This study examines translational regulation of erythropoietin (EPO). New findings show that EPO translation is controlled by an upstream small open reading frame (ORF). In unstressed cells, the ORF represses translation. However, under hypoxic stress, this repression is largely relieved by a mechanism of enhanced ribosome bypass of the small ORF.
Keywords: upstream open reading frame (uORF), eIF2α, phosphorylation, translational regulation, hypoxia, erythropoietin (EPO)
Abstract
Erythropoietin (EPO) is a key mediator hormone for hypoxic induction of erythropoiesis that also plays important nonhematopoietic functions. It has been shown that EPO gene expression regulation occurs at different levels, including transcription and mRNA stabilization. In this report, we show that expression of EPO is also regulated at the translational level by an upstream open reading frame (uORF) of 14 codons. As judged by comparisons of protein and mRNA levels, the uORF acts as a cis-acting element that represses translation of the main EPO ORF in unstressed HEK293, HepG2, and HeLa cells. However, in response to hypoxia, this repression is significantly released, specifically in HeLa cells, through a mechanism that involves processive scanning of ribosomes from the 5′ end of the EPO transcript and enhanced ribosome bypass of the uORF. In addition, we demonstrate that in HeLa cells, hypoxia induces the phosphorylation of eukaryotic translation initiation factor 2α (eIF2α) concomitantly with a significant increase of EPO protein synthesis. These findings provide a framework for understanding that production of high levels of EPO induced by hypoxia also involves regulation at the translational level.
INTRODUCTION
Regulation of mRNA translation is a key mechanism by which cells and organisms can rapidly change their gene expression patterns in response to extra- and intracellular stimuli. Translational control can occur on a global basis by modifications of the basic translation machinery or by selectively targeting defined subsets of messenger RNAs (mRNAs) to maintain the synthesis of certain proteins required either for the stress response or for aiding recovery from stress. These pathways are evolutionarily conserved and have been shown to impact translation significantly in organisms as diverse as yeast and humans. In many cases, features in the 5′-leader sequences of the corresponding mRNAs, such as internal ribosome entry sites (IRESs) and/or regulatory upstream open reading frames (uORFs), are important for them to evade global repression of translation (Sonenberg and Hinnebusch 2009).
uORFs are regulatory cis-acting elements present in the 5′-leader sequence of a transcript that are spread among different species and throughout the genome, but their prevalence has been difficult to calculate (Mignone et al. 2002). The most recent studies estimate that ∼49% of the human transcripts contain at least one uORF (Calvo et al. 2009), being conspicuously common in certain classes of genes, including oncogenes and genes involved in the control of cellular growth and differentiation (Morris and Geballe 2000; Wethmar et al. 2010). The fact that mutations that introduce or disrupt a uORF can cause human diseases illustrates their role in translational regulation (Cazzola and Skoda 2000; Chatterjee and Pal 2009; Barbosa et al. 2013).
For a uORF to function as a translational regulatory element, its initiation codon (upstream AUG [uAUG]) must be recognized, at least at certain times, by the scanning 40S ribosomal subunit and associated initiation factors (Hernández et al. 2010). When uORF recognition is regulated by a so-called leaky scanning mechanism, ribosomes either scan through the uAUG codon or recognize it, initiating translation. Indeed, the recognition of an AUG can be affected by its context, the AUG proximity to the cap site and the presence of nearby secondary structures. The optimal context is GCC(A/G)CCAUGG, with positions −3 and +4 being functionally the most important (Kozak 2002). In the case that the uORF is recognized and translated by a scanning ribosome, multiple alternative fates are available to the ribosome: It may (1) terminate and leave the mRNA, resulting in down-regulation of translation of the downstream main ORF; (2) translate the uORF and stall during either the elongation or termination phase of uORF translation, creating a blockade to additional ribosome scanning; or (3) terminate and reinitiate (Meijer and Thomas 2002; Poyry et al. 2004). When the option is for the ribosome to remain associated with the mRNA, it continues scanning and reinitiates further downstream at either a proximal or distal AUG codon. The potential of a ribosome to reinitiate further downstream depends on a number of factors, including the length of the uORF and the time it took to translate the uORF (Kozak 2001; Rajkowitsch et al. 2004). It has been shown that it is not so much the length per se that is the critical parameter but, rather, the time taken for the ribosome to translate the uORF (Poyry et al. 2004). The reason why uORF translation needs to be completed rapidly is related to the initiation factors necessary to remain associated with the 40S subunit to promote 40S scanning from the uORF termination codon to the downstream initiation site (Poyry et al. 2004). However, other initiation factors have to be acquired de novo. One initiation factor that has to be reacquired is eIF2 in the form of an eIF2/GTP/Met-tRNAi ternary complex (Hinnebusch 1997), because the Met-tRNAi in the ternary complex associated with the 40S subunit as it scans to the uORF initiation codon has been used to initiate the uORF translation (Poyry et al. 2004; Kozak 2005; Sachs 2006).
When mammalian cells encounter stress conditions such as during differentiation, development, pathogenic infection, chemical exposure, nutrient deprivation, and hypoxia, a family of protein kinases is activated to phosphorylate eIF2α. Phosphorylation of the α subunit of eIF2α on Ser 51 prevents the exchange of GDP for GTP by sequestering eIF2B, lowering the available pool of eIF2/GTP that is required for binding of initiator tRNA to the small ribosomal subunit and thus repressing protein synthesis (Sonenberg and Hinnebusch 2009). Concomitantly with the general inhibition of translation, phosphorylation of eIF2α selectively promotes translational up-regulation of a subset of mRNAs. Such mRNAs include those containing uORFs.
Human erythropoietin (EPO) is a circulating 34.4-kDa glycoprotein hormone that controls erythropoiesis by stimulating the proliferation of erythroid precursors (Jelkmann 1992; Ebert and Bunn 1999; Mole and Ratcliffe 2007). Indeed, its major action is the prevention of apoptosis in EPO-dependent, colony-forming unit–erythroid cells and erythroblasts that have not begun hemoglobin synthesis. Expression of the EPO gene is tightly controlled, and in the adult organism, kidneys produce ∼90% of circulating EPO, with its production being markedly up-regulated by hypoxia (Jelkmann 1992). Apart from the kidney and liver being the two major sites of synthesis, EPO mRNA expression has also been detected in the brain (neurons and glial cells), lung, heart, bone marrow, spleen, hair follicles, and the reproductive tract (Fandrey and Bunn 1993; Dame et al. 1998; Yasuda et al. 1998; Ghezzi and Brines 2004; Weidemann and Johnson 2009; Hoch et al. 2011). EPO synthesized in these organs appears to act locally, modulating, for example, regional angiogenesis and cellular viability, and does not seem to contribute to erythropoiesis (Maiese et al. 2008; Gassmann and Soliz 2009).
As mentioned above, EPO expression under normoxic conditions is low but increases during exposure to hypoxia; however, other stimuli, such as estrogen in the uterus, can also increase EPO expression (Yasuda et al. 1998; Chin et al. 2000; Besarab et al. 2009). Hypoxia inducible factor (HIF) 1 is the transcriptional activator responsible for the hypoxic induction of EPO that binds to the hypoxia-responsive element located in 3′ of the EPO gene, augmenting the EPO transcriptional rate (Wang et al. 1995; Noguchi et al. 2008).
Although the effect of many transcriptional mechanisms involved in EPO gene expression regulation has been well established, there is also an important contribution of post-transcriptional mechanisms that is less characterized (Goldberg et al. 1991). Alignment of the human and mouse EPO 5′-leader sequences has revealed high identity and the presence of a 14-codon uORF located upstream of the main AUG (Shoemaker and Mitsock 1986). These observations led us to investigate the potential role of this naturally occurring uORF in the translational control of human EPO expression. In this study, we report that the single uORF located in the human EPO mRNA inhibits translation in unstressed cells. However, this repression is significantly released by hypoxia in HeLa cells, via eIF2α phosphorylation. These findings provide a framework for understanding that production of high levels of EPO induced by hypoxia also involves regulation at the translational level.
RESULTS
The human EPO 5′-leader sequence comprises a conserved uORF
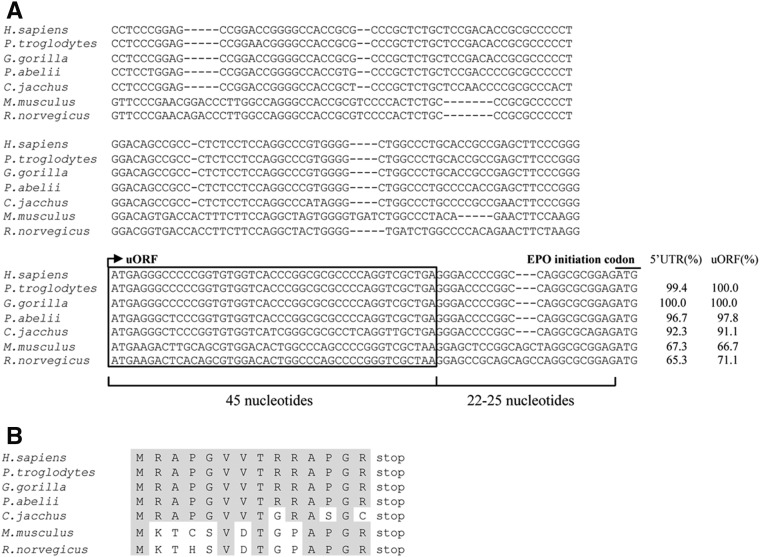
It has been shown that the 5′-leader sequence of the human EPO transcript has a significant homology with the murine EPO 5′ UTR, with both sequences having a high percentage of GC content (Shoemaker and Mitsock 1986). These investigators also reported the presence of a 14-codon ORF located upstream of the main AUG in both human and murine EPO transcripts. Figure 1A shows a broader alignment of the EPO 5′-leader sequences from human, chimpanzee, gorilla, orangutan, common marmoset, mouse, and rat, which all exhibit a high degree of similarity and conservation of the 14-codon uORF. These uORFs and the corresponding 5′-leader sequences have similar percentages of homology, with both structures equally conserved among species (Fig. 1A). The uORFs have a Kozak match of G/A at position −3 of the A(+1)UG. In addition, the position of the EPO uORF relative to the main AUG shows significant similarity among species, with the intercistronic region spanning 22 or 25 nucleotides (nt) (Fig. 1A). The alignment of the amino acid sequences for the EPO uORF in these species also shows a high degree of similarity (Fig. 1B). The conservation of the EPO uORF among the species may reflect an important evolutionary selection pressure and may suggest a potential regulatory function in EPO expression. These observations led us to investigate the role of the human EPO uORF in the translational control of the downstream main ORF.
FIGURE 1.
