Abstract
Lipidomics is a subspecialty of metabolomics that focuses on water insoluble metabolites that form membrane barriers. Most lipidomic databases catalog lipids from common model organisms, like humans or Escherichia coli. However, model organisms’ lipid profiles show surprisingly little overlap with that of specialized pathogens, creating the need for organism-specific lipidomic databases. Here we review rapid progress in lipidomic platform development with regard to chromatography, detection and bioinformatics. We emphasize new methods of comparative lipidomics, which use aligned datasets to identify lipids changed after introducing a biological variable. These new methods provide an unprecedented ability to broadly and quantitatively describe lipidic change during biological processes and identify changed lipids with low error rates.
1. Introduction
Lipids act as membrane barriers, sources of energy and signal transducers in ways that influence outcomes in Alzheimer’s disease [1], cardiovascular disease [2] as well as viral [3] and bacterial infection [4]. As contrasted with classical methods of analytical chemistry, which focus on detailed structures of individual lipids, lipidomics seeks to broadly measure the presence and abundance of most or all lipids in a cell. Supporting high throughput approaches, the LIPID MAPS consortium (http://www.lipidmaps.org/) [5] and the European Lipidomics Initiative (ELIfe) [6] are building databases and analytical methods for scanning and cataloguing cellular lipids.
A central issue for lipidomic development relates to the molecular diversity of compounds studied. Proteins or nucleic acids are polymers that vary in sequence, but the overall differences in the chemical behavior of individual RNA, DNA or peptide molecules in solvents and chromatography are relatively small. In contrast, the chemical diversity of metabolites is atomic, rather than molecular. Individual metabolites are composed of markedly different ratios of carbon, hydrogen, oxygen, sulfur and phosphorous, so they span an extremely broad spectrum of polarity, which influences their solubility in extraction solvents and behavior in chromatography. This review focuses on conceptual and practical approaches to handling lipid diversity as a key issue for developing lipidomics as an experimental discipline.
2. Lipidomics is a distinct field of metabolomics
The small molecule metabolites in cells can be divided into two groups based on one essential property: solubility in aqueous solutions. Conventional metabolomic methods extract cytosolic metabolites and so emphasize amino acids, nucleotides and other water-soluble molecules. In contrast, lipids are insoluble in aqueous solutions and naturally form barriers that separate intracellular and extracellular compartments. Thus, lipidomics can be understood as a specialized field of metabolomics that uses hydrophobic solvents to extract membranes and study barrier function.
The cytosol-soluble metabolites involved in central metabolism are somewhat well conserved across genetically diverse organisms. However, many lipids, especially secondary metabolites that form lipid barriers are often specialized for the cell type, so that lipid profiles from common eukaryotic cells and pathogens with specialized niches can be quite distinct. As contrasted with cytosolic metabolites, lipids tend to have larger size (600–3000 atomic mass units), lower polarity and slower turnover. These distinct chemical properties translate into a need for distinct extraction solvents, quenching reagents and chromatographic methods for separating lipids into classes. In fact, many classical metabolomics methods start with ethyl acetate to extract monoacylglycerols and compounds that are more hydrophilic benchmark molecules. In contrast, many lipidomic methods extract with combinations of chloroform and methanol molecules that are less polar than monoacylglycerols.
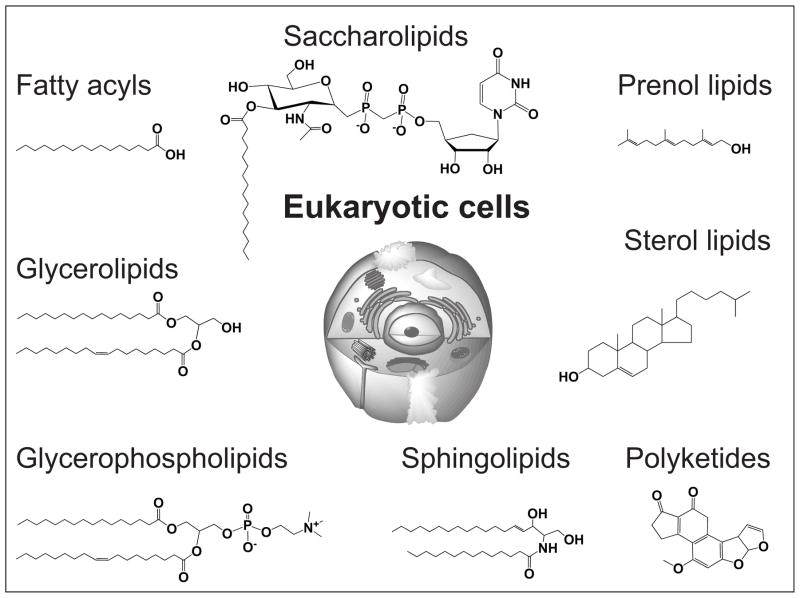
If lipids are described in a general way as molecules composed of a polar head group of variable size linked to aliphatic hydrocarbon chains, then there are many lipid classes with extremely diverse aliphatic hydrocarbon chains (Figures 1 and 2). Moreover, each lipid class is often comprised of slightly different combination of aliphatic chains and is produced as a family of acylforms or alkylforms, which posses the same core structure, but differ in the length, saturation state or substitution on individual alkyl chains. These fine molecular structures add another level of molecular diversity. Thus, a eukaryotic lipidome might contain up to 100,000 individual molecular lipid species [7–9]. This molecular diversity generates a need for developing a systematic ontology that can rationally name and catalog the molecules. Individual molecules with various trivial names and nomenclature systems are coalescing into one chemically comprehensive and widely used system that can be considered a lipidome. The LIPID MAPS project and other efforts seek to standardize nomenclature by describing each lipid with a unique 12-digit identifier, based on 8 lipid classes: fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids, prenol lipids, saccharolipids, and polyketides (Figure 1).
