This paper examines the function of the mammalian protein ZC3H14, which is a zinc finger polyadenosine binding protein that may be orthologous to the yeast protein Nab2. Studies in a neuronal cell line demonstrate that the protein controls poly(A) tail length. Furthermore, complementation analyses in Drosophila indicate that ZC3H14 plays an evolutionarily conserved role in controlling poly(A) tail length in neurons.
Keywords: ZC3H14/dNab2/Nab2, polyadenosine RNA-binding protein, poly(A) tail, intellectual disability
Abstract
The ZC3H14 gene, which encodes a ubiquitously expressed, evolutionarily conserved, nuclear, zinc finger polyadenosine RNA-binding protein, was recently linked to autosomal recessive, nonsyndromic intellectual disability. Although studies have been carried out to examine the function of putative orthologs of ZC3H14 in Saccharomyces cerevisiae, where the protein is termed Nab2, and Drosophila, where the protein has been designated dNab2, little is known about the function of mammalian ZC3H14. Work from both budding yeast and flies implicates Nab2/dNab2 in poly(A) tail length control, while a role in poly(A) RNA export from the nucleus has been reported only for budding yeast. Here we provide the first functional characterization of ZC3H14. Analysis of ZC3H14 function in a neuronal cell line as well as in vivo complementation studies in a Drosophila model identify a role for ZC3H14 in proper control of poly(A) tail length in neuronal cells. Furthermore, we show here that human ZC3H14 can functionally substitute for dNab2 in fly neurons and can rescue defects in development and locomotion that are present in dNab2 null flies. These rescue experiments provide evidence that this zinc finger-containing class of nuclear polyadenosine RNA-binding proteins plays an evolutionarily conserved role in controlling the length of the poly(A) tail in neurons.
INTRODUCTION
Post-transcriptional regulation of gene expression is critical for determining the spatial and temporal gene expression pattern of a cell. Such regulation is particularly critical in neurons where RNA trafficking and local translation play key roles in neuronal function (Sinnamon and Czaplinski 2011; Goldie and Cairns 2012). This post-transcriptional regulation of gene expression is mediated by numerous RNA-binding proteins. The functional importance of these RNA-binding proteins is highlighted by the growing number of proteins in this family that have been linked to human diseases (Cooper et al. 2009).
One key class of RNA-binding proteins, called polyadenosine RNA-binding proteins (Pabs), bind specifically to the poly(A) tails of mRNA transcripts and play important roles in the post-transcriptional control of gene expression (Eckmann et al. 2011). Previous studies demonstrate that Pabs can modulate transcript stability, export from the nucleus, and translation, highlighting their functions as key regulators of gene expression (Mangus et al. 2003). Recently, mutations in the human ZC3H14 gene, which encodes a zinc finger polyadenosine RNA-binding protein (Leung et al. 2009), were linked to inherited, nonsyndromic, intellectual disability revealing the critical role of this Pab protein in the brain (Pak et al. 2011; Kelly et al. 2012). The ZC3H14 protein (also termed mSUT-2) (Guthrie et al. 2009, 2011), belongs to an evolutionarily conserved, ubiquitously expressed family of Cys3His tandem zinc-finger (ZnF) polyadenosine (poly(A)) RNA-binding proteins (Anderson et al. 1993; Hector et al. 2002; Kelly et al. 2007; Leung et al. 2009; Pak et al. 2011). The mammalian ZC3H14 locus encodes three long protein isoforms (isoforms 1–3) that at steady state localize to the nucleus and one short isoform (isoform 4) that localizes to the cytoplasm (Leung et al. 2009; Kelly et al. 2012). Isoforms 1–3 are ubiquitously expressed and share a common domain structure (Fig. 1A) with both Drosophila melanogaster Nab2 (dNab2) and Saccharomyces cerevisiae Nab2, while the shorter ZC3H14 isoform 4, which is highly expressed only in testes, has a unique first exon but retains the C-terminal tandem ZnF RNA-binding domain (Leung et al. 2009). Interestingly, one of the ZC3H14 mutations in intellectually disabled patients (R154->termination codon) leads to loss of ZC3H14 isoforms 1–3 but leaves isoform 4 intact (Pak et al. 2011), suggesting that these longer isoforms are necessary for cognitive function in humans, while isoform 4 may perform a distinct function.
FIGURE 1.
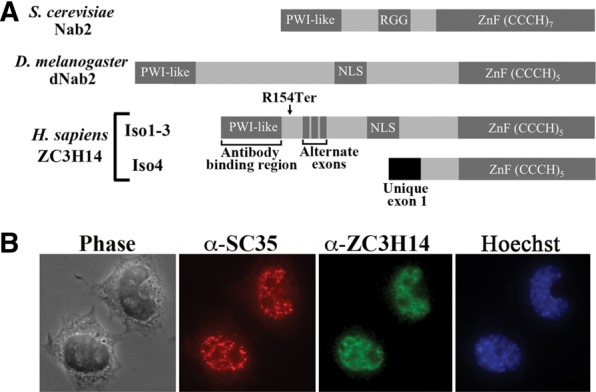
Human ZC3H14 isoforms 1–3 are localized to the nucleus of N2A cells. (A) Domain alignment of S. cerevisiae Nab2, D. melanogaster dNab2, and human ZC3H14 isoforms 1–3 and isoform 4. The conserved N-terminal PWI-like fold, Q-rich, RGG/predicted classical nuclear localization signal (cNLS), and C-terminal tandem CCCH zinc finger RNA-binding motif (ZnF CCCH) domains are indicated. The zinc finger domains of human ZC3H14 and fly dNab2 share significant homology with 48% amino acid similarity in the zinc fingers as determined by analysis with Clustal W (Larkin et al. 2007). The location of the Arginine-to-termination codon (R154Ter) mutation that eliminates isoforms 1–3 in intellectual disability patients is shown as is the location for the Alternate exons that are included/excluded to generate ZC3H14 isoforms 1–3. The PWI-like domain used to raise polyclonal ZC3H14 antibodies, which recognize ZC3H14-iso1-3, (Leung et al. 2009) is also indicated (Antibody binding region). (B) ZC3H14 localization in cultured N2A cells was assessed using an antibody directed against the PWI-like domain of ZC3H14 (α-ZC3H14) and thus isoforms 1–3 are visualized (green). Cells are costained with the RNA speckle marker SC-35 (α-SC-35) to indicate the position of nuclear speckles (red). Corresponding Hoescht staining (blue) is shown to indicate the position of the nucleus. A phase image is also shown.
