Abstract
The Arabidopsis accelerated cell death 6-1 (acd6-1) mutant shows constitutive defense, cell death, and extreme dwarf phenotypes. In a screen for acd6-1 suppressors, we identified a mutant that was disrupted by a T-DNA in the PHOSPHATE TRANSPORTER 4;1 (PHT4;1) gene. The suppressor mutant pht4;1-1 is dominant, expresses truncated PHT4;1 transcripts, and is more susceptible to virulent Pseudomonas syringae strains but not to several avirulent strains. Treatment with a salicylic acid (SA) agonist induced a similar level of resistance in Col-0 and pht4;1-1, suggesting that PHT4;1 acts upstream of the SA pathway. Genetic analysis further indicates that PHT4;1 contributes to SID2-dependent and -independent pathways. Transgenic expression of the DNA fragment containing the PHT4;1-1 region or the full-length PHT4;1 gene in wild-type conferred enhanced susceptibility to Pseudomonas infection. Interestingly, expression of PHT4;1 is regulated by the circadian clock. Together, these data suggest that the phosphate transporter PHT4;1 is critical for basal defense and also implicate a potential role of the circadian clock in regulating innate immunity of Arabidopsis.
Keywords: Biological clock, disease resistance, signal transduction, Pseudomonas syringae, phosphate transporter, salicylic acid
INTRODUCTION
Successful control of plant diseases depends on a thorough understanding of the mechanism of disease resistance in plants. In response to pathogen attacks, plants actively reprogram expression of thousands of genes (Maleck et al., 2000; Tao et al., 2003; Katagiri, 2004), among which only a few are known to play a direct role in regulating plant defense while most of them are diagnostic of defense responses. Thus, the major challenge in the field remains to identify components in the defense signaling networks and to understand their functions in regulating disease resistance.
One of the key nodes in the defense signaling networks is centered on the small phenolic compound salicylic acid (SA). SA is required for establishment of basal defense induced by pathogen elicitors, strong local resistance in the infected region induced by pathogen effector proteins as well as systemic acquired resistance (SAR) at the whole plant level (Hammond-Kosack and Jones, 1996; Ryals et al., 1996; Tsuda et al., 2008). Several genes that are important for SA-mediated defense have been identified in Arabidopsis and they can be grouped into three interconnected subgroups. The type I SA regulatory genes include SA INDUCTION-DEFICIENT 2 (SID2), encoding isochorismate synthase contributing to the bulk SA biosynthesis (Wildermuth et al., 2001). The type II SA regulatory genes are generally not considered to directly participate in SA biosynthesis because the protein products of these genes lack distinct enzymatic motifs. Examples of the type II SA regulators include ACCELERATED CELL DEATH 6 (ACD6), AGD2-LIKE DEFENSE 1 (ALD1), ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1), PHYTOALEXIN DEFICIENT 4, (PAD4) SID1/EDS5, HOPW1-1-INTERACTING/AVRPPHB SUSCEPTIBLE/GH3-LIKE DEFENSE GENE 1, and the MODIFIER OF SNC1 genes (Falk et al., 1999; Jirage et al., 1999; Nawrath et al., 2002; Lu et al., 2003; Song et al., 2004; Palma et al., 2005; Zhang et al., 2005; Zhang and Li, 2005; Goritschnig et al., 2007; Jagadeeswaran et al., 2007; Lee et al., 2007b; Nobuta et al., 2007; Palma et al., 2007). However, how some of these genes influence SA accumulation still remains to be determined (Lu, 2009). Genes acting downstream of SA signaling comprise the type III SA regulatory genes. The best-characterized defense gene in this group is NONEXPRESSOR OF PR GENES 1 (NPR1), the protein product of which translocates from the cytoplasm to the nucleus in response to redox changes to control defense gene expression and SAR activation (Cao et al., 1997; Ryals et al., 1997; Shah et al., 1997; Mou et al., 2003; Dong, 2004; Tada et al., 2008). To increase the complexity of defense signaling networks, SA is also known to cross-talk with signals derived from several phytohormones (Feys and Parker, 2000; Kunkel and Brooks, 2002; Wang et al., 2007; Koornneef and Pieterse, 2008; de Torres Zabala et al., 2009).
The type II SA regulator ACD6 is an ankyrin-repeat protein with a transmembrane domain and was recently shown to be a major determinant of fitness in Arabidopsis (Todesco et al., 2010). Loss-of-function mutation in the ACD6 gene leads to reduced SA accumulation and compromised defense against Pseudomonas syringae infection. In contrast, a gain-of-function mutant, acd6-1, caused by one amino acid substitution in the transmembrane domain of ACD6, exhibits extreme dwarfism and constitutive resistance to broad-spectrum pathogens, including P. syringae, Hyaloperonospora arabidopsidis, and Botrytis cinerea (Rate et al., 1999; Lu et al., 2003; Song et al., 2004; Wang and Lu, unpublished data). acd6-1 also accumulates high levels of SA and camalexin (an anti-fungal metabolite) and displays severe cell death. Interestingly, the small size of acd6-1 inversely correlates with the defense levels in the plant (Song et al., 2004; Lu et al., 2009). We took advantage of this unique feature of acd6-1 in a mutant screen for acd6-1 suppressors (sups), which are larger plants with potential disruptions in novel defense genes (Lu et al., 2009). T-DNA mutagenesis was used to introduce second site mutations in the acd6-1 background and to facilitate the subsequent cloning of the disrupted gene. Among 30 sup mutants isolated, we identified an allele of SID2 and cloned the SUP6 gene, encoding a predicted transmembrane protein with an N-terminal peptidase domain (Lu et al., 2009). Therefore, we have validated that acd6-1 suppressor screen is powerful in uncovering novel genes important for defense responses.
In this study, we report the isolation and characterization of a suppressor mutant that harbors a T-DNA insertion in the PHOSPHATE TRANSPORTER 4;1 (PHT4;1) gene (Guo et al., 2008a, 2008b). PHT4;1, also named ANTR1 (Roth et al., 2004; Pavon et al., 2008), belongs to a six-gene family in Arabidopsis. Only PHT4;6 was shown to regulate plant response to salt stress (Cubero et al., 2009); the biological functions of other members in the PHT4 family are largely unknown. We showed here that the suppressor mutant pht4;1-1 expressed truncated PHT4;1 transcripts and was dominant. pht4;1-1 conferred enhanced disease susceptibility to virulent Pseudomonas strains and this susceptibility could be suppressed by the treatment of an SA agonist. In addition, we showed that transgenic Col-0 plants carrying one or more copies of the truncated PHT4;1-1 genomic fragment or the full-length PHT4;1 gene were more susceptible to Pseudomonas infection. Thus, we provided the first evidence to implicate a member in the PHT4 family in regulating plant innate immunity. Interestingly, we found that expression of PHT4;1 was regulated by the biological clock, suggesting a role for the biological clock in control of disease resistance in plants.
