 2C).
Figure 2.
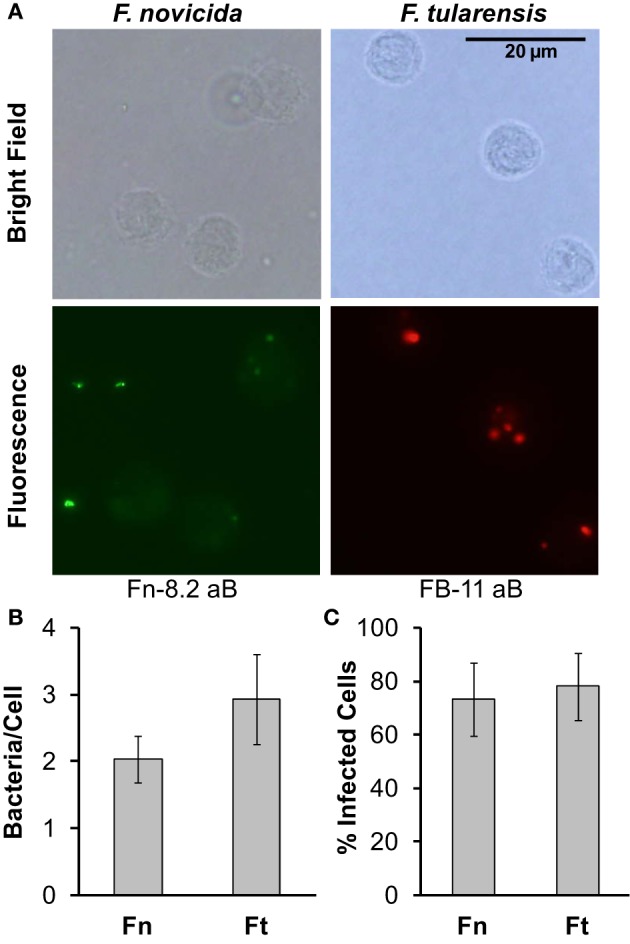
F. novicida and F. tularensis associate similarly with monocytes during infection. Primary human monocytes were infected with F. novicida (Fn) or F. tularensis Schu S4 (Ft) at an MOI of 50 for 5 h. (A) Representative images from cells fixed in paraformaldehyde and then stained with Fn-82 antibody specific for F. novicida or FB-11 antibody specific for F. tularensis. The corresponding secondary antibody for Fn-82 was Alexa Fluor 488 rabbit anti-goat IgG (green) and for FB-11 it was AlexaFluor 594 goat anti-mouse IgG (red). Graphs represent the number of bacteria per cell (B) or the number of infected cells (C). Graphs represent the mean ± s.e.m. from 1 donor incorporating a minimum of 4 frames. Data were analyzed by Student's t-test. No significant differences were found.
F. tularensis active suppression is dose-dependent
Mixed-infection experiments described above were performed at a 1:1 ratio between F. novicida and F. tularensis to allow equal opportunity for both bacteria to evoke an immune response. Since F. tularensis was found to suppress the normal monocyte responses to F. novicida, we next asked whether a smaller ratio of F. tularensis to F. novicida could still lead to suppression. To test this we infected human monocytes with F. tularensis at an MOI of 50, 25, or 5 in conjunction with F. novicida at a constant MOI of 50. Our results indicated that the active suppression of F. tularensis was dose-dependent (Figure 3).
Figure 3.
F. tularensis mediates immune suppression in a dose-dependent manner. Primary human monocytes were left untreated (NT) or infected overnight (16 h) with F. novicida (Fn), F. tularensis (Ft), or a combination of the two. Ft MOI was administered at decreasing levels (50, 25, and 5 MOI) while keeping Fn MOI constant at 50. Cell-free supernatants were assayed by sandwich ELISAs for TNF-α, IL-6, and IL-8. Graphs represent the mean ± s.e.m. from 4 independent donors. Data were analyzed by ANOVA. *p < 0.05 (Fn vs. selected groups), †p < 0.01 compared to Ft.
Bacterial viability is necessary for F. tularensis mediated cytokine suppression
Having established that F. tularensis could actively suppress host cell cytokine responses in a dose-dependent manner, we next tested whether or not bacterial viability played a role. For this, monocytes were infected overnight with F. novicida along with live, paraformaldehyde-killed or heat-killed F. tularensis. Both paraformaldehyde and heat-killed Francisella were unable to induce a pro-inflammatory response from human monocytes (Figure 5). Monocytes co-infected with live F. novicida and killed F. tularensis showed cytokine responses similar to those infected with live F. novicida alone (Figure 4). These results are in agreement with Ireland et al. (2013), who found that bacterial viability was required for suppression of NF-κB and interferon responses. The requirement for viability suggests that rather than suppressing through contact alone, F. tularensis is producing and/or secreting one or more factors in order to effect the dampening.
Figure 5.