Figure 1. Eukaryotic lipid classes.
Representative structures of the 8 eukaryotic lipid classes of the LIPID MAPS classification system.
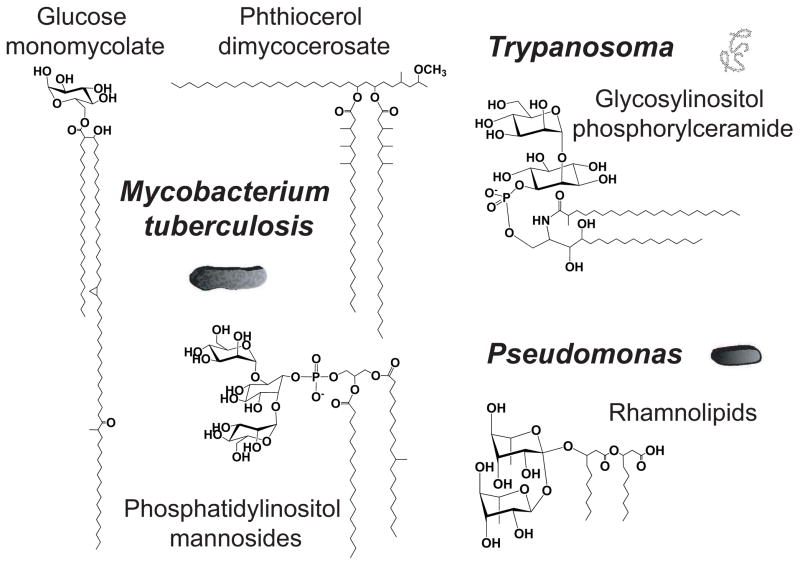
Figure 2. Pathogen-specific lipid classes.
Representative structures of pathogen-specific lipids.
3. Lipidomics strategies
3.1 Emergence of mass spectrometry (MS)-based methodology
Due to their high complexity and their heterogeneous physiochemical properties, measuring all types of lipids within a total lipid extract in a direct, comprehensive and quantitative manner creates analytical challenges. The earliest attempts consisted of conventional analytical methods such as high performance liquid chromatography, thin-layer chromatography [10] or gas chromatography of molecules modified with chromophores or optical density-based detection. Such approaches are time-consuming and require relatively large amounts of material. Furthermore, in the absence of radioactive or fluorescent labeling, these methodologies are mainly suitable for the characterization of global changes among lipid classes, rather than detailed remodeling of individual lipid species in one class. Yet, biological activities of a given lipid class often depend on the structure and distribution of molecular species [11, 12].
Nuclear magnetic resonance (NMR) is also an analytical method for lipidomics [13]. NMR-based strategies allow quantitative analyses of native molecules when present in complex mixtures and provide stereochemical information. Due to complexities in assigning signals to individual molecules in a mixture, this approach is often a fingerprint analysis, but can also identify certain individual lipids if they produce unique resonances [14, 15]. However, complexities related to interpretation of dozens or hundreds of mixed lipids limit analysis of individual compounds, and NMR requires relatively high input mass of lipids. Therefore, based on its high sensitivity and broad detection of most charged molecules in the mass range of naturally occurring lipids, mass spectrometry has emerged as a more widely used lipidomic methodology. Mass spectrometry has pushed the field forward so that thousands of distinct lipid species can be separately detected in one experiment [16].
Diverse lipids can be detected using electrospray ionization (ESI), atmospheric pressure chemical ionization, atmospheric pressure photoionization or matrix-assisted laser desorption/ionization. Currently, the most commonly employed configurations employ electrospray ionization, which is a relatively gentle and sensitive ionization method that can be combined with mass analyzers such as triple quadrupole, ion trap, Fourier transform or time of flight (Tof) devices, used in full scan mode or in collision induced dissociation mode (MS/MS). MS approaches can use simple shotgun methods with direct infusion of unfractionated lipid extract into the mass spectrometer [9], resulting in nearly simultaneous ionization of all analytes. Shotgun methods are commonly performed on tandem quadrupole instruments allow high throughput introduction of samples. However, shotgun methods are less broadly applicable to quantitative analysis of highly diverse lipids, due to cross-suppression of ionization by chemically diverse compounds [17]. Alternatively, LC-MS strategies partially separate lipid classes using liquid chromatography (LC) before mass spectrometry detection to reduce cross-suppression and gain other analytical information.
