Abstract
Waist circumference (WC) and waist-to-hip ratio (WHR) are surrogate measures of central adiposity that are associated with adverse cardiovascular events, type 2 diabetes and cancer independent of body mass index (BMI). WC and WHR are highly heritable with multiple susceptibility loci identified to date. We assessed the association between SNPs and BMI-adjusted WC and WHR and unadjusted WC in up to 57 412 individuals of European descent from 22 cohorts collaborating with the NHLBI's Candidate Gene Association Resource (CARe) project. The study population consisted of women and men aged 20–80 years. Study participants were genotyped using the ITMAT/Broad/CARE array, which includes ∼50 000 cosmopolitan tagged SNPs across ∼2100 cardiovascular-related genes. Each trait was modeled as a function of age, study site and principal components to control for population stratification, and we conducted a fixed-effects meta-analysis. No new loci for WC were observed. For WHR analyses, three novel loci were significantly associated (P < 2.4 × 10−6). Previously unreported rs2811337-G near TMCC1 was associated with increased WHR (β ± SE, 0.048 ± 0.008, P = 7.7 × 10−9) as was rs7302703-G in HOXC10 (β = 0.044 ± 0.008, P = 2.9 × 10−7) and rs936108-C in PEMT (β = 0.035 ± 0.007, P = 1.9 × 10−6). Sex-stratified analyses revealed two additional novel signals among females only, rs12076073-A in SHC1 (β = 0.10 ± 0.02, P = 1.9 × 10−6) and rs1037575-A in ATBDB4 (β = 0.046 ± 0.01, P = 2.2 × 10−6), supporting an already established sexual dimorphism of central adiposity-related genetic variants. Functional analysis using ENCODE and eQTL databases revealed that several of these loci are in regulatory regions or regions with differential expression in adipose tissue.
INTRODUCTION
Excess adiposity, especially central or visceral adiposity, is often a precursor to cardiovascular disease (CVD), type 2 diabetes (T2D) and cancer (1,2). The rising prevalence of obesity is becoming an increasing global concern (3,4). While major contributors leading to obesity such as diet and nutrition need to be further studied to inform better interventions, the biologic pathways that influence susceptibility to obesity are poorly understood. In an attempt to identify underlying genetic variants that affect adiposity traits and their distribution, many researchers have performed genome-wide association studies (GWAS) (5).
While measurements of obesity such as body mass index (BMI) and waist circumference (WC) represent a crude measurement of adiposity, they have been shown to be highly associated with cardiovascular disease-related outcomes and mortality (1,6–9). In addition, measurements of the distribution of adiposity such as waist-to-hip ratio (WHR), WC and visceral adipose tissue (VAT) have been associated with these adverse events, independent of BMI (10,11) indicating that WHR may be capturing overlapping and/or different etiologic pathways leading to poor health (12). Abdominal fat is thought to be more metabolically active and has been shown to confer a more adverse metabolic profile, in addition to increasing risk of cancer (13–17).
Several studies have also shown high heritability of adiposity measurements, indicating genetic contributions to variation in fat deposition (18). One such adiposity measure, WHR, has been shown to have ∼30–60% heritability (19,20) and shows large variation by sex (21). Previous GWAS have successfully highlighted a number of genetic loci and pathways that underpin obesity (22). Although there have been numerous GWAS of abdominal fat and adiposity-related traits (5,23–25), only one large meta-analysis by Heid et al. (26) focused on WHR adjusted for BMI (WHRadjBMI, henceforth referred to as WHR), which identified 14 associated loci. Of the 14 loci associated with WHR, 7 indicated heterogeneity by sex, and all 14 showed stronger effects among females in comparison with males. These findings are suggestive of distinct genetic effects on body shape and argue for the importance of sex stratification in the interrogation of genetic effects for adiposity-related traits.
Using meta-analysis, significant advancements have been made in identifying genetic loci, although only a small portion of the estimated heritability has to date been explained by the identified loci. Only 1.03% of the variance in WHR could be explained by the 14 loci identified by Heid et al. (26) from the Genetic Investigation of Anthropometric Traits (GIANT) WHR meta-analysis, indicating that the genetic contributions have not been fully identified or explained (27).
In this study, we used the ITMAT/Broad/CARe (IBC) array, designed to capture ∼50K SNPs across ∼2100 metabolic and cardiovascular-related loci, with the majority of these loci captured at equal or greater density when compared with conventional GWAS arrays (28). We employed this platform in a large meta-analysis of European descent individuals in an attempt to identify additional novel loci for WC and WHR and further investigate loci that affect these phenotypes in a sex-specific manner.
RESULTS
Population characteristics
A total of 22 studies, including 57 412 participants for WC analyses (49 380 for WHR analyses) of European descent met all criteria and were included in this meta-analysis (Supplementary Material, Tables S1–S3). The majority of participants were female (63.6%), with a mean age ranging between 32 and 69 years of age. Mean anthropometry measurements were similar between cohorts including WC, hip circumference (HC), height, weight, WHR and BMI (Supplementary Material, Table S2) with the exception of Look AHEAD which was selected for obesity. WC and BMI measurements were consistently higher among men in comparison with women in almost all cohorts. HC was on average higher in females in comparison with males amongst almost all cohorts (Supplementary Material, Table S3).
Meta-analysis
Sex-combined results
The sex-combined meta-analysis revealed six loci that reached array-wide significance (P < 2.4 × 10−6) for WHR adjusted for BMI (Table 1). Three loci were previously observed in the GIANT WHR meta-analysis greater than genome-wide significance thresholds: RSPO3, ADAMTS9 and ITPR2. SNPs rs2811337 (inTMCC1), rs7302703 (downstream of HOXC10) and rs936108 [in phosphatidylethanolamine N-methyltransferase (PEMT)], which reached genome-wide or array-wide significance (P = 7.65 × 10−9, 2.88 × 10−7, and 1.9 × 10−6, respectively), however, have not been reported before for association with WHR. The I2 values indicated mostly low levels of heterogeneity across studies. Regional association and Manhattan plots of detected loci are provided in Supplementary Material, Figures S1 and S2.
Table 1.
IBC array-wide significant SNPs associated with WHR in individuals of European descent
| Nearest gene | Chr | SNP | Variant type | CA | IBC |
GIANT (26) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Na | CAF | Effect (SE) | P-value | I2b | CAF | Effect (SE) | P-value | |||||
| RSPO3 | 6 | rs11154383 | Flanking 5UTR | G | 42 927 | 0.29 | 0.047 (0.007) | 3.0E−10 | 27.1 | 0.29 | 0.034 (0.005) | 1.3E−10c |
| TMCC1 | 3 | rs2811337 | Intronic | G | 48 549 | 0.82 | 0.048 (0.008) | 7.6E−09 | 21.1 | 0.82 | 0.018 (0.006) | 0.0036 |
| ADAMTS9 | 3 | rs4607103 | Intronic | C | 42 936 | 0.75 | 0.044 (0.008) | 2.4E−08 | 1.2 | 0.75 | 0.023 (0.006) | 4.3E−05c |
| ITPR2 | 12 | rs1049376 | Untranslated | C | 46 717 | 0.26 | 0.041 (0.007) | 5.6E−08 | 0.0 | 0.27 | 0.029 (0.005) | 7.6E−08c |
| HOXC10 | 12 | rs7302703 | 3′ downstream | G | 48 548 | 0.83 | 0.044 (0.008) | 2.9E−07 | 0.0 | 0.84 | 0.014 (0.008) | 0.067 |
| PEMT | 17 | rs936108 | Intronic | C | 35 827 | 0.46 | 0.035 (0.007) | 1.9E−06 | 23.5 | 0.47 | 0.014 (0.005) | 0.0041 |
Chr, chromosome; Pos, position; CA, coded allele; CAF, coded allele frequency; WHR, waist-to-hip ratio (adjusted for BMI, age, age2, center site and ancestry); SE, standard error.
aApproximately 63.2% female (exact percentage depending on SNP).
bHeterogeneity statistic (58).
cThese three loci were previously reported by Heid et al. (2010)26 but the most significant SNPs for each locus in Heid et al. were rs9491696 for RSPO3 (P = 1.8E−40, R2 = 0.28 with rs11154383); rs6795735 for ADAMTS9 (P = 9.8E−14, R2 = 0.27 with rs4607103) and rs718314 for ITPR2 (P = 1.14E−17; R2 = 0.64 with rs1049376). R2 (LD) was determined by 1000 Genomes Pilot 1 CEU.