The 5′-leader sequence of the EPO transcript includes a highly conserved uORF. (A) Nucleotide sequence alignment of the human (Homo sapiens), chimpanzee (Pan troglodytes), gorilla (Gorilla gorilla), orangutan (Pongo abelii), common marmoset (Callithrix jacchus), mouse (Mus musculus), and rat (Rattus norvegicus) EPO mRNA 5′-leader regions. The uORF sequences of the different species are framed, where the arrow indicates position of the upstream initiation AUG codon (in gray). The uORF termination codon (UGA or UAA), as well as the main EPO AUG codon (EPO initiation codon), is also shown in gray. The uORF and the intercistronic region lengths (in nucleotides) are indicated below. The percentage of homology relative to the human 5′-leader sequence and to the uORF is indicated on the right. (B) uORF-encoded amino acid sequence alignment of the human, chimpanzee, gorilla, orangutan, common marmoset, mouse, and rat EPO transcripts. The conserved amino acids are indicated in gray.
The EPO uORF represses translation of a downstream main ORF
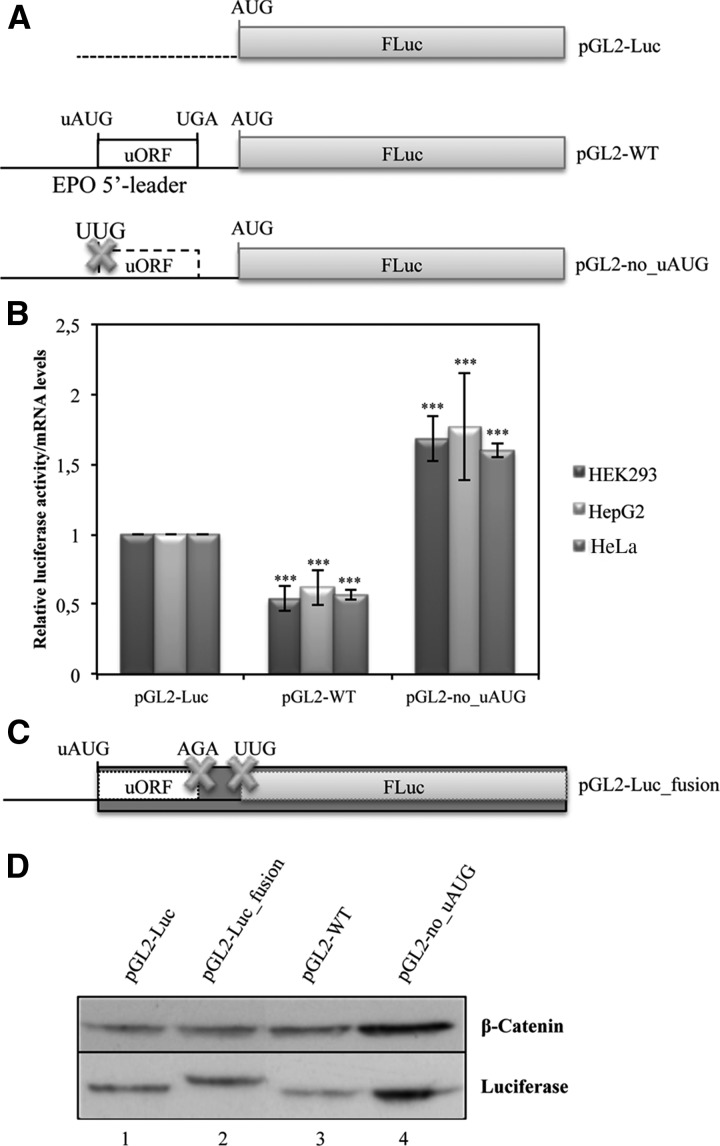
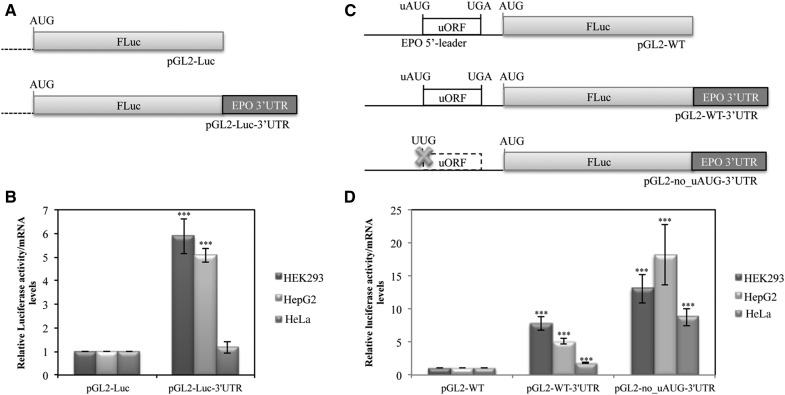
To determine the importance of the human EPO uORF in modulating translation efficiency of the downstream main ORF, the 181-bp sequence corresponding to the intact human EPO 5′-leader sequence was cloned into the cloning site of pGL2 expression vector, flanking the FLuc reporter gene to create the pGL2-WT construct (Fig. 2A). In addition, the EPO uORF was disrupted by site-directed mutagenesis of the uAUG (ATG→TTG), using the previous pGL2-WT construct as a template, originating the pGL2-no_uAUG construct (Fig. 2A). Expression of each of these reporter gene constructs was studied after transient transfection into a panel of cell lines: human embryonic kidney 293 (HEK293) cells, human hepatoma (HepG2) cells, and human cervix adenocarcinoma (HeLa) cells. For that, cellular extracts were prepared and assayed for luciferase activity, and total RNA was isolated to quantify the relative luciferase mRNA levels by reverse transcription–quantitative PCR (RT-qPCR). FLuc activity of each construct was normalized to the activity units from Renilla luciferase (RLuc) expressed from the cotransfected pRL-TK plasmid. The relative luciferase activity was then normalized to the corresponding mRNA levels and compared to the levels obtained with the empty pGL2-Luc vector, arbitrarily defined as one (Fig. 2B). Results show that in all cell lines studied, the human EPO 5′-leader sequence with the intact uORF induces a threefold repression of translation of the reporter transcript, compared with the data from the pGL2-no_uAUG construct without uORF (Fig. 2B). Thus, the intact EPO uORF induces a repression of gene expression at the translational level.
FIGURE 2.
The EPO uORF represses translation of the downstream main ORF. (A) Schematic representation of reporter constructs. The human EPO 5′-leader sequence encompassing its uORF (open box) with the intact initiation (uAUG) and termination (UGA) codons was cloned into the empty vector (pGL2-Luc), upstream of the firefly luciferase coding region (FLuc; gray boxes) to create the pGL2-WT construct. In the pGL2-no_uAUG construct, the uORF initiation codon is mutated (AUG→UUG; the cross represents the point mutation, and the dashed lined box represents the nonfunctional uORF). (B) The EPO 5′-leader sequence represses protein expression of the downstream reporter. HEK293, HepG2, and HeLa cells were transiently cotransfected with each one of the constructs described in A and with the pRL-TK plasmid encoding the Renilla luciferase (RLuc). Cells were lysed 24 h later, and the luciferase activity and mRNA levels were measured by luminometry assays and RT-qPCR, respectively. The graph represents the data as translational efficiency (relative luciferase activity/mRNA levels). Expression levels obtained from pGL2-Luc construct were defined as one. Average values and SD of three independent experiments are shown. Statistical analysis was performed using Student's t-test (unpaired, two-tailed): (**) P<0.01; (***) P<0.001. (C) Schematic representation of additional reporter constructs. The uORF stop codon was mutated in cis with a deletion of 1 nt downstream 4 nt from the stop codon (TGAgggac→AGAggg-c), so that both initiation codons (uAUG and AUG) are in frame. Also the AUG of the FLuc coding region was mutated (AUG→UUG) in the pGL2-Luc_fusion construct, encoding a fusion protein represented by the darker box. Crosses represent the point mutations. (D) Translation initiation can occur at the EPO uAUG. The constructs specified above each lane were transiently transfected in HEK293 cells. Twenty-four hours later, lysates were prepared and analyzed by Western blot. Immunoblotting was performed by using a firefly luciferase (FLuc)–specific antibody and a human β-catenin–specific antibody as a control for variations in protein loading.
To confirm that the uAUG of the EPO uORF is recognized by the ribosome, we cloned a construct in which the EPO uORF was fused in-frame to the luciferase ORF. This was achieved by site-directed mutagenesis of the pGL2-WT construct to introduce a mutation at the uORF stop codon in cis with a base pair deletion 4 nt downstream from the stop codon (TGAgggac→AGAggg-c) and also by mutating the FLuc main AUG (ATG→TTG; see Materials and Methods; pGL2-Luc_fusion construct) (Fig. 2C). This construct, as well as the pGL2-Luc, pGL2-WT, and pGL2-no_uAUG constructs, was transiently transfected into HEK293 cells. Twenty-four hours later, cell extracts were purified and analyzed by Western blot using a specific antibody that recognizes FLuc, with β-catenin as a loading control (Fig. 2D). As shown in Figure 2D (lane 2), the pGL2-Luc_fusion construct expresses a protein with higher molecular weight than that at lanes 1, 3, or 4; the larger band detected in lane 2 (Fig. 2D) corresponds to the uORF-FLuc fusion protein, while the other lanes (Fig. 2D, lanes 1,3,4) only show a smaller band that corresponds to the native form of the FLuc protein. These data demonstrate that indeed the human EPO uORF is recognized and translated by the ribosome and is thus functional.
