Abstract
Mammalian target of rapamycin (mTOR) is a conserved Ser/Thr kinase that is part of mTOR complex 1 (mTORC1), a master regulator that couples amino acid availability to cell growth and autophagy. Multiple cues modulate mTORC1 activity, such as growth factors, stress, energy status and amino acids. Although amino acids are key environmental stimuli, exactly how they are sensed and how they activate mTORC1 is not fully understood. Recently, a model has emerged whereby mTORC1 activation occurs at the lysosome and is mediated through an amino acid sensing cascade involving RAG GTPases, Ragulator and vacuolar H+-ATPase (v-ATPase).
Cells and organisms need to integrate information from the environment to ensure that they only grow when conditions are favourable. The highly conserved Ser/Thr protein kinase target of rapamycin (TOR) is a key integrator of environmental cues, including nutrient and growth factor availability as well as stress. Under nutrient-rich conditions, TOR promotes cell growth by stimulating biosynthetic pathways, including protein synthesis, and by inhibiting cellular catabolism such as through repression of the autophagy pathway (BOX 1). Therefore, understanding how the different stimuli are detected and how they signal to TOR is integral to elucidating how the cell or organism grows.
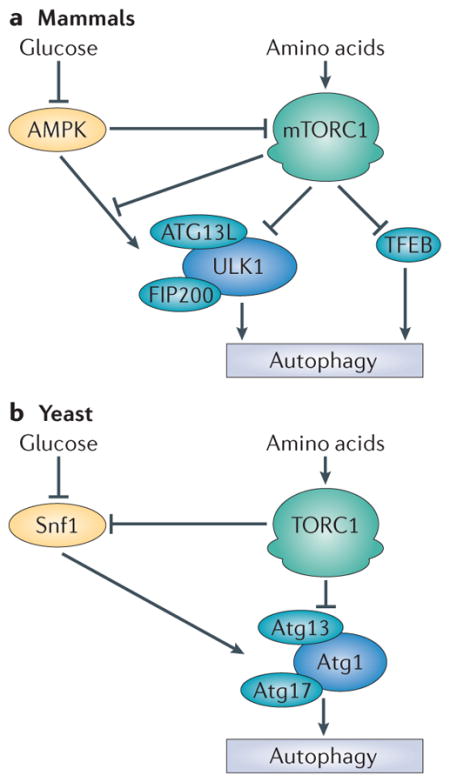
Box 1. mTORC1 and autophagy.
Macroautophagy, hereafter referred to as autophagy, is the primary catabolic cellular degradation process that generates nutrients and energy to maintain essential cellular activities upon nutrient starvation42. Target of rapamycin complex 1 (TORC1) is a potent repressor of autophagy in all eukaryotes43. Mammalian TORC1 (mTORC1) is inhibited under starvation conditions by the energy sensor AMP-activated protein kinase (AMPK) and by amino acid signalling. Under these conditions, ULK1 (unc-51-like kinase 1) undergoes autophosphorylation and phosphorylates ATG13L (ATG13-like) and FIP200 (FAK family kinase-interacting protein of 200 kDa), thus forming an active kinase complex to initiate autophagy. Under conditions of nutrient sufficiency, mTORC1 is active and inhibits autophagy by phosphorylating ATG13L and ULK1 subunits, thus repressing ULK1 kinase activity44–47 (see the figure, part a). The ULK1 complex is also activated by AMPK, and this regulation was recently shown to be antagonized by mTORC1-mediated phosphorylation of ULK1 on Ser757 (REF. 44). mTORC1 also indirectly regulates autophagy by controlling lysosome biogenesis through phosphorylation of transcription factor EB (TFEB)48,49, which drives the transcription of several lysosome- and autophagy-specific genes. mTORC1 and TFEB colocalize to the lysosomal membrane, where mTORC1-mediated phosphorylation promotes TFEB cytoplasmic sequestration48.
Like mammalian mTORC1, yeast TORC1 inhibits autophagy through the regulation of the Atg1 kinase complex (see the figure, part b). Atg1 (the homologue of mammalian ULK1) forms an active complex with Atg13 and Atg17. The presence of amino acids induces TORC1-mediated phosphorylation of Atg13. This phosphorylation event prevents its association with Atg17 and thus blocks autophagy. Inhibition of TORC1 activity by amino acid starvation or rapamycin treatment results in a de-repression of Atg13 and promotion of the active trimeric Atg1–Atg13–Atg17 complex. Another difference between yeast and mammals is that TORC1 inhibits the yeast orthologue of AMPK, termed Snf1. However, it remains unclear whether Snf1 regulates TORC1 (REF. 50). In yeast, Atg1 has been proposed to act downstream of Snf1, although the underlying mechanism remains to be elucidated51.
Metabolic homeostasis is orchestrated by mTORC1 through the promotion of biosynthetic pathways and the repression of catabolic autophagy in response to energy and amino acid sufficiency. Collectively, the regulation of these processes is strictly determined by energy and amino acid sensing pathways within the cell. Further research, particularly in mammals, is needed to determine whether mTORC1 regulates any aspects of the autophagy machinery in addition to the ULK1 complex. Although it seems that amino acids generated by autophagy have a role in mTORC1 activation, additional studies detailing mechanistic action will be of great importance.