3.2 Targeted analyses
The comprehensive identification and quantitation of the different lipid classes and their molecular diversity at an organism-wide level is challenging. One approach to dealing with the diversity is to carry out several experiments, which separately target pre-defined subsets of lipid classes. Such targeted approaches take advantage of multiple extraction and chromatography methods, each of which is optimized for the class of lipids analyzed. For example, the Lipid Maps consortium uses a targeted approach for the study of all major membrane lipids in which lipid classes are separately analyzed using distinct LC-MS and MS/MS protocols [18]. As an illustration, atmospheric pressure chemical ionization favors the ionization of the most hydrophobic lipids like triglycerides, whereas ESI with negative ion mode scanning is favored for anionic membrane phospholipids. Ionization of specific lipids can be favored in shotgun approaches by modifying solvent and spraying conditions for each class [19]. Further, lipidome coverage by shotgun analysis can be improved by achieving separate analyses in positive and negative modes, with different solvents or sample concentrations [20]. Such shotgun approaches are most effective when analyzing mixtures of structurally related lipids. The targeted approach provides class-specific method optimization, but necessarily offers a limited view of the lipidome or requires reconciling multiple datasets into one lipidome.
3.3 Global Lipidomics
When searching for a new virulence factor, antigen or carrier, the identity of the biological substance, among all lipids in an organism, is not known at the outset, so the design imperative is broad lipidomic coverage of the organism using a single method. Such untargeted studies seek detection of the broadest number of lipids, including potentially new molecules whose names and chemical formulas are unknown, but whose masses can be reliably detected. In this situation, comparing or quantitatively reconciling data from different chromatographic, ionization or detection methods is difficult, and in some cases, invalid. Global analysis requires reproducible analytical methods for the analysis of total lipid extract in one LC-MS system, generated with one set of detection conditions.
Although more complex and requiring detailed validation, LC-MS methods provide several advantages over shotgun methods for the analysis of complex mixtures derived from the total lipid extract of an organism [21, 22]. Chromatographic separation increases homogeneity of lipids at the ionization interface at any moment in time, which reduces cross suppression for more sensitive and quantitative detection of minor or poorly ionizing lipid species within complex mixtures. For example, chromatographic separation has allowed the analysis of eicosanoids generated after cellular activation and present in a biological sample at subpicomolar levels [23]. Also, LC provides additional retention time information that is useful for lipid identification. Thus, for global lipidomics, new LC-MS methods must be optimized against a broad spectrum of lipids derived from one type of cell, tissue or body fluid, often containing thousands of distinct compounds in a mixture. One approach in method development is to provide a series of focused benchmark lipids against which new analytical approaches can be optimized [21]. This approach differs fundamentally from the usual methods of analytical chemistry, which seek to optimize detection of one type of lipids. Instead, global lipdomics method development requires that new method changes are evaluated as to whether they increase or decrease the total number of lipids detected, as well as its effects on diverse, defined benchmark lipids.
4. Lipidomic data processing
4.1 Data processing
Lipidomics experiments generate vast amounts of data. As in other systems biology platforms, computational tools are necessary to bypass overwhelming work of manual translation into practically useful data using stepwise conversion. Typically, raw MS data are organized into individual molecular features, whereby mass, retention time and intensity are calculated for thousands of ions, measured in replicate. Computational pipelines for lipidomic data analysis vary among laboratories, based on shotgun or HPLC-MS analytical approaches and the type of mass analyzer used. However, most approaches share certain common features:
extraction of mass specific signals from noise to reduce false positive detection,
detection of discrete peaks corresponding to each detected m/z value, which results in list of three-dimensional datapoints, described here as molecular features, with linked m/z, retention time and intensity coordinates,
aligning datasets by matching peaks across multiple sample runs,
measuring variance among replicate datasets.
In comparative lipidomics, replicate datasets are first combined to give variance data for each molecular feature, and then separate datasets are generated under distinct biological conditions. Two or more datasets are aligned and systematically compared to provide an overview of changes. Software packages that support these analyses include XCMS (R) [24], MZmine (Java) [25] or Lipid Data Analyzer [26]. For shotgun lipidomic datasets that include fragmentation information, data processing includes spectral averaging and isotopic correction as well as algorithms for simultaneous identification and quantification of lipids.
4.2 MS signals assignment to described or known lipid structures
Over the years, targeted experiments have provided accurate mass, fragmentation patterns and polarity information for thousands lipid species. In addition, studies of ion chemistry fragmentation upon collision-induced dissociation have highlighted differential fragmentation kinetics and establish regiospecific fragmentation rules. Indeed, relative intensities of fragments reflect the position of fatty acids on several backbone structures [27, 28]. Experimental and theoretical information has been compiled in databases for automatic identification of previously described or “known” lipids in newly acquired datasets [29].
Mass spectrometers with high resolution (105) and mass accuracy (5–10 parts per million) enable elemental composition determination based on detected m/z values with the help of programs such as the Fatty Acid Analysis Tool [30]. Recent high mass resolution shotgun lipidomics, known as “top-down” lipidomics have been developed for the acquisition of exact masses [31, 32]. However, m/z information does not differentiate two molecules of the same nominal masses, also called isobars, within a mixture of lipids. An advantage of compound separation in LC-MS includes minimizing inter-class isobaric overlap. Intra-class isobars such as two phosphatidylcholine species with different fatty acid combinations, but the same formula (C18:2, C18:0 or C16:0, C20:2), can be differentiated by tandem mass spectrometry (MS/MS). The measurement of specific molecules within a mixture can be targeted for quantitative measurements using a multi-sector analyzer, such as a triple quadrupole, by searching for specific fragmentation patterns in parent ion scanning, daughter ion scanning or neutral loss scanning [9]. A sensitive and specific scanning mode is called selected reaction monitoring, whereby both specified parent and daughter ions are monitored. For LC-MS strategies performed on quadrupole or Q-Tof analyzers, specific measurements of targeted lipids eluting from chromatographic column at any time is achieved by repeatedly scanning for selected reactions, a method called multiple reaction monitoring [33]. These multidimensional mass spectrometry analyses have been applied for detailed analysis of glycerolipids, phospholipids and sphingolipids [34–37]. In parallel, techniques have enabled more detailed structural characterization like double bond position in fatty acyl groups by collision-induced dissociation in the presence of ozone [38] or the location of fatty acyl groups on the glycerol backbone by chiral LC-MS methods [39, 40].