RESULTS
pht4;1-1 Suppresses acd6-1-Conferred Phenotypes
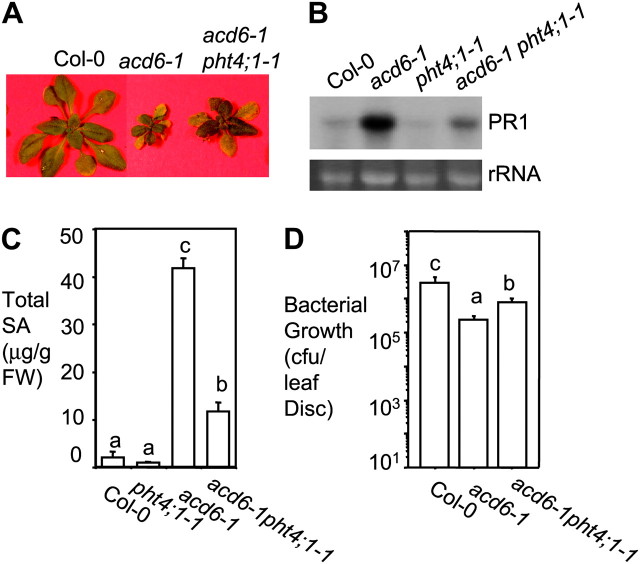
We previously showed that the small size of acd6-1 is grossly in inverse correlation with defense levels in the plant. We took advantage of this unique feature of acd6-1 in a suppressor screen in order to discover novel defense genes. Among the suppressor (sup) mutants isolated, acd6-1sup3-1, later designated acd6-1pht4;1-1, has an intermediate size compared with acd6-1 and Col-0 (Figure 1A). The size phenotype of acd6-1pht4;1-1 was confirmed in progenies of two backcrosses. Consistent with the change in plant size, pht4;1-1 partially suppressed acd6-1 for the expression of the defense marker gene PATHOGENESIS RELATED 1 (PR1) and SA accumulation (Glazebrook et al., 1997) (Figure 1B and 1C). When challenged with the virulent bacterium, Pseudomonas syringae pv. maculicola strain DG3 (PmaDG3), pht4;1-1 partially suppressed constitutive defense in acd6-1 (Figure 1D).
Figure 1.
The pht4;1-1 Mutant Suppresses acd6-1-Conferred Phenotypes.
(A) Picture of 25-day-old plants.
(B) Northern blot analysis of PR1 expression. Total RNA was isolated from 25-day-old uninfected plants. rRNA was used as a loading control.
(C) SA quantification. Uninfected 25-day-old plants were harvested for SA extraction followed by HPLC analysis.
(D) Bacterial growth assay. 25-day-old plants were infected with Pseudomonas syringae pv. maculicola strain DG3 (PmaDG3) (OD600 = 0.0001) and leaf discs were collected for bacterial growth assay 3 d after infection. CFU, colony forming unit.
Different letters in (C) and (D) indicate significant difference among samples (P < 0.05; n = 3 in (C) and n = 6 in (D)). These experiments were repeated two times, with similar results.
pht4;1-1 Is Dominant and Confers Enhanced Disease Susceptibility to Virulent Pseudomonas Strains
To further investigate the role of pht4;1-1 in defense regulation, we crossed acd6-1pht4;1-1 with Col-0 and obtained the pht4;1-1 homozygous mutant in the absence of acd6-1. We challenged pht4;1-1 and Col-0 with both virulent and avirulent Pseudomonas strains. We found that pht4;1-1 showed significantly more growth of two virulent strains (PmaDG3 and P. syringae pv. tomato strain DC3000 (DC3000)), compared to Col-0 (Figure 2A and 2B). pht4;1-1 also showed more severe disease symptoms than Col-0 with PmaDG3 infection (Supplemental Figure S1). However, similar susceptibility was found in pht4;1-1 and Col-0 to the avirulent strains, Pma avrRpt2 or Pma avrRpm1 (Figure 2C). These data suggest that PHT4;1 is involved in basal defense but not in defense mediated by R genes, such as RPS2 and RPM1. Surprisingly, we found a single copy of the pht4;1-1 mutation conferred enhanced disease susceptibility to PmaDG3 (Figure 2D), suggesting a dominant nature of the pht4;1-1 mutation.
Figure 2.
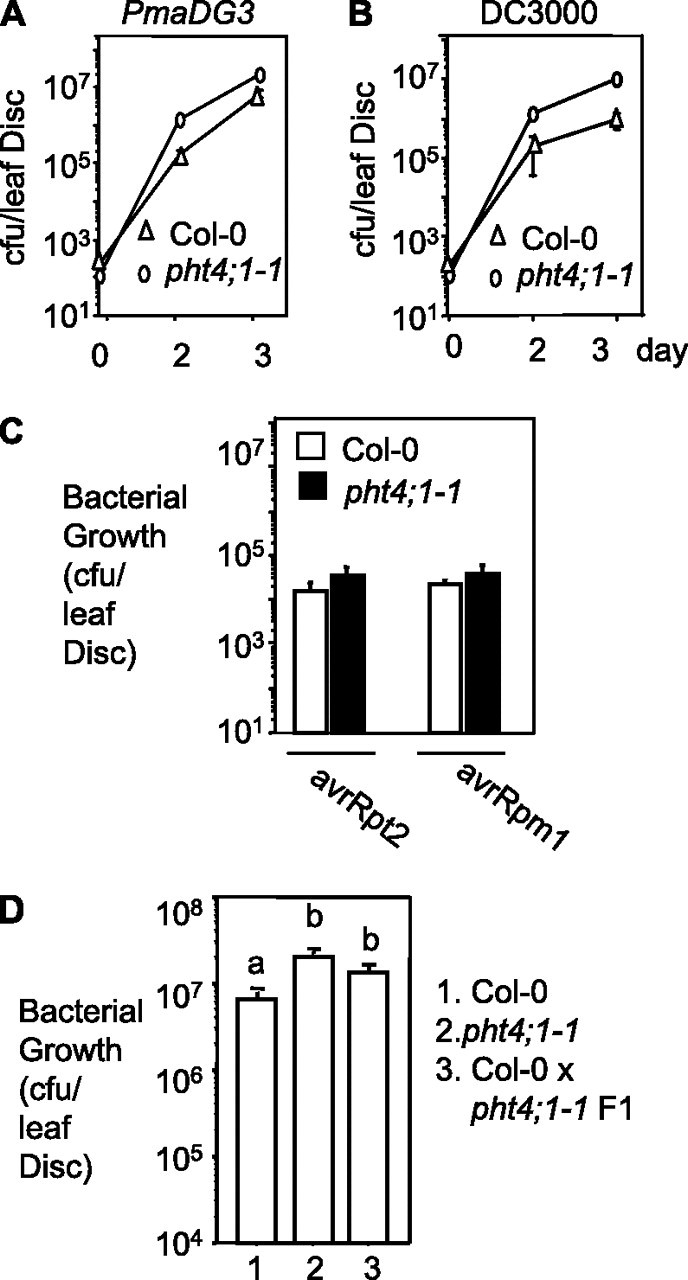
pht4;1-1 Is Dominant and Confers Enhanced Disease Susceptibility to Virulent Pseudomonas Strains.
(A) Infection with PmaDG3 (OD600 = 0.0001).
(B) Infection with P. syringae pv. tomato DC3000 (DC3000) (OD600 = 0.0001).
(C) Infection with Pma avrRpt2 or Pma avrRpm1 (OD600 = 0.0002).
(D) Infection of heterozygous pht4;1-1 with PmaDG3 (OD600 = 0.0001).
25-day-old plants were infected with the indicated Pseudomonas strains and assayed for bacterial growth. Significant difference between pht4;1-1 and Col-0 was observed at 2 and 3 days after infection in (A) and (B) and was indicated with different letters in (D) (P < 0.05; n = 6). No difference was observed in the two samples in (C) 3 d after infection with either avirulent strain. These experiments were repeated two times, with similar results.
PHT4;1 Encodes a Phosphate Transporter
Using the TAIL–PCR method (Liu et al., 1995), we identified a T-DNA insertion in the fifth exon of the PHT4;1 gene (At2g29650) in the pht4;1-1 mutant (Figure 3A). Instead of abolishing gene expression, the T-DNA insertion led to the production of a truncated transcript from the 5’ end of this gene (Figure 3B), which presumably encodes a truncated PHT4;1 protein with the first 347 amino acids. PHT4;1, previously also named ANTR1 (Roth et al., 2004; Guo et al., 2008b; Pavon et al., 2008), encodes a transmembrane protein that belongs to a small family with six members. Recently, members of the PHT4 family were demonstrated to have phosphate uptake activity (Guo et al., 2008b; Pavon et al., 2008; Cubero et al., 2009). One of the PHT4 family members (PHT4;6) was shown to regulate plant response to salt stress (Cubero et al., 2009). However, the biological functions of PHT4;1 and other members in the family are largely unknown.
