Earlier studies showed that tRNA isoacceptor bodies have evolved to reduce differences between natural aminoacyl-tRNAs in their affinity to EF-Tu. Simply put, “weaker” amino acids tend to be paired with “stronger” tRNAs, and vice versa. This study explores the coupling of different tRNA bodies with unnatural amino acids to determine their biochemical behavior during tRNA selection on the ribosome. The results generally argue that the coupling of a strong tRNAAla body with several unnatural amino acids (allyl-glycine, methyl-serine, and biotinyl-lysine) can increase the overall extent of incorporation by increasing the affinity of these aminoacyl-tRNAs for EF-Tu. The same strong tRNAAla body failed to increase the efficiency of N-methyl-phenylalanine incorporation, arguing that its deficits are not related to EF-Tu affinity. These results have broad implications for unnatural amino acid incorporation during translation elongation.
Keywords: unnatural amino acid, tRNAAla, EF-Tu affinity, ribosome, translation kinetics
Abstract
There is evidence that tRNA bodies have evolved to reduce differences between aminoacyl-tRNAs in their affinity to EF-Tu. Here, we study the kinetics of incorporation of L-amino acids (AAs) Phe, Ala allyl-glycine (aG), methyl-serine (mS), and biotinyl-lysine (bK) using a tRNAAla-based body (tRNAAlaB) with a high affinity for EF-Tu. Results are compared with previous data on the kinetics of incorporation of the same AAs using a tRNAPheB body with a comparatively low affinity for EF-Tu. All incorporations exhibited fast and slow phases, reflecting the equilibrium fraction of AA-tRNA in active ternary complex with EF-Tu:GTP before the incorporation reaction. Increasing the concentration of EF-Tu increased the amplitude of the fast phase and left its rate unaltered. This allowed estimation of the affinity of each AA-tRNA to EF-Tu:GTP during translation, showing about a 10-fold higher EF-Tu affinity for AA-tRNAs formed from the tRNAAlaB body than from the tRNAPheB body. At ∼1 µM EF-Tu, tRNAAlaB conferred considerably faster incorporation kinetics than tRNAPheB, especially in the case of the bulky bK. In contrast, the swap to the tRNAAlaB body did not increase the fast phase fraction of N-methyl-Phe incorporation, suggesting that the slow incorporation of N-methyl-Phe had a different cause than low EF-Tu:GTP affinity. The total time for AA-tRNA release from EF-Tu:GDP, accommodation, and peptidyl transfer on the ribosome was similar for the tRNAAlaB and tRNAPheB bodies. We conclude that a tRNA body with high EF-Tu affinity can greatly improve incorporation of unnatural AAs in a potentially generalizable manner.
INTRODUCTION
Ribosomal incorporation of unnatural amino acids (AAs) into polypeptides has become a useful tool in numerous studies of protein structure and function (Xie and Schultz 2005; Doi et al. 2007) and bears promise to facilitate drug discovery by peptidomimetic evolution (Yamagishi et al. 2011; Guillen Schlippe et al. 2012; Watts and Forster 2012). Unnatural AA-tRNAs are typically prepared from unmodified transcripts by ligation or charging with activated AAs (Hecht et al. 1978; Bain et al. 1989; Robertson et al. 1989; Murakami et al. 2006). Translation with unnatural AAs is often inefficient, even in purified in vitro systems (Forster et al. 2003), which limits its utility. Much effort has been spent during the last two decades to improve the incorporation of unnatural AAs in cell-free protein synthesis. It was found that the incorporation yield of a nonproteinogenic L-AA in a crude translation system could be significantly improved by swapping tRNA bodies (Cload et al. 1996). Furthermore, tRNA bodies based on E. coli tRNAAsn or tRNAAla instead of tRNAPhe increased the yield of incorporation of single N-methyl-Ala and N-methyl-Phe about twofold in a purified translation system (Zhang et al. 2007). Also, tRNA bodies based on tRNAAsn instead of tRNAPhe substantially increased the yield of incorporation of multiple adjacent unnatural non-N-alkyl-AAs (Forster 2009). However, a mechanistic understanding awaits investigation of the step(s) in translation affected by these body swaps.
Incorporation yields of unnatural AAs have also been improved in vitro (Doi et al. 2007) and in vivo (Park et al. 2011; Mittelstaet et al. 2013) by the use of EF-Tu mutants with increased affinity to the corresponding AA-tRNAs. In a recent rapid kinetics study on translation incorporation of members of the most commonly used unnatural AA class, the non-N-alkyl-L-AAs, it was demonstrated that inefficient incorporations were mainly due to weak binding of these unnatural AA-tRNAs to EF-Tu, not slow peptide bond formation per se. Individual incorporations of two small AAs (allyl-glycine [aG] and methyl-serine [mS]) and one bulky AA (biotinyl-lysine [bK]) from this class, attached to tRNAPheC3G,G70C (tRNAPheB), were markedly increased in response to the increased fractions of AA-tRNA in ternary complex resulting from increased EF-Tu concentration or lowered incubation temperature (Ieong et al. 2012).
Here, we have used rapid kinetics techniques to explore the effect on incorporation of these three unnatural AAs and also N-methyl-Phe by using a tRNA body with comparatively high affinity for EF-Tu, tRNAAlaB (Fig. 1B). We studied the time of EF-Tu-dependent delivery of the corresponding AA-tRNAs to the ribosomal A/T site as well as the total time of subsequent steps leading to peptide bond formation. We have found that the kinetics of incorporation improved significantly for the small aG and mS and dramatically for the bulky bK by the swap from the tRNAPheB to the tRNAAlaB. Moreover, we found that the accommodation/peptidyl-transfer reactions on the ribosome proceeded with comparable rates for natural and unnatural amino acids with short or long side chains.
FIGURE 1.
A tRNA based on tRNAAla (tRNAAlaB) charged with natural and unnatural AAs. (A) Natural and unnatural L-AAs used in our kinetics studies. (B) Synthetic tRNAAlaB (Zhang et al. 2007), which is an unmodified tRNA based on natural E. coli tRNAAla (black with purple anticodon; tRNA modifications are in green) with changes in blue.
RESULTS
Rapid kinetics of incorporation of natural and unnatural L-AAs: the method
In our prior quench-flow studies of unnatural AA incorporation into peptide in an E. coli translation system with components of high purity (Pavlov et al. 2009; Ieong et al. 2012), we used tRNAPheB, a tRNA body with intermediate affinity for EF-Tu (Asahara and Uhlenbeck 2002). We previously observed that incorporation rates of unnatural non-N-alkyl-L-AAs were limited by poor ternary complex formation (Ieong et al. 2012). Here, we compared incorporation from a tRNAAla-based unmodified body (tRNAAlaB) (Zhang et al. 2007), which we predicted would have a high affinity for EF-Tu (Asahara and Uhlenbeck 2002). Apart from its ease of synthesis by T7 RNA polymerase, we selected tRNAAlaB because we reasoned it might have the advantage of a tight EF-Tu binder but not the disadvantage of a too-tight binder (Schrader et al. 2011). We studied dipeptide bond formation with the natural L-AAs Phe and Ala as well as with the unnatural L-AAs aG, mS, the bulky bK, and N-methyl-Phe (Fig. 1).