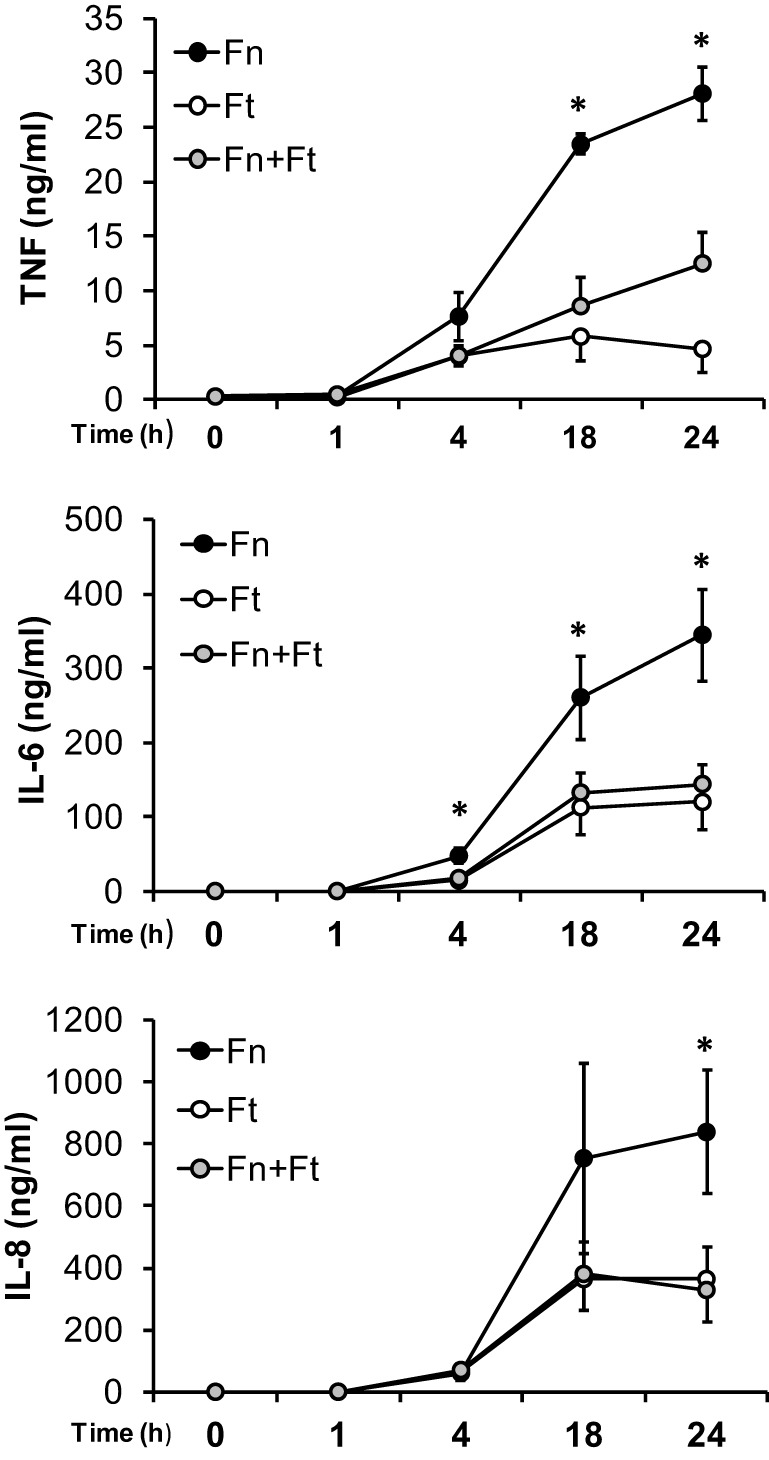
F. tularensis suppression is observed during early infection time points. Primary human monocytes were infected at for 1, 4, 18, or 24 h with F. novicida (Fn), F. tularensis (Ft) or both at an MOI of 50 for each bacteria. Cell-free supernatants were collected and assayed using sandwich ELISAs for TNF-α, IL-6, and IL-8. Graphs represent the mean ± s.e.m. from 4 independent donors. Data were analyzed by ANOVA. *p < 0.05 (Fn vs. Ft and Fn + Ft).
Figure 4.
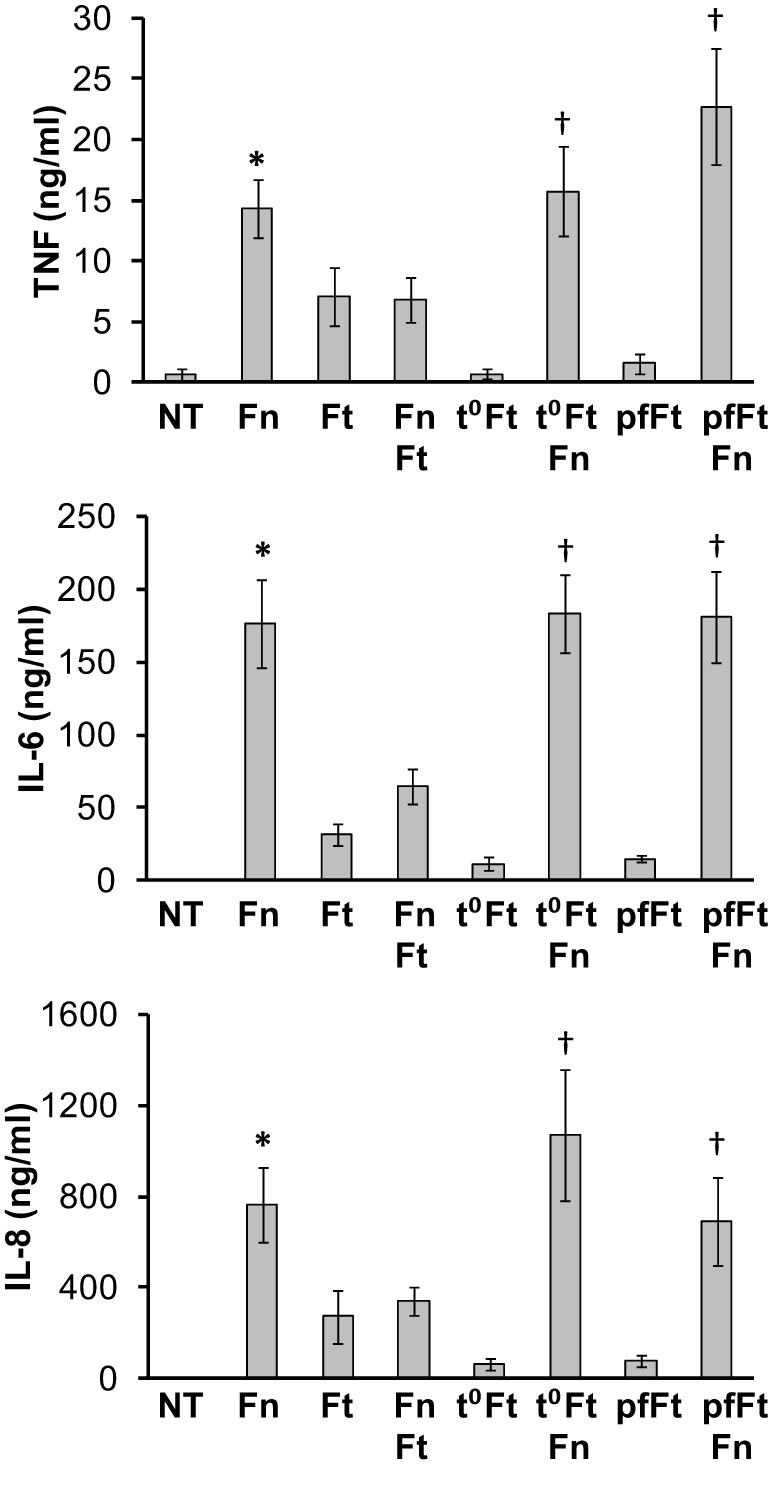
Viability of F. tularensis is essential to mediate immune suppression. Primary human monocytes were left untreated (NT) or infected overnight (16 h) with viable F. novicida (Fn), F. tularensis (Ft), or a combination of the two at an MOI of 50 for each bacteria. Additionally, monocytes were infected with heat-killed (t°Ft) or paraformaldehyde-fixed (pfFt) F. tularensis alone or in combination with live F. novicida (Fn). Cell-free supernatants from infected monocytes were assayed by sandwich ELISAs for TNF-α, IL-6, and IL-8. Graphs represent the mean ± s.e.m. from 4 independent donors. Data were analyzed by ANOVA. *p < 0.05 (Fn vs. Ft and Fn + Ft); †p < 0.05 compared to Ft, t°Ft, pfFt.
Time course of F. tularensis-mediated suppression
Previous in vivo studies reported that mice showed impaired pulmonary inflammatory responses to secondary stimuli when first challenged with F. tularensis (Bosio et al., 2007). These findings, combined with our above results showing that bacterial viability (and presumably production of immunosuppressive factors) was needed for the effects brought about by F. tularensis, led us to examine the length of time required for this suppression. Hence, we performed time course studies in monocytes infected with F. novicida, F. tularensis or both for 1, 4, 18, and 24 h. Supernatants were collected and analyzed by ELISA as above. Our results showed that although the greatest suppression occurred following overnight infection (18 and 24 h), co-infection with F. tularensis led to significant decreases in IL-6 cytokine suppression as early as 4 h after infection (Figure 5).
F. tularensis can attenuate ongoing immune responses
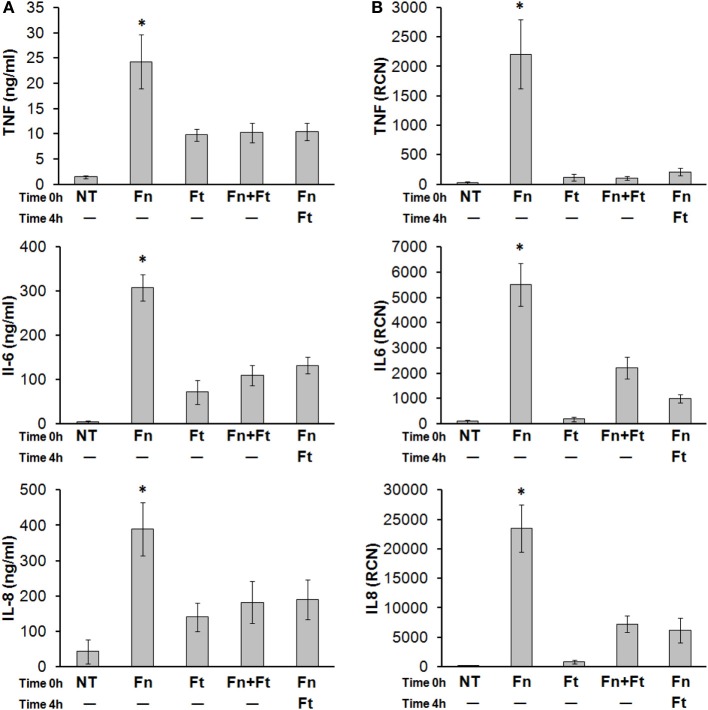
F. tularensis can begin dampening immune responses within just several hours of infection but its ability to inhibit an already-existing inflammatory response has not yet been demonstrated (Mares et al., 2008). To test this, we infected monocytes overnight with F. novicida, along with F. tularensis either concurrently or 4 h after F. novicida. Cells were lysed in TRIzol® and cleared supernatants were collected to measure cytokine transcript and secretion levels, respectively. Results showed that F. tularensis led to attenuated cytokine / chemokine responses even when added 4 h following F. novicida infection (Figures 6A,B). These results suggest that F. tularensis is likely interfering directly with one or more pro-inflammatory response pathways, as the bacteria are able to modulate responses already in progress.
Figure 6.