Figure 3.
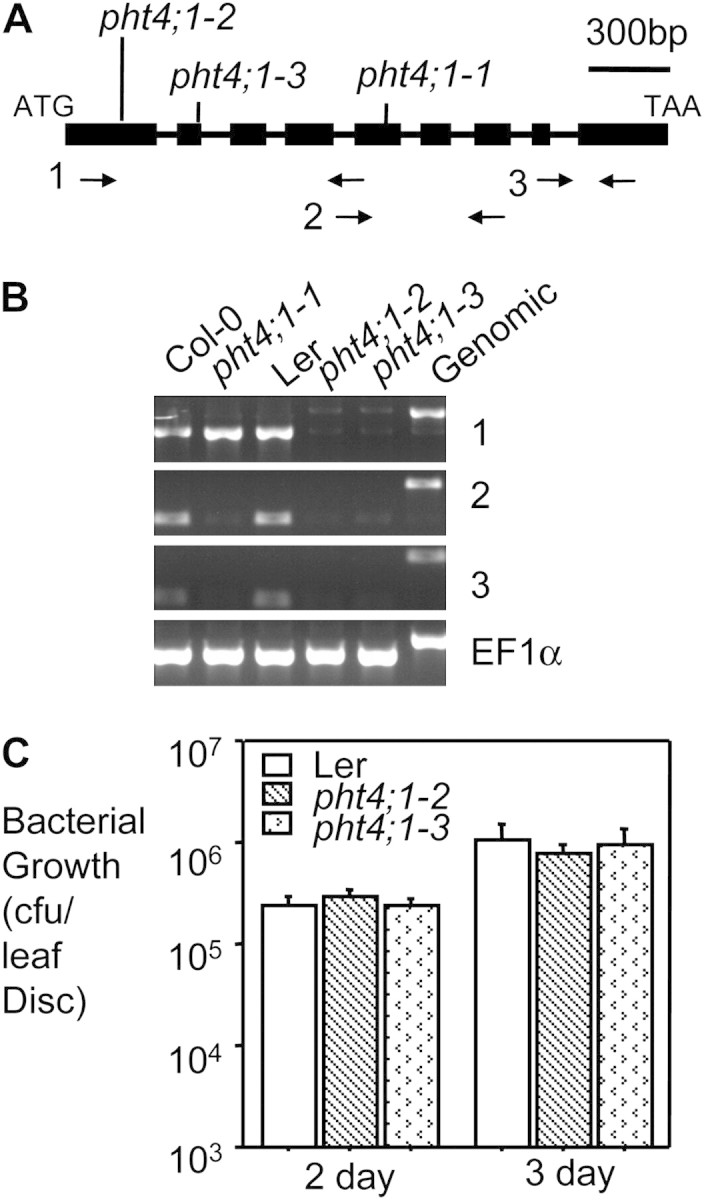
Two PHT4;1 Null Alleles Are Not Compromised in Disease Resistance.
(A) Structure of the PHT4;1 gene and positions of the mutant alleles. Filled boxes indicate exons and horizontal lines indicate introns. Vertical lines indicate the positions of PHT4;1 alleles. The arrow pairs are primer sets used in RT–PCR in (B).
(B) RT–PCR analysis. EF1α was used as a loading control. Note the PCR products amplified from a genomic DNA template included introns and therefore were larger than their corresponding RT–PCR products.
(C) Bacterial growth assay. 25-day-old plants were infected with PmaDG3 (OD600 = 0.0002). Six leaf discs from infected individual plants of each genotype were collected for bacterial growth assay 3 d after infection. No significant difference was observed among the genotypes. Experiments in (B) and (C) were repeated three times, with similar results.
We identified two loss-of-function alleles of PHT4;1 in Landsberg accession—pht4;1-2 (GT_5_110509) and pht4;1-3 (CSHL_GT22119)—both of which were disrupted by transposon insertions (Figure 3A). RT–PCR analysis indicated that PHT4;1 expression was completely abolished in these two alleles (Figure 3B), suggesting that they are null mutants. However, unlike pht4;1-1, pht4;1-2 and pht4;1-3 showed similar disease resistance to Pseudomonas as wild-type (WT) (Figure 3C). Since there are five other PHT4;1-like genes in Arabidopsis, it is possible that some of these genes share redundant function with PHT4;1.
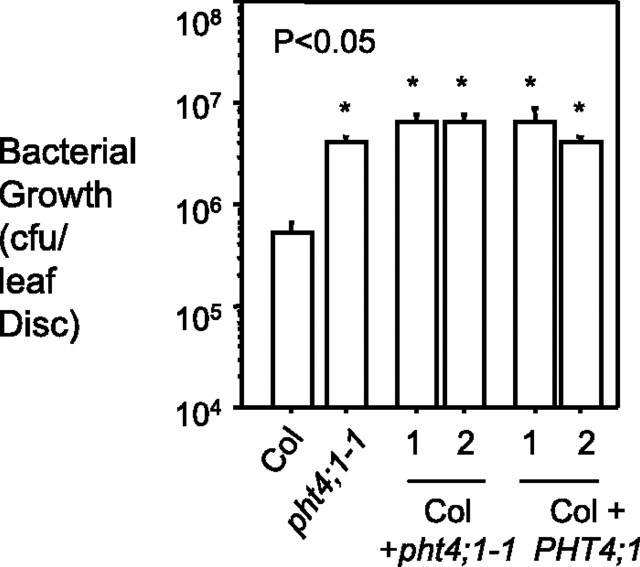
To confirm that pht4;1-1-conferred phenotypes are due to the perturbation of the PHT4;1 gene, we made transgenic Col-0 plants carrying one or more copies of the genomic fragment of PHT4;1-1 (encoding the N-terminal 1–347 amino acid of the protein) driven by the native PHT4;1 promoter. Like pht4;1-1, most transgenic plants were more susceptible than the control to PmaDG3 infection. In addition, we generated transgenic Col-0 plants expressing extra copies of the full-length PHT4;1 gene. Five out of seven such transgenic plants showed increased susceptibility to PmaDG3 infection. Bacterial growth results for two representative lines from each transformation are shown in Figure 4. Together, these data indicate that T-DNA disruption of the PHT4;1 gene is responsible for the pht4;1-1-conferred phenotypes and that PHT4;1 is a negative regulator of plant defense.
Figure 4.
Transgenic Plants Expressing PHT4;1-1 or PHT4;1 Are More Susceptible to PmaDG3 Infection.