In addition to the WHR analysis, we conducted a meta-analysis for WC and WC adjusted for BMI (WCadjBMI). Loci significantly associated with WC were FTO and APOE, and MFAP2 was significantly associated in this study with WC after adjusting for BMI (Supplementary Material, Table S4).
Sex-specific associations
Given that WHR has been previously reported to have significantly heterogeneous genetic effects by sex, we conducted a sex-specific analysis in addition to our combined meta-analysis. The female-only meta-analysis revealed two more array-wide significant associations: rs12076073-A in SHC1 (frequency = 0.96) increased WHR by 0.101 units (SE = 0.021) among females (P = 2.2 × 10−6) and had a slight association with WHR among males in the opposite direction (β = −0.066 ± 0.032, P = 0.040); and rs1037575-A in ATBDB4 (frequency = 0.79), which was associated with increased WHR in females only (β = 0.046 ± 0.010, P = 2.2 × 10−6) (Table 2). In males, the same allele had a null association with WHR (β = −0.002 ± 0.014, P = 0.89). No novel SNPs were identified in the male-specific meta-analysis for WHR (Table 3). Of all array-wide significant SNPs, the magnitude of absolute effect was stronger among females in comparison with males except for the rs7302703 SHC1 signal.
Table 2.
IBC array-wide significant SNPs associated with WHR among females
| Nearest gene | Chr | IBC |
GIANT, female (26) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | N | CA | CAF | Effect (SE) | P-value | I2 | SNPa | R2b | CA | CAF | Effect (SE) | P-value | ||
| RSPO3 | 6 | rs11154383 | 28 916 | G | 0.30 | 0.064 (0.009) | 1.5E−12 | 0.0 | rs9491696 | 0.28 | G | 0.48 | 0.050 (0.004) | 1.9E−32 |
| ADAMTS9 | 3 | rs4607103 | 28 927 | C | 0.75 | 0.055 (0.010) | 8.5E−09 | 0.0 | rs6795735 | 0.27 | C | 0.59 | 0.038 (0.005) | 1.9E−16 |
| TMCC1 | 3 | rs2811337 | 32 165 | G | 0.82 | 0.055 (0.010) | 1.7E−07 | 3.3 | rs2811337 | N/A | G | 0.82 | 0.038 (0.009) | 1.1E−05 |
| ITPR2 | 12 | rs1049376 | 31 254 | C | 0.26 | 0.046 (0.009) | 3.8E−07 | 0.0 | rs718314 | 0.64 | G | 0.26 | 0.042 (0.005) | 2.4E−17 |
| SHC1 | 1 | rs12076073 | 30 191 | A | 0.96 | 0.101 (0.021) | 1.9E−06 | 0.0 | rs12076073 | N/A | A | 0.96 | 0.087 (0.025) | 5.6E−04 |
| ATPBD4 | 15 | rs1037575 | 31 255 | A | 0.79 | 0.046 (0.010) | 2.2E−06 | 0.0 | rs1037575 | N/A | A | 0.79 | 0.017 (0.008) | 0.042 |
| SNPs that were significant in sex combined analysis (Table 1) but did not reach significance in female-specific analysis | ||||||||||||||
| HOXC10 | 12 | rs7302703 | 32 166 | G | 0.83 | 0.040 (0.010) | 1.3E−04 | 3.9 | rs7302703 | N/A | G | 0.84 | 0.007 (0.011) | 0.52 |
| PEMT | 17 | rs936108 | 24 807 | C | 0.46 | 0.037 (0.009) | 4.4E−05 | 38.9 | rs936108 | N/A | C | 0.47 | 0.020 (0.007) | 3.2E−03 |
Chr, chromosome; Pos, position; CA, coded allele; CAF, coded allele frequency; WHR, waist-to-hip ratio (adjusted for BMI, age, age2, center site and ancestry), SE, standard error.
aSame SNP used where available; otherwise closest proxy SNP was used (highest R2 in 1000 Genomes Pilot 1 CEU).
bR2 with IBC SNP in 1000 Genomes Pilot 1 CEU. The coding allele was selected based on consistency of MAF with the IBC SNP.
Table 3.
SNPs associations and effects with WHR among males for SNPs that reached significance in the sex-combined and female-specific analyses
| Nearest gene | Chr | IBC |
GIANT, male (26) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | N | CA | CAF | Effect (SE) | P-value | I2 | SNPa | R2b | CA | CAF | Effect (SE) | P-value | ||
| SNPs that were significant in sex combined or female-specific analysis but did not reach significance in male-specific analysis | ||||||||||||||
| RSPO3 | 6 | rs11154383 | 12 935 | G | 0.29 | 0.012 (0.014) | 0.39 | 31.9 | rs9491696 | 0.28 | G | 0.48 | 0.031 (0.005) | 1.1E−11 |
| ADAMTS9 | 3 | rs4607103 | 12 933 | C | 0.74 | 0.024 (0.014) | 0.088 | 5.3 | rs6795735 | 0.27 | C | 0.60 | 0.011 (0.005) | 0.27 |
| TMCC1 | 3 | rs2811337 | 15 213 | G | 0.82 | 0.035 (0.015) | 0.020 | 20.8 | rs2811337 | N/A | G | 0.83 | −0.002 (0.009) | 0.79 |
| ITPR2 | 12 | rs1049376 | 15 211 | C | 0.26 | 0.025 (0.013) | 0.057 | 0.0 | rs718314 | 0.64 | G | 0.26 | 0.017 (0.005) | 0.0014 |
| SHC1 | 1 | rs12076073 | 13 532 | A | 0.96 | −0.066 (0.032) | 0.040 | 4.2 | rs12076073 | N/A | A | 0.97 | −0.004 (0.026) | 0.86 |
| ATPBD4 | 15 | rs1037575 | 15 207 | A | 0.78 | −0.002 (0.014) | 0.91 | 0.0 | rs1037575 | N/A | A | 0.79 | 0.002 (0.009) | 0.81 |
| HOXC10 | 12 | rs7302703 | 15 211 | G | 0.83 | 0.058 (0.015) | 1.3E−04 | 0.0 | rs7302703 | N/A | G | 0.84 | 0.021 (0.011) | 0.054 |
| PEMT | 17 | rs936108 | 9026 | C | 0.47 | 0.035 (0.015) | 0.018 | 22.5 | rs936108 | N/A | C | 0.47 | 0.011 (0.007) | 0.13 |
Chr, chromosome; Pos, position; CA, coded allele; CAF, coded allele frequency; WHR, waist-to-hip ratio (adjusted for BMI, age, age2, center site and ancestry); SE, standard error.
aSame SNP used where available; otherwise closest proxy SNP was used (highest R2 in 1000 Genomes Pilot 1 CEU).
bR2 with IBC SNP in 1000 Genomes Pilot 1 CEU. The coding allele was selected based on consistency of MAF with the IBC SNP.