F. tularensis administered after F. novicida infection can still suppress the pro-inflammatory cytokine response elicited by F. novicida. Primary human monocytes were left untreated (NT) or infected overnight (16 h) with F. novicida (Fn), F. tularensis (Ft), or both at an MOI of 50 for each bacteria. In other samples, F. tularensis infection was performed 4 h following F. novicida infection. Cell-free supernatants were assayed by (A) sandwich ELISAs and (B) RT-qPCR, for TNF-α, IL-6, and IL-8. “RCN” represents Relative Copy Number for the Y-axis. Graphs represent the mean ± s.e.m. from 4 independent donors. Data were analyzed by ANOVA. *p < 0.05 (Fn vs. all groups).
Discussion
Here, we provide evidence that F. tularensis can actively suppress host cell immune responses, including those already in progress. We chose to examine human peripheral blood monocytes, since Francisella predominantly targets these cells in the blood stream. Our results showed that human monocytes infected with F. novicida demonstrate robust pro-inflammatory responses. In contrast, co-infected with F. tularensis and F. novicida produced cytokines at low levels, similar to those seen with F. tularensis alone. Furthermore, F. tularensis was able to dampen monocyte responses even if administered several hours following infection with F. novicida. The cytokines TNF-α and IL-6 were reduced, and we also observed a significant reduction in the neutrophil-attracting chemokine IL-8 at 4 h after F. tularensis infection. This would lead to the prediction that neutrophil responses might be compromised at the whole-organism level after infection, but Hall et al. found a substantial neutrophil influx in mice infected with F. tularensis (Hall et al., 2008). Additional studies are needed to determine the degree to which the IL-8 reduction we observed would influence neutrophil responses in vivo.
F. tularensis has pleiotropic effects on individual cell types as well as whole organisms, and one of these is manipulation of cytokine profile. For example, Periasamy et al. (2011) showed that Th1 pro-inflammatory cytokines were absent within the first 72 h of pulmonary infection despite an intense neutrophil infiltrate and high bacterial burden. However, Th2 (Singh et al., 2013) and Th17 (Woolard et al., 2008) cytokines have been observed in mouse models of respiratory tularemia. It was postulated that the lack of Th1 pro-inflammatory response during the early phase of infection was mediated by such Th2 and Th17 cytokines (Periasamy et al., 2011).
Regarding our findings in human monocytes, it is unlikely that F. tularensis-induced shifts toward Th2 and Th17 responses explain its suppressive effect, either alone or during co-infection with F. novicida. For example, we found that F. tularensis SchuS4 supresses IL10 gene expression in a pattern similar to Th1 cytokines (Supplementary Figure 1). In addition, TGFB1 and IL17RA expression was suppressed in human monocytes by both F. tularensis and F. novicida (Supplementary Figure 1). In contrast to significant IL-17 response following respiratory Francisella LVS infection (Woolard et al., 2008), we were unable to detect reliable expression of IL17A in human monocytes infected with Francisella (data not shown), which is in agreement with the finding by Periasamy et al. in mouse lung macrophages (Periasamy et al., 2011). Thus, observed suppression of robust human monocyte pro-inflammatory responses for F. novicida by F.tularensis co-infection may not be explained by only Th2/Th17 activation as we did not detect this activation within the timeframe of our experiments. However, a Th2/Th17 response may be a potent regulator of the pro-inflammatory response during tularemia at the whole-organism level as other cells such as dendritic cells may contribute by releasing Th2 anti-inflammatory cytokines (Periasamy et al., 2011; Singh et al., 2013). Also, the difference in Francisella recognition between mice and men should not be ignored (Gavrilin and Wewers, 2011).
There is a possibility that host cell death rather than active suppression by F. tularensis is responsible for the reduced proinflammatory responses. Indeed, we observed this in the present study wherein infection by F. tularensis led to greater LDH release (Supplementary Figure2). Although cell death may contribute toward the observed suppression of cytokine production, reduced cytokine transcripts were also observed via RT-qPCR, which compares against 2 endogenous housekeeping genes (Figure 3). Moreover, we were able to observe differential increases in some genes such as RELA and NFKBIA following F. tularensis infection (Supplementary Figure 3).
Our results also showed that inhibition of monocyte cytokine production was dependent on the viability of F. tularensis, since heat- or paraformaldehyde-killed F. tularensis showed no effect. This is in agreement with the work by the Bosio group (Telepnev et al., 2003; Bosio and Dow, 2005; Bosio et al., 2007; Chase et al., 2009), who showed that live Francisella exposure could lead to an attenuation of responses to subsequent innate immune stimuli.
Numerous earlier studies have shown that Francisella is capable of evading host immune detection and eliciting suboptimal pro-inflammatory cytokine responses (Telepnev et al., 2003; Bosio and Dow, 2005; Andersson et al., 2006; Sjostedt, 2006; Chase and Bosio, 2010; Medina et al., 2010; Melillo et al., 2010; Zarrella et al., 2011). This phenomenon is not unique to F. tularensis, since other virulent pathogens such as the Ebola virus and Mycobacterium leprae show similar characteristics (Bosio et al., 2003; Sinsimer et al., 2010). Further examinations of the mechanisms underlying host cell responses to such immunosuppressive pathogens will likely uncover additional commonalities that may ultimately lead to new host-directed therapeutic strategies.
It has often been suggested that Francisella could, at least to some degree, directly antagonize pro-inflammatory responses (Metzger et al., 2007) as well as escape detection. Discerning between the two possibilities has been problematic, and further complicated by the possibility that F. tularensis-mediated suppression of immune responses could be a reflection of endotoxin tolerance, wherein immune cells become refractory (early phase) and desensitized (later phase) to immune stimuli following exposure to an initial stimulus such as mycobacterium, LPS or TNF-α (Greisman and Hornick, 1975; West and Heagy, 2002; Bosio et al., 2007; Morris and Li, 2012; Dai et al., 2013). However, recent work (Bosio et al., 2007; Dai et al., 2013) along with work detailed in this study provides strong evidence that a component of active suppression exists. Also, the dependence of suppression on F. tularensis viability in this study suggests that F. tularensis is producing one or more immunosuppressive agents that act on the host cell and that this effect is a dose-dependent manner. Alternatively we cannot rule out that the lower MOIs of F. tularensis resulted in the infection of fewer monocytes with this bacterium (vs. F. novicida at 50 MOI presumably infecting a greater number of monocytes).