Both translation reinitiation and uAUG leaky scanning are involved in the translational initiation at the main AUG codon
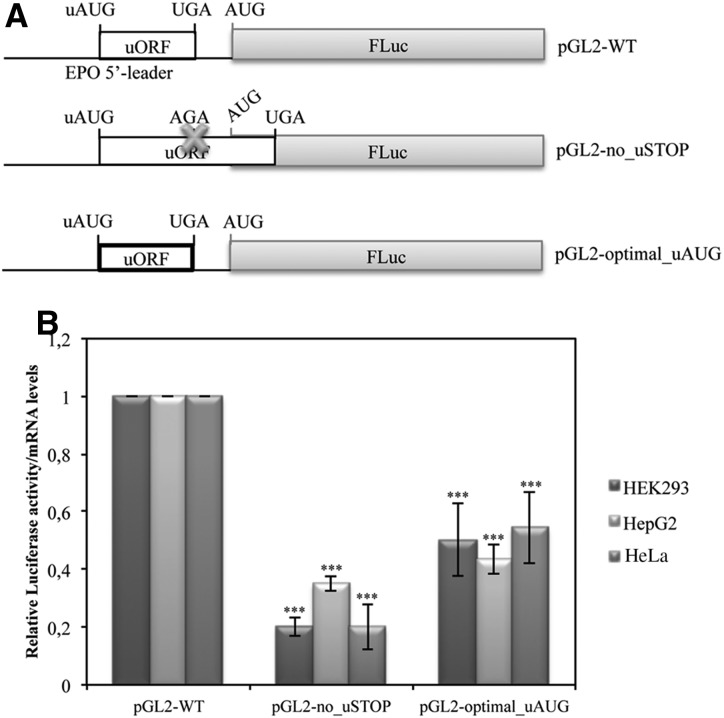
Since the AUG codon of the EPO uORF is in a good, but not optimal, context for initiation (gggAUGa), we expected that some ribosomes that load onto the EPO mRNA would initiate at the uAUG codon but that others could leak past the uAUG codon and initiate at the main AUG. Nevertheless, a few ribosomes that translate the uORF may reinitiate at the main AUG codon. To evaluate these possibilities, we first mutated the stop codon of the uORF (TGA→AGA; pGL2-no_uSTOP construct), creating an extended uORF that terminates at the next in-frame stop codon, 83 nt downstream from the FLuc initiation codon (pGL2-no_uSTOP construct) (Fig. 3A). This mutation completely abrogates the possibility that FLuc can be made by reinitiation after translation of the uORF, thereby allowing evaluation of the efficiency of leaky ribosome scanning. In addition, in the pGL2-WT vector we mutated the context of the uAUG codon (gggAUGa→gccAUGg) to obtain the pGL2-optimal_uAUG construct (Fig. 3A) with a uAUG sequence context shown by Kozak to yield maximum initiation frequency in higher eukaryotes (Kozak 1997; Wang 2004; Loughran et al. 2011). In this case, the majority of the ribosomes that load on the EPO mRNA 5′-leader sequence are unable to leak past the uAUG codon, and most likely they translate the uORF and may reinitiate at the downstream AUG codon. To estimate the relative contribution of leaky scanning and translation reinitiation, HEK293, HepG2, and HeLa cells were transiently transfected with the pGL2-no_uSTOP or pGL2-optimal_uAUG constructs or with the pGL2-WT construct, and translational efficiencies were monitored by dual luciferase assays normalized to the corresponding mRNA levels, as before. Results were compared to those obtained from the pGL2-WT construct (Fig. 3B). As shown in Figure 3B, mutation of the uORF stop codon (pGL2-no_uSTOP construct) reduces the translational efficiency to ∼20% of that of the pGL2-WT construct, suggesting that the percentage of ribosomes that leak past the uORF is low, and thus translation of the main ORF mostly occurs by reinitiation of the ribosomes after translation termination of the uORF. The analysis of pGL2-optimal_uAUG expression allowed us to observe that translation reinitiation at the main ORF can account for ∼60%, in comparison to the translation efficiency of the pGL2-WT construct (Fig. 3B). Our data demonstrate that a minority of ribosomes gain access to the main initiation codon by leaky scanning past the uAUG codon, while the majority does so by reinitiating translation after having translated the uORF.
FIGURE 3.
Both translation reinitiation and uAUG leaky scanning are involved in the translational initiation at the main AUG codon. (A) Schematic representation of reporter constructs. The pGL2-WT plasmid contains the wild-type human EPO 5′-leader sequence; the pGL2-no_uSTOP construct presents the EPO 5′-leader sequence with a mutation (UGA→AGA) at the uORF translation termination codon, which makes the uORF overlap with the luciferase ORF (the cross represents the point mutation); and the pGL2-optimal_uAUG contains the EPO 5′-leader sequence with an optimal uAUG sequence context (gggAUGa→gccAUGg; represented by a bold lined box). (B) HEK293, HepG2, and HeLa cells were transiently cotransfected with each one of the constructs described in A and with a plasmid encoding RLuc (pRL-TK) and analyzed as described in the legend to Figure 2B.
Translational repression exerted by the EPO uORF is peptide sequence–independent
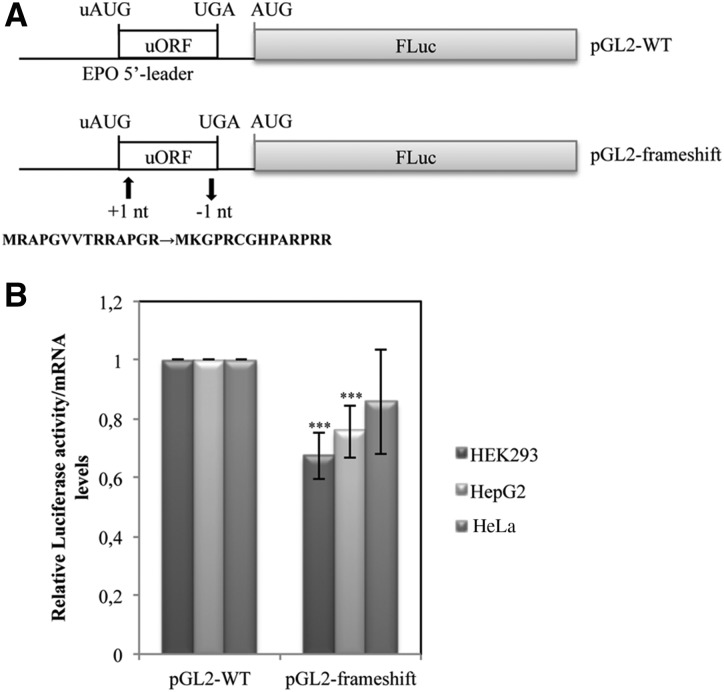
Translational inhibition by uORFs in some eukaryotic transcripts is dependent upon the peptide-coding sequence of the uORF (Karagyozov et al. 2008; Wei et al. 2012). Based on these data, and knowing that the peptide encoded by the EPO uORF is conserved among mammalian species (Fig. 1B), which may indicate its functional role, we next examined whether this peptide is required for inhibition of downstream translation. For that, we cloned the pGL2-frameshift construct (Fig. 4A) in which the uORF was modified by shifting the reading frame to generate a different amino acid sequence while preserving the uAUG context and most of the nucleotide sequence (see Materials and Methods). The pGL2-frameshift construct was used to transiently transfect the same panel of cells, and the analysis of translational efficiency was assessed as before; results were compared with those of the pGL2-WT construct (Fig. 4B). Our data show that in HeLa cells, the mutant uORF of pGL2-frameshift construct allows for a translational efficiency slightly lower than that of the pGL2-WT construct with the normal uORF. This difference is significant in HEK293 and HepG2 cells (Fig. 4B). The lower level of translation obtained by the pGL2-frameshift construct may reflect the effect of a rare codon encoded by the uORF of pGL2-frameshift construct that may decrease the efficiency of translational reinitiation. Since frameshifting the EPO uORF does not derepress translation, we can conclude that the native EPO uORF functions in a peptide sequence–independent manner to inhibit translation.
FIGURE 4.
Translational repression exerted by the EPO uORF is peptide sequence–independent. (A) Schematic representation of the expression constructs. The pGL2-WT plasmid contains the human normal EPO 5′-leader transcript sequence, and the pGL2-frameshift vector carries a EPO uORF sequence modified by frameshift mutations, which consist in the insertion of 1 nt in the second codon (+1 nt) and the deletion of 1 nt in 13th codon (−1 nt). The resulting uORF-encoded peptide sequence is shown below. (B) HEK293, HepG2, and HeLa cells were transiently cotransfected with each one of the constructs described in A and with a plasmid encoding RLuc (pRL-TK) and analyzed as described in the legend to Figure 2B.
The 3′ UTR of the EPO mRNA has no impact on the relative inhibitory effect of the uORF
It has been shown that translational repression exerted by a uORF present in a transcript can be modulated by the corresponding 3′ UTR, through protein interactions between both UTRs of the mRNA (Mehta 2006). On the other hand, it has been shown that a pyrimidine-rich region within the human EPO 3′ UTR is implicated in regulation of EPO mRNA stability and shown to bind two isoforms of a 40-kD poly(C) binding protein (PCBP), PCBP1 and PCBP2 (Wang et al. 1995; McGary et al. 1997; Czyzyk-Krzeska and Bendixen 1999; Ohigashi et al. 1999). Since these data show that the 3′ UTR of the EPO transcript influences its expression, we hypothesized that the EPO 3′ UTR could affect the translational inhibition exerted by the EPO uORF. To address this question, we first tested the effect of the EPO 3′ UTR on the translational efficiency of the FLuc. For that, the EPO 3′ UTR was cloned into the pGL2-Luc vector downstream from the FLuc cistron (pGL2-Luc-3′ UTR construct) (Fig. 5A). Expression of this reporter gene construct was studied following transfection into HEK293, HepG2, and HeLa cell lines. Translational efficiency was analyzed as before and compared with that of the empty pGL2-Luc construct. Results show that the EPO 3′ UTR alone induces about a fivefold increase of the translation efficiency in HEK293 and HepG2 cells (Fig. 5B); nevertheless in HeLa cells, the translational efficiency is unaltered (Fig. 5B). Therefore, it seems that the EPO 3′ UTR affects expression of the reporter gene through different mechanisms in the three cell lines.
FIGURE 5.
The 3′ UTR of the EPO mRNA has no influence in the relative inhibitory effect of the uORF. (A) Schematic of the FLuc reporter constructs containing the native luciferase 3′ UTR (pGL2-Luc) or the 3′ UTR sequence (dark gray box) of the human EPO transcript (pGL2-Luc-3′ UTR). (B) HEK293, HepG2, and HeLa cells were transiently cotransfected with each one of the constructs described in A and with a plasmid encoding RLuc (pRL-TK) and analyzed as described in the legend to Figure 2B. (C) Schematic of the FLuc reporter constructs containing the human EPO 5′-leader sequence with the intact uORF and the luciferase 3′ UTR (pGL2-WT), pGL2-WT-3′ UTR (dark gray box), or the EPO 5′-leader sequence with a disrupted uORF due to the uAUG→UUG mutation (represented by a cross) and the EPO 3′ UTR sequence (dark gray box; pGL2-no_uAUG-3′ UTR). (D) HEK293, HepG2, and HeLa cells were transiently cotransfected with each one of the constructs described in C and with a plasmid encoding RLuc (pRL-TK) and analyzed as described in the legend to Figure 2B.
We then monitored the translational efficiency of the pGL2-WT-3′ UTR construct that harbors both EPO 5′ and 3′ UTRs (Fig. 5C) and compared it to that of the corresponding construct with the disrupted uORF (pGL2-no_uAUG-3′ UTR construct) after normalization of both to the translational efficiency of the pGL2-WT construct (Fig. 5C). For that, each one of these constructs was cotransfected with pRL-TK in the same cell lines as above, and translational efficiency was measured as previously. Results show that insertion of the EPO 3′ UTR into pGL2-WT construct significantly increases expression of the main ORF; however, the fold increase is much lower in HeLa cells, which might indicate a tissue-specific effect (Fig. 5D). Of note, the intact EPO 5′-leader and 3′ UTR sequences in the pGL2-WT-3′ UTR construct still allow a significant threefold decrease in translational efficiency compared with that observed from the pGL2-no_uAUG-3′ UTR construct (Fig. 5D). Thus, the EPO 3′ UTR fails to overcome relative translational repression induced by the EPO uORF.