Statistical tests for heterogeneity by sex
Of the three SNPs identified in the sex-combined meta-analysis, two (rs7302703, downstream of HOXC10 and rs936108-PEMT) had similar effect sizes in males and females. There was a larger observed effect and stronger association among females compared with males for rs2811337 (TMCC1), although a Wald chi-square test for heterogeneity did not yield any statistical evidence of differences between sexes (Pheterogeneity = 0.29). Of the signals observed in females only (rs12076073-SHC1 and rs1037575-ATBDB4) the direction of effect was positive among females with strong association levels with WHR, while in comparison the direction of effect was negative for males with low association with WHR. The Wald test for heterogeneity was significant for both SNPs (Pheterogeneity = 1.0 × 10−5 and 0.01, respectively). The adjusted P-value allowing for heterogeneity by sex did not indicate an array-wide significant association with WHR for rs1037575 (P = 1.3 × 10−5) but allowing for heterogeneity by sex resulted in a stronger association for rs12076073 (P = 1.4 × 10−6).
Corroboration of findings using the GIANT central adiposity studies
Of the 14 significant WHR loci identified within the GIANT consortium, five loci were represented on the IBC array, including RSPO3, ADAMTS9 and ITPR2, which were array-wide significant in this study. The GIANT index SNPs were different from the sentinel SNPs from our analysis, but were in linkage disequilibrium (LD) (r2 > 0.2) with one another in each of the three loci, suggesting the SNPs are tagging the same causal variants. The remaining two loci, LYPLAL1 and NISCH-STAB1, displayed nominally significant associations (P = 1.32 × 10−5 and 0.0019, respectively) with WHR in our study (Supplementary Material, Table S5) with both having an r2 >0.6 with the previous GIANT index signal.
To substantiate our five novel findings in other large European descent populations, we attempted to corroborate our top signals in the WHR association results from the GIANT study (26). Although rs2811337 in TMCC1, rs730273 near HOXC10, rs936108 in PEMT, rs12076073 in SHC1 and rs936108 in PEMT were not genome-wide significant in GIANT, the effect allele frequencies and the direction of effect were consistent with our findings in both the sex-combined and sex-specific results (Tables 1–3; Supplementary Material, Fig. S4a–e). The observed significance was P = 3.6 × 10−3 for rs2811337, P = 0.067 for rs730273 and P = 4.1 × 10−3 for rs936108 in the sex-combined GIANT results. Consistent with our findings, rs12076073 in SHC1 showed a larger positive direction of effect among females and no significant effect among males (P = 5.6 × 10−4 and P = 0.86, respectively). Also consistent with our study, the A allele of rs1037575 (ATPBD4) was associated with increased WHR in GIANT among females (P = 0.042) but not males (P = 0.81). Although the HOXC10 locus was not strictly corroborated in GIANT (P = 0.067), we include it in this table because of the consistent direction of effect across the studies and noted the stronger effect among males in both the GIANT study and our study (Tables 2 and 3).
Shared genetic effects across other anthropometry phenotypes
To describe potential shared or pleiotropic effects of our significant WC, WCadjBMI and WHR signals on other anthropometric traits, we examined effect sizes, standard errors and P-values for the same SNPs across the anthropometric traits BMI, height, WC, WCadjBMI and WHR both in sex-combined and in sex-stratified groups.
The top three independent WC SNPs (in FTO, APOE and CD19) were also associated with BMI in the sex-combined, female-only and male-only meta-analyses (Pbinomial = 1.25 × 10−4) and also showed consistency in the direction of effect. However, none of the SNPs identified for WC were also associated with WCadjBMI, height or WHR. The MFAP2 signal, identified for WCadjBMI, was associated with height (P < 0.05) but not with other anthropometric traits.
The six loci associated with WHR revealed a slightly more complex pattern of shared effects across various anthropometry phenotypes. In the sex-combined analyses, WHR signals were also associated with WCadjBMI, height and BMI. Stratification by sex indicated that the WHR signals may also have effects on WCadjBMI among females with generally consistent directions of effect. Among males, WHR signals may also have effects on height but with the opposite direction of effect (Supplementary Material, Tables S6–S8).
Pleiotropic effects across other CVD-related phenotypes
To the extent that the data were available, we investigated the effects of our novel SNPs across cardiovascular-related phenotypes published in other GWAS meta-analyses (Supplementary Material, Table S9). Although there were a few nominally significant P-values (between 0.03 and 0.05), none of the SNPs replicated across the other phenotypes including height, blood pressure, cholesterol, glucose levels, HOMA-B or HOMA-IR after accounting for the number of tests.
An additional review of all variants in LD (r2 > 0.2) with associated SNPs through the NIH GWAS catalog did not reveal other central adiposity-related phenotypes such as lipids, blood pressure, BMI and insulin resistance. The locus on Chr17 has some very weak evidence of pleiotropy: the SNP rs12936587 has previously been associated with coronary artery disease (29), but the SNP shows low LD with our association (r2 = 0.26) and no evidence of association in imputation analysis.
Functionality analysis of variants and genes in novel associated loci
Network analysis
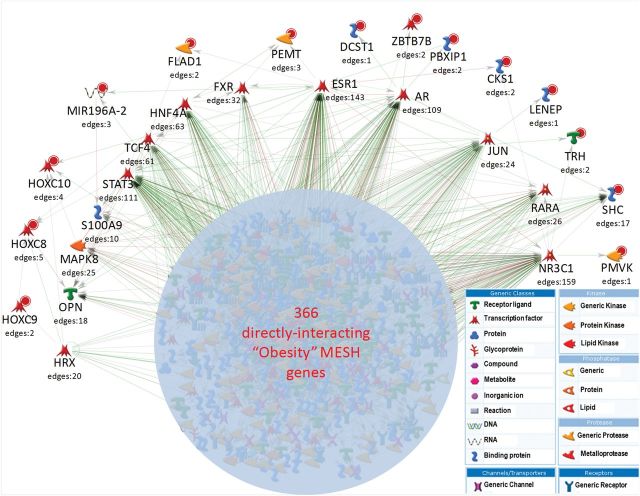
A network analysis of the extended LD (r2 > 0.2) region surrounding the five novel reported loci, resulted in 22 genes showing evidence of association. In order to evaluate the biological rationale of these 22 genes in adiposity, we used genego metacore to evaluate them for direct interaction with the 726 genes previously linked to adiposity and obesity in the literature by Medical Subject Headings (MeSH) terms. Fourteen genes in the five associated loci showed direct interaction with genes previously linked to adiposity by MESH in a single network of 631 directly interacting genes (Fig. 1). In the network, 158 genes were defined as hubs, based on eight or more interactions within the network, including SHC1, which underlies one of our index adiposity SNP findings.
Figure 1.
A medical subject headings (MESH)-defined obesity gene network including interacting genes reported in this study. GWAS genes reported here are indicated by a red dot. Key network hubs connecting with GWAS genes including SHC1 are shown. The number of reported gene–gene interactions (‘edges’) is indicated below the gene. All 631 genes are not shown for clarity; however, they are listed in Supplementary Materials, Table S11. Network was prepared using GeneGO metacore (Thomson Reuters).
Functional analysis of the five novel loci
Several bioinformatic databases were queried to highlight variants and genes of interest within the novel loci. The full genomic context of the five novel loci reported here are shown in Supplementary Material, Fig. S3a–e. The figures include associated SNPs, LD relationships, imputation results, Encyclopedia of DNA Elements (ENCODE) and tissue specific RNA-seq data, including adipose tissue. Functional analysis of all variants showing LD with associated SNPs is reported in Supplementary Material, Table S12. All genes reported in the loci are listed in Supplementary Material, Table S10. Of the five novel loci with array-wide significance, all contained some evidence to support involvement in adipogenic or obesogenic pathways, explained in more detail below.
