Abstract
The elaborate cytoarchitecture of the mammalian neocortex requires the timely production of its constituent pyramidal neurons and interneurons and their disposition in appropriate layers. Numerous chemotropic factors present in the forebrain throughout cortical development play important roles in the orchestration of these events. The Roundabout (Robo) family of receptors and their ligands, the Slit proteins, are expressed in the developing forebrain, and are known to play important roles in the generation and migration of cortical interneurons. However, few studies have investigated their function(s) in the development of pyramidal cells. Here, we observed expression of Robo1 and Slit genes (Slit1, Slit2) in cells lining the telencephalic ventricles, and found significant increases in progenitor cells (basal and apical) at embryonic day (E)12.5 and E14.5 in the developing cortex of Robo1−/−, Slit1−/−, and Slit1−/−/Slit2−/−, but not in mice lacking the other Robo or Slit genes. Using layer-specific markers, we found that both early- and late-born pyramidal neuron populations were significantly increased in the cortices of Robo1−/− mice at the end of corticogenesis (E18.5). The excess number of cortical pyramidal neurons generated prenatally appears to die in early postnatal life. The observed increase in pyramidal neurons was due to prolonged proliferative activity of their progenitors and not due to changes in cell cycle events. This finding, confirmed by in utero electroporation with Robo1 short hairpin RNA (shRNA) or control constructs into progenitors along the ventricular zone as well as in dissociated cortical cell cultures, points to a novel role for Robo1 in regulating the proliferation and generation of pyramidal neurons.
Keywords: corticogenesis, progenitors, proliferation, Robo
Introduction
The formation of the mammalian neocortex requires the timely production of its constituent pyramidal neurons and interneurons, and their migration to appropriate layers. Pyramidal neurons are generated by radial glial (RG) cells (or apical progenitors), present in the proliferative ventricular zone (VZ) and by intermediate progenitor cells (IPCs; basal progenitors) found predominantly in the subventricular zone (SVZ; (Kriegstein and Alvarez-Buylla, 2009). Throughout corticogenesis, RG cells undergo stereotypical patterns of symmetric and asymmetric divisions, continually generating diverse subtypes of neurons while maintaining a pool of progenitor cells (Miyata et al., 2004; Noctor et al., 2004). Basal progenitors arise in the ventricular surface and migrate to the upper part of the VZ to create the SVZ. Unlike their RG counterparts, they divide symmetrically to produce two pyramidal neurons or two additional IPCs.
The molecular mechanisms that govern cortical neuronal production and migration have recently been the focus of intense research, as defects in these processes can lead to neurological and psychiatric disorders (Bielas and Gleeson, 2004; Gressens, 2005; Guerrini and Filippi, 2005). Numerous chemotropic factors present in the developing forebrain play important roles in the orchestration of neurogenesis and migration. One such factor is Roundabout (Robo), first identified as a receptor for the chemorepulsive ligand Slit in Drosophila (Kidd et al., 1998). In the mammalian forebrain, it has been established that Slit–Robo signaling plays key roles in axonal pathfinding (Bagri et al., 2002; López-Bendito et al., 2007). Other studies have shown that inhibition of Robo1-mediated signaling alters the proliferation and migration of neocortical interneurons (Andrews et al., 2006; Hernández-Miranda et al., 2011), and more recently, Robo4 and Robo1 receptors have been shown to play crucial roles in the radial migration of pyramidal neurons (Zheng et al., 2012; Gonda et al., 2013). These findings support the notion that Robo receptors function beyond axonal pathfinding in the developing neocortex.
Here, we investigated the functional roles of Robo and Slit genes in the generation and disposition of pyramidal neurons. We first established expression of Robo1 and Slit genes (Slit1, Slit2) in cells lining the telencephalic ventricles. We, then, discovered significant increases in both apical and basal progenitor cells at E12.5 and E14.5 in the developing cortex of Robo1−/−, Slit1−/− and Slit1−/−/Slit2−/− mice, but not in those lacking the other Robo or Slit genes. Further, analysis of Robo1−/−/Robo2−/− double mutants indicated that Robo2 is unlikely to be involved in these proliferative events. These results are in contrast to the recently reported decrease in proliferating apical progenitors and the abnormal increase in basal progenitors in Robo1−/−/Robo2−/− mice at the onset of corticogenesis, with the phenotypic changes due to loss of Robo2 (Borrell et al., 2012).
Using layer-specific markers, we found that both early- and late-born pyramidal neuron populations were significantly increased in the cortices of Robo1−/− mice at the end of corticogenesis. The observed increase in pyramidal neurons was likely due to prolonged proliferative activity of progenitors, and not due to changes in their cell cycle parameters. This finding, confirmed by in utero electroporation with Robo1 shRNA or control constructs into progenitor cells along the VZ as well as in dissociated cortical cell cultures, points to a novel role for Robo1 in regulating the proliferation and generation of pyramidal neurons. Microarray analysis revealed that the observed changes in the generation of pyramidal neurons resulting from deletion of Robo1 are accompanied by the differential expression of a number of proliferation and apoptotic genes. The excess number of pyramidal neurons generated prenatally appears to die in early postnatal life, but the lamination of the cortex is altered, especially in the upper layers.
Materials and Methods
Transgenic mice.
All experimental procedures were performed in accordance with the UK Animals (Scientific Procedures) Act, 1986 and institutional guidelines. Wild-type C57BL/6J mice were obtained from Charles River, and time-mated Sprague-Dawley albino rats were provided by UCL Biological Services. Transgenic mouse lines used in this study included Robo1+/− (Andrews et al., 2008), Robo2+/− (Lu et al., 2007), Robo3+/− (Sabatier et al., 2004), Robo1+/−;Robo2+/− (Long et al., 2004), Slit1+/− (Plump et al., 2002), Slit2+/− (Plump et al., 2002), and Slit3+/− (Yuan et al., 2003). All mouse strains were maintained on the C57BL/6J background, with the exception of Slit3 transgenic mice which had a mixed genetic background. The day the vaginal plug was found was considered as embryonic day (E)0.5. Animals of both sexes were used in our experiments.
In situ hybridization and immunohistochemistry.
Probes and protocols for the nonradioactive in situ hybridization and immunohistochemistry methods were described previously (Moorman et al., 2001; Mommersteeg et al., 2013). Briefly, embryos were fixed overnight in 4% paraformaldehyde made in phosphate buffer saline (PFA), embedded in paraffin, and sectioned at 7–10 μm for immunohistochemistry or 12 μm for in situ hybridization. Sections were immunostained using one of the following antibodies: mouse monoclonal anti-BrdU (1:200, ProGen), mouse monoclonal anti-Iba1 (1:200, Abcam), rat monoclonal anti-Ctip2 (1:500, Abcam), goat polyclonal raised against Robo1 (1:250, BD Biosciences), Robo2 (1:250, BD Biosciences), or Robo3 (1:250, BD Biosciences), chicken polyclonal raised against GFP (1:500, Aves Laboratories), rabbit polyclonal raised against Cux1 (1:100, Santa Cruz Biotechnology), phosphohistone H-3 (PH-3; 1:1000, Millipore), Ki-67 (1:1000, Novocastra), Tbr2 (1:2000, gift from Professor R. Hevner, Seattle Children's Research Institute, Seattle, WA; 1:300, Millipore) or cleaved caspase-3 (CC3; 1:250, Cell Signaling Technology). After incubation in primary antibodies, sections were washed in PBS, incubated in biotinylated anti-species (1:250; Vector Laboratories) for 2 h, and processed using conventional immunohistochemistry protocols described previously (Andrews et al., 2006). Fluorescent secondary antibodies used were AlexaFluor 568 donkey anti-goat, 488 and 568 goat anti-rabbit, and 488 goat anti-mouse (1:250, Invitrogen), biotinylated horse anti-goat and goat anti-rabbit (1:250, Vector Laboratories). Nuclei were counterstained with 4′, 6-diamidino-2-phenylindole (DAPI; 2.5 μg/ml, Sigma-Aldrich).
Quantification of PH-3-positive cells.
All PH-3-positive cells present in embryonic coronal sections, along the entire VZ/SVZ, from the corticostriatal junction to the cortical hem (CH), and throughout the rostral-caudal extent of the cortex were included in all measurements (minimum of 8 sections from each of 4 animals for each condition). The extent of the layers was determined by methyl green counterstaining (Vector Laboratories). Quantification of apical progenitors lining the VZ was presented as PH-3-labeled cells per mm. Basal progenitors in the SVZ were presented as PH-3-labeled cells per 104 μm2. Basal progenitors here were defined as any cell more than three cells width away from ventricle surface.
Quantification of pyramidal neurons.
Pyramidal neurons were counted in coronal strips (400 μm wide) spanning the thickness of the middle (along the rostrocaudal axis) regions of the cortex at E18.5 (minimum of 8 sections from each of 3 animals for each condition). Strips were divided into the different layers of the developing cortex: VZ, SVZ, intermediate zone (IZ), subplate (SP), and cortical plate (CP) for the purpose of accurately quantifying pyramidal neuron numbers and distribution as labeled with Emx-1, Ctip2, or Cux-1. The extent of the layers was determined by methyl green counterstaining.
Dissociated cortical cell cultures.
Dissociated cell cultures were prepared from either E13.5 mouse or E15.5 Sprague-Dawley albino rat brains as described previously (Cavanagh et al., 1997). Briefly, cortices were dissected out in cold artificial CSF (ACSF) under a stereo microscope. They were incubated in trypsinization medium [0.05% trypsin (Sigma-Aldrich) with 100 μg/ml DNaseI (Roche) in neurobasal medium (Invitrogen)] for 15 min at 37°C. Trypsinization was quenched with neutralization medium (10% of FBS, Invitrogen, in neurobasal medium) for 5 min at 37°C. Cortices were then triturated by pipetting until no cellular aggregates were visible. The homogenous cell suspensions were subsequently pelleted by centrifugation at 1000 × g for 3 min. Cells were resuspended in dissociated culture medium [neurobasal media containing 10% of FBS and N2 supplement (Invitrogen), 100 μg/ml penicillin/streptomycin (Invitrogen), and 2 mm l-glutamine (Invitrogen)] and 1,000,000 cells were seeded on to 13 mm coverslips coated previously with 10 μg/ml poly-l-lysine and 10 μg/ml laminin and incubated in a the humidified incubator at 37°C. The next day, the culture medium was changed and the cell cultures were used for proliferation assays (see below).
Proliferation rate.
The rate of cell proliferation was established using tissue taken from Robo1+/+ and Robo1−/− embryos. Specifically, we determined the proportion of cells that incorporated bromodeoxyuridine (BrdU) after either a 2 h exposure, a period long enough to label as many cells as possible in S-phase without allowing them to enter mitosis, or a 24 h exposure to assess the number of cells that have left S-phase (Cavanagh et al., 1997). In these experiments, sections through the cortex were immunostained for BrdU and Ki67, and the number of double-positive cells was determined in an area measuring 104 μm2, encompassing the VZ and SVZ.
In dissociated cortical cell cultures, 10 mm BrdU was added to the medium for either 2 or 24 h after which, cells were washed, fixed with 4% PFA, immunostained for BrdU and Tbr2 or Pax6 to identify different progenitor types. The percentage of cells immunoreactive for BrdU and Tbr2 or Pax6 was counted using a ×40 objective lens in nine fields of view for each coverslip. We evaluated at least three coverslips from each animal for each time point and treatment, and significance was established using one-way ANOVA.
To determine the effect of Slit, Robo1, and Nrp1 on proliferation, we prepared E15.5 rat and E13.5 Robo1+/+ and Robo1−/− cortical cell cultures. These were incubated overnight in the presence or absence of 1 μg/ml (final concentration) Slit1, Slit2, Slit1/Slit2, Robo1-Fc, or Nrp1-Fc (R&D Systems). Cells were washed and fixed with PFA, immunostained for BrdU and Tbr2 or Pax6, and counterstained with DAPI. Cell counts were made with a ×40 objective in nine fields-of-view for each sample performed in quadruplicate.
Apoptosis and microglia labeling.
Sections taken through the brains of Robo1+/+ and Robo1−/− embryos at E12.5–E18.5, or mice at postnatal (P) days P8 and P21 were immunostained for CC3 and microglia marker Iba1. The number of labeled cells was counted in an area measuring 4 × 104 μm2, encompassing VZ and SVZ, and assessed for differences.
Plasmid construction.
For plasmid-based RNA inhibition of Robo1, the complementary oligonucleotides for the following target sequences (Robo1-shRNA1: 5′-ACTCAAACCTAACGCCATTTA-3′; Mt-Robo1-shRNA:5′-ACTCAAACCATTCGCCATTTA-3′) were annealed and inserted into the BamHI/HindIII sites of pSilencer 3.0-H1 (Ambion) as described previously (Gonda et al., 2013).
In utero electroporation.
In utero electroporation was performed as previously described (Gonda et al., 2013). Briefly, E12.5 timed-pregnant ICR mice were deeply anesthetized with sodium pentobarbitone (somnopentyl; Kyoritsu Pharmaceuticals), and their uterine horns exposed. Approximately 1–2 μl of plasmid DNA solution, dissolved to a final concentration of 5 mg/ml in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid-buffered saline, was injected into a lateral ventricle of each embryo with a glass micropipette, using electric pulses (30 V, 50 ms) that were discharged four times at intervals of 950 ms with an electroporator (CUY21E; Nepa Gene). The uterine horns were then replaced in the abdominal cavity to allow the embryos to continue normal development until E14.5. To assess proliferation in these mice, BrdU was administered through intraperitoneal injections 2 h before kill.
Laser-capture microdissection.
Embryonic brains (E12.5) from Robo1+/+ and Robo1−/− mice (n = 3 both groups) were dissected in RNase-free PBS, placed in cryostat molds, and frozen in Tissue-Tek OCT (Sakura Finetek). Forebrains were sectioned at 20 μm and adhered to laser-capture microdissection (LCM) membrane-mounted slides (Molecular Machines), and stored at −80°C until use. For LCM, slides were individually thawed for 30 s, fixed in cold methanol for 1 min, and rinsed rapidly in PBS. Slides were dehydrated through 70–100% ethanol, and allowed to dry thoroughly (30 s to 1 min). Cortical derived VZ/SVZ cells were excised within 15 min of drying using a PixCell II Laser Capture Microdissection System (Arcturus). VZ/SVZ cells were adhered to capture tube lids (Molecular Machines). Tubes were placed on dry ice and kept at −80°C until RNA extraction.