A genomic fragment containing the PHT4;1-1 region or the full-length PHT4;1 gene was PCR amplified and cloned into the binary vector pGreenII 0029 (Hellens et al., 2000) for plant transformation. The PHT4;1 promoter (1329 bp) was used to drive the expression of these two constructs. 25-day-old plants were infected with PmaDG3 (OD600 = 0.0001) and leaf discs were collected for bacterial growth assay 3 d after infection. Asterisks indicate significant difference between Col-0 and the transgenic plants (P < 0.05; n = 6). These experiments were repeated two times, with similar results.
pht4;1-1 Contributes to SA-Mediated Defense
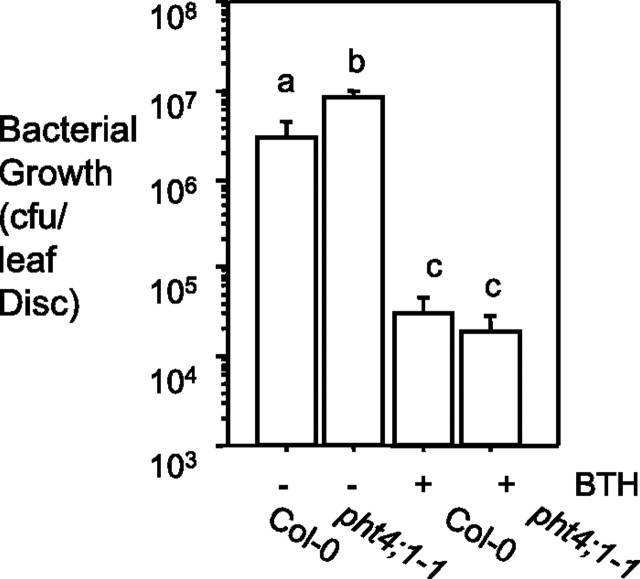
SA plays a key role in signaling plant defense (Hammond-Kosack and Jones, 1996; Ryals et al., 1996; Tsuda et al., 2008). To test whether pht4;1-1-conferred susceptibility is SA-related, we pre-treated Col-0 and pht4;1-1 with 300 μM benzo(1,2,3)thiadiazole-7-carbothioic acid (BTH), an agonist of SA (Lawton et al., 1996), and subsequently challenged the plants with PmaDG3. We found that BTH pretreatment induced a similar level of resistance in Col-0 and pht4;1-1 (Figure 5), suggesting that the pht4;1-1 mutation does not impair the ability of the plant to induce SA-mediated defense in response to exogenous BTH. Thus, PHT4;1 likely acts upstream of SA signaling.
Figure 5.
pht4;1-1-Conferred Susceptibility Can Be Suppressed by BTH Treatment.
Plants were treated with 300 μM BTH for 36 h before PmaDG3 infection (OD600 = 0.0001). Different letters indicate significant difference among samples (P < 0.05; n = 6). These experiments were repeated two times, with similar results.
To further investigate how PHT4;1 affects SA-mediated defense, we crossed acd6-1pht4;1-1 to acd6-1sid2-1. SID2 is known to contribute to bulk SA biosynthesis and the sid2-1 mutation abolishes most SA accumulation in acd6-1 (Lu et al., 2009). The fact that acd6-1pht4;1-1 accumulated intermediated SA levels compared with Col-0 and acd6-1 (Figure 1C) suggests that pht4;1-1 partially affects SID2-mediated SA biosynthesis. If pht4;1-1 also impairs SID2-independent SA biosynthesis, we expect that the triple mutant acd6-1pht4;1-1sid2-1 would be larger than the parental double mutants. Otherwise, if pht4;1-1 impairs only SID2-dependent SA biosynthesis, the triple mutant should be similar to the two parental double mutants. We found that the former case was true for acd6-1pht4;1-1sid2-1 (Figure 6A).
Figure 6.
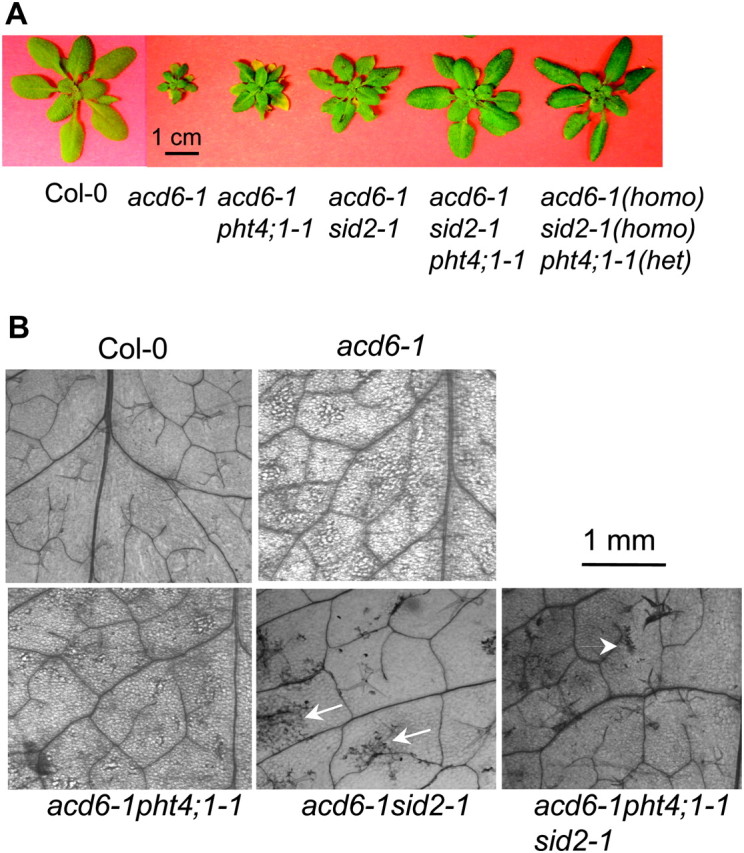
The pht4;1-1 Mutation Contributes to SID2-Dependent and -Independent Pathways.
(A) Picture of 25-day-old plants. Note the triple mutants are larger than the double mutants.
(B) Cell death phenotypes. The fifth leaves of the indicated genotypes were stained with trypan blue and photographed with a Photometrics CCD camera connected to a Leica dissecting microscope. Arrows indicate minor cell death in acd6-1sid2-1 and acd6-1pht4; 1-1sid2-1. These experiments were repeated two times, with similar results.
acd6-1 exhibited severe cell death, which could be suppressed by the sid2-1 mutation (Figure 6B; Lu et al., 2009). We found that the pht4;1-1 mutant also suppressed the severity of cell death in acd6-1. In addition, consistent with its increased size, acd6-1pht4;1-1sid2-1 had much reduced cell death compared with acd6-1pht4;1-1 and acd6-1sid2-1 (Figure 6B). Furthermore, we found that acd6-1pht4;1-1sid2-1 accumulated lower total SA (0.07 ± 0.05 μg gFW−1) than acd6-1pht4;1-1 (5.73 ± 0.64 μg gFW−1) and acd6-1sid2-1 (0.22 ± 0.14 μg gFW−1). Since the protein product of PHT4;1 lacks distinct motifs for being SA biosynthetic enzyme, we concluded from our studies that PHT4;1 is a type II SA regulator contributing to both SID2-dependent and -independent SA accumulation.
Consistent with pht4;1-1 being dominant (Figure 2D), we found that, among 114 progenies of a heterozygous triple mutant, acd6-1/acd6-1;sid2-1/sid2-1;pht4;1-1/PHT4;1, 33 plants were smaller in size, while 81 plants were larger (a 1:3 ratio; X2 = 0.45; P > 0.5). The smaller plants were genotyped to be acd6-1sid2-1 homozygous, had no pht4;1-1 allele, and were BASTA-sensitive. The larger ones were indistinguishable in size and were BASTA-resistant (Figure 6A). They segregated into 24 homozygous triple mutants and 57 heterozygotes (acd6-1/acd6-1;sid2-1/sid2-1;pht4;1-1/PHT4;1). These observations indicate that pht4;1-1 is due to a single T-DNA insertion, which is associated with the increased size phenotype and BASTA resistance. In addition, the fact that disrupting one copy of PHT4;1-1 is sufficient to suppress acd6-1sid2-1 phenotypes further supports that the pht4;1-1 mutation is dominant.