In each case, ternary complex consisting of AA-tRNA, EF-Tu, and GTP was assembled by preincubating various concentrations of EF-Tu:GTP at a fixed concentration of chemoenzymatically produced AA-tRNAs. Ternary complex was then rapidly mixed with preinitiated 70S ribosomal initiation complexes (ICs) containing the initiator tRNA (fMet-tRNAfMet) in the P site and the A site programmed with a GCA (Ala) or UUC (Phe) codon. In all experiments, the ternary complex concentration was limited by the fixed concentration of AA-tRNA, and the rate of ternary complex binding to the A site was determined by the concentrations of preinitiated ribosomes, always present in large molar excess over the AA-tRNA.
Swapping tRNAPheB for tRNAAlaB speeds up kinetics of incorporation of natural and small unnatural AAs
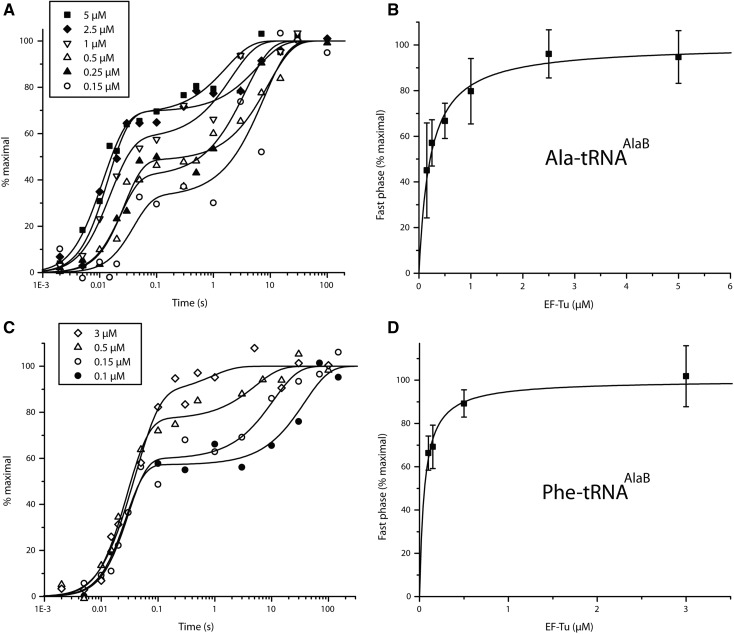
We first monitored fMet-Ala dipeptide formation on 70S IC with a GCA(Ala) codon at the A site and fixed (0.1 µM) Ala-tRNAAlaB substrate concentration at EF-Tu:GTP concentrations varying from 0.15 to 5 µM (Fig. 2A). At all EF-Tu concentrations, the incorporation reactions displayed biphasic kinetics with varying amplitudes of fast and slow phases. The amplitude of the fast phase increased with increasing EF-Tu concentration but the intrinsic rates of the two phases did not change significantly (Table 1). In this case, the amino acid (Ala) had weak affinity for EF-Tu, whereas tRNAAlaB had strong affinity for the factor. Therefore, the affinity of Ala-tRNAAlaB was optimal according to the thermodynamic compensation hypothesis that tRNA bodies are paired with their cognate (natural) amino acids for near-uniform affinity to EF-Tu (LaRiviere et al. 2001; Schrader et al. 2011).
FIGURE 2.
Effects of EF-Tu concentration on the kinetics of dipeptide synthesis from fMet-tRNAifMet and Ala-tRNAAlaB (A,B) or Phe-tRNAAlaB (C,D). Left: Time course (normalized) of f[3H]Met-Ala (A) or f[3H]Met-Phe (C) dipeptide formation at different EF-Tu concentrations in the reaction mixture. Right: (B,D) Hyperbolic fitting of the dependence of the fast phase fraction of the dipeptide formation (A,C) on EF-Tu concentrations. Kd for the binding of Ala-tRNAAlaB and Phe-tRNAAlaB to EF-Tu:GTP were estimated as 0.22 and 0.056 µM, respectively, from the fit. Experiments were done in LS3 buffer at 37°C (see Materials and Methods).
TABLE 1.
Kinetics values for dipeptide synthesis from fMet-tRNAfMet and different AA-tRNAs
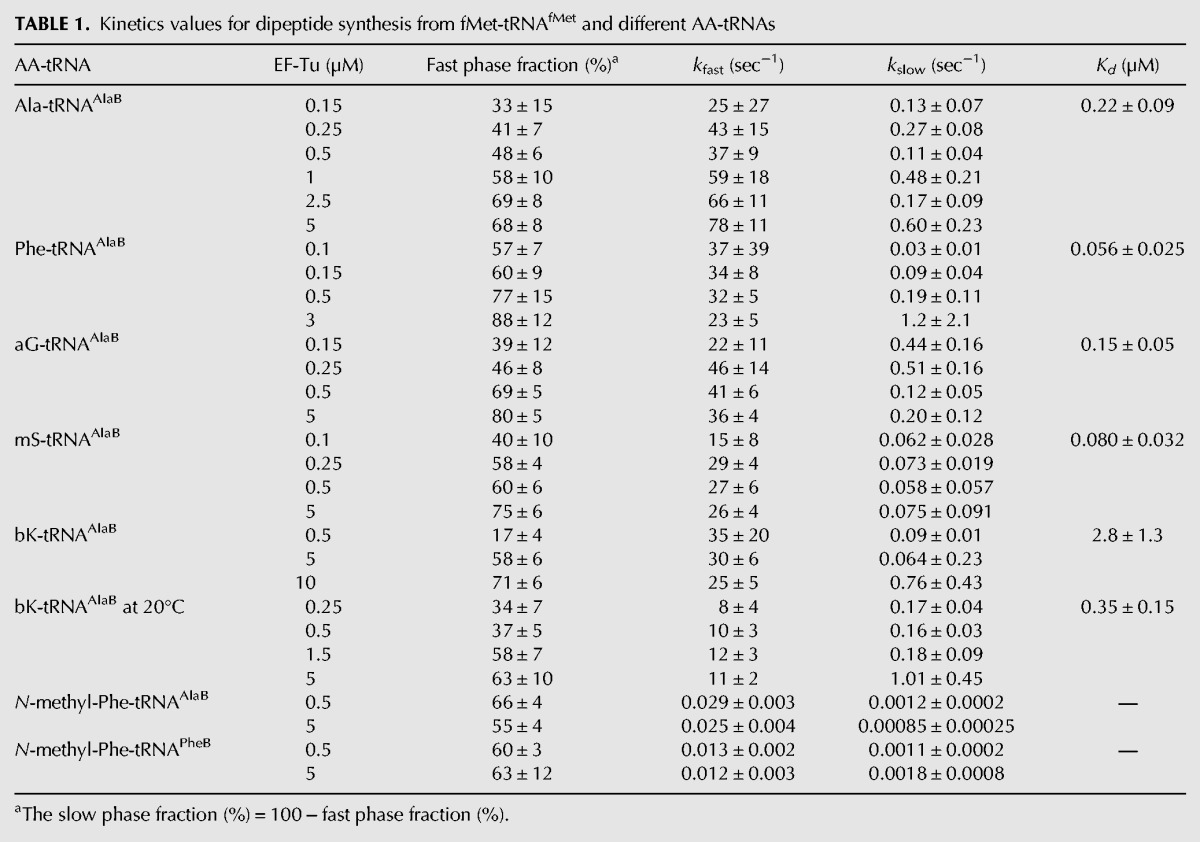
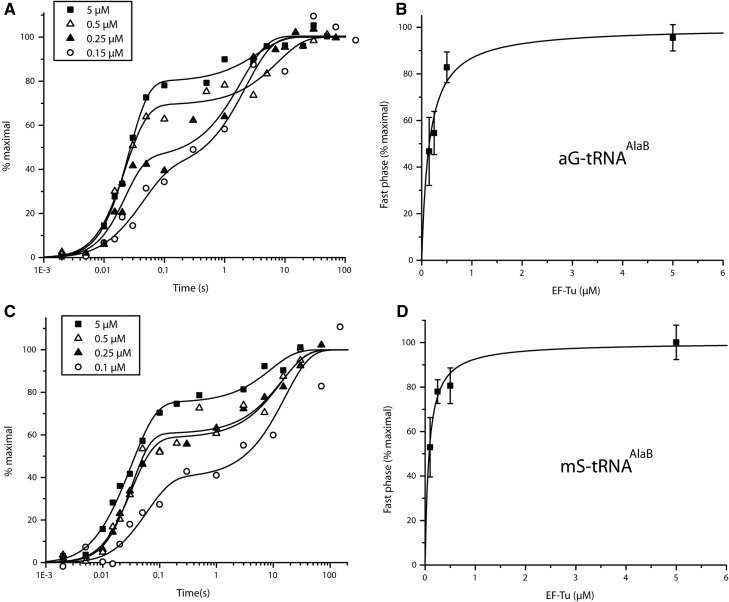