RNA extraction and microarray analysis.
Total RNA from cortical VZ/SVZ derived cells was extracted immediately after collection using the Qiagen RNeasy Miniplus kit. RNA was sent to the Wolfson Institute for Biomedical Research (UCL Genomics) for cDNA production, hybridization and scanning. The quality of the RNA was assessed using an Agilent bioanalyzer nanochip. All RNA had 18S and 28S ribosomal RNA bands. RNA (100 ng per chip) was converted to single-strand, sense-strand cDNA using the Affymetrix Sense target labeling protocol and the Mouse Gene 1.0ST Array kit. After fragmenting and end labeling, the cDNA were hybridized to Mouse Gene Gene-1_0-st-v1 Array's (Affymetrix) for 16 h at 45°C according to the manufacturer's instructions. The arrays were then washed and stained on the Fluidics station 450 using the hybridization, wash and stain kits and scanned on the GeneChip Scanner 3000. Analyses of microarray data were performed using the Genespring GX software package (Agilent Technologies). The p values were corrected for multiple testing using the Benjamini-Hochberg correction and a corrected p value threshold of 0.05, together with a fold cutoff of >2 was used to select differentially expressed genes.
qPCR validation.
For validation of the differentially expressed genes, quantitative PCR (qPCR) was performed. E12.5 and E14.5 Robo1+/+ and Robo1−/− (n = 3, both groups) mouse embryos were dissected and RNA extraction were performed as described above. For qPCR analysis, whole cortex was used, as opposed to the microarray analysis, where VZ and SVZ were dissected using LCM. RNA was treated with DNaseI (amplification grade; Invitrogen) to remove any remaining trace amounts of DNA. cDNA was generated with 450 ng of RNA by use of the Qiagen Whole Transcriptome Amplification Kit as described in the manufacturer's protocol. Primers for qPCR were designed using the Harvard Medical School Primer bank web facility (): Bcl2 (forward: GTCGCTACCGTCGTGACTTC; reverse: CAGACATGCACCTACCCAGC), Casp2 (forward: TACTCCCACCGTTGAGCTGT; reverse: CCGTAGCATCTGTGGATAGGC), Casp8 (forward: TGCTTGGACTACATCCCACAC; reverse: TGCAGTCTAGGAAGTTGACCA), Gapdh (forward: ATGACATCAAGAAGGTGGTG; reverse: CATACCAGGAAATGAGCTTG), Hey1 (forward: GCGCGGACGAGAATGGAAA; reverse: TCAGGTGATCCACAGTCATCTG), Itga6 (forward: TGCAGAGGGCGAACAGAAC; reverse: GCACACGTCACCACTTTGC), Itga9 (forward: AAGTGTCGTGTCCATACCAAC; reverse: GGTCTGCTTCGTAGTAGATGTTC), Ki67 (forward: ATCATTGACCGCTCCTTTAGGT; reverse: GCTCGCCTTGATGGTTCCT), Nfyc (forward: GAAGCGCATGTCGAAAGAAGA; reverse: GGCGGAGGCAGTCAATTCTC), Notch2 (forward: GAGAAAAACCGCTGTCAGAATGG; reverse: GGTGGAGTATTGGCAGTCCTC), Pax6 (forward: TACCAGTGTCTACCAGCCAAT; reverse TGCACGAGTATGAGGAGGTCT), Ppia (forward: GAGCTGTTTGCAGACAAAGTTC; reverse: CCCTGGCACATGAATCCTGG), Rb1cc1 (forward: GACACTGAGCTAACTGTGCAA; reverse: GCGCTGTAAGTACACACTCTTC), Robo1 (forward: GAGCCTGCTCACTTTTACCTC; reverse: GGTCTGAAGGGTGTTCAACAAT), TGFβ2 (forward: CTTCGACGTGACAGACGCT; reverse: GCAGGGGCAGTGTAAACTTATT), Top2a (forward: CAACTGGAACATATACTGCTCCG; reverse: GGGTCCCTTTGTTTGTTATCAGC), Tpx2 (forward: GATGCCCCCACCGACTTTATC; reverse: CTTGTTCTCCAAGTTGGCCTT), and Zfp423 (forward: TGGCCTGGGATTCCTCTGT; reverse: CTCTTGACTTGTCACGCTGTT). The qPCR was performed with SYBR Green reagent on a Chromo4 PTC-200 Real-Time PCR Detector system (Bio-Rad). PCR conditions were 94°C for 2 min, followed by 40 three-step cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) and peptidylprolylisomerase A (Ppia) were used for endogenous reference gene controls. Each primer set amplified a single PCR product of predicted size as determined by melt-curve analysis following PCR and by agarose gel electrophoresis, and had approximately equal amplification efficiencies when validated with a serial dilution of representative cDNA. Each qPCR was performed in triplicate, and relative quantification was determined according to the ΔΔC(t) method (Livak and Schmittgen, 2001; Faux et al., 2010).
Digital image acquisition and processing.
Optical and fluorescent images were collected using a Leica Microsystems light microscope (DM5000B). Images were reconstructed and digitized with Photoshop CS4 software (Adobe Systems).
Statistics.
Statistical analyses were performed by GraphPad 3 (GraphPad Software). All data are reported as mean number and SEM. The statistical significance between group means was tested by one-way ANOVA, followed by Bonferroni's post hoc test (for multiple-comparison tests). Significance was set at p ≤ 0.05.
Results
Expression of Slit-Robo in the proliferative zones of the telencephalon
The expression of Slit and Robo families of genes have been extensively studied in the rat forebrain (Marillat et al., 2002; Whitford et al., 2002; Camurri et al., 2004; Zheng et al., 2012), but although some members have been shown to be expressed in the developing mouse forebrain (Bagri et al., 2002; Andrews et al., 2008; Barber et al., 2009), detailed information is lacking. Here, we performed a comprehensive expression study of Slit (Slit1–3) and Robo (Robo1–3) mRNAs during early (E12.5) and middle (E14.5) stages of embryonic development, as the role of Robo4 in the development of cortical pyramidal neurons has recently been reported (Zheng et al., 2012).
During early development (E12.5) of the ventral telencephalon, Slit1 was strongly expressed in the mantle zones of the medial ganglionic eminence (MGE) and lateral ganglionic eminence (LGE), and in the preoptic area (POA); whereas Slit2 was present in the MGE and POA (Fig. 1A,B); and Slit3 was absent from both the ventral and dorsal telencephalon (data not shown). In the dorsal telencephalon, Slit1 exhibited strong expression along the preplate layer (PPL) and weak expression in the VZ, whereas Slit2 only showed weak expression in medial VZ (Fig. 1A,B). The punctate appearance of Slit1 and Slit2 in the forebrain suggested either cellular localization or expression along cerebral vasculature.
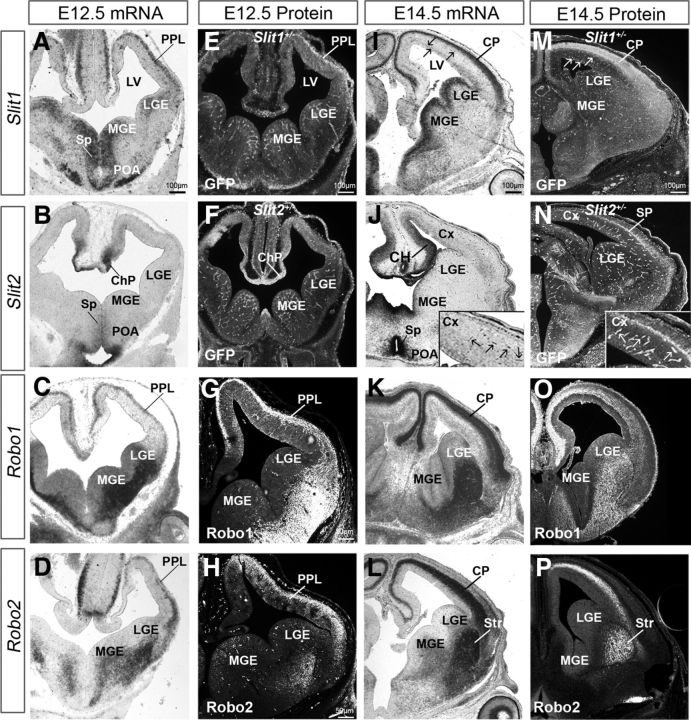
Figure 1.
Slit/Robo expression in the developing forebrain. Coronal sections of the middle forebrain at E12.5 (A–H) and E14.5 (I–P) showing localization by in situ hybridization (A–D, I–L) expression of Slit1 (A, I), Slit2 (B, J), Robo1 (C, K), and Robo2 (D, L). A, I, In the dorsal forebrain, strong Slit1 mRNA expression can be seen along the PPL at E12.5 and in the CP at E14.5, while considerably weaker expression is present in the VZ at both ages. In the ventral forebrain, strong expression is predominantly localized in the MGE, LGE, POA, and Sp. B, J, Slit2 mRNA is restricted mainly in the POA and ChP at E12.5, whereas at E14.5 is strongly expressed in the CH and weakly in the VZ of the cortex, MGE, and LGE. C, K, At E12.5, Robo1 mRNA is expressed strongly in the PPL and weakly in the VZ of the dorsal telencephalon. In the ventral telencephalon, it is localized predominantly in the MGE and LGE. This pattern is maintained at E14.5, with additional strong expression in the CP. D, L, Robo2 mRNA is abundantly expressed in the PPL, but absent from the proliferative zones of the cortex. E, F, M, N, Immunohistochemistry for TauGFP-labeling in Slit1+/− (E, M) and Slit2+/− (F, N) mice show GFP protein expression in the forebrain, in a pattern similar to that of Slit1 and Slit2 mRNA. I, M (arrows) and J, N (insets) point to Slit-expressing cortical vasculature. Protein expression patterns for Robo1 (G, O) and Robo2 (H, P) mirror mRNA expression. ChP, Choroid plexus; LV, lateral ventricle; Sp, septum; Str, striatum.
Labeling for the tau-GFP fusion protein in Slit1+/− and Slit2+/− mice allowed us to visualize in greater detail the cellular elements that express Slit1 or Slit2 in the murine forebrain (Fig. 1E,F). Thus, GFP immunohistochemical analysis of Slit1+/− mice at E12.5 (Fig. 1E) showed comparable expression to Slit1 mRNA, with strong staining in the PPL and POA (Fig. 1A,E). Similar analysis in Slit2+/− mice at E12.5 revealed robust staining in the choroid plexus and septum (Fig. 1F). GFP+ cells were also noted in the cortical VZ and SVZ and in the cerebral vasculature of both groups of mice (Fig. 1E,F), confirming recent evidence which points to a role for Slit2 in blood vessel formation (Han and Geng, 2011).
Robo1 and Robo2 mRNA localization was prominent in the ventral telencephalon, as previously described (Bagri et al., 2002). However, contrary to earlier reports, we observed low levels of Robo1 mRNA along the ventricular surfaces of the dorsal and ventral telencephalon (Fig. 1C), which was confirmed with Robo1 immunohistochemistry (Fig. 1G, Fig. 2A–F), suggesting a potential role for this receptor in regulating the proliferative activity of progenitor cells at this early stage of forebrain development. Robo2 mRNA was expressed abundantly laterally in the forebrain, occupying the mantle zones of the ventral telencephalon in a similar pattern to that of Robo1 (Fig. 1D). At the corticostriatal junction, Robo2 expression formed a decreasing gradient as it extended dorsally along the PPL, and appeared nearly absent in the CH (Fig. 1D). Robo3 mRNA was expressed weakly in the POA, LGE, and sparsely distributed throughout the dorsal telencephalon including the CH (data not shown). Neither Robo2 nor Robo3 mRNA or proteins were observed in the proliferative zones of the cortex (Fig. 1D,H; data not shown for Robo3).
Figure 2.
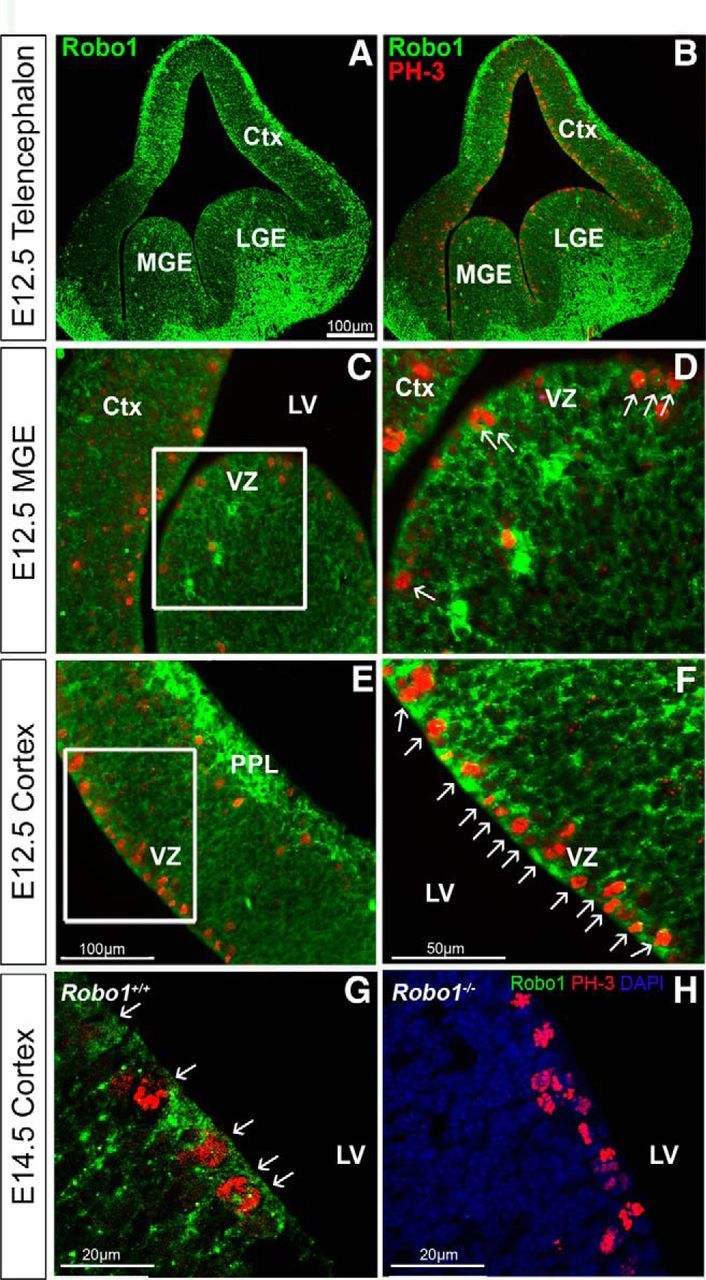
Coexpression of Robo1 protein with the proliferation marker, PH-3. Immunohistochemical localization of Robo1 (green) and PH-3 (red) in coronal sections through the telencephalon (A, B), and through the VZ of the MGE (C, D) and cortex of C57BL/6J mice at E12.5 (E, F), and in Robo1+/+ (G) and Robo1−/− littermate (H) at E14.5. D, F, Higher-magnification of the boxed areas in C and E, respectively, and illustrate coexpression of the two markers (arrows) in individual cells of the VZ of the MGE (D) and in the majority of the cells in the VZ of the cortex at both ages (F, G). Ctx, Cortex; LV, lateral ventricle.