PHT4;1 Expression Is Regulated by Light and the Circadian Clock
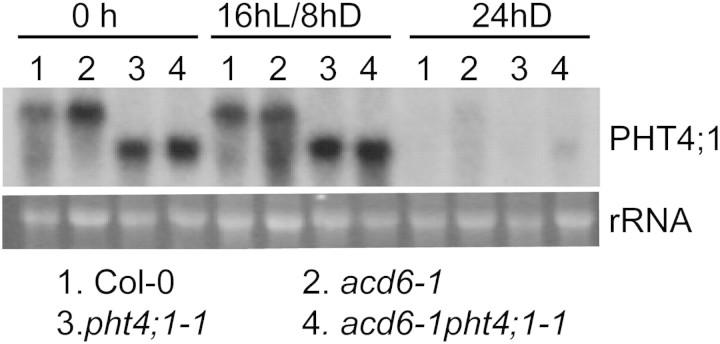
Bioinformatics analysis of publicly available microarray database (Genevesitgator; (Zimmermann et al., 2004)) and a previous study from Guo et al. (2008a) suggest that light regulates PHT4;1. To further investigate the role of light in regulating PHT4;1 expression, we performed northern blotting with total RNA extracted from Col-0, acd6-1, pht4;1-1, and acd6-1pht4; 1-1 in the presence or absence of light. Figure 7 shows that 24 h dark treatment completely abolished expression of PHT4;1 in Col-0 and acd6-1 and the truncated version PHT4;1-1 in the pht4;1-1 background. These observations suggest that light is required for PHT4;1 expression while dark suppresses its expression.
Figure 7.
Expression of PHT4;1 and PHT4;1-1 Is Light-Regulated.
25-day-old plants grown in a chamber with a 16-h light/8-h dark cycle were kept in the same condition or dark-treated for 24 h. Leaves were harvested for total RNA extraction followed by northern blotting. rRNA was used as a loading control. These experiments were repeated two times, with similar results.
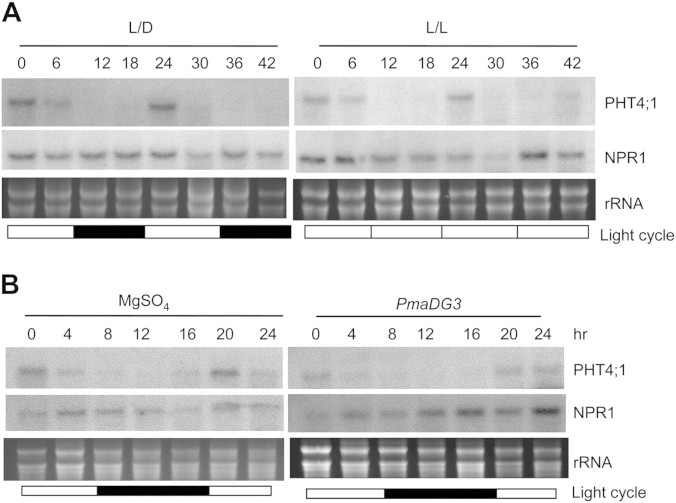
The circadian clock was also shown to regulate PHT4;1 expression (Guo et al., 2008a). In particular, two copies of CIRCADIAN CLOCK ASSOCIATED 1 (CCA1)-binding site (CBS) were found in the PHT4;1 promoter. CBS is a cis-element important for morning-specific circadian expression regulated by the central oscillator component of the circadian clock, CCA1 (Wang et al., 1997; Michael and McClung, 2002). To further investigate circadian regulation of PHT4;1 expression, we harvested leaf tissues from Col-0 and pht4;1-1 at every 4-h interval in 32 h under a light/dark cycle. RNA analysis revealed that both PHT4;1 and the truncated version PHT4;1-1 exhibited a similar diurnal expression pattern, with peaks in the daytime and troughs in the nighttime (Supplemental Figure S2). Such an expression pattern of PHT4;1 was also observed in the acd6-1 background under a light/dark cycle and persisted under a continuous light condition (Figure 8A). In contrast, expression of NPR1 was constant in acd6-1. In addition, we found that the diurnal expression of PHT4;1 was unaffected by PmaDG3 infection, although a slight suppression of PHT4;1 abundance was observed at 16 h and 20 h after PmaDG3 infection. As a control, the level of NPR1 transcripts was induced 12 h after the infection (Figure 8B). These observations strongly suggest that expression of PHT4;1 is under the control of the circadian clock.
Figure 8.
Expression of PHT4;1 Is Regulated by the Circadian Clock.
(A) Circadian expression of PHT4;1 in acd6-1. 25-day-old acd6-1 plants grown in a chamber with a 12-h light/12-h dark cycle were kept in the same chamber or moved to a chamber with continuous light conditions. Starting from time 0 (9:00 am), plants were harvested every 6 h for 42 h.
(B) Circadian expression of PHT4;1 during Pseudomonas infection. 25-day-old Col-0 plants grown in a chamber with a 12-h light/12-h dark cycle were infiltrated with PmaDG3 (OD = 0.0001) or 10 mM MgSO4 as control. The infiltrated leaves were collected at 4-h interval for 24 h.
Total RNA were extracted from the above samples and analyzed by northern blotting. White boxes indicate light periods and black boxes indicate dark periods. rRNA was used as a loading control. These experiments were repeated two times, with similar results.
DISCUSSION
From the acd6-1 suppressor screen, we isolated a mutant that harbors a T-DNA insertion in PHT4;1, the protein product of which was demonstrated to have phosphate transport activity (Roth et al., 2004; Guo et al., 2008b; Pavon et al., 2008). PHT4;1, also named ANTR1, belongs to a small gene family with six members, functions of which are largely unknown. In this study, we provided evidence to implicate a role of PHT4;1 in regulating plant defense. We showed that pht4;1-1 was dominant, possibly due to expression of the truncated PHT4;1. pht4;1-1 suppresses acd6-1-conferred phenotypes and is more susceptible to virulent Pseudomonas strains but not to two avirulent strains. The pht4;1-1-conferred susceptibility could be suppressed by treatment with BTH, an SA agonist and recapitulated by transgenically expressing a PHT4;1-1 or PHT4;1 genomic fragment in Col-0. Together, our data suggest that PHT4;1 is a negative regulator of basal defense in Arabidopsis. Interestingly, expression of PHT4;1 was under circadian control, suggesting a potential role of the biological clock in regulating plant innate immunity.
The Role of PHT4;1-Mediated Phosphate Transport in Plant Defense
Phosphorus (P) is an essential element for all living organisms and plays a crucial role in many physiological processes. Plant cells obtain their P via absorbing inorganic phosphate (Pi; the assimilable form of P) directly from the soil and/or from neighboring cells. Intracellular Pi can also be reallocated to different organelles of the cell. These processes are mediated by Pi transporters. The PHT4 family is one of the four Pi transporter families in Arabidopsis. Additional non-transporter genes are also known to regulate Pi sensing and signaling, leading to altered Pi transport and subsequently affecting Pi homeostasis in plants (Poirier and Bucher, 2002; Lin et al., 2009). Although mutations in many genes are known to influence Pi homeostasis in plants, only a few of such genes are known to be involved in defense regulation (Miura et al., 2005; Lee et al., 2007a; Murphy et al., 2008), suggesting that defense activation is not due to a general perturbation of Pi homeostasis in the cell.
pht4;1-1 and its two loss-of-function alleles are not known to have altered responses to Pi starvation (Lu, unpublished data and Wayne Versaw, personal communication), suggesting that PHT4;1 does not contribute to major Pi homeostasis in the cell. Evidence from our studies implicates a role of PHT4;1 in regulating plant defense, possibly by acting upstream of SA. Thus, we hypothesize that, rather than disrupting the Pi homeostasis in the cell, Pi reallocation mediated by PHT4;1 may generate a signal to regulate SA-mediated plant defense. A candidate downstream signaling could be derived from the second messenger molecule inositol (1, 4, 5) phosphate (InsP3), which is involved in regulating both biotic and abiotic stress responses in plants. InsP3 is generated by phospholipase C (PLC) catalyzing the hydrolysis of phosphatidylinositol-4,5-bisphosphate, a process activated by biotic and abiotic stimuli (Mueller-Roeber and Pical, 2002). Inositol 5-phosphatases (5PTases) terminate InsP3 signaling by hydrolyzing InsP3 (Erneux et al., 1998). One previous study showed that expression of PHT4;1 is co-regulated with InsP3 accumulation in the 5ptase mutant (Chen et al., 2008). This observation suggests a possibility that Pi transport by PHT4;1 is important for InsP3 signaling, which subsequently leads to defense responses. Further studies from metabolite analysis and genetics should reveal whether the pht4;1-1 mutant affects IP3 levels and, if so, how PHT4;1-mediated IP3 signaling is involved in regulating plant disease resistance.