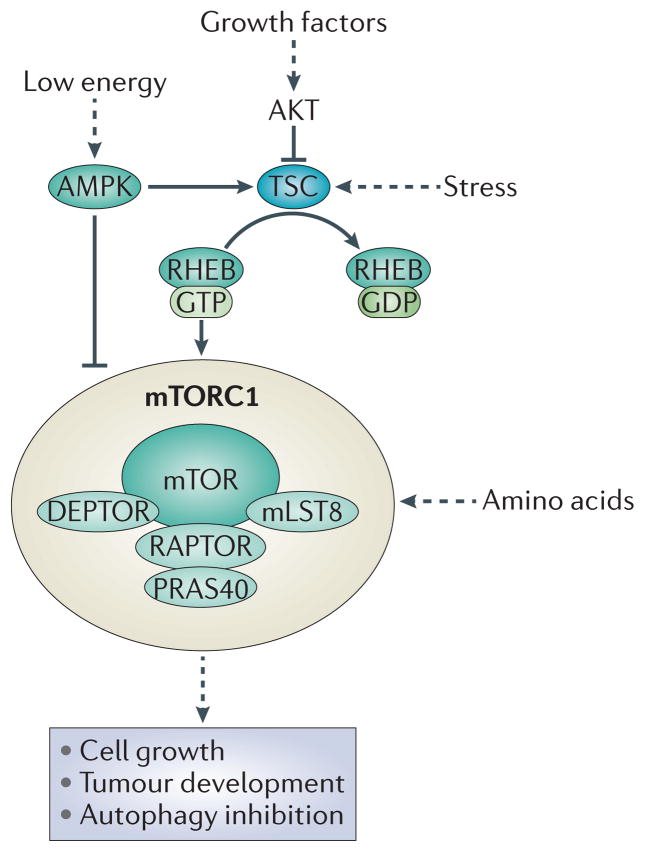
Mammalian TOR (mTOR) is part of a complex known as mTORC1, comprising: regulatory-associated protein of mTOR (RAPTOR), which aids in substrate recognition; 40 kDa Pro-rich AKT substrate (PRAS40; also known as AKT1S1) and DEP domain-containing mTOR-interacting protein (DEPTOR), both of which are negative regulators of mTORC1; and mammalian lethal with SEC13 protein 8 (mLST8; also known as GβL), which positively regulates mTORC1. Some mTORC1-activating stimuli, such as growth factors, signal through the tuberous sclerosis complex (TSC; comprising TSC1 and TSC2) (FIG. 1). TSC is a GTPase-activating protein (GAP) for the small GTPase RHEB (RAS homologue enriched in brain) and negatively regulates mTORC1 by promoting RHEB·GTP hydrolysis, converting RHEB into its inactive GDP-bound state. As a result, inhibition of TSC by growth factors gives rise to GTP-bound RHEB, which is a potent activator of mTORC1 kinase activity (reviewed in REFS 1,2). mTORC1 is inhibited directly and indirectly through AMP-activated protein kinase (AMPK)-mediated phosphorylation of RAPTOR and TSC2, respectively, in response to low energy3,4.
Figure 1. The mTORC1 signalling pathway.
Through a multiple-step process, growth factors stimulate AKT, which in turn phosphorylates and inhibits the tuberous sclerosis complex (TSC; comprising TSC1 and TSC2). TSC acts as a GTPase-activating protein (GAP) for the small GTPase RAS homologue enriched in brain (RHEB), promoting hydrolysis of its bound GTP and thus inhibiting RHEB. Upon inhibition of TSC, GTP-bound RHEB levels increase and can potently activate mammalian target of rapamycin complex 1 (mTORC1), presumably at the lysosome. In addition to growth factors, mTORC1 can be regulated by stress, energy status and amino acids. Stress and energy status regulate mTORC1 through TSC. Moreover, AMPK (AMP-activated protein kinase) is activated in response to a low energy status and phosphorylates RAPTOR (regulatory- associated protein of mTOR) and TSC2. This results in the inhibition of mTORC1. Amino acids regulate mTORC1 in a TSC-independent pathway. Collectively multiple stimuli modulate mTORC1 to control cell growth and autophagy. DEPTOR, DEP domain-containing mTOR-interacting protein; mLST8, mammalian lethal with SEC13 protein 8; PRAS40, 40 kDa Pro-rich AKT substrate.
Unlike the other stimuli, amino acids do not signal through TSC–RHEB, and it is currently unclear how amino acid sufficiency or limitation is sensed to modulate mTORC1 activity. It seems that amino acids are the most crucial signals for mTORC1 activation, as growth factors cannot efficiently activate mTOR when amino acids are limiting5–7. Although Leu5,6, Gln and Arg6,8–12 have been implicated in mTORC1 activation, which specific amino acids are sensed to activate mTORC1 is currently uncertain. Moreover, it remains elusive where the amino acids are first detected, but multiple studies have recently been published that implicate components residing at the lysosome surface.
In this Progress article, we summarize the current understanding of the protein cascade that senses amino acids and leads to mTORC1 activation at the lysosome, ultimately resulting in cell growth or inhibition of autophagy. In addition, we highlight what outstanding questions remain to be addressed.