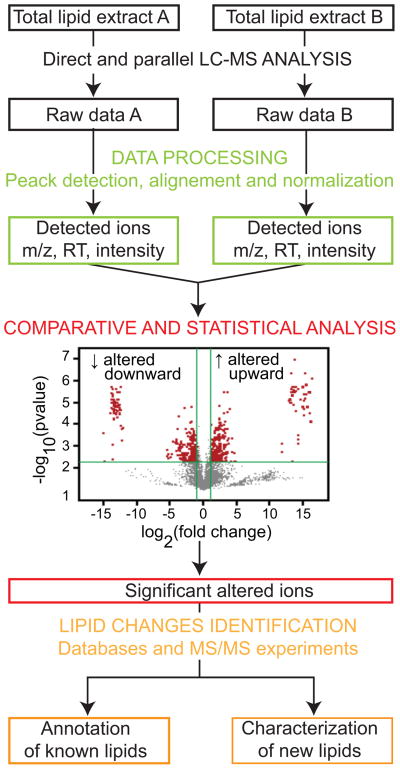
Software has been optimized for automatic identification, including Lipid Profiler/LipidView [33], Lipid Inspector [41], Fatty Acid Analysis Tool [30], and LipidQA [29]. In contrast to software, which relies on databases of reference MS/MS spectra, LipidXplorer [42] carries out cross-platform interpretation of shotgun lipidomics datasets and identifies lipids via user-defined queries formulated in the molecular fragmentation query language, allowing new queries for the identification of new lipids. For automatic identification based on accurate mass retention time in the absence of fragmentation information, platform-specific databases, most often created by the user, are required. In broad and untargeted LC-MS analysis, the lipid identification step is not essential for data processing of unnamed compounds of known mass for statistical analysis (Figure 3) [21]. XCMS and MZmine are widely used open-source tools to achieve untargeted data processing, and other programs are commercially available. Subsequent to statistical analyses, selected ions of interest are assigned to lipid structures by off-line MS/MS experiments. Manual collision induced dissociation methods have lower throughput than automated comparisons to databases, but can discover new types of lipids (Figure 3).
Figure 3. Global and unbiased comparative lipidomics.
Lipid extracts (dark) enter a workflow involving direct LC-MS detection using a steep gradient that separates diverse molecules in a single run (black), software-assisted raw curve finding and quantitation (green) and alignment for statistical comparative analysis (red). Finally, significantly changed features meeting predefined criteria provide a targeted list of compounds that correlated with the biological variable. This smaller list of candidate compounds can be identified through database interrogation or collisional mass spectrometry (orange). RT: retention time.
5. Comparative lipidomics
Comparative lipidomic analysis uses aligned datasets from samples obtained in at least two biological conditions, which differ by one biological variable such as growth condition, gene knockout or cell type. Much of the software described above, which is used for the lipid identification task, enables quantitation of targeted lipids. Quantitation is achieved by metabolic stable isotope labeling [30] or by addition of standards before extraction, which are often deuterated similar lipid class or with odd-carbon chain that are naturally absent in the studied organism [43]. Using calibration curves and internal standards, programs perform automatic conversion of intensity values of targeted lipids to absolute mass values [29, 44]. Such bioinformatics tools have successfully allowed the simultaneous absolute quantitation of hundreds lipid species using shotgun lipidomics [20]. For untargeted and global readouts, programs such as Lipid Array perform semi-quantitative analyses in the absence of standards using peak intensities [45]. Multiple testing corrections limit the false discovery rate [46].
Once all datapoints are assigned quantitative mean intensity or concentration values with calculated variance, lipidomes are systemically aligned to identify the number of changed features (Figure 3). This is the area where lipidomic analysis benefits from statistical procedures and visualization methods previously developed for use in transcriptomics (reviewed in [47]) to obtain correlation maps and volcano plots that describe fold-changes and p-values for all molecular events (Figure 3). As an illustration, metaXCMS (R) allows comparison of multiple datasets from different biological conditions for the identification of metabolites associated with biological variables [48]. After very large datasets with thousands of molecular features are compared to identify the smaller number of changed lipids, the final stage involves compound naming using databases or MS/MS that can be carried out on those lipids most significantly changed (Figure 3). The lipidomic community has undertaken recent efforts to standardize lipid naming and computational strategies. However, file formats are not yet universally standardized. Also, dataset analysis typically requires several bioinformatic tools that separately focus on quantitation, alignment, lipid identification and comparative display.