Multiple mechanisms have been discovered to date by which Francisella defeats host immune responses (Bosio et al., 2007; Cremer et al., 2011; Jones et al., 2012). These include an unconventional LPS that is poorly recognized by TLR4 (Duenas et al., 2006; Bosio, 2011), surface lipopeptides such as Tul4 that induce TLR2 signaling but fail to elicit a strong cytokine response (Thakran et al., 2008), OmpA that prevents nuclear translocation of NF-κB p65 (Mahawar et al., 2012), the pathogen's interactions with CR3 (Balagopal et al., 2006; Ben Nasr et al., 2006; Ben Nasr and Klimpel, 2008; Barker et al., 2009; Dai et al., 2013), and its ability to avoid both serum-mediated killing and antibody detection (Bosio et al., 2007; Ben Nasr and Klimpel, 2008; Clay et al., 2008). F. tularensis also leads to host cell transcriptional changes such that immune response pathways such as IFNγ, PI3K, Erk and TLR2 may be weakened (Butchar et al., 2008). Earlier results from our microarray study showed that expression of the Akt-inactivating phosphatase PTEN was higher in monocytes infected with Schu S4 than with F. novicida (Butchar et al., 2008), and it has been shown in human monocyte-derived macrophages (MDM) that Schu S4 induces higher levels of PTEN (Melillo et al., 2010). Interestingly, as well as inducing increased PTEN levels, F. tularensis Schu S4 also inhibited the inactivation of PTEN in human MDM via antioxidant activity, again leading to dampened Akt phosphorylation during infection (Melillo et al., 2010). Our group also confirmed that the Akt antagonist, PTEN, is induced in Schu S4 infections. Collectively, these findings point out the multifaceted nature of Francisella with regard to overcoming immune responses and successfully infecting the host organism. As a facultative bacterium, F. tularensis possesses the ability to respond to changes in its immediate environment, which includes host cells and host organisms. Indeed, our group has shown that suppression can occur as early as during phagocytosis (Dai et al., 2013) and it has been shown that F. tularensis rapidly alters its own transcriptional profile during the course of host cell infection (Wehrly et al., 2009). As such, it is clear that F. tularensis employs a battery of methods in order to actively suppress host responses. Novel additional mechanisms will almost certainly be uncovered as genetic and biochemical studies become increasingly sophisticated
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the NIH/NIAID Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (RCE) Program. The authors wish to acknowledge membership within and support from the Region V “Great Lakes” RCE (NIH award 1-U54-AI-057153). MAG was partly supported by Public Health Preparedness for Infectious Diseases (PHPID) grant from OSU. We thank Tatiana Gavrilina for help with statistical analyses.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fcimb.2014.00045/abstract
F. tularensis suppresses expression of anti-inflammatory genes. Primary human monocytes infected overnight (16 h) with F. novicida (Fn), F. tularensis (Ft), or both at an MOI of 50 for each bacteria were analyzed for expression levels of IL10, TGFB1 and IL17RA genes. Data are expressed as mean ± s.e.m., n = 3 independent experiments.
LDH release from monocytes followed by infection with Francisella. Primary human monocytes were left untreated (NT) or infected for 8 h (A) or 18 h (B) with F. novicida (Fn), F. tularensis (Ft), or both at an MOI of 50 for each bacteria. Cell-free supernatants were assayed for LDH release as a signature of cell death. Data are expressed as mean ± s.e.m., n = 10 independent experiments.
Differential effect of Francisella on expression of selected genes in human monocytes. Primary human monocytes infected overnight (16 h) with F. novicida (Fn), F. tularensis (Ft), or both at an MOI of 50 for each bacteria were analyzed for expression levels of RELA, IKBIA, PSTPIP1 and PYCARD genes. Data are expressed as mean ± s.e.m., n = 3 independent experiments.