Although little is known about the molecular function of human ZC3H14, both the S. cerevisiae and D. melanogaster presumptive orthologs of ZC3H14 bind polyadenosine RNA and are required to restrict poly(A) tail length in vivo (Anderson et al. 1993; Hector et al. 2002; Pak et al. 2011). As with ZC3H14 isoforms 1–3, the budding yeast and fly Nab2 proteins both localize to the nucleus at steady state (Anderson et al. 1993; Pak et al. 2011). Genetic studies reveal interactions between Nab2 and genes encoding components of the nuclear polyadenylation machinery (Kelly et al. 2010; Pak et al. 2011) suggesting that one principal Nab2 function is control of polyadenylation. Consistent with a critical role for Nab2 in post-transcriptional regulation, S. cerevisiae and D. melanogaster Nab2 are essential for cell viability and development, respectively (Anderson et al. 1993; Pak et al. 2011).
Our previous work demonstrating a requirement for D. melanogaster Nab2 in normal locomotor behavior (Pak et al. 2011) led to the hypothesis that neurons are exquisitely sensitive to defects in nuclear poly(A) tail length control. The subsequent findings that neuron-specific expression of wild-type dNab2 rescues dNab2 null developmental and locomotor defects, and that neuron-specific depletion of dNab2 in otherwise wild-type animals can elicit similar defects, confirmed this hypothesis (Pak et al. 2011). These studies, coupled with the finding that humans with ZC3H14 mutations display nonsyndromic intellectual disability (Pak et al. 2011), suggest that ZC3H14 and dNab2 may perform functionally analogous roles in RNA metabolism in invertebrate and vertebrate neurons. However, no studies have been performed previously to assess the molecular function of ZC3H14.
Here we demonstrate that depletion of ZC3H14 leads to extended poly(A) tails in a cultured mouse neuronal cell line without significant accumulation of poly(A) RNA in the nucleus, supporting a model in which ZC3H14 plays a primary role in control of poly(A) tail length in mammalian cells. To extend these molecular results and assess the function of ZC3H14 in neurons in vivo, we exploit Drosophila mutant for Nab2. Our results show that neuron-specific transgenic expression of a nuclear isoform of human ZC3H14, but not the shorter, cytoplasmic isoform retained in human ZC3H14-associated intellectual disability patients (Pak et al. 2011), rescues developmental and behavioral phenotypes of Nab2 null D. melanogaster. Critically, this overt rescue by a nuclear isoform of ZC3H14 is accompanied by repair of an underlying defect in poly(A) tail length in the fly brain. These data indicate that human ZC3H14 is a functional ortholog of fly Nab2 that plays a critical role in the control of poly(A) tail length of neuronal RNAs in vivo.
RESULTS
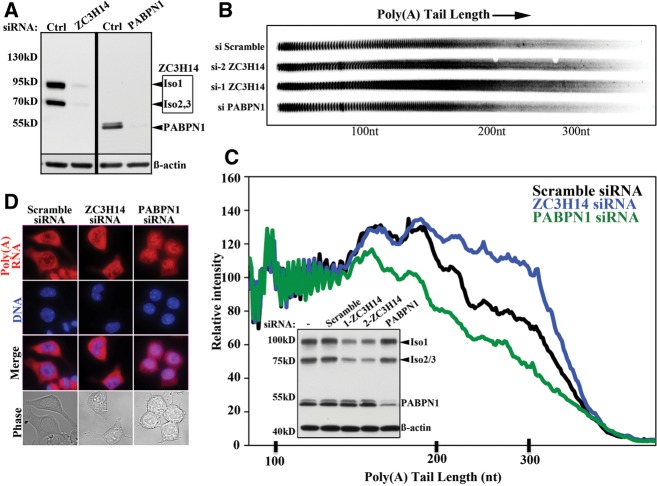
To examine the molecular function of ZC3H14, we utilized neuro 2A (N2A) cells (Olmsted et al. 1970). Consistent with previous results (Leung et al. 2009; Guthrie et al. 2011), ZC3H14 isoforms 1–3 (Fig. 1A) are primarily localized to the nucleus in this neuronal cell line (Fig. 1B). To assess the functional consequences of loss of ZC3H14, we used siRNA-mediated depletion. The siRNAs targeted to ZC3H14 were designed to deplete all transcript variants. As a control for a protein implicated in regulation of poly(A) tail length, we also examined the consequences of siRNA-mediated depletion of the nuclear poly(A)-binding protein, PABPN1. PABPN1 modulates the processivity of poly(A) polymerase to achieve optimal poly(A) tail length (Kuhn and Wahle 2004) and previous work has shown that depletion of PABPN1 leads to shortening of bulk poly(A) tails (Apponi et al. 2010). As shown in Figure 2A, both ZC3H14-iso1-3 and PABPN1 could be efficiently depleted from N2A cells. We then examined bulk poly(A) tail length in N2A cells treated with either of two independent ZC3H14 siRNAs, PABPN1 siRNA, or control scrambled siRNA (Fig. 2B,C). Depletion of ZC3H14 resulted in an increase in bulk poly(A) tail length compared with cells treated with control siRNA (Fig. 2B,C). As expected, depletion of PABPN1 led to shortened poly(A) tails (Apponi et al. 2010). Similar results were obtained with both siRNAs directed against ZC3H14 ensuring that results obtained were not due to off-target effects of the siRNA used. As the S. cerevisiae Nab2 protein has been implicated in both control of poly(A) tail length and poly(A) RNA export from the nucleus (Hector et al. 2002), in parallel with the bulk poly(A) tail length assays, we used fluorescence in situ hybridization (FISH) to assess the localization of bulk poly(A) RNA in cells depleted of ZC3H14. Consistent with what has been reported for dNab2 mutant flies (Pak et al. 2011), little or no accumulation of poly(A) was detected in the nuclei of cells treated with ZC3H14 siRNA (Fig. 2D). In contrast, as expected (Apponi et al. 2010), PABPN1 depletion led to significant nuclear accumulation of poly(A) RNA. These results provide molecular evidence that ZC3H14 plays a role in poly(A) tail length control but do not directly address the function of ZC3H14 in vivo.