Functional Redundancy among Members in the PHT4 Family
We showed that extra copies of PHT4;1 confer enhanced disease susceptibility, suggesting that PHT4;1 is a negative regulator of plant defense. However, we did not observe altered defense phenotypes in the two loss-of-function alleles (Figure 3). This is probably due to functional redundancy between PHT4;1 and other members in the family. Indeed, PHT4;1 shares high homology (from 27 to 65% identity) with five other family members. Except PHT4;6, all other members were localized or predicted to be in the plastid (Roth et al., 2004; Guo et al., 2008a; Pavon et al., 2008; Cubero et al., 2009). Thus, it is tempting to speculate that, like PHT4;1, some members in the PHT4;1 family also play a role in regulating plant defense. Further mutant analysis with multiple PHT4 family members being disrupted should reveal this possibility.
Bioinformatics analysis also revealed that PHT4;1-like proteins are present in diverse organisms, including plants and animals. Interestingly, a high degree of conservation (about 30%) is also shared between PHT4;1 and proteins from evolutionarily distant organisms, such as human sialin, rat VGLUT1, and VGLUT2 proteins, mouse Npt1, rabbit NaPi-1, and EAT-4 of Caenorhabditis elegans (Werner et al., 1991; Ni et al., 1994; Chong et al., 1995; Lee et al., 1999; Verheijen et al., 1999; Aihara et al., 2000). Notably, mutations in VGLUT1, VGLUT2, and EAT-4 result in severe neuronal diseases associated with aberrant cell death in rat and C. elegans (Raizen and Avery, 1994; Verheijen et al., 1999; Wojcik et al., 2004; Tordera et al., 2007; Garcia-Garcia et al., 2009). Consistent with the function of its animal homologs, we show here that PHT4;1 regulates disease resistance and cell death in Arabidopsis. Like some animal homologs, PHT4;1 family members were also demonstrated to have Pi transport activities (Guo et al., 2008a; Pavon et al., 2008; Cubero et al., 2009). Together, these observations suggest a common mechanism for the action of PHT4;1 and its homologs from plants and animals.
Like introducing extra copies of PHT4;1, the pht4;1-1 mutation also confers enhanced disease susceptibility, suggesting that the predicted PHT4;1-1 protein is gain-of-function in nature. For instance, the PHT4;1-1 protein might have an altered biochemical activity by changing the kinetics of substrate uptake and/or transporting Pi in a different direction at the organelle level, compared with the WT protein. It is also possible that PHT4;1-1 might be permeable to a different substrate profile compared with the WT protein. Further elucidation of the biochemical activity of PHT4;1 and PHT4;1-1 should facilitate the understanding of mechanisms of action of these proteins in plant defense and also reveal whether pht4;1-1 is a neomorphic or hypermorphic mutation.
A Potential Role of the Circadian Clock in Regulating Plant Defense
The circadian clock is the intrinsic time-measuring machinery that plays a central role in regulating plant growth and development and responses to environmental stimuli, such as light. Although light has long been implicated in regulating plant innate immunity (for review, see Roden and Ingle, 2009), whether or not the circadian clock regulates plant defense has not been well understood. However, previous studies that showed expression of some pathogen-inducible genes oscillated in a circadian manner indeed suggest such a possibility (Wang et al., 2001; Sauerbrunn and Schlaich, 2004; Weyman et al., 2006). To further corroborate this notion, we show here that expression of PHT4;1 is regulated by both the circadian clock and light (this study and Guo et al., 2008a). PHT4;1 is likely a target of the central oscillator component CCA1, since two copies of CBS were identified in the PHT4;1 promoter. CBS was also identified in the promoters of several key SA regulatory genes, such as SID2, suggesting a potential role of CCA1 in defense regulation. Further genetic and molecular analysis should reveal whether CCA1 regulates expression of PHT4;1 and other defense regulators and subsequently leads to altered defense responses.
METHODS
Plant Growth and Mutant Isolation
Unless otherwise indicated, all plants used in this paper are in Columbia background. Plants were grown in growth chambers with 200 μmol m−2 s−1 light intensity, 16-h light/8-h dark cycle, and 60% humidity. The mutants, acd6-1 and acd6-1sid2-1, and acd6-1 suppressor screen were described before (Lu et al., 2009). The acd6-1pht4;1-1sid2-1 triple mutant was made by crossing acd6-1pht4;1-1 and acd6-1sid2-1 and confirmed with a cleaved amplified polymorphic sequence (dCAPS) marker for acd6-1 and sid2-1 (Lu et al., 2009) and PCR markers to detect T-DNA insertion in pht4;1-1. pht4;1-2 (GT_5_110509) and pht4;1-3 (CSHL_GT22119), both disrupted by a transposon tag, were in Landsberg accession and were obtained, respectively, from Arabidopsis Biological Resource Center (ABRC) at Ohio State University and from the Martienssen's Laboratory at Cold Spring Harbor Laboratory in New York. Both mutants were confirmed with corresponding PCR markers for the DNA insertions. Primers used to detect pht4;1 mutants are listed in Supplemental Table S1.
Bacterial Infection
Bacterial strains Pseudomonas syringae pv. maculicola (Pma) ES4326 strain DG3 (PmaDG3), Pma avrRpt2, Pma avrRpm1, and P. syringae pv. tomato strain DC3000 (DC3000) were described previously (Guttman and Greenberg, 2001; Lee et al., 2007b). The fifth to seventh leaves of 25-day-old plants were infected with Pseudomonas strains by infiltration with a 1-ml needleless syringe. Details for bacterial culturing, infection, and growth analysis were described before (Greenberg et al., 2000; Lu et al., 2003). For benzo(1,2,3) thiadiazole-7-carbothioic acid (BTH) treatment, 25-day-old plants were sprayed with 300 μM BTH or water for 36 h before Pseudomonas infection. BTH was kindly provided by Robert Dietrich (Syngenta).
SA Measurement
Total SA were extracted from 25-day-old plants as previously described (Lu et al., 2003; Song et al., 2004) and quantified with a Dionex AS50 HPLC instrument with Acclaim® 120 C18 reverse column (4.6 × 250 mm) and RF2000 fluorescence detector. O-anisic acid was used as an internal standard. SA and o-anisic acid were eluted with a gradient of methanol and 0.5% acetic acid, with o-anisic acid being detected at 4.9 min with 301-nm excitation/365-nm emission and SA being detected at 6.5 min with 301-nm excitation/412-nm emission. The final concentration of each sample was calculated based on the average of three replicates, using a standard curve made from quantification of o-anisic acid and SA at concentrations of 50, 100, 250, 500, and 1000 ng ml−1.