At midembryonic stage (E14.5), Slit1 mRNA expression appeared more robust in the entire forebrain and formed distinct patterns in the VZ, SVZ, and CP, dorsally, and in the entire MGE and LGE, ventrally (Fig. 1I). Slit2 mRNA expression appeared sparse in the VZ and SVZ of the cortex, LGE, and MGE (Fig. 1J), but strong levels were evident in the septum, POA, and CH (Fig. 1J). Unlike Slit1, Slit2 mRNA was absent from the CP (Fig. 1I,J). Both Slit1 and Slit2 mRNA appeared to be expressed in the vasculature (Fig. 1I,J, arrows). Last, Slit3 mRNA showed scant expression and was largely absent from the proliferative zones dorsally and ventrally (data not shown).
GFP immunohistochemical analysis of Slit1+/− mice at E14.5 (Fig. 1M) showed comparable expression to Slit1 mRNA (Fig. 1I). Similar analysis in Slit2+/− mice at the same age revealed a thin, but distinct, band at the level of the SP (Fig. 1N). GFP+ cells were also noted in the cortical VZ and SVZ of both groups of mice and in the cerebral vasculature (Fig. 1M,N), in agreement with the mRNA expression patterns of Slit1 and Slit2.
Robo1 expression in the dorsal telencephalon at E14.5 was particularly robust in the CP and less so in the VZ/SVZ (Fig. 1K). Robo2 mRNA in the developing cortex was restricted to the lower region of the CP and the entire IZ (Fig. 1L). In the ventral telencephalon, Robo1 and Robo2 displayed equally strong expression patterns in the LGE and MGE (Fig. 1K,L). Robo3 mRNA expression was significantly down-regulated by E14.5, being only weakly expressed in the CP, in agreement with previous findings (Camurri et al., 2005; Barber et al., 2009), and totally absent by E16.5 (data not shown). Immunohistochemistry for Robo1 and Robo2 proteins generally confirmed the mRNA expression patterns in the dorsal telencephalon (Fig. 1O,P). Together, our expression patterns of Robo and Slit genes in the developing cortex show that Robo1 as well as Slit1 and Slit2 are expressed, among other areas, in the proliferative zones during early- and mid-phases of corticogenesis, suggesting that they may play a role in the generation of pyramidal neurons.
Robo1−/− mice display an increase in progenitors in the developing cortex
We have previously demonstrated a role for Robo1 in cortical interneuron generation and migration (Andrews et al., 2006; Hernández-Miranda et al., 2011). The observed expression of Robo1 mRNA and protein in the VZ of the developing cortex suggested that it might also play a role in the generation of pyramidal neurons. To assess this possibility, we first colabeled sections for PH-3, a marker of mitotically active cells, and Robo1 at E12.5 (Fig. 2). These sections showed the majority of PH-3-positive (PH-3+) cells lined the ventricular wall (presumptive apical progenitors), and nearly all expressed Robo1 (Fig. 2F, arrows). At E14.5 we observed similar colabeling for Robo1 protein and PH-3 in Robo1+/+ animals (Fig. 2G), but no staining for Robo1 in the cortices of Robo1−/− littermates (Fig. 2H), thus confirming the specificity of our Robo1 antibody. We then quantified the number of PH-3+ cells in the cortices of Robo1+/+ and Robo1−/− mice at E12.5, E14.5 (Fig. 3G), and E16.5 (data not shown). Specifically, we counted the number of PH-3+ apical progenitors that line the ventricular surface, and basal progenitors situated away from the ventricular surface, both of which give rise to pyramidal neurons (Miyata et al., 2004; Noctor et al., 2004).We performed similar counts in the proliferative zones of the GEs. At E12.5, we observed a significant increase in the number of PH-3+ apical progenitor cells in the cortex (Robo1−/−28.82 ± 1.29; Robo1+/+ 24.88 ± 1.41; p < 0.03) and MGE (Robo1−/− 27.04 ± 1.73; Robo1+/+ 24.29 ± 2.02; p < 0.05; Fig. 3G), and in basal progenitors in the cortex, MGE, and LGE of Robo1−/− mice compared with control littermates (n = 4 for each genotype; cortex: Robo1−/− 2.95 ± 0.21; Robo1+/+ 1.83 ± 0.20; p < 0.0008; LGE: Robo1−/− 3.57 ± 0.16; Robo1+/+ 2.58 ± 0.21; p < 0.0009; MGE: Robo1−/−3.10 ± 0.10; Robo1+/+ 2.44 ± 0.11; p < 0.0003; Fig. 3G).
Figure 3.
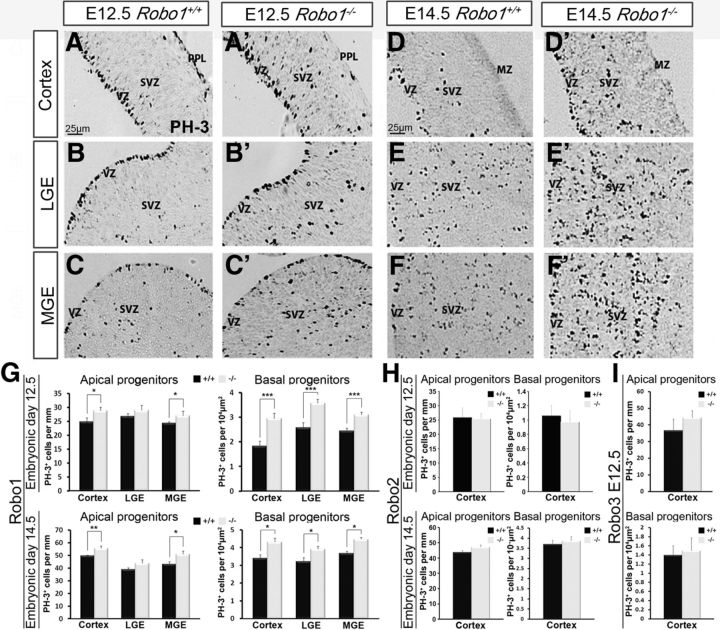
Increased PH-3-labeling in the telencephalon in Robo1−/−, but not Robo2−/− or Robo3−/− mice during corticogenesis. (A–F′) Images of coronal sections through the cortex (A, A′, D, D′), LGE (B, B′, E, E′) and MGE (C, C′, F, F′) at E12.5 (A–C′) and E14.5 (D–F′) showing mitotically active cells labeled with PH-3 in the VZ and SVZ of Robo1+/+ (A–F) and Robo1−/− (A′–F′) mice. G, Histograms indicate the presence of a significantly greater number of mitotically active apical and basal progenitors at E12.5 and E14.5 in Robo1−/− mice. H, Similar analysis in Robo2−/− mice at E12.5, E14.5, and in Robo3−/− mice at E12.5 (I) did not show any significant differences in PH-3-labeling of apical or basal progenitor cells in the dorsal cortex. Histograms show means and error bars represent SEM; *p < 0.05, **p < 0.007, ***p < 0.0009. MZ, Marginal zone.
At E14.5, the cortices of Robo1−/− mice exhibited a significant increase in PH-3+ apical (n = 4 for each genotype; Robo1−/− 55.65 ± 1.73; Robo1+/+ 49.57 ± 0.69; p < 0.002) and basal progenitor cells (Robo1−/− 4.32 ± 0.31; Robo1+/+ 3.39 ± 0.18; p < 0.01; Fig. 3G) compared with wild-type littermates. Within the ventral forebrain at E14.5, the increase in basal progenitors persisted both in the LGE (Robo1−/− 3.92 ± 0.19; Robo1+/+ 3.22 ± 0.19; p < 0.02) and MGE (Robo1−/− 4.47 ± 0.25; Robo1+/+ 3.68 ± 0.19; p < 0.02; Fig. 3G). In agreement with an earlier study (Andrews et al., 2008), we also found an increase in PH-3+ apical progenitors within the LGE (Robo1−/− 43.90 ± 2.70; Robo1+/+ 39.01 ± 1.21; p < 0.10) and MGE (Robo1−/− 51.03 ± 2.53; Robo1+/+ 43.01 ± 2.06; p < 0.02) at E14.5 (Fig. 3G). Interestingly, there was no significant increase in either apical or basal progenitor pools (cortex, LGE, MGE) in Robo1−/− mice compared with wild-type littermates at later stages of forebrain development (E16.5). This coincided with the observed decrease in Robo1 expression in the proliferative zones at this age (data not shown).
Although we failed to detect expression of either Robo2 or Robo3 receptor in the proliferative zones of the cortex, we cannot exclude that they are expressed at very low levels and that, like Robo1, they may play a role in the proliferation of pyramidal cell progenitors. However, PH-3+ cell counts in the proliferative zones of the cortices of Robo2−/− and Robo3−/− mice at E12.5 did not reveal significant changes in proliferation of either apical (n = 3 for each genotype; Robo2−/− 25.47 ± 1.84; Robo2+/+ 25.84 ± 3.46; p < 0.91; Robo3−/− 44.86 ± 4.15; Robo3+/+ 36.69 ± 6.69; p < 0.29) or basal (Robo2−/− 0.97 ± 0.15; Robo2+/+ 1.06 ± 0.14; p < 0.67; Robo3−/− 1.49±.029; Robo3+/+ 1.40 ± 0.20; p < 0.80) progenitor pools (Fig. 3H,I). Similarly, at E14.5, Robo2−/− mice did not display any significant changes in apical (Robo2−/− 47.14 ± 1.45; Robo2+/+ 43.82 ± 1.30; p < 0.07) or basal (Robo2−/− 3.86 ± 0.22; Robo2+/+ 3.69 ± 0.20; p < 0.60) progenitor cells (Fig. 3H). The reduction in Robo3 mRNA expression between early and middle stages of corticogenesis suggested that Robo3 does not play an active role in regulating cell proliferation during cortical development. Accordingly, PH-3+ cell counts at E15.5 in Robo3 knock-out mice and wild-type littermates showed no significant differences along the VZ or SVZ (data not shown).
To assess the possibility of receptor redundancy or antagonism, we quantified PH-3-labeling in Robo1−/−/Robo2−/− double-knock-out mice at E12.5 and E15.5 (Fig. 4A,B). Consistent with our data for Robo1−/− forebrain at E12.5, we observed an increase, albeit statistically insignificant, in basal progenitors in the cortex (n = 3 for each genotype; Robo1−/−/Robo2−/− 1.94 ± 0.18; Robo1+/+/Robo2+/+ 1.52 ± 0.09; p < 0.57), and in the LGE and MGE (LGE: Robo1−/−/Robo2−/− 2.30 ± 0.15; Robo1+/+/Robo2+/+ 1.91 ± 0.12; p < 0.10; MGE: Robo1−/−/Robo2−/− 2.23 ± 0.07; Robo1+/+/Robo2+/+ 1.80 ± 0.13; p < 0.56; Fig. 4A). In addition, apical progenitor cells along the cortex and LGE did not show a significant increase in Robo1−/−/Robo2−/− double-mutants compared with wild-type littermates at this stage (Fig. 4A). However, apical progenitor cells along the MGE did show a statistically significant increase in Robo1−/−/Robo2−/− double-mutants compared with wild-type littermates (Robo1−/−/Robo2−/− 20.45 ± 4.06; Robo1+/+/Robo2+/+ 17.52 ± 3.86; p < 0.001).
Figure 4.