Amino acid sensing at the lysosome
RAG GTPases
RHEB is required for mTORC1 activation by all stimuli, including amino acids. However, Tsc2-null mouse embryonic fibroblasts (MEFs) are still sensitive to amino acid starvation, indicating that other components that are not part of the TSC–RHEB pathway are likely to be involved in this process13,14 (FIG. 1). The discovery of the small RAG GTPases, through screening in Drosophila melanogaster and in mammalian cells, was an important breakthrough in understanding amino acid signalling to mTORC1 (REFS 5,15). RAG GTPases belong to the RAS superfamily; however, they have unique characteristics that distinguish them from other small GTPases, such as a long carboxyl-terminal domain, the lack of a membrane-targeting sequence and the ability to form heterodimers16,17. In mammals, there are four RAG proteins: RAGA and RAGB (Gtr1 in yeast52), which have high sequence similarity and are functionally redundant; and RAGC and RAGD (Gtr2 in yeast53), which are also highly related in sequence and are functionally equivalent. RAGA or RAGB forms a heterodimer with RAGC or RAGD, with the possibility of forming four distinct complex arrangements17. The crystal structure of the yeast Gtr1–Gtr2 heterodimer was recently solved. The structure reveals an amino-terminal GTPase domain on each protein where GTP binding and hydrolysis occur, and a carboxy-terminal domain on each protein with multiple contacts required for Gtr1·GTP–Gtr2·GDP dimerization18,19. Dimerization of RAG GTPases is important for mTORC1 activation and RAG protein stability5,15.
Like other small GTPases, the activation state of RAG GTPases is reflected by their guanine nucleotide state, and this is regulated by amino acids. Specifically, the presence of amino acids promotes the formation of the active complex configuration, in which RAGA and RAGB are GTP-bound and RAGC and RAGD are GDP-bound (for simplicity, RAGA/B·GTP–RAGC/D·GDP denotes the active complex and RAGA/B·GDP–RAGC/D·GTP the inactive complex). Likewise, GTP-bound Gtr1 in complex with GDP-bound Gtr2 is the active form in yeast when amino acids are available (FIG. 2).
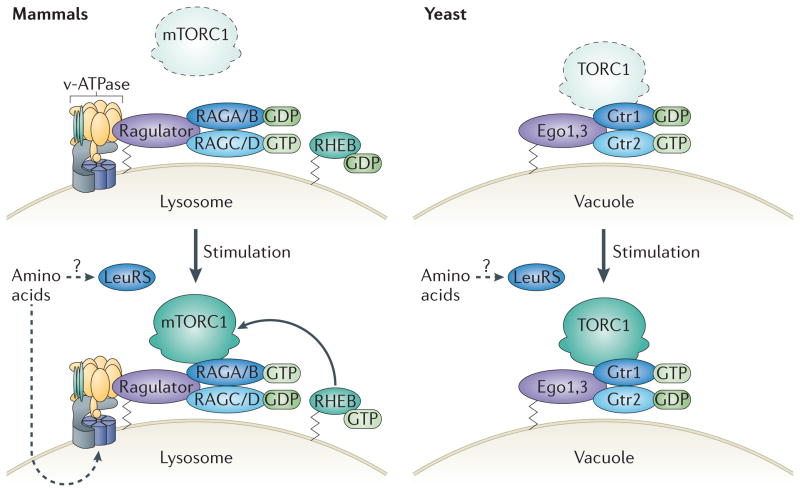
Figure 2. Amino acid-induced mTORC1 activation in mammals and yeast.
Amino acids promote the formation of the active configuration of the RAG GTPase complex in mammals at the lysosome (left) and of Gtr1–Gtr2 at the vacuole in yeast (right). Under amino acid starvation conditions in mammals, inactive mammalian target of rapamycin complex 1 (mTORC1) is diffuse throughout the cytosol. The RAG GTPase complex is inactive, with RAGA or RAGB loaded with GDP (RAGA/B·GDP) and RAGC or RAGD loaded with GTP (RAGC/D·GTP) (left, top). During amino acid deficiency in yeast, inactive TORC1 and Gtr1–Gtr2 remain localized at the vacuolar membrane but do not physically interact. Similarly to RAG GTPases in mammals under amino acid-deficient conditions, Gtr1 is bound to GDP and Gtr2 is bound to GTP, which results in an inactive complex (right, top). Amino acid stimulation signals to vacuolar H+-ATPase (v-ATPase), which is required to induce the guanine exchange factor (GEF) activity of Ragulator (right, bottom). Ragulator acts as a GEF for RAGA/B, promoting the conversion of RAGA/B·GDP to RAGA/B·GTP. Amino acids also facilitate the formation of RAGC/D·GDP, giving rise to the active RAG complex, RAGA/B·GTP–RAGC/D·GDP. The mechanisms involved in switching the guanine nucleotide state of RAGC/D are not clear. mTORC1 binds to the RAG complex and is recruited to the lysosome through an unknown mechanism, where it becomes activated. Leucyl-tRNA synthetase (LeuRS) may act as a direct sensor for the amino acid Leu in the cytoplasm and might be involved in the activation of mTORC1. In yeast under amino acid sufficiency, TORC1, already bound to the vacuole, is activated when Gtr1 is loaded with GTP and Gtr2 is loaded with GDP (left, bottom). LeuRS has also been reported to have a role in TORC1 activation in yeast.