Cell Death Staining
The fifth and sixth leaves of 25-day-old plants were collected for trypan blue staining for cell death (Rate et al., 1999). Stained leaves were examined with a Leica dissecting microscope and images were captured with a Photometrics CCD camera connected to the microscope and analyzed by MetaMorph image software.
RNA Analysis
Total RNA from 25-day-old plants was isolated using TRIzol reagent (Invitrogen). Northern blotting was performed as previously described (Lu et al., 2003). To make radioactive probes, polymerase chain reaction (PCR) was performed with primers and a DNA template specific for each gene in the presence of [32P]dCTP. For reverse-transcriptase PCR (RT–PCR), total RNA was reverse-transcribed into cDNAs, using the First Strand cDNA Synthesis kit (Fermentas) according to the manufacturer's instructions. Gene-specific primers were designed to span two exons sandwiched with an intron and were used in PCR to amplify the cDNA fragment of the corresponding gene, using RT products as templates. PCR products amplified from genomic DNA from Col-0 were different in size from their corresponding RT–PCR products and were used as controls. PCR products at 25 and 30 cycles were collected and separated on a 1.5% agarose gel. Primers used to make radioactive probes and in RT–PCR are listed in Supplemental Table S1.
DNA Construction and Plant Transformation
The PHT4;1-1 genomic DNA, including 1329-bp promoter region, was PCR amplified with At2g29650longST_493F and SUP3_T-DNA5' primers and cloned into the binary vector pGreenII 0029 (Hellens et al., 2000). The construct was transferred to Agrobacterium GV3101 for plant transformation, according to the floral dipping method (Clough and Bent, 1998). The full-length PHT4;1 genomic fragment was similarly cloned, using primers At2g29650longST_493F and At2g29650longST_4617R, and transferred to Col-0. Plant transformants were selected on soil by spraying with BASTA at a dilution of 1:4000 (AgrEvo USA, Wilmington, DE). At the T2 generation, homozygous lines were selected on MS agar plates containing the herbicide BASTA and further planted on soil for resistance test with Pseudomonas infection. Primers used to make DNA constructs are listed in Supplemental Table S1.
Accession Number
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession number At2g29650.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by startup funds from University of Maryland Baltimore County and a grant from National Science Foundation (NSF RIG-0818651) to H.L. and a scholarship from China Scholarship Council to J.L.S.
Supplementary Material
Acknowledgments
We thank Dr Stephen Miller at UMBC for valuable advice on the manuscript, Dr Robert McClung at Dartmouth College for discussions on the circadian work, and Dr Wayne Versaw at TAMU for sharing unpublished data. We thank Dr William LaCourse for his assistance in using the HPLC instrument and Tim Ford for taking pictures for this publication. Microscopic images were taken at Keith R. Porter Core Imaging Facility at UMBC. No conflict of interest declared.
References
- Aihara Y, et al. Molecular cloning of a novel brain-type Na+-dependent inorganic phosphate cotransporter. J. Neurochem. 2000;74:2622–2625. doi: 10.1046/j.1471-4159.2000.0742622.x. [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- Chen X, Lin WH, Wang Y, Luan S, Xue HW. An inositol polyphosphate 5-phosphatase functions in PHOTOTROPIN1 signaling in Arabidopsis by altering cytosolic Ca2+ Plant Cell. 2008;20:353–366. doi: 10.1105/tpc.107.052670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong SS, Kozak CA, Liu L, Kristjansson K, Dunn ST, Bourdeau JE, Hughes MR. Cloning, genetic mapping, and expression analysis of a mouse renal sodium-dependent phosphate cotransporter. Am. J. Physiol. 1995;268:F1038–F1045. doi: 10.1152/ajprenal.1995.268.6.F1038. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cubero B, Nakagawa Y, Jiang X, Miura K, Li F, Raghothama KG, Bressan RA, Hasegawa PM, Pardo JM. The phosphate transporter PHT4;6 is a determinant of salt tolerance that is localized to the Golgi apparatus of Arabidopsis. Mol. Plant. 2009;2:535–552. doi: 10.1093/mp/ssp013. [DOI] [PubMed] [Google Scholar]
- de Torres Zabala M, Bennett MH, Truman WH, Grant MR. Antagonism between salicylic and abscisic acid reflects early host–pathogen conflict and moulds plant defence responses. Plant J. 2009;59:375–386. doi: 10.1111/j.1365-313X.2009.03875.x. [DOI] [PubMed] [Google Scholar]
- Dong X. NPR1, all things considered. Curr. Opin. Plant Biol. 2004;7:547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Erneux C, Govaerts C, Communi D, Pesesse X. The diversity and possible functions of the inositol polyphosphate 5-phosphatases. Biochim. Biophys. Acta. 1998;1436:185–199. doi: 10.1016/s0005-2760(98)00132-5. [DOI] [PubMed] [Google Scholar]
- Falk A, Feys BJ, Frost LN, Jones JD, Daniels MJ, Parker JE. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl Acad. Sci. U S A. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Parker JE. Interplay of signaling pathways in plant disease resistance. Trends Genet. 2000;16:449–455. doi: 10.1016/s0168-9525(00)02107-7. [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia AL, Elizalde N, Matrov D, Harro J, Wojcik SM, Venzala E, Ramirez MJ, Del Rio J, Tordera RM. Increased vulnerability to depressive-like behavior of mice with decreased expression of VGLUT1. Biol. Psychiatry. 2009;66:275–282. doi: 10.1016/j.biopsych.2009.02.027. [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. Use of Arabidopsis for genetic dissection of plant defence responses. Annu. Rev. Genet. 1997;31:547–569. doi: 10.1146/annurev.genet.31.1.547. [DOI] [PubMed] [Google Scholar]
- Goritschnig S, Zhang Y, Li X. The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J. 2007;49:540–551. doi: 10.1111/j.1365-313X.2006.02978.x. [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Silverman FP, Liang H. Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics. 2000;156:341–350. doi: 10.1093/genetics/156.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Irigoyen S, Fowler TB, Versaw WK. Differential expression and phylogenetic analysis suggest specialization of plastid-localized members of the PHT4 phosphate transporter family for photosynthetic and heterotrophic tissues. Plant Signal. Behav. 2008;3:784–790. doi: 10.4161/psb.3.10.6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Jin Y, Wussler C, Blancaflor EB, Motes CM, Versaw WK. Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 2008;177:889–898. doi: 10.1111/j.1469-8137.2007.02331.x. [DOI] [PubMed] [Google Scholar]
- Guttman DS, Greenberg JT. Functional analysis of type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single copy genomic integration system. Mol. Plant–Microbe Interact. 2001;14:145–155. doi: 10.1094/MPMI.2001.14.2.145. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JD. Resistance gene-dependent plant defense responses. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran G, Raina S, Acharya BR, Maqbool SB, Mosher SL, Appel HM, Schultz JC, Klessig DF, Raina R. Arabidopsis GH3-LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae. Plant J. 2007;51:234–246. doi: 10.1111/j.1365-313X.2007.03130.x. [DOI] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl Acad. Sci. U S A. 1999;96:13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F. A global view of defense gene expression regulation: a highly interconnected signaling network. Curr. Opin. Plant Biol. 2004;7:506–511. doi: 10.1016/j.pbi.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CM. Cross talk in defense signaling. Plant Physiol. 2008;146:839–844. doi: 10.1104/pp.107.112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 2002;5:325–331. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 1996;10:71–82. doi: 10.1046/j.1365-313x.1996.10010071.x. [DOI] [PubMed] [Google Scholar]
- Lee J, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007;49:79–90. doi: 10.1111/j.1365-313X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- Lee MW, Lu H, Jung HW, Greenberg JT. A key role for the Arabidopsis WIN3 protein in disease resistance triggered by Pseudomonas syringae that secrete AvrRpt2. Mol. Plant–Microbe Interact. 2007;20:1192–1200. doi: 10.1094/MPMI-20-10-1192. [DOI] [PubMed] [Google Scholar]