FIGURE 2.
ZC3H14 is required for proper poly(A) tail length control. (A) An immunoblot shows that both ZC3H14 (isoforms 1–3) and a control polyadenosine RNA-binding protein, PABPN1, are robustly depleted from N2A cells by treatment with specific siRNA with no effect of a scrambled control RNA (Ctrl). ZC3H14-iso4 is not detected as the antibody was raised to the N-terminal PWI-like domain not present in isoform 4 (see Fig. 1A). The double band observed for PABPN1 is likely due to the presence of phosphorylated PABPN1. The corresponding β-actin band is shown as a loading control. (B) ZC3H14 was depleted from N2A cells using two independent siRNAs (C, inset). As controls, cells were also treated with PABPN1 siRNA to deplete PABPN1 or scrambled control siRNA (siScramble). Total RNA samples were prepared and subjected to bulk poly(A) tail length analysis as described in Materials and Methods. A gel of the resolved bulk poly(A) samples is shown. Approximate positions of nucleotide size markers (nt) are indicated. (C) To provide a measure of poly(A) tail length, the length of each lane of an independent poly(A) tail length assay was scanned for N2A samples treated with Control scramble siRNA, ZC3H14 siRNA, or PABPN1 siRNA as indicated. The relative intensity of the signal is plotted vs. poly(A) tail length as determined by size markers. The positions on the scan corresponding to 100, 200, and 300 nt are indicated. (Inset) An immunoblot shows the level of ZC3H14 (isoforms 1–3) knockdown obtained with two independent siRNAs for this experiment. The knockdown obtained for the control polyadenosine RNA-binding protein, PABPN1, is also shown. No treatment (−) or a control Scramble siRNA has no effect on levels of ZC3H14 or PABPN1. β-actin serves as a loading control. (D) The siRNA-treated samples shown in the inset in C were used for fluorescence in situ hybridization to assess poly(A) RNA localization. Total poly(A) RNA is visualized with an oligo dT probe. The corresponding DAPI and phase images are shown. Representative cell fields are shown. Results are typical of more than 50 cells analyzed for each siRNA where the distribution of poly(A) RNA in cells treated with ZC3H14 siRNA was indistinguishable from cells treated with scramble control siRNA.
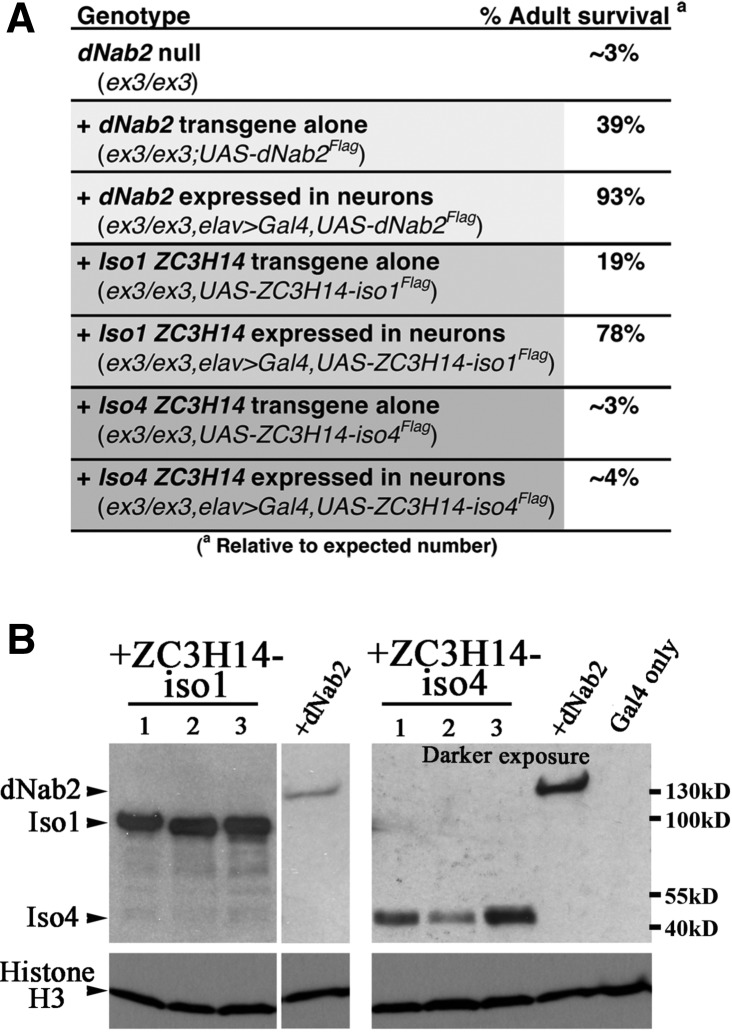
Given that patients with mutations that eliminate the nuclear forms of ZC3H14 display intellectual disability without other reported symptoms (Pak et al. 2011; Kelly et al. 2012), we sought to assess the function of ZC3H14 in neurons. For these studies, we exploited a Drosophila model that is null for Nab2 and shows neuronal-specific defects in both development and behavior (Pak et al. 2011). As shown in Figure 1A, the sole isoform of Drosophila Nab2 (dNab2), which is localized to the nucleus (Pak et al. 2011), shares a similar domain structure with the nuclear isoforms of ZC3H14, isoforms 1–3. To directly assess whether H. sapiens ZC3H14 and D. melanogaster Nab2 are functional orthologs, we used tissue-specific transgenic expression. Multiple transgenes encoding either human ZC3H14 isoform 1 (UAS-ZC3H14-iso1Flag), which is localized to the nucleus (Leung et al. 2009), or human ZC3H14 isoform 4 (UAS-ZC3H14-iso4Flag), which is localized to the cytoplasm (Leung et al. 2009), were tested for the ability to rescue developmental and behavioral defects in flies homozygous for the protein and mRNA null allele dNab2ex3 (hereafter referred to as dNab2null) (Pak et al. 2011). Only a small percentage (∼3%) of homozygous dNab2null flies eclose as adults (Fig. 3A; Pak et al. 2011). Transgenic expression of ZC3H14-iso1 specifically in D. melanogaster neurons using a pan-neuronal driver (elav-Gal4,UAS-ZC3H14-Iso1Flag) rescues the eclosion rate of these dNab2null animals from ∼3% to ∼78% (Fig. 3A). This rescue by ZC3H14-iso1 is comparable to the rescue conferred by transgenic, neuronal re-expression of dNab2, which restores the eclosion rate to ∼93% (Fig. 3A). In contrast, pan-neuronal expression of UAS-ZC3H14-iso4Flag had no ameliorating effect on eclosion rates of dNab2null animals (elav-GAL4/+;UAS-ZC3H14-iso4Flag/+;dNab2ex3/dNab2ex3) (Fig. 3A). Immunoblot analysis confirms expression of Flag-tagged dNab2 and ZC3H14 isoforms in head lysates of transgenic animals (Fig. 3B); however, the relative level of expression of ZC3H14-iso4 attained was always significantly lower than ZC3H14-iso1 even in several independent transgenic lines. Taken together, these results provide evidence that ZC3H14-iso1 may substitute for dNab2 in key developmental functions.