Chromosome 1 locus analysis
The 185 kb LD region (r2 > 0.2) surrounding the SNP rs12076073 includes KCNN3, CKS1B, PMVK, FLAD1, PBXIP, ZBTB7B, SHC1, LENEP, DCST1 and another microRNA precursor, mir4258. SHC1, containing this index SNP, is involved in a complex network of gene and protein pathways, and in our network analysis SHC1 is a hub protein interacting with 17 other proteins in the obesity network (Fig. 1). Review of published RNA-seq expression data from adipose tissue shows that SHC1 is one of the most highly adipose-expressed genes in the region (Supplementary Materials, Fig. S3a). Additionally, the sentinel SNP is in complete LD with a non-synonymous Met410Val variant (rs8191979). Although this is a conservative amino acid substitution which is predicted benign by Annovar analysis, the methionine residue is conserved in all available mammalian sequences. Seven other genes showed interaction with the obesity MeSH network (PBXIP1, CKS1B, PMVK, FLAD1, ZBTB7B, LENEP and DCST1); however, investigation of functionality did not reveal any to be strong candidates in this region, apart from PBXIP1 which may interact in known hyperglycemia pathways.
Chromosome 3 locus analysis
The 342 kb LD region surrounding the SNP rs2811337 spans the genes TMCC1, PLXND1, TRH and a hypothetical gene, LOC100507032. A review of ENCODE information identified several variants with potential for regulatory impact. Biological literature resulting from pathway analysis implicates TRH in hypothyroidism and hyperglycemia. No other gene in the region was an obvious obesity candidate gene, although RNA-seq data identifies PLXND1 as the most highly adipose expressed gene in the region (Supplementary Material, Fig. S3b).
Chromosome 12 locus analysis
The 17 kb LD block surrounding the SNP rs7302703 encompasses a well-preserved family of homeobox genes (HOXC9, HOXC10, HOXC8 and the micro RNA precursor, mir196A2). A review of expression data for the genes within the region highlighted HOXC8 and HOXC9 in relation to adipose studies. Possible functionality of variants across the region showing direct or indirect evidence of association with adiposity indicated a variant within HOXC10 may influence regulation. The HOXC10 signal (rs7302703) is also in a binding site of the second most highly connected hub in our obesity network, the estrogen receptor (ESR1). Another variant (rs12822416) 2.7 kb upstream of HOXC8 is also located in an ESR1-binding site: this SNP is in strong LD (r2 = 0.85) with the index signal and also shows highly significant association by imputation (P = 1.65 × 10−7).
Chromosome 15 locus analysis
The 202 kb LD region surrounding rs1037575 spans the ATPBD4 gene and its antisense transcript ATPBD4-AS1. Evaluation of ENCODE data finds strong evidence to support rs1037575 as the most putative causal variant in the region. The SNP is located in both CEBPB and STAT3 transcription factor-binding sites and functional disruption of ATPBD4 appears most likely at a transcriptional level, via disruption of CEBPB and STAT3 transcription factor-binding sites. As the rs1037575 association is female specific, we reviewed DNase I data across 125 ENCODE cell types (derived from both males and females), using the UCSC genome browser. The only cell types showing DNase I hypersensitivity at the rs1037575 locus were female derived, including HELA cells (Cervical Carcinoma), HMEC (human mammary epithelial cells) and GM12878-XiMat (B-lymphocytes derived from a female subject). This information is not sufficient to conclusively support a sex-specific effect of rs1037575, but it does highlight the locus as a candidate for further investigation. The CEBPB and STAT3 genes were the third and fourth most highly connected hubs in our obesity network.
Chromosome 17 locus analysis
The 133 kb LD region surrounding rs936108 spans the PEMT and an antisense transcript, RP11-524F11.2. Analysis of associated variants identifies two variants in the PEMT gene that may be of interest. First, rs897453 encodes a Val58Leu variant in the second transmembrane domain of the protein. The variant is predicted benign in impact and valine is not highly conserved at this position in mammals. Secondly, ENCODE data indicates a strongly associated SNP (rs750546, R2 = 0.67 in 1000 Genomes CEU) with a putative regulatory impact in an ESR1 binding site, linking PEMT into our extended obesity network.
DISCUSSION
In a large meta-analysis of ∼2100 cardiovascular-related genes in nearly 50 000 individuals of European descent from 22 cohorts, variation in 5 additional novel loci (3 in sex-combined and 2 in sex-specific analyses) was associated with WHR. Consistent with previous literature, analysis of the sexes separately showed larger effects in females, with two loci only identifiable in females. Association results, in silico corroboration efforts, examination of the effect of identified variants on multiple anthropometric measures, network analysis, transcriptional analysis and biological plausibility suggest the identified genes play a role in the biology of WHR.
Most of the heritability of central adiposity traits has yet to be explained by variants identified through genetic meta-analyses to date. Analysis conducted by Heid et al. (26), in the GIANT (Genetic Investigation of Anthropometric Traits) consortium, employing a genome-wide genotyping platform, identified loci with strong adiposity-related phenotypic associations. The candidate gene-centric CARe IBC SNP array was designed before the report by Heid et al. and thus did not explicitly include subsequently index SNPs or loci unveiled for central adiposity. Moreover, while their results were used as in silico corroboration to validate our findings, it should be noted that either a fraction or total number of participants in 7 of the 22 cohorts (up to 38% of the individuals) included in the current study were also genotyped in GIANT. However, a different genotyping platform was used in GIANT, and even with a different platform and different index SNPs for some loci, the GIANT GWAS results showed consistency in the direction of effect with the IBC array results. The forest plots (Supplementary Materials, Fig. 4A-e) also demonstrate the lack of heterogeneity across the studies used in this analysis, regardless of whether the study was also part of GIANT. Thus, it does not appear that the overlapping cohorts alone are driving the effects observed in this analysis.
Despite its larger sample size, the effect sizes in GIANT were smaller than those detected in the IBC discovery cohorts. Varying LD patterns in different populations with lower LD between the index SNP and the functional variant in some studies could diminish the detected effect. Though corroboration with GIANT findings substantiated our findings, further replication analyses will be needed to label our novel SNPs as lying within established loci. The 14 loci identified in the GIANT study explained 1.03% of the variance in WHR, and the 3 novel SNPs identified here in the sex-combined analysis explain an additional 0.18% of variance. However, as we did not conduct a two-stage study design, our effect estimates may be inflated, though our large sample size would reduce such a bias (30). The large sample size employed in our analysis also resulted in the identification of a low frequency variant associated with central adiposity [rs12076073 in SHC1 which has a minor allele frequency (MAF) of ∼0.04].
Pleiotropic effects across other anthropometric and CVD-related phenotypes
Comparison of our top identified SNPs across other anthropometric traits suggests that many SNPs have shared effects across multiple measures of anthropometry. Not surprisingly, SNPs associated with WC were also associated with BMI, affirming that the SNPs are capturing overall body size. The WCadjBMI identified SNP in MFAP2 was originally identified in a previous height GWAS study (31). It can be reasoned that when WC, which is highly correlated with weight, is adjusted for BMI, a composite measure of weight and height, the resulting SNPs may capture the residual variances in height. This highlights the importance of understanding what aspects of anthropometry are captured when using measures that are supposed to be proxies of adiposity.
Potential biologic significance of newly discovered loci
There is growing evidence to indicate that non-coding regions of the genome may have a regulatory function which can impact disease phenotypes at a gene or pathway level (32). We have taken advantage of the increased accessibility to large data sets that have cataloged genetic regions with potential functionality, allowing us to perform a comprehensive evaluation of the impact of direct and indirectly associated variants on the adiposity phenotype (32–34). By combining this with the rich structured annotation afforded by MeSH (http://www.ncbi.nlm.nih.gov/mesh/), we have been able to construct a conservative network of direct gene interactions that have been reproducibly linked to obesity in the literature. We carried out a network analysis of the 22 genes in the 5 novel loci reported here and found that 14 genes interact in the obesity gene network presented. These interactions are not enriched above the level expected by chance (data not shown); however, at a qualitative level they highlight a number of potential novel interactions with known obesity-related pathways.