6. Model organisms versus major pathogens
Most efforts to catalogue lipidomes emphasize eukaryotic cells [49] or common model organisms like humans or E. coli [50]. In contrast to cytosol-soluble, central metabolites studied in conventional metabolomics, membrane lipids are secondary metabolites that show substantial variation from organism to organism. Medically important pathogens with genomes that diverge substantially from these two model organisms are beginning to be systematically categorized, including Candida albicans [51], Toxoplasma gondii [52], Tripanosoma species [53], Leishmania donovani [54] and Mycobacterium tuberculosis [21, 55, 56]. The degree of overlap of lipid profiles among model organisms and medically important organisms can be surprisingly limited. Therefore, databases for individual pathogens need to be created and hosted in a systematic way.
This general point is well illustrated for M. tuberculosis, a major human pathogen responsible for more than 1.5 million deaths each year [57]. Members of the genus Mycobacterium produce one of the most complex and unique bacterial lipidomes. It is estimated that 8% of the mycobacterial genome encodes around 250 enzymes for the biosynthesis of its lipid repertoire, as contrasted to 50 lipid modifying genes in E. Coli [58]. These enzymes produce apolar lipids, glycolipids, lipoproteins and lipoglycans, which account up to 40 percent of the bacterial dry weight [59]. Complete Freund’s adjuvant is a mixture of cell wall components from which trehalose dimycolate and acylated lipoproteins contribute strongly to host response [60, 61]. Lipids can interfere in a stimulatory or inhibitory manner on human immune system. Lipids are T cell antigens presented by CD1 proteins, adjuvants and virulence factors that modulate the host cytokines pattern [4, 62, 63].
The widespread nature of the tuberculosis epidemic, as well as the plethora of immunologic and virulence determining properties found into mycobacterial lipids, have initiated systematic efforts to study M. tuberculosis lipidome on a global level [21, 55, 56]. The question arises as to whether highly specialized eukaryotic or bacterial pathogens require distinct approaches or can instead rely on databases and methods from model organisms. The MycoMass database catalogs more than 5000 theoretical lipid species into 58 lipid families of which 40 families are not found in databases emphasizing model organisms. With the exception of some glycerophospholipid classes found in the inner mycobacterial membrane, most of the lipid structures produced by mycobacteria are only found in mycobacteria and related actinobacteria (Figure 2). This basic finding, which to a lesser extent is reflected in new lipidomic datasets for toxoplasma or trypanosome species, reflects the need to catalog and organize lipids from specialized cell types with high biomedical significance.
Further, the relatively high mass and low polarity of mycobacterial polyketides and mycolic acids contrasts with the lower mass and higher polarity of anionic phospholipids that dominate in mammalian cells. Therefore, developing a lipidomics platform for M. tuberculosis required hydrophobic run solvents, high ionization energy and lower countercurrent gas at the electrospray interface, as compared to those previously optimized for phospholipids [21]. Last, in contrast to anionic phospholipids found in most cells, which are preferentially detected in the negative ion mode, mycolyl glycolipids and polyketides in the mycobacterial outer membrane are neutral lipids. Because these lipids are diverse and are more readily detected as cation adducts, we achieved broader lipidomic coverage in the positive ion mode. These examples illustrate why lipidomic methods must be optimized for this medically important bacterial parasite [21, 55, 64].
These basic observations lead to several general insights relating to new directions for lipidomic study. First, given the lack of coverage of lipids of pathogens among databases designed for model organisms, automated naming of ions based on accurate mass is currently limited. Separate databases for specialized pathogens, or one database that is separately interrogated by cell or origin, need to be established along with new resources for hosting this information. The differing lipid content of these pathogens opens new avenues for investigation of comparative lipidomics. For example, systematic investigation of the particular lipids that distinguish pathogens from related species or strains could provide information about biomarkers of the strain for use in diagnosis. Also, systematic comparisons of lipid profiles in pathogen strains with genetically related, but non-pathogenic strains, represents a strategy to identify virulence factors.