Next, we used the same type of experimental conditions to monitor incorporation from tRNAAlaB linked to Phe and small unnatural AAs (Fig. 1A) on the GCA-programmed 70S IC. Biphasic kinetics was observed in all cases, with the fast and slow rates of incorporation for Phe, aG, and mS (Figs. 2C, 3A,C) similar to those for Ala (Fig. 2A; Table 1) and other small AAs on tRNAPheB (Ieong et al. 2012). The fast phase amplitudes for Phe, aG, and mS on tRNAAlaB increased with the EF-Tu concentration and were consistently larger than those on tRNAPheB (about twofold larger at 0.5 µM EF-Tu) (Table 2). The amplitudes of the fast phases increased hyperbolically, but the corresponding rates did not change significantly with increasing concentration of EF-Tu:GTP, as observed above in the Ala-tRNAAlaB case and previously in similar experiments with tRNAPheB (Table 1; Ieong et al. 2012). Accordingly, we infer that the pronounced biphasic kinetics observed at low EF-Tu concentration (Figs. 2, 3) was primarily due to the existence of a mixture of free and EF-Tu:GTP-bound AA-tRNA with slow peptide bond formation from the free and fast peptide bond formation from the EF-Tu:GTP-bound fraction of AA-tRNA (see Discussion). In the next section, we use the free and bound fractions to estimate the constants of binding of the various AA-tRNAs to EF-Tu:GTP. Also, the apparent constancy of the rate of the slow phase of AA-incorporation (Table 1) is intriguing, since a simple model for binding of AA-tRNA would predict a linear relation between the rate and the EF-Tu concentration (Ieong et al. 2012) (see Discussion).
FIGURE 3.
Effects of EF-Tu concentration on the kinetics of dipeptide synthesis from fMet-tRNAifMet and aG-tRNAAlaB (A,B) or mS-tRNAAlaB (C,D).
TABLE 2.
Comparison between two different tRNA bodies of fast phase fractions at 37°C at various EF-Tu concentrations
EF-Tu:GTP displays higher affinity to the tRNAAlaB than to the tRNAPheB body during translation
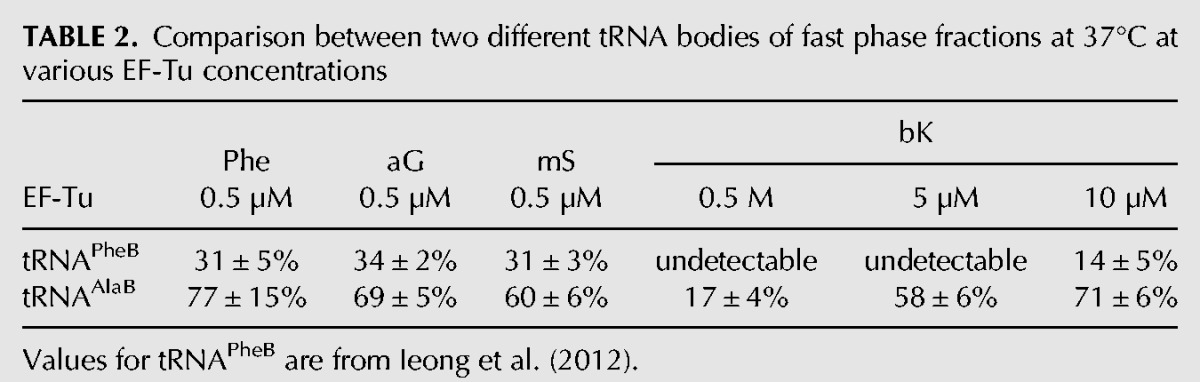
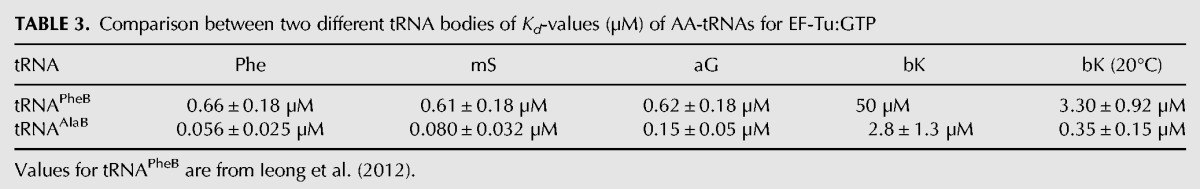
To understand the speeding up of the incorporation kinetics by swapping tRNA bodies, equilibrium dissociation constants (Kd-values) for the binding of the AA-tRNAAlaBs to EF-Tu:GTP were estimated and compared to those of the AA-tRNAPheBs. For this, the amplitudes of the fast phase in the dipeptide kinetics were plotted as functions of the EF-Tu:GTP concentration at fixed AA-tRNAAlaB concentration as shown in Figure 2B (Ala-tRNAAlaB), Figure 2D (Phe-tRNAAlaB), Figure 3B (aG-tRNAAlaB), and Figure 3D (mS-tRNAAlaB). Compared to our previous data with small AAs attached to the tRNAPheB body (Ieong et al. 2012), the present use of tRNAAlaB increased the binding affinity of AA-tRNAs for EF-Tu:GTP 4–18 times (Table 3).
TABLE 3.