We found strongest evidence linking a gene to adiposity in the chromosome 1 locus, with evidence for regulation of metabolic and adipose transport pathways. Although the locus contains 10 genes, the Src homology 2 domain containing transforming protein (SHC1) gene is a strong adiposity candidate. The gene is one of the strongest functional candidates in the locus, based on the association at a Met410Val variant, which although is a conservative amino acid substitution predicted to be benign, the methionine residue is conserved in all available mammalian sequences. But perhaps most significantly, it is also a key hub in the obesity MeSH network, indicating its potential role in the regulation of metabolic and adipose pathways. Mice lacking the SHC1 protein live longer and are leaner than wild-type animals, suggesting that this molecule may have a role in metabolic derangement and premature senescence by over-nutrition (35). SHC1 is known to activate the insulin receptor, by release of phospho-SHC, triggering a cascade of signaling events via SOS, RAF and the MAP kinases (http://www.ncbi.nlm.nih.gov/biosystems/106423) (36). The SHC1 gene also potentially represents a highly novel target opportunity in adiposity: Choi et al. (37) described 64 small molecule peptide mimetic molecules with antagonistic-binding activity at SHC1 (full activities reported in ChEMBL). These molecules could represent important tool compounds for functional studies of SHC1 in animal models and could certainly represent a starting point for further medicinal chemistry. In addition to the SHC1 gene, the literature from our functionality analysis shows that the pre-B-cell leukemia homeobox interacting protein 1 (PBXIP1) gene within the chromosome 1 locus may also be relevant, as it is known to interact with PBX1, which has developmental roles in the pancreas and can affect hyperglycemia and weight loss (38,39). However, considering the strong evidence linking SHC1 to obesity we presently consider it the most likely candidate in this region.
In the chromosome 3 locus, thyrotropin-releasing hormone (TRH) is the most promising biological candidate with a role in hypothyroidism in mouse models, which also affects pancreas function, resulting in hyperglycemia (40). However, no strong candidate functional variants were observed in TRH. Another candidate in the region on the basis of functional variation is transmembrane and coiled coil domain 1 (TMCC1), a variant in which [rs12494774 (r2 = 0.43)] is reported to mediate a TMCC1 eQTL and is also located in a binding site for HNF4A which showed substantial connectivity in the obesity MESH network. The role of TMCC1 is largely unclear, although it appears to function in cell signaling and cell regulation and has homology to plectin proteins, which have been shown to be important during cytoskeletal remodeling in the adipogenesis (41).
In the chromosome 12 locus a cluster of HOX family genes showed a range of support in adiposity, including altered expression in adipose tissue. Yamamoto et al. (42) reported differential expression of both HOXC8 and HOXC9 in subcutaneous and intra-abdominal adipose tissue. Fasting in both lean and ob/ob mice systematically decreased the expression of HOXC8 but produced only variable changes in the expression of other developmental and adipogenic genes. In a study of depot- and sex-dependent differences in gene expression in human abdominal and gluteal adipose tissues, Karastergiou et al. (43) reported differential expression of many HOX genes between the abdominal and gluteal depot, including highly significant down-regulation of HOXC8 in the gluteal depot in both sexes. Several variants across the region showed evidence of functional impact on binding sites for the estrogen receptor (ESR1), possibly suggesting a haplotype influence on the expression of multiple HOX genes via an ESR1 mediated regulatory mechanism. Though the results of observational studies have been mixed, studies with model organisms indicate that ER-alpha has an effect on obesity (ER-alpha is encoded by the gene ESR1 in humans). ER-alpha knockout mice have been shown to develop increased abdominal tissue, insulin resistance and glucose intolerance (44,45). This mounting evidence for the association of adiposity traits with the HOX cluster gene may indicate a combined influence of multiple HOX genes, possibly via a common regulatory mechanism through ESR1. Several HOX genes, including HOXC9 and HOXC8, are also known to dimerize with the PBX1 protein described above (46). Finally, we note that rs7302703 is downstream of HOXC10. The GIANT WHR meta-analysis previously reported an SNP within HOXC13, which is near HOXC10 but no LD is observed between the two SNPs, indicating that these variants may be independent but may function similarly on WHR.
Our final two loci reported have less clear established rationale in adiposity, but may offer an important new insight into the phenotype. In the chromosome 15 locus, ATPBD4, recently identified as a diphthamide amidase, is a critical component of the metabolic pathway targeted by diphtheria toxin and may play a role in adiposity (47). Although pathway and literature-based exploration of ATPBD4 and ATPBD4-AS1 has to date not revealed any clear involvement in adipogenesis, the metabolic role of the protein is still under investigation and the protein family are known to act on a range of endogenous lipid messengers, including oleamide and the endocannabinoid anandamide, which are known to modulate a number of neurobehavioral processes in mammals, including pain, sleep, feeding and locomotor activity (48). We performed a structure-based evaluation of the druggability of the ATPBD4 protein using the dogsitescorer tool (49) and identified two potentially druggable-binding pockets in the protein (data not shown). If further evidence is found to support this gene in adiposity, it could represent a highly novel therapeutic target. Finally in the chromosome 17 locus, we found a non-synonymous variant and a regulatory variant that could mediate a role for PEMT gene in adiposity, which is also supported by mouse knockout studies that have demonstrated that PEMT-deficient mice are protected from diet-induced obesity, supporting the likelihood that this gene and, possibly the antisense transcript, may have a role(s) in adiposity (50).
In conclusion, our study has identified five novel signals associated with WHR. Like previous studies, the effect size and strength of association is larger among females for most of the SNPs in comparison with males. Our investigation of the potential biological function of these novel loci suggest that many of these SNPs play a functional role that may disrupt the biological pathways that affect adiposity development.
MATERIALS AND METHODS
Studies
Studies with participants that had at least baseline anthropometry measurements of European ancestry and typed using the IBC array were invited to participate in the meta-analysis. Twenty-two studies (15 population-based, 6 case–control and 1 randomized controlled trial) provided summary level data for all SNPs on the IBC array. Detailed information regarding recruitment and phenotype measurement methods are provided in Supplementary Material, Table S1, and basic study level characteristics including average age and average anthropometry measurements of all 22 studies inclusive of CARe are outlined in Supplementary Material, Tables S2–S3.
Phenotypic measurements
Individual studies
Individuals <20 years of age were excluded. All waist-related measurements were measured either by trained personnel or health practitioners. WC and HC measurements were measured in each participating cohort and were converted to centimeters as necessary. WHR was calculated by dividing waist (cm) by hip (cm). Individuals provided informed consent according to each of the contributing studies' protocol. Individuals who did not consent to genetic analyses were excluded.
Genotyping
All participants were genotyped using the ITMAT/Broad/CARE (IBC) chip, which includes up to 49 240 cosmopolitan tagged SNPs across ∼2100 candidate genes (28). As several of the cohorts did not have imputed variants available, only directly genotyped SNPs were included for the analysis. Quality control of the genotypic data was maintained by removing SNPs with call rates <90% or not in Hardy–Weinberg equilibrium (P < 1 × 10−6) and removing samples with call rates <95%. An array-wide Bonferroni correction was applied taking LD into account for the number of independent tests using CEPH HapMap1 data (P = 2.4 × 10−6) (51), which has been employed in a number of IBC meta-analyses to date (52–54). Genotype imputation for a subset of the cohorts was performed using the software MACH (55). HapMap phase 2 data sets (www.hapmap.org) were used for the reference panel (build 36 release 22) consisting of CEU HapMap-phased haplotypes. Genotype imputation resulted in allelic-dosage data, representing the expected number of copies of the minor allele a subject carries, on ∼2.2 million autosomal SNPs. Imputation results were only available for the six cohorts from the CARe consortium (ARIC, CARDIA, CHS, MESA, CFS and FHS) and were used to investigate regions around the top loci for functional analysis.