Under amino acid-sufficient conditions, the active RAGA/B·GTP–RAGC/D·GDP complex has been shown to bind directly to the mTORC1 component RAPTOR and redistribute mTORC1 to the lysosome. Interestingly, mTORC1 is dispersed throughout the cell under amino acid starvation conditions but is redistributed to vesicles containing lysosome-associated membrane protein 2 (LAMP2) and RAB7 (which are markers of lysosomes and late endosomes, respectively) in response to amino acid stimulation5,20. This suggests that RAG GTPases have a role in transducing amino acid signals to mTORC1. Consistent with this, knockdown of RAG GTPases ablates mTORC1 relocalization to the lysosome in response to amino acid stimulation5. This relocalization has been proposed to promote the interaction of mTORC1 with RHEB, which is itself thought to be targeted and anchored to the lysosome through its C-terminal CAAX (in which C is Cys, A is an aliphatic residue and X the terminal amino acid) motif5,20, leading to mTORC1 activation through an unknown mechanism. Although RHEB lysosomal localization has been demonstrated in fluorescent microscopy studies using overexpressed fluorescently tagged RHEB, this has yet to be confirmed for endogenous RHEB owing to the lack of a quality RHEB antibody. A RHEB homologue in budding yeast does not seem to be an upstream activator of TORC1, suggesting divergent evolution of TORC1 regulation21. Because RHEB responds to growth factor signalling (FIG. 1), it is possible that mTORC1 regulation by RHEB evolved in organisms in which growth factor signalling occurs.
In contrast to mammals, yeast TORC1 remains at the vacuole, the yeast equivalent of the lysosome, under both starvation and normal conditions. This suggests that the amino acid-induced shuttling mechanism evolved in higher eukaryotes22 (FIG. 2). However, like the active RAG complex in mammals, Gtr1·GTP–Gtr2·GDP physically binds activated TORC1 in a manner dependent on nucleotide loading and amino acid availability22.
The nucleotide-bound state of the RAG heterodimer is crucial for mTORC1 activation. Under starvation conditions, overexpression of RAGA/B·GTP alone is sufficient to activate mTORC1, which indicates that it is the nucleotide-bound state of RAGA/B, rather than of RAGC/D, that primarily regulates mTORC1 activation22,23. However, co-expression of RAGC/D enhances this effect, in part because RAGC/D stabilizes RAGA/B. Furthermore, an inactive mutant RAGA/B·GDP–RAGC/D·GTP complex that does not respond to amino acids restrains the activity of mTORC1 even under amino acid sufficiency5,15. These findings reveal that the presence of amino acids determines the guanine nucleotide state of RAG GTPases and consequently controls the recruitment of mTORC1 to the lysosome and thus its activation. This model suggests a mechanism by which mTORC1 senses multiple stimuli at the lysosome, such as amino acids through RAG GTPases and growth factors through RHEB; amino acids and growth factors seem to converge at the lysosome in order to efficiently activate mTORC1.
Ragulator
RAG GTPases localize at the lysosomal surface but lack a lipid-anchoring motif, which suggests that RAG-binding proteins may tether them to the lysosome. Indeed, mass spectrometry analysis using RAG GTPases as ‘bait’ identified a complex termed Ragulator that acts as a scaffold for the heterodimeric active RAG complex at the lysosome. Originally, Ragulator was defined as a trimeric complex consisting of p18 (encoded by LAMTOR1 (late endosomal/lysosomal adaptor, MAPK and mTOR activator 1)), p14 (encoded by LAMTOR2) and MP1 (MEK-binding partner 1; encoded by LAMTOR3)20. Recently, two additional components, C7orf59 and HBXIP (hepatitis B virus X-interacting protein; encoded by LAMTOR4 and LAMTOR5, respectively) have been described and, together with p18, p14 and MP1, form a pentameric Ragulator complex24.
But how does Ragulator tether RAG GTPases, and thus mTORC1, to the lysosome? Ragulator and RAG GTPases are tethered at the lysosome by p18, which associates with the membrane via its myristoylated and palmitoylated residues20,24. Depletion of Ragulator components disrupts RAG GTPase and mTORC1 lysosomal localization and thus mTORC1 activation, which confirms the importance of this complex in tethering RAG and mTORC1 to the lysosome20,24.
Ragulator orthologues have not been identified in yeast; however, TORC1 is a component of the Ego complex (EGOC), which is located at the vacuolar membrane (FIG. 2). EGOC consists of Ego1, Ego3, Gtr1 and Gtr2. Although the precise function of Ego3 is not well defined, studies have shown that, like p18, Ego1 is a palmitoylated and myristoylated protein that maintains Gtr1–Gtr2 and TORC1 at the vacuole membrane25,26. An important point to keep in mind is that amino acids regulate TORC1 binding to EGOC, similar to mTORC1–RAG–Ragulator binding in mammals. However, unlike in mammals, in which mTORC1 is dispersed across the cell in the absence of amino acids, in yeast TORC1 remains at the vacuole even under starvation conditions. Regardless of the different anchoring machinery, it seems that mTORC1 localization at the lysosome in mammals, or TORC1 at the vacuole in yeast, is crucial in facilitating its function and activation1,27.
Despite the lack of sequence similarity between EGOC and Ragulator, overall structure conservation is apparent, as Ego3 is structurally similar to MP1 and p14 (REFS 26,28). High-resolution crystal structures of MP1, p14, HBXIP and Gtr1–Gtr2 also identified the presence of a roadblock domain in each protein29–31. Structural predictions indicate roadblock domains in the C-terminal domain of RAG GTPases and of C7orf59 (REFS 18,24). Therefore, each component of the RAG–Ragulator complex contains a roadblock domain, six in total, except for the anchoring protein p18, which acts as a scaffold for the roadblock domain-containing proteins. The function of this domain is currently unknown; however, it may provide a specific architectural element for protein–protein interactions during amino acid signalling to mTORC1. It will be interesting to determine whether other proteins that may potentially be involved in amino acid signalling to mTORC1 contain this domain.