Comparison between two different tRNA bodies of Kd-values (µM) of AA-tRNAs for EF-Tu:GTP
tRNAAlaB improves the kinetics of incorporation of bK, a bulky unnatural AA
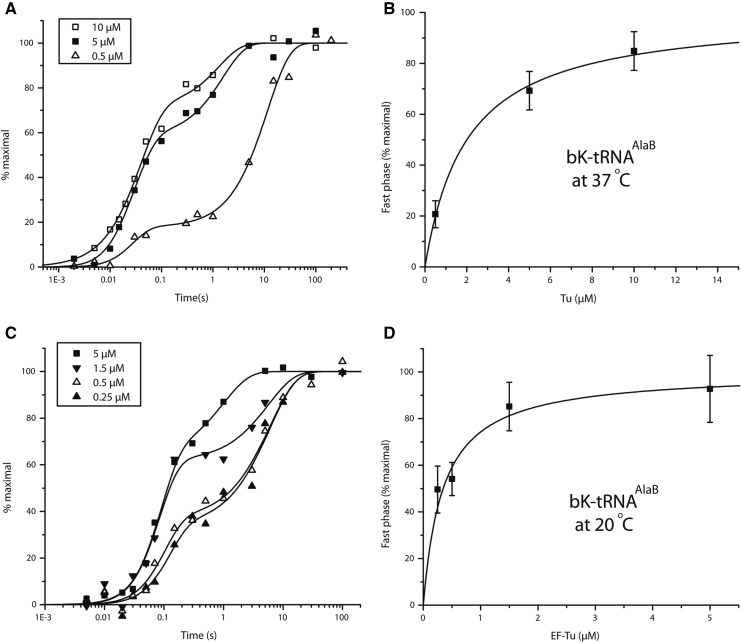
Only a slow phase of incorporation of the bulky unnatural AA, bK (Fig. 1A) from tRNAPheB could previously be observed at 37°C with EF-Tu concentrations up to 5 µM (Ieong et al. 2012). The lack of a fast phase under those conditions means that formation of an active ternary complex during preincubation was negligible. However, a fast phase could be detected by increasing the EF-Tu concentration to 10 µM or by decreasing the incubation temperature to 20°C. In contrast, the kinetics of dipeptide formation from bK-tRNAAlaB at 37°C exhibited a significant fast phase amplitude for bK incorporation even at EF-Tu concentrations down to 0.5 µM (Fig. 4A). Furthermore, at EF-Tu concentrations >5 µM, the fast phase amplitude dominated over that of the slow phase in the bK incorporation curves, and the specific rates of the fast and slow phases were similar to those for incorporations of Ala, Phe, aG, and mS (Fig. 4A). The kinetics data obtained for bK-tRNAAlaB and bK-tRNAPheB at the two temperatures could readily be explained by the about 18-fold (37°C) or ninefold (20°C) smaller Kd-values for EF-Tu:GTP binding to bK-tRNAAlaB than to bK-tRNAPheB (Fig. 4B,D; Table 3). The similarity of the rate of the fast phase of bK with the fast phases of the Ala, Phe, aG, and mS incorporations suggests that the major kinetic deficiency of the bulky bK was its very weak binding to EF-Tu:GTP and not A-site accommodation or the chemistry of peptide bond formation (see below).
FIGURE 4.
Effects of EF-Tu concentration on the kinetics of dipeptide synthesis from fMet-tRNAifMet and bK-tRNAAlaB. Experiments were done at 37°C (A,B) and 20°C (C,D).
Body swap from tRNAPheB to tRNAAlaB moderately hastens incorporation of N-methyl-Phe
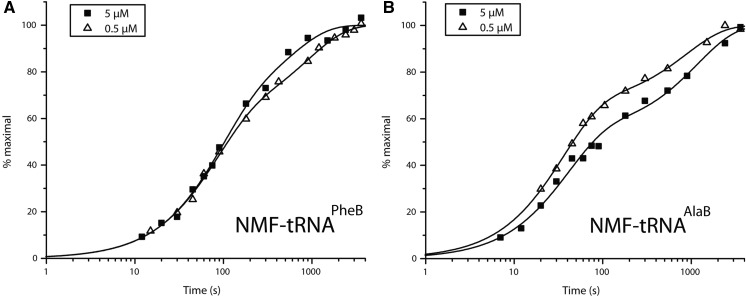
We previously found that N-methylation of Phe decreased its rate of incorporation from tRNAPheB by four orders of magnitude (Pavlov et al. 2009), and body swapping from tRNAPheB to the tRNAAlaB increased the incorporation yield of N-methyl-AAs twofold in 40-min assays (Zhang et al. 2007). Here, we monitored dipeptide formation with N-methyl-Phe-tRNAPheB using EF-Tu at 0.5 µM, as in previous work (Pavlov et al. 2009), and at 5 µM (Fig. 5A, filled squares). The kinetics was biphasic at both concentrations of EF-Tu, with a very slow and a super slow phase, but the amplitudes of the phases remained unchanged at varying EF-Tu concentrations. Furthermore, swapping of the tRNAPheB body with tRNAAlaB did not alter the amplitudes of the fast and slow phases (Fig. 5A,B), but the rate constant of the fast phase increased about twofold at both concentrations of EF-Tu (Table 1). These data suggest that neither the slow incorporation of N-methyl-Phe nor its biphasic nature can be rationalized by inefficient ternary complex formation but must have other explanations (see Discussion).
FIGURE 5.
Effects of EF-Tu concentration on the kinetics of dipeptide synthesis from fMet-tRNAifMet and N-methyl-Phe-tRNAPheB (A) or N-methyl-Phe-tRNAAlaB (B).
Impact of AA-tRNA binding affinity for EF-Tu on intra-ribosomal kinetics
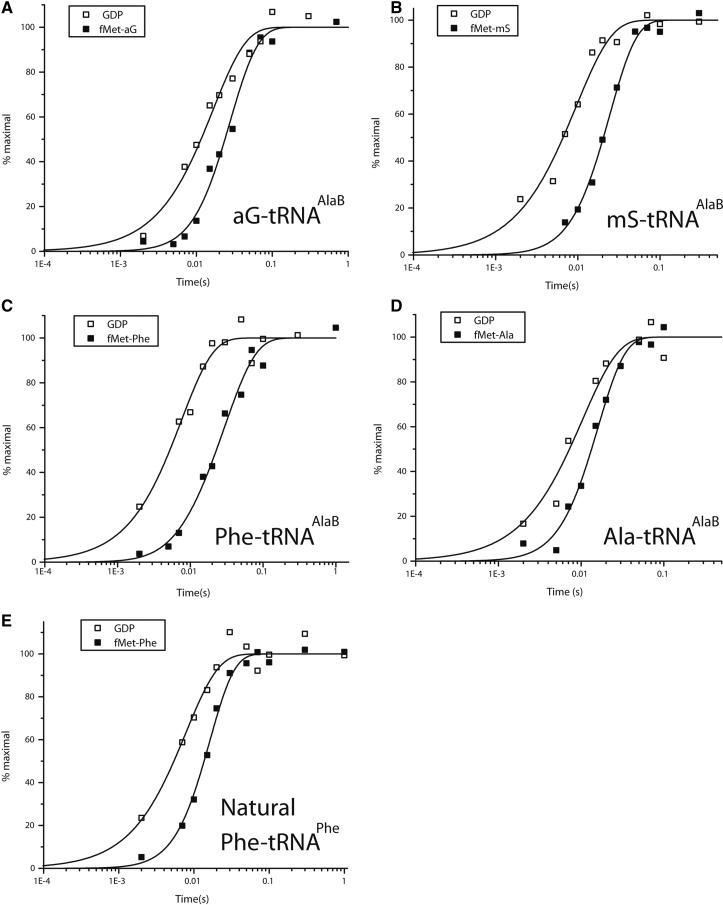
Very little is known about the rates of individual steps on the ribosome for incorporation of non-N-alkyl unnatural AAs from any tRNA body. In this section, we only consider the fast phase of the incorporation reaction and describe how different natural and unnatural L-AAs on the tRNAAlaB body affected the average times of GTP hydrolysis on EF-Tu (τGTP) and the average times of dipeptide formation (τdip). τGTP is the sum of times for ternary complex binding to the ribosome, GTPase activation, and GTP hydrolysis, whereas τdip is the sum of τGTP and the average time, τacc,pep, for AA-tRNA release from EF-Tu:GDP, its A-site accommodation and the time of peptidyl transfer (Pavlov et al. 2009; Johansson et al. 2011). By measuring GTP hydrolysis and dipeptide formation in the very same experiment, we estimated τacc,pep from the difference τdip− τGTP (Pavlov et al. 2009; Johansson et al. 2011). We used this assay to estimate τacc,pep for all aminoacyl-tRNAs (Fig. 6) except bK-tRNAAlaB, which required a different type of experiment due to its particularly low affinity (high Kd-value) for EF-Tu:GTP (see below). A major discriminating feature among these AA-tRNAs was their significantly different τacc,pep-values (Table 4), which were smallest for Ala-tRNAAlaB and Phe-tRNAPheB (the two naturally acylated AA-tRNAs), intermediate for aG- and mS-tRNAAlaB (the two small unnatural AAs), and largest for Phe-tRNAAlaB (the highest affinity to EF-Tu:GTP) (Table 3).