EPO is regulated at the translational level in response to hypoxia, but not nutrient deprivation, specifically in HeLa cells
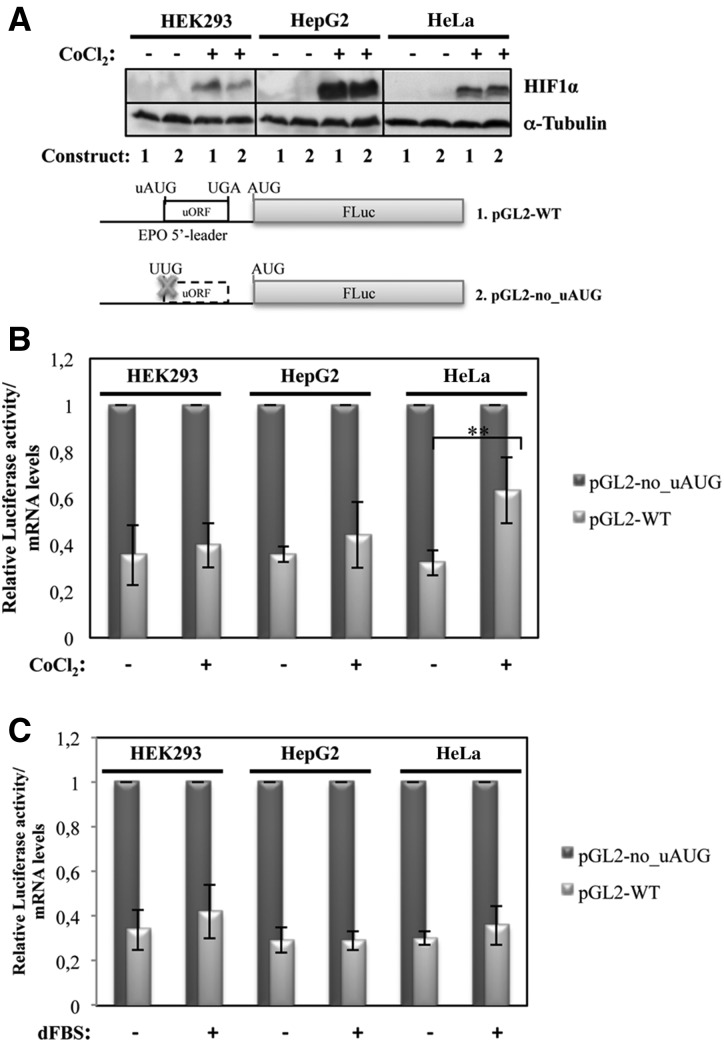
A number of stresses, including temperature shock, DNA damage, nutrient stress, and hypoxia, can lead to changes in gene expression patterns caused by a general shutdown and reprogramming of protein synthesis. This process of translational control decreases global protein synthesis rates and increases synthesis of selective subsets of mRNAs, such as those carrying IRESs and/or uORFs, which encode stress-response proteins (Blais et al. 2004; Barbosa et al. 2013). Given that EPO is widely known to respond to hypoxia (Ebert and Bunn 1999; Haase 2013), we decided to investigate whether the EPO uORF directs translational control in response to this cellular stress. Therefore, HEK293, HepG2, and HeLa cells were transiently transfected with the pGL2-WT and pGL2-no_uAUG constructs that carry the intact or disrupted EPO uORF, respectively, and the cells were treated with 200 µM cobalt chloride (CoCl2) to mimic hypoxia. Twenty-four hours after exposure, cells were lysed, and protein and RNA were extracted and analyzed as previously described. As the transcription factor HIF complex is the key regulator of hypoxia-inducible EPO gene expression in HepG2 cells (Fandrey and Bunn 1993), the cellular hypoxic stimulus was monitored by Western blot against HIF1α. This analysis demonstrated an increase in HIF1α protein levels induced by hypoxia, compared with results obtained in untreated cells. Detection of α-tubulin was used to control the amount of loaded protein (Fig. 6A). Under normoxic and hypoxic conditions, we observed that the translational efficiency of the pGL2-WT construct, normalized to that of the pGL2-no_uAUG construct under corresponding conditions, does not significantly change in response to hypoxia in HEK293 and HepG2 cells (Fig. 6B). In contrast, the pGL2-WT expression increased 1.7-fold in the HeLa cells in response to hypoxia (Fig. 6B), consistent with the intact 5′-leader sequence of the EPO mRNA directing translational derepression in response to hypoxia, specifically in HeLa cells.
FIGURE 6.
The EPO uORF responds to hypoxia but not to nutrient starvation, specifically in HeLa cells. The pGL-WT (construct 1) and pGL2-no_uAUG (construct 2) vectors represented as in Figure 2 were separately cotransfected with a plasmid encoding RLuc (pRL-TK) in HEK293, HepG2, and HeLa cells. Six hours after transfection, cells were untreated (−) or treated (+) for 24 h with 200 µM CoCl2 to mimic hypoxic conditions or with medium supplemented with 10% (v/v) dialyzed fetal bovine serum (dFBS) to induce nutrient starvation. (A) Representative Western blot analysis of HEK293, HepG2, and HeLa cell extracts untreated (−) or treated (+) with CoCl2 as indicated. Immunoblotting was performed using a human HIF1α–specific antibody to control the stress conditions and a human α-tubulin–specific antibody to control for variations in protein loading. (B) Normoxic (CoCl2 −) and hypoxic (CoCl2 +) transfected cells were lysed and analyzed as described in the legend to Figure 2B. (C) Cells cultured in nutrient deprivation (dFBS +) or in control conditions (dFBS −) were lysed and analyzed as described in the legend to Figure 2B.
Thereafter, we investigated if this effect is observed in stimuli other than hypoxia. To test this hypothesis, HEK293, HepG2, and HeLa cells transiently transfected with the pGL2-WT or pGL2-no_uAUG constructs were cultured under conditions of nutrient deprivation using dialyzed fetal bovine serum (dFBS) in the media, as described in the Materials and Methods. Translational efficiency of the pGL2-WT construct was compared with that of the pGL2-no_uAUG construct as before. Our data have shown that translational efficiency of pGL2-WT does not significantly change in response to nutrient deprivation in all cell lines studied (Fig. 6C). Taken together, this set of data shows that the EPO uORF controls translation specifically in response to hypoxia in HeLa cells.
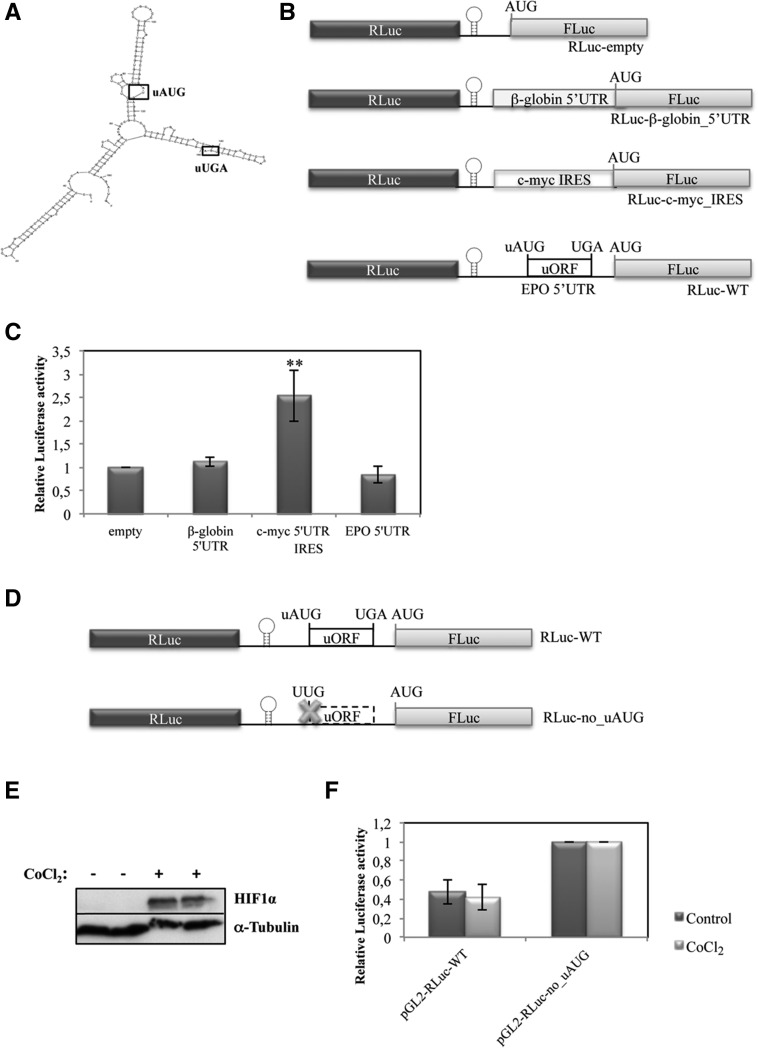
EPO translational derepression in response to hypoxia in HeLa cells is not mediated by an IRES
As already stated, translational control can be regulated by elements within the 5′ and 3′ UTRs of mRNAs, including uORFs and IRESs, among others. These elements can act singly or in combination. The 5′-leader sequence of the EPO mRNA is relatively long, with a high CG content (81%) forming a secondary Y structure with strong and structured hairpins, as predicted by Mfold (ΔG = −93.18 kcal/mol) (Fig. 7A), which are common features of the 5′ UTRs of cellular mRNAs reported to have IRES activity (Bastide et al. 2008; Tahiri-Alaoui et al. 2009; Komar and Hatzoglou 2011). Based on these data, we first aimed to test whether the 5′-leader sequence of the human EPO mRNA exhibits IRES activity. For that, we cloned a dicistronic reporter construct in which the native EPO 5′-leader sequence was inserted between the Renilla and FLuc cistrons but downstream from a stem–loop structure (ΔG value of −55.50 kcal/mol) to impede ribosomal reinitiation (Fig. 7B). In parallel, the β-globin (HBB) 5′ UTR and the c-myc IRES sequence were also inserted in the dicistronic reporter system to serve as negative and positive controls for IRES activity, respectively (Fig. 7B; Stoneley et al. 1998; Cobbold et al. 2007). The resulting plasmids, called RLuc-β-globin_5′ UTR, RLuc-c-myc_IRES, and RLuc-WT, were each transiently transfected into HeLa cells. Twenty-four hours post transfection, cells were harvested and lysates subjected to a dual luciferase reporter assay, and the subsequent ratio of FLuc to RLuc was compared with that from the control RLuc-empty plasmid, which contains a short linker sequence between the two luciferase cistrons (Fig. 7B). Results show that the c-myc sequence significantly increased the relative luciferase activity by 2.5-fold (Fig. 7C), which shows its capacity to drive cap-independent FLuc expression, in accordance to what is expected for an mRNA containing IRES activity (Stoneley et al. 1998; Cobbold et al. 2007). However, the EPO 5′ UTR behaved similarly to the β-globin 5′ UTR, and each maintained relative luciferase activity at levels similar to those of the RLuc-empty construct, showing that neither of them allows detectable internal translation initiation (Fig. 7C). These results illustrate that the EPO 5′-leader sequence does not exhibit IRES activity in normoxic HeLa cells.
FIGURE 7.