FIGURE 3.
Neuron-specific expression of ZC3H14-iso1 can rescue viability defects of homozygous dNab2nullflies. (A) The percentage of adult flies eclosed (relative to expected Mendelian ratios) for each of the indicated genotypes is summarized. Genotypes are as follows: dNab2 null (dNab2ex3/dNab2ex3), +dNab2 transgene alone (UAS-dNab2Flag/+;;dNab2ex3/dNab2ex3), +dNab2 expressed in neurons (UAS-dNab2Flag/elav-GAL4;;dNab2ex3/dNab2ex3), +ZC3H14-iso1 transgene alone (UAS-ZC3H14-iso1Flag/+;;dNab2ex3/dNab2ex3), +ZC3H14-iso1 expressed in neurons (UAS-ZC3H14-iso1Flag/elav-GAL4;;dNab2ex3/dNab2ex3), +ZC3H14-iso4 transgene alone (+/+ or +/Y;UAS-ZC3H14-iso4Flag/+;;dNab2ex3/dNab2ex3), +ZC3H14-iso4 expressed in neurons (elav-GAL4/+ or elav-GAL4/Y;UAS-ZC3H14-iso4Flag/+;dNab2ex3/dNab2ex3). (B) Anti-Flag immunoblots to detect pan-neuronal (using elav-GAL4) expression of the UAS-dNab2Flag, UAS-ZC3H14-iso1Flag, and UAS-ZC3H14-iso4Flag transgenes in fly heads. The blot on the right corresponding to ZC3H14 iso4 is a darker exposure (as indicated) due to lower-level expression of ZC3H14 iso4 relative to ZC3H14 iso1. The dNab2 band, which is the same band but shown at the different exposures, for both ZC3H14-iso1 and ZC3H14-iso4, illustrates the relative levels of exposure. Control flies expressing Gal4 only are also shown. The approximate position of molecular weight markers is indicated to the right.
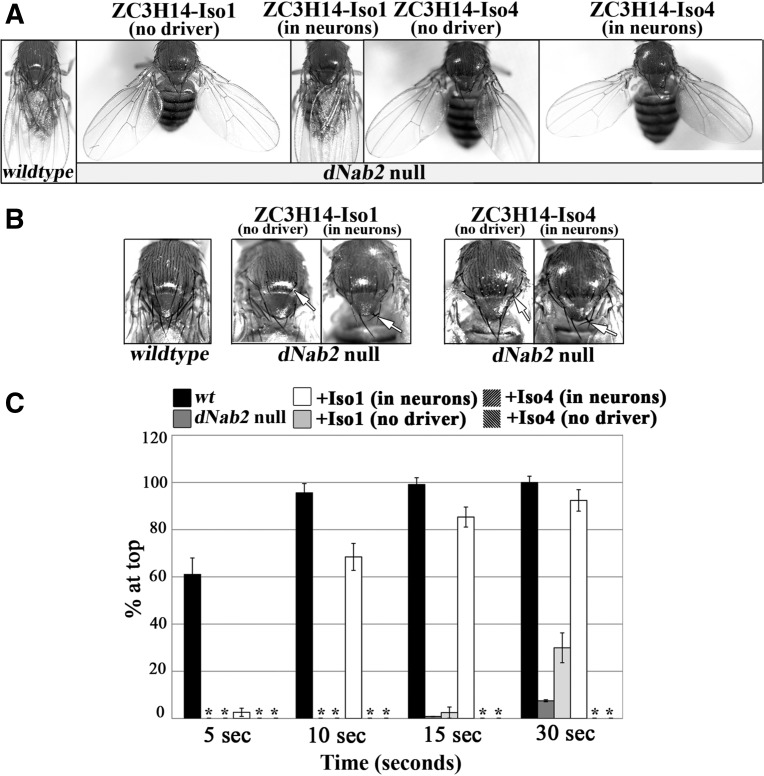
While 97% of dNab2null homozygotes die as pharate adults, the 3% that eclose display a variety of organismal phenotypes including impaired locomotor behavior, fully penetrant kinking of thoracic bristles, and a wings-held-out posture (Fig. 4; Pak et al. 2011). Importantly, neuronal expression of H. sapiens ZC3H14-iso1 restores normal wing posture (Fig. 4A) to a degree comparable to that achieved by the transgenic re-expression of wild-type dNab2 (Fig. 4A; Pak et al. 2011). Thoracic bristle kinking is not rescued by pan-neuronal expression of either wild-type dNab2 or ZC3H14-iso1 (Fig. 4B, white arrows), indicating a non-neuronal role for dNab2/ZC3H14 in bristle development. Two additional ZC3H14-iso1 transgenes displayed similar degrees of rescue when expressed in fly neurons (data not shown), indicating that rescuing activity is linked to expression of ZC3H14 and not the site of transgene insertion. Consistent with the failure of ZC3H14-iso4 to rescue eclosion, three separate transgenic lines expressing this isoform showed no rescue of wing posture when expressed in neurons (Fig. 4A).
FIGURE 4.