Statistical analysis
Regression analysis
Studies used genotype strand conversion scripts when applicable to ensure uniform comparisons between studies in the meta-analysis phase. Each study created the phenotype variable by modeling WHR and WC as a function of age, age2, BMI where applicable and center site (if the study was composed of multiple centers) to create residuals. The residuals were then regressed on each SNP and 10 principal components to account for population stratification (principle components identified with Eigenstrat or PLINK (56)). For each study we assumed an additive genetic model and analyses were stratified by sex and combined for joint analysis where possible.
Meta-analysis
We conducted a fixed-effect meta-analysis using the regression results from each study with an inverse variance-weighted approach in METAL (57). Meta-analysis results were filtered to remove monomorphic alleles and SNPs, and those with missing strand information. Heterogeneity across studies was quantified using the I2 statistic (58).
Sex-specific analysis
Given previous findings on sex dimorphism of the WHR trait, we conducted a sex-specific analysis by running the meta-analysis on males and females separately. Pheterogeneity values were calculated using a Wald chi-square test using the effect size (β) and stand error (SE) specific to males (M) and females (F) to detect heterogeneity driven by sex differences. A two degree of freedom chi-square test was used to identify any significant SNPs once allowing for the heterogeneity between sexes (59).
Test for heterogeneity =
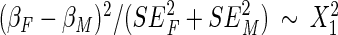 .
.Test allowing for heterogeneity =
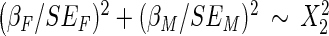 .
.
Pleiotropic analysis
Shared genetic effects across other anthropometry phenotypes
To assess whether our strongest signals in each of our reference phenotypes (WC, WCadjBMI or WHR) have effects across other anthropometric phenotypes, we first examined the associations of the same SNPs with other anthropometric phenotypes with IBC array meta-analysis data (BMI, height, WC, WCadjBMI and WHR). We then employed a binomial test to identify the comparator phenotypes that indicate shared effects.
For the binomial test P-value calculation, we treated the number of independent SNPs that were array-wide significant with the reference phenotype as the number of tests. To ensure the tests were independent, we selected only one SNP with the strongest P-value from each independent locus. Of those SNPs, the number of SNPs that also have a P-value of <0.05 with the comparator phenotype was the number of ‘successes’. We treated a binomial test resulting in a P-value of <0.05 as significant.
Shared genetic effects across other CVD-related phenotypes
We investigated whether the five novel loci showed evidence of association with related phenotypes using publicly available data from the GIANT consortium [BMI (23) and height (31)]; the MAGIC consortium [fasting glucose, fasting insulin, HOMA-B and HOMA-IR (60), and 2 h glucose (61)]; the ICBP-GWAS consortium [(SBP and DBP) (62)] and a genome-wide association scan for lipids [(HDL, LDL, triglycerides and total cholesterol) (63)] and glycemic traits from www.magicinvestigators.org. We reported the P-value for association for the same SNP and also the direction of effect where available.
In addition, potential pleiotropic effects of all variants in LD (r2 > 0.2) with the index SNPs were reviewed in the NIH GWAS catalog (http://www.genome.gov/gwastudies/) presented in the UCSC genome browser (64).
Functional analyses
Investigation of potential functionality and biologic role of variants and genes within resulting loci was evaluated based on known gene annotations, literature information, pathway analysis and functional analysis of variants as described below. Data collected from the bioinformatic methods described below were used to make an overall qualitative assessment of genes with the highest degree of support in adiposity-related traits in the identified loci.
The extended locus around each index SNP was defined by identification of all SNPs showing an r2 > 0.2, and a core locus within this region was defined by SNPs showing r2 > 0.5. We used an inclusive strategy for our LD analysis, reviewing all SNPs with r2 LD > 0.2. We used a low threshold for LD to allow for weak LD that may exist between variants of widely differing allele frequencies, as might be expected between higher frequency SNPs on genotyping panels and lower frequency SNPs with potential for disease causality that are likely to be subject to negative selection pressure. We considered the more widely used r2 > 0.5 as moderate to good evidence of LD, again depending on the difference in allele frequencies. LD was defined using the HaploReg tool which includes LD data derived from Phase I of the 1000 genomes project. The full genomic context of the loci defined by this process is shown in Supplementary Material, Figure S3a–e and the genes are listed in Supplementary Material, Table S10.
Network analysis: exploration of involvement in adiposity related pathways
We undertook a comprehensive approach to investigate the involvement of genes in associated loci with adiposity-related traits. First, we performed a text mining approach, querying genes in Medline with obesity and diabetes Medical subject headings (MeSH). Next all 726 genes linked to obesity by MeSH term were combined with the 22 genes present in the five novel loci reported here. The combined list was used to build a network of direct interactions in GeneGo (Supplementary Material, Table S11).
Exploration of regulation and expression
Loci of interest were investigated at both the gene and variant levels using a range of bioinformatics tools and databases. Variants showing evidence of LD with associated variants were explored for impact on gene function using ANNOVAR (65) and regulatory function using a combination of HaploReg (34) and Regulomedb (33), which both draw on comprehensive data from the Encyclopedia of DNA Elements (ENCODE) (32) and published eQTL studies. We mainly focused on three functional elements provided through ENCODE. ChIP-seq provides information on transcription-factor-binding sites. DNase-seq provides information on both transcription-factor-binding sites and chromatin structure. FAIRE-seq provides information on chromatin structure through exploitation of differences in cross-linking efficiency of nucleosomes to regulatory factors. Loci including ENCODE data and RNA-seq expression data were visualized in the UCSC genome browser.
Pathway analysis
Genes showing evidence of association were reviewed for evidence of involvement in adiposity at a pathway level using Ingenuity Pathway Analysis (Ingenuity Systems, Inc.) and GeneGo Metacore (Thomson Reuters). By combining comprehensive data on gene and variant function we were able to build up a view of genes with the highest level of support in WHR.
Druggability annotation and analysis
We annotated genes in loci of interest with information concerning potential druggability, defined as the potential modulation of a protein target by a water-soluble small molecule drug. Druggable proteins usually contain a defined-binding pocket or active site, which could act as a site of action (pharmacophore) for a small molecule drug. We grouped proteins into four druggability classes based on a collation of complementary published annotations of the potentially druggable genome and publically available databases of small molecules (66–68). Targets in class 1 have a known drug recorded in drugbank (www.drugbank.ca); class 2 have small molecules recorded in chembl (www.ebi.ac.uk/chembl) and may be in current development within pharmaceutical companies; class 3 are homologous to class 1 or class 2 targets; class 4 are predicted to contain a potentially druggable pharmacophore based on de novo structure-based druggability prediction using the dogsitescorer tool (dogsite.zbh.uni-hamburg.de) (49).
SUPPLEMENTARY MATERIAL
FUNDING
Funding for this collaborative work is also provided for each cohort in the Supplementary Materials.
Supplementary Material
ACKNOWLEDGEMENTS
Acknowledgements are provided for each cohort in the Supplementary Materials.
Conflict of Interest statement: No conflicts of interest have been reported by any of the authors.