References
- Andersson H., Hartmanova B., Kuolee R., Ryden P., Conlan W., Chen W., et al. (2006). Transcriptional profiling of host responses in mouse lungs following aerosol infection with type A Francisella tularensis. J. Med. Microbiol. 55, 263–271 10.1099/jmm.0.46313-0 [DOI] [PubMed] [Google Scholar]
- Balagopal A., Macfarlane A. S., Mohapatra N., Soni S., Gunn J. S., Schlesinger L. S. (2006). Characterization of the receptor-ligand pathways important for entry and survival of Francisella tularensis in human macrophages. Infect. Immun. 74, 5114–5125 10.1128/IAI.00795-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J. H., McCaffrey R. L., Baman N. K., Allen L. A., Weiss J. P., Nauseef W. M. (2009). The role of complement opsonization in interactions between F. tularensis subsp. novicida and human neutrophils. Microbes. Infect. 11, 762–769 10.1016/j.micinf.2009.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauler T. J., Chase J. C., Bosio C. M. (2011). IFN-beta mediates suppression of IL-12p40 in human dendritic cells following infection with virulent Francisella tularensis. J. Immunol. 187, 1845–1855 10.4049/jimmunol.1100377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Nasr A., Haithcoat J., Masterson J. E., Gunn J. S., Eaves-Pyles T., Klimpel G. R. (2006). Critical role for serum opsonins and complement receptors CR3 (CD11b/CD18) and CR4 (CD11c/CD18) in phagocytosis of Francisella tularensis by human dendritic cells (DC): uptake of Francisella leads to activation of immature DC and intracellular survival of the bacteria. J. Leukoc. Biol. 80, 774–786 10.1189/jlb.1205755 [DOI] [PubMed] [Google Scholar]
- Ben Nasr A., Klimpel G. R. (2008). Subversion of complement activation at the bacterial surface promotes serum resistance and opsonophagocytosis of Francisella tularensis. J. Leukoc. Biol. 84, 77–85 10.1189/jlb.0807526 [DOI] [PubMed] [Google Scholar]
- Bosio C. M. (2011). The subversion of the immune system by francisella tularensis. Front. Microbiol. 2:9 10.3389/fmicb.2011.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio C. M., Aman M. J., Grogan C., Hogan R., Ruthel G., Negley D., et al. (2003). Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J. Infect. Dis. 188, 1630–1638 10.1086/379199 [DOI] [PubMed] [Google Scholar]
- Bosio C. M., Bielefeldt-Ohmann H., Belisle J. T. (2007). Active suppression of the pulmonary immune response by Francisella tularensis Schu4. J. Immunol. 178, 4538–4547 [DOI] [PubMed] [Google Scholar]
- Bosio C. M., Dow S. W. (2005). Francisella tularensis induces aberrant activation of pulmonary dendritic cells. J. Immunol. 175, 6792–6801 [DOI] [PubMed] [Google Scholar]
- Bublitz D. C., Noah C. E., Benach J. L., Furie M. B. (2010). Francisella tularensis suppresses the proinflammatory response of endothelial cells via the endothelial protein C receptor. J. Immunol. 185, 1124–1131 10.4049/jimmunol.0902429 [DOI] [PubMed] [Google Scholar]
- Butchar J. P., Cremer T. J., Clay C. D., Gavrilin M. A., Wewers M. D., Marsh C. B., et al. (2008). Microarray analysis of human monocytes infected with Francisella tularensis identifies new targets of host response subversion. PLoS ONE 3:e2924 10.1371/journal.pone.0002924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchar J. P., Rajaram M. V., Ganesan L. P., Parsa K. V., Clay C. D., Schlesinger L. S., et al. (2007). Francisella tularensis induces IL-23 production in human monocytes. J. Immunol. 178, 4445–4454 [DOI] [PubMed] [Google Scholar]
- Celli J., Zahrt T. C. (2013). Mechanisms of Francisella tularensis intracellular pathogenesis. Cold Spring Harb. Perspect. Med. 3:a010314 10.1101/cshperspect.a010314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase J. C., Bosio C. M. (2010). The presence of CD14 overcomes evasion of innate immune responses by virulent Francisella tularensis in human dendritic cells in vitro and pulmonary cells in vivo. Infect. Immun. 78, 154–167 10.1128/IAI.00750-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase J. C., Celli J., Bosio C. M. (2009). Direct and indirect impairment of human dendritic cell function by virulent Francisella tularensis Schu S4. Infect. Immun. 77, 180–195 10.1128/IAI.00879-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Ivashkiv L. B. (2010). IFN-gamma abrogates endotoxin tolerance by facilitating Toll-like receptor-induced chromatin remodeling. Proc. Natl. Acad. Sci. U.S.A. 107, 19438–19443 10.1073/pnas.1007816107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay C. D., Soni S., Gunn J. S., Schlesinger L. S. (2008). Evasion of complement-mediated lysis and complement C3 deposition are regulated by Francisella tularensis lipopolysaccharide O antigen. J. Immunol. 181, 5568–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens D. L., Horwitz M. A. (2007). Uptake and intracellular fate of Francisella tularensis in human macrophages. Ann. N.Y. Acad. Sci. 1105, 160–186 10.1196/annals.1409.001 [DOI] [PubMed] [Google Scholar]
- Crane D. D., Griffin A. J., Wehrly T. D., Bosio C. M. (2013a). B1a cells enhance susceptibility to infection with virulent Francisella tularensis via modulation of NK/NKT cell responses. J. Immunol. 190, 2756–2766 10.4049/jimmunol.1202697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane D. D., Ireland R., Alinger J. B., Small P., Bosio C. M. (2013b). Lipids derived from virulent Francisella tularensis broadly inhibit pulmonary inflammation via toll-like receptor 2 and peroxisome proliferator-activated receptor alpha. Clin. Vaccine Immunol. 20, 1531–1540 10.1128/CVI.00319-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T. J., Butchar J. P., Tridandapani S. (2011). Francisella subverts innate immune signaling: focus on PI3K/Akt. Front. Microbiol. 5:13 10.3389/fmicb.2011.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T. J., Fatehchand K., Shah P., Gillette D., Patel H., Marsh R. L., et al. (2012). MiR-155 induction by microbes/microbial ligands requires NF-kappaB-dependent de novo protein synthesis. Front. Cell. Infect. Microbiol. 