Neuron-specific expression of ZC3H14-iso1 but not ZC3H14-iso4 is sufficient to rescue organismal and functional defects of dNab2null flies. (A) Normal wing posture is shown in control (denoted “wildtype”) adult flies. dNab2 null mutants with UAS-ZC3H14 transgenes but lacking a Gal4 driver (UAS-ZC3H14-iso1Flag/+;;dNab2ex3/dNab2ex3, or UAS-ZC3H14-iso4Flag/+;;dNab2ex3/dNab2ex3 denoted as “no driver”) display a fully penetrant wings-held-out phenotype, which is rescued in dNab2 null flies transgenically expressing ZC3H14-iso1 in all neurons (elav-GAL4/UAS-ZC3H14-iso1Flag;;dNab2ex3/dNab2ex3, denoted as “in neurons”) but not in dNab2null flies expressing ZC3H14-iso4 in all neurons (elav-Gal4/UAS-ZC3H14-iso4Flag;;dNab2ex3/dNab2ex3, denoted as “in neurons”). (B) Thoracic bristle kinking in dNab2nulladults is not rescued by neuronal expression of ZC3H14 isoforms. Dorsal views of the thorax are shown. Genotypes are the same as listed in A. Wild-type (wildtype) flies have straight extended bristles, but dNab2null flies containing UAS-ZC3H14-iso1 (ZC3H14-Iso1) or UAS-ZC3H14-iso4 (ZC3H14-Iso4) with (in neurons) or without (no driver) the neuronal elav-Gal4 driver still show bristle kinking (denoted by white arrows). Thus, neuron-specific expression of ZC3H14-iso1 or ZC3H14-iso4 does not rescue bristle kinking. (C) A negative geotaxis assay (Ganetzky and Flanagan 1978) was used to assess locomotor activity in flies with the following genotypes (wt=+/+, dNab2 null=ex3/ex3, “+iso1 [in neurons]”= elav-Gal4/UAS-iso1;;ex3/ex3, “+iso1 [no driver]”= UAS-iso1/+;;ex3/ex3, “+iso4 [in neurons]”= elav-Gal4/UAS-iso4;;ex3/ex3, “+iso4 [no driver]”= UAS-iso4/+;;ex3/ex3). Data are presented as the average percentage of flies reaching the top of a cylinder (% at top) at indicated time points across all trials. At least 10 independent trials each containing 10 flies were tested for all genotypes, except ZC3H14-iso4 expressing flies in which two groups of 10 flies were analyzed due to the low frequency of adults obtained for this genotype. Error bars, SEM. Asterisks denote genotypes in which no flies reached the top of the vial within the parameters of the assay.
Impaired locomotion of Nab2null animals in a negative geotaxis assay (Ganetzky and Flanagan 1978) can be rescued by re-expressing wild-type dNab2 specifically in neurons (Pak et al. 2011). This paradigm provides an opportunity to assess the functional equivalence of fly Nab2 and human ZC3H14-iso1 in supporting a complex behavior. dNab2null flies (Fig. 4C, dark-gray bars) show severely reduced locomotion compared with wild-type control flies (Fig. 4C, black bars). Neuron-specific expression of ZC3H14-iso1 (Fig. 4C, white bars) significantly rescued this behavioral defect. A low level of rescue was also observed in the absence of the GAL4 driver (Fig. 4C, light-gray bars) likely due to leaky transgene expression (Pak et al. 2011). As with eclosion rates and organismal phenotypes, the ZC3H14-iso4 transgene failed to rescue the dNab2null locomotor phenotype either in the absence or presence of the pan-neuronal GAL4 driver (Fig. 4C). Specific rescue of the locomotor deficit by ZC3H14-iso1 further supports the hypothesis that D. melanogaster Nab2 and human ZC3H14-iso1 are functional orthologs at a step of RNA metabolism that is critical for nervous system function.
If ectopic lengthening of poly(A) tails in neurons underlies the functional defects in dNab2null flies, then rescue of functional defects should be accompanied by rescue of this molecular phenotype. To test whether rescue of dNab2null phenotypes by human ZC3H14-iso1 is accompanied by restoration of proper RNA poly(A) tail length control, bulk poly(A) tail length was compared among control wild-type flies, dNab2null animals, and dNab2null animals with wild-type dNab2 or human ZC3H14-iso1 transgenically expressed specifically in neurons (Fig. 5). To enrich for neuronal RNAs, samples were harvested from a pool of ∼250–300 fly heads per genotype and analyzed in triplicate. As reported previously (Pak et al. 2011), dNab2 mutant flies show extended poly(A) tails among a subset of the total pool of RNAs. Expression of either dNab2 or H. sapiens ZC3H14-iso1 specifically in neurons conferred significant and comparable rescue of this poly(A) tail-length defect (Fig. 5), indicating that ZC3H14 can replace dNab2 in the molecular machinery that restricts poly(A) length in D. melanogaster CNS neurons.
FIGURE 5.
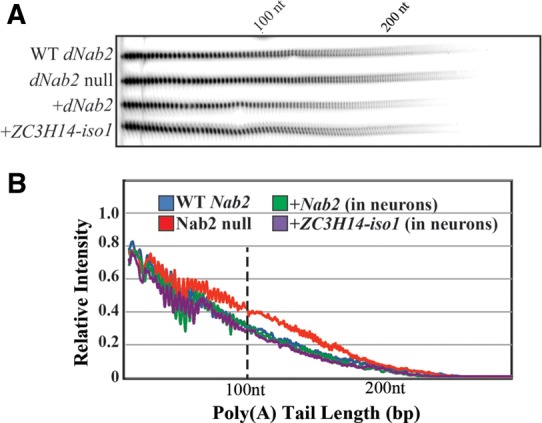
Neuronal expression of ZC3H14-iso1 rescues the poly(A) tail length defect of homozygous dNab2null flies. (A) Total RNA was isolated from flies of the indicated genotypes (“WT dNab2”=control, “dNab2 null”=ex3/ex3, “+dNab2” or +“ZC3H14-iso1” = the corresponding protein expressed in dNab2null neurons with elav-Gal4), end-labeled with [32P]pCp, and non-poly(A) tracts were digested with RNAses A and T1. Labeled RNA was processed as described in Materials and Methods. The position of a 100-nt marker is indicated on the gel. (B) Bulk poly(A) tail length from fly heads of the indicated genotypes corresponding to the samples shown in the gel in A was analyzed by densitometric quantification of poly(A) tracts using ImageJ and plotted as normalized signal intensity (Relative Intensity) as a function of Poly(A) Tail Length in base pairs using Microsoft Excel. The approximate positions of the nucleotide size markers (nt) are indicated.