Recently, pentameric Ragulator was determined as a guanine exchange factor (GEF) for RAGA/B, promoting the exchange of GDP for GTP and the consequent activation of the RAG complex, which is crucial for mTORC1 lysosomal localization and activation24. The identification of Ragulator as the GEF of RAG GTPases provided a key link between amino acid sensing and RAG guanine nucleotide loading (FIG. 3). The entire pentameric Ragulator complex is necessary for GEF activity towards RAGA/B; the originally identified trimeric Ragulator, or each individual Ragulator component on its own, displays no GEF activity towards RAGA/B. Surprisingly, Ragulator does not contain a domain that is homologous to any of the known GEF catalytic domains. Nucleotide-free rather than nucleotide-bound RAGA/B preferentially interacts with Ragulator, a common feature found in other GEF–GTPase interactions24. Unlike its effects on RAGA/B, Ragulator does not display any GEF activity towards RAGC/D, perhaps owing to the differences in switch I and switch II regions among the GTPases, which are known to act as recognition motifs for GEF–GTPase interactions. Rapid intrinsic dissociation of GDP from RAGC/D was also demonstrated, possibly suggesting that no GEF is required24.
Figure 3. mTORC1 activation at the lysosome.
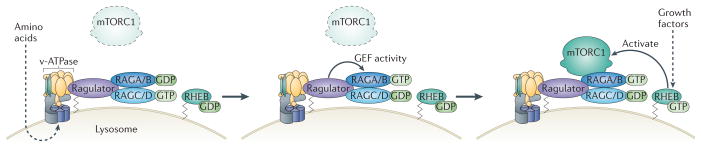
Amino acids are thought to accumulate within the lysosomal lumen and to signal to vacuolar H+-ATPase (v-ATPase) through an ‘inside–out’ mechanism. v-ATPase controls RAG GTPase–Ragulator binding, and therefore Ragulator guanine exchange factor (GEF) activity and RAGA and RAGB guanine nucleotide loading (RAGA/B·GTP). The active RAG complex (RAGA/B·GTP–RAGC/D·GDP) binds to mammalian target of rapamycin complex 1 (mTORC1) and recruits it to the lysosome, through an unknown mechanism, possibly in close proximity to RHEB (RAS homologue enriched in brain). Downstream of growth factor signalling, GTP-bound RHEB potently activates mTORC1.
As mentioned previously, there are no clear orthologues of the Ragulator components in yeast. Instead, in yeast, Vam6 (a homologue of the mammalian vacuolar protein sorting 39 (VPS39), which promotes late endosome to lysosome fusion) was identified as the GEF for Gtr1 (REFS 22,32). However, VPS39 does not seem to bind to and function as the GEF for RAGA/B in mammals24. It is possible that the GEF for Gtr1 in yeast and RAGA in higher eukaryotes has diverged, although this is surprising considering the high degree of sequence similarity between RAGA/B and Gtr1. Mammalian cells express homologues of VPS39, such as TGFβ receptor- associated protein 1 (TGFβRAP1), which have not been assessed as potential GEFs33. Therefore, readers should be cautious about a proposed model in which VPS39 functions as a GEF for RAGA/B in mammals until it can be confirmed.
Vacuolar H+-ATPase
The findings described above suggest that, in response to the presence of amino acids, Ragulator tethers RAG GTPases to the lysosome, which in turn relocalize mTORC1 to this organelle, leading to mTORC1 activation. What stimulates Ragulator itself in response to changes in amino acid levels? Vacuolar H+-ATPase (v-ATPase) interacts with RAG GTPases and Ragulator on the lysosomal membrane, thus regulating mTORC1 activation in response to amino acids both in D. melanogaster and mammalian cells34 (FIG. 3). The highly conserved v-ATPase pumps protons into intracellular organelles, such as the lysosome, to acidify them and across the plasma membrane, thereby maintaining cytosolic pH in numerous cell types. v-ATPase is a multicomponent complex composed of two domains: the peripheral cytosolic V1 domain, which contains eight subunits (subunits A–H); and the integral membrane V0 domain, which consists of five subunits (subunits a, d, c, c′ and c″). The V1 domain hydrolyses ATP to fuel proton translocation through the V0 domain channel from the cytoplasm into the lysosomal lumen, resulting in its acidification35.
The v-ATPase V1 domain interacts with RAG GTPases, whereas both the V1 and V0 domains interact with Ragulator. The interaction of the V1 domain with RAG GTPases and Ragulator is regulated by amino acids, with the interactions being strengthened by amino acid starvation and weakened in response to amino acid stimulation. Moreover, inhibition of v-ATPase with the macrolide salicylihalamide A renders these interactions unresponsive to amino acids34. Similarly to V1 domain–RAG and V1 domain–Ragulator interactions, the RAG GTPase–Ragulator interaction also strengthens when cells are deprived of amino acids and weakens with amino acid stimulation24. v-ATPase is thought to reside upstream of Ragulator GEF activity. Moreover, chemical inhibition of v-ATPase renders the Ragulator–RAG interaction insensitive to amino acids, implicating Ragulator as a bridge between v-ATPase and RAG in the amino acid cascade24. Furthermore, knockdown of individual v-ATPase subunits, or the use of chemical inhibitors, has been shown to not affect RAG localization to the lysosome34. It is currently unclear whether RAG GTPases dissociate from the lysosome or whether they are always anchored to this organelle by Ragulator. If RAG GTPases relocalize to another cellular compartment, which signals regulate this movement remains unclear.