2:73 10.3389/fcimb.2012.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T. J., Ravneberg D. H., Clay C. D., Piper-Hunter M. G., Marsh C. B., Elton T. S., et al. (2009). MiR-155 induction by F. novicida but not the virulent F. tularensis results in SHIP down-regulation and enhanced pro-inflammatory cytokine response. PLoS ONE. 4:e8508 10.1371/journal.pone.0008508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S., Rajaram M. V., Curry H. M., Leander R., Schlesinger L. S. (2013). Fine tuning inflammation at the front door: macrophage complement receptor 3-mediates phagocytosis and immune suppression for Francisella tularensis. PLoS Pathog. 9:e1003114 10.1371/journal.ppat.1003114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duenas A. I., Aceves M., Orduna A., Diaz R., Sanchez C. M., Garcia-Rodriguez C. (2006). Francisella tularensis LPS induces the production of cytokines in human monocytes and signals via Toll-like receptor 4 with much lower potency than E. coli LPS. Int. Immunol. 18, 785–795 10.1093/intimm/dxl015 [DOI] [PubMed] [Google Scholar]
- Edwards J. A., Rockx-Brouwer D., Nair V., Celli J. (2010). Restricted cytosolic growth of Francisella tularensis subsp. tularensis by IFN-gamma activation of macrophages. Microbiology 156, 327–339 10.1099/mic.0.031716-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestal C. A., Malik M., Catlett S. V., Savitt A. G., Benach J. L., Sellati T. J., et al. (2007). Francisella tularensis has a significant extracellular phase in infected mice. J. Infect. Dis. 196, 134–137 10.1086/518611 [DOI] [PubMed] [Google Scholar]
- Gavrilin M. A., Abdelaziz D. H., Mostafa M., Abdulrahman B. A., Grandhi J., Akhter A., et al. (2012). Activation of the pyrin inflammasome by intracellular Burkholderia cenocepacia. J. Immunol. 188, 3469–3477 10.4049/jimmunol.1102272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilin M. A., Bouakl I. J., Knatz N. L., Duncan M. D., Hall M. W., Gunn J. S., et al. (2006). Internalization and phagosome escape required for Francisella to induce human monocyte IL-1beta processing and release. Proc. Natl. Acad. Sci. U.S.A. 103, 141–146 10.1073/pnas.0504271103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilin M. A., Mitra S., Seshadri S., Nateri J., Berhe F., Hall M. W., et al. (2009). Pyrin critical to macrophage IL-1beta response to Francisella challenge. J. Immunol. 182, 7982–7989 10.4049/jimmunol.0803073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilin M. A., Wewers M. D. (2011). Francisella Recognition by Inflammasomes: differences between Mice and Men. Front. Microbiol. 2:11 10.3389/fmicb.2011.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greisman S. E., Hornick R. B. (1975). The nature of endotoxin tolerance. Trans. Am. Clin. Climatol. Assoc. 86, 43–50 [PMC free article] [PubMed] [Google Scholar]
- Greisman S. E., Hornick R. B., Carozza F. A., Jr., Woodward T. E. (1963). The role of endotoxin during typhoid fever and tularemia in man. I. Acquisition of tolerance to endotoxin. J. Clin. Invest. 42, 1064–1075 10.1172/JCI104792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. D., Woolard M. D., Gunn B. M., Craven R. R., Taft-Benz S., Frelinger J. A., et al. (2008). Infected-host-cell repertoire and cellular response in the lung following inhalation of Francisella tularensis Schu S4, LVS, or U112. Infect. Immun. 76, 5843–5852 10.1128/IAI.01176-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland R., Wang R., Alinger J. B., Small P., Bosio C. M. (2013). Francisella tularensis SchuS4 and SchuS4 lipids inhibit IL-12p40 in primary human dendritic cells by inhibition of IRF1 and IRF8. J. Immunol. 191, 1276–1286 10.4049/jimmunol.1300867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. L., Napier B. A., Sampson T. R., Llewellyn A. C., Schroeder M. R., Weiss D. S. (2012). Subversion of host recognition and defense systems by Francisella spp. Microbiol. Mol. Biol. Rev. 76, 383–404 10.1128/MMBR.05027-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. J., Park S. H., Choi Y. S., Shim S. K., Park M. Y., Park M. S., et al. (2008). Cytokine response in Balb/c mice infected with Francisella tularensis LVS and the Pohang isolate. J. Vet. Sci. 9, 309–315 10.4142/jvs.2008.9.3.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahawar M., Atianand M. K., Dotson R. J., Mora V., Rabadi S. M., Metzger D. W., et al. (2012). Identification of a novel Francisella tularensis factor required for intramacrophage survival and subversion of innate immune response. J. Biol. Chem. 287, 25216–25229 10.1074/jbc.M112.367672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mares C. A., Ojeda S. S., Morris E. G., Li Q., Teale J. M. (2008). Initial delay in the immune response to Francisella tularensis is followed by hypercytokinemia characteristic of severe sepsis and correlating with upregulation and release of damage-associated molecular patterns. Infect. Immun. 76, 3001–3010 10.1128/IAI.00215-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina E. A., Morris I. R., Berton M. T. (2010). Phosphatidylinositol 3-kinase activation attenuates the TLR2-mediated macrophage proinflammatory cytokine response to Francisella tularensis live vaccine strain. J. Immunol. 185, 7562–7572 10.4049/jimmunol.0903790 [DOI] [PubMed] [Google Scholar]
- Melillo A. A., Bakshi C. S., Melendez J. A. (2010). Francisella tularensis antioxidants harness reactive oxygen species to restrict macrophage signaling and cytokine production. J. Biol. Chem. 285, 27553–27560 10.1074/jbc.M110.144394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D. W., Bakshi C. S., Kirimanjeswara G. (2007). Mucosal immunopathogenesis of Francisella tularensis. Ann. N.Y. Acad. Sci. 1105, 266–283 10.1196/annals.1409.007 [DOI] [PubMed] [Google Scholar]