DISCUSSION
Here we present the first functional characterization of the mammalian polyadenosine RNA-binding protein, ZC3H14. The data couple evidence from a cultured neuronal cell line with functional studies exploiting a Drosophila model and support a role for ZC3H14 in modulating poly(A) tail length. Significantly, we also find evidence that members of this class of zinc finger polyadenosine RNA-binding proteins are functional orthologs in metazoan neurons. Results presented here reveal that the ZC3H14 protein, which is lost in cases of human intellectual disability (Pak et al. 2011), is required for proper control of poly(A) tail length. Consistent with the observation that human patients lacking ZC3H14 display brain-specific defects, expression of human ZC3H14 solely in the neurons of dNab2 mutant flies is sufficient to rescue both behavioral defects and an underlying molecular defect in poly(A) tail length control. These data identify a conserved cell type, neurons, and a corresponding molecular process, poly(A) tail length control, which are specifically affected by ZC3H14 loss. A key question that remains is to understand the functional consequences of extended poly(A) tails on neuronal development and function, as well as whether the observed neuronal phenotypes are due to changes in specific RNAs or classes of RNAs.
To assess the function of human ZC3H14 in vivo, we have taken advantage of a Drosophila model in which functional rescue conferred by tissue-specific expression can be readily assessed. D. melanogaster has been utilized to identify conserved genes and signaling pathways that function in a variety of human diseases ranging from cancer (Pan 2010) to muscular dystrophy (Chartier et al. 2006; Lloyd and Taylor 2010) and has proven particularly useful in defining the genetic requirements of higher cognitive function (Narro et al. 2007). Genetic and molecular data presented here support a model in which the human ZC3H14-iso1 and Drosophila Nab2 proteins function at a common step in RNA processing, and that human ZC3H14 protein can maintain molecular interactions with critical RNAs and proteins in Drosophila neurons, and consequently can elicit similar downstream effects on cellular physiology and development.
Several lines of evidence suggest that the ZC3H14-iso1-3 proteins may have a function distinct from ZC3H14-iso4. First, the protein isoforms localize to different cellular compartments with ZC3H14-iso1-3 localized to the nucleus at steady state, while ZC3H14-iso4 is located in the cytoplasm (Leung et al. 2009). Second, expression studies suggest that ZC3H14-iso4 is highly expressed only in the testes (Leung et al. 2009). Finally, the most well-characterized ZC3H14 mutation in intellectual disability patients leads to loss of ZC3H14-iso1-3, while ZC3H14-iso4 is likely unaffected (Pak et al. 2011). Consistent with the suggestion that the nuclear and cytoplasmic isoforms of ZC3H14 perform distinct functions, this study shows that ZC3H14-iso4 does not replace the function of dNab2. However, a caveat to this conclusion is that levels of transgenic ZC3H14-iso4 attained are not comparable to those obtained for transgenic expression of ZC3H14-iso1 (see Fig. 3B). Furthermore, the transgenic proteins are both N-terminally epitope-tagged which could interfere with protein function. Thus, our results suggest that ZC3H14-iso4 is not a functional ortholog of dNab2 but further studies would be required to directly address this point.
Our analysis provides evidence that rescue of both organismal and behavioral defects in the dNab2 mutant flies correlates with a rescue of a molecular defect in polyadenylation. However, this work does not directly demonstrate that altered polyadenylation underlies the Drosophila phenotypes. Given the interdependent nature of transcription, RNA processing, export, and translation, dNab2/ZC3H14 could play direct roles in functions other than control of poly(A) tail length. Indeed, recent work has defined roles for the S. pombe Nab2 protein in regulated RNA turnover (Grenier St-Sauveur et al. 2013) and other studies identified the corresponding C. elegans protein, termed Sut-2, as a suppressor of a Tau toxicity model (Guthrie et al. 2009). Understanding the precise molecular functions of RNA-binding proteins, as well as defining their target RNAs, is a significant challenge and a major focus of our ongoing studies of dNab2/ZC3H14.
Although most mRNA transcripts are polyadenylated at the 3′-end, we infer from our analysis that the shared molecular properties of the Nab2/ZC3H14 nuclear poly(A)-binding proteins are nonetheless more critical in neurons than in other cell types. This inference is congruent with the fact that mutation of ZC3H14 in humans causes nonsyndromic intellectual disability (Pak et al. 2011), a form of disease where brain function is impaired without other detectable symptoms. The enhanced requirement for dNab2/ZC3H14 in neurons relative to other cell types also parallels an existing body of evidence showing that regulated polyadenylation is an important mechanism that guides the localized translation of mRNAs involved in synaptic development and plasticity (Keleman et al. 2007; Udagawa et al. 2012). Altered polyadenylation, as occurs in Drosophila and mammalian neurons lacking dNab2/ZC3H14, could perturb or override this cytoplasmic coupling between poly(A) tail length and translation and thus alter expression of RNAs that are subject to both temporal and spatial control.
The present study demonstrating that human ZC3H14 is a functional ortholog of Drosophila Nab2 provides a validated genetic model that can be used to understand how defects in otherwise ubiquitously expressed RNA-binding proteins give rise to neuron-specific dysfunction. ZC3H14 joins a growing class of RNA-binding proteins that when disrupted or silenced result in neuron-specific diseases (Cooper et al. 2009). Other members of this class of proteins include the Fragile-X Syndrome (FXS) protein, FMRP, and the spinal muscular atrophy (SMA) protein, SMN1 (Lukong et al. 2008; Ibrahim et al. 2012). The specific dependence of neurons on Nab2/ZC3H14 fits well with an emerging model that the 3′ UTRs of neuronal RNAs are subject to more extensive regulation than their non-neuronal counterparts (An et al. 2008; Hilgers et al. 2011; Smibert et al. 2012). Future studies investigating the precise molecular role of ZC3H14 and the neuronal target RNAs that it binds and regulates will be critical in defining how defects in Nab2/ZC3H14-dependent post-transcriptional regulation of gene expression contribute to neuronal dysfunction.