FIGURE 6.
Kinetics of the fast phase of GTP hydrolysis and dipeptide synthesis with different AA-tRNA combinations. Time course of GTP hydrolysis (□) and dipeptide synthesis (▪) for dipeptide formation reaction from fMet-tRNAifMet and aG-tRNAAlaB (A), mS-tRNAAlaB (B), Phe-tRNAAlaB (C), Ala-tRNAAlaB (D), or natural Phe-tRNAPhe (E) are shown. Preinitiated ribosomes had Ala codon GCA (A–D) or Phe codon UUC (E) in the A site. All kinetics were measured at 37°C in LS3 buffer. Representative plots are shown for each assay.
TABLE 4.
Rates for dipeptide synthesis from fMet-tRNAifMet and different AA-tRNAs at 37°C and dissociation constant (Kd) of the corresponding AA-tRNAs for EF-Tu:GTP
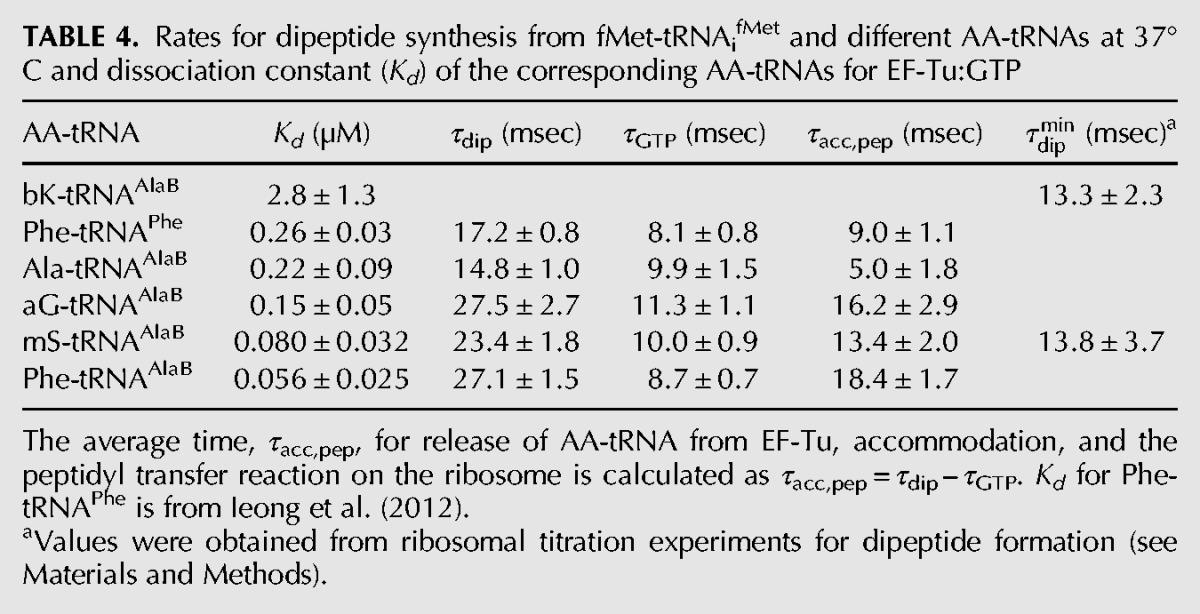
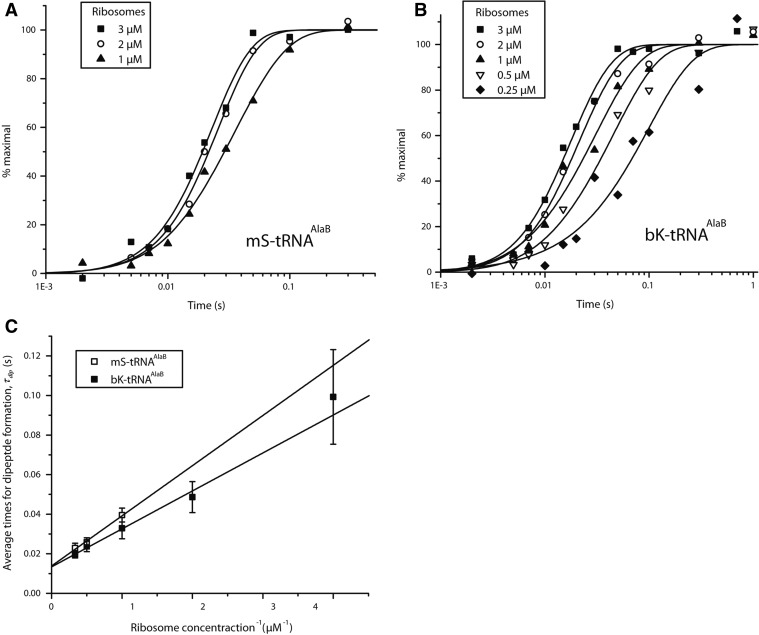
The relatively low affinity of bK-tRNAAlaB to EF-Tu:GTP (Table 3) made the measurement of τGTP technically unfeasible (Pavlov et al. 2009). However, the tRNAAlaB body did provide sufficient affinity for EF-Tu to allow for a precise estimate of the minimal τdip time, τdipmin, operationally defined as the τdip-value at saturating 70S ribosome concentration (Pape et al. 1998). By definition, therefore, τdipmindid not contain the time for binding of ternary complex to the ribosome but all subsequent steps leading to peptide bond formation. Since the time of GTP hydrolysis is expected to be very short (Johansson et al. 2008), τdipmin approximated τacc,pep. (Indeed, in a calibration experiment we estimated τdipmin for incorporation of mS from tRNAAlaB as 13.8 msec [Fig. 7A,C], a value close to the 13.4 msec estimated for the τacc,pep-value of the same incorporation [Table 4].) The finding that bK incorporation from tRNAAlaB had aτdipmin-value of 13.3 msec, virtually identical with the τdipmin-value of 13.8 msec estimated for the mS incorporation with the same method (Fig. 7B,C; Table 4), was surprising (see Discussion).
FIGURE 7.
Dependence of the rate of fast phase of dipeptide synthesis on ribosome concentration in the reaction mixture. The normalized time course of f[3H]Met-mS (A) or f[3H]Met-bK (B) dipeptide formation for different concentrations of preinitiated ribosomes with the Ala codon (GCA) in the A site (see Materials and Methods). (C) The average times of dipeptide formation τdip, estimated from the experiments in A (□) and B (▪), were plotted versus the inverse of ribosome concentration (Lineweaver-Burke plot); τdipmin- values for dipeptide formation, which is equal to the y-intercept, were estimated from the fits as 13.8 ± 3.7 msec for mS-tRNAAlaB (□) and 13.3 ± 2.3 msec for bK-tRNAAlaB (▪).