EPO translational derepression in response to hypoxia in HeLa cells is not mediated by an IRES. (A) Representation of the secondary structure of the 5′-leader sequence of the human EPO transcript as predicted by the Mfold webserver. (B) Schematic of the dicistronic luciferase vectors. The 5′ UTR sequence of the human EPO transcript (EPO 5′ UTR) to be tested for IRES activity, as well as the 5′ UTR sequence of the human β-globin transcript (β-globin 5′ UTR), or the c-myc IRES sequence previously described by Stoneley et al. (1998), was inserted between the Renilla (RLuc) and firefly (FLuc) luciferase cistrons, downstream from a hairpin structure (represented by a stem–loop) in the multiple cloning site spacer of the RLuc-empty vector to create the RLuc-WT, RLuc-β-globin_5′ UTR, and the RLuc-c-myc_IRES constructs, respectively. (C) HeLa cells were transiently transfected with each one of the constructs described in B and with a plasmid encoding RLuc (pRL-TK) and analyzed as described in the legend to Figure 2B. (D) Schematic of the dicistronic reporter constructs used to test if the EPO 5′-leader sequence contains an IRES activated during hypoxia. RLuc-WT contains the human EPO 5′-leader sequence with the intact uORF, as defined in A, and the RLuc-no_uAUG contains the EPO 5′-leader sequence with a disrupted uORF due to the uAUG→UUG mutation (represented by a cross). HeLa cells were transfected with these constructs. Six hours later, cells were untreated or treated with 200 µM CoCl2 for 24 h. (E) Representative Western blot analysis of HeLa cell extracts untreated (−) or treated (+) with CoCl2. Immunoblotting was performed using a human HIF1α–specific antibody to control the stress conditions and a human α-tubulin–specific antibody to control for variations in protein loading. (F) Relative luciferase activity was quantified as described in the legend to Figure 2B.
Knowing that in response to hypoxia, some transcripts may increase efficiency of translation by facilitating internal translation initiation through a process such as IRES-mediated initiation (Schepens 2005), we subsequently tested if EPO translational derepression in response to hypoxia in HeLa cells is mediated by IRES. Thus, we analyzed the expression of the RLuc-WT construct with the intact EPO uORF, relative to that of the RLuc-no_uAUG construct carrying the disrupted EPO uORF, both cloned in the dicistronic reporter plasmid (Fig. 7D) as before, in normoxic and hypoxic HeLa cells. To mimic hypoxia, cells were treated with CoCl2, and induction of hypoxia was monitored, as previously, by Western blot using an antibody against HIF1α. As expected, results show high accumulation of HIF1α protein during hypoxic incubation (Fig. 7E). In addition, data show that relative luciferase activity from the RLuc-WT construct in normoxic cells is at ∼45% of the relative luciferase activity of the RLuc-no_uAUG construct. Under hypoxia, the same relative luciferase activity was observed (Fig. 7F). This result supports the idea that the EPO 5′-leader sequence does not allow for internal translation initiation irrespective of stress conditions. Thus, in both normoxic and hypoxic HeLa cells, EPO translation involves the processive scanning of ribosomes from the 5′-end of the EPO transcript.
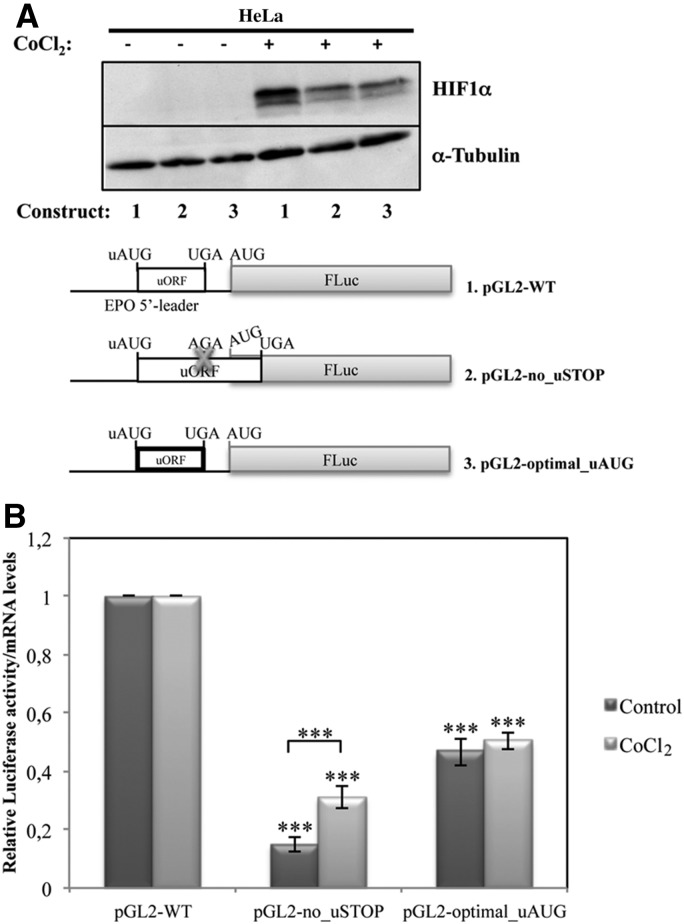
EPO translational derepression in response to hypoxia is mediated by leaky scanning of ribosomes through the inhibitory uORF
Based on our data showing that both translation reinitiation and uAUG leaky scanning are involved in the translational initiation at the main AUG codon of the pGL2-WT construct, we next addressed which of these two mechanisms of initiation occur to overcome the inhibitory function of the EPO uORF in response to hypoxia in HeLa cells. Thus, as previously (see Fig. 3), protein expression of the pGL2-WT (construct carrying the wild type EPO uORF), pGL2-no_uSTOP (construct carrying a mutation at the uORF stop codon [TGA→AGA]), and pGL2-optimal_uORF (construct carrying the uAUG in an optimal context) constructs was analyzed in HeLa cells under normoxic and hypoxic conditions. To mimic hypoxia, cells were treated with CoCl2, and induction of hypoxia was monitored, as before, by Western blotting using an antibody against HIF1α. As expected, hypoxia conditions led to the accumulation of HIF1α (Fig. 8A). Furthermore, our results show that the translational efficiency from the pGL2-no_uSTOP construct with the extended uORF increases 1.6-fold in hypoxic conditions, compared with its levels in normoxia, meaning that, under hypoxia, the uAUG is less efficiently recognized (Fig. 8B). In contrast, increasing the translation initiation sequence context to an optimal start codon context at the pGL2-optimal_uAUG construct does not affect the corresponding translational efficiency under hypoxic versus normoxic conditions (Fig. 8B). These results are consistent with a model in which derepression of translation in hypoxic HeLa cells occurs by an increase in leaky scanning of ribosomes through the inhibitory EPO uORF.
FIGURE 8.
EPO translational derepression in response to hypoxia of HeLa cells is mediated by leaky scanning of ribosomes through the inhibitory uORF. The pGL-WT (construct 1), pGL2-no_STOP (construct 2), and pGL2-optimal_uAUG (construct 3) vectors, represented as in Figure 3, were separately cotransfected with a plasmid encoding RLuc (pRL-TK) in HeLa cells. Six hours after transfection, cells were untreated or treated for 24 h with 200 µM CoCl2 to mimic hypoxic conditions. (A) Representative Western blot analysis of transfected HeLa cell extracts untreated (−) or treated (+) with CoCl2 as shown. Immunoblotting was performed using a human HIF1α–specific antibody to control for hypoxia and a human α-tubulin–specific antibody to control for variations in protein loading. (B) Relative luciferase activity was quantified as described in the legend to Figure 2B.
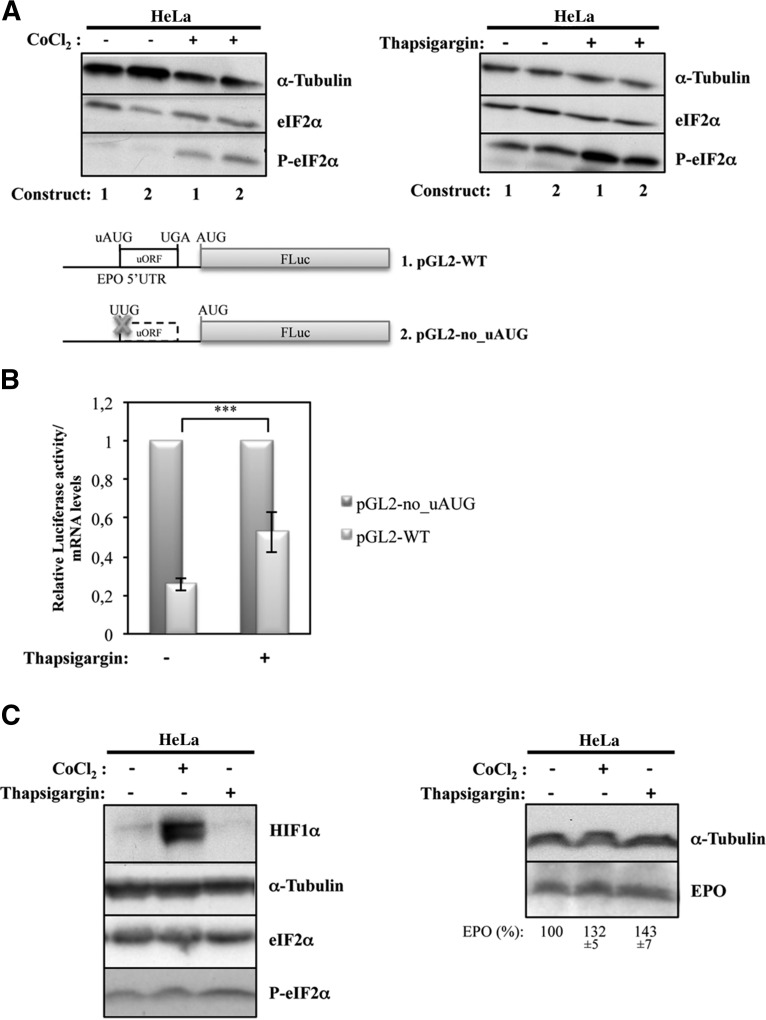
EPO translational regulation in response to hypoxia occurs with concomitant eIF2α phosphorylation
Phosphorylation of eIF2α is a rapid consequence of hypoxic stress, reducing the availability of competent initiation complexes. Indeed, when eIF2α is phosphorylated, the ternary complex becomes scarce and global translation compromised (Sonenberg and Hinnebusch 2009). Despite eIF2α phosphorylation, the presence of uORFs can promote the increased expression of certain stress-related mRNAs (Koritzinsky et al. 2007; Dang Do et al. 2009; Barbosa et al. 2013). Based on these data, we next addressed whether the mechanism by which EPO uORF significantly derepresses translation in hypoxic HeLa cells occurs through eIF2α phosphorylation. For that, the state of eIF2α phosphorylation was examined by immunoblotting using anti-phospho-eIF2α antibody, in HeLa cells transiently transfected with pGL2-WT or pGL2-no_uAUG constructs, and treated with 200 µM CoCl2 for 24 h. Results show that the extent of eIF2α phosphorylation in this cell line is increased by induction of hypoxia (Fig. 9A). Taking advantage of these data, we next tested whether the treatment of these cells with thapsigargin, a potent ER stress agent that directly activates eIF2α kinases without activating any other signaling pathway (Harding et al. 2001; Koumenis et al. 2002), would induce translational derepression of the luciferase reporter. HeLa cells transiently transfected with pGL2-WT or pGL2-no_uAUG constructs were treated with 1 µM thapsigargin. Twenty-four hours later, cells were lysed, and protein levels were measured by luminometry assays and normalized to the corresponding mRNA levels quantified by RT-qPCR, as previously. The extent of eIF2α phosphorylation in thapsigargin-treated cells was examined by immunoblotting, as before. Figure 9A shows that the extent of eIF2α phosphorylation was increased by thapsigargin treatment. Figure 9B shows that phosphorylation of eIF2α effectively induces a significant increase of translation of the pGL2-WT mRNA relative to that of pGL2-no_uAUG (twofold increase), specifically in treated compared with untreated HeLa cells. From these findings, it seems that eIF2α phosphorylation is involved in the regulation of translation of the pGL2-WT reporter mRNA via the EPO uORF in HeLa cells in response to hypoxia. Together, these results and those shown in Figure 8 indicate that in HeLa cells exposed to hypoxia, eIF2α is phosphorylated, which up-regulates the translation of EPO mRNA by increasing the rate of ribosomal bypass of the inhibitory uORF.