MATERIALS AND METHODS
Cell culture
Neuro 2A (N2A) cells (Olmsted et al. 1970) were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 0.1 mg/mL streptomycin, and 100 units/mL penicillin. DNA plasmids were transfected into cultured cells using Lipofectamine2000 (Invitrogen) according to manufacturer's protocol.
D. melanogaster stocks and genetics
All crosses and stocks were maintained in standard conditions unless otherwise noted. The Nab2ex3 null allele (referred to here as Nab2null has been described previously (Pak et al. 2011). Complete open reading frames of ZC3H14 isoform 1 (NCBI #NM_024824.4) and ZC3H14 isoform 4 (NCBI #NM_207662) were cloned into a Gateway (Invitrogen) entry vector. The Gateway system was then used to transfer either ZC3H14-iso1 or ZC3H14-iso4 into a plasmid encoding an N-terminal Flag epitope (plasmid pTFW). Plasmids were then sequenced to ensure that the correct reading frame was maintained and that no mutations had been introduced during the cloning process. Sequenced plasmids were then injected by BestGene, Inc. into white− embryos. Integrated transgenes were then mapped and balanced using standard genetic techniques.
siRNA-mediated depletion
Stealth RNAi (Invitrogen) was used to knockdown ZC3H14 and PABPN1 expression in cultured N2A cells. Cells were plated in GM at a density of 1 × 105 cells per well on collagen-coated six-well plates and after 6 h, duplexed siRNAs at the final concentration of 80 nM were used to transfect cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions in GM. After overnight incubation, medium containing transfection complexes were replaced by fresh GM. PABPN1 knockdown was assessed by immunoblotting using either anti-ZC3H14 (Leung et al. 2009) or anti-PABPN1 (Apponi et al. 2010) antibody after 40 h of transfection. Cells were transfected with scrambled control or one of two ZC3H14 siRNAs (Invitrogen). ZC3H14-1, TGACTGACCTGAGTGTGGCACAGAA; ZC3H14-2, GAAGAGCCTCGATACTGACTCCAAA. PABPN1 siRNAs were previously described (Apponi et al. 2010).
Immunoblotting
Analyses of protein levels were performed by immunoblotting using standard procedures (Green and Sambrook 2012). For N2A cells, cells were lysed with RIPA-2 (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS) containing protease inhibitors (Mini Complete, Roche) and equal amounts of total protein (5–20 μg) were resolved by SDS-PAGE, transferred to nitrocellulose, and the desired protein was detected by immunoblotting with appropriate antibody (polyclonal ZC3H14) (Leung et al. 2009) or PABPN1 (Apponi et al. 2010) diluted 1:5000. For flies, 20 D. melanogaster heads were collected from each genotype being analyzed and homogenized in sample loading buffer. Samples were resolved by SDS-PAGE and transferred to PVDF membranes. Membranes were probed for 1 h at room temperature with anti-FLAG (Sigma M2, 1:1000) antibody to detect Nab2/ZC3H14 or overnight with anti-histone H3 (Abcam, 1:10,000) to detect the histone control.
Determination of bulk poly(A) tail length
Bulk poly(A) tails were analyzed using a standard PAT assay (Chekanova et al. 2001) as described previously (Apponi et al. 2010). Total RNA was isolated from either cultured N2A cells or 250–300 fly heads of 5–6 d old flies of the selected genotype. Total RNA was 3′-end labeled with [32P]pCp using T4 RNA ligase, and digested with a cocktail of RNAse A/T1. The resulting tracts of poly(A) RNA were resolved by gel electrophoresis and imaged by film or PhosphorImager. At least three biological replicates were performed for each fly genotype analyzed. Using ImageJ software, densitometry analysis of each experiment was performed in order to obtain profile curves for each sample that was analyzed. Profiles of the relative signal intensity at a given location on the gel were plotted in Microsoft Excel and a moving average (period = 25) calculated.
Fluorescence in situ hybridization (FISH)
The protocol used for poly(A) RNA localization was previously described (Farny et al. 2008). Briefly, cells were allowed to adhere to ECL-coated coverslips in DM for 6 h. Cells were fixed with 4% paraformaldehyde in PBS for 10 min, permeabilized with 100% cold methanol for 10 min and 70% ethanol at 4°C overnight. Cells were hybridized for 1–2 h at 37°C with DIG-Oligo-dT(50) probe (Integrated DNA Technologies) at 0.2 mM in hybridization buffer (25% formamide, 10% dextran sulfate, 0.005% BSA, 1 mg/mL yeast tRNA in 2X SSC). Following washes with 2X SSC, cells were incubated with FITC-conjugated anti-DIG antibody (Molecular Probes) at RT for 1 h and with Hoechst 33325 dye (Sigma) for 5 min. Images were obtained using an Olympus IX81 microscope with a 0.3 NA 100X Zeiss Plan-Neofluor objective. Images were captured using a Hamamatsu digital camera with Slidebook software (version 5.0). At least three independent experiments were performed.
Microscopy
D. melanogaster were imaged with a Leica DFC500 CCD digital camera. Post acquisition image processing was performed using Adobe Photoshop. N2A Cells were visualized for fluorescence in situ hybridization by indirect immunofluorescence microscopy. Cells were fixed with 2% formaldehyde (EM Science) for 10 min, permeabilized with 0.1%Triton X-100 for 5 min, and incubated with Hoechst to mark nuclear DNA. Images were obtained using an Olympus IX81 microscope with a 0.3 NA 100X Zeiss Plan-Neofluor objective unless otherwise stated. Images were captured using a Hamamatsu digital camera with Slidebook software (version 1.63) and globally processed for brightness and contrast using Adobe Photoshop.
D. melanogaster behavioral assays
The negative-geotaxis assay was performed as previously described (Ganetzky and Flanagan 1978) with some modifications. Briefly, newly eclosed flies (day 0) were collected daily, divided into groups of 10, and aged in separate vials for 5 d. Cohorts of age-matched flies were transferred to a 25-mL graduated cylinder, tapped to the bottom, and analyzed for a climbing response. The number of flies reaching the top after 5, 10, 15, and 30 sec was recorded. A minimum of 10 groups were tested for each genotype except for elav>GAL4/+;UAS-ZC3H14-iso4Flag/+;Nab2ex3/Nab2ex3.