What is the role of v-ATPase in amino acid-induced mTORC1 activation? Knockdown of v-ATPase subunits inhibited mTORC1 lysosomal localization and activation, highlighting a key role of v-ATPase in this pathway. Hydrolysis of ATP by v-ATPase is essential for the interaction of RAG GTPases with mTORC1 and consequently for mTORC1 activation, but the reason for this is unknown. v-ATPase-mediated ATP hydrolysis does not seem to drive a proton gradient required for amino acid transport and accumulation within the lysosome, as freely diffusible alcohol ester derivatives of amino acids could still collect within the lysosome regardless of whether the v-ATPase was inhibited. Furthermore, an ionophore that disrupts the lysosomal proton gradient without affecting v-ATPase function had no effect on the RAG GTPase–mTORC1 interaction under amino acid sufficient conditions34. Other reports have revealed that a lower cytoplasmic pH correlates with inhibition of mTORC1 activity36,37, with impaired v-ATPase function increasing cytoplasmic acidification, resulting in mTORC1 inhibition36,37. These findings are complimentary to studies showing that chemical inhibition of v-ATPase is important in regulating mTORC1. It thus remains to be determined whether the role of v-ATPase in amino acid signalling to mTORC1 is keeping the lysosome acidic or whether it ensures that the pH within the cytoplasm is optimal for cell growth. Interestingly, in a cell-free system with isolated RAG GTPase-bound lysosomes and purified mTORC1, the addition of amino acids promoted RAG GTPase–mTORC1 binding, indicating that the lysosome contains all of the necessary machinery that is required for amino acid-induced mTORC1 activation34.
Although the precise amino acid sensor that stimulates v-ATPase is unknown, a recent model has proposed that amino acid signalling to mTORC1 begins within the lysosomal lumen communicating to v-ATPase through an ‘inside–out’ mechanism34 (that is, the build-up of amino acids inside the lysosomal lumen signals to and activates mTORC1 that resides outside of lysosomes). Stimulation of starved cells with radiocarbon (14C)-labelled amino acids leads to the rapid accumulation of these amino acids in isolated lysosomes34. How the amino acids are transported into the lysosome and what particular transporters facilitate this process is currently unknown. However, it has been observed that overexpression of the lysosome-localized amino acid transporter PAT1 (proton-assisted transporter 1), which is known to export amino acids out of the lysosome, inhibits mTORC1 activation34. Moreover, lysosomal membrane permeabilization, which allows amino acids to escape, blocked RAG GTPase–mTORC1 interaction34. Together, these observations suggest that the buildup of amino acids within the lysosomal lumen seems to be required for downstream mTORC1 activation.
Whether the amino acids that are transported into the lysosome originate from within the cell or are transported from the extracellular environment remains unclear. However, an increase in intracellular amino acid concentrations mediated by the protein synthesis inhibitor cycloheximide enhanced mTORC1 activity, which indicates that intracellular amino acids can accumulate within the lumen of the lysosome34.
On the basis of these findings, it has been proposed that amino acids, whether intracellular or extracellular, accumulate within the lysosomal lumen and signal to v-ATPase. This enhances the GEF activity of Ragulator, promoting the active RAG GTPase conformation (RAGA/B·GTP–RAGC/D·GDP), which can then recruit mTORC1 to the lysosome, ultimately leading to mTORC1 activation. Whether v-ATPase has a role in amino acid sensing and TOR activation in yeast remains to be determined.
Activation by leucyl-tRNA synthetase
Two independent groups showed in mammals and yeast that leucyl-tRNA synthetase (LeuRS) acts as a direct sensor for the amino acid Leu and is involved in the activation of mTORC1, although the mechanistic details differ considerably between the two studies38,39 (FIG. 2). LeuRS is a cytoplasmic enzyme that catalyses the ATP-dependent ligation of l-Leu to its corresponding tRNA and is required for protein synthesis. LeuRS has previously been reported to be a key amino acid sensor upstream of mTOR40. In mammals, in response to Leu, LeuRS was found to translocate to the lysosomal membrane, where it bound to and acted as a GAP for RAGD. LeuRS interacted via its C terminus with the C terminus of RAGD, which is likely to be important for heterodimerization with RAGA/B based on the structure of Gtr1–Gtr2 (REF. 38) (see above). Surprisingly, the Arg residue in human LeuRS that is required for the proposed GAP activity is not conserved in the D. melanogaster LeuRS homologue, which is unexpected because amino acid-induced mTORC1 activation is highly conserved in all eukaryotes.
In yeast, LeuRS was shown to regulate the activation of Gtr1 (the RAGA/B homologue), in this case by blocking GTP hydrolysis rather than promoting it. Specifically, the LeuRS editing domain (which hydrolyses mischarged tRNA Leu) interacts with GTP-bound Gtr1 and blocks GTP hydrolysis, and this interaction is facilitated by a conformational change in LeuRS triggered by binding of Leu. The domain in LeuRS that interacts with Gtr1·GTP is different from the one suggested to mediate the interaction with RAGD in mammals. It was proposed that the binding between LeuRS and Gtr1·GTP blocks the access of an unidentified GAP, thus inhibiting GTP hydrolysis39.
Further work is needed to verify the exact mechanism of action of LeuRS in this context and also to confirm that it does indeed have a role in mTORC1 activation. Also, in contrast to the amino acid sensing inside–out model, LeuRS detects the availability of Leu in the cytoplasm. It will be important to further examine how the two mechanisms (the lysosomal inside–out amino acid sensing mechanism involving v-ATPase and the cytoplasmic amino acid sensing mechanism involving LeuRS) are integrated. Radioactively labelled amino acids accumulate in the lysosome within 10 minutes of addition, whereas mTORC1 has been reported to be activated after only 3 minutes5,34. It is still not clear whether amino acids are required to accumulate within the lysosome before mTORC1 is being recruited there, or if there is also a parallel signal, such as the one mediate by LeuRS, aiding mTORC1 activation.