Finally, elegant work on Bacillus subtilis has integrated multidisciplinary platforms that profile, transcripts, proteins and metabolites in parallel for one organism in defined conditions [65]. The integration of results from genomics, proteomics, metabolomics and lipidomics supports a new approach whereby genetic regulators, enzymes and small molecules are measured in parallel during one experiment.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sagin FG, Sozmen EY. Lipids as key players in Alzheimer disease: alterations in metabolism and genetics. Curr Alzheimer Res. 2008;5(1):4–14. doi: 10.2174/156720508783884648. [DOI] [PubMed] [Google Scholar]
- 2.Tselepis AD, John Chapman M. Inflammation, bioactive lipids and atherosclerosis: potential roles of a lipoprotein-associated phospholipase A2, platelet activating factor-acetylhydrolase. Atheroscler Suppl. 2002;3(4):57–68. doi: 10.1016/s1567-5688(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 3.Marsh M, Helenius A. Virus entry: open sesame. Cell. 2006;124(4):729–40. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neyrolles O, Guilhot C. Recent advances in deciphering the contribution of Mycobacterium tuberculosis lipids to pathogenesis. Tuberculosis (Edinb) 2011;91(3):187–195. doi: 10.1016/j.tube.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Schmelzer K, Fahy E, Subramaniam S, Dennis EA. The lipid maps initiative in lipidomics. Methods Enzymol. 2007;432:171–183. doi: 10.1016/S0076-6879(07)32007-7. [DOI] [PubMed] [Google Scholar]
- 6.van Meer G, Leeflang BR, Liebisch G, Schmitz G, Goni FM. The European lipidomics initiative: enabling technologies. Methods Enzymol. 2007;432:213–232. doi: 10.1016/S0076-6879(07)32009-0. [DOI] [PubMed] [Google Scholar]
- 7.Murphy RC, Fiedler J, Hevko J. Analysis of nonvolatile lipids by mass spectrometry. Chem Rev. 2001;101(2):479–526. doi: 10.1021/cr9900883. [DOI] [PubMed] [Google Scholar]
- 8.van Meer G. Cellular lipidomics. Embo J. 2005;24(18):3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005;24(3):367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 10.Fontell K, Holman RT, Lambertsen G. Some new methods for separation and analysis of fatty acids and other lipids. J Lipid Res. 1960;1:391–404. [PubMed] [Google Scholar]
- 11.Guiard J, Collmann A, Garcia-Alles LF, Mourey L, Brando T, Mori L, Gilleron M, Prandi J, De Libero G, Puzo G. Fatty acyl structures of mycobacterium tuberculosis sulfoglycolipid govern T cell response. J Immunol. 2009;182(11):7030–7037. doi: 10.4049/jimmunol.0804044. [DOI] [PubMed] [Google Scholar]
- 12.Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, Yamashita E, Tsukihara T, Yoshikawa S. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. Embo J. 2007;26(6):1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reo NV. NMR-based metabolomics. Drug Chem Toxicol. 2002;25(4):375–382. doi: 10.1081/dct-120014789. [DOI] [PubMed] [Google Scholar]
- 14.Mahrous EA, Lee RB, Lee RE. A rapid approach to lipid profiling of mycobacteria using 2D HSQC NMR maps. J Lipid Res. 2008;49(2):455–463. doi: 10.1194/jlr.M700440-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Fernando H, Bhopale KK, Kondraganti S, Kaphalia BS, Shakeel Ansari GA. Lipidomic changes in rat liver after long-term exposure to ethanol. Toxicol Appl Pharmacol. 2011;255(2):127–137. doi: 10.1016/j.taap.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLuckey SA, Wells JM. Mass analysis at the advent of the 21st century. Chem Rev. 2001;101(2):571–606. doi: 10.1021/cr990087a. [DOI] [PubMed] [Google Scholar]
- 17.Han X, Yang K, Gross RW. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom Rev. 2012;31(1):134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreyev AY, Fahy E, Guan Z, Kelly S, Li X, McDonald JG, Milne S, Myers D, Park H, Ryan A, Thompson BM, Wang E, Zhao Y, Brown HA, Merrill AH, Raetz CR, Russell DW, Subramaniam S, Dennis EA. Subcellular organelle lipidomics in TLR-4-activated macrophages. J Lipid Res. 2010;51(9):2785–2797. doi: 10.1194/jlr.M008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han X, Yang K, Yang J, Fikes KN, Cheng H, Gross RW. Factors influencing the electrospray intrasource separation and selective ionization of glycerophospholipids. J Am Soc Mass Spectrom. 2006;17(2):264–274. doi: 10.1016/j.jasms.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci USA. 2009;106(7):2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Layre E, Sweet L, Hong S, Madigan CA, Desjardins D, Young DC, Cheng TY, Annand JW, Kim K, Shamputa IC, McConnell MJ, Debono CA, Behar SM, Minnaard AJ, Murray M, Barry CE, 3rd, Matsunaga I, Moody DB. A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem Biol. 2011;18(12):1537–1549. doi: 10.1016/j.chembiol.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandra K, Pereira Ados S, Vanhoenacker G, David F, Sandra P. Comprehensive blood plasma lipidomics by liquid chromatography/quadrupole time-of-flight mass spectrometry. J Chromatogr A. 2010;1217(25):4087–4099. doi: 10.1016/j.chroma.2010.02.039. [DOI] [PubMed] [Google Scholar]
- 23.Murphy RC, Barkley RM, Zemski Berry K, Hankin J, Harrison K, Johnson C, Krank J, McAnoy A, Uhlson C, Zarini S. Electrospray ionization and tandem mass spectrometry of eicosanoids. Anal Biochem. 2005;346(1):1–42. doi: 10.1016/j.ab.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 24.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78(3):779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 25.Katajamaa M, Miettinen J, Oresic M. MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics. 2006;22(5):634–636. doi: 10.1093/bioinformatics/btk039. [DOI] [PubMed] [Google Scholar]