DISCUSSION
Fast and slow phases of ribosomal incorporation of natural and unnatural AA-tRNAs
In the present work, we studied the kinetics of ribosome-catalyzed dipeptide formation (fMet-AA) with the natural L-AAs Phe and Ala, the small unnatural AAs aG and mS, the bulky bK, and N-methyl-Phe charged on a tRNAAla body (tRNAAlaB) of comparatively high affinity to EF-Tu (Asahara and Uhlenbeck 2002). Results were compared to those obtained for the same AAs charged on a tRNAPhe body (tRNAPheB) of comparatively intermediate affinity to EF-Tu:GTP (Asahara and Uhlenbeck 2002). In this, as well as in the previous study (Ieong et al. 2012), the incorporation kinetics invariably displayed two phases: one with fast and one with slow incorporation (Figs. 2–5). For all AAs except N-methyl-Phe, the fast phase was due to rapid delivery of activated ternary complex (unnatural AA-tRNA:EF-Tu:GTP) to the ribosome. The slow phase was primarily due to de novo formation of ternary complex from free AA-tRNA and free EF-Tu at the start of the translation incubation (Ieong et al. 2012). This interpretation is supported by the observation that the fast phase amplitude increased hyperbolically, and the fast phase rate remained unaltered with increasing EF-Tu:GTP concentration at a constant concentration of AA-tRNA in the reaction mixture. Accordingly, the biphasic kinetics made it possible to estimate the dissociation constant for each aminoacyl-tRNA to EF-Tu:GTP from the rapid translation kinetics experiments themselves (Table 4). Intriguing aspects of these and previous (Ieong et al. 2012) observations are that the rate constant ascribed to the slow phase did not increase linearly with the concentration of free EF-Tu:GTP, and the slow phase did not vanish even at a very high concentration of EF-Tu:GTP (Figs. 2–4). This suggests that an active ternary complex was formed according to a two-step mechanism: first AA-tRNA bound to EF-Tu:GTP in the formation of an inactive form of ternary complex which, subsequently, slowly transformed to its fully active conformation. Although the physiological role of this unexpectedly complex kinetics of ternary complex formation remains to be clarified, it might provide an explanation for the vastly different estimates that have been obtained for the rate constant of AA-tRNA association to EF-Tu:GTP (Gromadski et al. 2002; Ling et al. 2009).
Faster delivery of unnatural AA-tRNAs to the ribosomal A/T site by increased affinity of the tRNA body to EF-Tu
From our rapid kinetics experiments, we found that AAs Phe, Ala, aG, mS, or bK charged on the tRNAAlaB body had about ten times higher affinity to EF-Tu:GTP than those with the same AAs linked to the tRNAPheB body (Table 2). The smallest Kd-value (highest affinity) was observed for the combination of the tRNAAlaB body with Phe, in line with the thermodynamic compensation hypothesis (Asahara and Uhlenbeck 2002), suggesting that the affinity to EF-Tu:GTP of a tRNA body correlated negatively with that of its cognate, natural AA so that the binding affinities of natural AA-tRNAs to EF-Tu:GTP were near-uniform in the living cell (LaRiviere et al. 2001). In the present case, the noncognate combination of “tight” tRNAAlaB body with “intermediate” Phe AA led to an “unnaturally strong” binding of Phe-tRNAAlaB to EF-Tu:GTP. By an extension of this nomenclature to unnatural AAs based on the data in Tables 1 and 3, bK was a “very weak” AA, whereas aG and mS were similar to “intermediate” Phe. The very weak binding of the bulky unnatural bK to EF-Tu:GTP may be due to steric exclusion of this unnatural side chain from the AA binding pocket on EF-Tu (Doi et al. 2007).
For aG and mS, the amplitude of the fast phase of dipeptide formation was significantly increased by the swap from the tRNAPheB to the tRNAAlaB body and, for bulky bK, the amplitude increase due to the swap was dramatic (Table 2; Ieong et al. 2012). Our previous experiments revealed very inefficient ribosomal incorporation of bK from tRNAPheB (Ieong et al. 2012), but the present experiments show the rate of the fast phase of bK incorporation from tRNAAlaB to be close to those rates observed for the much smaller unnatural AAs, aG, and mS from the same tRNA body (Table 1). The explanation for the greatly increased amplitude of the fast phase can be traced to the 18-fold higher affinity of EF-Tu:GTP to bK-tRNAAlaB than to bK-tRNAPheB (Table 2). This result suggests that efficient incorporation of many other unnatural AAs with low affinity to EF-Tu can be achieved by attaching them to tRNAAlaB or other tRNA bodies that are rationally designed for high affinity to EF-Tu (Roy et al. 2007; Schrader et al. 2009). Such design is greatly facilitated by the finding that the affinity between EF-Tu:GTP and a tRNA body can be tuned over a wide range by the choice of three base pairs in the T-stem of the tRNA (Roy et al. 2007; Schrader et al. 2009). The tightest of these tRNA binders, which are tighter than tRNAAlaB, may be optimal for the weakest AAs, like bK; but it should be noted that if EF-Tu binding is too tight, as in the case of Val-tRNAVal T1 (Table 1 in Schrader et al. 2009), the incorporation can actually be slower, not faster. Increasing the efficiencies of incorporation of unnatural AA-tRNAs at their cognate codons may also help reduce unwanted side reactions, like cross-reading of codons by noncognate tRNAs (Subtelny et al. 2011).
Incorporation of N-methylated amino acids displays unexpected features
Concerning incorporation of N-methyl-Phe from tRNAAlaB and tRNAPheB, kinetics was again biphasic, but the “faster” phases (60% amplitudes) were about 1000 times slower than for the non-N-alkylated AAs (Table 1). The amplitudes and rate constants remained unaltered when the EF-Tu:GTP concentration was increased from 0.5 to 5 µM. There was also a poorly defined super slow phase with 40% amplitude and a rate constant of ∼0.001 sec−1 for both tRNA bodies and EF-Tu concentrations. From these data it follows that the incorporation defect of N-methyl-Phe was not caused by poor ternary complex formation but must have had another reason.
We previously observed that a ternary complex with EF-Tu:GTP and N-methyl-Phe-tRNAPhe had rapid GTP hydrolysis when entering the A site (Pavlov et al. 2009). Taken together, these data sets suggest that N-methylation inhibits a function late in the chain of events, leading to peptide bond formation, most likely the chemistry of peptidyl transfer itself. That would lead to the interesting option that the tRNAAlaB body placed N-methyl-Phe in a more favorable context for peptidyl transfer than did tRNAPheB. Definitive answers to these types of questions, relevant not only to the efficiency of incorporation of unnatural AAs but also to mechanistic aspects of A-site accommodation and peptidyl-transfer, require further experimental work.