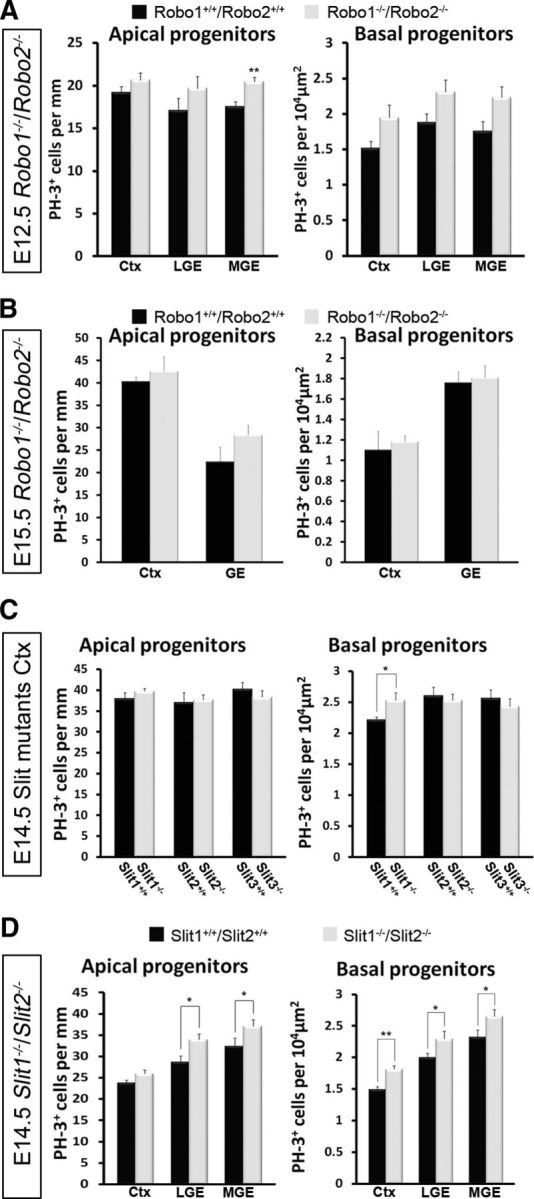
Increased PH-3-labeling in the cortices of Robo1−/−/Robo2−/− and Slit1−/− mice at E14.5. Histograms show number of PH-3+ apical and basal progenitor cells in Robo1−/−/Robo2−/− mice at E12.5 (A) and E15.5 (B), Slit single (Slit1−/−, Slit2−/−, and Slit3−/−) mutants (C) and Slit1−/−/Slit2−/− double mutants at E14.5 (D), compared with control littermates. A, At E12.5, histograms show statistically significant increases in apical progenitor cells in the MGE of Robo1−/−/Robo2−/− mice compared with control littermates. B, At E15.5, no significant increase in either progenitor cell population was observed. C, Similarly, no significant increases were found in Slit single mutants, with the exception of basal progenitors in the cortices of Slit1−/− mice. D, However, in Slit1−/−/Slit2−/− mice, an increase in apical and basal progenitors was observed in the cortex, LGE, and MGE; *p < 0.05, **p < 0.004.Ctx, Cortex.
At E15.5, Robo1−/−/Robo2−/− mice showed an increase, albeit statistically insignificant, in the cortex and GEs for both apical (n = 3 for each genotype; cortex: Robo1−/−/Robo2−/− 42.56 ± 3.31; Robo1+/+/Robo2+/+ 40.27 ± 1.01; p < 0.53; GE: Robo1−/−/Robo2−/− 28.35 ± 2.12; Robo1+/+/Robo2+/+ 22.38 ± 3.30; p < 0.16) and basal (n = 3 for each genotype; cortex: Robo1−/−/Robo2−/− 1.18 ± 0.05; Robo1+/+/Robo2+/+ 1.09 ± 0.18; p < 0.69; GE: Robo1−/−/Robo2−/− 1.81 ± 0.12; Robo1+/+/Robo2+/+ 1.76 ± 0.11; p < 0.76) progenitors (Fig. 4B), similar to that observed in E16.5 Robo1−/− mice. This observation suggests that loss of Robo2 function in Robo1−/−/Robo2−/− mice did not significantly alter the strength of the phenotype observed in Robo1−/− mice, pointing to the specific requirement for Robo1 and a redundant role for Robo2 in cortical proliferation events. This concurs with our observations in Robo2−/− mice, which indicated no significant differences in PH-3+ cell number compared with controls (Fig. 3H).
Increase in cortical progenitors in Slit1−/− and Slit1−/−/Slit2−/− double knock-out mice
We previously demonstrated weak expression of Slit mRNA in the telencephalon at E12.5, becoming stronger at E14.5, especially in the proliferative zones of the developing dorsal cortex (Fig. 1A,B,I,J). To determine whether the Robo1−/− phenotype is recapitulated in Slit mutants, we assessed the number of PH-3+ cells in the cortices of all three single Slit mutants (Slit1–3) at E14.5, when proliferation appears to be most affected in Robo1−/− mice. Interestingly, we only observed a small, but significant increase in basal progenitors in the cortices of Slit1−/− mice (n = 4 for each genotype; Slit1−/− 2.53 ± 0.12; Slit1+/+ 2.21 ± 0.05; p < 0.02), and no marked differences in either progenitor cell population in Slit2−/− or Slit3−/− animals (Fig. 4C).
Our analysis of Slit1−/−/Slit2−/− double knock-out mice at E14.5 uncovered a statistically significant increase in apical and basal progenitors in the LGE (n = 3 for each genotype: apical: Slit1−/−/Slit2−/− 33.93 ± 1.36; Slit1+/+/Slit2+/+ 28.67 ± 1.43; p < 0.01; basal: Slit1−/−/Slit2−/− 2.29 ± 0.12; Slit1+/+/Slit2+/+ 2.00 ± 0.06; p < 0.04) and MGE (n = 3 for each genotype; apical: Slit1−/−/Slit2−/− 37.05 ± 1.62; Slit1+/+/Slit2+/+ 32.37 ± 1.96; p < 0.04; basal: Slit1−/−/Slit2−/− 2.65 ± 0.10; Slit1+/+/Slit2+/+ 2.32 ± 0.11; p < 0.04). However, in the cortex, there was an increase only in basal progenitors (n = 3 for each genotype; Slit1−/−/Slit2−/− 1.81 ± 0.06; Slit1+/+/Slit2+/+ 1.49 ± 0.04; p < 0.002). Thus, it appears that removal of both Slit1 and Slit2 results in a slightly stronger phenotype compared with Slit1−/− mice, which suggests these two ligands are not redundant but, instead, are likely to interact synergistically (Fig. 4C,D). Also, the E14.5 Slit1−/−/Slit2−/− data are similar to that acquired from Robo1−/− single mutants at this age (Figs. 3G,4D), which strongly suggests that Slit1 and Slit2, in addition to Robo1, have a function in the regulation of the proliferative activity during the mid-stages of corticogenesis.
Our data support the notion that a Slit-mediated Robo signaling mechanism is involved in proliferation since we observed very similar phenotypes in both Slit and Robo mutants. Given that absence of Slit or Robo leads to increased proliferation, we were interested to know whether the converse was true, i.e., does the addition of Slit or Robo lead to a reduction in proliferation. To test this, we added recombinant Slit1, Slit2, Robo1-Fc, or Neuropilin-Fc (Nrp1), at concentrations which have been shown to affect proliferation of GE cells (Andrews et al., 2008), to E15.5 rat dissociated cortical cultures overnight. The next day, cultures were pulsed with BrdU for 2 h, fixed, and stained for BrdU and either Tbr2 or Pax6 to assess proliferation in the basal and apical progenitors pools, respectively. Addition of either Slit1 or Slit2 alone, did not have an effect on proliferation of either apical or basal progenitor populations (Fig. 5A,B). However, addition of Slit1 and Slit2 together (Slit1/Slit2), and Robo1-Fc resulted in a significant decrease in proliferation of both apical (control: 55.19%±0.69; Slit1/Slit2: 35.48%±0.70, p < 0.005; Robo1-Fc: 40.35%±0.90; p < 0.003) and basal (control: 39.15%±0.49; Slit1/Slit2: 25.16%±0.50, p < 0.004; Robo1-Fc: 30.75%±0.64, p < 0.003) progenitor pools, whereas addition of Nrp1-Fc had no effect (Fig. 5A,B).
Figure 5.
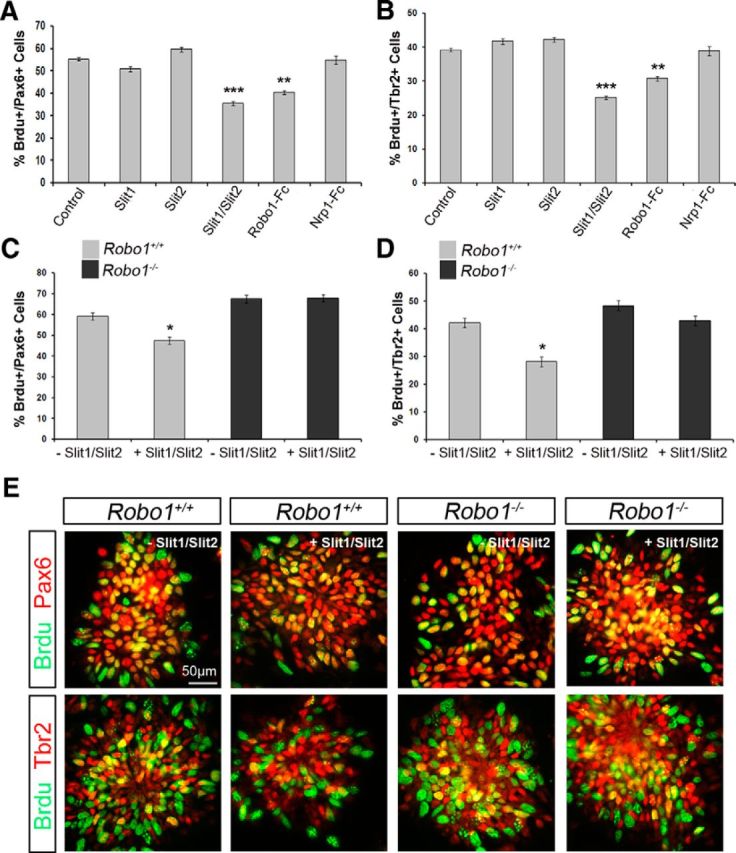
Reduced proliferation in Robo1+/+, but not in Robo1−/− dissociated cortical cell cultures following Slit1/Slit2 treatment. A, B, Histograms show percentage of BrdU+ E15.5 rat apical (Pax6+; A) and basal (Tbr2+; B) progenitor cells following treatment with either: Control, Slit1, Slit2, Slit1/Slit2, Robo1-Fc, or Nrp1-Fc overnight. Treatment with Slit1/Slit2 and Robo1-Fc caused a significant decrease in proliferation of both progenitor pools. C, D, Dissociated cortical cell cultures prepared from E13.5 Robo1+/+ and Robo1−/− mice were incubated overnight in the presence or absence of Slit1/Slit2, and the percentages of proliferating apical (C) and basal (D) progenitors were assessed following a 2 h BrdU pulse. Wild-type cultures showed a significant decrease in proliferation following Slit1/Slit2 treatment. Robo1−/− dissociated cortical cell cultures were unaffected by Slit treatment, but showed increased proliferation compared with wild-type cultures (E); *p < 0.05, **p < 0.004, ***p < 0.0004.
To confirm these findings, we tested the effects of exogenous Slit1/Slit2 on proliferation in E13.5 Robo1+/+ and Robo1−/− (n = 4) dissociated cortical cell cultures. Thus, addition of Slit1/Slit2 to Robo1+/+ cell cultures overnight caused a significant decrease in proliferation of apical (Robo1+/+-Slit: Pax6/BrdU 59.19% ± 1.68; +Slit 47.42% ± 1.06; p < 0.04) and basal progenitors (Robo1+/+-Slit: Tbr2/BrdU 43.03% ± 1.67; +Slit 33.21% ± 1.79; p < 0.04), which is in agreement with our rat cortical cultures of similar age (Fig. 5C–E). However, while we observed a significant increase in the level of proliferation of both progenitor populations in Robo1−/− cultures compared with Robo1+/+ in the absence of exogenous Slit protein, we did not note any alteration following the addition of Slit1/Slit2 proteins to either apical (Robo1−/−-Slit: Pax6/BrdU 67.61% ± 0.35; +Slit 67.88% ± 0.84; p < 0.87) or basal (Robo1−/−-Slit: Tbr2/BrdU 48.42% ± 1.87; +Slit 42.95% ± 1.78; p < 0.56) progenitor populations. Together, these findings clearly show a role for Slit1/Slit2-Robo1-mediated signaling transduction mechanism in regulating proliferation in cortical progenitor pools.
Interestingly, addition of Robo1-Fc to Robo1+/+ and Robo1−/− cortical cell cultures produced a decrease in the number of apical Pax6+/BrdU+ cells (Robo1+/+-Robo1-Fc: 57.62% ± 1.43; +Robo1-Fc 41.38% ± 1.18; p < 0.04; Robo1−/−-Robo1-Fc: Pax6/BrdU 67.61% ± 0.35; +Robo1-Fc 50.842% ± 0.48; p < 0.03) and basal Tbr2+/BrdU+ progenitor cell types (Robo1+/+-Robo1-Fc: 45.16% ± 1.87; +Robo1-Fc 35% ± 1.95; p < 0.04; Robo1−/−-Robo1-Fc: 54.61% ± 0.35; +Robo1-Fc 42.84% ± 0.48; p < 0.03). This suggests that Robo1-Fc may be acting independently of Robo1 function in these cells, possibly by interacting heterophilically with other Robo receptors (Liu et al., 2004; Camurri et al., 2005) or with other receptors, such as neuropilin-plexins (Hernández-Miranda et al., 2011).