FIGURE 9.
Hypoxia stimulates phosphorylation of eIF2α, which induces EPO translational regulation in HeLa cells. The pGL-WT (construct 1) and pGL2-no_uAUG (construct 2) vectors represented as in Figure 2 were separately cotransfected with a plasmid encoding RLuc (pRL-TK) in HeLa cells. Six hours after transfection, cells were untreated or treated for 24 h with 200 µM CoCl2 to mimic hypoxic conditions or with 1 µM thapsigargin. (A) Representative Western blot analyses of HeLa cell extracts untreated (−) or treated (+) as indicated. Immunoblotting was performed using human eIF2α–specific and human phosphorylated eIF2α–specific antibodies to control for stress conditions and human α-tubulin–specific antibody to control for variations in protein loading. (B) Relative luciferase activity was quantified as described in the legend to Figure 2B. (C) Endogenous EPO protein expression responds to CoCl2 and thapsigargin cellular treatment. (Left) Panel is representative of Western blot analyses of HeLa cell extracts untreated (−) or treated (+) with CoCl2 or thapsigargin, as indicated. Immunoblotting was performed using human HIF1α–specific, human eIF2α–specific, and human phosphorylated eIF2α–specific antibodies to control for stress conditions and human α–specific antibody to control for variations in protein loading. (Right) A representative Western blot of HeLa cell extracts untreated or treated, as in the left panel. Immunoblotting was performed using a human EPO–specific antibody and α-tubulin–specific antibody to control for variations in protein loading. The percentage (%) of endogenous EPO protein expressed in the HeLa cells after treatment, in comparison to that before treatment, is indicated below each lane and was achieved by densitometric analysis using ImageJ software.
Next, we investigated whether CoCl2 and thapsigargin treatments show a parallel effect on the expression of the endogenous human EPO protein. For that, HeLa cells were treated with each one of these drugs, and the stimuli were monitored by immunoblotting against HIF1α and phosphorylated eIF2α, as before. This analysis demonstrated an increase in HIF1α protein levels induced by hypoxia and an increase of phosphorylated eIF2α after both treatments (Fig. 9C, left). Under these conditions, immunoblotting against the endogenous human EPO protein revealed an increase of its levels as a result of both of these stimuli (Fig. 9C, right). This leads to the conclusion that, indeed, human EPO protein expression is regulated by hypoxia via eIF2α phosphorylation, recapitulating our previous results from reporter constructs. Furthermore, the response of endogenous EPO expression to thapsigargin treatment unequivocally shows that its expression is controlled at the translational level.
DISCUSSION
EPO is one of many examples of genes that present several layers of expression regulation. The most well characterized mechanisms are at the transcriptional level and are correlated with the increase of EPO mRNA levels under hypoxic conditions (Jelkmann 2011). Human EPO mRNA (NM_000799), which encodes a 166-amino-acid hormone, possesses a 5′-leader sequence with 181 nt that encompasses a uORF with 14 codons, located 22 nt upstream of the EPO AUG codon (Fig. 1). The 5′-leader sequences of the EPO mRNAs of human, chimpanzee, gorilla, orangutan, common marmoset, mouse, and rat show elevated sequence similarity, with the uORF also highly conserved among these species (Fig. 1). Knowing that a high percentage of transcripts encoding hormones have uORFs involved in their translational control, responding to cell type and external stimuli (Sachs 2006; Wethmar et al. 2010; Medenbach et al. 2011), we investigated the function of the human EPO uORF in its translational control. We found that the EPO uORF is translatable and that the presence of the intact uORF significantly inhibits the translation of the downstream ORF in different cell lines (Fig. 2). The preservation of this effect in different cell types suggests that the uORF is a major determinant of EPO protein expression.
In the context of questioning how ribosomes gain access to the main EPO AUG codon, the results shown in Figure 3 suggest that a small percentage of ribosomes bypass the uAUG codon and the corresponding uORF and that additional ribosomes are also able to reinitiate at the EPO start site after translation of the uORF. Knowing that the presence of a purine at the −3 position relative to the AUG codon is usually thought to be sufficient for efficient initiation (Kozak 2001), it is really not too surprising that some ribosomes leak past the EPO uORF AUG codon, but the majority of them recognize the uAUG, translate the uORF, and may reinitiate at the downstream main ORF, even though the uORF AUG codon has the nonoptimal gggAUGa sequence context. As an A at position −3 can be superior to G (Kozak 2001), which occurs in the mouse and rat sequences (Fig. 1), we hypothesize that in these species, EPO uORF may be even more efficiently recognized than in humans. In this work, we also show that the EPO uORF-encoded peptide, although highly conserved among species (Fig. 1), is not involved in the mechanism by which the uORF represses translation (Fig. 4). Thus, we do not currently understand the significance of the conservation of the peptide sequence. In addition, using reporter constructs where the EPO 5′-leader sequence and/or the EPO 3′ UTR flank the FLuc cistron, we substantiate that the relative translational repression exerted by the uORF is not released in the presence of the EPO 3′ UTR sequence (Fig. 5). These results suggest that the cis-acting elements present in the EPO 3′ UTR involved in increasing EPO gene expression do not affect the mechanism by which the uORF represses translation. Thus, these two regions may act at different layers of EPO gene expression regulation.
Trying to understand the mechanism by which EPO translational derepression occurs in response to hypoxia in HeLa cells, we have observed that EPO translation does not implicate the induction of IRES activity despite the high CG content of the EPO 5′-leader sequence forming a secondary Y structure with strong and structured hairpins, characteristics of IRES sequences (Fig. 7). Instead, it involves the processive scanning of ribosomes from the 5′-end of the EPO transcript whether in normoxic or hypoxic HeLa cells (Figs. 3, 8), suggesting that EPO translation is not controlled via multiple elements located in its 5′-leader region, a condition that occurs in some transcripts such as the VEGF-A isoform, in which an uORF is located within an IRES (Bastide et al. 2008).
EPO is the primary regulator of mammalian erythropoiesis and is produced by the kidney and the liver in an oxygen-dependent manner. However, it is now clear that EPO is a multifunctional molecule produced and utilized by many tissues that rapidly responds to different cell stress stimuli and tissue injuries (Arcasoy 2008; Brines et al. 2008; Ruifrok et al. 2008; Ryou et al. 2012). Based on these data, we have investigated the role of the EPO uORF in three different cell lines derived from embryonic kidney, liver, and cervical cancer in the response to chemical hypoxia or nutrient deprivation. We found that the protein expression from the construct with the intact EPO 5′-leader sequence is significantly increased, specifically in HeLa cells in response to hypoxia, but not during nutrient limitation (Fig. 6). On the other hand, no effect was observed in HEK293 and HepG2 cells (Fig. 6), suggesting that a tissue-specific factor might be involved in the different EPO uORF-mediated translational control observed in the different cell lines. This factor might be, for example, a kinase that specifically phosphorylates eIF2α in HeLa cells in response to hypoxia, inducing EPO translational derepression in these cells specifically. These results show that the translational control mediated by the EPO uORF is another layer in the already complex control of EPO gene expression in response to hypoxia.
To understand how scanning ribosomes better reach the main ORF to increase translation when HeLa cells are hypoxic, we have observed that besides the reinitiation mechanism, more ribosomes bypass the EPO uORF in response to hypoxia, and thus, the uORF decreases its barrier function to scanning ribosomes and translational rate of the main ORF is significantly increased (Fig. 8). What is the biochemical mechanism by which scanning ribosomes bypass EPO uORF and reach the main AUG in hypoxic HeLa cells? It is known that hypoxia activates eIF2α phosphorylation (Sonenberg and Hinnebusch 2009), which is also in accordance with our data shown in Figure 9. On the other hand, stress-induced eIF2α phosphorylation significantly increases translation of the main reporter ORF in HeLa cells (Fig. 9); these data may indicate that different tissues evolved differently in order to increase EPO expression locally, independently of the renal transcription induction. Taken together, our results indicate that in basal conditions where eIF2α phosphorylation is low, translation of the EPO uORF serves as a barrier that inhibits translation of the main downstream EPO ORF in different cell types. During hypoxia, enhanced eIF2α phosphorylation significantly increases ribosome bypass of the uORF in HeLa cells, probably due to the nonoptimal uAUG sequence context, and translation of the downstream main ORF occurs with higher efficiency. A similar mechanism has been previously described for two other human transcripts: ATF4/5 and CCAAT/enhancer-binding protein homologous protein (CHOP) mRNAs (Watatani et al. 2007; Zhou et al. 2008; Palam et al. 2011). Indeed, the nonoptimal sequence context of the uAUG is a feature conserved among each of the species illustrated in Figure 1, which may reflect its functional role in translational control. On the other hand, the tissue specificity observed in the translational control mediated by the EPO uORF in response to hypoxia may suggest the involvement of potential tissue-specific regulator(s) that would facilitate the bypass of the EPO uORF, specifically in HeLa cells during hypoxia. In the future, it will be interesting to determine whether EPO responds to other stress stimuli in combination with eIF2α phosphorylation through the uORF bypass mechanism in different cell types.
We do not yet completely understand the biochemical basis for the ribosomal bypass of the uORF in our model of EPO translational control in response to hypoxia specifically in HeLa cells. Lowered eIF2-GTP levels may contribute to the reduced recognition of the EPO uORF. Additional contributors to this bypass may be the eIF2α phosphorylation-mediated expression regulation of other critical translation factors, or the tissue-specific regulators that would then facilitate the bypass of the EPO uORF during hypoxia. Also, this mechanism may involve specific sequences or conditions, such as cancerigenesis, that have not yet been identified but will be challenged to investigate.
Overall, the present results report an additional mechanism involved in the regulation of human EPO gene expression. The translational control by the EPO uORF and its response to hypoxia might present a new target for therapeutic interventions in diseases, such as cancer, related to the nonhematopoietic functions of EPO.