Passage time from A/T site to peptidyl transfer for unnatural AA-tRNAs
When an AA-tRNA is bound in the A/T site of the ribosome, it is still in ternary complex with EF-Tu:GTP (Voorhees et al. 2010). Eventual incorporation of the AA requires (1) hydrolysis of GTP; (2) release of the esterified CCA end from EF-Tu:GDP; (3) movement of the CCA end into the peptidyl transfer center (PTC) of the large ribosomal subunit (accommodation); and (4) participation in the peptidyl transfer reaction. It has been shown that the total time for these four steps can be significantly prolonged by greatly increased AA-tRNA affinity to EF-Tu:GTP, probably due to extended time of execution of Step 2 above (Schrader et al. 2011; Mittelstaet et al. 2013). In line with this, the longest time monitored for the execution of Steps 2–4 in the present work (τacc,pep = 18 msec) was observed for peptidyl transfer to Phe-tRNAAlaB, which had the highest affinity (Kd = 0.056 µM) to EF-Tu:GTP (Table 3). Shorter τacc,pep estimates were obtained for peptidyl transfer to aG-tRNAAlaB (16 msec) and mS-tRNAAlaB (13 msec), two aminoacyl-tRNAs with somewhat higher Kd estimates for EF-Tu:GTP binding (0.08 µM and 0.15 µM, respectively) than Phe-tRNAAlaB. The smallest τacc,pep estimates were obtained for Phe-tRNAPhe (9 msec) and Ala-tRNAAlaB (5 msec), both with their AAs cognate to their tRNA bodies. An interesting case is that of the bulky bK. In the previous section, we observed that its fast phase amplitude increased greatly when the tRNAPheB body was exchanged for the tRNAAlaB body due to the much higher affinity of EF-Tu:GTP to bK-tRNAAlaB (Kd = 2.8 µM) than to bK-tRNAPheB (Kd = 50 µM). Here, we see that τacc,pep (approximated by τdipmin) for bK-tRNAAlaB was 13 msec, a value slightly larger than the τacc,pep-value of 9 msec for Phe-tRNAPheB and significantly shorter than the τacc,pep-value of 18 msec for Phe-tRNAAlaB (Table 4). This means that the bulky side chain of bK, which caused the low affinity of bK-tRNAAlaB to EF-Tu:GTP compared to that of Phe-tRNAAlaB (Table 3), had but a small effect on the τacc,pep-value. If the latter was dominated by accommodation of tRNA in the A site (Bieling et al. 2006), this would mean that accommodation was virtually unhindered by the bulky bK side chain. If, in contrast, the τacc,pep-value was dominated by the chemistry of peptidyl transfer (Pavlov et al. 2009; Johansson et al. 2011), this would mean that peptide bond formation itself was virtually unobstructed by the very bulky side chain of bK.
CONCLUSION
We have found that the tight tRNAAlaB body significantly improved the incorporation kinetics of (1) unnatural small and bulky AAs by greatly increasing their affinity to EF-Tu:GTP, thereby increasing the fast phase fractions; and (2) N-methyl-Phe by increasing the rate of dipeptide formation by a mechanism yet to be characterized. Our results open a kinetic window to understanding known effects of misacylated tRNA body and unnatural AA in translation and to rational and potentially generalizable improvement of incorporation of unnatural AAs. Our observations suggest that carefully selected unnatural AAs can be used as tools to monitor kinetic steps late into the chain of events leading to peptide bond formation on the translating ribosome.
MATERIALS AND METHODS
Materials
The purified E. coli translation system and N-NVOC-AA-pdCpAs were prepared in our laboratory and characterized according to Ieong et al. (2012) and references therein. 3′CA-truncated tRNAAlaB and tRNAPheB were prepared as described (Zhang et al. 2007). AA-tRNAs were prepared by ligating the N-NVOC-AA-pdCpA derivatives of Phe, Ala, aG, mS, bK, and N-methyl-Phe (Fig. 1A) to unmodified 3′CA-truncated tRNAAlaB (Fig. 1B) or tRNAPheB (only for N-methyl-Phe) using T4 RNA ligase, followed by removal of the NVOC group by photolysis prior to translation experiments as previously described (Ieong et al. 2012). LS3 buffer (Pavlov et al. 2008) (pH was adjusted to 7.5) contains 95 mM KCl, 5 mM NH4Cl, 0.5 mM CaCl2, 8 mM putrescine, 1 mM spermidine, 30 mM HEPES, 1 mM dithioerythritol, 2 mM phosphoenolpyruvate (PEP), 5 mM Mg(OAc)2, 1 mM ATP, and 1 mM GTP, supplemented with 1 µg/mL pyruvate kinase and 0.1 µg/mL myokinase for energy regeneration. LS3A buffer (Pavlov et al. 2009) was identical to LS3 buffer except containing 2 mM ATP and lacking GTP.
Kinetics measurements
All kinetics assays were performed in a temperature-controlled quench-flow apparatus (RQF-3; KinTeck Corp.), where the ribosome mixture and ternary complex mixture were rapidly mixed and stopped by quenching with 50% formic acid (17% final concentration) at different incubation times. Compositions of the ribosome mixture and ternary complex mixture are given below for different types of measurements. The rate of dipeptide formation was determined by the concentration of ribosome complexes, which was always in excess over ternary complexes. Unless specified otherwise, all experiments were at 37°C.
Dipeptide synthesis at different EF-Tu concentrations
Experiments were performed in LS3 buffer as previously described (Ieong et al. 2012) for AA-tRNAPheB. For dipeptide synthesis with AA-tRNAAlaB, mRNA encoding fMet-Ala-Phe-Stop (codons AUGGCAUUCUAA) was used in the ribosome mixture. EF-Tu was present at equal concentrations in both mixtures at the concentration indicated in each experiment and was in excess over AA-tRNA. Thus, the concentration of ternary complexes was limited by the concentration of AA-tRNA (Ieong et al. 2012). For each dipeptide reaction, time points were taken until the yield of dipeptide formed was saturated (e.g., up to 100 sec for non-N-akyl-AA or 1 h for N-akyl-AA). Yields at saturation (within 20% experimental error) always corresponded to the amount of active AA-tRNA input to the reaction, which was limiting. The active concentration of the AA-tRNA was determined by O.D.260nm multiplied by the ligation yield. Ligation yields were at least 50% of the total tRNAAlaB based on urea polyacrylamide gel electrophoresis at pH 5.
Simultaneous measurement of GTP hydrolysis and dipeptide synthesis
In this paragraph, final concentrations are given after mixing equal volumes of ribosome mixture and ternary complex mixture. The ribosome mixture was prepared by incubating 1 µM 70S ribosomes, 1.5 µM IF1, 0.5 µM IF2, 1.5 µM IF3, 2 µM mRNA, and 1.2 µM f[3H]Met-tRNAifMet in buffer LS3 for 15 min at 37°C. The ternary complex mixture, containing 0.7 µM EF-Tu and 0.7 µM [3H]GDP in LS3A buffer, was first preincubated for 15 min at 37°C (to convert GDP to GTP on EF-Tu and in solution); then, 0.3 µM AA-tRNAAlaB was added to the mixture, and the incubation continued for another 15 min. For natural Phe-tRNAPhe, the ternary complex mixture containing 0.3 µM EF-Tu and 0.3 µM [3H]GDP in LS3A buffer was first preincubated for 15 min at 37°C; then, 0.2 mM phenylalanine, 0.1 unit/µL PheRS, and 1.5 µM E. coli tRNAPhe was added to the mixture, and the incubation continued for another 15 min.
Ribosome titration experiments
The ribosome mixture was prepared by incubating 70S ribosomes (variable concentrations), IF1 (1.5× ribosome concentration), IF2 (0.5× ribosome concentration), IF3 (1.5× ribosome concentration), mRNA (2× ribosome concentration), and f[3H]Met-tRNAifMet (1.2× ribosome concentration) in buffer LS3 for 15 min at 37°C. The ternary complex mixture was prepared in LS3 buffer as previously described (Ieong et al. 2012), in which 10 µM EF-Tu (concentration in ternary complex mixture before translation reaction) was used to ensure a high fraction of preformed ternary complex. Here, only the fast phase was measured, and it showed the rapid peptide bond formation on the ribosome.