Increased proliferation in Robo1−/− mice alters pyramidal neuron numbers
To assess whether the increased number of mitotically active progenitor cells in the cortex of Robo1−/− mice results in an increase in pyramidal neurons, we examined their number and distribution in the CP of Robo1−/− mice and wild-type littermates (n = 3, Robo1−/−; n = 3, Robo1+/+) using the specific pyramidal neuron marker, Emx1 (Chan et al., 2001). At E18.5, Robo1−/− mice displayed a significant increase in Emx1-positive cells in the CP compared with control mice (Robo1−/− 321.83 ± 24.19; Robo1+/+ 256.17 ± 10.95; p < 0.0005). There was also a concomitant increase in the thickness of the CP in Robo1−/− mice (Robo1−/− 440.23 μm ± 55.72; Robo1+/+ 369.70 μm ± 22.65; p < 0.0005). We then asked whether lack of Robo1 signaling differentially affected the number and distribution of one or more pyramidal neuron subtypes. According to the “inside-out” pattern of pyramidal neuron generation and cortical layer formation, early-born neurons occupy deeper layers while late-born cells are disposed more superficially (Angevine and Sidman, 1961; Rakic, 1974). Thus, we assessed the number of Cux1- and Ctip2-positive neurons, which are predominantly located in layers II/III and V/VI of the adult cortex, respectively (Arlotta et al., 2005; Chen et al., 2005; Cubelos et al., 2008, 2010). In the cortices of E18.5 Robo1−/− mice, Cux1-labeled cells were significantly increased compared with wild-type controls, both in the upper cortical plate (UCP) and throughout the thickness of the cortex (UCP: Robo1−/− 309.00 ± 67.80; Robo1+/+ 209.33 ± 33.02; p < 0.03; cortex: Robo1−/− 344.40 ± 26.81; Robo1+/+ 264.76 ± 30.35; p < 0.0005; Fig. 6A′,B′). Concurrently, Ctip2-labeled cells increased in the lower layers and throughout the thickness of Robo1−/− cortices compared with wild-type controls (Robo1−/− 182.08 ± 4.21; Robo1+/+ 156.00 ± 3.38; p < 0.0008; Fig. 6A,B). When the developing cortex, including the IZ, SVZ, and VZ, was divided into 10 equal bins, we were able to determine that the overall distribution of Cux1- and Ctip2-labeled pyramidal neurons was not altered in the absence of Robo1, despite an increase in the overall numbers of both subtypes (Fig. 6C,D). The observed increase in the thickness of the CP and the overall higher number of both upper- and lower-layer pyramidal neurons in the cortices of Robo1 knock-out mice at E18.5 contributed to an overall increase in the thickness of the entire cortex. Indeed, measurements in the region of the presumptive somatosensory cortex showed a small, but significant increase at E18.5 (n = 4 for each genotype; Robo1−/− 495.79 μm ± 11.73; Robo1+/+ 463.43 μm ± 6.28; p < 0.03).
Figure 6.
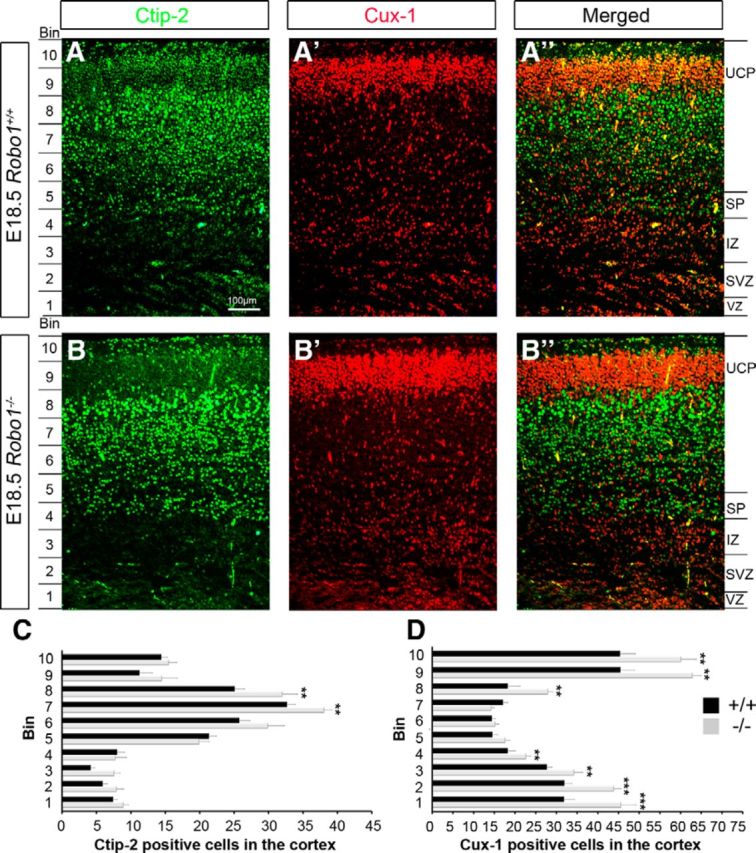
Increase in upper and lower layer pyramidal neurons in the cortices of Robo1−/− mice at E18.5. (A–B″) Immunohistochemistry in the cortex of Robo1+/+ (A–A″) and Robo1−/− (B–B″) mice for the early- (Ctip2, green; A, A″, B, B″), and late-born (Cux1, red; A′, A″, B′, B″) pyramidal neuron markers. Counts revealed a significant increase in both Ctip2+ and Cux1+ cells in the cortex of Robo1−/− mice. C, D, No distribution defects were observed despite the increase in cell density of either Ctip2+ or Cux1+ cells in Robo1 mutants; **p < 0.004, ***p < 0.0002.
The robust expression of Robo2 mRNA in the cortex during early and mid-corticogenesis (Fig. 1D,L) prompted us to examine the number and distribution of pyramidal neurons in Robo2−/− mice. Using Ctip2 and Cux1 to label early- and late-born neurons, respectively, we did not observe a significant change in either Cux1- (n = 3 for each genotype; Robo2−/− 238.58 ± 2.82; Robo2+/+ 244.40 ± 2.90; p < 0.17) or Ctip2-labeled (Robo2−/− 166.87 ± 2.14; Robo2+/+ 167.29 ± 2.53; p < 0.90) pyramidal neurons compared with wild-type littermates. In addition, there was no significant difference in the thickness of the cortex in Robo2−/− mice (Robo2+/+ 546.34 μm ± 4.33; Robo2−/− 541.44 μm ± 7.01; p < 0.58). Together with our PH-3 data, these results suggest that, contrary to a recent report (Borrell et al., 2012), Robo2 on its own does not play a role in the generation and disposition of cortical pyramidal neurons.
Cell cycle parameters are not altered in cortical progenitors in Robo1−/− mice
The process of self-renewal and neurogenesis performed by apical and basal progenitors during corticogenesis is tightly regulated. The observed increase in proliferation resulting in larger progenitor populations may be due to shortening of the cell cycle, causing cells to divide more rapidly in the absence of the Robo1 receptor, or to prolonged period of proliferation. To assess the proliferative activity of cortical progenitor cells in Robo1−/− mice and their wild-type littermates at E14.5 (n = 4, Robo1−/−; n = 4, Robo1+/+), animals were pulse-labeled with BrdU, a thymidine analog that becomes incorporated into DNA during S-phase of the cell cycle, for 2 h. Sections were colabeled with Ki67, a marker of proliferating cells from S-phase through M-phase of the cell cycle (Fig. 7A,B‴). This enabled us to quantify the S-phase fraction, or proliferative activity, determined by dividing the number of BrdU+ cells by the number of Ki67+ cells within the proliferative zones (VZ and SVZ) as described by Tanaka et al. (2011). Further, the same ratio was used to determine the percentage of cells exiting S-phase by pulse-labeling embryos for 24 h at E13.5 and colabeling coronal sections with Ki67.
Figure 7.
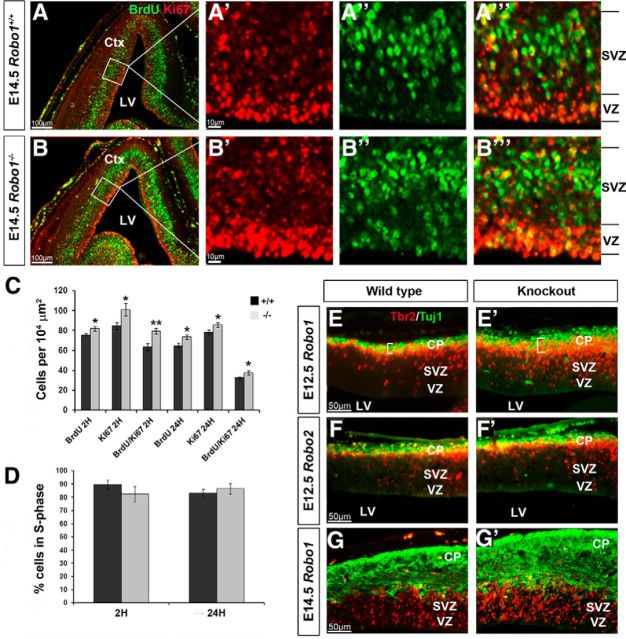
Increased expression of proliferation markers in the dorsal cortices of Robo1−/− mice at E14.5. (A–B‴) Immunohistochemistry in the cortices of Robo1+/+ (A–A‴) and Robo1−/− (B–B‴) mice for BrdU (green) and Ki67 (red). C, D, Counts revealed a significant increase in BrdU+, Ki67+, and BrdU+/Ki67+ cells in the proliferative zones in Robo1−/− mice after a 2 and 24 h BrdU pulse (C), and no significant differences in the percentage of cells in S-phase (D). E–G′, Immunohistochemistry in the cortices of Robo1+/+ (E, G) and Robo1−/− (E′,G′) mice at E12.5 (E, E′) and E14.5 (G, G′), and of Robo2+/+ (F) and Robo2−/− (F′) mice at E12.5, for the basal progenitor marker Tbr2 (red) and immature neuronal marker Tuj1 (green); *p < 0.04, **p < 0.004. Ctx, Cortex; LV, lateral ventricle.
In the VZ/SVZ zones of the Robo1−/− cortex, we observed significantly more BrdU+ cells after 2 h compared with wild-type controls (Robo1−/− 81.83 ± 2.37; Robo1+/+ 75.00 ± 1.77; p < 0.04; Fig. 7A″,B″,C). These animals also showed a significant increase in the number of Ki67+ cells (Robo1−/− 101.00 ± 6.13; Robo1+/+ 84.43 ± 3.39; p < 0.03; Fig. 7A′,B′,C) and in the number of double-positive cells (BrdU+/Ki67+; Robo1−/− 79.33 ± 2.84; Robo1+/+ 63.71 ± 3.26; p < 0.005; Fig. 7C). However, the ratio of BrdU+/Ki-67+-positive cells in Robo1−/− cortex was slightly lower, but not significantly different from wild-type littermates (Robo1−/− 0.8255 ± 0.0566; Robo1+/+ 0.8953 ± 0.0349; p < 0.30) suggesting that, although there is a general increase in the number of dividing cells in Robo1−/− mice, there is no change in the length of the S-phase of the cell cycle (Fig. 7D).
Our observations at E14.5 after a 24 h BrdU pulse further reinforced our results obtained from Robo1−/− 2 h pulse-labeled sections, showing a significant increase in BrdU+ cells (Robo1−/− 73.67 ± 2.19; Robo1+/+ 64.89 ± 2.46; p < 0.02), Ki67+ cells (Robo1−/− 85.67 ± 2.18; Robo1+/+ 78.22 ± 2.26; p < 0.03), and BrdU+/Ki67+ cells (Robo1−/− 37.67 ± 2.02; Robo1+/+ 32.56 ± 0.97; p < 0.04) in Robo1 knock-out mice compared with wild-type littermates (Fig. 7C). Using the same ratio to obtain the S-phase fraction after a 2 h BrdU pulse, we were able to calculate the percentage of progenitor cells in the VZ and SVZ exiting from the cell cycle over a 24 h period. Our results indicate that Robo1−/− mice exhibit a slightly higher, albeit insignificant, percentage of progenitors exiting S-phase (Robo1−/− 86.55% ± 3.85; Robo1+/+ 83.20% ± 2.99; p < 0.50; Fig. 7D). This is in line with our previous observations, which indicated that there is no significant change in the length of the cell cycle of progenitor cells at E14.5 in the Robo1−/− cortex.
Typically, basal progenitors in the dorsal cortex will terminally divide to produce newborn neurons (Haubensak et al., 2004; Noctor et al., 2004; Huttner and Kosodo, 2005). We have observed a substantial increase in the number of progenitor cells in early (E12.5) and middle (E14.5) stages of corticogenesis. To further reinforce our hypothesis that cortical progenitors in Robo1−/−, but not Robo2−/− knock-out mice, undergo a prolonged period of proliferation compared with wild-type controls independently of altered cell cycle parameters, we performed colabeling experiments for the newborn neuronal marker Tuj1 and Tbr2, which label basal progenitor cells in Robo1−/−, Robo2−/− and control littermates at E12.5 and E14.5 (Fig. 7E–G′). At E12.5, we observed a statistically significant increase in Tbr2+ cells (Robo1−/− 17.46 ± 0.70; Robo1+/+ 12.81 ± 0.48; p < 0.0002) and thickness of Tuj1 labeling (Robo1−/−25.07 μm ± 0.02; Robo1+/+ 19.93 μm ± 0.56; p < 0.0004) in the cortex of Robo1−/− mice compared with wild-type littermates (Fig. 7E,E′). However, no difference was observed in either Tbr2+ cells (Robo2−/− 13.52 ± 0.27; Robo2+/+ 13.55 ± 0.41; p < 0.95) or Tuj1 thickness (Robo2−/− 20.36 μm ± 0.31; Robo2+/+ 19.61 μm ± 0.47; p < 0.18) in Robo2−/− mice during early forebrain development (Fig. 7F,F′). As expected, the increase in basal progenitors and immature neurons persisted at E14.5 in Robo1−/− knock-out mice (Fig. 7G,G′), indicating that progenitor cells continue to proliferate, creating a larger progenitor pool and thus, more neurons in the cortex.
In light of cell cycle parameters playing no or only a marginal role in the increased proliferative activity in Robo1-null mice in early and middle stages of cortical development, we next investigated the role of programmed cell death (PCD) in the formation of cortical pyramidal neurons. Previous studies have linked PCD with a morphogenetic function during the development of the CNS (Thomaidou et al., 1997). Immunohistochemical staining of the dorsal telencephalon at E12.5–E18.5 for the apoptotic marker CC3 revealed significantly fewer labeled cells in the cortex of Robo1−/− mice compared with wild-type littermates (n = 3 for both groups at all ages) in the early stages of corticogenesis. Thus, at E12.5 (Robo1−/− 2.72 ± 0.15; Robo1+/+ 3.69 ± 0.05; p < 0.03), E13.5 (Robo1−/− 3.4 ± 0.06; Robo1+/+ 4.08 ± 0.04; p < 0.046), and E14.5 (Robo1−/− 4.05 ± 0.16; Robo1+/+ 5.46 ± 0.19; p < 0.032) we observed fewer CC3+ cells in the cortices of Robo1−/− mice than in control littermates (Fig. 8A–F,H). However, at the later stages of E15.5 (Robo1−/− 7.45 ± 0.06; Robo1+/+ 7.35 ± 0.05; p < 0.71) and E18.5 (Robo1−/− 16.32 ± 0.121; Robo1+/+ 17.22 ± 0.14; p < 0.73,) there were no significant differences between the two groups (Fig. 8A–F,H).