REFERENCES
- 1.McGee D.L. Body mass index and mortality: a meta-analysis based on person-level data from twenty-six observational studies. Ann. Epidemiol. 2005;15:87–97. doi: 10.1016/j.annepidem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Renehan A.G., Tyson M., Egger M., Heller R.F., Zwahlen M. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet. 2008;371:569–578. doi: 10.1016/S0140-6736(08)60269-X. [DOI] [PubMed] [Google Scholar]
- 3.Obesity: preventing and managing the global epidemic. Report of a WHO consultation. World Health Organ. Tech. Rep. Ser. 2000;894:16–97. [PubMed] [Google Scholar]
- 4.Wang Y., Beydoun M.A., Liang L., Caballero B., Kumanyika S.K. Will All Americans Become Overweight or Obese? Estimating the Progression and Cost of the US Obesity Epidemic. Obesity. 2008;16:2323–2330. doi: 10.1038/oby.2008.351. [DOI] [PubMed] [Google Scholar]
- 5.Fox C.S., Liu Y., White C.C., Feitosa M., Smith A.V., Heard-Costa N., Lohman K., Johnson A.D., Foster M.C., Greenawalt D.M., et al. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012;8:e1002695. doi: 10.1371/journal.pgen.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dagenais G.R., Yi Q., Mann J.F.E., Bosch J., Pogue J., Yusuf S. Prognostic impact of body weight and abdominal obesity in women and men with cardiovascular disease. Am. Heart J. 2005;149:54–60. doi: 10.1016/j.ahj.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Poirier P., Giles T.D., Bray G.A., Hong Y., Stern J.S., Pi-Sunyer F.X., Eckel R.H. Obesity and cardiovascular disease pathophysiology, evaluation, and effect of weight loss. Arterioscler. Thromb. Vasc. Biol. 2006;26:968–976. doi: 10.1161/01.ATV.0000216787.85457.f3. [DOI] [PubMed] [Google Scholar]
- 8.Cornier M.-A., Després J.-P., Davis N., Grossniklaus D.A., Klein S., Lamarche B., Lopez-Jimenez F., Rao G., St-Onge M.-P., Towfighi A., et al. Assessing adiposity: a scientific statement from the American Heart Association. Circulation. 2011;124:1996–2019. doi: 10.1161/CIR.0b013e318233bc6a. [DOI] [PubMed] [Google Scholar]
- 9.Faeh D., Braun J., Bopp M. Body mass index vs cholesterol in cardiovascular disease risk prediction models. Arch. Intern. Med. 2012;172:1766–1768. doi: 10.1001/2013.jamainternmed.327. [DOI] [PubMed] [Google Scholar]
- 10.Bodenant M., Kuulasmaa K., Wagner A., Kee F., Palmieri L., Ferrario M.M., Montaye M., Amouyel P., Dallongeville J. Measures of Abdominal Adiposity and the Risk of Stroke. Stroke. 2011;42:2872–2877. doi: 10.1161/STROKEAHA.111.614099. [DOI] [PubMed] [Google Scholar]
- 11.Khalili S., Hatami M., Hadaegh F., Sheikholeslami F., Azizi F. Prediction of Cardiovascular Events with Consideration of General and Central Obesity Measures in Diabetic Adults: Results of the 8.4-Year Follow-Up. Metab. Syndr. Relat. Disord. 2012;10:218–224. doi: 10.1089/met.2011.0070. [DOI] [PubMed] [Google Scholar]
- 12.Seidell J.C., Pérusse L., Després J.-P., Bouchard C. Waist and hip circumferences have independent and opposite effects on cardiovascular disease risk factors: the Quebec Family Study. Am. J. Clin. Nutr. 2001;74:315–321. doi: 10.1093/ajcn/74.3.315. [DOI] [PubMed] [Google Scholar]
- 13.Kershaw E.E., Flier J.S. Adipose tissue as an endocrine organ. J. Clin. Endocrinol. Metab. 2004;89:2548–2556. doi: 10.1210/jc.2004-0395. [DOI] [PubMed] [Google Scholar]
- 14.Vohl M.-C., Sladek R., Robitaille J., Gurd S., Marceau P., Richard D., Hudson T.J., Tchernof A. A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obes. Res. 2004;12:1217–1222. doi: 10.1038/oby.2004.153. [DOI] [PubMed] [Google Scholar]
- 15.Galic S., Oakhill J.S., Steinberg G.R. Adipose tissue as an endocrine organ. Mol. Cell. Endocrinol. 2010;316:129–139. doi: 10.1016/j.mce.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Després J.-P., Lemieux I. Abdominal obesity and metabolic syndrome. Nature. 2006;444:881–887. doi: 10.1038/nature05488. [DOI] [PubMed] [Google Scholar]
- 17.Doyle S.L., Donohoe C.L., Lysaght J., Reynolds J.V. Visceral obesity, metabolic syndrome, insulin resistance and cancer. Proc. Nutr. Soc. 2012;71:181–189. doi: 10.1017/S002966511100320X. [DOI] [PubMed] [Google Scholar]
- 18.Clark P.J. The heritability of certain anthropometric characters as ascertained from measurements of twins. Am. J. Hum. Gen. 1956;8:49–54. [PMC free article] [PubMed] [Google Scholar]
- 19.Maes H.H., Neale M.C., Eaves L.J. Genetic and environmental factors in relative body weight and human adiposity. Behav. Genet. 1997;27:325–351. doi: 10.1023/a:1025635913927. [DOI] [PubMed] [Google Scholar]
- 20.Rose K.M., Newman B., Mayer-Davis E.J., Selby J.V. Genetic and behavioral determinants of waist-hip ratio and waist circumference in women twins. Obes. Res. 1998;6:383–392. doi: 10.1002/j.1550-8528.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 21.Zillikens M., Yazdanpanah M., Pardo L., Rivadeneira F., Aulchenko Y., Oostra B., Uitterlinden A., Pols H., van Duijn C. Sex-specific genetic effects influence variation in body composition. Diabetologia. 2008;51:2233–2241. doi: 10.1007/s00125-008-1163-0. [DOI] [PubMed] [Google Scholar]
- 22.Wang J., Thornton J.C., Kolesnik S., Pierson R.N., Jr. Anthropometry in Body Composition: An Overview. Ann. N.Y. Acad. Sci. 2000;904:317–326. doi: 10.1111/j.1749-6632.2000.tb06474.x. [DOI] [PubMed] [Google Scholar]
- 23.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Allen H.L., Lindgren C.M., Luan J., Magi R., et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyre D., Delplanque J., Chevre J.-C., Lecoeur C., Lobbens S., Gallina S., Durand E., Vatin V., Degraeve F., Proenca C., et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat. Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 25.Thorleifsson G., Walters G.B., Gudbjartsson D.F., Steinthorsdottir V., Sulem P., Helgadottir A., Styrkarsdottir U., Gretarsdottir S., Thorlacius S., Jonsdottir I., et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 26.Heid I.M., Jackson A.U., Randall J.C., Winkler T.W., Qi L., Steinthorsdottir V., Thorleifsson G., Zillikens M.C., Speliotes E.K., Mägi R., et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musani S.K., Erickson S., Allison D.B. Obesity—still highly heritable after all these years. Am. J. Clin. Nutr. 2008;87:275–276. doi: 10.1093/ajcn/87.2.275. [DOI] [PubMed] [Google Scholar]
- 28.Keating B.J., Tischfield S., Murray S.S., Bhangale T., Price T.S., Glessner J.T., Galver L., Barrett J.C., Grant S.F.A., Farlow D.N., et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schunkert H., König I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F.R., Barbalic M., Gieger C., et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraft P. Curses—winner's and otherwise—in genetic epidemiology. Epidemiology. 2008;19:649–651. doi: 10.1097/EDE.0b013e318181b865. [DOI] [PubMed] [Google Scholar]