ACKNOWLEDGMENTS
We are most grateful to members of the Corbett and Moberg laboratories for helpful discussions and comments. This work was supported grants from the NIH to A.H.C. (GM058728) and K.H.M. (CA123368) and a Development Award from the MDA to A.B. (255856).
REFERENCES
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, et al. 2008. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134: 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Wilson SM, Datar KV, Swanson MS 1993. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol Cell Biol 13: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apponi LH, Leung SW, Williams KR, Valentini SR, Corbett AH, Pavlath GK 2010. Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum Mol Genet 19: 1058–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier A, Benoit B, Simonelig M 2006. A Drosophila model of oculopharyngeal muscular dystrophy reveals intrinsic toxicity of PABPN1. EMBO J 25: 2253–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Shaw RJ, Belostotsky DA 2001. Analysis of an essential requirement for the poly(A) binding protein function using cross-species complementation. Curr Biol 11: 1207–1214 [DOI] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G 2009. RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann CR, Rammelt C, Wahle E 2011. Control of poly(A) tail length. Wiley Interdiscip Rev RNA 2: 348–361 [DOI] [PubMed] [Google Scholar]
- Farny NG, Hurt JA, Silver PA 2008. Definition of global and transcript-specific mRNA export pathways in metazoans. Genes Dev 22: 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganetzky B, Flanagan JR 1978. On the relationship between senescence and age-related changes in two wild-type strains of Drosophila melanogaster. Exp Gerontol 13: 189–196 [DOI] [PubMed] [Google Scholar]
- Goldie BJ, Cairns MJ 2012. Post-transcriptional trafficking and regulation of neuronal gene expression. Mol Neurobiol 45: 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR, Sambrook J 2012. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Grenier St-Sauveur V, Soucek S, Corbett AH, Bachand F 2013. Poly(A) tail-mediated gene regulation by opposing roles of Nab2 and Pab2 nuclear poly(A)-binding proteins in pre-mRNA decay. Mol Cell Biol 33: 4718–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie CR, Schellenberg GD, Kraemer BC 2009. SUT-2 potentiates τ-induced neurotoxicity in Caenorhabditis elegans. Hum Mol Genet 18: 1825–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie CR, Greenup L, Leverenz JB, Kraemer BC 2011. MSUT2 is a determinant of susceptibility to τ neurotoxicity. Hum Mol Genet 20: 1989–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector RE, Nykamp KR, Dheur S, Anderson JT, Non PJ, Urbinati CR, Wilson SM, Minvielle-Sebastia L, Swanson MS 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J 21: 1800–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Perry MW, Hendrix D, Stark A, Levine M, Haley B 2011. Neural-specific elongation of 3′ UTRs during Drosophila development. Proc Natl Acad Sci 108: 15864–15869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim F, Nakaya T, Mourelatos Z 2012. RNA dysregulation in diseases of motor neurons. Annu Rev Pathol 7: 323–352 [DOI] [PubMed] [Google Scholar]
- Keleman K, Kruttner S, Alenius M, Dickson BJ 2007. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci 10: 1587–1593 [DOI] [PubMed] [Google Scholar]
- Kelly SM, Pabit SA, Kitchen CM, Guo P, Marfatia KA, Murphy TJ, Corbett AH, Berland KM 2007. Recognition of polyadenosine RNA by zinc finger proteins. Proc Natl Acad Sci 104: 12306–12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SM, Leung SW, Apponi LH, Bramley AM, Tran EJ, Chekanova JA, Wente SR, Corbett AH 2010. Recognition of polyadenosine RNA by the zinc finger domain of nuclear poly(A) RNA-binding protein 2 (Nab2) is required for correct mRNA 3′-end formation. J Biol Chem 285: 26022–26032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S, Pak C, Garshasbi M, Kuss A, Corbett AH, Moberg K 2012. New kid on the ID block: neural functions of the Nab2/ZC3H14 class of Cys3His tandem zinc-finger polyadenosine RNA binding proteins. RNA Biol 9: 555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U, Wahle E 2004. Structure and function of poly(A) binding proteins. Biochim Biophys Acta 1678: 67–84 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Leung SW, Apponi LH, Cornejo OE, Kitchen CM, Valentini SR, Pavlath GK, Dunham CM, Corbett AH 2009. Splice variants of the human ZC3H14 gene generate multiple isoforms of a zinc finger polyadenosine RNA binding protein. Gene 439: 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd TE, Taylor JP 2010. Flightless flies: Drosophila models of neuromuscular disease. Ann N Y Acad Sci 1184: e1–e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Chang KW, Khandjian EW, Richard S 2008. RNA-binding proteins in human genetic disease. Trends Genet 24: 416–425 [DOI] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narro ML, Yang F, Kraft R, Wenk C, Efrat A, Restifo LL 2007. NeuronMetrics: software for semi-automated processing of cultured neuron images. Brain Res 1138: 57–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted JB, Carlson K, Klebe R, Ruddle F, Rosenbaum J 1970. Isolation of microtubule protein from cultured mouse neuroblastoma cells. Proc Natl Acad Sci 65: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak C, Garshasbi M, Kahrizi K, Gross C, Apponi LH, Noto JJ, Kelly SM, Leung SW, Tzschach A, Behjati F, et al. 2011. Mutation of the conserved polyadenosine RNA binding protein, ZC3H14/dNab2, impairs neural function in Drosophila and humans. Proc Natl Acad Sci 108: 12390–12395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D 2010. The hippo signaling pathway in development and cancer. Dev Cell 19: 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnamon JR, Czaplinski K 2011. mRNA trafficking and local translation: the Yin and Yang of regulating mRNA localization in neurons. Acta Biochim Biophys Sin (Shanghai) 43: 663–670 [DOI] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads BD, Carlson J, Brown JB, et al. 2012. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep 1: 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, Lorenz LJ, Zukin RS, Bassell GJ, Richter JD 2012. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol Cell 47: 253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]