Figure 8.
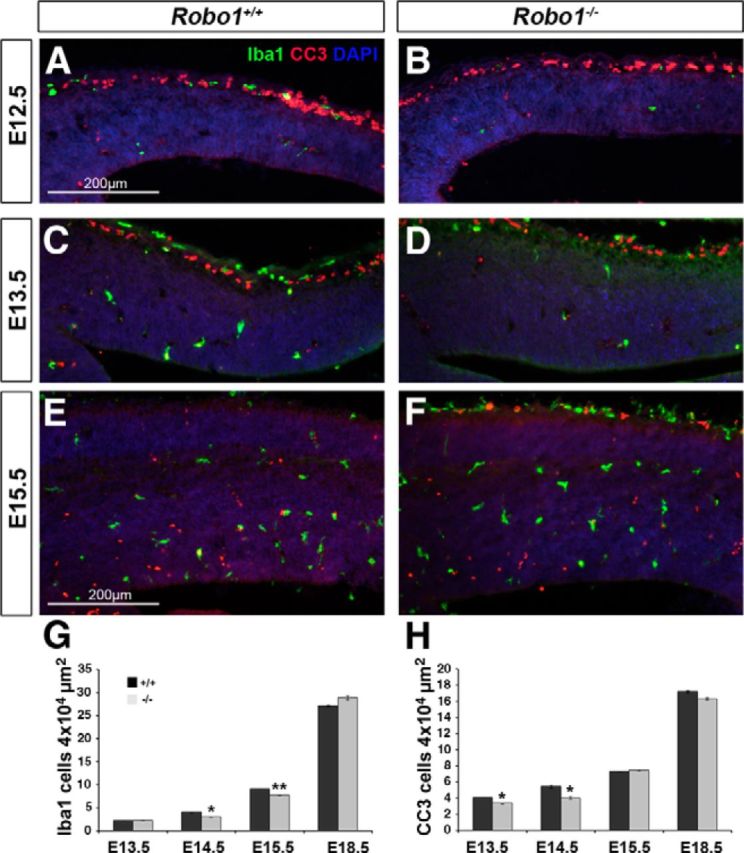
Reduced number of microglia and apoptotic cells in early and middle stages of corticogenesis in Robo1−/− mice. A–F, Immunohistochemistry in the cortices of Robo1+/+ (A, C, E) and Robo1−/− (B, D, F) mice for Iba1 (green) and CC3 (red) at E12.5 (A, B), E13.5 (C, D), and E15.5 (E, F). G, H, Counts revealed significantly reduced numbers of Iba1- (G) and CC3-positive (H) cells in the cortices of Robo1−/− at E13.5–E15.5 compared with control littermates; *p < 0.04, **p < 0.004.
Recent work by Cunningham et al. (2013) have provided compelling evidence that microglia play a fundamental role in regulating the size of the progenitor pool in the developing cerebral cortex. Specifically, these authors have demonstrated that the presence of microglia in the cortical proliferative zone results in a reduction in the size of the progenitor cell pool, whereas elimination of microglia has the opposite effect on the number of progenitors. Thus, we examined sections taken from the cortices of Robo1−/− mice and the corresponding controls at E12.5–E18.5 and stained for Iba1, a marker of microglia (n = 3 for both groups at all ages). These sections revealed relatively few microglia before E14.5, making it difficult to quantify accurately (Fig. 8A–F,G). However, at E14.5 (Robo1−/− 3.05 ± 0.06; Robo1+/+ 4.1 ± 0.09; p < 0.04) and E15.5 (Robo1−/− 7.71 ± 0.06; Robo1+/+ 27.43 ± 0.27; p < 0.006), we observed a small, but significant decrease in the number of microglia in the cortex of Robo1−/− mice compared with control littermates, but not at E18.5 (Robo1−/− 28.9 ± 0.44; Robo1+/+ 27.15 ± 0.19; p < 0.285). Together, these studies demonstrated that the increased presence of progenitor cells in the developing cortex and the generation of higher number of neurons in the absence of Robo1 may be due, at least in part, to a lower incidence of PCD and reduced number of microglia during mid-stages of corticogenesis.
The effect of Robo1 on proliferation is cell autonomous
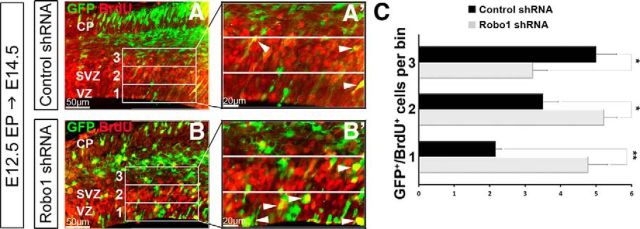
To determine whether the effect of loss of Robo1 function on proliferation is cell autonomous, we introduced Robo1 small hairpin RNA (shRNA) by in utero electroporation into cells along the VZ at E12.5 and killed the dam 48 h later (Gonda et al., 2013). To assess proliferation, BrdU was administered 2 h before kill. We speculate that Robo1 knockdown progenitor cells in the cortex could continue to proliferate longer than control progenitors, thus remaining in the VZ and SVZ and ultimately giving rise to a greater number of pyramidal neurons. To test this hypothesis, we quantified the number of electroporated cells (GFP+), which were also BrdU+ (GFP+/BrdU+) within the VZ/SVZ (Fig. 9A–C). To further assess their position within the VZ/SVZ, we divided the proliferative zone of the dorsal cortex into three 50-μm-thick bins, each spanning 200 μm (Fig. 9A′,B′). We found significantly more Robo1 knockdown cells colabeled with BrdU in Bin 1 (n = 3 for each construct; Robo1shRNA: 4.78 ± 0.54; control shRNA: 2.16 ± 0.16; p < 0.002) and Bin 2 (Robo1shRNA: 5.22 ± 0.36; control shRNA: 3.50 ± 0.01; p < 0.01), and fewer in the third bin (Robo1shRNA: 3.22 ± 0.40; control shRNA: 5.00 ± 0.03; p < 0.01) compared with control knockdown cells (Fig. 9C). This suggests that progenitor cells that lack the Robo1 receptor continue to proliferate for a longer period in the lower part of the VZ/SVZ than progenitor cells electroporated with the control construct.
Figure 9.
Increase in proliferative activity in the absence of Robo1 is cell autonomous. (A–B′) Immunohistochemistry in the dorsal cortex of C57BL/6J mice at E14.5, 48 h after in utero electroporation of either control shRNA (A, A′) or Robo1shRNA (B, B′) for GFP+ (green, electroporated) and BrdU+ (red) cells, following a 2 h BrdU pulse before kill. C, Histogram shows a significant increase in proliferating Robo1 shRNA electroporated cells (GFP+/BrdU+) present in lower bins within the VZ and SVZ of the cortex compared with control shRNA electroporated embryos (A′, B′); *p < 0.05, **p < 0.004.
Similar results were obtained in dissociated cortical cell cultures of Robo1−/− and wild-type littermates at E14.5 (n = 4, Robo1−/−; n = 4, Robo1+/+). These cultures were pulsed with BrdU for 2 h before labeling for apical (Pax6+) and basal (Tbr2+) progenitor cell markers. After quantifying the percentage of Pax6+ and Tbr2+ cells colabeled with BrdU, we found an increase in the number of BrdU-positive apical (Pax6+; Robo1−/− 61.97% ± 0.09; Robo1+/+ 49.00% ± 0.16; p < 0.005) and basal progenitors (Tbr2+; Robo1−/− 35.54% ± 0.08; Robo1+/+ 23.64% ± 0.28; p < 0.03) compared with control cultures. In experiments performed in dissociated cortical cell cultures from Robo2−/− and wild-type littermates at E14.5 (n = 5, Robo2−/−; n = 3, Robo2+/+), we found no significant differences in the number of BrdU-positive apical (Robo2−/− 57.98% ± 0.83; Robo2+/+ 53.63% ± 0.63; p < 0.089) or basal progenitors (Robo2−/− 39.21% ± 0.74; Robo2+/+ 37.93% ± 0.54; p < 0.527) compared with control cultures. These results further suggest a cell-autonomous role for Robo1, and not Robo2, in regulating the proliferative activity of progenitor cells in the cortical VZ and SVZ.
Postnatal ages
A recent study by Gonda et al. (2013) reported a reduction in cortical thickness, especially of layers II/III, but no alterations in the number of Cux1-positive cells in the cortices of adult Robo1−/− mice compared with controls. This phenotype differs from our findings at the end of corticogenesis (E18.5), which included significantly increased cortical thickness and markedly increased number of pyramidal neurons of both upper (Cux1+) and lower layers (Citip2+). This would suggest that significant alterations take place postnatally in both the number and disposition of pyramidal neurons in Robo1−/− mice compared with controls.
We first set out to confirm the observations of Gonda et al. (2013) by examining the cortices of adult animals (P56). Specifically, we found a small, but significant decrease in the thickness of the cortex in Robo1−/− mice compared with wild-type controls (Robo1−/− n = 4; 1169.26 μm ± 12.66; Robo1+/+ n = 3; 1223.69 μm ± 16.32; p < 0.02). Further, using subtype-specific markers, Cux1 and Ctip2, we noted no significant decrease in the overall number of upper- or lower-layer pyramidal neurons, respectively (Cux1:; Robo1−/− 254.00 ± 12.79; Robo1+/+ 268.16 ± 14.16; p < 0.49; Ctip2: Robo1−/− 166.00 ± 3.49; Robo1+/+ 175.67 ± 5.12; p < 0.11). Instead, there was a decrease in the thickness of layers II/III in mice lacking Robo1 (Robo1−/− 434.33 μm ± 7.00; Robo1+/+ 524.33 μm ± 28.61; p < 0.03), but no significant change in layers IV/V (Robo1−/− 604.48 μm ± 13.19; Robo1+/+ 640.87 μm ± 12.40; p < 0.06).
To assess when during the first postnatal 8 weeks the observed phenotypic changes between the late embryonic and adult stages occurred, we analyzed cortical thickness, apoptosis, and the number of neurons in layers II/III (Cux1+) at two stages in the first 3 weeks of postnatal life. At P21, similar to the adult, we observed no differences in the number of Cux1+ cells in layers II/III of Robo1−/− mice compared with wild-type littermates (Robo1−/− 326.00 ± 14.00; Robo1+/+ 320.00 ± 6.85; p < 0.69), but we found a significant decrease in the thickness of the layers (Robo1−/− 418.83 μm ± 13.32; Robo1+/+ 488.98 μm ± 16.89; p < 0.02). However, at P8, we found no significant difference in overall cortical thickness between Robo1−/− mice and wild-type littermates (Robo1−/− 1086.28 μm ± 13.54; Robo1+/+ 1089.84 μm ± 25.90; p < 0.90), yet the significant decrease in the thickness of layers II/III noted in the adults was already evident (Robo1−/− 379.71 μm ± 8.19; Robo1+/+ 406.44 μm ± 4.70; p < 0.01). This suggests that the observed change in phenotype between late embryonic stage and the adult is most likely occurring at around the start of the second postnatal week. Interestingly, we found an almost double the number of CC-3-positive cells in the cortices of Robo1−/− mice compared with their wild-type littermates at this time (P8; Robo1−/− 53.00 ± 2.46; Robo1+/+ 28.00 ± 2.01; p < 0.0002), suggesting that increased cell death contributes, at least in part, to the reduction in the number of pyramidal neurons and decrease in cortical thickness, especially of the upper cortical layers, in postnatal life.
Robo1 regulates activity of genes involved in proliferation, apoptosis, and tumorogenesis during development
In an attempt to identify novel downstream signaling cascades for Robo1 to regulate proliferation and apoptosis within the neurogenic zones of the cortex, we performed a microarray analysis of RNA extracted from VZ/SVZ of Robo1+/+ and Robo1−/−littermates (n = 3 for both groups) at E12.5 using laser capture microscope dissection. Our analyses identified 312 genes that were differentially expressed in the cortical proliferative zones, which may have a role in neurogenesis. Genes were selected only if they showed a fold-change >2, with a corrected p < 0.05.
Validation by qPCR was performed at E14.5, when the proliferative effect in the absence of Robo1 was the strongest (Table 1). In addition to the genes selected from our microarray analysis, we performed qPCR for additional genes known to be involved in proliferation (Ki67, Pax6, Zfp423) and apoptosis (Bcl2, Casp2, Casp8, Top2a, and Tpx2). QPCR analysis for Ki67 (3.03 ± 1.24), Pax6 (4.95 ± 1.58), and Zfp423 (2.35 ± 0.47) showed positive fold-changes, whereas Bcl2 (−4.24 ± 1.96), Casp2 (−3.31 ± 0.06), Casp8 (−2.18 ± 0.43), Top2a (−3.50 ± 0.80), and Tpx2 (−3.19 ± 0.61) revealed negative fold-changes, indicating a general increase in expression of proliferation genes and a concomitant decrease in apoptotic genes. These results corroborated our immunohistochemistry experiments, which showed increased numbers of Ki67+ and Pax6+ cells and reduced numbers of apoptotic (CC-3+) cells in the cortex of Robo1−/− mice.