- 31.Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S., et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.A., Doyle F., Epstein C.B., Frietze S., Harrow J., Kaul R., et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S., et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranieri S.C., Fusco S., Panieri E., Labate V., Mele M., Tesori V., Ferrara A.M., Maulucci G., De Spirito M., Martorana G.E., et al. Mammalian life-span determinant p66shcA mediates obesity-induced insulin resistance. Proc. Natl. Acad. Sci. USA. 2010;107:13420–13425. doi: 10.1073/pnas.1008647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geer L.Y., Marchler-Bauer A., Geer R.C., Han L., He J., He S., Liu C., Shi W., Bryant S.H. The NCBI BioSystems database. Nucleic Acids Res. 2010;38:D492–D496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi W.J., Kim S.-E., Stephen A.G., Weidlich I., Giubellino A., Liu F., Worthy K.M., Bindu L., Fivash M.J., Nicklaus M.C., et al. Identification of Shc Src homology 2 domain-binding peptoid-peptide hybrids. J. Med. Chem. 2009;52:1612–1618. doi: 10.1021/jm800789h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muharram G., Beucher A., Moerman E., Belaïch S., Gmyr V., Vandewalle B., Pattou F., Kerr-Conte J. Endocrine pancreatic tissue plasticity in obese humans is associated with cytoplasmic expression of PBX-1 in pancreatic ductal cells. Biochem. Biophys. Res. Commun. 2005;333:1153–1159. doi: 10.1016/j.bbrc.2005.05.199. [DOI] [PubMed] [Google Scholar]
- 39.Kim S.K., Selleri L., Lee J.S., Zhang A.Y., Gu X., Jacobs Y., Cleary M.L. Pbx1 inactivation disrupts pancreas development and in Ipf1-deficient mice promotes diabetes mellitus. Nat. Genet. 2002;30:430–435. doi: 10.1038/ng860. [DOI] [PubMed] [Google Scholar]
- 40.Yamada M., Saga Y., Shibusawa N., Hirato J., Murakami M., Iwasaki T., Hashimoto K., Satoh T., Wakabayashi K., Taketo M.M., et al. Tertiary hypothyroidism and hyperglycemia in mice with targeted disruption of the thyrotropin-releasing hormone gene. Proc. Natl. Acad. Sci. USA. 1997;94:10862–10867. doi: 10.1073/pnas.94.20.10862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verstraeten V.L.R.M., Renes J., Ramaekers F.C.S., Kamps M., Kuijpers H.J., Verheyen F., Wabitsch M., Steijlen P.M., van Steensel M.A.M., Broers J.L.V. Reorganization of the nuclear lamina and cytoskeleton in adipogenesis. Histochem. Cell Biol. 2011;135:251–261. doi: 10.1007/s00418-011-0792-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto Y., Gesta S., Lee K.Y., Tran T.T., Saadatirad P., Kahn C.R. Adipose depots possess unique developmental gene signatures. Obesity. 2010;18:872–878. doi: 10.1038/oby.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karastergiou K., Fried S.K., Xie H., Lee M.-J., Divoux A., Rosencrantz M.A., Chang R.J., Smith S.R. Distinct developmental signatures of human abdominal and gluteal subcutaneous adipose tissue depots. J. Clin. Endocrinol. Metab. 2013;98:362–371. doi: 10.1210/jc.2012-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heine P.A., Taylor J.A., Iwamoto G.A., Lubahn D.B., Cooke P.S. Increased adipose tissue in male and female estrogen receptor-alpha knockout mice. Proc. Natl. Acad. Sci. USA. 2000;97:12729–12734. doi: 10.1073/pnas.97.23.12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohlsson C., Hellberg N., Parini P., Vidal O., Bohlooly-Y M., Bohlooly M., Rudling M., Lindberg M.K., Warner M., Angelin B., et al. Obesity and disturbed lipoprotein profile in estrogen receptor-alpha-deficient male mice. Biochem. Biophys. Res. Commun. 2000;278:640–645. doi: 10.1006/bbrc.2000.3827. [DOI] [PubMed] [Google Scholar]
- 46.Calvo K.R., Knoepfler P., McGrath S., Kamps M.P. An inhibitory switch derepressed by pbx, hox, and Meis/Prep1 partners regulates DNA-binding by pbx1 and E2a-pbx1 and is dispensable for myeloid immortalization by E2a-pbx1. Oncogene. 1999;18:8033–8043. doi: 10.1038/sj.onc.1203377. [DOI] [PubMed] [Google Scholar]
- 47.Uthman S., Bär C., Scheidt V., Liu S., ten Have S., Giorgini F., Stark M.J.R., Schaffrath R. The amidation step of diphthamide biosynthesis in yeast requires DPH6, a gene identified through mining the DPH1-DPH5 interaction network. PLoS Genet. 2013;9:e1003334. doi: 10.1371/journal.pgen.1003334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Labar G., Michaux C. Fatty acid amide hydrolase: from characterization to therapeutics. Chem. Biodivers. 2007;4:1882–1902. doi: 10.1002/cbdv.200790157. [DOI] [PubMed] [Google Scholar]
- 49.Volkamer A., Kuhn D., Grombacher T., Rippmann F., Rarey M. Combining global and local measures for structure-based druggability predictions. J. Chem. Inf. Model. 2012;52:360–372. doi: 10.1021/ci200454v. [DOI] [PubMed] [Google Scholar]
- 50.Jacobs R.L., Zhao Y., Koonen D.P.Y., Sletten T., Su B., Lingrell S., Cao G., Peake D.A., Kuo M.-S., Proctor S.D., et al. Impaired de novo choline synthesis explains why phosphatidylethanolamine N-methyltransferase-deficient mice are protected from diet-induced obesity. J. Biol. Chem. 2010;285:22403–22413. doi: 10.1074/jbc.M110.108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lo K.S., Wilson J.G., Lange L.A., Folsom A.R., Galarneau G., Ganesh S.K., Grant S.F.A., Keating B.J., McCarroll S.A., Mohler E.R., III., et al. Genetic association analysis highlights new loci that modulate hematological trait variation in Caucasians and African Americans. Hum. Genet. 2011;129:307–317. doi: 10.1007/s00439-010-0925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ganesh S.K., Tragante V., Guo W., Guo Y., Lanktree M.B., Smith E.N., Johnson T., Castillo B.A., Barnard J., Baumert J., et al. Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum. Mol. Genet. 2013;22:1663–1678. doi: 10.1093/hmg/dds555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asselbergs F.W., Guo Y., van Iperen E.P.A., Sivapalaratnam S., Tragante V., Lanktree M.B., Lange L.A., Almoguera B., Appelman Y.E., Barnard J., et al. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am. J. Hum. Genet. 2012;91:823–838. doi: 10.1016/j.ajhg.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo Y., Lanktree M.B., Taylor K.C., Hakonarson H., Lange L.A., Keating B.J. Gene-centric meta-analyses of 108 912 individuals confirm known body mass index loci and reveal three novel signals. Hum. Mol. Genet. 2013;22:184–201. doi: 10.1093/hmg/dds396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higgins J.P.T., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 59.Magi R., Lindgren C.M., Morris A.P. Meta-analysis of sex-specific genome-wide association studies. Genet. Epidemiol. 2010;34:846–853. doi: 10.1002/gepi.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saxena R., Hivert M.-F., Langenberg C., Tanaka T., Pankow J.S., Vollenweider P., Lyssenko V., Bouatia-Naji N., Dupuis J., Jackson A.U., et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat. Genet. 2010;42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C., Hwang S.-J., et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hopkins A.L., Groom C.R. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 67.Russ A.P., Lampel S. The druggable genome: an update. Drug Discov. Today. 2005;10:1607–1610. doi: 10.1016/S1359-6446(05)03666-4. [DOI] [PubMed] [Google Scholar]
- 68.Costa P.R., Acencio M.L., Lemke N. A machine learning approach for genome-wide prediction of morbid and druggable human genes based on systems-level data. BMC Genomics. 2010;11:S5–S9. doi: 10.1186/1471-2164-11-S5-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.