Conclusions and perspectives
Recent work identifying new lysosomal components has provided a model whereby amino acids that have accumulated in the lysosome signal to mTORC1 through an inside–out mechanism. Moreover, new work involving LeuRS suggests that mTORC1 may also be activated by amino acids in the cytoplasm38,39. Although the precise amino acid sensor at the lysosome is still unknown, the first downstream target discovered thus far is v-ATPase34. v-ATPase promotes the GEF activity of Ragulator, resulting in RAG GTPase nucleotide exchange and activation24. The activated RAGA/B·GTP– RAGC/D·GDP complex binds to and recruits mTORC1 from an undefined cellular location to the lysosome, possibly in close proximity to the potent mTORC1 activator RHEB5,20. Amino acids are thought to relay messages to RAG GTPases, which directly bind to and redistribute mTORC1 to lysosomes. RHEB, which is activated by growth factors downstream of TSC, is also thought to reside on the lysosome. Together, these stimuli coordinate to activate mTORC1 and thus promote cell growth and inhibit autophagy. This model demonstrates how multiple stimuli may coordinately activate mTORC1 at the lysosome.
The identification of Ragulator as the RAGA/B GEF is an important advance, although the GAP has yet to be found. Future work is now needed to identify other components of the pathway. The further characterization of mTORC1 negative regulators, such as SH3 domain-binding protein 4 (SH3BP4), PAT1 and the small GTPases ARF and RAB5, will provide important insight23,34,41. The precise amino acid sensor and its location have yet to be identified. If the sensor is located in the lysosomal lumen, is the acidification of the lysosome required for its activation? Indeed, the functions of many lumenal proteins depend on an acidic pH. Furthermore, hydrolysis of ATP by the V1 subunit of v-ATPase promotes acidification of the lysosome and activation of mTORC1 through undefined mechanisms. Other reports have shown that an increase in cytoplasm acidity inhibits mTORC1 (REFS 36,37). Although this is consistent with v-ATPase having a role in mTORC1 activation, is it lysosomal acidification or the pH of the cytoplasm that regulates mTORC1? Future work will undoubtedly be focused on understanding these mechanisms and how v-ATPase is regulated. For example, signals that regulate v-ATPase could potentially modulate mTORC1 activity and cell growth. It will also be important to understand how this pathway integrates with the one that is activated by LeuRS in the cytoplasm.
If the inside–out mechanism holds true and amino acids accumulate within the lysosome to signal to mTORC1, where are the amino acids coming from? Are intracellular or extracellular amino acid pools filling the lumen of the lysosome? How are the amino acids being transported into the lysosome? The identification of the lysosomal amino acid transporter will be a significant advancement.
Moreover, the precise details or unidentified intermediates involved in mTORC1 translocation to the lysosome require further investigation. Do RAG GTPases dissociate from the lysosome, perhaps in response to amino acids, and shuttle mTORC1 to the lysosome? Or are there additional, unidentified intermediates that translocate mTORC1 to the RAG GTPases at the lysosome in response to amino acids?
Amino acids are fundamental to life and essential in facilitating mTORC1 activation. Therefore, determining the molecular mechanisms involved in mTORC1 activation will lead to a better understanding of cell growth and autophagy, enhance our basic understanding of biology and also clarify the role of this pathway in disease. For example, although numerous oncogenes and tumour suppressors have been identified in the mTORC1-activating pathway involving TSC, such proteins have yet to be characterized in the amino acid cascade. Identification of additional regulators could shed light onto the role of this signalling pathway in human pathogenesis, in particular cancer.
Acknowledgments
The authors are grateful to their colleague R. Gong and the rest of the Guan laboratory for valuable discussions and insightful comments. In addition, the authors would like to thank V. S. Tagliabracci for critical reading of this manuscript. The authors would like to apologize to their colleagues whose work could not be cited owing to space limitations. The work in the Guan laboratory was supported by a National Institutes of Health (NIH) grant (CA108941) and a grant from the Department of Defense (W81XWH-09-1-0279) to K.L.G. J.L.J is supported by a grant from the National Cancer Institute (T32CA121938), and R.C.R is supported by a grant from the Canadian Institute of Health Research (CIHR).