MATERIALS AND METHODS
Plasmid constructs
The plasmid pGL2-enhancer (Promega) encoding the FLuc was digested with BglII/HindIII, into which was inserted the BglII/HindIII-digested CMV promoter sequence from pcDNA3.1/hygro+ (Invitrogen), originating the so-called pGL2-Luc construct. Then, the 181-bp fragment corresponding to the human EPO 5′ UTR was inserted into the HindIII/XbaI sites of the pGL2-Luc plasmid, originating the pGL2-WT construct. The fragment corresponding to the 5′ UTR of the human EPO transcript was previously obtained by overlap-extension PCR. For that, two PCR products were obtained: one corresponding to the human EPO 5′ UTR amplified from human genomic DNA with the primer 1 (with the HindIII linker) (Table 1) and the overlapping reverse primer 3 (Table 1), and another corresponding to the amplification of the pGL2-Luc plasmid with the overlapping primer 4 (Table 1) and the reverse flanking primer 2 that produce a 160-bp fragment beginning at the FLuc AUG codon through downstream. Then, the flanking primers 1 and 2 and the two PCR-amplified products were used to amplify the final overlap-extension fragment encompassing the human EPO 5′ UTR and the 5′ part of the FLuc cistron, which was further digested with HindIII and XbaI enzymes (the fragment sequence includes a XbaI restriction site) before ligation to the pGL2-Luc vector to create the pGL2-WT construct.
TABLE 1.
DNA oligonucleotides used in the current work

The pGL2-no_uAUG variant, carrying a mutation at the uORF AUG codon (ATG→TTG), was created by replacing the native HindIII/XbaI fragment by the corresponding fragment carrying the ATG→TTG mutation. For that, the same overlap-extension approach was used, in which the two first PCR reactions were performed to amplify fragments from the pGL2-WT plasmid with primers 1 and 5 (reverse primer 5 carries the mutation) (Table 1) and with primers 6 and 2 (forward primer 6 carries the mutation) (Table 1), respectively. Then, primers 1 and 2 were used to amplify the overlapped fragment, using the previous fragments as template. The resulting DNA fragment was digested with HindIII/XbaI and then ligated to the pGL2-WT plasmid previously digested with the same enzymes.
The pGL2-Luc_fusion carrying a mutation of the uORF stop codon in cis with a deletion of 1 nt downstream 4 nt from the stop codon (TGAgggac→AGAggg-c), so that both AUG codons (the uAUG and the main AUG) are in frame, and a mutation (ATG→TTG) at the main AUG, the pGL2-no_uSTOP carrying a mutation (TGA→AGA) at the uORF stop codon, and the pGL2-optimal_uAUG carrying a mutation of the uAUG sequence context (gggATGa→gccATGg) were all created by site-directed mutagenesis, using the pGL2-WT plasmid as template and the mutagenic primers 7 to 14, respectively (Table 1).
The pGL2-frameshift construct was obtained by site-directed mutagenesis, insertion of one A nucleotide at the beginning of the uORF (5′-ATGAAGG…-3′), and deletion of one T nucleotide at the 3′ end (5′-…GGCGCTGA-3′), using the primers 15 to 18 (Table 1) and the pGL2-WT as template.
The sequence corresponding to the 3′ UTR of the human EPO transcript was inserted into the EcoRI/PflMI sites of the pGL2-Luc, pGL2-WT, and the pGL2-no_uAUG constructs to obtain the pGL2-Luc-3′ UTR, pGL2-WT-3′ UTR, and pGL2-no_AUG-3′ UTR constructs, respectively. For this purpose, overlap-extension PCR was performed as above, with the flanking primers 19 and 20 (primer 20 with PflMI linker) (Table 1), using as templates the PCR amplification of the 3′ part of the FLuc cistron (450-bp fragment located upstream of the FLuc stop codon, which encompasses a EcoRI site) from the pGL2-Luc construct with primers 19 and 22 (Table 1) and the PCR amplification of the 3′ UTR fragment of the human EPO gene with primers 20 and 21 (Table 1).
The dicistronic constructs carrying both RLuc and FLuc cistrons were derived from the psiRF vector previously described (Tahiri-Alaoui et al. 2009). In order to prevent ribosome read-through, the sequence encompassing a stable hairpin structure was PCR amplified from the plasmid p53 “construct A” (Candeias et al. 2006), with the primers 23 and 24. This PCR product was digested with XhoI and cloned into the XhoI site of the psiRF vector. The resulting construct was named RLuc-empty. To obtain the RLuc-β-globin_5′ UTR, the same overlap-extension approach described above was used. In this case, the two first PCR reactions were performed to amplify the following fragments: The human β-globin 5′ UTR was amplified from genomic DNA with the primers 25 (with the XmaI linker) and 26 (Table 1), and the 663-bp fragment located downstream from the FLuc AUG codon, encompassing one AccI site, was amplified from the RLuc-empty plasmid with the primers 27 and 28 (Table 1). Then, primers 25 and 28 were used to amplify the two overlapped fragments. The resulting DNA fragment was digested with XmaI/AccI and then ligated to the RLuc-empty plasmid previously digested with the same enzymes. The RLuc-c-myc_IRES construct was generated using the same approach. The two first PCR reactions were performed to amplify the c-myc IRES 340-bp sequence (Stoneley et al. 1998) from human genomic DNA with the primers 29 (with the EcoRI linker) and 30 (Table 1) and to amplify a 663-bp fragment from the AUG codon through downstream into the FLuc coding sequence of the RLuc-empty construct, with the primers 31 and 28 (Table 1). Then primers 29 and 28 were used to amplify the two overlapped fragments. The resulting DNA fragment was digested with EcoRI/AccI and then ligated to the RLuc-empty plasmid previously digested with the same enzymes.
To obtain the RLuc-WT and the RLuc-no_uAUG dicistronic constructs, the RLuc-empty plasmid was digested with BglII/PmeI and the RLuc cistron sequence along with the SV40 promoter sequence, and the hairpin structure was inserted into the BglII/PmeI sites of pGL2-WT and pGL2-no_uAUG plasmids, creating the RLuc-WT and the RLuc-no_uAUG constructs, respectively.
Cell culture and plasmid transfection
HEK293 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum. HepG2 cells and HeLa cells were grown in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% (v/v) fetal bovine serum (Gibco). Cells were grown at 37°C in a humidified incubator containing 5% CO2. Transient transfections were performed using Lipofectamine 2000 transfection reagent (Invitrogen), following the manufacturer's instructions, in 35-mm plates. Cells were cotransfected with 750 ng of the test DNA construct corresponding to pGL2-Luc, pGL2-WT, or its derivative plasmids and 500 ng of the pRL-TK plasmid (Promega), which encodes RLuc as an internal control, and then harvested after 24 h. Dicistronic plasmids were single transfected at the same conditions but using 1 μg of test DNA construct. To mimic hypoxia, 6 h post-transfection the cultures were changed to fresh medium supplemented with 200 µM CoCl2 (Sigma-Aldrich). To induce nutrient starvation, 6 h post-transfection the cultures were changed to fresh medium supplemented with 10% (v/v) of dFBS (Gibco), and then cells were harvested 24 h post-treatment. Cells were treated with 1 μM thapsigargin (Sigma) to activate eIF2α kinases and induce eIF2α phosphorylation.
SDS-PAGE and Western blotting
Protein lysates were resolved, according to standard protocols, in 10% SDS-PAGE and transferred to PVDF membranes (Bio-Rad). Membranes were probed using mouse monoclonal anti-α-tubulin (Sigma) at 1:10000 dilution, rabbit polyclonal anti-eIF2α (Cell Signaling) at 1:500 dilution, rabbit polyclonal anti-phospho Ser 51 eIF2α (Life Technologies) at 1:500 dilution, mouse monoclonal β-catenin (Transduction Laboratories) at 1:3000 dilution, rabbit polyclonal anti-HIF1α (BD Biosciences) at 1:500, or rabbit polyclonal anti-EPO (Sigma) at 1:100. Detection was carried out using secondary peroxidase–conjugated anti-mouse IgG (Bio-Rad), anti-rabbit IgG (Bio-Rad), or anti-goat IgG (Sigma) antibodies followed by chemoluminescence. For densitometric analysis, films from at least three independent experiments were digitalized and analyzed using ImageJ software.
Luminometry assay
Lysis was performed in all cell lines with passive lysis buffer (Promega). The cell lysates were used to determine luciferase activity with the dual-luciferase reporter assay system (Promega) and a Lucy 2 luminometer (Anthos Labtec), according to the manufacturers’ standard protocols. One microgram of extract was assayed for firefly and RLuc activities. The ratio is the unit of FLuc after normalized with RLuc, and each value was derived from three independent experiments.
RNA isolation
Total RNA from transfected cells was isolated using the Nucleospin RNA extraction II kit (Macherey-Nagel), following the manufacturer's instructions. Then, all RNA samples were treated with RNase-free DNase I (Ambion) and purified by phenol:chloroform extraction.
Reverse transcription-quantitative PCR
Synthesis of cDNA was carried out using 1 µg of total RNA and Superscript II reverse transcriptase (Invitrogen), according to the manufacturer's instructions. Real-time PCR was performed in ABI Prism 7000 sequence detection system, using SybrGreen master mix (Applied Biosystems). Primers specific for the FLuc cDNA (primers 32 and 33) (Table 1) and RLuc cDNA (primers 34 and 35) (Table 1) were designed using the ABI Primer Express software. Primers specific for human EPO cDNA (primers 36 and 37) (Table 1) were according to the method previously described (Frede et al. 2011). Quantification was performed using the relative standard curve method (ΔΔCt; Applied Biosystems). The following cycling parameters were used: 10 min at 95°C and 40 cycles of 15 sec at 95°C and 1 min at 61°C. Technical triplicates from three to four independent experiments were assessed in all cases.
Statistical analysis
Results are expressed as mean ± SD. Student's t-test was used for estimation of statistical significance. Significance for statistical analysis was defined as a P < 0.05.
ACKNOWLEDGMENTS
We thank Margarida Gama Carvalho, Marco Candeias, and Abdessamad Tahiri-Alaoui for supplying the pRL-TK, p53 “A,” and psiRF plasmids, respectively. We thank Ana Ramos and Rafaela Lacerda for cloning the pGL2-RLuc-empty, pGL2-RLuc-β-globin_5′ UTR, and pGL2-RLuc-c-myc_IRES plasmids. Also, we thank Isabel Peixeiro and Alexandre Teixeira for critical reading of the manuscript and Luka Clarke for English editing. This research was partially supported by Fundação para a Ciência e a Tecnologia (PEst-OE/BIA/UI4046/2011, PTDC/BIM-MED/0352/2012 and SFRH/BD/63581/2009 to C.B.).