- 26.Hartler J, Trotzmuller M, Chitraju C, Spener F, Kofeler HC, Thallinger GG. Lipid Data Analyzer: unattended identification and quantitation of lipids in LC-MS data. Bioinformatics. 2011;27(4):572–577. doi: 10.1093/bioinformatics/btq699. [DOI] [PubMed] [Google Scholar]
- 27.Hsu FF, Turk J. Charge-remote and charge-driven fragmentation processes in diacyl glycerophosphoethanolamine upon low-energy collisional activation: a mechanistic proposal. J Am Soc Mass Spectrom. 2000;11(10):892–899. doi: 10.1016/S1044-0305(00)00159-8. [DOI] [PubMed] [Google Scholar]
- 28.Hsu FF, Turk J. Charge-driven fragmentation processes in diacyl glycerophosphatidic acids upon low-energy collisional activation. A mechanistic proposal. J Am Soc Mass Spectrom. 2000;11(9):797–803. doi: 10.1016/S1044-0305(00)00151-3. [DOI] [PubMed] [Google Scholar]
- 29.Song H, Hsu FF, Ladenson J, Turk J. Algorithm for processing raw mass spectrometric data to identify and quantitate complex lipid molecular species in mixtures by data-dependent scanning and fragment ion database searching. J Am Soc Mass Spectrom. 2007;18(10):1848–1858. doi: 10.1016/j.jasms.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leavell MD, Leary JA. Fatty acid analysis tool (FAAT): An FT-ICR MS lipid analysis algorithm. Anal Chem. 2006;78(15):5497–5503. doi: 10.1021/ac0604179. [DOI] [PubMed] [Google Scholar]
- 31.Graessler J, Schwudke D, Schwarz PE, Herzog R, Shevchenko A, Bornstein SR. Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS One. 2009;4(7):e6261. doi: 10.1371/journal.pone.0006261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwudke D, Hannich JT, Surendranath V, Grimard V, Moehring T, Burton L, Kurzchalia T, Shevchenko A. Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra. Anal Chem. 2007;79(11):4083–4093. doi: 10.1021/ac062455y. [DOI] [PubMed] [Google Scholar]
- 33.Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, Thiele C, Ekroos K, Shevchenko A. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal Chem. 2006;78(17):6202–6214. doi: 10.1021/ac060545x. [DOI] [PubMed] [Google Scholar]
- 34.Yang K, Zhao Z, Gross RW, Han X. Systematic analysis of choline-containing phospholipids using multi-dimensional mass spectrometry-based shotgun lipidomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(26):2924–36. doi: 10.1016/j.jchromb.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gross RW, Han X. Shotgun lipidomics of neutral lipids as an enabling technology for elucidation of lipid-related diseases. Am J Physiol Endocrinol Metab. 2009;297(2):E297–303. doi: 10.1152/ajpendo.90970.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han X, Gross RW. Shotgun lipidomics: multidimensional MS analysis of cellular lipidomes. Expert Rev Proteomics. 2005;2(2):253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- 37.Han X, Yang K, Cheng H, Fikes KN, Gross RW. Shotgun lipidomics of phosphoethanolamine-containing lipids in biological samples after one-step in situ derivatization. J Lipid Res. 2005;46(7):1548–1560. doi: 10.1194/jlr.D500007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas MC, Mitchell TW, Harman DG, Deeley JM, Nealon JR, Blanksby SJ. Ozone-induced dissociation: elucidation of double bond position within mass-selected lipid ions. Anal Chem. 2008;80(1):303–311. doi: 10.1021/ac7017684. [DOI] [PubMed] [Google Scholar]
- 39.Deng L, Nakano H, Iwasaki Y. Direct separation of monoacylglycerol isomers by enantioselective high-performance liquid chromatography. J Chromatogr A. 2008;1198–1199:67–72. doi: 10.1016/j.chroma.2008.03.095. [DOI] [PubMed] [Google Scholar]
- 40.Lee SH, Blair IA. Targeted chiral lipidomics analysis of bioactive eicosanoid lipids in cellular systems. BMB Rep. 2009;42(7):401–410. doi: 10.5483/bmbrep.2009.42.7.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzchalia T, Shevchenko A. Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal Chem. 2006;78(2):585–595. doi: 10.1021/ac051605m. [DOI] [PubMed] [Google Scholar]
- 42.Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011;12(1):R8. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore JD, Caufield WV, Shaw WA. Quantitation and standardization of lipid internal standards for mass spectroscopy. Methods Enzymol. 2007;432:351–367. doi: 10.1016/S0076-6879(07)32014-4. [DOI] [PubMed] [Google Scholar]
- 44.Song H, Ladenson J, Turk J. Algorithms for automatic processing of data from mass spectrometric analyses of lipids. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(26):2847–2854. doi: 10.1016/j.jchromb.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanova PT, Milne SB, Forrester JS, Brown HA. LIPID arrays: new tools in the understanding of membrane dynamics and lipid signaling. Mol Interv. 2004;4(2):86–96. doi: 10.1124/mi.4.2.6. [DOI] [PubMed] [Google Scholar]