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inoki K, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 4.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hara K, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Campbell LE, Miller CM, Proud CG. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem J. 1998;334:261–267. doi: 10.1042/bj3340261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauchart-Thevret C, Cui L, Wu G, Burrin DG. Arginine-induced stimulation of protein synthesis and survival in IPEC-J2 cells is mediated by mTOR but not nitric oxide. Am J Physiol Endocrinol Metab. 2010;299:e899–e909. doi: 10.1152/ajpendo.00068.2010. [DOI] [PubMed] [Google Scholar]
- 9.Nicklin P, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duran RV, et al. Glutaminolysis activates Rag–mTORC1 signaling. Mol Cell. 2012;47:349–358. doi: 10.1016/j.molcel.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 11.van der Vos KE, Coffer PJ. Glutamine metabolism links growth factor signaling to the regulation of autophagy. Autophagy. 2012;8:1862–1864. doi: 10.4161/auto.22152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Vos KE, et al. Modulation of glutamine metabolism by the PI(3)K–PKB–FOXO network regulates autophagy. Nature Cell Biol. 2012;14:829–837. doi: 10.1038/ncb2536. [DOI] [PubMed] [Google Scholar]
- 13.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 14.Long X, Ortiz-Vega S, Lin Y, Avruch J. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem. 2005;280:23433–23436. doi: 10.1074/jbc.C500169200. [DOI] [PubMed] [Google Scholar]
- 15.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nature Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakashima N, Noguchi E, Nishimoto T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 1999;152:853–867. doi: 10.1093/genetics/152.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekiguchi T, et al. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 18.Gong R, et al. Crystal structure of the Gtr1p–Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev. 2011;25:1668–1673. doi: 10.1101/gad.16968011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong JH, et al. Crystal structure of the Gtr1pGTP–Gtr2pGDP protein complex reveals large structural rearrangements triggered by GTP-to-GDP conversion. J Biol Chem. 2012;287:29648–29653. doi: 10.1074/jbc.C112.384420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sancak Y, et al. Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Virgilio C, Loewith R. Cell growth control: little eukaryotes make big contributions. Oncogene. 2006;25:6392–6415. doi: 10.1038/sj.onc.1209884. [DOI] [PubMed] [Google Scholar]
- 22.Binda M, et al. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 23.Li L, et al. Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem. 2010;285:19705–19709. doi: 10.1074/jbc.C110.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the Rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashrafi K, Farazi TA, Gordon JI. A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J Biol Chem. 1998;273:25864–25874. doi: 10.1074/jbc.273.40.25864. [DOI] [PubMed] [Google Scholar]
- 26.Kogan K, Spear ED, Kaiser CA, Fass D. Structural conservation of components in the amino acid sensing branch of the TOR pathway in yeast and mammals. J Mol Biol. 2010;402:388–398. doi: 10.1016/j.jmb.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 27.Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang T, Peli-Gulli MP, Yang H, De Virgilio C, Ding J. Ego3 functions as a homodimer to mediate the interaction between Gtr1–Gtr2 and Ego1 in the EGO complex to activate TORC1. Structure. 2012;20:2151–2160. doi: 10.1016/j.str.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Saez I, Lacroix FB, Blot D, Gabel F, Skoufias DA. Structural characterization of HBXIP: the protein that interacts with the anti-apoptotic protein survivin and the oncogenic viral protein HBx. J Mol Biol. 2011;405:331–340. doi: 10.1016/j.jmb.2010.10.046. [DOI] [PubMed] [Google Scholar]
- 30.Kurzbauer R, et al. Crystal structure of the p14/MP1 scaffolding complex: how a twin couple attaches mitogen-activated protein kinase signaling to late endosomes. Proc Natl Acad Sci USA. 2004;101:10984–10989. doi: 10.1073/pnas.0403435101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lunin VV, et al. The structure of the MAPK scaffold, MP1, bound to its partner, p14. A complex with a critical role in endosomal map kinase signaling. J Biol Chem. 2004;279:23422–23430. doi: 10.1074/jbc.M401648200. [DOI] [PubMed] [Google Scholar]
- 32.Valbuena N, Guan KL, Moreno S. The Vam6–Gtr1/Gtr2 pathway activates TORC1 in response to amino acids in fission yeast. J Cell Sci. 2012;125:1920–1928. doi: 10.1242/jcs.094219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Messler S, et al. The TGF-β signaling modulators TRAP1/TGFBRAP1 and VPS39/Vam6/TLP are essential for early embryonic development. Immunobiology. 2011;216:343–350. doi: 10.1016/j.imbio.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside–out mechanism that requires the vacuolar H+-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishi T, Forgac M. The vacuolar (H+)-ATPases — nature’s most versatile proton pumps. Nature Rev Mol Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- 36.Fonseca BD, et al. Structure-activity analysis of niclosamide reveals potential role for cytoplasmic pH in control of mammalian target of rapamycin complex 1 (mTORC1) signaling. J Biol Chem. 2012;287:17530–17545. doi: 10.1074/jbc.M112.359638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balgi AD, et al. Regulation of mTORC1 signaling by pH. PLoS ONE. 2011;6:e21549. doi: 10.1371/journal.pone.0021549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han JM, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 39.Bonfils G, et al. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell. 2012;46:105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Avruch J, et al. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab. 2009;296:e592–e602. doi: 10.1152/ajpendo.90645.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim YM, et al. SH3BP4 is a negative regulator of amino acid–Rag GTPase–mTORC1 signaling. Mol Cell. 2012;46:833–846. doi: 10.1016/j.molcel.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 43.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 44.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ganley IG, et al. ULK1·ATG13·FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung CH, et al. ULK–Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martina JA, Chen Y, Gucek M, Puertollano R. mTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Settembre C, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orlova M, Kanter E, Krakovich D, Kuchin S. Nitrogen availability and TOR regulate the Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:1831–1837. doi: 10.1128/EC.00110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z, Wilson WA, Fujino MA, Roach PJ. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol Cell Biol. 2001;21:5742–5752. doi: 10.1128/MCB.21.17.5742-5752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirose E, Nakashima N, Sekiguchi T, Nishimoto T. RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1–GTPase pathway. J Cell Sci. 1998;111:11–21. doi: 10.1242/jcs.111.1.11. [DOI] [PubMed] [Google Scholar]
- 53.Nakashima N, Noguchi E, Nishimoto T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 1999;152:853–867. doi: 10.1093/genetics/152.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]