Table 1.
| Gene name | Symbol | RefSeq | Fold-change array | Fold-change qPCR | Function | Reference |
|---|---|---|---|---|---|---|
| Caspase 2 | Casp2 | NM_007610 | −2.86 | −2.4 | Role in apoptosis | Li et al., 1997 |
| Hairy/enhancer-of-Split related with YRPW motif 1 | Hey1 | NM_010423 | −2.23 | −4.22 | Negatively regulates neuronal bHLH genes, promote maintenance of neural precursor cells | Sakamoto et al., 2003 |
| Integrin α-6 | Itga6 | NM_030691 | −2.38 | −7.07 | Involved in tumorigenicity | Cariati et al., 2008 |
| Integrin α-9 | Itga9 | NM_133721 | −2.006 | −4.04 | Downregulated or lost in 44% of breast tumors | Mostovich et al., 2011 |
| Notch gene homolog 2 (Drosophila) | Notch2 | NM_010928 | −2.29 | −1.38 | Negative regulator of glial differentiation in mammalian brain development | Tanaka et al., 1999 |
| Nuclear transcription factor-Y gamma | Nfyc | NM_008692 | −2.82 | −1.26 | Role in Hox gene regulation | Deng et al., 2007 |
| RB1-inducible coiled-coil 1 | Rb1cc1 | NM_009826 | −4.06 | −2.67 | Candidate human tumor suppressor gene and a modulator of TGF-β signaling | Koinuma et al., 2011 |
| Roundabout homolog 1 (Drosophila) | Robo1 | NM_019413 | −7.81 | −8.4 | Involved in interneuron generation and migration | Andrews et al., 2006; Andrews et al., 2008 |
| Transforming growth factor, β-2 | Tgfb2 | NM_009367 | −2.28 | −1.35 | Proliferation and differentiation of hippocampal granule neurons | Lu et al., 2005 |
| Zinc-finger protein 423 | Zfp423 | NM_033327 | 2.63 | 2.34 | Coordinates Notch and bone morphogenetic protein signaling | Masserdotti et al., 2010 |
Recently, Borrell et al. (2012) demonstrated an association between Robo2 and members of the Notch family of genes. In our microarray analysis, we also observed differential expression of members of this family, including Notch2 and Hey1, which were significantly downregulated in Robo1−/− compared with Robo1+/+ controls. This was confirmed by qPCR at E14.5, which showed negative fold-changes for Notch2 (−1.38 ± 0.55) and Hey1 (−4.22 ± 0.06).
In addition, our microarray analysis and qPCR showed that members of the Integrin family of cell-surface proteins, Itga6 and Itga9, were downregulated in Robo1−/− mice at E14.5 (Table 1). Disruption in the expression and function of these genes has been implicated in the growth and spread of different types of breast cancers (Cariati et al., 2008; Mostovich et al., 2011). Likewise, Tgfβ2, which has been identified as a key factor in the progression and suppression of malignant gliomas (Hau et al., 2011), showed a negative fold-change in both microarray analysis and qPCR at E14.5 (Table 1). This is important not only because it reinforces the importance of Robo1 in governing proliferative activity, but because it suggests that mechanisms involved in cancer growth or suppression may also play a role in proliferation during cortical development.
Discussion
The maintenance of the proportions of pyramidal neurons and interneurons is essential for normal cortical function. Disruption of this delicate ratio may result in wide-ranging neurological and cognitive defects. Thus, the previously reported increase in the number of interneurons in prenatal and postnatal mice lacking Robo1 (Andrews et al., 2006; Hernández-Miranda et al., 2011) disrupts this balance, with hitherto unknown functional and behavioral deficits, and brings into question the role of this receptor in the development of their pyramidal counterparts.
Cortical pyramidal neurons arise from RG cells that line the dorsal aspect of the lateral ventricles. Throughout corticogenesis, these cells migrate radially from the VZ toward the pial surface before taking up positions in the CP in a temporally guided inside-out manner (Rakic, 1990). There has been considerable interest over the years on the molecular mechanisms that underlie their radial migration (Nadarajah et al., 2001; Marín and Rubenstein, 2003; Jossin, 2004). However, more recently, attention has shifted to cortical interneurons, and an ever-increasing list of molecules that play important roles in their generation, differentiation, and migration has come to light, including Robo1 (Faux et al., 2010; Hernández-Miranda et al., 2011). In experiments aimed to understand how mice lacking this receptor show increased number of cortical interneurons, we used proliferation markers and found that this phenotype is, at least in part, due to increased proliferation in the MGE, thus implicating Robo1 in the process of proliferation in the forebrain. Abundant evidence already exists in other systems, which points to a role for Slit-Robo signaling in cell proliferation (Sundaresan et al., 1998; Xian et al., 2001, 2004). The aim of the present study was to investigate whether Robo1 is also involved in the proliferation of pyramidal neuron progenitors, as any changes in their generation are likely to have an effect on the laminar and functional organization in the cortex.
We first used in situ hybridization to expand previous studies performed in rat (Marillat et al., 2002) and encompass Robo1/2/3 as well as Slit1/2/3 mRNA expression throughout the developing mouse forebrain. More importantly, these experiments revealed the presence of Robo1 in progenitor cells lining the ventricular surface of the cortex and GEs as early as E12.5. This observation is compatible with the notion that Robo1 influences the proliferative activity of MGE cells that generate cortical interneurons (Andrews et al., 2008). The presence of Robo1 in RG cells in the ventricular surface of the dorsal telencephalon and the observed increase in PH-3-labeled cells in this zone and in the SVZ at early and middle stages of corticogenesis (E12.5 and E14.5) implicate Robo1 in the generation of cortical pyramidal neurons. Robo1 expression was not detected in the proliferative zones of the developing cortex as corticogenesis progressed through to the later stages. Interestingly, we found that in Robo1−/−/Robo2−/− double-mutants, the increase in basal progenitors at E12.5 was less than in Robo1−/− mice, suggesting that Robo2 did not compensate for the loss of Robo1 signaling and is likely to have a redundant role in cortical proliferation events. This is consistent with our observation that neither Robo2 nor Robo3 were expressed in the proliferative zones at E12.5 or E14.5, nor were there any significant changes in mitotically active cells in Robo2 or Robo3 knock-out mice.
The present findings differ significantly from results of a very similar study on the role of Slit/Robo signaling in progenitor cell proliferation in the developing cortex (Borrell et al., 2012). First, these authors reported, using in situ hybridization and immunohistochemistry, expression of Robo2 and barely detectable Robo1 in the VZ at E12.5. Using the same techniques, we did observe Robo1 expression in cells lining the telencephalic ventricle at this age, but failed to localize Robo2 and Robo3 in this proliferating cell population at the same age. We can only suggest that differences in the protocols and antibodies used by the two groups could account for the different results. Second, Borrell et al. (2012) reported a decrease in the density of dividing VZ cells (apical progenitors), but an abnormal increase in basal progenitors at the early stages (E11.5 and E12.5) of corticogenesis in Robo1−/−/Robo2−/− mutants; these phenotypic changes were primarily due to the loss of Robo2. In contrast, using single- (Robo1, Robo2, Robo3) and double-mutants(Robo1−/−/Robo2−/−), we found a significant increase in both apical and basal progenitors during early and middle stages of corticogenesis only in the absence of Robo1; analysis of Robo1−/−/Robo2−/− mutants showed a milder phenotype than Robo1−/− mice. It may be suggested that the contrasting results reported by the two groups are due to differences in the genetic strains and background of the animals. It has been reported that the Robo knock-out strains used in the Borrell et al. (2012) study are hypomorphs (Long et al., 2004), whereas our stains are complete nulls (Lu et al., 2007; Andrews et al., 2008). Further, Robo1−/−/Robo2−/− mutants used here were raised in a different background than those reported in the study of Borrell et al. (2012). Importantly, our finding of increased proliferation in cortical progenitor cells in the absence of Robo1 function was further confirmed in dissociated cortical cell cultures taken from Robo1−/− mice and wild-type littermates at E14.5 and by in utero electroporation of RNA interference and control constructs into cells in the VZ at E12.5.
Unlike earlier ages, PH-3-positive cells did not show a significant increase in Robo1-null mice at E16.5. However, the VZ was thicker in these mice compared with wild-type littermates as well as at the end of corticogenesis (E18.5) when visualized in Emx1- and Cux1-stained sections. We may venture to suggest that this could be due to delay in the onset of migration of the late generated upper-layer pyramidal neurons as demonstrated in a recent study by Gonda et al. (2013).
In view of the increase in both apical and basal progenitors in early- and mid-corticogenesis, we hypothesized that pyramidal neuron numbers must be affected. We used markers of pyramidal neuron subtypes, and found that both upper- and lower-layer populations increased in the cortices of Robo1-null mice at E18.5. Further, the CP was thicker in these animals. Similar experiments performed on Robo2 knock-out mice at this age did not show any difference in the number and disposition of pyramidal neurons. The observations that loss of Robo1 function leads to an increase in both cortical pyramidal cells and interneurons indicates that its role in cell proliferation is at a level different from that of neuronal specification and differentiation.
The presence of larger progenitor populations in Robo1−/− mice may be due to changes in cell cycle time or to prolonged period of proliferation. We assessed proliferative activity, defined as the S-phase fraction of actively cycling cells, by using BrdU in conjunction with Ki-67 as described by Tanaka et al. (2011). These experiments showed no statistical differences in the length of S-phase of the cell cycle, nor in the number of progenitors exiting S-phase in mice lacking Robo1 compared with wild-type controls. However, it is possible that a small change in cell-cycle parameters could have a large effect on cell output.
To test the hypothesis that cortical progenitor cells undergo a prolonged period of proliferation in Robo1 knock-out animals we used Tuj1, a marker of young neurons, and Tbr2. We found that the increase in basal progenitors in the early and middle stages of corticogenesis was accompanied by concomitant increase in young cortical neurons in Robo1, but not in Robo2, knock-out mice. This experiment clearly indicated that removal of Robo1 function results in a larger progenitor pool in the proliferative zones and, consequently, more neurons in the cortex at the end of corticogenesis. However, the presence of a larger progenitor pool in these animals may be due, at least in part, to the observed reduction in cell death in these animals. Previous studies have highlighted the importance of programmed cell death in shaping the cortex throughout development (Thomaidou et al., 1997; Haydar et al., 1999). Alternatively, loss of Robo1 function could lead to perturbations in downstream signaling cascades (Ypsilanti et al., 2010), which could potentially result in altered cell proliferation as observed in Robo1−/− mice. Indeed, our microarray analysis seems to support a role for a number of proliferation and apoptotic genes in Robo signaling events during corticogenesis.
In summary, we report here that the majority of progenitor cells lining the dorsal telencephalic ventricle express Robo1, but not other members of the Robo receptor family. Absence of this receptor leads to increase in the size of progenitor pools in the developing cortex and, consequently, to increased production of pyramidal neurons during corticogenesis. The increase in progenitor pools is likely due to prolonged period of proliferation resulting in delay in the onset of migration of pyramidal neurons and altered lamination, especially of the supragranular layers. However, reduction in apoptotic cells and microglia in the early stages of corticogenesis may also contribute to the observed increase in progenitor cells. These results, together with our earlier finding of significant increase of interneurons in the cortices of Robo1−/− mice, implicate Robo1 in the regulation of progenitor cell dynamics in the developing forebrain, and suggest that the same Robo1 signal transduction mechanisms may operate within the dorsal and ventral telencephalon during neurogenesis.
Footnotes
The work was supported by a Wellcome Trust Programme Grant (089775) to J.G.P. and W.D.A., and by Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to C.H. We thank Drs Alain Chedotal and Pavol Zelina (Institut de la Vision, Paris) for providing the brains from Robo3−/−, Robo1−/−/Robo2−/−, and Slit3−/− mice used in the present study, and Mary Rahman for technical assistance.
The authors declare no competing financial interests.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution and reproduction in any medium provided that the original work is properly attributed.