- Mohapatra N. P., Soni S., Rajaram M. V., Strandberg K. L., Gunn J. S. (2013). Type A Francisella tularensis acid phosphatases contribute to pathogenesis. PLoS ONE. 8:e56834 10.1371/journal.pone.0056834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M., Li L. (2012). Molecular mechanisms and pathological consequences of endotoxin tolerance and priming. Arch. Immunol. Ther. Exp. (Warsz) 60, 13–18 10.1007/s00005-011-0155-9 [DOI] [PubMed] [Google Scholar]
- Oyston P. C., Sjostedt A., Titball R. W. (2004). Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2, 967–978 10.1038/nrmicro1045 [DOI] [PubMed] [Google Scholar]
- Periasamy S., Singh A., Sahay B., Rahman T., Feustel P. J., Pham G. H., et al. (2011). Development of tolerogenic dendritic cells and regulatory T cells favors exponential bacterial growth and survival during early respiratory tularemia. J. Leukoc. Biol. 90, 493–507 10.1189/jlb.0411197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick L. C., Wu T. H. (2007). Animal models of Francisella tularensis infection. Ann. N.Y. Acad. Sci. 1105, 238–265 10.1196/annals.1409.003 [DOI] [PubMed] [Google Scholar]
- Sharma J., Mares C. A., Li Q., Morris E. G., Teale J. M. (2011). Features of sepsis caused by pulmonary infection with Francisella tularensis Type A strain. Microb. Pathog. 51, 39–47 10.1016/j.micpath.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Rahman T., Malik M., Hickey A. J., Leifer C. A., Hazlett K. R., et al. (2013). Discordant results obtained with Francisella tularensis during in vitro and in vivo immunological studies are attributable to compromised bacterial structural integrity. PLoS ONE 8:e58513 10.1371/journal.pone.0058513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinsimer D., Fallows D., Peixoto B., Krahenbuhl J., Kaplan G., Manca C. (2010). Mycobacterium leprae actively modulates the cytokine response in naive human monocytes. Infect. Immun. 78, 293–300 10.1128/IAI.00816-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostedt A. (2006). Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes. Infect. 8, 561–567 10.1016/j.micinf.2005.08.001 [DOI] [PubMed] [Google Scholar]
- Sjostedt A. (2007). Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann. N.Y. Acad. Sci. 1105, 1–29 10.1196/annals.1409.009 [DOI] [PubMed] [Google Scholar]
- Sly L. M., Rauh M. J., Kalesnikoff J., Song C. H., Krystal G. (2004). LPS-induced upregulation of SHIP is essential for endotoxin tolerance. Immunity 21, 227–239 10.1016/j.immuni.2004.07.010 [DOI] [PubMed] [Google Scholar]
- Telepnev M., Golovliov I., Grundstrom T., Tarnvik A., Sjostedt A. (2003). Francisella tularensis inhibits Toll-like receptor-mediated activation of intracellular signalling and secretion of TNF-alpha and IL-1 from murine macrophages. Cell. Microbiol. 5, 41–51 10.1046/j.1462-5822.2003.00251.x [DOI] [PubMed] [Google Scholar]
- Telepnev M., Golovliov I., Sjostedt A. (2005). Francisella tularensis LVS initially activates but subsequently down-regulates intracellular signaling and cytokine secretion in mouse monocytic and human peripheral blood mononuclear cells. Microb. Pathog. 38, 239–247 10.1016/j.micpath.2005.02.003 [DOI] [PubMed] [Google Scholar]
- Thakran S., Li H., Lavine C. L., Miller M. A., Bina J. E., Bina X. R., et al. (2008). Identification of Francisella tularensis lipoproteins that stimulate the toll-like receptor (TLR) 2/TLR1 heterodimer. J. Biol. Chem. 283, 3751–3760 10.1074/jbc.M706854200 [DOI] [PubMed] [Google Scholar]
- Walters K. A., Olsufka R., Kuestner R. E., Cho J. H., Li H., Zornetzer G. A., et al. (2013). Francisella tularensis subsp. tularensis induces a unique pulmonary inflammatory response: role of bacterial gene expression in temporal regulation of host defense responses. PLoS ONE 8:e62412 10.1371/journal.pone.0062412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrly T. D., Chong A., Virtaneva K., Sturdevant D. E., Child R., Edwards J. A., et al. (2009). Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol. 11, 1128–1150 10.1111/j.1462-5822.2009.01316.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M. A., Heagy W. (2002). Endotoxin tolerance: a review. Crit. Care Med. 30, S64–S73 10.1097/00003246-200201001-00009 [DOI] [PubMed] [Google Scholar]
- Woolard M. D., Hensley L. L., Kawula T. H., Frelinger J. A. (2008). Respiratory Francisella tularensis live vaccine strain infection induces Th17 cells and prostaglandin E2, which inhibits generation of gamma interferon-positive T cells. Infect. Immun. 76, 2651–2659 10.1128/IAI.01412-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrella T. M., Singh A., Bitsaktsis C., Rahman T., Sahay B., Feustel P. J., et al. (2011). Host-adaptation of Francisella tularensis alters the bacterium's surface-carbohydrates to hinder effectors of innate and adaptive immunity. PLoS ONE. 6:e22335 10.1371/journal.pone.0022335 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
F. tularensis suppresses expression of anti-inflammatory genes. Primary human monocytes infected overnight (16 h) with F. novicida (Fn), F. tularensis (Ft), or both at an MOI of 50 for each bacteria were analyzed for expression levels of IL10, TGFB1 and IL17RA genes. Data are expressed as mean ± s.e.m., n = 3 independent experiments.
LDH release from monocytes followed by infection with Francisella. Primary human monocytes were left untreated (NT) or infected for 8 h (A) or 18 h (B) with F. novicida (Fn), F. tularensis (Ft), or both at an MOI of 50 for each bacteria. Cell-free supernatants were assayed for LDH release as a signature of cell death. Data are expressed as mean ± s.e.m., n = 10 independent experiments.
Differential effect of Francisella on expression of selected genes in human monocytes. Primary human monocytes infected overnight (16 h) with F. novicida (Fn), F. tularensis (Ft), or both at an MOI of 50 for each bacteria were analyzed for expression levels of RELA, IKBIA, PSTPIP1 and PYCARD genes. Data are expressed as mean ± s.e.m., n = 3 independent experiments.