REFERENCES
- Arcasoy MO 2008. The non-haematopoietic biological effects of erythropoietin. Br J Haematol 141: 14–31 [DOI] [PubMed] [Google Scholar]
- Barbosa C, Peixeiro I, Romão L 2013. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet 9: e1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide A, Karaa Z, Bornes S, Hieblot C, Lacazette E, Prats H, Touriol C 2008. An upstream open reading frame within an IRES controls expression of a specific VEGF-A isoform. Nucleic Acids Res 36: 2434–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besarab A, Frinak S, Yee J 2009. What is so bad about a hemoglobin level of 12 to 13 g/dL for chronic kidney disease patients anyway? Adv Chronic Kidney Dis 16: 131–142 [DOI] [PubMed] [Google Scholar]
- Blais JD, Filipenko V, Bi M, Harding HP, Ron D, Koumenis C, Wouters BG, Bell JC 2004. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol 24: 7469–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brines M, Patel NSA, Villa P, Brines C, Mennini T, De Paola M, Erbayraktar Z, Erbayraktar S, Sepodes B, Thiemermann C, et al. 2008. Nonerythropoietic, tissue-protective peptides derived from the tertiary structure of erythropoietin. Proc Natl Acad Sci 105: 10925–10930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK 2009. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci 106: 7507–7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candeias MM, Powell DJ, Roubalova E, Apcher S, Bourougaa K, Vojtesek B, Bruzzoni-Giovanelli H, Faahraeus R 2006. Expression of p53 and p53/47 are controlled by alternative mechanisms of messenger RNA translation initiation. Oncogene 25: 6936–6947 [DOI] [PubMed] [Google Scholar]
- Cazzola M, Skoda RC 2000. Translational pathophysiology: a novel molecular mechanism of human disease. Blood 95: 3280–3288 [PubMed] [Google Scholar]
- Chatterjee S, Pal J 2009. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol Cell 101: 251–262 [DOI] [PubMed] [Google Scholar]
- Chin K, Yu X, Beleslin-Cokic B, Liu C, Shen K, Mohrenweiser HW, Noguchi CT 2000. Production and processing of erythropoietin receptor transcripts in brain. Mol Brain Res 81: 29–42 [DOI] [PubMed] [Google Scholar]
- Cobbold LC, Spriggs KA, Haines SJ, Dobbyn HC, Hayes C, de Moor CH, Lilley KS, Bushell M, Willis AE 2007. Identification of internal ribosome entry segment (IRES)–trans-acting factors for the myc family of IRESs. Mol Cell Biol 28: 40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyzyk-Krzeska MF, Bendixen AC 1999. Identification of the poly(C) binding protein in the complex associated with the 3′ untranslated region of erythropoietin messenger RNA. Blood 93: 2111–2120 [PubMed] [Google Scholar]
- Dame C, Fahnenstich H, Freitag P, Hofmann D, Abdul-Nour T, Bartmann P, Fandrey J 1998. Erythropoietin mRNA expression in human fetal and neonatal tissue. Blood 92: 3218–3225 [PubMed] [Google Scholar]
- Dang Do AN, Kimball SR, Cavener DR, Jefferson LS 2009. eIF2α kinases GCN2 and PERK modulate transcription and translation of distinct sets of mRNAs in mouse liver. Physiol Genomics 38: 328–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert BL, Bunn HF 1999. Regulation of the erythropoietin gene. Blood 94: 1864–1877 [PubMed] [Google Scholar]
- Fandrey J, Bunn H 1993. In vivo and in vitro regulation of erythropoietin mRNA: measurement by competitive polymerase chain reaction. Blood 81: 617–623 [PubMed] [Google Scholar]
- Frede S, Freitag P, Geuting L, Konietzny R, Fandrey J 2011. Oxygen-regulated expression of the erythropoietin gene in the human renal cell line REPC. Blood 117: 4905–4914 [DOI] [PubMed] [Google Scholar]
- Gassmann M, Soliz J 2009. Erythropoietin modulates the neural control of hypoxic ventilation. Cell Mol Life Sci 66: 3575–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi P, Brines M 2004. Erythropoietin as an antiapoptotic, tissue-protective cytokine. Cell Death Differ 11: S37–S44 [DOI] [PubMed] [Google Scholar]
- Goldberg M, Gaut C, Bunn H 1991. Erythropoietin mRNA levels are governed by both the rate of gene transcription and posttranscriptional events. Blood 77: 271–277 [PubMed] [Google Scholar]
- Haase VH 2013. Regulation of erythropoiesis by hypoxia-inducible factors. Blood Rev 27: 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D 2001. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell 7: 1153–1163 [DOI] [PubMed] [Google Scholar]
- Hernández G, Altmann M, Lasko P 2010. Origins and evolution of the mechanisms regulating translation initiation in eukaryotes. Trends Biochem Sci 35: 63–73 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG 1997. Translational regulation of yeast GCN4. J Biol Chem 272: 21661–21664 [DOI] [PubMed] [Google Scholar]
- Hoch M, Fischer P, Stapel B, Missol-Kolka E, Sekkali B, Scherr M, Favret F, Braun T, Eder M, Schuster-Gossler K, et al. 2011. Erythropoietin preserves the endothelial differentiation capacity of cardiac progenitor cells and reduces heart failure during anticancer therapies. Cell Stem Cell 9: 131–143 [DOI] [PubMed] [Google Scholar]
- Jelkmann W 1992. Erythropoietin: structure, control of production, and function. Physiol Rev 72: 449–489 [DOI] [PubMed] [Google Scholar]
- Jelkmann W 2011. Regulation of erythropoietin production. J Physiol 589: 1251–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagyozov L, Godfrey R, Böhmer SA, Petermann A, Hölters S, Östman A, Böhmer FD 2008. The structure of the 5′-end of the protein-tyrosine phosphatase PTPRJ mRNA reveals a novel mechanism for translation attenuation. Nucleic Acids Res 36: 4443–4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Hatzoglou M 2011. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 10: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koritzinsky M, Rouschop KMA, van den Beucken T, Magagnin MG, Savelkouls K, Lambin P, Wouters BG 2007. Phosphorylation of eIF2α is required for mRNA translation inhibition and survival during moderate hypoxia. Radiother Oncol 83: 353–361 [DOI] [PubMed] [Google Scholar]
- Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, Koromilas A, Wouters BG 2002. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2α. Mol Cell Biol 22: 7405–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 1997. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J 16: 2482–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 2001. Constraints on reinitiation of translation in mammals. Nucleic Acids Res 29: 5226–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 2002. Pushing the limits of the scanning mechanism for initiation of translation. Gene 299: 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361: 13–37 [DOI] [PubMed] [Google Scholar]
- Loughran G, Sachs MS, Atkins JF, Ivanov IP 2011. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res 40: 2898–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiese K, Chong ZZ, Shang YC 2008. Raves and risks for erythropoietin. Cytokine Growth Factor Rev 19: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGary EC, Rondon IJ, Beckman BS 1997. Post-transcriptional regulation of erythropoietin mRNA stability by erythropoietin mRNA-binding protein. J Biol Chem 272: 8628–8634 [DOI] [PubMed] [Google Scholar]
- Medenbach J, Seiler M, Hentze MW 2011. Translational control via protein-regulated upstream open reading frames. Cell 145: 902–913 [DOI] [PubMed] [Google Scholar]
- Mehta A 2006. Derepression of the Her-2 uORF is mediated by a novel post-transcriptional control mechanism in cancer cells. Genes Dev 20: 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, Thomas AAM 2002. Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem J 367: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignone F, Gissi C, Liuni S, Pesole G 2002. Untranslated regions of mRNAs. Genome Biol 3: previews0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole DR, Ratcliffe PJ 2007. Cellular oxygen sensing in health and disease. Pediatr Nephrol 23: 681–694 [DOI] [PubMed] [Google Scholar]
- Morris DR, Geballe AP 2000. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 20: 8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi CT, Wang L, Rogers HM, Teng R, Jia Y 2008. Survival and proliferative roles of erythropoietin beyond the erythroid lineage. Expert Rev Mol Med 10: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohigashi T, Mallia CS, McGary E, Scandurro AB, Rondon I, Fisher JW, Beckman BS 1999. Protein kinase C α protein expression is necessary for sustained erythropoietin production in human hepatocellular carcinoma (Hep3B) cells exposed to hypoxia. Biochim Biophys Acta 1450: 109–118 [DOI] [PubMed] [Google Scholar]
- Palam LR, Baird TD, Wek RC 2011. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J Biol Chem 286: 10939–10949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyry TAA, Kaminski A, Jackson RJ 2004. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev 18: 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L, Vilela C, Berthelot K, Ramirez CV, McCarthy JEG 2004. Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J Mol Biol 335: 71–85 [DOI] [PubMed] [Google Scholar]
- Ruifrok WP, de Boer RA, Westenbrink BD, van Veldhuisen DJ, van Gilst WH 2008. Erythropoietin in cardiac disease: new features of an old drug. Eur J Pharmacol 585: 270–277 [DOI] [PubMed] [Google Scholar]
- Ryou M-G, Liu R, Ren M, Sun J, Mallet RT, Yang S-H 2012. Pyruvate protects the brain against ischemia-reperfusion injury by activating the erythropoietin signaling pathway. Stroke 43: 1101–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs MS 2006. Downstream control of upstream open reading frames. Genes Dev 20: 915–921 [DOI] [PubMed] [Google Scholar]
- Schepens B 2005. The polypyrimidine tract-binding protein stimulates HIF-1 IRES-mediated translation during hypoxia. Nucleic Acids Res 33: 6884–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CB, Mitsock LD 1986. Murine erythropoietin gene: cloning, expression, and human gene homology. Mol Cell Biol 6: 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE 1998. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene 16: 423–428 [DOI] [PubMed] [Google Scholar]
- Tahiri-Alaoui A, Smith LP, Baigent S, Kgosana L, Petherbridge LJ, Lambeth LS, James W, Nair V 2009. Identification of an intercistronic internal ribosome entry site in a Marek's disease virus immediate-early gene. J Virol 83: 5846–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-Q 2004. 5′-Untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation. Nucleic Acids Res 32: 1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Jiang B-H, Rue EA, Semenza GL 1995. Hypoxia-inducible factor 1 is a basic–helix–loop–helix–PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci 92: 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watatani Y, Ichikawa K, Nakanishi N, Fujimoto M, Takeda H, Kimura N, Hirose H, Takahashi S, Takahashi Y 2007. Stress-induced translation of ATF5 mRNA is regulated by the 5′-untranslated region. J Biol Chem 283: 2543–2553 [DOI] [PubMed] [Google Scholar]
- Wei J, Wu C, Sachs MS 2012. The arginine attenuator peptide interferes with the ribosome peptidyl transferase center. Mol Cell Biol 32: 2396–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidemann A, Johnson RS 2009. Nonrenal regulation of EPO synthesis. Kidney Int 75: 682–688 [DOI] [PubMed] [Google Scholar]
- Wethmar K, Smink JJ, Leutz A 2010. Upstream open reading frames: molecular switches in (patho)physiology. BioEssays 32: 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda Y, Masuda S, Chikuma M, Inoue K, Nagao M, Sasaki R 1998. Estrogen-dependent production of erythropoietin in uterus and its implication in uterine angiogenesis. J Biol Chem 273: 25381–25387 [DOI] [PubMed] [Google Scholar]
- Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC 2008. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem 283: 7064–7073 [DOI] [PubMed] [Google Scholar]