References
- Andrews W, Liapi A, Plachez C, Camurri L, Zhang J, Mori S, Murakami F, Parnavelas JG, Sundaresan V, Richards LJ. Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development. 2006;133:2243–2252. doi: 10.1242/dev.02379. [DOI] [PubMed] [Google Scholar]
- Andrews W, Barber M, Hernadez-Miranda LR, Xian J, Rakic S, Sundaresan V, Rabbitts TH, Pannell R, Rabbitts P, Thompson H, Erskine L, Murakami F, Parnavelas JG. The role of Slit-Robo signaling in the generation, migration and morphological differentiation of cortical interneurons. Dev Biol. 2008;313:648–658. doi: 10.1016/j.ydbio.2007.10.052. [DOI] [PubMed] [Google Scholar]
- Angevine JB, Jr, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Bagri A, Marín O, Plump AS, Mak J, Pleasure SJ, Rubenstein JL, Tessier-Lavigne M. Slit proteins prevent midline crossing and determine the dorsoventral position of major axonal pathways in the mammalian forebrain. Neuron. 2002;33:233–248. doi: 10.1016/S0896-6273(02)00561-5. [DOI] [PubMed] [Google Scholar]
- Barber M, Di Meglio T, Andrews WD, Hernandez-Miranda LR, Murakami F, Chédotal A, Parnavelas JG. The role of Robo3 in the development of cortical interneurons. Cereb Cortex. 2009;19:i22–31. doi: 10.1093/cercor/bhp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas SL, Gleeson JG. Cytoskeletal-associated proteins in the migration of cortical neurons. J Neurobiol. 2004;58:149–159. doi: 10.1002/neu.10280. [DOI] [PubMed] [Google Scholar]
- Borrell V, Cárdenas A, Ciceri G, Galcerán J, Flames N, Pla R, Nóbrega-Pereira S, García-Frigola C, Peregrín S, Zhao Z, Ma L, Tessier-Lavigne M, Marín O. Slit/Robo signaling modulates the proliferation of central nervous system progenitors. Neuron. 2012;76:338–352. doi: 10.1016/j.neuron.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camurri L, Mambetisaeva E, Sundaresan V. Rig-1 a new member of Robo family genes exhibits distinct pattern of expression during mouse development. Gene Expr Patterns. 2004;4:99–103. doi: 10.1016/S1567-133X(03)00142-X. [DOI] [PubMed] [Google Scholar]
- Camurri L, Mambetisaeva E, Davies D, Parnavelas J, Sundaresan V, Andrews W. Evidence for the existence of two Robo3 isoforms with divergent biochemical properties. Mol Cell Neurosci. 2005;30:485–493. doi: 10.1016/j.mcn.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Cariati M, Naderi A, Brown JP, Smalley MJ, Pinder SE, Caldas C, Purushotham AD. Alpha-6 integrin is necessary for the tumourigenicity of a stem cell-like subpopulation within the MCF7 breast cancer cell line. Int J Cancer. 2008;122:298–304. doi: 10.1002/ijc.23103. [DOI] [PubMed] [Google Scholar]
- Cavanagh JF, Mione MC, Pappas IS, Parnavelas JG. Basic fibroblast growth factor prolongs the proliferation of rat cortical progenitor cells in vitro without altering their cell cycle parameters. Cereb Cortex. 1997;7:293–302. doi: 10.1093/cercor/7.4.293. [DOI] [PubMed] [Google Scholar]
- Chan CH, Godinho LN, Thomaidou D, Tan SS, Gulisano M, Parnavelas JG. Emx1 is a marker for pyramidal neurons of the cerebral cortex. Cereb Cortex. 2001;11:1191–1198. doi: 10.1093/cercor/11.12.1191. [DOI] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci U S A. 2005;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubelos B, Sebastián-Serrano A, Kim S, Moreno-Ortiz C, Redondo JM, Walsh CA, Nieto M. Cux-2 controls the proliferation of neuronal intermediate precursors of the cortical subventricular zone. Cereb Cortex. 2008;18:1758–1770. doi: 10.1093/cercor/bhm199. [DOI] [PubMed] [Google Scholar]
- Cubelos B, Sebastián-Serrano A, Beccari L, Calcagnotto ME, Cisneros E, Kim S, Dopazo A, Alvarez-Dolado M, Redondo JM, Bovolenta P, Walsh CA, Nieto M. Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron. 2010;66:523–535. doi: 10.1016/j.neuron.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CL, Martínez-Cerdeño V, Noctor SC. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J Neurosci. 2013;33:4216–4233. doi: 10.1523/JNEUROSCI.3441-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Sun Y, Zhang Y, Luo X, Hou W, Yan L, Chen Y, Tian E, Han J, Zhang H. Transcription factor NFY globally represses the expression of the C. elegans Hox gene Abdominal-B homolog egl-5. Dev Biol. 2007;308:583–592. doi: 10.1016/j.ydbio.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Faux C, Rakic S, Andrews W, Yanagawa Y, Obata K, Parnavelas JG. Differential gene expression in migrating cortical interneurons during mouse forebrain development. J Comp Neurol. 2010;518:1232–1248. doi: 10.1002/cne.22271. [DOI] [PubMed] [Google Scholar]
- Gonda Y, Andrews WD, Tabata H, Namba T, Parnavelas JG, Nakajima K, Kohsaka S, Hanashima C, Uchino S. Robo1 regulates the migration and laminar distribution of upper-layer pyramidal neurons of the cerebral cortex. Cereb Cortex. 2013;23:1495–1508. doi: 10.1093/cercor/bhs141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressens P. Neuronal migration disorders. J Child Neurol. 2005;20:969–971. [PubMed] [Google Scholar]
- Guerrini R, Filippi T. Neuronal migration disorders, genetics, and epileptogenesis. J Child Neurol. 2005;20:287–299. doi: 10.1177/08830738050200040401. [DOI] [PubMed] [Google Scholar]
- Han HX, Geng JG. Over-expression of Slit2 induces vessel formation and changes blood vessel permeability in mouse brain. Acta Pharmacol Sin. 2011;32:1327–1336. doi: 10.1038/aps.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hau P, Jachimczak P, Schlaier J, Bogdahn U. TGF-beta2 signaling in high-grade gliomas. Curr Pharm Biotechnol. 2011;12:2150–2157. doi: 10.2174/138920111798808347. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydar TF, Kuan CY, Flavell RA, Rakic P. The role of cell death in regulating the size and shape of the mammalian forebrain. Cereb Cortex. 1999;9:621–626. doi: 10.1093/cercor/9.6.621. [DOI] [PubMed] [Google Scholar]
- Hernández-Miranda LR, Cariboni A, Faux C, Ruhrberg C, Cho JH, Cloutier JF, Eickholt BJ, Parnavelas JG, Andrews WD. Robo1 regulates semaphorin signaling to guide the migration of cortical interneurons through the ventral forebrain. J Neurosci. 2011;31:6174–6187. doi: 10.1523/JNEUROSCI.5464-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner WB, Kosodo Y. Symmetric versus asymmetric cell division during neurogenesis in the developing vertebrate central nervous system. Curr Opin Cell Biol. 2005;17:648–657. doi: 10.1016/j.ceb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Jossin Y. Neuronal migration and the role of reelin during early development of the cerebral cortex. Mol Neurobiol. 2004;30:225–251. doi: 10.1385/MN:30:3:225. [DOI] [PubMed] [Google Scholar]
- Kidd T, Brose K, Mitchell KJ, Fetter RD, Tessier-Lavigne M, Goodman CS, Tear G. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell. 1998;92:205–215. doi: 10.1016/S0092-8674(00)80915-0. [DOI] [PubMed] [Google Scholar]
- Koinuma D, Shinozaki M, Nagano Y, Ikushima H, Horiguchi K, Goto K, Chano T, Saitoh M, Imamura T, Miyazono K, Miyazawa K. RB1CC1 protein positively regulates transforming growth factor-beta signaling through the modulation of Arkadia E3 ubiquitin ligase activity. J Biol Chem. 2011;286:32502–32512. doi: 10.1074/jbc.M111.227561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Bergeron L, Cryns V, Pasternack MS, Zhu H, Shi L, Greenberg A, Yuan J. Activation of caspase-2 in apoptosis. J Biol Chem. 1997;272:21010–21017. doi: 10.1074/jbc.272.34.21010. [DOI] [PubMed] [Google Scholar]
- Liu Z, Patel K, Schmidt H, Andrews W, Pini A, Sundaresan V. Extracellular Ig domains 1 and 2 of Robo are important for ligand (Slit) binding. Mol Cell Neurosci. 2004;26:232–240. doi: 10.1016/j.mcn.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (−δδCT) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long H, Sabatier C, Ma L, Plump A, Yuan W, Ornitz DM, Tamada A, Murakami F, Goodman CS, Tessier-Lavigne M. Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron. 2004;42:213–223. doi: 10.1016/S0896-6273(04)00179-5. [DOI] [PubMed] [Google Scholar]
- López-Bendito G, Flames N, Ma L, Fouquet C, Di Meglio T, Chedotal A, Tessier-Lavigne M, Marín O. Robo1 and Robo2 cooperate to control the guidance of major axonal tracts in the mammalian forebrain. J Neurosci. 2007;27:3395–3407. doi: 10.1523/JNEUROSCI.4605-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Wu Y, Sousa N, Almeida OF. SMAD pathway mediation of BDNF and TGF beta 2 regulation of proliferation and differentiation of hippocampal granule neurons. Development. 2005;132:3231–3242. doi: 10.1242/dev.01893. [DOI] [PubMed] [Google Scholar]
- Lu W, van Eerde AM, Fan X, Quintero-Rivera F, Kulkarni S, Ferguson H, Kim HG, Fan Y, Xi Q, Li QG, Sanlaville D, Andrews W, Sundaresan V, Bi W, Yan J, Giltay JC, Wijmenga C, de Jong TP, Feather SA, Woolf AS, et al. Disruption of ROBO2 is associated with urinary tract anomalies and confers risk of vesicoureteral reflux. Am J Hum Genet. 2007;80:616–632. doi: 10.1086/512735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillat V, Cases O, Nguyen-Ba-Charvet KT, Tessier-Lavigne M, Sotelo C, Chédotal A. Spatiotemporal expression patterns of slit and Robo genes in the rat brain. J Comp Neurol. 2002;442:130–155. doi: 10.1002/cne.10068. [DOI] [PubMed] [Google Scholar]
- Marín O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- Masserdotti G, Badaloni A, Green YS, Croci L, Barili V, Bergamini G, Vetter ML, Consalez GG. ZFP423 coordinates Notch and bone morphogenetic protein signaling, selectively up-regulating Hes5 gene expression. J Biol Chem. 2010;285:30814–30824. doi: 10.1074/jbc.M110.142869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Mommersteeg MT, Andrews WD, Ypsilanti AR, Zelina P, Yeh ML, Norden J, Kispert A, Chédotal A, Christoffels VM, Parnavelas JG. Slit-roundabout signaling regulates the development of the cardiac systemic venous return and pericardium. Circ Res. 2013;112:465–475. doi: 10.1161/CIRCRESAHA.112.277426. [DOI] [PubMed] [Google Scholar]
- Moorman AF, Houweling AC, de Boer PA, Christoffels VM. Sensitive nonradioactive detection of mRNA in tissue sections: novel application of the whole-mount in situ hybridization protocol. J Histochem Cytochem. 2001;49:1–8. doi: 10.1177/002215540104900101. [DOI] [PubMed] [Google Scholar]
- Mostovich LA, Prudnikova TY, Kondratov AG, Loginova D, Vavilov PV, Rykova VI, Sidorov SV, Pavlova TV, Kashuba VI, Zabarovsky ER, Grigorieva EV. Integrin alpha9 (ITGA9) expression and epigenetic silencing in human breast tumors. Cell Adh Migr. 2011;5:395–401. doi: 10.4161/cam.5.5.17949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajah B, Brunstrom JE, Grutzendler J, Wong RO, Pearlman AL. Two modes of radial migration in early development of the cerebral cortex. Nat Neurosci. 2001;4:143–150. doi: 10.1038/83967. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Plump AS, Erskine L, Sabatier C, Brose K, Epstein CJ, Goodman CS, Mason CA, Tessier-Lavigne M. Slit1 and Slit2 cooperate to prevent premature midline crossing of retinal axons in the mouse visual system. Neuron. 2002;33:219–232. doi: 10.1016/S0896-6273(01)00586-4. [DOI] [PubMed] [Google Scholar]
- Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183:425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- Rakic P. Principles of neural cell migration. Experientia. 1990;46:882–891. doi: 10.1007/BF01939380. [DOI] [PubMed] [Google Scholar]
- Sabatier C, Plump AS, Le Ma, Brose K, Tamada A, Murakami F, Lee EY, Tessier-Lavigne M. The divergent Robo family protein rig-1/Robo3 is a negative regulator of slit responsiveness required for midline crossing by commissural axons. Cell. 2004;117:157–169. doi: 10.1016/S0092-8674(04)00303-4. [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Hirata H, Ohtsuka T, Bessho Y, Kageyama R. The basic helix-loop-helix genes Hesr1/Hey1 and Hesr2/Hey2 regulate maintenance of neural precursor cells in the brain. J Biol Chem. 2003;278:44808–44815. doi: 10.1074/jbc.M300448200. [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Chung G, Heppell-Parton A, Xiong J, Grundy C, Roberts I, James L, Cahn A, Bench A, Douglas J, Minna J, Sekido Y, Lerman M, Latif F, Bergh J, Li H, Lowe N, Ogilvie D, Rabbitts P. Homozygous deletions at 3p12 in breast and lung cancer. Oncogene. 1998;17:1723–1729. doi: 10.1038/sj.onc.1202103. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Kadokawa Y, Hamada Y, Marunouchi T. Notch2 expression negatively correlates with glial differentiation in the postnatal mouse brain. J Neurobiol. 1999;41:524–539. doi: 10.1002/(SICI)1097-4695(199912)41:4<524::AID-NEU8>3.3.CO%3B2-9. [DOI] [PubMed] [Google Scholar]
- Tanaka R, Tainaka M, Ota T, Mizuguchi N, Kato H, Urabe S, Chen Y, Fustin JM, Yamaguchi Y, Doi M, Hamada S, Okamura H. Accurate determination of S-phase fraction in proliferative cells by dual fluorescence and peroxidase immunohistochemistry with 5-bromo-2′-deoxyuridine (BrdU) and Ki67 antibodies. J Histochem Cytochem. 2011;59:791–798. doi: 10.1369/0022155411411090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomaidou D, Mione MC, Cavanagh JF, Parnavelas JG. Apoptosis and its relation to the cell cycle in the developing cerebral cortex. J Neurosci. 1997;17:1075–1085. doi: 10.1523/JNEUROSCI.17-03-01075.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford KL, Marillat V, Stein E, Goodman CS, Tessier-Lavigne M, Chédotal A, Ghosh A. Regulation of cortical dendrite development by Slit-Robo interactions. Neuron. 2002;33:47–61. doi: 10.1016/S0896-6273(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Xian J, Clark KJ, Fordham R, Pannell R, Rabbitts TH, Rabbitts PH. Inadequate lung development and bronchial hyperplasia in mice with a targeted deletion in the Dutt1/Robo1 gene. Proc Natl Acad Sci U S A. 2001;98:15062–15066. doi: 10.1073/pnas.251407098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian J, Aitchison A, Bobrow L, Corbett G, Pannell R, Rabbitts T, Rabbitts P. Targeted disruption of the 3p12 gene, Dutt1/Robo1, predisposes mice to lung adenocarcinomas and lymphomas with methylation of the gene promoter. Cancer Res. 2004;64:6432–6437. doi: 10.1158/0008-5472.CAN-04-2561. [DOI] [PubMed] [Google Scholar]
- Ypsilanti AR, Zagar Y, Chédotal A. Moving away from the midline: new developments for Slit and Robo. Development. 2010;137:1939–1952. doi: 10.1242/dev.044511. [DOI] [PubMed] [Google Scholar]
- Yuan W, Rao Y, Babiuk RP, Greer JJ, Wu JY, Ornitz DM. A genetic model for a central (septum transversum) congenital diaphragmatic hernia in mice lacking Slit3. Proc Natl Acad Sci U S A. 2003;100:5217–5222. doi: 10.1073/pnas.0730709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Geng AQ, Li PF, Wang Y, Yuan XB. Robo4 regulates the radial migration of newborn neurons in developing neocortex. Cereb Cortex. 2012;22:2587–2601. doi: 10.1093/cercor/bhr330. [DOI] [PMC free article] [PubMed] [Google Scholar]