- Lee RY, Sawin ER, Chalfie M, Horvitz HR, Avery L. EAT-4, a homolog of a mammalian sodium-dependent inorganic phosphate cotransporter, is necessary for glutamatergic neurotransmission in Caenorhabditis elegans. J. Neurosci. 1999;19:159–167. doi: 10.1523/JNEUROSCI.19-01-00159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WY, Lin SI, Chiou TJ. Molecular regulators of phosphate homeostasis in plants. J. Exp. Bot. 2009;60:1427–1438. doi: 10.1093/jxb/ern303. [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Lu H. Dissection of salicylic acid-mediated defense signaling networks. Plant Signal. Behav. 2009;4:1–5. doi: 10.4161/psb.4.8.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Rate DN, Song JT, Greenberg JT. ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell. 2003;15:2408–2420. doi: 10.1105/tpc.015412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Salimian S, Gamelin E, Wang G, Fedorowski J, LaCourse W, Greenberg JT. Genetic analysis of acd6-1 reveals complex defense networks and leads to identification of novel defense genes in Arabidopsis. Plant J. 2009;58:401–412. doi: 10.1111/j.1365-313X.2009.03791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- Michael TP, McClung CR. Phase-specific circadian clock regulatory elements in Arabidopsis. Plant Physiol. 2002;130:627–638. doi: 10.1104/pp.004929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl Acad. Sci. U S A. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- Mueller-Roeber B, Pical C. Inositol phospholipid metabolism in Arabidopsis: characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 2002;130:22–46. doi: 10.1104/pp.004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy AM, Otto B, Brearley CA, Carr JP, Hanke DE. A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J. 2008;56:638–652. doi: 10.1111/j.1365-313X.2008.03629.x. [DOI] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Metraux JP. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell. 2002;14:275–286. doi: 10.1105/tpc.010376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B, Rosteck PR, Jr., Nadi NS, Paul SM. Cloning and expression of a cDNA encoding a brain-specific Na+-dependent inorganic phosphate cotransporter. Proc. Natl Acad. Sci. U S A. 1994;91:5607–5611. doi: 10.1073/pnas.91.12.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K, Okrent RA, Stoutemyer M, Rodibaugh N, Kempema L, Wildermuth MC, Innes RW. The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 2007;144:1144–1156. doi: 10.1104/pp.107.097691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma K, Zhang Y, Li X. An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr. Biol. 2005;15:1129–1135. doi: 10.1016/j.cub.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Palma K, Zhao Q, Cheng YT, Bi D, Monaghan J, Cheng W, Zhang Y, Li X. Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 2007;21:1484–1493. doi: 10.1101/gad.1559607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavon LR, Lundh F, Lundin B, Mishra A, Persson BL, Spetea C. Arabidopsis ANTR1 is a thylakoid Na+-dependent phosphate transporter: functional characterization in Escherichia coli. J. Biol. Chem. 2008;283:13520–13527. doi: 10.1074/jbc.M709371200. [DOI] [PubMed] [Google Scholar]
- Poirier Y, Bucher M. Phosphate transport and homeostasis in Arabidopsis. 2002 doi: 10.1199/tab.0024. in The Arabidopsis Book, pp.1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Avery L. Electrical activity and behavior in the pharynx of Caenorhabditis elegans. Neuron. 1994;12:483–495. doi: 10.1016/0896-6273(94)90207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate DN, Cuenca JV, Bowman GR, Guttman DS, Greenberg JT. The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell. 1999;11:1695–1708. doi: 10.1105/tpc.11.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden LC, Ingle RA. Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant–pathogen interactions. Plant Cell. 2009;21:2546–2552. doi: 10.1105/tpc.109.069922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C, Menzel G, Petetot JM, Rochat-Hacker S, Poirier Y. Characterization of a protein of the plastid inner envelope having homology to animal inorganic phosphate, chloride and organic-anion transporters. Planta. 2004;218:406–416. doi: 10.1007/s00425-003-1121-5. [DOI] [PubMed] [Google Scholar]
- Ryals J, et al. The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell. 1997;9:425–439. doi: 10.1105/tpc.9.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD. Systemic acquired resistance. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerbrunn N, Schlaich NL. PCC1: a merging point for pathogen defence and circadian signalling in Arabidopsis. Planta. 2004;218:552–561. doi: 10.1007/s00425-003-1143-z. [DOI] [PubMed] [Google Scholar]
- Shah J, Tsui F, Klessig DF. Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA- inducible expression of the tms2 gene. Mol. Plant–Microbe Interact. 1997;10:69–78. doi: 10.1094/MPMI.1997.10.1.69. [DOI] [PubMed] [Google Scholar]
- Song JT, Lu H, McDowell JM, Greenberg JT. A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 2004;40:200–212. doi: 10.1111/j.1365-313X.2004.02200.x. [DOI] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Dong X. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science. 2008;321:952–956. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell. 2003;15:317–330. doi: 10.1105/tpc.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M, et al. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature. 2010;465:632–636. doi: 10.1038/nature09083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordera RM, Totterdell S, Wojcik SM, Brose N, Elizalde N, Lasheras B, Del Rio J. Enhanced anxiety, depressive-like behaviour and impaired recognition memory in mice with reduced expression of the vesicular glutamate transporter 1 (VGLUT1) Eur. J. Neurosci. 2007;25:281–290. doi: 10.1111/j.1460-9568.2006.05259.x. [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F. Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 2008;53:763–775. doi: 10.1111/j.1365-313X.2007.03369.x. [DOI] [PubMed] [Google Scholar]
- Verheijen FW, et al. A new gene, encoding an anion transporter, is mutated in sialic acid storage diseases. Nat. Genet. 1999;23:462–465. doi: 10.1038/70585. [DOI] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 2007;17:1784–1790. doi: 10.1016/j.cub.2007.09.025. [DOI] [PubMed] [Google Scholar]
- Wang ZX, Yamanouchi U, Katayose Y, Sasaki T, Yano M. Expression of the Pib rice-blast-resistance gene family is up-regulated by environmental conditions favouring infection and by chemical signals that trigger secondary plant defences. Plant Mol. Biol. 2001;47:653–661. doi: 10.1023/a:1012457113700. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell. 1997;9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A, Moore ML, Mantei N, Biber J, Semenza G, Murer H. Cloning and expression of cDNA for a Na/Pi cotransport system of kidney cortex. Proc. Natl Acad. Sci. U S A. 1991;88:9608–9612. doi: 10.1073/pnas.88.21.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyman PD, Pan Z, Feng Q, Gilchrist DG, Bostock RM. A circadian rhythm-regulated tomato gene is induced by Arachidonic acid and Phythophthora infestans infection. Plant Physiol. 2006;140:235–248. doi: 10.1104/pp.105.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- Wojcik SM, Rhee JS, Herzog E, Sigler A, Jahn R, Takamori S, Brose N, Rosenmund C. An essential role for vesicular glutamate transporter 1 (VGLUT1) in postnatal development and control of quantal size. Proc. Natl Acad. Sci. U S A. 2004;101:7158–7163. doi: 10.1073/pnas.0401764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li X. A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell. 2005;17:1306–1316. doi: 10.1105/tpc.104.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cheng YT, Bi D, Palma K, Li X. MOS2, a protein containing G-patch and KOW motifs, is essential for innate immunity in Arabidopsis thaliana. Curr. Biol. 2005;15:1936–1942. doi: 10.1016/j.cub.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.