Analysis of kinetics measurements
The samples quenched at different time points in the quench-flow apparatus were first centrifuged at 20,000g for 15 min. For analysis of dipeptide synthesis, the extent of dipeptide formation in the pellets was analyzed by RP-HPLC as described (Ieong et al. 2012). For analysis of GTP hydrolysis, the [3H]GDP and [3H]GTP in the supernatants were analyzed by MonoQ HPLC as described (Pavlov et al. 2009). The data were analyzed by the nonlinear regression program Origin 7.5 (OriginLab Corp.). The rates and fractions for the fast phase (kfast and Afast) and slow phase (kslow and Aslow) in dipeptide formation at different EF-Tu concentration, and Kd-values for AA-tRNA binding to EF-Tu:GTP were estimated as described (Ieong et al. 2012). The biphasic kinetics can be described as
where dip(t) is the normalized amount of dipeptide formed. τGTP was estimated by fitting the data of GTP hydrolysis to a single-step exponential model, and τdip was estimated by fitting the data of dipeptide formation to a two-step kinetic model (Johansson et al. 2008), τacc,pep = τdip − τGTP. For ribosome titration experiments, the average times of dipeptide formation (τdip) were plotted versus the inverse of ribosome concentration (Lineweaver-Burke plot) and fitted by linear regression; τdipmin-values for dipeptide formation were obtained by the y-axis intercept.
ACKNOWLEDGMENTS
This work was supported by grants from the Swedish Research Council (project grants and Linnaeus Uppsala RNA Research Centre to M.E. and A.C.F.) and the Knut and Alice Wallenberg Foundation (to M.E.).
Footnotes
Freely available online through the RNA Open Access option.
REFERENCES
- Asahara H, Uhlenbeck OC 2002. The tRNA specificity of Thermus thermophilus EF-Tu. Proc Natl Acad Sci 99: 3499–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain JD, Glabe CG, Dix TA, Chamberlin AR, Diala ES 1989. Biosynthetic site-specific incorporation of a non-natural amino acid into a polypeptide. J Am Chem Soc 111: 8013–8014 [Google Scholar]
- Bieling P, Beringer M, Adio S, Rodnina MV 2006. Peptide bond formation does not involve acid-base catalysis by ribosomal residues. Nat Struct Mol Biol 13: 423–428 [DOI] [PubMed] [Google Scholar]
- Cload ST, Liu DR, Froland WA, Schultz PG 1996. Development of improved tRNAs for in vitro biosynthesis of proteins containing unnatural amino acids. Chem Biol 3: 1033–1038 [DOI] [PubMed] [Google Scholar]
- Doi Y, Ohtsuki T, Shimizu Y, Ueda T, Sisido M 2007. Elongation factor Tu mutants expand amino acid tolerance of protein biosynthesis system. J Am Chem Soc 129: 14458–14462 [DOI] [PubMed] [Google Scholar]
- Forster AC 2009. Low modularity of aminoacyl-tRNA substrates in polymerization by the ribosome. Nucleic Acids Res 37: 3747–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster AC, Tan Z, Nalam MNL, Lin H, Qu H, Cornish VW, Blacklow SC 2003. Programming peptidomimetic syntheses by translating genetic codes designed de novo. Proc Natl Acad Sci 100: 6353–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Wieden HJ, Rodnina MV 2002. Kinetic mechanism of elongation factor Ts-catalyzed nucleotide exchange in elongation factor Tu. Biochemistry 41: 162–169 [DOI] [PubMed] [Google Scholar]
- Guillen Schlippe YV, Hartman MC, Josephson K, Szostak JW 2012. In vitro selection of highly modified cyclic peptides that act as tight binding inhibitors. J Am Chem Soc 134: 10469–10477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht SM, Alford BL, Kuroda Y, Kitano S 1978. “Chemical aminoacylation” of tRNA's. J Biol Chem 253: 4517–4520 [PubMed] [Google Scholar]
- Ieong KW, Pavlov MY, Kwiatkowski M, Forster AC, Ehrenberg M 2012. Inefficient delivery but fast peptide bond formation of unnatural L-aminoacyl-tRNAs in translation. J Am Chem Soc 134: 17955–17962 [DOI] [PubMed] [Google Scholar]
- Johansson M, Bouakaz E, Lovmar M, Ehrenberg M 2008. The kinetics of ribosomal peptidyl transfer revisited. Mol Cell 30: 589–598 [DOI] [PubMed] [Google Scholar]
- Johansson M, Ieong KW, Trobro S, Strazewski P, Åqvist J, Pavlov MY, Ehrenberg M 2011. pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc Natl Acad Sci 108: 79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRiviere FJ, Wolfson AD, Uhlenbeck OC 2001. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 294: 165–168 [DOI] [PubMed] [Google Scholar]
- Ling J, So BR, Yadavalli SS, Roy H, Shoji S, Fredrick K, Musier-Forsyth K, Ibba M 2009. Resampling and editing of mischarged tRNA prior to translation elongation. Mol Cell 33: 654–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelstaet J, Konevega AL, Rodnina MV 2013. A kinetic safety gate controlling the delivery of unnatural amino acids to the ribosome. J Am Chem Soc 135: 17031–17038 [DOI] [PubMed] [Google Scholar]
- Murakami H, Ohta A, Ashigai H, Suga H 2006. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat Methods 3: 357–359 [DOI] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV 1998. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J 17: 7490–7497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Hohn MJ, Umehara T, Guo LT, Osborne EM, Benner J, Noren CJ, Rinehart J, Soll D 2011. Expanding the genetic code of Escherichia coli with phosphoserine. Science 333: 1151–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov MY, Antoun A, Lovmar M, Ehrenberg M 2008. Complementary roles of initiation factor 1 and ribosome recycling factor in 70S ribosome splitting. EMBO J 27: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC 2009. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci 106: 50–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SA, Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG 1989. The use of 5′-phospho-2 deoxyribocytidylylriboadenosine as a facile route to chemical aminoacylation of tRNA. Nucleic Acids Res 17: 9649–9660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy H, Becker HD, Mazauric MH, Kern D 2007. Structural elements defining elongation factor Tu mediated suppression of codon ambiguity. Nucleic Acids Res 35: 3420–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader JM, Chapman SJ, Uhlenbeck OC 2009. Understanding the sequence specificity of tRNA binding to elongation factor Tu using tRNA mutagenesis. J Mol Biol 386: 1255–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader JM, Chapman SJ, Uhlenbeck OC 2011. Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. Proc Natl Acad Sci 108: 5215–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Hartman MC, Szostak JW 2011. Optimal codon choice can improve the efficiency and fidelity of N-methyl amino acid incorporation into peptides by in-vitro translation. Angew Chem Int Ed Engl 50: 3164–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V 2010. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330: 835–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts RE, Forster AC 2012. Update on pure translation display with unnatural amino acid incorporation. Methods Mol Biol 805: 349–365 [DOI] [PubMed] [Google Scholar]
- Xie J, Schultz PG 2005. An expanding genetic code. Methods 36: 227–238 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y, Shoji I, Miyagawa S, Kawakami T, Katoh T, Goto Y, Suga H 2011. Natural product-like macrocyclic N-methyl-peptide inhibitors against a ubiquitin ligase uncovered from a ribosome-expressed de novo library. Chem Biol 18: 1562–1570 [DOI] [PubMed] [Google Scholar]
- Zhang B, Tan Z, Dickson LG, Nalam MNL, Cornish VW, Forster AC 2007. Specificity of translation for N-alkyl amino acids. J Am Chem Soc 129: 11316–11317 [DOI] [PMC free article] [PubMed] [Google Scholar]