- 46.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125(1–2):279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 47.Niemela PS, Castillo S, Sysi-Aho M, Oresic M. Bioinformatics and computational methods for lipidomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(26):2855–2862. doi: 10.1016/j.jchromb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 48.Tautenhahn R, Patti GJ, Kalisiak E, Miyamoto T, Schmidt M, Lo FY, McBee J, Baliga NS, Siuzdak G. metaXCMS: second-order analysis of untargeted metabolomics data. Anal Chem. 2011;83(3):696–700. doi: 10.1021/ac102980g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dennis EA, Deems RA, Harkewicz R, Quehenberger O, Brown HA, Milne SB, Myers DS, Glass CK, Hardiman G, Reichart D, Merrill AH, Jr, Sullards MC, Wang E, Murphy RC, Raetz CR, Garrett TA, Guan Z, Ryan AC, Russell DW, McDonald JG, Thompson BM, Shaw WA, Sud M, Zhao Y, Gupta S, Maurya MR, Fahy E, Subramaniam S. A mouse macrophage lipidome. J Biol Chem. 2010;285(51):39976–39985. doi: 10.1074/jbc.M110.182915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oursel D, Loutelier-Bourhis C, Orange N, Chevalier S, Norris V, Lange CM. Lipid composition of membranes of Escherichia coli by liquid chromatography/tandem mass spectrometry using negative electrospray ionization. Rapid Commun Mass Spectrom. 2007;21(11):1721–1728. doi: 10.1002/rcm.3013. [DOI] [PubMed] [Google Scholar]
- 51.Singh A, Prasad R. Comparative lipidomics of azole sensitive and resistant clinical isolates of Candida albicans reveals unexpected diversity in molecular lipid imprints. PLoS One. 2011;6(4):e19266. doi: 10.1371/journal.pone.0019266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welti R, Mui E, Sparks A, Wernimont S, Isaac G, Kirisits M, Roth M, Roberts CW, Botte C, Marechal E, McLeod R. Lipidomic analysis of Toxoplasma gondii reveals unusual polar lipids. Biochemistry. 2007;46(48):13882–13890. doi: 10.1021/bi7011993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richmond GS, Gibellini F, Young SA, Major L, Denton H, Lilley A, Smith TK. Lipidomic analysis of bloodstream and procyclic form Trypanosoma brucei. Parasitology. 2010;137(9):1357–1392. doi: 10.1017/S0031182010000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng L, T’Kind R, Decuypere S, von Freyend SJ, Coombs GH, Watson DG. Profiling of lipids in Leishmania donovani using hydrophilic interaction chromatography in combination with Fourier transform mass spectrometry. Rapid Commun Mass Spectrom. 2010;24(14):2074–2082. doi: 10.1002/rcm.4618. [DOI] [PubMed] [Google Scholar]
- 55.Madigan CA, Cheng TY, Layre E, Young DC, McConnell MJ, Debono CA, Murry JP, Wei JR, Barry CE, 3rd, Rodriguez GM, Matsunaga I, Rubin EJ, Moody DB. Lipidomic discovery of deoxysiderophores reveals a revised mycobactin biosynthesis pathway in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2012;109(4):1257–1262. doi: 10.1073/pnas.1109958109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sartain CV, Cui J, Meisel RP, Wolfner MF. The poly(A) polymerase GLD2 is required for spermatogenesis in Drosophila melanogaster. Development. 2011;138(8):1619–1629. doi: 10.1242/dev.059618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.W.H.O. Global tuberculosis control. 2011 http://www.who.int/tb/publications/global_report/2011/gtbr11_full.pdf.
- 58.Camus JC, Pryor MJ, Medigue C, Cole ST. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148(Pt 10):2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- 59.Goren MB, Brennan PJ. Mycobacterial lipids: chemistry and biologic activities. In: Youmans GP, editor. Tuberculosis (Edinb) W. B. Saunders Co; Philadelphia: 1979. pp. 63–193. [Google Scholar]
- 60.Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, Kinoshita T, Akira S, Yoshikai Y, Yamasaki S. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med. 2009;206(13):2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thoma-Uszynski S, Stenger S, Takeuchi O, Ochoa MT, Engele M, Sieling PA, Barnes PF, Rollinghoff M, Bolcskei PL, Wagner M, Akira S, Norgard MV, Belisle JT, Godowski PJ, Bloom BR, Modlin RL. Induction of direct antimicrobial activity through mammalian toll-like receptors. Science. 2001;291(5508):1544–1547. doi: 10.1126/science.291.5508.1544. [DOI] [PubMed] [Google Scholar]
- 62.Moody DB, Besra GS, Wilson IA, Porcelli SA. The molecular basis of CD1-mediated presentation of lipid antigens. Immunol Rev. 1999;172:285–296. doi: 10.1111/j.1600-065x.1999.tb01373.x. [DOI] [PubMed] [Google Scholar]
- 63.Young DC, Moody DB. T-cell recognition of glycolipids presented by CD1 proteins. Glycobiology. 2006;16(7):103R–112R. doi: 10.1093/glycob/cwj111. [DOI] [PubMed] [Google Scholar]
- 64.Sartain MJ, Dick DL, Rithner CD, Crick DC, Belisle JT. Lipidomic analyses of Mycobacterium tuberculosis based on accurate mass measurements and the novel “Mtb LipidDB”. J Lipid Res. 2011;52(5):861–872. doi: 10.1194/jlr.M010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buescher JM, Liebermeister W, Jules M, Uhr M, Muntel J, Botella E, Hessling B, Kleijn RJ, Le Chat L, Lecointe F, Mader U, Nicolas P, Piersma S, Rugheimer F, Becher D, Bessieres P, Bidnenko E, Denham EL, Dervyn E, Devine KM, Doherty G, Drulhe S, Felicori L, Fogg MJ, Goelzer A, Hansen A, Harwood CR, Hecker M, Hubner S, Hultschig C, Jarmer H, Klipp E, Leduc A, Lewis P, Molina F, Noirot P, Peres S, Pigeonneau N, Pohl S, Rasmussen S, Rinn B, Schaffer M, Schnidder J, Schwikowski B, Van Dijl JM, Veiga P, Walsh S, Wilkinson AJ, Stelling J, Aymerich S, Sauer U. Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science. 2012;335(6072):1099–1103. doi: 10.1126/science.1206871. [DOI] [PubMed] [Google Scholar]